Fitness consequences of a regulatory polymorphism in a seasonal environment (original) (raw)
Abstract
Gene regulation is commonly assumed to have evolved in response to environmental variability. Although tightly regulated in Escherichia coli strain K12, transcriptional control of arginine biosynthesis is deregulated in strain B. Caused by a single amino acid replacement in the arginine repressor, these contrasting regulatory strategies result in a fitness tradeoff. The K12 repressor is selectively favored in the presence of arginine and disfavored in its absence. In environments that cycle between high and low arginine, short seasons favor the K12 allele, whereas long seasons favor the B allele. Unexpectedly then, deregulated expression is adaptive in some seasonal habitats.
Determining the course of adaptation in variable environments remains a fundamental issue in evolutionary biology (1–4). One obvious outcome is the evolution of regulatory mechanisms that control development and metabolism (5). Another is the appearance of balanced polymorphisms where natural selection maintains functional variation in changeable habitats (5–7). Control of arginine biosynthesis in Escherichia coli does not fit neatly into either of these categories: the mechanism of regulation itself differs among strains, with arginine strongly repressing gene expression in strain K12, but increasing expression in strain B (8–10).
These contrasting regulatory strategies arise as a consequence of a single amino acid replacement at site 70 of the _argR_-encoded repressor (11, 12). X-ray crystallographic structures reveal that site 70 lies at the critical interface between the arginine-binding domain and the DNA-binding domain (13, 14). Replacing the proline in ArgRK12 by the leucine in ArgRB affects communication between these domains, preventing arginine from acting as a potent corepressor of arginine biosynthesis. Consequently, ArgRB allows elevated expression whenever arginine is available (10, 15).
It has been hypothesized (10) that these dissimilar patterns of regulation might reflect adaptations to different natural habitats, and even that the greater potential for enzyme synthesis of strain K12 is an adaptation to survival under conditions of arginine starvation. However, no data to support this conjecture have been proffered. Also unclear are how and whether a fully functional regulatory system, the result of adaptation to a variable environment, might persist with a deregulated system, the expected result of adaptation to a constant environment. These issues make ArgR a suitable model system for exploring the fitness consequences of two contrasting regulatory strategies.
Materials and Methods
Media. Minimal medium is Davis salts [MD: 7 g of K2HPO4, 2 g of KH2PO4, 1 g of (NH4)2SO4, and 0.5 g of trisodium citrate in 1 liter of distilled deionized water with 1 ml of 1 M MgSO4·7H2O and 0.5 ml of 1% thiamine added after autoclaving] supplemented with 2 g/liter glucose and 15 g/liter Bacto agar for plates. Rich medium is Luria broth (LB: 5 g of yeast extract, 10 g of tryptone, and 10 g of NaCl in 1 liter of distilled deionized water with 1 g of glucose and 2.5 mM CaCl2 added after autoclaving). Arginine-free medium (AF) is MD supplemented with an assortment of amino acids (other than arginine) and vitamins (AF is MD with the following added from a sterile 25-fold concentrated stock: 580 mg of dl-alanine, 60 mg of l-aspartatic acid, 70 mg of l-cysteine, 400 mg of l-glutamic acid, 160 mg of glycine, 50 mg of l-histidine, 320 mg of dl-isoleucine, 270 mg of l-leucine, 360 mg of l-lysine, 120 mg of l-methionine, 560 mg of dl-phenylalanine, 140 mg of l-proline, 330 mg of dl-serine, 55 mg of l-tryptophan, 60 mg of l-tyrosine, 200 mg of l-threonine, 330 mg of l-valine, 200 mg of l-asparagine, 20 mg of guanosine, 20 mg of adenosine, 20 mg of uracil, 1 mg of pantothenic acid, 1 mg of riboflavin, 1 mg of thiamine, 1 mg of nicotinic acid, 1 mg of biotin, and 1 mg of pyridoxine). MD was supplemented with 55 mg of l-tryptophan, 60 mg of l-tyrosine, 560 mg of dl-phenylalanine, and 100 mg/liter shikimic acid to support growth of AroE– strains. Antibiotics were added to media at the following concentrations: 20 mg/liter chloramphenicol, 100 mg/ liter streptomycin, 15 mg/liter tetracycline, 5 mg/liter trimethoprim, and 100 mg/liter canavanine.
