RAP74 induces promoter contacts by RNA polymerase II upstream and downstream of a DNA bend centered on the TATA box (original) (raw)
Abstract
RAP74, the large subunit of transcription factor IIF, associates with a preinitiation complex containing RNA polymerase II (pol II) and other general initiation factors. We have mapped the location of RAP74 in close proximity to promoter DNA at similar distances both upstream and downstream of a DNA bend centered on the TATA box. Binding of RAP74 induces a conformational change that affects the position of pol II relative to that of the DNA. This reorganization of the preinitiation complex minimally requires the N-terminal region of RAP74 containing both its RAP30-binding domain and another region necessary for accurate transcription in vitro. We propose a role for RAP74 in controlling the topological organization of the pol II preinitiation complex.
Initiation of mRNA synthesis by mammalian RNA polymerase II (pol II) is a complex biochemical process controlled by a set of general transcription factors including TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH (1–5). Transcription initiation is preceded by the assembly of a preinitiation complex consisting of pol II and the general transcription factors on promoter DNA. For genes containing a TATA box, the first step in preinitiation complex assembly is the binding of TATA box-binding protein (TBP), the TATA box-binding subunit of TFIID, to the TATA element (6). The other general transcription factors and pol II can assemble, mostly through protein-protein interactions, onto the TBP–promoter complex (1–6).
Human TFIIF, which is composed of two subunits known as RAP30 and RAP74, is involved at both the initiation and elongation stages of transcription (7). It is required for accurate initiation of transcription at all promoters tested except the IgH promoter (8–10). RAP30 is directly involved in recruiting pol II to a preinitiation complex containing TBP and TFIIB (11, 12). TFIIF binds to several general initiation factors, including TFIIB (13–15), TFIID (16), and TFIIE (17), as well as to pol II (10, 14, 15, 18). It is also an integral component of pol II holoenzymes isolated from both yeast and mammalian cells (19–21). These findings suggest that TFIIF plays a central role in preinitiation complex assembly. TFIIF is a target for at least some transcriptional activators, since the interaction of RAP74 with the serum response factor that binds the c-fos promoter is required for transcriptional activation (22, 23). In addition, TFIIF can stimulate RNA chain elongation (7, 24, 25).
Over the past few years, both high and low resolution techniques have been utilized to analyze the structure of the pol II preinitiation complex (26). NMR and x-ray crystallography studies revealed surprising features of TBP–promoter (27, 28), TBP–TFIIB–promoter (29, 30), and TBP–TFIIA–promoter (31, 32) complexes. Binding of TBP to the TATA element induces bending of the promoter DNA (27, 28). TFIIA and TFIIB both interact with TBP in the preinitiation complex, and TFIIB primarily by clamping onto the C-terminal stirrup of TBP (29, 30). Alanine scanning mutagenesis has identified regions on the surface of TBP that are required for interactions with the various components of the complex (33). Protein–DNA crosslinking has been used to obtain information on the relative positions of the general transcription factors and pol II along promoter DNA (34–37). Using 5-[_N_-(_p_-azidobenzoyl)-aminoallyl] photocrosslinking (38), we have previously mapped the locations of two subunits of TFIIA (A35 and A21), TBP, TFIIB, RAP30, RAP74, TFIIE34, and the two largest subunits of pol II along the adenovirus major late promoter (34, 35). This low resolution technique has the advantage of providing information on the topology of large complexes assembled using full-length or truncated polypeptides.
We have now mapped the location of RAP74 in close proximity to the major groove of DNA at similar distances both upstream and downstream of a DNA bend centered on the TATA box. We show that the association of RAP74 with the preinitiation complex induces a molecular reorganization of the complex that affects the position of pol II relative to that of the DNA.
MATERIALS AND METHODS
Protein Factors.
Recombinant TBP, TFIIB, RAP30, RAP74 (full-length and various deletion mutants), TFIIE34, and TFIIE56, as well as calf thymus pol II were purified as described (15, 34, 35). The various proteins, including the RAP74 deletion mutants, were analyzed for quality and quantity by SDS/PAGE followed by Coomassie blue staining.
Photocrosslinking.
The synthesis of deoxyuridine monophosphate (38), preparation of photoprobes, and the conditions for binding reactions were as described (34, 35). For each photoprobe, the concentration of poly(dI-dC) in binding reactions was optimized to favor specific over nonspecific binding, and was between 50 and 200 ng per reaction. A typical reaction with all factors present contained 200 ng each of TBP, TFIIB, RAP30, RAP74 (full-length or deletion mutants), pol II, TFIIE34, and TFIIE56. Ultraviolet irradiation, nuclease treatment, and SDS/PAGE analysis of radiolabeled crosslinking products were performed as described (34, 35). In some cases, crosslinking products were submitted to immunoprecipitation using either antibodies raised against RAP74 or a control antibody (34, 35). The calf thymus pol II used in our experiments was primarily in the IIb form, lacking the C-terminal domain of the largest subunit (pol180). Therefore, in the crosslinking experiments, we have followed pol180 for our analyses. Longer exposure of our gels has revealed that pol II containing the C-terminal domain (pol220) behaves exactly as pol180 in all of our experiments.
