Alternative genetic pathways in colorectal carcinogenesis (original) (raw)
Abstract
The comparative typing of matched tumor and blood DNAs at dinucleotide repeat (microsatellite) loci has revealed in tumor DNA the presence of alleles that are not observed in normal DNA. The occurrence of these additional alleles is possibly due to replication errors (RERs). Although this observation has led to the recognition of a subtype of colorectal cancer with a high incidence of RERs (caused by a deficiency in DNA mismatch repair), a thorough analysis of the RER frequency in a consecutive series of colorectal cancers had not been reported. It is shown here that the extensive typing of 88 colorectal tumors reveals a bimodal distribution for the frequency of RER at microsatellite loci. Within the major mode (75 tumors, RER− subtype), the probability that a locus exhibited instability did not differ significantly among loci and tumors, being 0.02. The subsequent development of a statistical test for an operational discrimination between the RER− and RER+ subtypes indicated that the probability of misclassification did not exceed 0.001 in this series. The frequency of K-ras mutation was found to be equivalent in the two subtypes. However, in the RER+ tumors, the p53 gene mutation was less frequently detected, the adenomatous polyposis coli (APC) mutation was rare, and the biallelic inactivation of either of these genes was not observed. Furthermore, the concomitant occurrence of APC and tumor growth factor β receptor type II gene alterations was found only once. These data suggest that the repertoires of genes that are frequently altered in RER+ and RER− tumors may be more different than previously thought.
Colorectal tumorigenesis has been associated with the progressive acquisition of a variety of genomic alterations in neoplastic cells. Genes of the ras family have been found activated by missense mutation in 45% of colorectal cancers (K-ras in 40% of cases, N-ras in 5%) (1, 2). The tumor suppressor genes p53 and adenomatous polyposis coli (APC) were each found mutated in about two-thirds of the tumors (3, 4). For these latter two genes, frequent association of point mutation in one allele and loss of the other allele has been observed, indicating that their functional inactivation takes place through a two-hit mechanism often involving chromosomal deletions (5–7).
In 1993, comparative analysis at microsatellite loci revealed, in the DNA of some tumors, the frequent presence of alleles that were not observed in the matched normal DNA. These new alleles are possibly generated as the result of errors occurring during replication (replication error, RER) (8). The tumors that exhibit the highest frequency of RER at microsatellite loci, thus termed RER+ (9), were shown to be impaired in the DNA mismatch-repair pathway. Among the genes involved in this pathway, MSH2 (10, 11), MLH1 (12, 13), PMS2 (14), GTBP/MSH6 (15–17), MSH3 (17), and perhaps PMS1 (14) and DNA polymerase δ (18) are the sites of somatic mutations.
RER+ tumors account for 10–15% of all colorectal cancers (19). They arise preferentially from the proximal colon, a site where tumors rarely exhibit losses of 17p, 18q, and 5q (2). Mismatch-repair deficiency may be an early event in tumorigenesis. This deficiency is expected in part to determine the subsequent genetic events associated with tumor progression (20, 21). Indeed, the high mutation rate of the HPRT gene in RER+ cell lines grown in the presence of 6-thioguanine (22, 23) is indicative of a genetic instability enabling the rapid accumulation of somatic mutations (24). This high rate may eventually alleviate the requirement of chromosomal loss in the biallelic inactivation of tumor suppressor genes. In support of the latter hypothesis are the observations that RER+ tumors or cell lines infrequently demonstrate allelic losses (25), exhibit normal or quasinormal karyotypes (26), and are the site of biallelic frame-shift mutations in the tumor growth factor β receptor type II (TGF-βRII) gene (27–29) and in the BAX gene (30).
With the exception of the TGF-βRII and BAX genes, conflicting information is available on the repertoire of genes which, when altered, contribute directly to the oncogenic properties of the RER+ tumors. Reported frequencies of mutations in the three critical genes initially recognized as being recurrently mutated in colorectal cancer, K-ras, p53, and APC, differ markedly among series of RER+ colorectal tumors (8, 31–36). To delineate more accurately the genetic mechanism involved in colorectal cancer we characterized with respect to microsatellite instability tumors that were calibrated for their DNA index and that were also screened for loss of alleles on 17p and 5q, and for point mutations in the APC, p53, K-ras, and TGF-βRII genes.
PATIENTS AND METHODS
Patients.
This study was based on two series of tumors. The first series was collected from 96 patients (mean age, 66 ± 11.3 years) undergoing surgery for colorectal cancer in our institution. Identification of Ki-ras mutations and determination of DNA index and allelic losses on chromosomes 17p and 5q have been in part previously reported in a larger series of 109 tumors (2, 37, 38).
A second, more recent, series of 219 colorectal tumors was made from surgical specimens provided to our laboratory by several collaborating institutions. Flow cytometric analysis revealed in this series 70 tumors with near diploid DNA index (0.95 < n < 1.05). These tumors were selected for further analysis.
Tumors collected from patients with familial adenomatous polyposis or with a family history of colorectal cancer suggestive of hereditary nonpolyposis colon cancer (HNPCC; Amsterdam criteria) were excluded. In all cases, the freshly removed tumor and normal mucosae samples were rapidly frozen and stored in liquid nitrogen.
Genotyping of Microsatellite Loci.
A total of 110 highly polymorphic microsatellite loci evenly distributed on the autosomes were selected from the Genethon database (list available on request). They were used to determine the RER status of 88 colorectal carcinomas. Each locus was scored for its stability status according to the absence or presence of mobility shifts or additional bands in the amplification product from the tumor as compared with that from normal DNA. A partial characterization of microsatellite instability has previously been reported for 46 tumors of the first series and for 44 tumors of the second series (39).