Strains and Alleles. The genetic background used in these experiments is derived from DD320, a K12 wild-type strain except for a small deletion spanning the lactose operon (16) that is often used for chemostat competition experiments (17). Allele argRK12 (72.91 min) comes from DD320 and allele argRB comes from wild-type B (18) strain CGSC no. 5365 obtained from the E. coli Genetic Stock Center (Yale University, New Haven, CT). Strains were constructed by using the generalized transducing bacteriophage P1(cml clr100) (19).
ArgR– strains DJS2AT and DJS53652AT [trimethoprim-resistant (Tmpr) Δ_argR_::fol_ derivatives of DD320 and CGSC no. 5365, respectively) were constructed by using P1(cml clr100) isolated from MG1655R (20) and selected on MD Tmp plates, the ArgR– phenotype being confirmed by growth on AF canavanine plates. An aromatic amino acid auxotrophy (aroE24,rpsL186 at 73.88 min and 74.84 min) from CGSC no. 5553 (21) was introduced into DJS2AT by cotransduction with the streptomycin resistance marker to produce DJS681, which was isolated on LB streptomycin plates and screened for the AroE– phenotype on minimal plates supplemented with arginine. DJS681 (Δ_argR::fol,aroE24,rpsL186) is the genetic background used in all experiments.
There is no convenient direct selection at argR. Consequently, argRB was moved into a K12 background by cotransduction with an adjacent tetracycline resistance cassette which was introduced specifically for this purpose. First zhc-6::Tn10 (72.5 min) from strain CGSC no. 7442 (22) was transduced into DJS53652AT and a Tetr,Tmpr transductant (DJS5A2A) isolated. Transduction of zhc-6::Tn10 from DJS5A2A into CGSC no 5365, selecting for Tetr and screening for Tmps (DJS102), brought the Tetr marker adjacent to argRB. Strain DJS249 was constructed by cotransduction of argRB with zhc-6::Tn10 from DJS102 into DJS2AT, selecting for Tetr and screening for Tmps. Strain DJS249 has the argRB allele placed the DD320 background.
Cotransduction of argR alleles, from DD320 and DJS249, with aroE+ into DJS681 (Δ_argR_::fol,aroE24,rpsL186), selecting for growth on minimal medium and screening for Tets and Tmps (Strr is retained), completes the strain constructions. Several independently constructed strains were isolated, DJS1204 and DJS1441 carry argRK12 and DJS1262 and DJS1319 carry argRB. Allele argR38, which carries a point mutation inactivating the repressor, was similarly cotransduced from CGSC no. 4519 (23) into DJS681 to produce strains MCS509 and MCS560. Sequencing of PCR argR amplicons confirmed their identities. Spontaneous mutants resistant to the bacteriophage T5 (_fhuA_–) were isolated on LB plates supplemented with 2.5 mM CaCl2. All strains were stored at –80°C in 16% glycerol.
Competition Experiments. Competition experiments are conducted by using standard methods (17). A peristaltic pump delivers fresh medium (MD salts, pH 7.3, supplemented with 5 μM FeSO4, 0.1 g/liter glucose and 10–3 g/liter thiamine with l-arginine added as required) into the chemostat growth chamber at a dilution rate D = 0.33 h–1. Spent medium and cells exit through an overflow siphon. Filtered humidified air, entering the base of the growth chamber, mixes and aerates the culture before escaping through a vent. Experiments are conducted in a 37°C warm room. Competitions in variable environments are conducted in modified chemostats having two media inlets, with the peristaltic pumps that deliver media to the chemostats being turned on and off at specific intervals by a timer.
