The Complement Factor H R1210C Mutation Is Associated With Atypical Hemolytic Uremic Syndrome (original) (raw)
Abstract
Mutations in the gene encoding complement factor H (CFH) that alter the C3b/polyanions-binding site in the C-terminal region impair the capacity of factor H to protect host cells. These mutations are also strongly associated with atypical hemolytic uremic syndrome (aHUS). Although most of the aHUS-associated CFH mutations seem “unique” to an individual patient or family, the R1210C mutation has been reported in several unrelated aHUS patients from distinct geographic origins. Five aHUS pedigrees and 7 individual aHUS patients were analyzed to identify potential correlations between the R1210C mutation and clinical phenotype and to characterize the origins of this mutation. The clinical phenotype of aHUS patients carrying the R1210C mutation was heterogeneous. Interestingly, 12 of the 13 affected patients carried at least one additional known genetic risk factor for aHUS. These data are in accord with the 30% penetrance of aHUS in R1210C mutation carriers, as it seems that the presence of other genetic or environmental risk factors significantly contribute to the manifestation and severity of aHUS in these subjects. Genotype analysis of CFH and CFHR3 polymorphisms in the 12 unrelated carriers suggested that the R1210C mutation has a single origin. In conclusion, the R1210C mutation of complement factor H is a prototypical aHUS mutation that is present as a rare polymorphism in geographically separated human populations.
HUS is characterized by thrombocytopenia, Coomb's test negative microangiopathic hemolytic anemia, and acute renal failure. The typical form of HUS follows a diarrheal prodrome and is associated with Shiga toxin-producing 0157:H7 Escherichia coli infections. However, 5% to 10% of HUS patients lack an association with infection. This atypical form of HUS (aHUS) occurs in both adults and in young children and has a poor prognosis. Recurrent episodes of aHUS are common with a mortality rate that approaches 30%. Endothelial cell injury appears to be the primary event in the pathogenesis of HUS. The endothelial damage triggers a cascade of events that result in the formation of platelet-fibrin hyaline microthrombi that occlude arterioles and capillaries. A hallmark of HUS is the presence of schistocytes (fragmented cells) that generate as the red blood cells traverse these partially occluded microvessels.1
aHUS is associated with mutations or polymorphisms in the genes encoding the complement regulatory proteins factor H (CFH),2–9 membrane cofactor protein (MCP, CD46),10–13 and factor I (IF, CFI),14,15 and with mutations in the complement activating components factor B (BF, CFB)16 and C3 (C3).17 Importantly, mutations in CFH, MCP (CD46), and IF (CFI) are loss-of-function mutations, whereas mutations in BF (CFB) are gain-of-function mutations. In addition, autoantibodies and deletion of the CFHR1-CFHR3 genes are associated with aHUS.18–20
Missense mutations in the exons encoding the C-terminal region of factor H are the commonest genetic alteration among aHUS patients. Carriers of these CFH mutations express factor H molecules that present normal regulatory activity in plasma but a limited capacity to protect cells from complement lysis.4,7,21 The combination of both an active complement system in plasma and a defective protection of cellular surfaces is critical in the pathogenesis of aHUS.22 In a situation that triggers complement activation, such as during bacterial or viral infections, exposure to drugs that cause endothelial activation and damage or the presence of immune complexes, deposition, and amplification of C3b on the microvasculature cellular surfaces cannot be controlled, and this results in tissue damage and destruction.
CFH R1210C is the prototypic aHUS-associated CFH mutation. Functional analysis demonstrated that R1210C mutant factor H has normal cofactor activity but defective binding to C3b, heparin, and endothelial cells.4,7 A unique feature of the R1210C factor H mutant is its capacity to form covalent heterodimers with serum albumin through the cysteine residue generated by the mutation.7 Interestingly, CFH R1210C is a common mutation that has been described in several unrelated aHUS patients from different countries.2,3,7,23,24
Here we have both examined whether there is a common clinical phenotype associated with CFH R1210C and analyzed the origins of this mutation. To this end, we evaluated in 13 aHUS patients and 16 healthy carriers the clinical phenotypes, determined the CFH genotypes associated with CFH R1210C, and analyzed the concurrence of other known aHUS risk factors. These include mutations and polymorphisms in the CFH, MCP (CD46), and IF (CFI) genes, and genomic rearrangements in the CFH-CFHR1–5 genomic region.