Electron Microscopy (EM).
Immediately after binding reactions, protein–DNA complexes were diluted and deposited on carbon grids using the amylamine method (39). The complexes were not submitted to a fixation procedure prior to deposition onto the grids. The samples were positively stained with uranyl acetate and rotary shadowed with tungsten prior to observation using a Philips Electrical Instruments (Mahwah, NJ) model 201 electron microscope at 60 kV. DNA length measurements were obtained from electron micrographs at a final magnification of ×190,000 by direct planimetry using a Map Videoplan-2 system (Zeiss). Micrographs of 35 different DNA fragments, either uncomplexed or carrying a preinitiation complex in the region of the TATA box, were analyzed. A Student’s t test was used for statistical analysis (Statistical Sciences, Seattle, version 3.3 for Windows).
RESULTS
The RAP74 Subunit of TFIIF and the 145-kDa Subunit of Pol II Crosslink to Nucleotides Located Both Upstream and Downstream of the TATA Box.
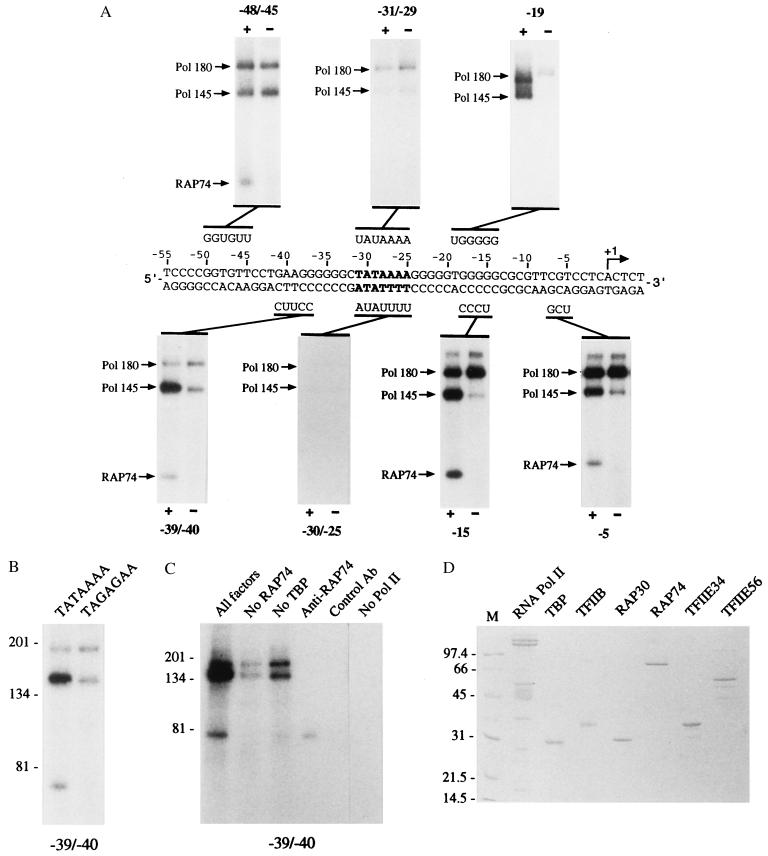
Our previous photocrosslinking data indicated that RAP30 and RAP74 occupy adjacent spaces on the same face of the DNA helix between the TATA box and the transcriptional initiation site of the adenovirus major late promoter, a region where pol II and TFIIE are also known to crosslink (34, 35). We have now extended our topological analysis to the promoter region located upstream of the TATA box. Interestingly, we have also obtained strong crosslinking of RAP74 and pol II to various positions upstream of the TATA element (Fig. 1A). In our experiments, we assess the specificity of crosslinking by comparing crosslinking reactions performed in the presence or in the absence of TBP. This is necessary because pol II and some general transcription factors bind to DNA in a nonspecific manner (34, 35, 40). A crosslinking signal that is significantly weaker in the absence of TBP is interpreted as promoter-specific because we always observed that assembling a reaction in the absence of TBP had the same effect as using a photoprobe with a mutated TATA element (refs. 34 and 35, and below). In the presence of TBP, TFIIB, TFIIF (RAP74 and RAP30), pol II, and TFIIE (p56 and p34), we obtained specific crosslinking of RAP74 and the 145-kDa subunit of pol II to nucleotides of the noncoding strand both downstream (positions −5 and −15) and upstream (positions −39/−40) of the TATA box. RAP74, but not pol II, crosslinked to additional positions on the coding strand upstream of the TATA element (positions −48/−45). We did not obtain specific crosslinking of RAP74 and pol II to nucleotides in the vicinity of the TATA element (positions −31/−29 on the coding strand and positions −30/−25 on the noncoding strand) where TBP and TFIIA have been shown (34) to crosslink. The two largest subunits of pol II specifically crosslinked to position −19 on the coding strand. Strong crosslinking of RAP74 and the 145-kDa subunit of pol II both upstream and downstream of the TATA element required the presence of TBP (Fig. 1A, positions −39/−40; compare lanes + and −). The absence of TBP had the same effect as the use of a photoprobe containing a mutated TATA element (TATAAAA to TAGAGAA) (Fig. 1B). The crosslinked polypeptide at ≈70 kDa was identified as being RAP74 because (i) the crosslinked band was only obtained when RAP74 was present in the reaction (Fig. 1C, compare “All factors” and “No RAP74”), and (ii) this polypeptide can be immunoprecipitated with specific antibodies raised against RAP74 (Fig. 1C), but not with control antibodies. Crosslinking of the 180-kDa and 145-kDa polypeptides was only obtained when pol II was present in the binding reactions (Fig. 1C, compare “All factors” and “No Pol II”). The factors we used in our crosslinking experiments were highly purified pol II and recombinant TBP, TFIIB, TFIIE34, TFIIE56, RAP74, and RAP30 expressed in Escherichia coli (Fig. 1D).