Competition experiments are conducted between pairs of strains: the first, carrying one argR allele, is sensitive to the bacteriophage T5, whereas the second, carrying the other argR allele, is a resistant isolate. The progress of competition is monitored by following the frequency of T5 resistance over time. Samples, stored overnight at 4°C, are enumerated by flow cytometry (17). Sensitive cells, their membranes depolarized by attachment of excess T5, accumulate YoPro-1-iodide (Molecular Probes) to fluoresce green when excited by a 25-mW air-cooled 488-nm argon laser in a FACScaliber flow (Becton Dickinson). Data acquisition is triggered by sideways-scattered light, with data collected for sideways-(SS) and forwards-(FS) scattered light, and fluorescence (FL) between 505 and 545 nm. The Log10_SS_ vs. Log10_FS_ plots are gated to remove points, such as the bacteriophage T5, that are too small to be E. coli cells. Cell counts are determined from the bimodal Log10_SS_ vs. Log10_FL_ plots that show the fluorescent T5-sensitive population well separated from the nonfluorescent T5-resistant population. Each sample is counted for ≈40 s (between 25,000 and 30,000 counted cells with noncellular “background” counts below 250 ± 20) at 30, 45, and 60 min after staining begins. Sample counts typically show little difference and are considered replicates.
Estimating Fitness. The selection coefficient, _s_12, is estimated as the slope of a plot of the Loge ratio of population densities (_N_i) against time in t (measured in _D_–1 generations),
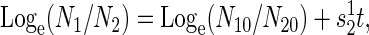 |
[1] |
|---|
where _N_i0 are the population densities at time 0. The mean selection coefficient among replicate experiments is estimated by pooled regression analysis (24). The mean selection coefficient in a cyclically varying environment is determined by least-squares nonlinear regression to
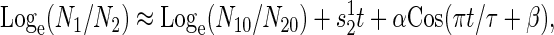 |
[2] |
|---|
where α is the amplitude of the oscillation, π is degrees in radians, τ is the period spent in an environment, and β is the phase of the wave with respect to time. The mean selection coefficient among replicate experiments is estimated as an arithmetic mean. The fitness of one strain relative to a second (_w_12) is estimated as
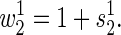 |
[3] |
|---|
Enzyme Assays. The amount of citrulline produced by the action of ornithine transcarbamylase on ornithine and carbamyl phosphate was determined spectrophotometrically after derivitization with diacetyl monoxime in toluenized extracts (25) from chemostat-grown cells. Cell lysates were added to 50 mM diethanolamine/acetic acid, pH 8.1, 5 mM ornithine, and 5 mM carbamyl phosphate and incubated at 37°C for 10 min. To a volume of 0.5 ml of the reaction was added 0.125 ml of 9% NH4Fe(SO4)2·12H2O, 11% (NH4)2Fe(SO4)2·6H2O in 0.5 M H2SO. To this were added 0.625 ml of 16% H2SO4, 42% H3PO4, and then 0.25 ml of 0.75% diacetylmonoxime, and the resulting mixture immediately heated at 100°C in the dark for 30 min. After rapid cooling on ice to room temperature, the A490 was determined in a Cary 300 spectrophotometer and the concentration of citrulline was determined by using a set of standards. Protein concentrations in toluenized extracts were determined by the method of Bradford by using IgG as a standard (26).
Results
We studied the patterns of regulation and the fitnesses conferred by three alternative arginine repressor alleles of E. coli. Two, argRK12 and argRB, occur naturally. The third, argR38, is a laboratory mutant that, in rendering arginine biosynthesis constitutive through lack of activity, serves as an experimental control for the fitness effects of argR regulation. Note that all fitnesses are expressed relative to the argRB allele.
Patterns of Regulation. We assayed the activity of ornithine transcarbamylase, an enzyme involved in arginine biosynthesis, to confirm that different ArgR alleles result in different patterns of regulation in the K12 genetic background during slow growth in chemostats. The expression seen with argRK12 is strongly repressed in the presence of arginine, whereas the weaker deregulated expression seen with argRB is slightly elevated (Table 1). The inactive repressor encoded by argR38 produces very high levels of expression. These results concur with earlier reports (9) that ArgR alleles alone determine the patterns of regulation, and that these same patterns of regulation appear under the competitive environment imposed by slow glucose-limited growth in chemostats.