RESULTS AND DISCUSSION
Our series of aHUS patients carrying the CFH R1210C mutation includes 13 affected individuals comprising 11 unrelated individuals (R016, S006, H29IV-1, D45348, D40308, S198, S197, S223, S196, FHUSA11, and 6089) and two brothers (F106 and F108). These patients were identified in routine CFH mutation analyses of aHUS patients. They belong to five aHUS cohorts, including approximately 800 unrelated patients, from France, Germany, Italy, Spain, and the United Kingdom (see “Concise Methods”). The overall number of patients who carry the CFH R1210C mutation ranges from 1% to 4% in the different cohorts, which represents an estimable 5% to 15% of all of the patients carrying mutations in CFH and illustrates the high prevalence of the CFH R1210C mutation among atypical HUS patients.
Age at onset ranges from 6 mo to 53 yr, with 6 patients developing the disease under 13 yr old and 7 patients over 21 yr old. Eight patients developed end-stage renal disease needing dialysis, following their initial presentation (R016 D45348, S198, S223, and FHUSA11) or following recurrent episodes (S006, S197, and F108). Three patients developed chronic renal insufficiency needing dialysis following their initial presentation (S196 and D40308) or several recurrences (H29IV-1). In contrast, patient 6089 had a single episode and recovered renal function without sequelae and patient F106 had two episodes that also resolved without any renal sequelae. Time interval to reach end-stage renal disease or chronic renal insufficiency from the initial episode ranged from 3 mo (H29IV-1) to 15 yr (R016 and F108). Four patients (S198, R016, D45348, and FHUSA11) have been transplanted, and three of them had a recurrence in the allograft. Transplantation was successful without recurrence of the disease in patient S198 and also in patient D45348 when he was transplanted a second time. In agreement with previous reports,1 there is an elevated frequency of recurrences in the allograft among CFH R1210C mutation carriers. Patient D45348 was transplanted twice and illustrates the risks associated with living related donors who share aHUS mutations with the receptor. D45348 had a recurrence in the allograft from her mother, with whom she shares two aHUS risk factors (the CFH R1210C mutation and the MCPggaac haplotype, Figure 1). In a second unrelated cadaver renal transplant (unlikely carrying the CFH R1210C mutation), she has had no episodes of recurrent HUS and the graft is functioning well.
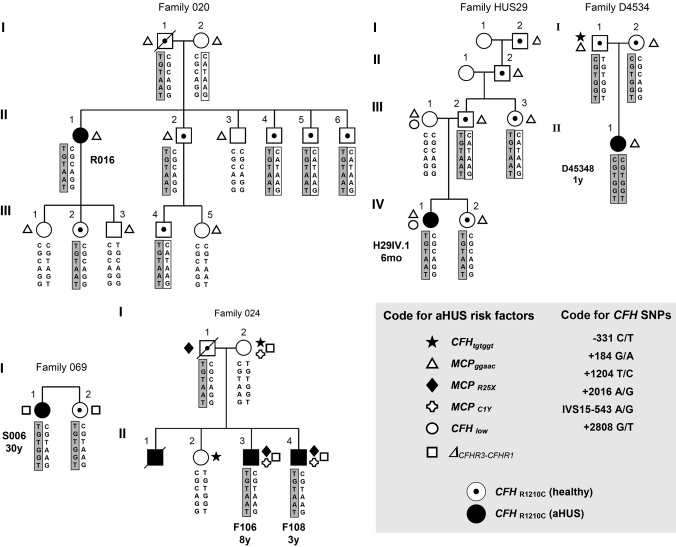
Figure 1.
aHUS pedigrees carrying CFH R1210C. Patient code and age of onset are indicated. The 6 SNPs that compose the CFH haplotypes are represented in the columns. CFH haplotypes that carry the R1210C mutation are squared and shaded. The protective CFH-H2 haplotype is squared and blank. A code for all symbols is included in the figure.
As a whole, data summarized in Table 1 illustrate significant heterogeneity in the clinical phenotype and severity of the disease in the 13 affected individuals. This does not support a correlation between clinical phenotype and the CFH R1210C mutation. Moreover, CFH R1210C mutation analysis in the relatives of patients R016, F106/F108, H29IV-1, D45348, and S006 demonstrated the presence of 16 asymptomatic CFH R1210C mutation carriers (Figure 1), suggesting that presence of other genetic or environmental risk factors contributes to the manifestation and severity of aHUS in CFH R1210C mutation carriers. Penetrance of the disease among CFH R1210C mutation carriers was estimated to be approximately 30%.
Table 1.