Figure 1.
Crosslinking of RAP74 and pol II along the adenovirus major late promoter. (A) Independent photoprobes that place the modified nucleotide deoxyuridine monophosphate (U) and radiolabeled nucleotides at various positions along the adenovirus major late promoter were used in crosslinking experiments. Binding reactions contained TFIIB, TFIIF (RAP74 and RAP30), pol II, and TFIIE (56- and 34-kDa subunits) in either the presence (+) or the absence (−) of TBP. The positions of RAP74 as well as those of the 145- and 180-kDa subunits of pol II are indicated. (B) Photocrosslinking experiments in the presence of TBP, TFIIB, TFIIF, pol II, and TFIIE were performed using photoprobe −39/−40 that contained either a wild-type (TATAAAA) or a mutated (TAGAGAA) TATA element. (C) Photocrosslinking experiments were performed using TBP, TFIIB, TFIIF, pol II, and TFIIE (All factors), or all of the above factors except TBP (No TBP), RAP74 (No RAP74), or pol II (No Pol II). In some cases, crosslinked polypeptides were immunoprecipitated using either antibodies directed against RAP74 (Anti-RAP74) or control antibodies (Control Ab). (D) Protein factors used in the crosslinking experiments.
Specific Crosslinking of the 145-kDa Subunit of Pol II to Nucleotides −39/−40 and the 180-kDa Subunit of Pol II to Nucleotide −19 Requires the Presence of RAP74.
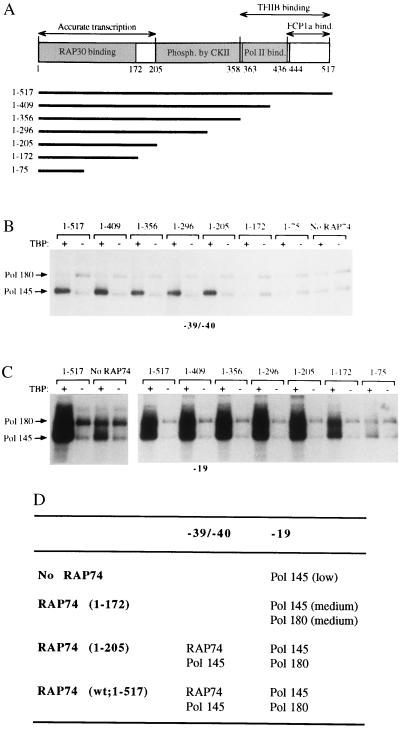
To analyze the role of RAP74 on the crosslinking of pol II upstream of the TATA element we used various RAP74 mutants with C-terminal deletions (15) in our experiments (Fig. 2A). As mentioned above, specific crosslinking was monitored by comparing the intensities of crosslinked polypeptides obtained in the presence of TBP with those obtained in its absence. When full-length RAP74 (amino acids 1–517) was added to reactions containing TBP, TFIIB, RAP30, pol II, and TFIIE (p34 and p56), we obtained specific crosslinking of the 145-kDa subunit of pol II to positions −39/−40 (Fig. 2B). RAP74 deletion mutants containing the N-terminal 205 amino acids (e.g., mutants 1–409, 1–356, 1-296, and 1-205) supported the specific crosslinking of pol II to position −39/−40 (Fig. 2B). Longer deletions abolished this crosslinking of the polymerase upstream of the TATA element (Fig. 2B, compare mutant 1–205 to mutants 1-172 and 1–75). In the absence of RAP74 (Fig. 2B), no specific crosslinking of pol II to positions −39/−40 was obtained. Crosslinking of pol II downstream of the TATA box has previously been shown to occur in the absence of RAP74 (refs. 34 and 35, and below). In Fig. 2, only the upper part of the crosslinking gels containing the two largest pol II subunits is shown to simplify the data, but the lower part of the gels indicated that only fragments of RAP74 able to support accurate transcription in vitro specifically crosslink to positions −39/−40 as well as to positions −15, −5, and −48/−45 (data not shown). Our data using photoprobe −39/−40 are summarized in Fig. 2D. They indicate that RAP74, through its N-terminal region (amino acids 1–205), induces a conformational change in the preinitiation complex that brings pol II closer to our photosensitive nucleotide upstream of the TATA element at positions −39/−40.