Identification of TGF-βRII Mutations.
Codons 110–134, including 10 repeating adenines of TGF-βRII gene, were amplified using the following primers: sense 5′-CTTTATTCTGGAAGATGCTG-3′ and antisense 5′-GAAGAAAGTCTCACCAGGC-3′. Each case was scored for mutation status according to the size of amplified products.
Identification of APC Mutations.
Search for somatic APC mutations was performed in 85 tumors of the first series by using sequentially two pre-screening methods based on denaturing gradient gel electrophoresis (DGGE) (40, 41). In addition, the PTT procedure was applied to exon 15, using 5 overlapping 1,800-bp fragments (Table 1) (42). All electrophoretic variants were sequenced directly from PCR-amplified products prepared as previously described (41) using Prism ready reaction dye primer cycle sequencing kits (Applied Biosystems) and an Applied Biosystems 373A sequencer. When a mutation was found in a tumor DNA sample, the corresponding normal DNA was systematically checked for the absence of this mutation.
Table 1.
PCR conditions for protein truncation test
| Fragment | Primer | Annealing temp; °C |
|---|---|---|
| I | 5′-(a)GGAACTTTGTGGAATCTCTC | 60 |
| 5′-TTCGGTTTTACTGCTTTGTCC | ||
| II | 5′-(a)GTTTCTCCATACAGGTCACG | 60 |
| 5′-TGTAGGAATGGTATCTCGTTT | ||
| III | 5′-(a)GCAGTAAATGCTGCAGTTCAGAGG | 50 |
| 5′-CTTTTTTGGCATTGCGGAGCTT | ||
| IV | 5′-(a)GATGATGTTGACCTTTCCAGGG | 58 |
| 5′-GTTGACTGGCGTACTAATACAG | ||
| V | 5′-(a)GCAAACATGCCTTCAATCTCTCG | 58 |
| 5′-CCCTCTAACAAGAATCAAACCT |
Identification of p53 Mutations.
A total of 87 tumors from the first series was screened for somatic mutations in p53 exons 4 to 8 (36).
Allelic Losses.
In addition to the previously reported determination of allelic losses on 17p and 5q by using restriction fragment length polymorphism (RFLP) loci and the Southern technique (2, 38), the tumors of patients that were heterozygous for intragenic polymorphisms in the p53 and APC genes or those that carried a somatic mutation in these genes could be directly monitored for the loss of alleles of the corresponding genes by examining the sequence profile of the relevent amplified product.
Statistics.
Qualitative variables were compared with respect to one another by using χ2 analysis with Yates’ correction when necessary. The FUM values (see below) and instability rate at each locus in RER− tumors were tested for a Poisson distribution by a Kolmogorov–Smirnov test.
The probability for observing m unstable microsatellite loci among n tested in RER− tumors was calculated from a binomial distribution using p and n as parameters. The p parameter was derived assuming a Poisson distribution for the frequency of unstable microsatellite loci (FUM value) in RER− tumors.
RESULTS
Classification of RER+ and RER− Tumors.
To investigate microsatellite stability in a series of colorectal tumors, an initial set of 110 microsatellite loci scattered over the entire genome was chosen. Using this set of markers, we characterized a group of 46 tumors by comparing the typing data of matched normal and tumor DNA samples (Fig. 1a). These abnormal bands correspond to the somatic generation of new alleles. Occasionally, multiple additional bands were observed, resulting in a ladder. Most tumors demonstrated little or no microsatellite instability in spite of the fact that more than 80 loci were informative for each tumor. In contrast, a small group of tumors demonstrated many instabilities.
Figure 1.
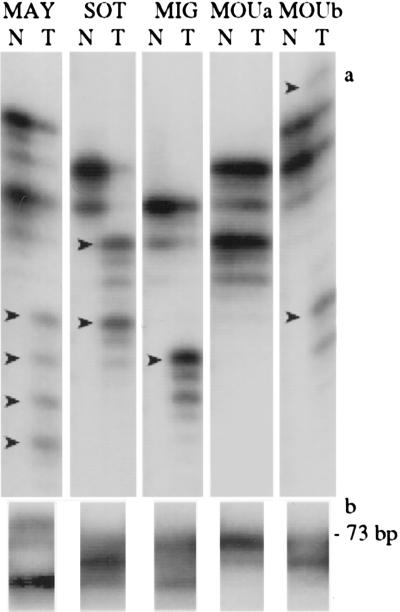
Detection of instability at microsatellite loci. (a) The PCR products generated at D9S178 from normal (N) and matched tumor (T) DNAs are compared for five cases. No difference was observed for sample MOUa. Arrows indicate the presence of additional alleles (RER) in the four other cases. (b) The PCR product generated by primers spanning the coding A10 tract of the TGF-βRII gene from the tumor DNA is shown for the same five cases. The product generated from normal DNA is 73 bp long. This is the only product observed for case MOUa. In the four other cases, additional product(s) demonstrate(s) the presence of frameshift mutation(s).