Clinical data in aHUS patients carrying the CFH_R1210C_ mutation
| ID | Gender | Biopsya | Onset | Triggering Factor | Time Interval to ESRD/CRI | Renal Statusb | Recurrences (no.) | Renal Transplant | Recurrence After Transplantc |
|---|---|---|---|---|---|---|---|---|---|
| S198 | M | — | 34 yr | Vomiting | During 1st episode | ESRD | No | Yes | No |
| S197 | F | Yes | 6 mo | ? | 3 yr | ESRD | Yes (several) | No | — |
| S196 | F | Yes | 53 yr | Treatment RAd | During 1st episode | CRI | No | No | — |
| S223 | M | — | 43 yr | ? | During 1st episode | ESRD | No | No | — |
| F106 | M | — | 8 yr | Bronchopneumonia | — | Normal | Yes (1) | No | — |
| F108 | M | Yes | 3 yr | Diarrhea/vomiting | 15 yr | ESRD | Yes (1) | No | — |
| S006 | F | Yes | 30 yr | Flu-like episode | 4 mo | ESRD | Yes (1) | No | — |
| R016 | F | Yes | 31 yr | Vomiting | 15 yr | ESRD | No | Yes | Yes (3 mo) |
| D40308 | F | — | 40 yr | Postpartum | During 1st episode | CRI | No | No | — |
| D45348 | F | — | 1 yr | ? | During 1st episode | ESRD | No | Twice | 1st yes (27 mo) 2nd no |
| 6089 | M | Yes | 13 yr | Infection | — | Normal | No | No | — |
| FHUSA11 | F | — | 21 yr | ? | During 1st episode | ESRD | No | Yes | Yes (6 d) |
| H29IV-1 | F | — | 6 mo | ? | 3 mo | CRI | Yes (3) | No | — |
To determine whether mutations or polymorphisms in other complement genes, previously associated with aHUS, influence the development and severity of aHUS in the CFH R1210C mutation carriers, we undertook mutation screening of MCP (CD46) and IF (CFI) and genotyped all samples for the MCPggaac, CFH TGTGGT, and Δ_CFRH3-CFRH1_ polymorphisms.10,20,22 Figures 1 and 2 and Table 2 summarize these data. Two of our patients are also compound heterozygotes for mutations in MCP (F106 and F108), and a third one carries a mutation in the second CFH allele (H29IV-1) (Table 2).
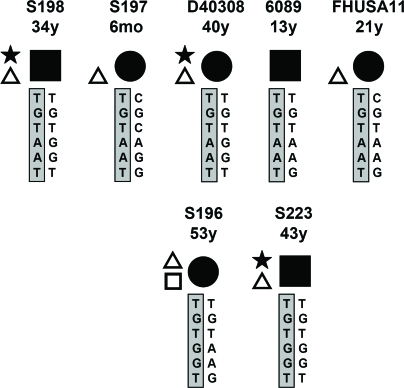
Figure 2.
Individual aHUS patients carrying CFH R1210C. Most probable CFH haplotypes indicated. Patient code and onset age are shown. The CFH haplotype that carries CFH R1210C is shaded. Code for symbols is as in Figure 1.
Table 2.
Complement and genetic data in aHUS patients carrying the CFH R1210C mutation
| ID | C3 Levels (80–177 mg/dl) | Additional aHUS Risk Factorsa | |||
|---|---|---|---|---|---|
| Mutations | MCP ggaac | CFH TGTGGT | Δ_CFHR3-CFHR1_ | ||
| S198 | 82 | No | Yes (homozygous) | Yes | ? |
| S197 | 94 | No | Yes (homozygous) | No | No |
| S196 | 101 | No | Yes (heterozygous) | No | Yes (heterozygous) |
| S223 | 92 | No | Yes (homozygous) | Yes | No |
| F106 | 61 | MCP R25X/MCP C1Y | No | No | Yes (heterozygous) |
| F108 | 61 | MCP R25X/MCP C1Y | No | No | Yes (heterozygous) |
| S006 | 75 | No | ? | No | Yes (heterozygous) |
| R016 | 108 | No | Yes (heterozygous) | No | No |
| D40308 | ? | No | Yes (homozygous) | Yes | No |
| D45348 | ? | CFH R1210C (homozygous) | No | No | No |
| 6089 | 90 | No | No | No | No |
| FHUSA11 | 73 | No | Yes (heterozygous) | No | No |
| H29IV-1 | 58 | CFH low | Yes (homozygous) | No | No |
Convalescent complement C3 levels are clearly decreased only in patients who also carry MCP mutations (F106 and F108) or CFH low expression alleles (H29IV-1), suggesting a constitutive dysregulation of the alternative complement pathway in these patients (Table 1). Interestingly, these three individuals and D45348, who is homozygous for CFH R1210C, have an early onset of the disease. All but one of the 13 aHUS patients included in this report carry additional aHUS risk factors and, in general, these affected individuals tend to have more aHUS risk factors than their asymptomatic relatives who are CFH R1210C carriers (Figure 1). With this in mind, it is interesting that patient 6089, who does not carry additional known aHUS risk factors, had only a single aHUS episode and recovered renal function without sequelae (Table 1). In addition to CFH haplotypes that predispose to aHUS, we have recently reported the existence of CFH haplotypes that protect from aHUS (_CFH_-H2 in Table 3).22 Interestingly, some of the asymptomatic CFH R1210C carriers also have the aHUS low-risk CFH CATAAG haplotype (Figure 1).