Figure 2.
Effect of RAP74 deletion mutants on the crosslinking of pol II upstream of the TATA element. (A) Functional domains of RAP74 and the sequences included in the various C-terminal deletion mutants are indicated. (B and C) Photocrosslinking experiments with either photoprobe −39/−40 (B) or −19 (C) were performed using TFIIB, RAP30, pol II, and TFIIE in the presence (+) or in the absence (−) of TBP and/or RAP74 (either full-length or various C-terminal deletion mutants). Some binding reactions were conducted in the absence of RAP74 (No RAP74). The positions of the 145- and 180-kDa subunits of pol II are indicated. (D) Summary of the photocrosslinking data shown in B and C.
We have previously shown (34, 35) that RAP74 and TFIIE strongly stimulate crosslinking of the two largest subunits of pol II and RAP30 to position −19 of promoter DNA. As shown in Fig. 2C, when full-length RAP74 (e.g., amino acids 1–517) was added to reactions containing TBP, TFIIB, RAP30, pol II, and TFIIE, we obtained specific crosslinking of the 180- and 145-kDa subunits of pol II to nucleotide −19. In the absence of RAP74, only the 145-kDa pol II subunit weakly crosslinked to this position (Fig. 2C, compare “1–517” and “No RAP74”). The 145-kDa subunit of pol II also crosslinks to nucleotides −15 and −5 in the absence of RAP74 (data not shown). RAP74 mutants containing the N-terminal 205 amino acids (e.g., mutants 1–409 to 1–205) fully supported the specific crosslinking of the two largest pol II subunits to nucleotide −19 (Fig. 2C). The RAP74 mutant containing the N-terminal 172 amino acids, which are sufficient to bind RAP30, but not for accurate transcription in vitro (15) only partly supported the specific crosslinking of both the 180- and 145-kDa pol II subunits to position −19. In the presence of RAP74 mutant 1–75, only the 145-kDa pol II subunit specifically crosslinked to position −19. Our data using photoprobe −19 are summarized in Fig. 2D. They indicate that RAP74 brings the 180-kDa pol II subunits closer to our photosensitive nucleotide at position −19, further supporting the notion that RAP74 induces a reorganization of the preinitiation complex.
The Preinitiation Complex Assembles on Promoter DNA That Forms a Bend Centered on the TATA Box.
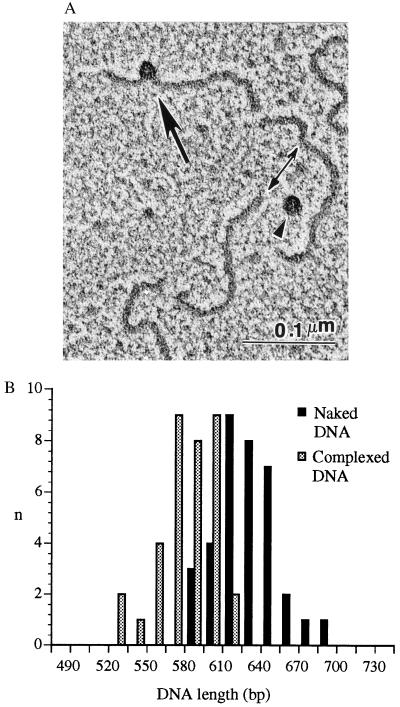
A possible explanation for the crosslinking of RAP74 and pol II to DNA regions both upstream and downstream of the TATA box, but not to the actual nucleotides of the TATA box itself, is that promoter DNA forms a bend centered approximately on the TATA element. Such a bending of the promoter DNA would place the nucleotides located between −15 and −5 closer to those located between −40 and −50. Previous studies (27, 28) indicate that the binding of TBP to the TATA box induces a bend in the promoter DNA. To visualize the structure of protein–DNA complexes assembled on promoter DNA in the presence of TBP, TFIIB, TFIIF, pol II, and TFIIE, we submitted promoter fragments, either naked or carrying a preinitiation complex, to EM (Fig. 3). Examination of several micrographs revealed that the complexes assembled in the region of the TATA box, which was 160 bp from one end of the 630-bp DNA fragment utilized, were generally placed slightly on the edge of the DNA helix (Fig. 3A). In most cases visualization clearly revealed that DNA forms a bend within the protein complex (for an example, see Fig. 3A). To clearly demonstrate the extent of the bent DNA within the core of the complex, we measured the lengths of DNA fragments that were either naked or carrying a preinitiation complex. Micrographs of 35 different fragments revealed that the lengths of fragments carrying a protein complex were significantly smaller than the lengths of naked fragments (579 ± 21 bp vs. 630 ± 21 bp; P < 0.0001) (Fig. 3B). From these data, we conclude that ≈51 bp of promoter DNA are included within the core of the complex. Taken together, our data support the concept that the preinitiation complex assembles onto promoter DNA sequences that form a bend centered approximately on the TATA element. The exact angle formed by the DNA helices on each side of the TATA box cannot be measured with precision from our experiments.
Figure 3.