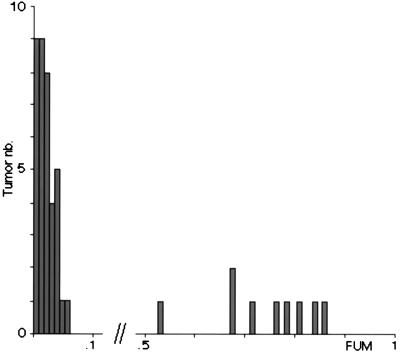
A quantitative estimate of the frequency of unstable microsatellite loci in each tumor was derived by computing the ratio of the number of unstable microsatellite loci to the total number of typed microsatellites. By analogy with the FAL parameter, which measures the fractional allelic losses (43), this ratio was termed FUM, for fractional unstable microsatellites. Analysis of the FUM values revealed a bimodal distribution (Fig. 2). The first subgroup, which included 37 tumors, demonstrated FUM below 0.06 (minimum 0; mean 0.02). These tumors were classified as RER−. In this subgroup, the numbers of microsatellite instabilities per tumor and per locus are compatible with a Poisson distribution (Kolmogorov–Smirnov test, p = 0.35 and p = 0.41, respectively), thus failing to reveal heterogeneity in the FUM values of the tumors or in the frequency with which microsatellite loci would demonstrate instabilities. The second subgroup included 9 tumors that had FUM values larger than 0.53 (maximum 0.86; mean 0.70). They were termed RER+. This group was too small to test for heterogeneity. No tumor had a FUM value between 0.06 and 0.53 (Fig. 2).
Figure 2.
FUM distribution in colorectal cancers. A total of 46 tumors that had been typed on 110 dinucleotide repeat loci are ordered according to their FUM values. The tumors that have a FUM value less than or equal to 0.06 were classified in the RER− subtype. Although about 4,000 elementary genotypic data on microsatellite loci were available for this subgroup, statistical analysis revealed heterogeneity neither for FUM values nor for the occurrence of RER at specific loci.
Taking into account the presence of two clearly distinct subtypes, the number of microsatellite loci to be typed was decreased to 20 for the classification of an additional group of 42 tumors. Again none of the tumors had a FUM value between 0.06 and 0.5. This second group was found to contain 4 RER+ and 38 RER− tumors. Thus the merged subgroups of this first series contained a total of 75 RER− and 13 RER+ tumors. All RER+ tumors were found to have a near-diploid DNA content (n < 1.3, Table 2).
Table 2.
Relation of RER phenotype with single genetic alterations in the first series of tumors
| Measurement | Criterion | Tumors, n | χ2 P | |
|---|---|---|---|---|
| RER+ | RER− | |||
| DNA index | n < 1.3 | 13 | 29 | <10−4 |
| n ≥ 1.3 | 0 | 46 | ||
| LOH | ||||
| 17p | Yes | 0 | 50 | <10−4 |
| No | 13 | 21 | ||
| 5q | Yes | 0 | 44 | 2 × 10−3 |
| No | 9 | 24 | ||
| Mutation | ||||
| _p_53 | Yes | 4 | 41 | NS |
| No | 9 | 33 | ||
| K-ras | Yes | 2 | 24 | NS |
| No | 7 | 39 | ||
| APC | Yes | 1 | 53 | 2 × 10−4 |
| No | 8 | 15 | ||
| TGF-βRII | Yes | 13 | 1 | <10−4 |
| No | 0 | 74 |
Frequency of p53, APC, K-ras, and TGF-βRII Mutations in RER+ and RER− Tumors.
The screening for p53 and APC mutations that was applied to this series of tumors was performed for exons 4 to 8 of the p53 gene and for all coding exons of the APC gene with the exception of the newly identified small alternative exon 10b, in which no mutation has been described in the literature (44, 45). For both genes, the intron/exon boundaries were also investigated. The details of the p53 mutations have been reported elsewhere (6). Of the 13 tumors that were classified as RER+, 4 (31%) demonstrated a p53 mutation (IVS3–1G → A, CCC152CC, AGG249TGG, and CGT273TGT). APC mutations were found in 59 different tumor samples. Fourteen tumors were found to carry two mutations (Table 3). A single RER+ tumor (11%) demonstrated an APC mutation (CGA1450TGA) (Table 2).
Table 3.
Somatic APC mutations
| Patient | Location | Nucleotide change | Consequence | LOH | Patient | Location | Nucleotide change | Consequence | LOH |
|---|---|---|---|---|---|---|---|---|---|
| DEM | EX5 | C607T | Q203X | − | SCH | EX15 | 3155insCGAC | Frameshift | |
| VER | EX5 | C637T | R213X | + | EX15 | 3926del5 | Frameshift | − | |
| GIM | EX6 | C646T | R213X | + | GAL | EX15 | 3191insT | Frameshift | |
| RUB | EX6 | C646T | R213X | − | EX15 | T4565A | L1522X | − | |
| MARb | EX6 | C667T | Q223X | − | BON | EX15 | C3340T | R1114X | − |
| DUT | EX6 | C694T | R232X | + | LEB | EX15 | G3466T | E1156X | + |
| MARc | EX6 | C694T | R232X | ELO | EX15 | G3625T | E1209X | + | |
| EX15 | C3581A | S1194X | − | BOG | EX15 | C3682T | Q1228X | + | |
| THE | EX8 | C847T | R283X | + | MOUa | EX15 | C2626T | R876X | |
| BOU | EX8 | C904T | R302X | EX15 | C3682T | Q1228X | − | ||
| EX9 | 983del7 | Frameshift | − | DEP | EX15 | G3856T | E1286X | + | |
| LAS | EX8 | A931T | K311X | + | KAT | EX15 | 3859del | Frameshift | + |
| FLE | IVS8a | IVS8a-2A → T | Splice mutation | − | SAU | EX15 | 3872insA | Frameshift | + |
| SER | EX9 | 1253del | Frameshift | COU | EX15 | G3883T | E1295X | + | |
