The UIM-containing protein RAP80 interacts with BRCA1 and functions in DNA damage repair response (original) (raw)
. Author manuscript; available in PMC: 2008 May 21.
Abstract
In this study, we examine the potential role of receptor-associated protein 80 (RAP80), a nuclear protein containing two ubiquitin-interacting motifs (UIMs), in DNA damage response and double-strand break (DSB) repair. We demonstrate that following ionizing radiation (IR) and treatment with DNA-damaging agents RAP80 translocates to discrete nuclear foci that co-localize with those of γ-H2AX. The UIMs and the region between aa 204–304 are critical for the re-localization of RAP80 to IR-induced foci (IRIF). These observations suggest that RAP80 becomes part of a DNA-repair complex at the sites of IRIF. We also demonstrate that RAP80 forms a complex with the tumor repressor BRCA1 and that this interaction is mediated through the BRCT repeats of BRCA1. The UIMs are not required for the interaction of RAP80 with BRCA1. Knockdown of RAP80 in HEK293 cells significantly reduced DSB-induced homology-directed recombination (HDR). Moreover, inhibition RAP80 expression by siRNA increased radiosensitivity, whereas increased radioresistance was observed in human breast cancer MCF-7 cells over-expression of RAP80. Taken together, our data suggest that RAP80 plays an important role in DNA damage response signaling and HDR-mediated DSB repair. We further demonstrate that RAP80 can function as a substrate of the ataxia-telangiectasia mutated (ATM) protein kinase in vitro which phosphorylates RAP80 at Ser205 and Ser402. We show that this phosphorylation is not required for the migration of RAP80 to IRIF.
Keywords: RAP80, UIMC1, ubiquitin-interacting motif, DNA damage, ATM, BRCA1, BRCT
INTRODUCTION
DNA double strand-breaks (DSB) are potential mutagenic/carcinogenic lesions that can lead to chromosomal aberrations, disruption of genomic integrity, and cause cancer (1–4). DSB trigger a series of cellular responses that are involved in the control of DNA damage repair and activation of cell cycle checkpoints. The regulation of these two processes is very closely coordinated (5–8). Early in DNA damage response signaling nuclear proteins are recruited into multiprotein complexes at the site of the lesion. Some of these proteins are engaged in DNA repair, while others coordinate DNA repair and the activation of cell cycle checkpoints. The latter results in a block in cell cycle progression at the G1/S, intra-S, or G2/M checkpoints until the damage is repaired. If the damage is not properly repaired, this may lead to permanent cell cycle arrest, cell death, or oncogenesis (1, 4, 5). Damage sensors, including Nijmegen Breakage Syndrome 1 (NBS1) and RAD50, play a role early in DNA damage responses and provide a scaffold for downstream proteins (5, 9). The nuclear protein kinase ataxia-telangiectasia mutated (ATM) also plays a critical role in the early responses to DNA damage and appears to be the primary player in response to ionizing radiation (IR) (10–12). IR and DNA damage-inducing agents induce dissociation of the inactive, dimeric ATM complex, auto-phosphorylation of Ser1981, and ATM activation. Subsequently, active ATM localizes to IR-induced foci (IRIF) and catalyzes the phosphorylation of many damage sensors, as well as signal transducer and effector proteins. The ATM substrates include NBS1, H2AX, breast cancer susceptibility gene 1 (BRCA1), 53BP1, checkpoint kinase 2 (CHK2), and the tumor suppressor p53 (5, 10, 11, 13, 14). These effector proteins regulate cell cycle arrest, apoptosis, DNA-repair and transcription (4, 5).
The receptor-associated protein 80 (RAP80) or ubiquitin-interacting motif containing 1 (UIMC1, as approved by the HUGO Gene Nomenclature Committee) is a nuclear protein containing two ubiquitin-interacting motifs (UIMs) at its amino terminus (15, 16). A recent study demonstrated the importance of these UIMs in the binding of K63-linked (poly)ubiquitin and the regulation of the stability and transcriptional activity of the estrogen receptor α by RAP80 (16). UIMs have been found in a variety of proteins with roles in endocytosis, (de)ubiquitination, replication, transcription, and DNA-repair (17, 18). The potential role of UIM-containing proteins in DNA repair (19, 20) and the observed association of RAP80 with several proteins involved in the activation of cell cycle checkpoints (Yan, J., unpublished observations) suggested a possible role for RAP80 in DNA damage repair responses. In this study, we provide evidence that support this hypothesis. We demonstrate that RAP80 translocates to IRIF after IR and that the UIMs are essential for this re-localization of RAP80. We provide evidence showing that RAP80 and BRCA1 are associated with the same protein complex and that this interaction is dependent on the BRCA1 C-terminal (BRCT) repeats of BRCA1. We further demonstrate that RAP80 is a target of ATM phosphorylation. Knock-down of RAP80 expression by small interfering RNA (siRNA) reduced DSB-induced HDR and the cells sensitivity to IR-induced cytotoxicity. These observations suggest that RAP80 may play a critical role in the regulation of BRCA1 function(s) and DNA damage response signaling.
Materials and Methods
Plasmids
pLXIN-3xFLAG-RAP80, pLXIN-3xFLAG-RAP80ΔUIM, pLXIN-3xFLAG-RAP80 mutants (aa 1–582, 1–504, 1–404, 1–204, and 1–122) and pEGFP-RAP80 were described previously (15, 16). Details on other mutant RAP80, ATM, and BRCA1 plasmids used in this study are provided in the Supplement.
Generation of MCF-7-RAP80
MCF-7-RAP80, which stably expresses 3xFLAG-RAP80, was obtained by transfecting MCF-7 cells with pLXIN-3xFLAG-RAP80, followed by selection with 750 µg/ml geneticin. Resistant clones were pooled and expression of 3xFLAG-RAP80 was verified by Western blot analysis and confocal microscopy.
Retroviral infection
PT67 cells were transfected with pLXIN-3xFLAG-RAP80, geneticin-resistant cells isolated, and virus-containing conditioned medium collected. Normal human fibroblasts (GM05757) and A–T cells (GM05823) (Coriell, Camden, NJ) were then infected with the retrovirus and 48 later examined by confocal microscopy.
siRNA knockdown
MCF-7 cells were transfected with control or RAP80 siRNA Smart Pool (Dharmacon, Lafayette, CO) following the manufacturer’s suggestions. MCF-7 cells were seeded at 105/ml in antibiotics free RPMI 1640 medium containing 10% FBS. On the second day, siRNA was mixed with Dharmafect 1 (Dharmacon) in antibiotics and serum free medium for 20 min and then added to the cells at a final concentration of 100 nM. Twenty-four hours later the transfection was repeated. After an additional 48 h incubation cells were harvested for western blot analysis or colony forming assay.
Confocal microscopy
MCF-7 cells were transiently transfected with wild type or mutant pEGFP-RAP80 plasmid DNA and 48 h later irradiated at the dose indicated. At different time intervals after irradiation cells were fixed for 20 min in 4% paraformaldehyde, and subsequently treated for 7 min with 0.2% Triton X-100. Cells were washed in PBS and then incubated for 15 min in Superblock Blocking Buffer (Pierce, Rockford, IL). Cells were subsequently incubated for 2 h with anti-γ-H2AX antibody (Upstate Biotech., Temecula, CA), and finally for 40 min with anti-mouse Alexa 595 (Molecular Probes, Eugene, OR). Endogenous RAP80 and BRCA1 were detected with an anti-RAP80 antibody (Bethyl, Montgomery, TX) and an anti-BRCA1 antibody (Calbiochem, San Diego, CA), respectively. FLAG-RAP80 in MCF-7-RAP80 cells was detected with a rabbit anti-3xFLAG antibody (gift from Dr. Yue Xiong, UNC, Chapel Hill, NC) and an anti-rabbit Alexa 488 antibody (Molecular Probes). Nuclei were stained with 4′,6-diamidino-2-phenyindole (DAPI). Cells were then covered with 80% glycerol and fluorescence observed in a Zeiss LSM 510 NLO confocal microscope (Zeiss, Thornwood, NY). The photographs shown are representative for each experiment.
Co-immunoprecipitation (co-IP) analysis
MCF-7 cells were treated with IR and 4 h later harvested and lysed for 1 h in RIPA buffer (Upstate Biotech.) containing protease inhibitor and phosphatase cocktails I and II (Sigma, St. Louis, Missouri). The cell lysates were centrifuged at 14,000 ×g at 4 °C for 10 min. The supernatants were then incubated with anti-RAP80 antibody (Bethyl) and protein-G Sepharose beads (Sigma) overnight to isolate RAP80 protein complexes. The beads were then washed three times with RIPA buffer. The bound protein complexes were then solubilized in sample buffer and analyzed by Western blot analysis using anti-BRCA1 (Calbiochem) and anti-RAP80 antibodies.
ATM kinase assay
Phosphorylation of GST-RAP80 proteins by ATM kinase was examined following a procedure described by Ziv et al. (21). For details see Supplement.
Colony-forming assay
MCF-7 parental, MCF-7-RAP80, or MCF-7 cells transfected with RAP80 siRNA were trypsinized and resuspended at 5 × 104 cells/ml. Cells were then irradiated at different doses, plated in triplicate dishes, and grown for 14 days at 37°C. Cells were then stained with 1% Commassie brilliant blue, 5% acetic acid and 30% methanol and the number of colonies counted.