Table 3.
CFH haplotype frequencies
| CFH Haplotypesa | CFH SNPs | Haplotype Frequencyb (n = 317) | |||||
|---|---|---|---|---|---|---|---|
| −331C>T | c.184G>A | c.1204C>T | c.2016A>G | IVS15–543 | c.2808G>T | ||
| H1 | C | G | C | A | G | G | 0.257 |
| H2 | C | A | T | A | A | G | 0.204 |
| H3 | T | G | T | G | G | T | 0.207 |
| H4a | C | G | T | A | A | G | 0.139 |
| H4b | T | G | T | A | A | G | 0.071 |
| H5 | T | G | C | A | G | G | 0.037 |
| H6 | C | G | T | A | G | G | 0.021 |
| H7 | T | G | T | A | G | G | 0.016 |
| H8 | C | G | T | G | G | T | 0.015 |
| H9 | C | A | T | G | G | T | 0.008 |
| H10 | T | G | T | G | G | G | 0.006 |
| H11 | C | A | C | A | G | G | 0.005 |
Therefore, the data presented here are consistent with previous reports showing that affected individuals carry more than one aHUS risk factor10,16,25 and strongly support the concept that susceptibility to aHUS results from the additive effect of multiple genetic factors involving plasma and membrane-associated complement regulators. Within the families included in this report there are, however, relatives who apparently share identical genetic susceptibility profiles with the aHUS patients and have not developed the disease (Figure 1). As indicated before, this suggests that there are other, as yet unidentified, genetic or environmental risk factors contributing to manifestation of the disease.
Haplotype analysis using six CFH and two CFHR3 intragenic SNPs enable us to identify three extended CFH haplotypes segregating with CFH R1210C in the affected individuals (Table 2). In families 020, 024, and HUS29, CFH R1210C was associated with a rare CFH haplotype (CFH TGTAAT, H4b* in Table 4) that was absent in a control population of 317 unrelated individuals (Figure 1; Table 3) and differs from haplotype H4b only at SNP 2808G>T. Interestingly, this unique CFH haplotype was also present in patients S198, S197, D40308, 6089, and FHUSA11, which suggests that CFH R1210C was also associated with the CFH TGTAAT haplotype in these individuals. Family D4534 illustrates that the CFH CGTGGT haplotype is also associated with the CFH R1210C mutation (Figure 1). Finally, patient S223, who is a CFH TGTGGT homozygote, unambiguously associates the CFH TGTGGT haplotype with CFH R1210C (Figure 2). Haplotype CFH TGTGGT is also likely associated with the CFH R1210C in patients S006 and S196 (Figures 1 and 2). This conclusion is supported by the observation that patients S006 and S196 lack one copy of the CFHR1 and CFHR3 genes, which is associated with the CFH CGTAAG and CFH TGTAAG haplotypes (Martinez-Barricarte et al., unpublished observations), making the most probable genotypes for S006 and S196, CFH TGTGGT/CFH CGTAAG and CFH TGTGGT/CFH TGTAAG, respectively.
Table 4.