| EX15 | 3926del5 | Frameshift | − | LAN | EX15 | G3916T | E1306X | + | |
| NOV | IVS9 | IVS9+2insTAT | Splice mutation | LIS | EX15 | 3926del5 | Frameshift | + | |
| EX15 | C3638A | S1213X | − | PIS | EX15 | 3926del5 | Frameshift | + | |
| LET | EX14 | 1944insA | Frameshift | ROB | EX15 | G3934T | G1312X | + | |
| EX15 | C4348T | R1450X | − | NOE | EX15 | 3950insG | Frameshift | + | |
| HAM | EX15 | G1972T | E658X | − | BOR | EX15 | C4031A | S1344X | + |
| VAS | EX15 | G2572T | E858X | − | MIG | EX15 | 4060insT | Frameshift | − |
| FRE | EX15 | C2626T | R876X | + | GAU | EX15 | C4099T | Q1367X | + |
| LOU | EX15 | C2626T | R876X | + | AVE | EX15 | 4146del | Frameshift | + |
| ARG | EX15 | C2626T | R876X | DESb | EX15 | 4147delAT | Frameshift | + | |
| EX15 | 4661insAA | Frameshift | − | JEA | EX15 | 4185insT | Frameshift | − | |
| LEC | EX15 | C2626T | R876X | GOD | EX15 | G4189T | E1397X | + | |
| EX15 | 4242del17 | Frameshift | − | DAU | EX15 | C4199A | S1400X | − | |
| DESa | EX15 | C2626T | R876X | DER | EX15 | C4202A | S1401X | + | |
| EX15 | 4243del | Frameshift | − | POU | EX15 | G4222T | E1408X | − | |
| PET | EX15 | C2626T | R876X | SOUa | EX15 | 4233del | Frameshift | + | |
| EX15 | C4348T | R1450X | − | LEF | EX15 | C4448T | R1450X | + | |
| JAN | EX15 | 2857del | Frameshift | FAL | EX15 | C4448T | R1450X | − | |
| EX15 | G4138T | E1380X | − | HUB | EX15 | G4381T | E1461X | + | |
| LEO | EX15 | G2950T | E984X | SOUb | EX15 | 4462del | Frameshift | − | |
| EX15 | C4285T | Q1429X | − | BRI | EX15 | 4462del | Frameshift | + | |
| MARa | EX15 | A2977T | E993X | + | LEG | EX15 | 4489del | Frameshift | − |
| WIT | EX15 | 4589del | Frameshift | + |
Mutation of K-ras was searched for in codons 12 and 13 by using oligonucleotide-specific hybridization. Mutations were found in 2 of 9 RER+ tumors (22%) and in 24 of 63 RER− tumors (38%).
Mutations for TGF-βRII were revealed by a deletion or insertion of 1 to 2 bp in the A10 tract (Fig. 1b). Mutations were found in all 13 RER+ tumors and in 1 of the 75 RER− tumors (Table 2). Because the presence of contaminating normal DNA may lead to erroneous conclusions when LOH at microsatellite loci is monitored by PCR, it was not possible in most cases to decide reliably whether one or both alleles were altered.
Search for Evidence of Biallelic Inactivation of p53 and APC.
This series of tumors had previously been screened for allelic losses on 17p and 5q by typing a set of restriction fragment length polymorphisms located at these two chromosomal segments. During the screening for point mutations in APC and p53 genes, a number of patients were identified as being heterozygous for intragenic polymorphic DNA variations that could be used to further document allelic losses. A final method to monitor LOH that was also used took advantage of the detection of somatic mutations. Sequencing of the DNA fragment containing the mutation, obtained by PCR amplification, from the tumor DNA sample permitted documentation of loss or conservation of the nonmutated allele. All three methods provided consistent data when used on the same tumor. Allelic losses on 17p and 5q were observed in 50 and 44, cases respectively. All tumors that demonstrated LOH in these chromosome segments belonged to the RER− group of tumors (Table 2). Tumors were classified according to the number of alterations (point mutation and LOH, counting each for one alteration) that had been detected at a given locus. None of the 42 tumors with two APC alterations, and only 1 of 22 tumors bearing a single APC alteration, was found to exhibit a RER+ phenotype. In contrast, 8 of 13 tumors (62%) in which no APC alteration was evidenced demonstrated a RER+ phenotype. This difference was highly significant (χ2 test, 2 degrees of freedom, P < 10−4). Similarly, 36 tumors inactivating both p53 alleles and 19 of 23 tumors bearing a single p53 alteration were of RER− phenotype; 9 of 26 tumors in which no p53 alteration was detected exhibited a RER+ phenotype. This difference was also found statistically significant (χ2 test, 2 degrees of freedom, P = 6 × 10−4). Biallelic inactivation of APC and/or p53 gene(s) was not observed in tumors with a TGF-βRII gene mutation (Table 4).
Table 4.
Classification of the first series of tumors according to the number of altered APC or p53 alleles for the RER tumor subtype or the presence of TGF-βRII mutation
| Gene | Mutated/deleted alleles, n | Tumors, n | χ2 P | No. with mutation in TGF-βRII | χ2 P | |
|---|---|---|---|---|---|---|
| RER+ | RER− | Yes | No | |||
| APC | 2 | 0 | 42 | 0 | 45 | |
| 1 | 1 | 21 | <10−4 | 2 | 22 | <10−4 |
| 0 | 8 | 5 | 8 | 8 | ||
| p53 | 2 | 0 | 36 | 0 | 39 | |
| 1 | 4 | 19 | 6 × 10−4 | 5 | 20 | 2 × 10−3 |
| 0 | 9 | 17 | 9 | 22 |
Search for APC Mutation in Tumors with TGF-βRII Gene Mutation.