(A) A typical EM field showing a preinitiation complex (arrow) assembled on the adenovirus major late promoter in the presence of TBP, TFIIB, TFIIF, pol II, and TFIIE. Two uncomplexed DNA fragments (double-headed arrow) and one free protein complex (arrowhead) are also shown. (B) Histograms of DNA lengths of promoter fragments either carrying a preinitiation complex or in the uncomplexed state.
DISCUSSION
The exact role of RAP74 in transcriptional initiation is not clear from previous studies. In this report, we have analyzed the topology of preinitiation complexes assembled in either the presence or the absence of RAP74 to define more precisely the role of the large subunit of TFIIF in preinitiation complex assembly.
Our photocrosslinking data indicate that RAP74 is located in close proximity to the major groove of promoter DNA in regions both upstream and downstream of the TATA element, but not in the vicinity of the TATA box itself (Fig. 1). A probable explanation for this observation is that promoter DNA forms a bend centered approximately on the TATA box. Such a bending of the promoter DNA would place the nucleotides between −15 and −5 closer to those located between −40 and −50. Binding of TBP to the TATA box has been reported to induce bending of ≈95° (41). Our EM data indicate that bending of the promoter DNA also occurs in the context of complexes assembled on the TATA box in the presence of TBP, TFIIB, TFIIF, pol II, and TFIIE, and that ≈51 bp of promoter DNA are included within the core of the complex (Fig. 3). In the context of a 95° bend in promoter DNA, it seems unlikely that a single molecule of RAP74 can make simultaneous contacts with the major groove of DNA at positions −5, −15, −39/−40, and −45/−48. However, it is possible that greater bending exists in the context of a preinitiation complex containing TBP, TFIIB, TFIIF, pol II, and TFIIE. For example, additional bends totaling ≈90° would bring the −50/−40 and −15/−5 regions into juxtaposition. Alternatively, it is possible that more than one molecule of RAP74 associate with the preinitiation complex. For example, in the context of a 95° bend centered on the TATA box, a heterotetramer composed of two molecules of each RAP30 and RAP74 would be more likely to make simultaneous contacts with the major groove at positions −5, −15, −39/−40, and −45/−48. Interestingly, it has been shown (32, 43) that TFIIF can exist as a heterotetramer in solution. Both possibilities would be consistent with our observations that either the full-length RAP74 (Fig. 1A) or mutant proteins with C-terminal deletions (data not shown) and the 145-kDa pol II subunit (Fig. 1A) crosslink to promoter sequences located both upstream and downstream of the TATA box and that the DNA is visually shortened by ≈50 bp (Fig. 3).
Previous studies have demonstrated that the RAP30 subunit of TFIIF is sufficient for recruiting pol II to a TBP–TFIIB–promoter complex (9, 11, 12). The RAP74 subunit of TFIIF was shown to stabilize a TBP–TFIIB–RAP30–pol II–promoter complex (11, 44). Consistent with these observations, we have shown (35) that RAP74 increases crosslinking of both pol II and RAP30 to nucleotide −19 of the adenovirus major late promoter. We now provide evidence that the stabilization of the preinitiation complex induced by RAP74 is accompanied by a reorganization of the complex that affects the position of pol II relative to that of the DNA. Our data indicate that binding of RAP74 to the preinitiation complex has two different effects on the crosslinking of pol II along promoter DNA. Firstly, the presence of RAP74 in our reactions increased the specific crosslinking of the 145-kDa subunit of pol II to nucleotides −19 (Fig. 2C), −15, and −5 (ref. 35 and data not shown). Consistent with previous observations (11, 35, 44), these data indicate that RAP74 can enhance the formation of stable preinitiation complexes. Secondly, and more importantly, the presence of RAP74 in our reactions induced two new crosslinking sites for pol II subunits along promoter DNA, one for the 180-kDa subunit at nucleotide −19 (Fig. 2C), and one for the 145-kDa subunit at nucleotides −39/−40 (Fig. 2B). This finding shows that RAP74 can induce a conformational change that repositions pol II relative to the DNA, bringing the 180-kDa subunit closer to position −19 and the 145-kDa subunit closer to positions −39/−40 (Fig. 4A). We do not yet know whether the conformational change affects the absolute position of the DNA, the polymerase, or both. Because RAP74 (15) and pol II (40) both contain DNA-binding domains, it is possible that contacts between these proteins and the DNA helix upstream of the TATA box sharpen the angle of the bend, thereby bringing the nucleotides at positions −39/−40 closer to the polymerase. It is equally possible that the association of RAP74 with the preinitiation complex repositions the polymerase, bringing the 145-kDa subunit closer to DNA in the vicinity of position −40. The N-terminal region of RAP74 (amino acids 1–205), which contains the RAP30-binding domain, is both necessary and sufficient for the reorganization of the preinitiation complex (Fig. 2). Interestingly, the same RAP74 region was found to be minimally required to support accurate transcription in vitro (15). This may be because the conformational change induced by RAP74 is important for transcription.