HDR assay
HDR assay was performed as described previously (22). HEK293 cells with a chromosomally integrated single copy of the repair substrate pDR-GFP (23) were used to test RPA’s role in HDR. Chromosomal DSBs were induced with the expression of I-SceI. DSB-induced HDR resulted in restoration and expression of GFP and was quantified by FACS.
Results
RAP80 re-localizes to nuclear foci after DNA damage
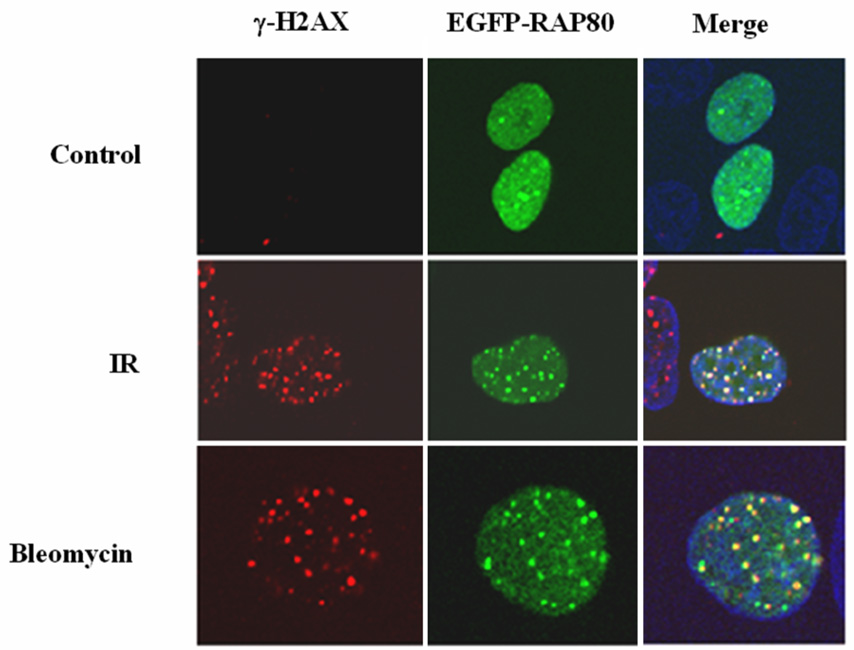
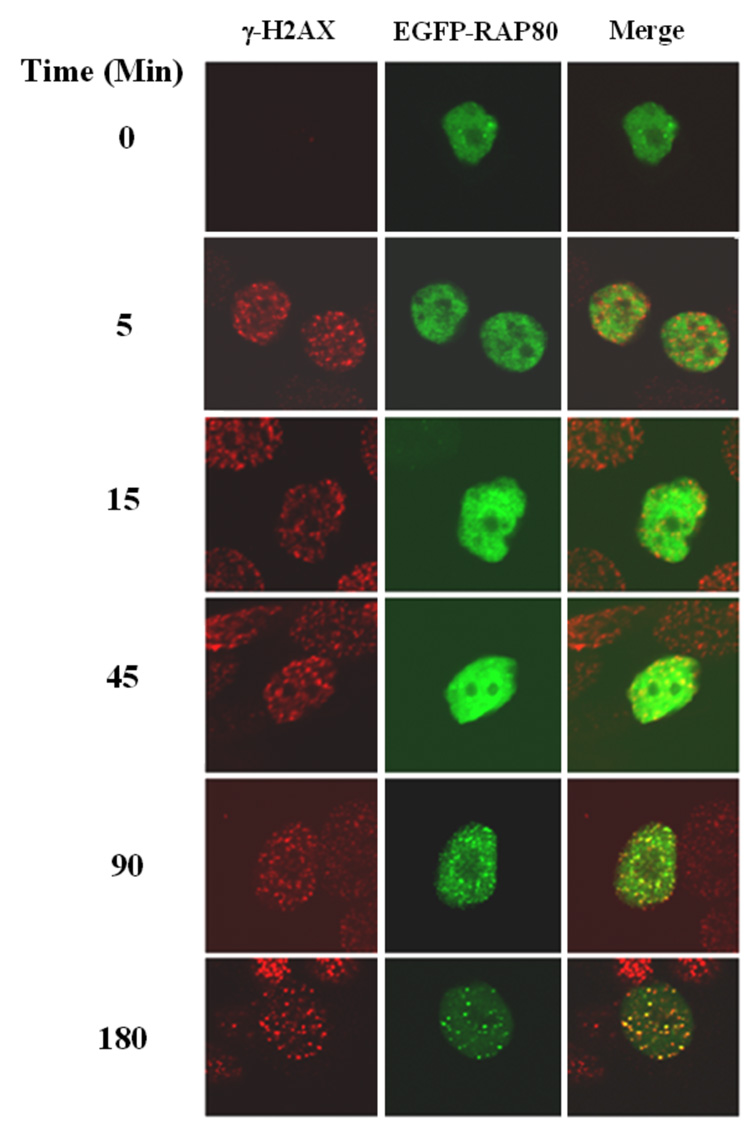
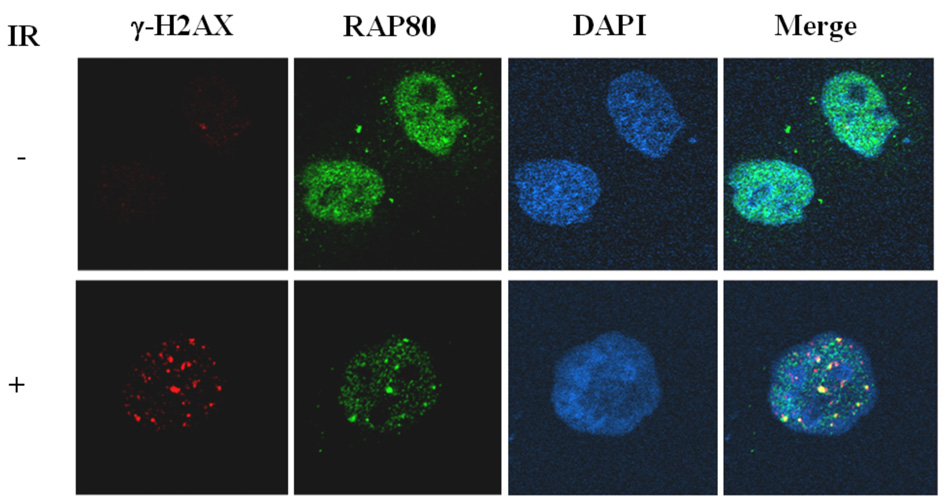
Many proteins involved in DNA damage responses translocate to nuclear foci after DNA damage (5, 24). To study the potential role of RAP80 in DNA damage responses, we first examined the effect of DNA damage on the potential re-localization of RAP80. MCF-7 cells were transiently transfected with pEGFP-RAP80 and the effect of γ-irradiated on the cellular distribution of RAP80 examined by confocal microscopy. As shown in Fig. 1_A_, RAP80 was rather homogeneously expressed in the nucleus of non-irradiated cells while after IR RAP80 was localized in discrete nuclear foci that co-localized with DSB-associated foci of γ-H2AX which served as an indicator for the sites of DNA damage. Treatment of the cells with bleomycin, a DNA DSB-inducing agent, induced a similar re-localization of RAP80 (Fig. 1_A_). These observations indicated that RAP80 translocates to DNA damage sites after IR or treatment with DSB-inducing agents. Previous studies have shown that after DNA damage proteins locate to foci at different time intervals. For example, γ-H2AX, 53BP1, and ATF2 localize to damage foci very rapidly in response to IR, while BRCA1 translocates to foci several hours after IR (25, 26). Examination of the time course of the translocation of RAP80 showed that RAP80 did not rapidly re-locate to DNA damage foci; however, translocation of RAP80 to the foci was clearly observed at 90 min after IR (Fig. 1_B_).
Figure 1.