A systematic screening for TGF-βRII gene was established in the laboratory and applied to a new set of 70 consecutive sporadic colorectal carcinomas with near diploid DNA content. An out-frame mutation in the A10 coding sequence was identified in 23 tumors. On 11 occasions, the absence of the normal PCR product derived from the A10 sequence indicated that both alleles were altered. For the remaining 12 tumors its persistence precluded any conclusion on the inactivation of the second allele because of the possible presence of nonneoplastic cells contaminating the tumor specimen. Of the 34 mutated TGF-βRII alleles that could be evaluated the A10 sequence was reduced to A8 in 11 cases, to A9 in 19 cases, and increased to A11 in 4 cases. Typing of 16 poly(CA) polymorphisms revealed for all 23 tumors more than 8 RERs. Thus these tumors met the criteria that had been defined for being classified as RER+. Search for APC mutation in this subgroup of tumors using the in vitro transcription–translation test failed to reveal any truncated APC protein. This observation contrasts with the detection, in 48 of 68 RER− tumors, of a mutation in exon 15 leading to a truncated protein (Fig. 3).
Figure 3.
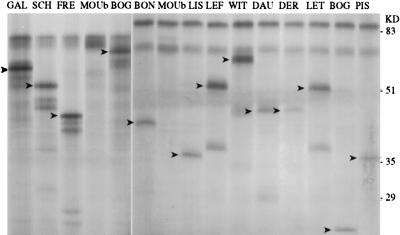
Detection of truncating mutation in exon 15 of the APC gene. Left and Right show polypeptides derived from the in vitro transcription–translation test performed on fragments I and II of APC exon 15, respectively. Fragment I is expected to give rise to a normal 72-kDa polypeptide that is observed in all lanes due to contamination of tumor fragments with nonneoplastic cells. Observation of shorter polypeptides (arrowheads) is indicative of the presence of an APC mutation that was in all cases confirmed by sequencing. Fragment II encodes a normal 85-kDa polypeptide. Note that in sample BOG shorter polypeptides are generated from both fragments I and II. This observation was suggestive of a localization of the truncating mutation on the DNA portion overlapping I and II, a hypothesis that was confirmed by sequencing.
DISCUSSION
The typing of at least 20 (more than 100 for half of the cases) dinucleotide repeat polymorphisms for each of 88 colorectal carcinomas has revealed, on the basis of their FUM values, two distinct groups of tumors, which were qualified RER+ and RER−. In each group the average FUM values were 0.7 and 0.02, respectively. Given the size of each group, it was not possible to distinguish within each of them subgroups for the FUM values.
Simulation studies were performed to derive, from the set of data collected on RER− tumors, the probability of finding RERs as a function of the number of tested loci (Table 5). The minimum FUM value that is to be observed for a tumor to exclude it from the RER− subtype (i.e., to classify it as RER+) was derived from these empirical probabilities. The resulting values are in close accordance with those derived assuming that RERs in RER− tumors were the result of stochastic events, following a Poisson distribution, which would occur with an equal probability of 0.02 on the different microsatellite loci tested and in the different tumors studied (Table 5).
Table 5.
Analysis of the criteria for the classification of colorectal tumors in the RER− subtypes
| Tested loci, n | Minimum FUM requested for classification of a tumor as not belonging to RER− subtype* | |||||||
|---|---|---|---|---|---|---|---|---|
| Probability of observation of RER at dinucleotide repeat loci in RER− tumors | Risk of misclassification = | |||||||
| None | 1 | 2 | 1 or 2 | ≥3 | P < 0.05 | P < 0.01 | P < 0.001 | |
| 2 | 0.960 | 0.039 | 0.000 | 0.040 | — | 0.50 | — | — |
| 0.967 | 0.033 | 0.000 | 0.033 | 0.50 | ||||
| 3 | 0.941 | 0.058 | 0.001 | 0.059 | 0.000 | 0.66 | 0.66 | — |
| 0.943 | 0.056 | 0.000 | 0.057 | 0.000 | 0.66 | 0.66 | 0.66 | |
| 4 | 0.922 | 0.075 | 0.002 | 0.078 | 0.000 | 0.50 | 0.50 | 0.75 |
| 0.933 | 0.065 | 0.002 | 0.067 | 0.000 | 0.50 | 0.05 | 0.75 | |
| 8 | 0.851 | 0.139 | 0.010 | 0.149 | 0.000 | 0.25 | 0.25 | 0.38 |
| 0.87 | 0.122 | 0.009 | 0.131 | 0.000 | 0.25 | 0.25 | 0.38 | |
| 12 | 0.785 | 0.192 | 0.022 | 0.214 | 0.002 | 0.17 | 0.25 | 0.33 |
| 0.806 | 0.175 | 0.017 | 0.192 | 0.001 | 0.17 | 0.25 | 0.33 | |
| 16 | 0.724 | 0.236 | 0.036 | 0.273 | 0.004 | 0.13 | 0.19 | 0.25 |
| 0.758 | 0.208 | 0.030 | 0.238 | 0.003 | 0.13 | 0.19 | 0.25 | |
| 20 | 0.668 | 0.272 | 0.053 | 0.325 | 0.007 | 0.15 | 0.15 | 0.20 |
| 0.700 | 0.247 | 0.048 | 0.295 | 0.005 | 0.10 | 0.15 | 0.20 |
None of the RER+ tumors demonstrated either allelic loss on 17p or loss on 5q, an observation that is compatible with previous cytogenetic (26) or allelic loss studies (25). These observations suggest that, if biallelic inactivation of p53 and APC were to occur in the RER+ tumors, an increased frequency of point mutations would be expected. However, the observed p53 and APC mutations were markedly less frequent in the RER+ tumors as compared with the RER− tumors. This unexpected observation may be due to a lower efficiency of our mutation screening methods in RER+ tumors as compared with RER− tumors or to a high incidence of mutations in RER+ tumors in regions that were not screened in this work. Alternatively, it may reflect a true low incidence of p53 and APC mutation in RER+ tumors. The recent observation of the presence of functional p53 mRNA in 4 of 4 RER+ cell lines derived from colorectal cancer (36) supports the proposal that development of RER+ tumors is frequently compatible with the continued presence of an intact p53 gene. No functional test is yet available for the APC gene. However, by far the most frequent mode of first-hit inactivation of the APC gene occurs through mutations causing protein truncation. The observation that full-length APC protein is present in RER+ cell lines indicates that this gene may also be frequently functional in these tumors (33).