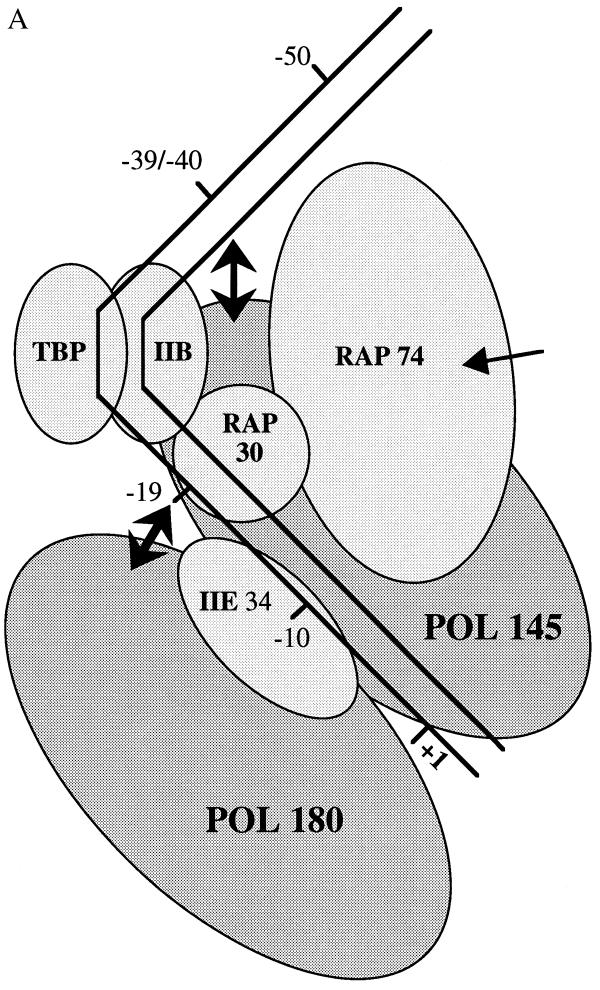
Figure 4.
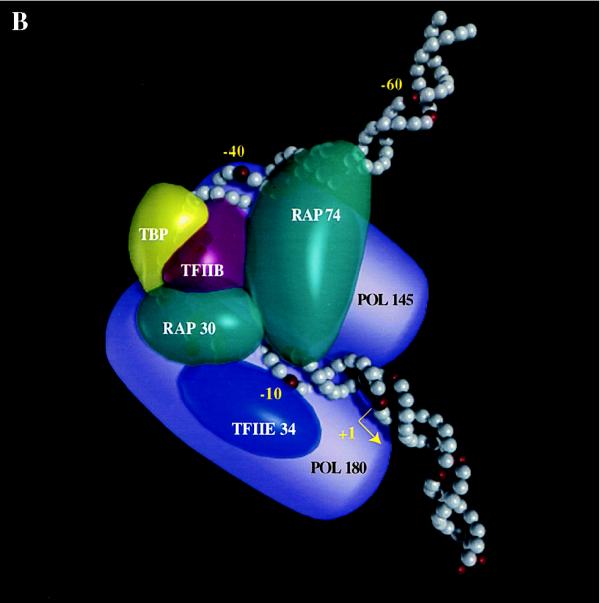
(A) Schematic representation of the conformational change induced by RAP74 in a TBP–TFIIB–RAP30–pol II–TFIIE–promoter complex. The entry of RAP74 in the complex modifies the position of the 145-kDa pol II subunit relative to positions −39/−40 upstream of the TATA box and that of the 180-kDa pol II subunit relative to position −19 downstream of the TATA box. Because we do not yet know whether the conformational change affects the absolute position of the DNA, the polymerase, or both, double-headed arrows are used to indicate the relative movement of pol II subunits. (B) Low resolution model of a TBP–TFIIB–TFIIF–pol II–TFIIE–promoter complex. The model accounts for the direct protein–protein interactions that have been reported between the various members of the general transcription machinery (1–6, 26). RAP74 is located in close proximity to the DNA on each side of a bend centered on the TATA box. RAP74, RAP30, and the 145-kDa subunit of pol II occupy a space on the same face of the DNA helix between positions −19 and −5. The placement of RAP74 in close proximity to pol145 is also consistent with the identification of a putative TFIIF interaction site in the IIc subunit (pol 140) of pol II in Drosophila (45). TFIIE34 and the 180-kDa subunit of pol II are placed on the other face of the helix between −19 and +1. The model is consistent with our EM data although no subunit structure can be resolved in the EM photo.
Our data suggest that the large subunit of TFIIF is involved in controlling the topological organization of the preinitiation complex. RAP74 was shown to bind directly to several members of the transcription machinery including RAP30 (10, 14, 15), pol II (15), TFIIB (13, 15), TBP-associated factorII250 (16), and TFIIE56 (17). RAP74 may orchestrate the organization of the preinitiation complex through these various protein-protein interactions. Proper molecular organization of the TBP–TFIIB–TFIIF–pol II–TFIIE complex (Fig. 4B) may be necessary for recruitment of TFIIH, formation of an open complex, initiation of RNA chain synthesis, stabilization of the RNA–DNA duplex during early elongation, and activation by some DNA-binding transcription factors. In addition, our results allow to speculate that TFIIF may play a central role in the molecular organization of the pol II holoenzyme. The previous finding that TFIIF is part of the so-called “mediator” in yeast (19) suggests that RAP30 and RAP74 may participate in the assembly of the holoenzyme by stabilizing the association of the mediator with the core pol II. As we have mentioned above, our data allow us to predict that TFIIF exists as a heterotetramer in the preinitiation complex. If this is indeed the case, a putative heterotetramer of TFIIF would contribute to a more extensive interface of interaction between the mediator and the core enzyme.