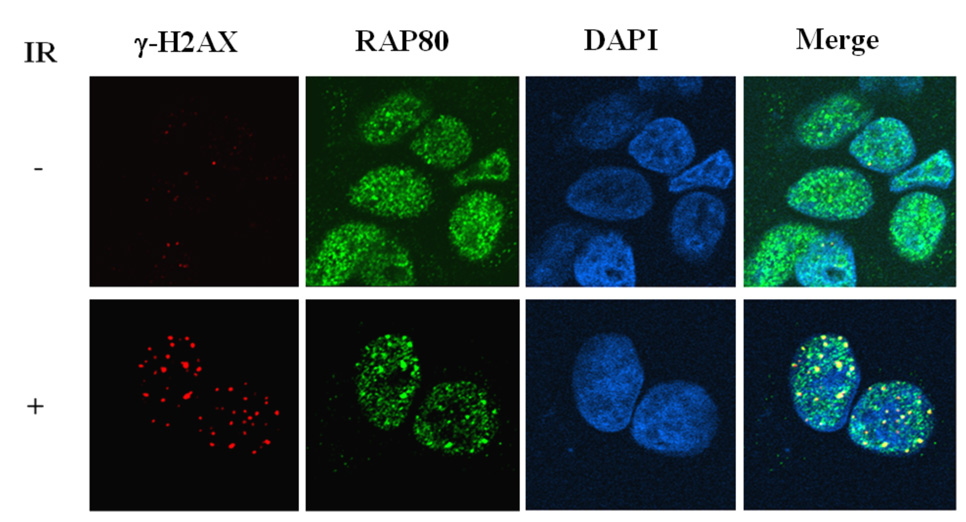
Irradiation and treatment with DNA-damage inducing chemicals induce translocation of RAP80 to IRIF. A. MCF-7 cells were transfected with pEGFP-RAP80 and 48 h later treated with IR (10 Gy) or bleomycin (0.06 U/ml). After 3 h incubation, cells were fixed and subsequently stained with anti-γ-H2AX and Alexa 595-conjugated mouse anti-rabbit IgG antibody. Nuclei were identified by DAPI staining. Localization of EGFP-RAP80 and γ-H2AX was examined by confocal microscopy. B. Time-dependent translocation of RAP80 to IRIF. MCF-7 cells were transfected with pEGFP-RAP80 and 48 h later γ-irradiated (10 Gy). At the times indicated cells were fixed and stained with anti-γ-H2AX antibody and DAPI. C. Translocation of endogenous RAP80 to IRIF. MCF-7 cells were treated with 10 Gy γ-irradiation and 3 h later the localization of endogenous RAP80 and γ-H2AX was examined by confocal microscopy with an anti-RAP80 and anti-γ-H2AX antibody, respectively.
To examine the translocation of endogenous RAP80 in response to DNA damage, MCF-7 cells were treated with IR and the localization of RAP80 examined by immunohistochemistry with anti-RAP80 antibody. Fig. 1_C_ shows that endogenous RAP80 translocated to DNA repair foci after IR in agreement with the observations obtained with exogenous RAP80.
Identification of domains important for the re-localization of RAP80
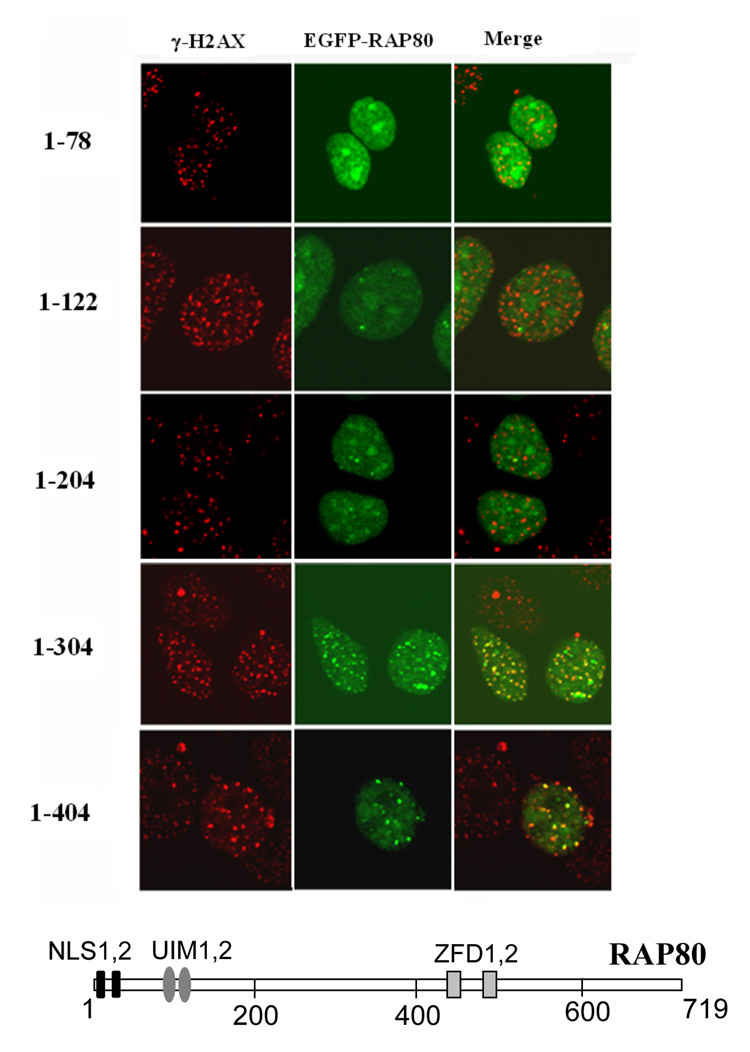
Previously, we reported that RAP80 contains two putative zinc finger motifs between aa 505 and 582 and two functional ubiquitin-interacting motifs (UIMs) between aa 80 and 124 (15, 16). To determine what regions within RAP80 are important for its translocation to IRIF, we examined the effect of a series of carboxyl-terminal deletions on its re-localization. This analysis showed that after IR the deletion mutants RAP80(1–404) and RAP80(1–304) still co-localized with γ-H2AX positive foci whereas RAP80(1–78), RAP80(1–122), and RAP80(1–204) did not (Fig. 2_A_). These observations suggest that the region between aa 204–304 is essential for the migration of RAP80 to IRIF and that the zinc finger domain in the C-terminal half of RAP80 is not required. The effect of several amino-terminal deletions in RAP80 could not be analyzed because they are rapidly degraded by the proteasome system.
Figure 2.
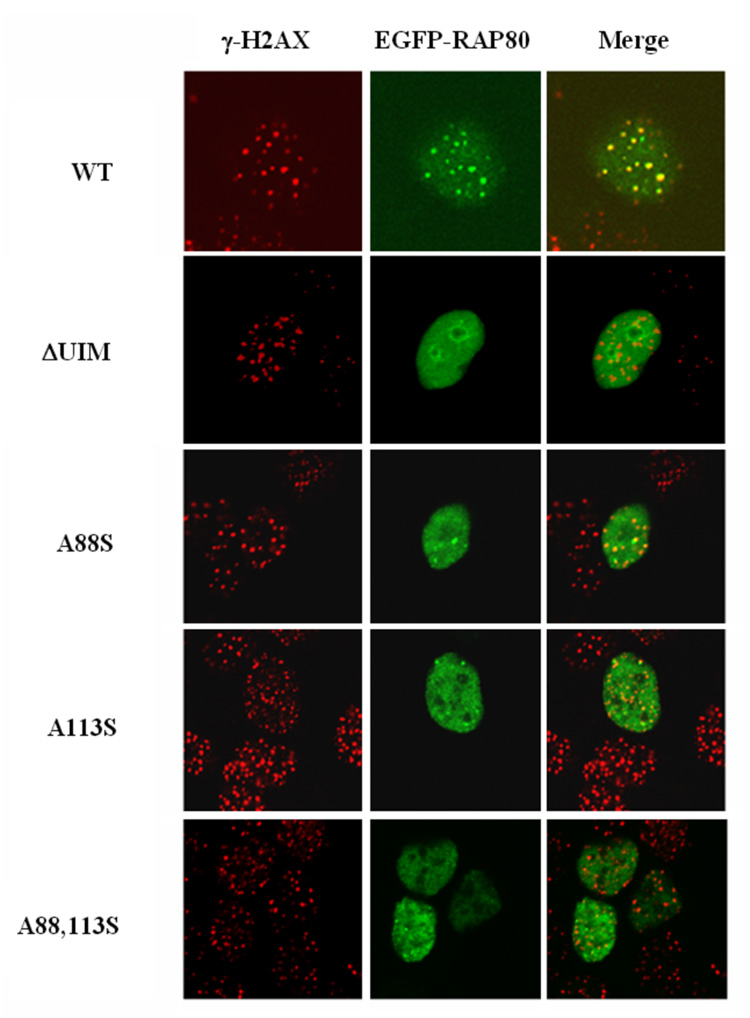
The region between aa 204–304 and the UIMs are required for the relocalization of RAP80 to IRIF. MCF-7 cells were transfected with several pEGFP-RAP80 mutants containing various C-terminal deletions as indicated (A), pEGFP-RAP80 or pEGFP-RAP80ΔUIM in which the two UIMs are deleted or pEGFP-RAP80 mutants containing point mutations within the UIMs as indicated (B). After 48 h cells were γ-irradiated (10 Gy) and 3 h later the subcellular distribution of EGFP-RAP80 and γ-H2AX examined by confocal microscopy. Schematic view of RAP80 is shown in A. NLS indicates nuclear localization signal; UIM, ubiquitin interacting motif; ZFD, zinc finger domain.
Previous studies have shown that the UIMs are important for the modulation of ERα-mediated transcriptional activation by RAP80 (16). We therefore examined these the effect of a UIM deletion on the IR-induced translocation of RAP80. In contrast to EGFP-RAP80, which co-localized with γ-H2AX following IR, EGFP-RAP80ΔUIM did not translocate to IRIF as efficiently as wild type RAP80 but remained rather diffusely distributed in the nucleus (Fig. 2_B_). Previous studies showed that Ala88 and Ala113 within the UIMs of RAP80 are highly conserved among UIMs and critical for the functions of UIMs (16, 27). Fig. 2C shows that the single point mutations Ala88Ser and Ala113Ser greatly impaired the migration of RAP80 to IRIF while the Ala88Ser/Ala113Ser double mutation totally abolished its translocation. These observations support the conclusion that the UIMs are of critical importance in the IR-induced re-localization RAP80 to IRIF.