The present study may help to resolve apparently conflicting conclusions recently reached by independent groups of investigators concerning the frequency of microsatellite instabilities in colorectal cancer and its relationship to APC mutation.
Konishi et al. (35) distinguish severe and mild RER+ phenotypes. The criterion for the severe RER+ phenotype (i.e., 3 to 5 instabilities of 5 tested loci) is, according to our statistical analysis, discriminative with a probability of misclassification lower than 10−4 (Table 5). In this subgroup the APC mutation prevalence is small (1 mutation in 10 cases) and compatible with that observed in our group of RER+ tumors. The criterion for their mild RER+ phenotype is the observation of 1 or 2 unstable loci among 5 typed dinucleotide repeats. In their series 11% of the tumors meet this criterion, a percentage that is comparable to the expected proportion of RER− tumors (9.6%) that would yield this observation (Table 5). It follows that most tumors classified by Konishi et al. in the mild RER+ subtype may correspond to tumors that we would have categorized RER−. In accord with this hypothesis, the mild RER+ tumors of Konishi et al. demonstrate a high frequency of APC mutation (35).
Huang et al. (34) suggest that APC mutation rates do not differ markedly in RER+ and RER− subtypes. In their report, the RER status determination was based on the typing of 4 or more microsatellite loci per tumor and tumors were entered in the RER+ subtype when at least half of the tested loci demonstrated RER. The RER+ subgroup thus defined may contain a minute proportion of tumors that would have been classified RER− in our study. This proportion, however, is not sufficient to account for the high APC mutation rate observed in this series of RER+ tumors. Although we cannot provide a specific explanation for this discrepancy with our present observation, it should be noted that the published data of Huang et al. (34) do suggest a significant difference in the rate of APC mutation in RER+ and RER− tumors (47 mutations in 63 RER− tumors versus 29 mutations in 52 RER+ tumors, χ2 test, 1 degree of freedom, P = 0.03).
It is suggested that to avoid future confusion, the classification of a colorectal tumor in the RER+ subgroup should rely on stringent criteria. Those advocated here may be provisionally adopted. We may not exclude, however, that subsequent larger series of data would reveal within each subtype or among microsatellite loci, smaller groups that would exhibit subtle differences in stability. Mononucleotide repeats such as Bat-26 have been shown to be systematically unstable in RER+ tumors (39). This locus also has demonstrated instability in the HCT 15 cell line, which has a low rate of alteration at dinucleotide repeats and an inactivation of the MSH6 gene.
When cell lines derived from RER+ tumors are grown under selective conditions, the genes that are placed under selective pressure exhibit a high rate of mutation (22, 23). It is therefore likely that the low incidence of mutation in p53 and APC in RER+ tumors reflects the lack of a strong requirement for mutation in these genes during tumor initiation and progression. It has been observed that genes that carry mononucleotide or dinucleotide repeats may be more readily mutated in RER+ tumors as compared with RER− tumors as exemplified by mutations of the A10 coding sequence of the TGF-βRII gene (27–29), the G8 coding sequence of the BAX and IGFIIR genes (30, 46), and the β2-microglobulin gene (47). Our data and those published by Konishi et al. indicate that mutation in the A10 tract of TGF-βRII and mutation in APC occur as alternative events in colorectal tumors. Interestingly, search for mutation in the entire coding sequence of the TGF-βRII gene for 30 colorectal tumors has revealed a mutation in a region distinct from the A10 tract. This single mutation had occurred in a tumor that displayed microsatellite instability (48). Taken together, these observations strongly suggest that the somatic mutagenesis associated with tumor initiation and progression of RER+ and RER− involves groups of target genes that are markedly more different than previously suspected. It prompts the search in RER+ tumors for additional genes that when mutated may contribute to the tumor phenotype.
Acknowledgments
This work was supported by the Ligue Nationale Contre le Cancer, the Association de la Recherche sur le Cancer, and the Ministère de l’Education, de la Santé et de la Recherche.