CFH-CFHR3 haplotypes associated with the CFH R1210C mutation
| Patient | Haplotype | CFH SNPs | CFHR3 SNPs | ||||||
|---|---|---|---|---|---|---|---|---|---|
| −331C>T | c.184G>A | c.1204C>T | c.2016A>G | IVS15–543 | c.2808G>T | IVS3–1106G>A | IVS3–1146T>A | ||
| S198 | H4b* | T | G | T | A | A | T | G | T |
| S197 | H4b* | T | G | T | A | A | T | G | T |
| D40308 | H4b* | T | G | T | A | A | T | ? | ? |
| 6089 | H4b* | T | G | T | A | A | T | ? | ? |
| FHUSA11 | H4b* | T | G | T | A | A | T | G | T |
| R016 | H4b* | T | G | T | A | A | T | G | T |
| F106 | H4b* | T | G | T | A | A | T | G | T |
| H29IV-1 | H4b* | T | G | T | A | A | T | G | T |
| S006 | H3 | T | G | T | G | G | T | G | T |
| S196 | H3 | T | G | T | G | G | T | G | T |
| S223 | H3 | T | G | T | G | G | T | G | T |
| D45348 | H8 | C | G | T | G | G | T | G | T |
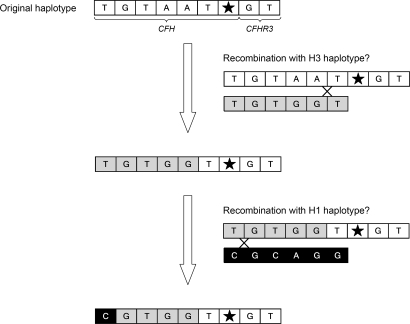
Alignment of the three extended CFH haplotypes associated with the CFH R1210C mutation illustrates that they are identical around the mutation site, suggesting that the three alleles may have a common origin and have been generated by recombination from a single original CFH chromosome carrying the CFH TGTAAT haplotype (Table 4; Figure 3). Because the CFH TGTAAT haplotype is strongly associated with the CFH R1210C mutation and is extremely rare in whites, we postulate that the CFH R1210C mutation originated in a different population, where the CFH TGTAAT haplotype is relatively frequent, and that was introduced in the European populations later on. However, we cannot rule out the possibility of a mutational event in this rare chromosome in whites.
Figure 3.
Model for the origin of the three CFH haplotypes associated with CFH R1210C. This model shows 2 putative recombination events that may have generated the 3 CFH haplotypes found associated with CFH R1210C in our patients. H1 and H3 are relatively frequent CFH haplotypes in the general population (Table 3).
In conclusion, this study shows that aHUS patients carrying CFH R1210C do not have a common clinical phenotype. This heterogeneity, the incomplete penetrance of the disease among the CFH R1210C mutation carriers and the identification of additional aHUS-associated risk factor in the affected individuals, is a common feature seen in all aHUS patients. The relatively high prevalence of the CFH R1210C mutation among aHUS patients suggests that CFH R1210C is a rare functional polymorphism in the general population that specifically predisposes to development of aHUS.
CONCISE METHODS
Patients
All participating centers have IRB approval for the studies included in this report. Our study includes five aHUS pedigrees and seven individual aHUS patients carrying the CFH R1210C mutation from Italy, United Kingdom, Australia, Germany, France, and the United States. These R1210C pedigrees and patients were identified in routine CFH mutation analysis of aHUS patients. aHUS was diagnosed because of the presence of one or more episodes of microangiopathic hemolytic anemia and thrombocytopenia defined on the basis of hematocrit <30%, hemoglobin <10 mg/dl, serum lactate dehydrogenase >460U/L, undetectable haptoglobin, fragmented erythrocytes in the peripheral blood smear, and platelet count <150,000/μl, associated with acute renal failure. Patients with Stx-HUS, defined as the presence of Shiga toxin in the stools (by the Vero cell assay) and/or of serum antibodies against Shiga toxin (by ELISA) and/or LPS (O157, O26, O103, O111, and O145, by ELISA) were excluded. None of the aHUS patients carrying the CFH R1210C had thrombotic thrombocytopenic purpura-like manifestations.
Family 024.
There are three affected individuals (F106, II-3 and F108, II-4) in this pedigree, two of whom are alive. Patients F106 and F108 first presented at the age of 8 and 3 yr, respectively. The family history revealed that a younger brother died of HUS at the age of 6 yr and the father, who had hepatitis C, was noted on various occasions to have thrombocytopenia and anemia. In addition, microscopic proteinuria/hematuria and a slightly elevated creatinine (1.5 mg/dl) were also reported. A paternal uncle died at the age of 28 yr from renal failure.
Family 020.
This pedigree included several asymptomatic carriers of the R1210C mutation but only one affected individual (II-1, R016), who was first admitted to hospital at the age of 31. Fifteen years later, after a progressive deterioration in renal function, she started peritoneal dialysis. She received a cadaveric renal transplant that was unsuccessful because of a recurrence of HUS. No relevant family history was reported. The same mutation has been found in the father, in four of five healthy siblings, and in one of her three children.
Family 069.
This pedigree includes a female (S006), who first presented at 30 yr old. After two recurrences, she started chronic hemodialysis. No relevant clinical history was reported in her family. The same mutation has been found in the healthy sister.
Family HUS29.
This pedigree has four generations of carriers and only one affected (H29IV-1) who first presented with HUS at 6 mo of age. She was treated with peritoneal dialysis and fresh plasma infusions (20 ml/kg 3 times a week for 2 wk) and completely recovered renal function. She had three recurrences in the following 3 mo and is currently treated with peritoneal dialysis and plasma infusions every 6 wk.