RAP80 is associated with a BRCA1 protein complex
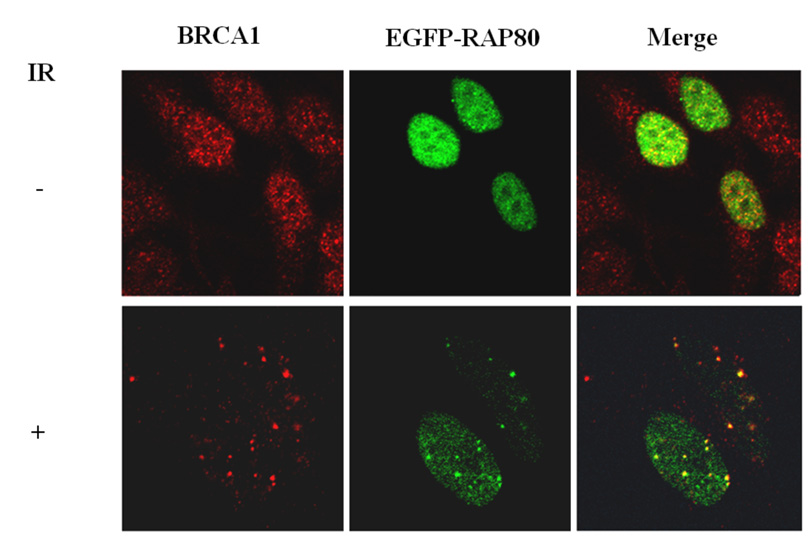
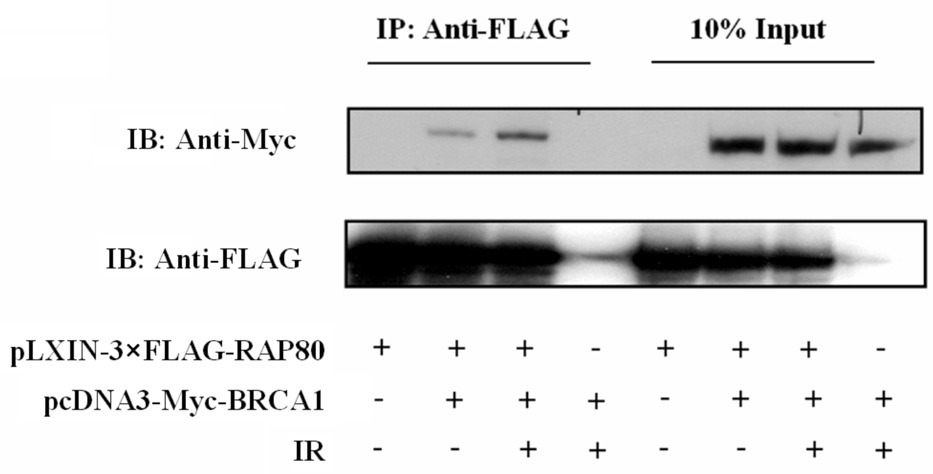
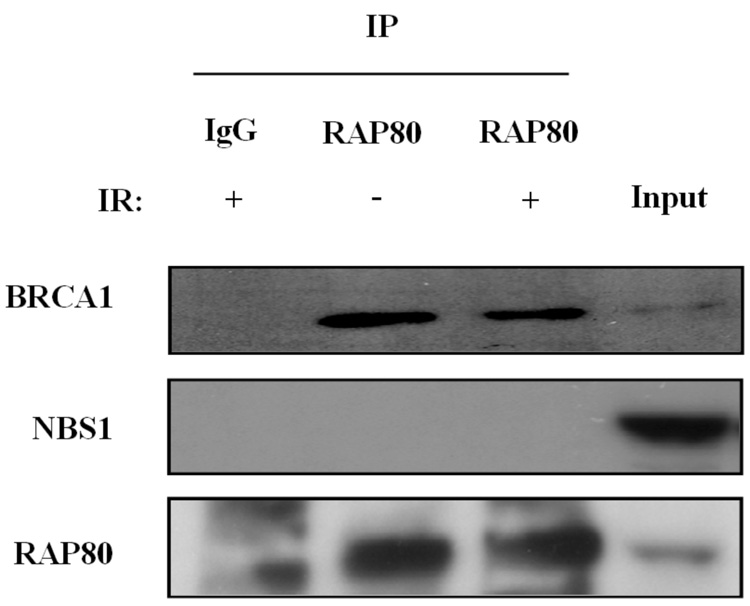
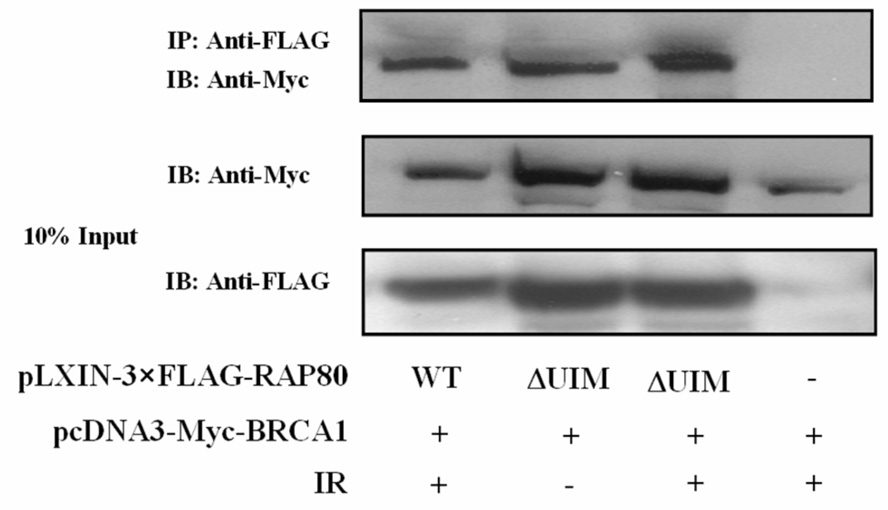
Proteins recruited to IRIF are part of large multiprotein complexes. These proteins migrate to IRIF at different time intervals (26, 28, 29). ATF2 is recruited very early after IR-induced damage, while RAP80, as reported for BRCA1, migrates to IRIF at a later time. RAP80 likely forms a complex with several other DNA response proteins. We therefore examined the association of RAP80 with several DNA damage repair proteins. To examine its association with BRCA1, HeLa cells were transfected with pEGFP-RAP80, irradiated and 4 h later the localization of RAP80 and BRCA1 examined by confocal microscopy. As shown in Fig. 3_A_, in non-irradiated cells both RAP80 and BRCA1 exhibited a rather diffuse pattern of nuclear distribution, while after irradiation both proteins co-localized to IRIF. The possible association of RAP80 and BRCA1 was further examined by co-IP analysis. Our data showed that 3xFLAG-RAP80 could co-IP Myc-BRCA1 from lysates isolated from both irradiated and non-irradiated cells (Fig. 3_B_). Moreover, co-IP analysis using an anti-RAP80 antibody endogenous RAP80 was able to pull-down endogenous BRCA1 (Fig 3_C_). These observations suggest that RAP80 and BRCA1 are associated with the same protein complex and that their association was independent of IR. In contrast, NBS1 was not immunoprecipitated by RAP80 antibody suggesting that it does not appear to be part of a RAP80 protein complex (Fig. 3_C_).
Figure 3.
RAP80 and BRCA1 are part of the same protein complex. A. HeLa cells were transfected with pEGFP-RAP80 and 48 h later treated with and without 15 Gy γ-irradiation. Four h later cells were stained with anti-BRCA1 antibody. Subsequently, the subcellular distribution of EGFP-RAP80 and BRCA1 was examined by confocal microscopy. B. HEK293T cells were transfected with pLXIN-3xFLAG-RAP80 and pcDNA3-Myc-BRCA1 and 48 h later treated with or without γ-irradiation. After 4 h incubation protein lysates were prepared, FLAG-RAP80 protein complexes isolated with anti-FLAG resin and examined by Western blot analysis with antibodies against Myc and FLAG. C. MCF-7 cells were treated with or without γ-irradiation. After 4 h incubation protein lysates were isolated and RAP80 protein complexes immunoprecipitated with an anti-RAP80 antibody or IgG (control). The immunoprecipitated protein complexes were then examined by Western blot analysis with antibodies against RAP80, BRCA1, or NBS1. D. HCC1937 cells containing mutant BRCA1 were γ-irradiated (10 Gy) and 3 h later the subcellular distribution of RAP80 and γ-H2AX was examined.
To study whether the translocation of RAP80 to IRIF was dependent on BRCA1, the migration of endogenous RAP80 was examined in HCC1937 cells, which express a truncated form of BRCA1 that does not migrate to IRIF after DNA damage (30). Confocal microscopy showed that RAP80 still translocated to IRIF after IR (Fig. 3_D_) suggesting that this function of BRCA1 was dispensable for this re-localization.