Table 1. ArgR-regulated ornithine transcarbamylase expression in chemostat grown cultures.
| Feed arginine, mM | ||
|---|---|---|
| Allele | 0 | 1 |
| argRB | 1.00 ± 0.02 | 2.06 ± 0.03 |
| argRK12 | 1.42 ± 0.14 | 0.14 ± 0.02 |
| argR38 | 67.35 ± 0.68 | 68.71 ± 0.68 |
Competition in Constant Environments. We determined the fitness of argRK12 relative to argRB by direct competition in the highly reproducible environment imposed by the chemostat (17), a continuous culture device that forces intense competition for a starvation diet of glucose. When present, arginine is incorporated directly into protein or metabolized to provide additional carbon and nitrogen for growth (27). However, E. coli cannot use arginine as the sole source of carbon and energy (27), and so glucose remains essential and growth limiting in all experiments. Abundant ammonium ensures nitrogen is never limiting. Placing argR alleles in a common genetic background eliminates any possibility that other genetic differences between strains K12 and B influence fitness.
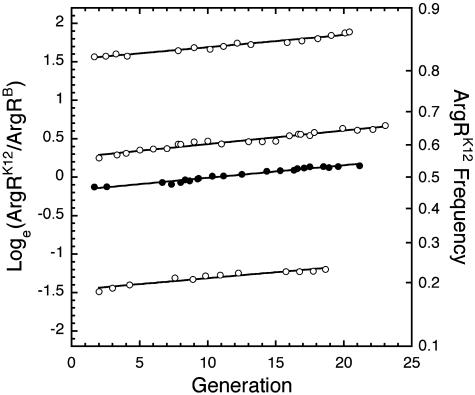
Fig. 1 presents data from four chemostat competition experiments. In these examples, the medium was supplemented with 0.1 mM arginine and the competitions were initiated across a range of allele frequencies. The selective advantage of argRK12 over argRB, estimated as the pooled slope of the lines, is s = 0.0154 ± 0.0008 per generation. T5 resistance, associated with argRK12 in one experiment and with argRB in the other three, has no selective effect and serves simply as a neutral genetic marker that hitchhikes along with the selected argR alleles.
Fig. 1.
Data from chemostat fitness assays reveal that ArgRK12 is fitter than ArgRB when given a constant feed supplemented with 0.1 mM arginine. Neither initial allele frequency nor T5 resistance affects the intensity of selection. Filled circles pertain to competitions where ArgRK12 is T5 resistant and ArgRB is T5 sensitive. Open circles designate competitions where ArgRK12 is T5 sensitive and ArgRB is T5 resistant.
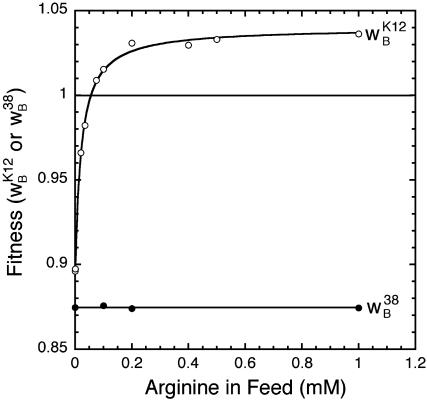
The relationship between fitness and arginine availability was determined in a series of experiments using the chemostat competition assay. Relative to argB, the selection against argRK12 seen in the absence of arginine diminishes rapidly as the concentration of feed arginine is increased (Fig. 2). Eventually, a broad fitness plateau is reached where argRK12 now has the competitive advantage over argRB.
Fig. 2.
The concentration of arginine in the feed medium strongly affects fitness (relative to ArgB) of the functional repressor ArgRK12 (open circles) but not the inactive repressor ArgR38 (filled circles). Fitnesses were calculated from the pooled selection coefficient estimated from at least four replicate experiments by pooled regression analysis. Standard errors lie inside the symbols.
Control competition experiments with the inactive argR38 allele show that the changes in fitness are caused by the differences in gene regulation. Isolated in the laboratory, argR38 encodes a repressor devoid of activity (hence the very high levels of expression seen in Table 1) that serves as a control for the effects on fitness of regulation by argRB and argRK12. Competitions between the inactive ArgR38 and the unresponsive ArgRB (Fig. 2) show that fitness is independent of arginine availability. Hence, the changes in fitness seen during competition between ArgRB and ArgRK12 are solely attributable to regulation by ArgRK12 in response to arginine availability.