Compared with high resolution techniques such as x-ray crystallography and NMR, the method of photocrosslinking has the advantage of providing information on the molecular organization of large complexes assembled using full-length proteins or deletion mutants. In this report, we have analyzed the topology of a protein–DNA complex composed of 15–20 polypeptides, namely a TBP–TFIIB–TFIIF–pol II–TFIIE–promoter complex. The photocrosslinking experiments can also provide information on the putative role of various factors in complex assembly. Here, we have obtained information on the role of RAP74. However, and as it is also the case for high resolution techniques, the recruitment of additional factors such as TFIIA and TBP-associated factors may affect the positions of the other factors bound to the complex. Similarly, the positions of human pol II subunits may be different than those presented for the calf thymus pol II used in this study. In the future, it will be relevant to analyze larger complexes that will resemble more closely the complexes likely to be found in vivo. These analyses will continue to provide useful information on the molecular organization and the mechanisms of assembly of transcription complexes on class II gene promoters.
Acknowledgments
We thank members of our laboratories for helpful discussions and for sharing some reagents, P. Vandenberghe for computer-generated models, P. Magny for expert assistance with EM, and W. Home, C. Déry, and C. Gagnon for critical reading of the manuscript. This work was supported by grants from the Medical Research Council of Canada (to B.C. and J.G.) and the American Cancer Society (to Z.B.). B.C. is a Junior Research Scholar of the Fonds de la recherche en santé du Québec. J.G. is a Medical Research Council Distinguished Scientist and an International Research Scholar of the Howard Hughes Medical Institute. Z.B. is also supported by the Michigan State University and the Michigan State University Agricultural Experiment Station. F.R. holds a studentship from the Fonds pour la formation de chercheurs et l’aide à la recherche.
ABBREVIATIONS
RAP
RNA polymerase II-associated protein
TF
transcription factor
pol II
RNA polymerase II
TBP
TATA box-binding protein
EM
electron microscopy
References
- 1.Orphanides G, Lagrange T, Reinberg D. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 2.Conaway R C, Conaway J W. Annu Rev Biochem. 1993;62:161–190. doi: 10.1146/annurev.bi.62.070193.001113. [DOI] [PubMed] [Google Scholar]
- 3.Ranish J A, Hahn S. Curr Opin Cell Biol. 1996;6:151–158. doi: 10.1016/s0959-437x(96)80044-x. [DOI] [PubMed] [Google Scholar]
- 4.Pugh B F. Curr Opin Cell Biol. 1996;8:303–311. doi: 10.1016/s0955-0674(96)80002-0. [DOI] [PubMed] [Google Scholar]
- 5.Roeder R G. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 6.Hernandez N. Genes Dev. 1993;7:1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- 7.Aso T, Conaway J W, Conaway R C. FASEB J. 1995;9:1419–1428. doi: 10.1096/fasebj.9.14.7589983. [DOI] [PubMed] [Google Scholar]
- 8.Parvin J D, Shykind B M, Meyers R E, Kim J, Sharp P A. J Biol Chem. 1994;269:18414–18421. [PubMed] [Google Scholar]
- 9.Tyree C M, George C P, Lira-Devito L M, Wampler S L, Dahmus M E, Zawel L, Kadonaga J T. Genes Dev. 1993;7:1254–1265. doi: 10.1101/gad.7.7a.1254. [DOI] [PubMed] [Google Scholar]
- 10.Burton Z F, Killeen M, Sopta M, Ortolan L G, Greenblatt J. Mol Cell Biol. 1988;8:1602–1613. doi: 10.1128/mcb.8.4.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flores O, Lu H, Killeen M, Greenblatt J, Burton Z F, Reinberg D. Proc Natl Acad Sci USA. 1991;88:9999–10003. doi: 10.1073/pnas.88.22.9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Killeen M, Coulombe B, Greenblatt J. J Biol Chem. 1992;267:9463–9466. [PubMed] [Google Scholar]
- 13.Ha I, Roberts S, Maldonado E, Sun X, Kim L-U, Green M, Reinberg D. Genes Dev. 1993;7:1021–1032. doi: 10.1101/gad.7.6.1021. [DOI] [PubMed] [Google Scholar]
- 14.Fang S M, Burton Z F. J Biol Chem. 1996;271:11703–11709. doi: 10.1074/jbc.271.20.11703. [DOI] [PubMed] [Google Scholar]