ABBREVIATIONS
APC
adenomatous polyposis coli
RER
replication error
TGF-βRII
tumor growth factor β receptor type II
FUM
fraction unstable microsatellites
LOH
loss of heterozygosity
References
- 1.Vogelstein B, Fearon E R, Hamilton S R, Kern S E, Preisinger A C, Leppert M, Nakamura Y, White R, Smits A M, Bos J L. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 2.Delattre O, Olschwang S, Law D J, Melot T, Remvikos Y, Salmon R J, Sastre X, Validire P, Feinberg A P, Thomas G. Lancet. 1989;ii:353–356. doi: 10.1016/s0140-6736(89)90537-0. [DOI] [PubMed] [Google Scholar]
- 3.Baker S J, Fearon E R, Nigro J M, Hamilton S R, Preisinger A C, Jessup J M, vanTuinen P, Ledbetter D H, Barker D F, Nakamura Y. Science. 1989;244:217–221. doi: 10.1126/science.2649981. [DOI] [PubMed] [Google Scholar]
- 4.Powell S M, Zilz N, Beazer-Barclay Y, Bryan T M, Hamilton S R, Thibodeau S N, Vogelstein B, Kinzler K W. Nature (London) 1992;359:235–237. doi: 10.1038/359235a0. [DOI] [PubMed] [Google Scholar]
- 5.Baker S J, Preisinger A C, Jessup J M, Paraskeva C, Markowitz S, Willson J K, Hamilton S, Vogelstein B. Cancer Res. 1990;50:7717–7722. [PubMed] [Google Scholar]
- 6.Hamelin R, Laurent-Puig P, Olschwang S, Jego N, Asselain B, Remvikos Y, Girodet J, Salmon R J, Thomas G. Gastroenterology. 1994;106:42–48. doi: 10.1016/s0016-5085(94)94217-x. [DOI] [PubMed] [Google Scholar]
- 7.Miyaki M, Konishi M, Kikuchi-Yanoshita R, Enomoto M, Igari T, Tanaka K, Muraoka M, Takahashi H, Amada Y, Fukayama M, Maeda Y, Iwama T, Mishima Y, Mori T, Koike M. Cancer Res. 1994;54:3011–3020. [PubMed] [Google Scholar]
- 8.Ionov Y, Peinado M A, Malkhosyan S, Shibata D, Perucho M. Nature (London) 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 9.Parsons R. Cell. 1993;75:1227–1236. doi: 10.1016/0092-8674(93)90331-j. [DOI] [PubMed] [Google Scholar]
- 10.Leach F S, Nicolaides N C, Papadopoulos N, Liu B, Jen J, et al. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 11.Fishel R, Lescoe M K, Rao M R, Copeland N G, Jenkins N A, Garber J, Kane M, Kolodner R. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 12.Bronner C E, Baker S M, Morrison P T, Warren G, Smith L G, Lescoe M K, Kane M, Earabino C, Lipford J, Lindblom A, Tannengard P, Bollag R, J, Godwin A R, Ward T C, Nordenskjold M, Fishel R, Kolodner R, Liskay R M. Nature (London) 1994;368:258–261. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- 13.Papadopoulos N, Nicolaides N C, Wei Y F, Ruben S M, Carter K C, et al. Science. 1994;263:1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 14.Nicolaides N C, Papadopoulos N, Liu B, Wei Y F, Carter K C, Ruben S M, Rosen C A, Haseltine W A, Fleischmann R D, Fraser C M, Adams M D, Venter J C, Dunlop M G, Hamilton S R, Petersen G M, de la Chapelle A, Vogelstein B, Kinzler K W. Nature (London) 1994;371:75–80. doi: 10.1038/371075a0. [DOI] [PubMed] [Google Scholar]
- 15.Papadopoulos N, Nicolaides N C, Liu B, Parsons R, Lengauer C, Palombo F, D’Arrigo A, Markowitz S, Willson J K, Kinzler K W, Jiricny J, Vogelstein B. Science. 1995;268:1915–1917. doi: 10.1126/science.7604266. [DOI] [PubMed] [Google Scholar]
- 16.Palombo F, Gallinari P, Iaccarino I, Lettieri T, Hughes M, D’Arrigo A, Truong O, Hsuan J J, Jiricny J. Science. 1995;268:1912–1914. doi: 10.1126/science.7604265. [DOI] [PubMed] [Google Scholar]
- 17.Malkhosyan S, Rampino N, Yamamoto H, Perucho M. Nature (London) 1996;382:499–500. doi: 10.1038/382499a0. [DOI] [PubMed] [Google Scholar]
- 18.da Costa L T, Liu B, el-Deiry W, Hamilton S R, Kinzler K W, Vogelstein B, Markowitz S, Willson J K, de la Chapelle A, Downey K M, So A G. Nat Genet. 1995;9:10–11. doi: 10.1038/ng0195-10. [DOI] [PubMed] [Google Scholar]
- 19.Liu B, Farrington S M, Petersen G M, Hamilton S R, Parsons R, Papadopoulos N, Fujiwara T, Jen J, Kinzler K W, Wyllie A H. Nat Med. 1995;1:348–352. doi: 10.1038/nm0495-348. [DOI] [PubMed] [Google Scholar]
- 20.Shibata D, Peinado M A, Ionov Y, Malkhosyan S, Perucho M. Nat Genet. 1994;6:273–281. doi: 10.1038/ng0394-273. [DOI] [PubMed] [Google Scholar]
- 21.Shibata D, Navidi W, Salovaara R, Li Z-H, Aaltonen L A. Nat Med. 1996;2:676–681. doi: 10.1038/nm0696-676. [DOI] [PubMed] [Google Scholar]
- 22.Eshleman J R, Lang E Z, Bowerfind G K, Parsons R, Vogelstein B, Willson J K, Veigl M L, Sedwick W D, Markowitz S D. Oncogene. 1995;10:33–37. [PubMed] [Google Scholar]