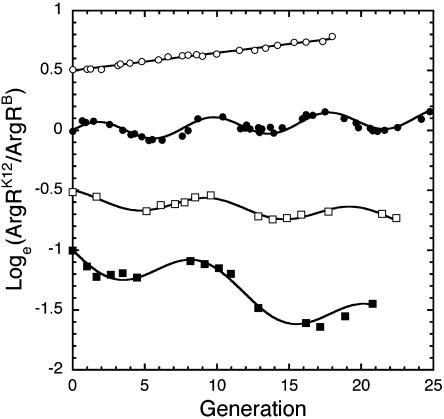
Competition in Variable Environments. The impact on competition of seasonal change was investigated by cycling between feed media supplemented with 0 and 0.2 mM arginine (Fig. 3). Two effects are apparent. First, the oscillations in allele frequency, which are undetectable with seasons two generations long, amplify as the length of the seasons increases (a season is defined as the length of time on 0 or 0.2 mM arginine). Second, a reversal in the overall direction of selection is evident as shorter seasons favoring argRK12 give way to longer seasons favoring argRB.
Fig. 3.
The direction of selection depends on the length of seasons when cycling between feeds supplemented with 0.0 and 0.2 mM arginine. Short seasons of two (open circles) and four (filled circles) generations favor ArgRK12 over ArgR_B_, whereas longer seasons of five (open squares) and six (filled squares) generations favor ArgRB over ArgRK1[supi]2. Each point represents the mean Loge ratio obtained from at least five experiments with standard errors lying inside the symbols. The y axis intercepts are spaced at 0.5 unit for clarity.
Both effects are expected. First, oscillations amplify during longer seasons simply because there is more time for selection to drag allele frequencies up and down. Second, the overall reversal in the direction of selection is a consequence of Jensen's Inequality, which ensures that, for a concave relationship of the sort depicted in Fig. 2, the function of the expectation will exceed the expectation of the function. With very short seasons, either of two situations pertains: (i) no longer having sufficient time to respond to very rapid changes organisms perceive the environment to be a constant average, or (ii) the environment is nearly constant, there being insufficient time when switching between media for arginine concentrations to diverge very far from the true average. In either event, the mean fitness (i.e., function) converges on 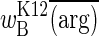 , where
, where 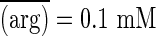 is the average (i.e., expected) arginine concentration. With long seasons, most of the time is spent at 0 and 0.2 mM arginine, and proportionally little time is spent in transit between them. The mean fitness now converges on
is the average (i.e., expected) arginine concentration. With long seasons, most of the time is spent at 0 and 0.2 mM arginine, and proportionally little time is spent in transit between them. The mean fitness now converges on 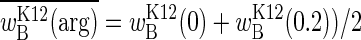 , which, as the average of two fitnesses, is the expectation of the function. That the two mean fitnesses are not equal is readily seen from Fig. 2: short seasons are predicted to yield
, which, as the average of two fitnesses, is the expectation of the function. That the two mean fitnesses are not equal is readily seen from Fig. 2: short seasons are predicted to yield 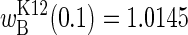 and argRK12 wins the competition, whereas long seasons are predicted to yield
and argRK12 wins the competition, whereas long seasons are predicted to yield 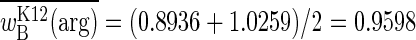 and argRB wins the competition. Experimental results concur with these predictions: the upper bound of
and argRB wins the competition. Experimental results concur with these predictions: the upper bound of 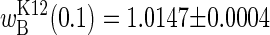 is attained with seasons two generations long, whereas the lower bound is approached with
is attained with seasons two generations long, whereas the lower bound is approached with 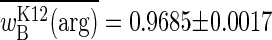 by seasons six generations long.
by seasons six generations long.
The data in Fig. 3 represent averages from at least five replicate experiments in which the initial argRK12 allele frequencies ranged between 0.4 and 0.6. As with competitions in constant environments, the evidence shows that initial allele frequencies have no effect on the long-term outcome of competition. Moreover, a deliberate attempt failed to detect evidence of frequency dependence when competitions with seasons four generations long were initiated with argRK12 allele frequencies of 0.2 and 0.8 (data not shown).