Family D4534.
This consanguineous pedigree is remarkable, comprising two healthy R1210C carrier parents and one affected R1210C homozygote daughter (D45348) who presented with HUS at the age of 1 yr. She received a live related transplant from her mother at the age of 4 yr. This was complicated by five episodes of recurrent HUS, each of which was treated with plasma exchange, but the graft lasted only 2 yr 3 mo. She then returned to hemodialysis. She received a cadaver renal transplant at the age of 10. There have been no episodes of recurrent HUS, and the graft is functioning well.
Patient D4038.
She presented at the age of 40 with postpartum HUS. She did not recover renal function and remains dialysis dependent.
Patient S198 is a man who first presented at the age of 34. He did not recover renal function and started hemodialysis. He received a cadaveric renal transplant. Initial immunosuppression was with steroids, cyclosporine, and mycophenolate mofetil. The outcome of the transplant has been good, with no episodes of recurrent disease. No relevant clinical history was reported in his family.
Patient S197 is a female who first presented at 6 mo of age. After several recurrences over the next 3 yr, she developed end-stage renal failure. She is currently on dialysis. There is no relevant family history.
Patient S223 is a man who had his first episode of HUS at the age of 43. He did not recover renal function and is currently on peritoneal dialysis. There is no relevant family history.
Patient S196 is a woman who had her first episode of HUS at the age of 53. Her renal function did not recover. She is currently on chronic hemodialysis.
Patient FHUSA11 was referred at the age of 21 with HUS. She did not recover renal function and started hemodialysis. At age 23, she received a cadaveric renal transplant. She lost the allograft within 6 d to recurrent HUS.
Patient 6089 is a male who developed HUS at 13 yr. His condition resolved spontaneously without either dialysis or plasma therapy. Eighteen months later, the only detectable abnormality was microscopic proteinuria.
CFH and MCP Genotyping
A set of six SNPs, representing a minimal informative set for genetic variations within CFH (c.-331C>T, c.184G>A, c.1204C>T, c.2016A>G, IVS15–543C>T, and c.2808G>T), and two additional SNPs in CFHR3 (rs370789 and rs389897) were genotyped on genomic DNA using Taqman probes (Applied Biosystems) and real-time PCR equipment (IQ5, Applied Biosystems), according to the manufacturer's specifications, or by automatic DNA sequencing of PCR derived amplicons in a sequencer (ABI 3730, Applied Biosystems, Fosters City, USA) using dye terminator cycle sequencing kit (Applied Biosystems, Fosters City, USA). CFH haplotypes in the pedigrees were determined by segregation analysis. CFH haplotypes in individual aHUS patients were assigned on the basis of the CFH haplotype frequencies in a sample of 317 Spanish controls (Table 3). The aHUS-associated MCPggaac haplotype was genotyped as described previously.10
CFHR3 Genotyping
Two SNPs in the intron 3 of CFHR3 (rs370789 IVS3–1106G>A and rs389897 IVS3–1046T>A) were genotyped on genomic DNA by automatic DNA sequencing of PCR-derived amplicons in a sequencer (ABI 3730, Applied Biosystems) using dye terminator cycle sequencing kit (Applied Biosystems). Copy number variation of CFHR1 and CFHR3 was analyzed by MLPA (Multiplex Ligation-Dependent Probe Amplification) as described previously.20
DISCLOSURES.
The authors declare that no financial conflict of interest exists.
Acknowledgments
The authors thank all patients and the collaborating clinicians for their participation in this study as well as the members of Secugen S.L. and the DNA sequencing laboratory at the CIB for invaluable technical assistance with patient genotyping. S.R.deC. is supported by the Spanish Ministerio de Educación y Cultura (SAF2005–00913); M.N. and G.R. by grants from Istituto Superiore della Sanità (Project no.526D/9) and NIH (Project no. DK71221); P.F.Z. by the Deutsche Forschungsgemeinschaft and KIDNEEDS Iowa; V.F.-B. by the Délégation Régionale à la Recherche Clinique, Assistance Publique-Hôpitaux de Paris (Grants PHRC AOM 05130); and T.G. by the Robin Davies Trust and the Foundation for Children with atypical HUS.
Published online ahead of print. Publication date available at www.jasn.org.