Mapping regions important for the association of RAP80 with BRCA1
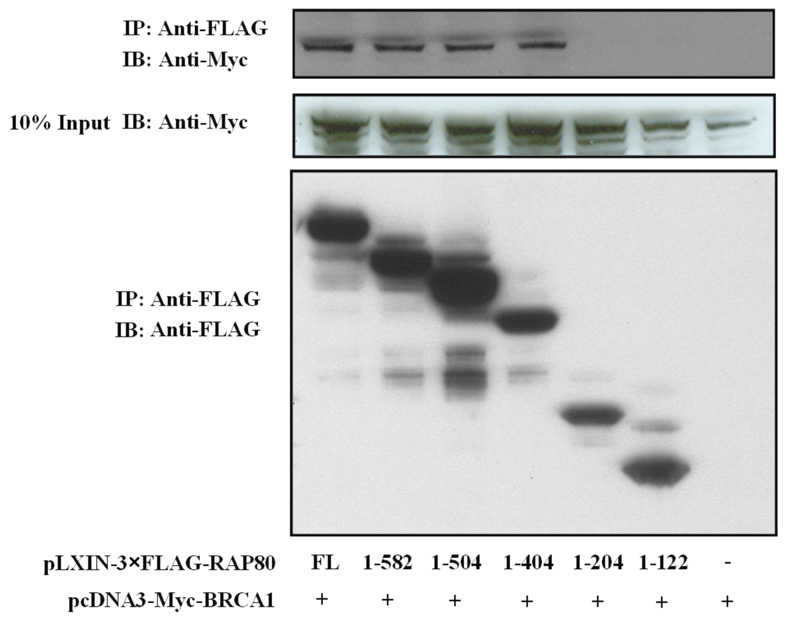
First, we examined whether the UIMs were required for the interaction of RAP80 with BRCA1. Co-IP analysis showed that, as RAP80, RAP80ΔUIM was able to pull-down BRCA1 in an IR-independent manner (Fig. 4_A_). Next, we examined the effect of a series of RAP80 C-terminal deletion mutants for their ability to co-immunoprecipitate BRCA1. Co-IP analysis demonstrated that the region between aa 204–404 was required for the interaction (Fig. 4_B_). Since the BRCT domain (aa 1528–1863) functions as a docking module for several BRCA1-interacting proteins (31–33), we determined its role in the interaction with RAP80. Fig. 4C shows that the BRCT domain was sufficient to mediate the interaction with RAP80.
Figure 4.
Identification of regions important for the association of RAP80 and BRCA1. HEK293T cells were transfected with pcDNA3-Myc-BRCA1, pcDNA3.1-HA-BRCT, and pLXIN-3xFLAG encoding wild type RAP80, RAP80ΔUIM or a C-terminal deletion mutant of RAP80 as indicated. Forty eight h later cells were γ-irradiated and after an additional 4 h incubation, FLAG-RAP80 protein complexes were isolated with anti-FLAG resin and examined by Western blot analysis with antibodies against FLAG, Myc or HA. A. Effect of the UIM deletion on the association of RAP80 and BRCA1. B. Effect of C-terminal deletions on the interaction of RAP80 with BRCA1. C. The BRCT domain is sufficient for the association of BRCA1 with RAP80.
RAP80 expression affects DSB-induced HDR and sensitivity of MCF7 to IR
Since RAP80 is in a protein complex with BRCA1, which plays a critical role in DSB-induced HDR, we examined the effect of RAP80 knockdown on DSB-induced HDR (22). HEK293 cells, stably expressing the HDR target vector pMJ-GFP, were transfected with control or RAP80 siRNA and pCMV-I-SceI, which encodes a product that induces DSB. Cells were subsequently examined by a flow cytometric analysis for HDR. As shown in Fig. 5_A_, cells in which RAP80 was depleted showed a significantly reduced HDR compared to cells transfected with control siRNA suggesting that RAP80 affects DSB repair through regulating HDR.
Figure 5.
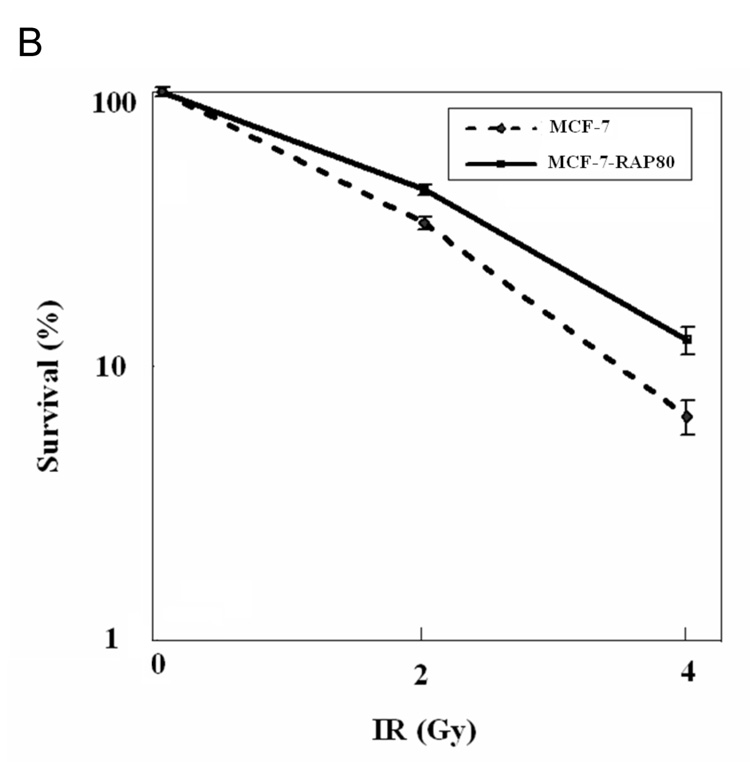
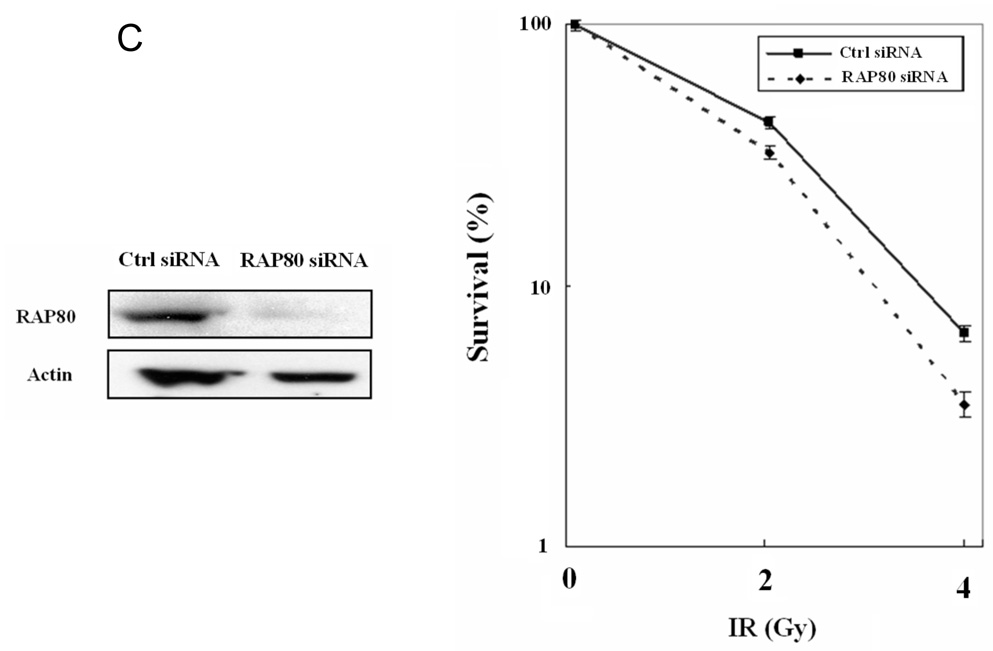
RAP80 expression affects DSB-induced HDR and sensitivity of MCF7 to IR. A. DSB-induced HDR in HEK293 cells transfected with scrambled or RAP80 siRNAs was assessed using flow cytometry as described in Materials and Methods. Data represent the mean of three independent experiments. Bars indicate standard deviation (SD). *p<0.025. Knockdown of RAP80 expression was examined by Western blot analysis with an anti-RAP80 antibody. B. MCF-7 and MCF-7-RAP80 cells were seeded at low density and irradiated with the indicated doses of IR. Cells were incubated for 14 days at 37°C to allow colonies to form. Colonies were then stained and counted. C. MCF-7 cells were transfected with scrambled or RAP80 siRNAs and then were treated as described under B. Right panel shows decreased survival of cells treated with RAP80 siRNA. Part of the cells was used in Western blot analysis to examine the down-regulation of RAP80 protein expression (left panel).