- 23.Bhattacharyya N P, Skandalis A, Ganesh A, Groden J, Meuth M. Proc Natl Acad Sci USA. 1994;91:6319–6323. doi: 10.1073/pnas.91.14.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu B, Nicolaides N C, Markowitz S, Willson J K, Parsons R E, Jen J, Papadopoulos N, Peltomaki P, de la Chapelle A, Hamilton S R, Kinzler K W, Vogelstein B. Nat Genet. 1995;9:48–55. doi: 10.1038/ng0195-48. [DOI] [PubMed] [Google Scholar]
- 25.Thibodeau S N, Bren G, Schaid D. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 26.Remvikos Y, Vogt N, Muleris M, Salmon R J, Malfoy B, Dutrillaux B. Genes Chromosomes Cancer. 1995;12:272–276. doi: 10.1002/gcc.2870120406. [DOI] [PubMed] [Google Scholar]
- 27.Parsons R, Myeroff L L, Liu B, Willson J K, Markowitz S D, Kinzler K W, Vogelstein B. Cancer Res. 1995;55:5548–5550. [PubMed] [Google Scholar]
- 28.Lu S L, Akiyama Y, Nagasaki H, Saitoh K, Yuasa Y. Biochem Biophys Res Commun. 1995;216:452–457. doi: 10.1006/bbrc.1995.2644. [DOI] [PubMed] [Google Scholar]
- 29.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan R S, Zborowska E, Kinzler K W, Vogelstein B, Brattain M, Willson J K V. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 30.Rampino N, Yamamoto H, Ionov Y, Li Y, Sawai H, Reed J C, Perucho M. Science. 1997;275:967–969. doi: 10.1126/science.275.5302.967. [DOI] [PubMed] [Google Scholar]
- 31.Aaltonen L A, Peltomaki P, Leach F S, Sistonen P, Pylkkanen L, Mecklin J P, Jarvinen H, Powell S M, Jen J, Hamilton S R, Petersen G M, Kinzler K W, Vogelstein B, de la Chapelle A. Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 32.Lazar V, Grandjouan S, Bognel C, Couturier D, Rougier P, Bellet D, Bressac-de Paillerets B. Hum Mol Genet. 1994;3:2257–2260. doi: 10.1093/hmg/3.12.2257. [DOI] [PubMed] [Google Scholar]
- 33.Heinen C D, Richardson D, White R, Groden J. Cancer Res. 1995;55:4797–4799. [PubMed] [Google Scholar]
- 34.Huang J, Papadopoulos N, McKinley A J, Farrington S M, Curtis L J, Wyllie A H, Zheng S, Willson J K V, Markowitz S D, Morin P, Kinzler K W, Vogelstein B, Dunlop M G. Proc Natl Acad Sci USA. 1996;93:9049–9054. doi: 10.1073/pnas.93.17.9049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Konishi M, Kikuchi-Yanoshita R, Tanaka K, Muraoka M, Onda A, Okumura Y, Kishi N, Iwama T, Mori T, Koike M, Ushio K, Chiba M, Nomizu S, Konishi F, Utsunomiya J, Miyaki M. Gastroenterology. 1996;111:307–317. doi: 10.1053/gast.1996.v111.pm8690195. [DOI] [PubMed] [Google Scholar]
- 36.Cottu P, Muzeau F, Estreicher A, Flijou J F, Iggo R, Thomas G, Hamelin R. Oncogene. 1996;13:2727–2730. [PubMed] [Google Scholar]
- 37.Laurent-Puig P, Olschwang S, Delattre O, Remvikos Y, Asselain B, Melot T, Validire P, Muleris M, Girodet J, Salmon R J, Thomas G. Gastroenterology. 1992;102:1136–1141. [PubMed] [Google Scholar]
- 38.Law D J, Olschwang S, Monpezat J P, Lefrancois D, Jagelman D, Petrelli N J, Thomas G, Feinberg A P. Science. 1988;241:961–965. doi: 10.1126/science.2841761. [DOI] [PubMed] [Google Scholar]
- 39.Hoang J M, Cottu P H, Thuille B, Salmon R J, Thomas G, Hamelin R. Cancer Res. 1997;57:300–303. [PubMed] [Google Scholar]
- 40.Olschwang S, Laurent-Puig P, Groden J, White R, Thomas G. Am J Hum Genet. 1993;52:273–279. [PMC free article] [PubMed] [Google Scholar]
- 41.Olschwang S, Tiret A, Laurent-Puig P, Muleris M, Parc R, Thomas G. Cell. 1993;75:959–968. doi: 10.1016/0092-8674(93)90539-3. [DOI] [PubMed] [Google Scholar]
- 42.Powell S M, Petersen G M, Krush A J, Booker S, Jen J, Giardiello F M, Hamilton S R, Vogelstein B, Kinzler K W. N Engl J Med. 1993;329:1982–1987. doi: 10.1056/NEJM199312303292702. [DOI] [PubMed] [Google Scholar]
- 43.Vogelstein B, Fearon E R, Kern S E, Hamilton S R, Preisinger A C, Nakamura Y, White R. Science. 1989;244:207–211. doi: 10.1126/science.2565047. [DOI] [PubMed] [Google Scholar]
- 44.Sulekova Z, Ballhausen W G. Hum Genet. 1995;96:469–471. doi: 10.1007/BF00191808. [DOI] [PubMed] [Google Scholar]
- 45.Xia L, St Denis K A, Bapat B. Genomics. 1995;28:589–591. doi: 10.1006/geno.1995.1195. [DOI] [PubMed] [Google Scholar]
- 46.Souza R F, Appel R, Yin J, Wang S, Smolinski K N, et al. Nat Genet. 1996;14:255–257. doi: 10.1038/ng1196-255. [DOI] [PubMed] [Google Scholar]
- 47.Bicknell D C, Rowan A, Bodmer W F. Proc Natl Acad Sci USA. 1994;91:4751–4756. doi: 10.1073/pnas.91.11.4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takenoshita S, Tani M, Nagashima M, Hagiwara K, Bennett W P, Yokota J, Harris C C. Oncogene. 1997;14:1255–1258. doi: 10.1038/sj.onc.1200938. [DOI] [PubMed] [Google Scholar]
- 49.Ad Hoc Committee on Mutation Nomenclature. Hum Mutat. 1996;8:197–202. doi: 10.1002/humu.1380080302. [DOI] [PubMed] [Google Scholar]