REFERENCES
- 1.Noris M, Remuzzi G: Hemolytic uremic syndrome. Lancet 16: 1035–1050, 2005 [DOI] [PubMed] [Google Scholar]
- 2.Caprioli J, Bettinaglio P, Zipfel PF, Amadei B, Daina E, Gamba S, Skerka C, Marziliano N, Remuzzi G, Noris M: The molecular basis of familial hemolytic uremic syndrome: mutation analysis of factor H gene reveals a hot spot in short consensus repeat 20. J Am Soc Nephrol 12: 297–307, 2001 [DOI] [PubMed] [Google Scholar]
- 3.Caprioli J, Castelletti F, Bucchioni S, Bettinaglio P, Bresin E, Pianetti G, Gamba S, Brioschi S, Daina E, Remuzzi G, Noris M: Complement factor H mutations and gene polymorphisms in haemolytic uraemic syndrome: the C-257T, the A2089G and the G2881T polymorphisms are strongly associated with the disease. Hum Mol Genet 12: 3385–3395, 2003 [DOI] [PubMed] [Google Scholar]
- 4.Manuelian T, Hellwage J, Meri S, Caprioli J, Noris M, Heinen S, Jozsi M, Neumann HPH, Remuzzi G, Zipfel PF: Mutations in factor H reduce binding affinity to C3b and heparin and surface attachment to endothelial cells in hemolytic uremic syndrome. J Clin Invest 111: 1181–1190, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perez-Caballero D, Gonzalez-Rubio C, Gallardo ME, Vera M, Lopez-Trascasa M, Rodriguez de Cordoba S, Sanchez-Corral P: Clustering of missense mutations in the C-terminal region of factor H in atypical hemolytic uremic syndrome Am J Hum Genet 68: 478–484, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richards A, Buddles M, Donne R, Kaplan B, Kirk E, Venning M, Tielemans C, Goodship JA, Goodship TH: Factor H mutations in hemolytic uremic syndrome cluster in exons 1820, a domain important for host cell recognition. Am J Hum Genet 68: 485–490, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sánchez-Corral P, Pérez-Caballero D, Huarte O, Simckes AM, Goicoechea de Jorge E, López-Trascasa M, Rodriguez de Córdoba S: Structural and functional characterization of factor H mutations associated with atypical hemolytic uremic syndrome. Am J Hum Genet 71: 1285–1295, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Venables JP, Strain L, Routledge D, Bourn D, Powell HM, Warwicker P, Diaz-Torres ML, Sampson A, Mead P, Webb M, Pirson Y, Jackson MS, Hughes A, Wood KM, Goodship JA, Goodship THJ: Atypical haemolytic uraemic syndrome associated with a hybrid complement gene. PLoS Med 3: 836–844, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warwicker P, Goodship THJ, Donne RL, Pirson Y, Nicholls A, Ward RM, Turnpenny P, Goodship JA: Genetic studies into inherited and sporadic hemolytic uremic syndrome. Kidney Int 53: 836–844, 1998 [DOI] [PubMed] [Google Scholar]
- 10.Esparza-Gordillo J, Goicoechea de Jorge E, Buil A, Berges LC, Lopez-Trascasa M, Sanchez-Corral P, Rodriguez de Cordoba S: Predisposition to atypical hemolytic uremic syndrome involves the concurrence of different susceptibility alleles in the regulators of complement activation gene cluster in 1q32. Hum Mol Genet 14: 703–712, 2005 [DOI] [PubMed] [Google Scholar]
- 11.Fremeaux-Bacchi V, Kemp EJ, Goodship JA, Dragon-Durey MA, Strain L, Loirat C, Deng HW, Goodship THJ: The development of atypical haemolytic-uraemic syndrome is influenced by susceptibility factors in factor H and membrane cofactor protein: evidence from two independent cohorts. J Med Genet 42: 852–856, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noris M, Brioschi S, Caprioli J, Todeschini M, Bresin E, Porrati F, Gamba S, Remuzzi G: Familial haemolytic uraemic syndrome and an MCP mutation. Lancet 362: 1542–1547, 2003 [DOI] [PubMed] [Google Scholar]