To investigate the potential role of RAP80 in DNA damage repair, we generated an MCF-7 cell line that stably expressed FLAG-RAP80. Confocal microscopy showed that FLAG-RAP80 co-localized with γ-H2AX after IR (Supplementary Fig. 1). To examine the effects of FLAG-RAP80 on DNA damage response, we examined the effect of various doses of IR on the colony-forming ability of MCF-7 and MCF-7-RAP80 cells. As shown in Fig. 5_B_, MCF-7-RAP80 cells exhibited a decreased sensitivity to IR, suggesting that overexpression of RAP80 enhances cell viability. We next examined the effect of RAP80 knockdown on cell survival. MCF-7 cells were transfected with control or RAP80 siRNA before the effect of various doses of IR on the colony-forming ability was examined. As shown in Fig. 5_C_, MCF7 cells in which RAP80 expression was reduced, formed fewer colonies than parental cells suggesting that RAP80 expression regulates cell survival after DNA damage. Although these effects are modest, they are in agreement with a regulatory role in DNA damage response signaling.
ATM phosphorylates RAP80 in vitro
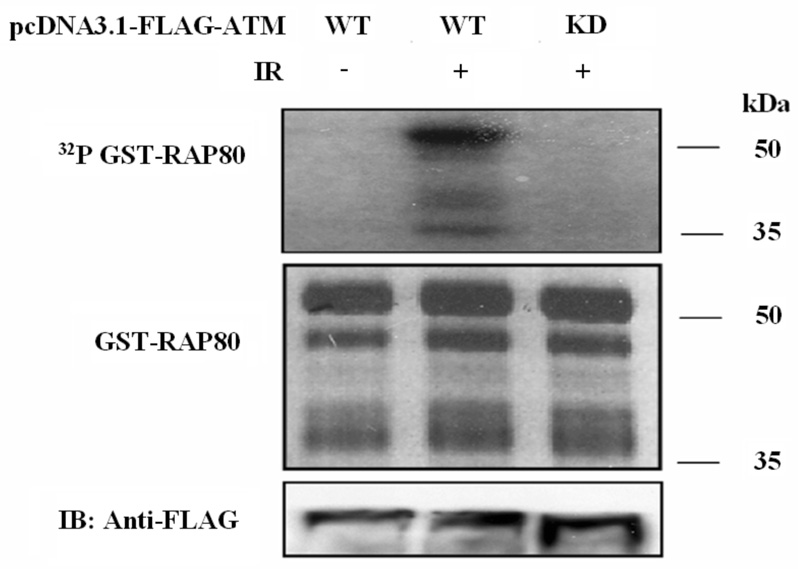
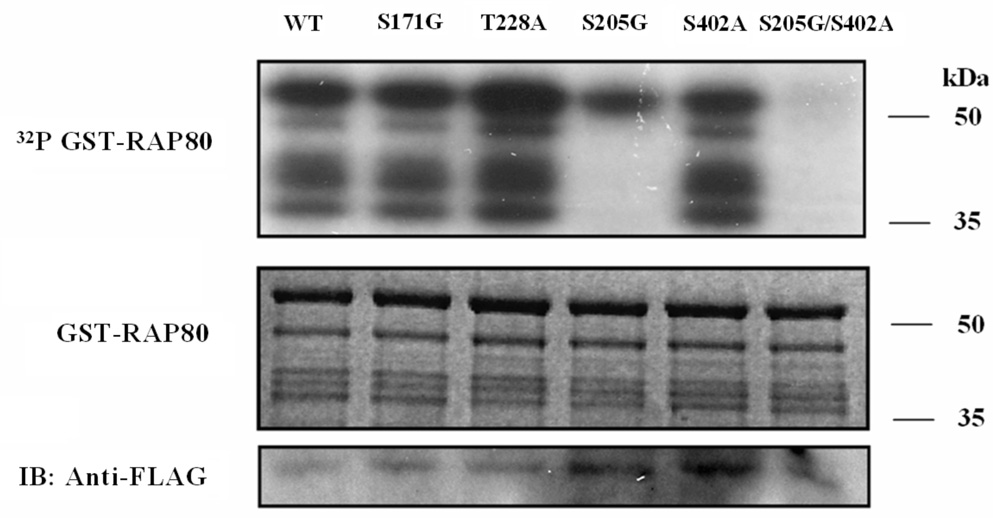
Many proteins involved in IR-induced DNA damage responses are phosphorylated by phosphatidylinositol 3-kinase-like (PIKK) family member ATM (5, 10, 11). In several instances the translocation of proteins to IRIF is dependent on their phosphorylation by ATM. We therefore examined whether the migration RAP80 to IRIF was dependent on its phosphorylation by ATM. Analysis of the RAP80 amino acid sequence identified several potential ATM phosphorylation sites (Fig. 6_A_). Based on their conservation between mouse and human RAP80 and their similarity to the consensus sequence of the ATM phosphorylation site ΦXΦS(T)Q (in which Φ is a hydrophobic amino acid, although there are some exceptions to this) (34), Ser171, Ser205, Thr228, and Ser402, Ser419, and Thr448 were identified as the most likely ATM phosphorylation sites. To determine whether RAP80 could function as a substrate of ATM, we examined the phosphorylation of GST-RAP80(413–500) and GST-RAP80(168–405) that containing the six potential ATM phosphorylation sites. In vitro kinase assays were performed using Flag-ATM immunoprecipitated from HEK293T cell lysates. In contrast to GST-RAP80(413–500) (not shown), RAP80(168–405) was efficiently phosphorylated by WT ATM (Fig. 6_B_) suggesting that the ATM phosphorylation site(s) reside(s) within the 168–405 region. We further demonstrate that this phosphorylation was dependent on the activation of ATM in HEK293 cells by IR and that “kinase-dead” ATM did not phosphorylate RAP80 (Fig. 6_B_). These results indicated that RAP80 might be a target for ATM phosphorylation in vivo. To determine which of the site(s) was/were phosphorylated, we examined the phosphorylation of several RAP80(168–405) mutants containing point mutations at the four potential sites. This analysis showed that mutation of Ser205 reduced RAP80 phosphorylation by ATM. Particularly the shorter GST-RAP80 fragments, which were generated by proteolytic cleavage of GST-RAP80(168–405) at its carboxyl-terminus, were no longer phosphorylated while mutation of Ser171 and Thr228 did not affect the phosphorylation of these shorter fragments. These data suggested that Ser205 and Ser402 function as ATM phosphorylation sites. This was confirmed by the data showing that the mutation of both Ser205 and Ser402 totally abolished the phosphorylation of RAP80(168–405) (Fig. 6_C_). These observations indicate that RAP80 functions as a substrate for ATM protein kinase in vitro and that ATM can phosphorylate RAP80 on both Ser205 and Ser402.
Figure 6.
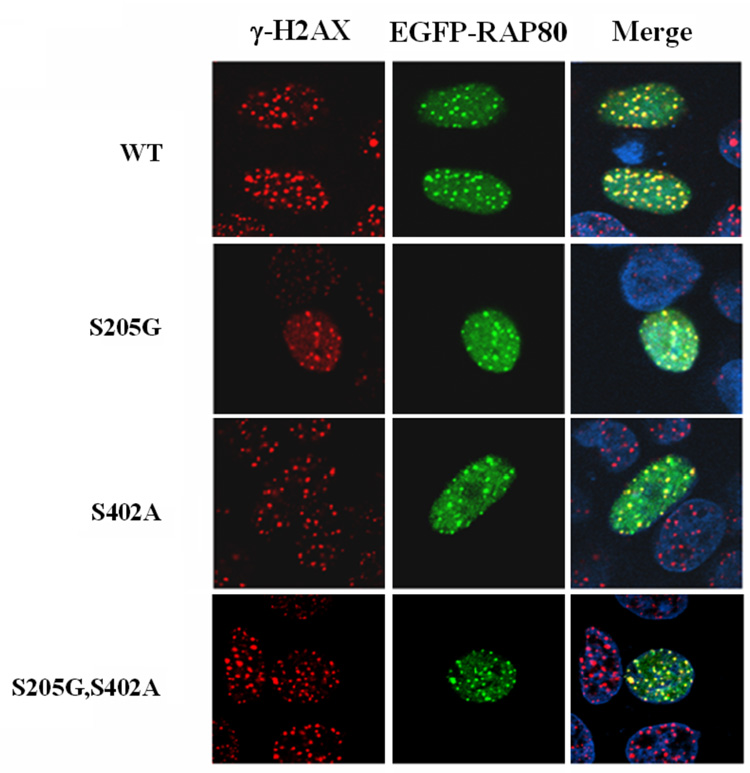
RAP80 is a substrate for phosphorylation by ATM kinase. A. Schematic view of the location the potential ATM phosphorylation sites (arrows; dashed arrows indicate sites not conserved between human and mouse) in human RAP80. Comparison of a partial human and mouse RAP80 amino acid sequence is shown below. The most likely ATM phosphorylation sites conserved between mouse and human are highlighted. B. Phosphorylation of GST-RAP80(168–405) protein by ATM kinase or ATM(KD) was examined as described in Materials and Methods. C. Identification of ATM phosphorylation sites in RAP80. Phosphorylation of WT and several mutant GST-RAP80(168–405) proteins by ATM kinase was examined. D. Phosphorylation of RAP80 by ATM is not required for its translocation to IRIF. MCF-7 cells were transfected with pEGFP-RAP80 plasmid DNA encoding wild type RAP80 or RAP80 in which the ATM-phosphorylation sites were mutated. After 48 h incubation cells were treated with γ-irradiation (10 Gy) and 3 h later stained with anti-γ-H2AX antibody and DAPI. Localization of EGFP-RAP80 and γ-H2AX was examined by confocal microscopy.