- 15.Wang B Q, Burton Z F. J Biol Chem. 1995;270:27035–27044. doi: 10.1074/jbc.270.45.27035. [DOI] [PubMed] [Google Scholar]
- 16.Dikstein R, Ruppert S, Tjian R. Cell. 1996;84:781–790. doi: 10.1016/s0092-8674(00)81055-7. [DOI] [PubMed] [Google Scholar]
- 17.Maxon M E, Goodrich J A, Tjian R. Genes Dev. 1994;8:515–524. doi: 10.1101/gad.8.5.515. [DOI] [PubMed] [Google Scholar]
- 18.Sopta M, Carthew R W, Greenblatt J. J Biol Chem. 1985;260:10353–10361. [PubMed] [Google Scholar]
- 19.Kim Y J, Bjorklund S, Li Y, Sayre M H, Kornberg R D. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 20.Koleske A J, Young R A. Nature (London) 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 21.Ossipow V, Tassan J-P, Nigg E A, Schibler U. Cell. 1995;83:137–146. doi: 10.1016/0092-8674(95)90242-2. [DOI] [PubMed] [Google Scholar]
- 22.Zhu H, Joliot V, Prywes R. J Biol Chem. 1994;269:3489–3497. [PubMed] [Google Scholar]
- 23.Joliot V, Demma M, Prywes R. Nature (London) 1995;373:632–635. doi: 10.1038/373632a0. [DOI] [PubMed] [Google Scholar]
- 24.Price D H, Sluder A E, Greenleaf A L. J Biol Chem. 1987;262:3244–3255. [PubMed] [Google Scholar]
- 25.Price D H, Sluder A E, Greenleaf A L. Mol Cell Biol. 1989;9:1465–1475. doi: 10.1128/mcb.9.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nikolov D B, Burley S K. Proc Natl Acad Sci USA. 1997;94:15–22. doi: 10.1073/pnas.94.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim Y, Geiger J H, Hahn S, Sigler P B. Nature (London) 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 28.Kim J L, Nikolov D B, Burley S K. Nature (London) 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 29.Bagby S, Kim S, Maldonado E, Tong K I, Reinberg D, Ikura M. Cell. 1995;82:857–867. doi: 10.1016/0092-8674(95)90483-2. [DOI] [PubMed] [Google Scholar]
- 30.Nikolov D B, Chen H, Halay E D, Usheva A A, Hisatake K, Lee D K, Roeder R G, Burley S K. Nature (London) 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- 31.Tan S, Hunziker Y, Sargent D F, Richmond T J. Nature (London) 1996;381:127–134. doi: 10.1038/381127a0. [DOI] [PubMed] [Google Scholar]
- 32.Geiger J H, Hahn S, Lee S, Sigler P B. Science. 1996;272:830–836. doi: 10.1126/science.272.5263.830. [DOI] [PubMed] [Google Scholar]
- 33.Tang H, Sun X, Reinberg D, Ebright R H. Proc Natl Acad Sci USA. 1996;93:1119–1124. doi: 10.1073/pnas.93.3.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coulombe B, Li J, Greenblatt J. J Biol Chem. 1994;269:19962–19967. [PubMed] [Google Scholar]
- 35.Robert F, Forget D, Li J, Greenblatt J, Coulombe B. J Biol Chem. 1996;271:8517–8520. doi: 10.1074/jbc.271.15.8517. [DOI] [PubMed] [Google Scholar]
- 36.Lagrange T, Kim T-K, Orphanides G, Ebright Y W, Ebright R H, Reinberg D. Proc Natl Acad Sci USA. 1996;93:10620–10625. doi: 10.1073/pnas.93.20.10620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oelgesclager T, Chiang C M, Roeder R G. Nature (London) 1996;382:735–738. doi: 10.1038/382735a0. [DOI] [PubMed] [Google Scholar]
- 38.Bartholomew B, Kassavetis G A, Braun B R, Geiduschek E P. EMBO J. 1990;9:2197–2205. doi: 10.1002/j.1460-2075.1990.tb07389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dubochet J, Ducommun M, Zollinger M, Kellenberger M. J Ultrastruct Res. 1971;35:145–167. doi: 10.1016/s0022-5320(71)80148-x. [DOI] [PubMed] [Google Scholar]
- 40.Killeen M T, Greenblatt J. Mol Cell Biol. 1992;12:30–37. doi: 10.1128/mcb.12.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guzikevich-Guerstein G, Shakked Z. Nat Struct Biol. 1996;3:32–37. doi: 10.1038/nsb0196-32. [DOI] [PubMed] [Google Scholar]
- 42.Conaway J W, Conaway R C. J Biol Chem. 1989;264:2357–2362. [PubMed] [Google Scholar]
- 43.Flores O, Ha I, Reinberg D. J Biol Chem. 1990;265:5629–5634. [PubMed] [Google Scholar]
- 44.Tan S, Aso T, Conaway R C, Conaway J W. J Biol Chem. 1994;269:25684–25691. [PubMed] [Google Scholar]
- 45.Skantar A M, Greenleaf A L. Gene Expression. 1995;5:49–69. [PMC free article] [PubMed] [Google Scholar]