- 13.Richards A, Kemp EJ, Liszewski MK, Goodship JA, Lampe AK, Decorte R, Muslumanogglu MH, Kavukcu S, Filler G, Pirson Y, Wen LS, Atkinson JP, Goodship THJ: Mutations in human complement regulator, membrane cofactor protein (CD46), predispose to development of familial hemolytic uremic syndrome. Proc Natl Acad Sci U S A 100: 12966–12971, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fremeaux-Bacchi V, Dragon-Durey MA, Blouin J, Vigneau C, Kuypers D, Boudailliez B, Loirat C, Rondeau E, Fridman, WH: Complement factor I: a susceptibility gene for atypical haemolytic uraemic syndrome. J Med Genet 41: e84, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kavanagh D, Kemp EJ, Mayland E, Winney RJ, Duffield JS, Warwick G, Richards A, Ward R, Goodship JA, Goodship THJ: Mutations in complement factor I predispose to development of atypical hemolytic uremic syndrome. J Am Soc Nephrol 16: 2150–2155, 2005 [DOI] [PubMed] [Google Scholar]
- 16.Goicoechea de Jorge E, Harris CL, Esparza-Gordillo J, Carreras L, Arranz EA, Garrido CA, Lopez-Trascasa M, Sanchez-Corral P, Morgan BP, Rodriguez de Cordoba S: Gain-of-function mutations in complement factor B are associated with atypical hemolytic uremic syndrome. Proc Natl Acad Sci U S A 104: 240–245, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fremeaux-Bacchi V, Regnier C, Blouin J, Dragon-Durey MA, Fridman WH, Janssen B, Loirat C: Protective or aggressive: paradoxical role of C3 in atypical hemolytic uremic syndrome. Mol Immunol 44: 172, 2007 [Google Scholar]
- 18.Dragon-Durey, M-A, Loirat C, Cloarec S, Macher, M-A, Blouin J, Nivet H, Weiss L, Fridman WH, Fremeaux-Bacchi V: Anti-factor H autoantibodies associated with atypical hemolytic uremic syndrome. J Am Soc Nephrol 16: 555–563, 2005 [DOI] [PubMed] [Google Scholar]
- 19.Jozsi M, Strobel S, Dahse, H-M, Liu, W-s, Hoyer PF, Oppermann M, Skerka C, Zipfel PF: Anti-factor H autoantibodies block C-terminal recognition function of factor H in hemolytic uremic syndrome. Blood 110: 1516–1518, 2007 [DOI] [PubMed] [Google Scholar]
- 20.Zipfel PF, Edey M, Heinen S, Józsi M, Richter H, Misselwitz J, Hoppe B, Routledge D, Strain L, Hughes AE, Goodship JA, Licht C, Goodship THJ, Skerka C: Deletion of complement factor H related genes CFHR1 and CFHR3 is associated with atypical hemolytic uremic syndrome. PLoS Genet 3: 387–392, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez-Corral P, Gonzalez-Rubio C, Rodriguez de Cordoba S, Lopez-Trascasa M: Functional analysis in serum from atypical hemolytic uremic syndrome patients reveals impaired protection of host cells associated with mutations in factor H. Mol Immunol 41: 81–84, 2004 [DOI] [PubMed] [Google Scholar]
- 22.Pickering MC, Goicoechea de Jorge E, Martinez-Barricarte R, Recalde S, Garcia-Layana A, Rose KL, Moss J, Walport MJ, Cook HT, Rodriguez de Cordoba S, Botto M: Spontaneous hemolytic uremic syndrome triggered by complement factor H lacking surface recognition domains. J Exp Med 204: 1249–1256, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neumann HPH, Salzmann M, Bohnert-Iwan B, Mannuelian T, Skerka C, Lenk D, Bender BU, Cybulla M, Riegler P, Konigsrainer A, Neyer U, Bock A, Widmer U, Male DA, Franke G, Zipfel PF: Haemolytic uraemic syndrome and mutations of the factor H gene: a registry-based study of German speaking countries. J Med Genet 40: 676–681, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perkins SJ, Goodship THJ: Molecular modelling of the C-terminal domains of factor H of human complement: a correlation between haemolytic uraemic syndrome and a predicted heparin binding site. J Mol Biol 316: 217–224, 2002 [DOI] [PubMed] [Google Scholar]
- 25.Esparza-Gordillo J, Goicoechea de Jorge E, Garrido CA, Carreras L, Lopez-Trascasa M, Sanchez-Corral P, Rodriguez de Cordoba S: Insights into hemolytic uremic syndrome: segregation of three independent predisposition factors in a large, multiple affected pedigree. Mol Immunol 43: 1769–1775, 2006 [DOI] [PubMed] [Google Scholar]
- 26.Hageman GS, Anderson DH, Johnson LV, Hancox LS, Taiber AJ, Hardisty LI, Hageman JL, Stockman HA, Borchardt JD, Gehrs KM, Smith RJH, Silvestri G, Russell SR, Klaver CCW, Barbazetto I, Chang S, Yannuzzi LA, Barile GR, Merriam JC, Smith RT, Olsh AK, Bergeron J, Zernant J, Merriam JE, Gold B, Dean M, Allikmets R: A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci U S A 102: 7227–7232, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]