To examine whether this phosphorylation could be involved in the translocation of RAP80 to DNA-damage foci, we examined the re-location of pEGFP-RAP80 proteins with point mutations at the different ATM phosphorylation sites. Confocal analysis showed that neither the single or double mutations affected the translocation of RAP80 to DNA repair foci after IR (Fig. 6_D_). The translocation of a pEGFP-RAP80 mutant (8MT) with all potential ATM phosphorylation sites mutated was also examined. This mutant also translocated to the IRIF (Supplementary Fig. 2_A_). The independency of RAP80 migration on phosphorylation by ATM was further confirmed by infecting normal and A–T cells with retrovirus encoding FLAG-RAP80. The data showed that RAP80 also translocated to IRIF in A–T cells (Supplementary Fig. 2_B_).
DISCUSSION
DNA damage initiates a series of cellular events that control DNA repair, gene transcription, and cell cycle checkpoints. These DNA signaling responses function to maintain genomic integrity and determine whether a cell survives or undergoes apoptosis. DNA damage triggers the recruitment of a large number of DNA damage signaling and repair proteins to the sites of genomic damage. In this study, we provide evidence indicating that RAP80 plays an important role in DNA damage repair signaling. Our study identifies RAP80/UIMC1 as a protein that migrates to IRIF after DNA damage. In untreated cells RAP80 is distributed rather diffusely within the nucleus while after treatment with IR or bleomycin RAP80 translocates to nuclear foci (IRIF) and where it co-localizes with γ-H2AX (Fig. 1A). The translocation of RAP80 to IRIF after DSB-induced damage suggests that RAP80 plays a role in DNA damage responses.
Proteins are recruited to IRIF at different times after genomic damage reflecting different functions of these proteins in DNA repair responses (5, 24). The initial migration of proteins to IRIF occurs very quickly in response to IR. For example, the appearance of ATF2 and 53BP1 occur within minutes while other proteins, including BRCA1, are recruited to IRIF hours after DNA damage (25, 26, 29). Our results show that IR-induced translocation of RAP80 to IRIF occurred after a substantial delay and was first observed 90 minutes after IR (Fig. 1B), a time course more similar to that reported for BRCA1 (28). These observations suggest that RAP80 may not be an initial sensor of genomic damage but function as a downstream mediator or modulator of DNA damage signals.
We reported previously that RAP80 contains two putative Cys-X2-Cys-X11-His-X3-Cys zinc finger-like motifs near its carboxyl-terminus and two UIMs near its amino-terminus that is able to bind K63-linked polyubiquitin chains (15, 16). To determine what region of RAP80 is important for its translocation to IRIF, the effects of several deletion and point mutations on this migration were examined. Our data showed that deletion of the carboxyl-terminus up to Gln304 had little effect on the translocation of RAP80 while RAP80(1–204) was unable to migrate to the foci. These results suggest that the zinc-finger domain is not required and the region between Ser204-Gln304 is critical for the migration of RAP80 to IRIF. We previously showed that the UIMs in RAP80 are functional and play a critical role in the function of RAP80 (16). UIMs consist of a short sequence motif of about 20 amino acids, that can bind ubiquitin and/or specific ubiquitinated proteins (17). UIM-containing proteins have been reported to direct (multi)monoubiquitination thereby regulating a broad range of cellular functions, including membrane protein trafficking, histone function, transcriptional regulation, DNA repair, replication, and (de)ubiquitination control (17, 18, 35, 36). Our results show that deletion of the UIMs (RAP80ΔUIM) abolished the ability of RAP80 to translocate to IRIF. In addition, point mutations A88S and A113S within the UIMs greatly impaired the migration of RAP80 to IRIF, while the double mutation totally abolished its translocation (Fig. 2). These data indicate that the UIMs play a critical role in the migration of RAP80 to IRIF. Since the UIMs in RAP80 bind K63-linked polyubiquitin, they might provide a binding site for a specific ubiquitinated protein(s) and a mechanism to recruit RAP80 to IRIF. Deletion of the UIMs would prevent such interaction and block the migration of RAP80 to IRIF. Alternatively, UIMs can promote mono-ubiquitination that result in a change in conformation and activity of certain UIM-containing proteins (17, 18). Such ubiquitination might be required for RAP80 to migrate to IRIF. Recently, ubiquitination of the BRCA1-interacting protein CtIP has been reported to be required for its association with chromatin and its participation in checkpoint control (37).
The time course of the translocation of RAP80 to IRIF shows similarities with that of BRCA1 and several other DNA repair signaling proteins (5, 38). In addition, we demonstrated that after DNA damage RAP80 co-localized with BRCA1. These observations suggested that RAP80 and BRCA1 might be part of the same protein complex. Co-immunoprecipitation analysis supported the conclusion that RAP80 is associated with BRCA1. This association appeared to be independent of the induction of DNA damage since RAP80 was also associated with a BRCA1 protein complex in non-irradiated cells (Fig. 3B). Deletion of the UIMs did not affect this association suggesting that this interaction was not regulated through the UIMs of RAP80. In addition, RAP80 was able to translocate to IRIF in HCC1937 cells which express a truncated BRCA1 that is unable to migrate to nuclear foci (30). These observations suggest that in contrast to the relocation of several other binding partners of BRCA1 (39), the translocation of RAP80 to IRIF is not dependent on the migration of BRCA1 to IRIF (Fig. 3D). Since the translocation of RAP80 is dependent on its UIMs, one might predict that the translocation of proteins associated with this RAP80 complex, including BRCA1, are also dependent on the UIMs of RAP80.
Many studies have demonstrated that altered expression or activity of DNA damage response proteins, including BRCA1 (22), ATF2 (26), MDC1 (40), and BRIT1 (41), can affect the sensitivity of cells to IR. Specific mutations in these genes or knockdown of their expression by siRNAs can result in aberrant control of DNA repair and cell cycle checkpoints, and subsequently reduce the survival of cells after IR. Our data demonstrate that knockdown of RAP80 expression by siRNAs caused a significant reduction in DSB-induced homology-directed recombination (HDR)(Fig. 4) which constitutes a major mechanism by which cells repair DSB (22, 42). In addition, our study shows that MCF-7 cells overexpressing RAP80 were more resistant to IR-induced cytotoxicity, while knockdown of the expression of endogenous RAP80 increased the sensitivity of MCF-7 cells to IR (Fig. 5). Although these effects were modest, they are in agreement with the concept that RAP80 functions as a facilitator of efficient DSB repair. Since HDR is regulated by BRCA1, the association of RAP80 with BRCA1 and its effect on HDR described in this study support our conclusion that RAP80 has a function in DNA damage repair signaling possibly by modulating a specific activity/function of BRCA1.
IR-induced DNA damage results in the activation of the ATM kinase, which in turn leads to the modulation of various signaling pathways through the phosphorylation of many target proteins (5, 11). Some of these proteins appear to function as scaffolds to recruit additional PIKK targets. Several of these target proteins are involved in the activation of cell cycle checkpoints. These include, p53, MDM2, and Chk2, which regulate the G1 checkpoint, and BRCA1 which regulates the G1 or G2 checkpoint depending on its site of phosphorylation. Our results demonstrate that RAP80 is an in vitro target of the ATM protein kinase. This phosphorylation was specific since the phosphorylation was dependent on the IR-activation of ATM while the “kinase-dead” ATM mutant did not phosphorylate RAP80. Analysis of the phosphorylation of several RAP80 mutants demonstrated that Ser205 and Ser402 are ATM phosphorylation sites (Fig. 6).
The translocation of many DNA damage response proteins to IRIF has been shown to be dependent on ATM phosphorylation (5, 26). In contrast, the migration of other proteins, including BRCA1 and NBS1 (43), occurs independently of their phosphorylation by ATM. We showed that a RAP80 mutant, in which both ATM phosphorylation sites are mutated, was still able to translocate to IRIF. This suggests that, as BRCA1 and NBS1, the migration of RAP80 to IRIF occurs independently of its phosphorylation by ATM. Phosphorylation by ATM could affect other functions of RAP80, including its interaction with other proteins. Moreover, phosphorylation of RAP80 at different sites may regulate different functions of RAP80 as has been reported for BRCA1. Phosphorylation at Ser1423 of BRCA1 is important for the G2/M checkpoint but not for the intra-S-checkpoint while the inverse is true for the phosphorylation at Ser1387 (44). Future studies have to determine the functional significance of in vivo phosphorylation of RAP80 by ATM.
In summary, in this study we demonstrate that DSB induce the relocation of the UIM-containing protein RAP80 to DNA damage foci. We provide evidence showing that RAP80 is associated with a protein complex containing BRCA1 but its translocation does not depend on the relocalization of BRCA1. The UIMs and a region between aa 204 and 304 are critical for the translocation of RAP80 and proteins, including BRCA1, associated with this RAP80 complex. We demonstrate that RAP80 knockdown significantly reduces DSB-induced HDR. In addition, overexpression or knockdown of RAP80 expression, respectively, reduces or increases IR-induced cytotoxicity. Its translocation to IRIF, its association with the tumor suppressor BRCA1, and its effects on DSB-induced damage repair suggest that RAP80 has a critical function in DNA damage repair signal transduction and promotes DSB repair. Therefore, mutations in the RAP80 gene that result in aberrant expression or function of RAP80, might have an impact on genome integrity and cancer development.
Supplementary Material
01
Acknowledgments
We would like to thank Drs. Richard Paules and Daniel Menendez for their comments on the manuscript. This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences.
References
- 1.Mills KD, Ferguson DO, Alt FW. The role of DNA breaks in genomic instability and tumorigenesis. Immunological reviews. 2003;194:77–95. doi: 10.1034/j.1600-065x.2003.00060.x. [DOI] [PubMed] [Google Scholar]
- 2.Nyberg KA, Michelson RJ, Putnam CW, Weinert TA. Toward maintaining the genome: DNA damage and replication checkpoints. Annual review of genetics. 2002;36:617–656. doi: 10.1146/annurev.genet.36.060402.113540. [DOI] [PubMed] [Google Scholar]
- 3.Hopfner KP, Putnam CD, Tainer JA. DNA double-strand break repair from head to tail. Curr Opin Struct Biol. 2002;12:115–122. doi: 10.1016/s0959-440x(02)00297-x. [DOI] [PubMed] [Google Scholar]
- 4.Deng CX. BRCA1: cell cycle checkpoint, genetic instability, DNA damage response and cancer evolution. Nucleic acids research. 2006;34:1416–1426. doi: 10.1093/nar/gkl010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Houtgraaf JH, Versmissen J, van der Giessen WJ. A concise review of DNA damage checkpoints and repair in mammalian cells. Cardiovasc Revasc Med. 2006;7:165–172. doi: 10.1016/j.carrev.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 6.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annual review of biochemistry. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 7.Rouse J, Jackson SP. Interfaces between the detection, signaling, and repair of DNA damage. Science (New York, NY. 2002;297:547–551. doi: 10.1126/science.1074740. [DOI] [PubMed] [Google Scholar]
- 8.Melo J, Toczyski D. A unified view of the DNA-damage checkpoint. Curr Opin Cell Biol. 2002;14:237–245. doi: 10.1016/s0955-0674(02)00312-5. [DOI] [PubMed] [Google Scholar]
- 9.Assenmacher N, Hopfner KP. MRE11/RAD50/NBS1: complex activities. Chromosoma. 2004;113:157–166. doi: 10.1007/s00412-004-0306-4. [DOI] [PubMed] [Google Scholar]
- 10.Kurz EU, Lees-Miller SP. DNA damage-induced activation of ATM and ATM-dependent signaling pathways. DNA repair. 2004;3:889–900. doi: 10.1016/j.dnarep.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 11.Kitagawa R, Kastan MB. The ATM-dependent DNA damage signaling pathway. Cold Spring Harbor symposia on quantitative biology. 2005;70:99–109. doi: 10.1101/sqb.2005.70.002. [DOI] [PubMed] [Google Scholar]
- 12.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 13.Banin S, Moyal L, Shieh S, et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science (New York, NY. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 14.Cortez D, Wang Y, Qin J, Elledge SJ. Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science (New York, NY. 1999;286:1162–1166. doi: 10.1126/science.286.5442.1162. [DOI] [PubMed] [Google Scholar]
- 15.Yan Z, Kim YS, Jetten AM. RAP80, a novel nuclear protein that interacts with the retinoid-related testis-associated receptor. The Journal of biological chemistry. 2002;277:32379–32388. doi: 10.1074/jbc.M203475200. [DOI] [PubMed] [Google Scholar]
- 16.Yan J, Kim S-K, Yang X-P, Albers M, Koegl M, Jetten AM. Ubiquitin interacting motifs of RAP80 are critical in its regulation of Estrogen Receptor α. Nucl Acids Res. 2007 doi: 10.1093/nar/gkl1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Fiore PP, Polo S, Hofmann K. When ubiquitin meets ubiquitin receptors: a signalling connection. Nature reviews. 2003;4:491–497. doi: 10.1038/nrm1124. [DOI] [PubMed] [Google Scholar]
- 18.Hoeller D, Crosetto N, Blagoev B, et al. Regulation of ubiquitin-binding proteins by monoubiquitination. Nat Cell Biol. 2006;8:163–169. doi: 10.1038/ncb1354. [DOI] [PubMed] [Google Scholar]
- 19.Wang H, Zhai L, Xu J, et al. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol Cell. 2006;22:383–394. doi: 10.1016/j.molcel.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 20.Huang TT, Nijman SM, Mirchandani KD, et al. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol. 2006;8:339–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- 21.Ziv Y, Banin S, Lim DS, Canman CE, Kastan MB, Shiloh Y. Expression and assay of recombinant ATM. Methods Mol Biol. 2000;99:99–108. doi: 10.1385/1-59259-054-3:99. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J, Willers H, Feng Z, et al. Chk2 phosphorylation of BRCA1 regulates DNA double-strand break repair. Mol Cell Biol. 2004;24:708–718. doi: 10.1128/MCB.24.2.708-718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakanishi K, Yang YG, Pierce AJ, et al. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:1110–1115. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lisby M, Rothstein R. Localization of checkpoint and repair proteins in eukaryotes. Biochimie. 2005;87:579–589. doi: 10.1016/j.biochi.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 25.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 26.Bhoumik A, Takahashi S, Breitweiser W, Shiloh Y, Jones N, Ronai Z. ATM-dependent phosphorylation of ATF2 is required for the DNA damage response. Mol Cell. 2005;18:577–587. doi: 10.1016/j.molcel.2005.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polo S, Sigismund S, Faretta M, et al. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature. 2002;416:451–455. doi: 10.1038/416451a. [DOI] [PubMed] [Google Scholar]
- 28.Celeste A, Fernandez-Capetillo O, Kruhlak MJ, et al. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol. 2003;5:675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- 29.Rai R, Dai H, Multani AS, et al. BRIT1 regulates early DNA damage response, chromosomal integrity, and cancer. Cancer Cell. 2006;10:145–157. doi: 10.1016/j.ccr.2006.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scully R, Ganesan S, Vlasakova K, Chen J, Socolovsky M, Livingston DM. Genetic analysis of BRCA1 function in a defined tumor cell line. Mol Cell. 1999;4:1093–1099. doi: 10.1016/s1097-2765(00)80238-5. [DOI] [PubMed] [Google Scholar]
- 31.Glover JN, Williams RS, Lee MS. Interactions between BRCT repeats and phosphoproteins: tangled up in two. Trends in biochemical sciences. 2004;29:579–585. doi: 10.1016/j.tibs.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 32.Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science (New York, NY. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- 33.Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science (New York, NY. 2003;302:639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 34.Kim ST, Lim DS, Canman CE, Kastan MB. Substrate specificities and identification of putative substrates of ATM kinase family members. The Journal of biological chemistry. 1999;274:37538–37543. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- 35.Fujiwara K, Tenno T, Sugasawa K, et al. Structure of the ubiquitin-interacting motif of S5a bound to the ubiquitin-like domain of HR23B. J Biol Chem. 2004;279:4760–4767. doi: 10.1074/jbc.M309448200. [DOI] [PubMed] [Google Scholar]
- 36.Huang TT, D'Andrea AD. Regulation of DNA repair by ubiquitylation. Nature reviews. 2006;7:323–334. doi: 10.1038/nrm1908. [DOI] [PubMed] [Google Scholar]
- 37.Yu X, Fu S, Lai M, Baer R, Chen J. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes & development. 2006;20:1721–1726. doi: 10.1101/gad.1431006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scully R, Xie A, Nagaraju G. Molecular functions of BRCA1 in the DNA damage response. Cancer Biol Ther. 2004;3:521–527. doi: 10.4161/cbt.3.6.842. [DOI] [PubMed] [Google Scholar]
- 39.Greenberg RA, Sobhian B, Pathania S, Cantor SB, Nakatani Y, Livingston DM. Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes & development. 2006;20:34–46. doi: 10.1101/gad.1381306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421:961–966. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- 41.Lin SY, Rai R, Li K, Xu ZX, Elledge SJ. BRIT1/MCPH1 is a DNA damage responsive protein that regulates the Brca1-Chk1 pathway, implicating checkpoint dysfunction in microcephaly. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15105–15109. doi: 10.1073/pnas.0507722102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhuang J, Zhang J, Willers H, et al. Checkpoint kinase 2-mediated phosphorylation of BRCA1 regulates the fidelity of nonhomologous end-joining. Cancer Res. 2006;66:1401–1408. doi: 10.1158/0008-5472.CAN-05-3278. [DOI] [PubMed] [Google Scholar]
- 43.Kitagawa R, Bakkenist CJ, McKinnon PJ, Kastan MB. Phosphorylation of SMC1 is a critical downstream event in the ATM-NBS1-BRCA1 pathway. Genes & development. 2004;18:1423–1438. doi: 10.1101/gad.1200304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu B, O'Donnell AH, Kim ST, Kastan MB. Phosphorylation of serine 1387 in Brca1 is specifically required for the Atm-mediated S-phase checkpoint after ionizing irradiation. Cancer Res. 2002;62:4588–4591. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01