Discussion
Deregulated expression sometimes provides a better adaptive strategy than regulated expression in certain variable environments. This result, so contrary to the prevailing view that regulation is necessarily a preferred adaptive strategy in variable environments, challenges us to test hypotheses of biological regulation, rather than simply accepting the plausible.
We have shown that both the direction and the intensity of selection in a cyclically variable environment can depend on the length of the seasons (Fig. 3). This result is a necessary consequence of the nonlinear relationship between fitness and resource availability (Fig. 2). An intuitive argument to explain why deregulated expression can be favored in a variable environment is as follows. With rapid fluctuations, organisms experience only the average environment. If that average environment contains sufficient arginine for growth, then organisms capable of repressing expression are favored because they do not waste energy and resources making unneeded enzymes and metabolites. With infrequent fluctuations, however, organisms experience each of the two environments (with and without arginine). Under these circumstances, mean fitness is determined by the average of the two fitnesses in each of the two environments, and this average happens to favor deregulated expression.
Our results demonstrate that competitive outcomes under variable conditions are not readily predicted from a few fitnesses measured under constant conditions. Even determining the relationship across a range of arginine environments is insufficient to predict competitive outcomes, except at the extremes of environmental seasonality where fitnesses approach the limits 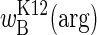 and
and 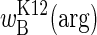 . Predicting fitnesses in seasons of finite length requires knowing the speed with which an organism's physiology can respond to environmental changes. In the absence of such knowledge, the direction of selection must be determined empirically for seasons that fluctuate at intermediate frequencies.
. Predicting fitnesses in seasons of finite length requires knowing the speed with which an organism's physiology can respond to environmental changes. In the absence of such knowledge, the direction of selection must be determined empirically for seasons that fluctuate at intermediate frequencies.
The possibility that argRB and argRK12 form a balanced polymorphism is suggested by each being favored under different circumstances, in both constant and variable environments. Indeed, E. coli B is a wild-type isolate obtained by Bronfebrenner (see ref. 18) and stored under nonmutagenic and grown under nonselective conditions during its entire laboratory history. However, DD320 [strain RV of Jacob (16) and derived from Hfr3300 (28)], like most derivatives of E. coli K12, has been exposed to mutagens and selective media. Nevertheless, the argR of DD320 is identical in sequence to that of MG1655 (GenBank accession no. NP_417704) (29), a wild-type K12 strain with an independent laboratory history (28), although its peptide sequence is identical to those of CFT073 and 0157:H7 (GenBank accession nos. NP_755858 and NP_312137), both recently isolated pathogenic isolates (30, 31). Because argRB and argRK12 occur naturally, it seems worthwhile to enquire whether either is protected from loss.
Theory suggests, and experiments confirm, that competing populations may coexist in an unvarying environment provided that the growth of each is limited to different extents by different resources (17, 32–34), or provided that the growth of one is limited by the availability of a metabolite excreted by an otherwise inferior competitor (35, 36). For example, growth might be limited to different extents by glucose and arginine, or one strain (argRK12) might be limited by the availability of arginine excreted by the second (argRB). Either ecological mechanism produces frequency-dependent selection, where strains are favored when rare and disfavored when common. However, we find no evidence that the intensity and direction of selection depends on allele frequencies across the entire range of arginine concentrations investigated (e.g., see Fig. 1). Frequency-dependent selection, generated by differential resource utilization or by metabolite cross-feeding, does not operate at argR.
Another means to maintain the polymorphism is in a patchy environment, with argRB favored where arginine is absent and argRK12 favored where it is present. Such a mechanism can only be invoked for the short term because, in the long term, regulation will be lost. Where arginine is absent, argRB will rapidly attain dominance, and where arginine is present, the entire metabolic pathway is superfluous and may be lost without affecting fitness, a victim of mutational decay and random genetic drift. Only if organisms are exposed to temporal variation in arginine availability is a regulatory system needed (5).
Theory (37, 38) suggests that two competing populations may coexist on a single seasonally variable resource provided that (i) the direction of selection changes with resource abundance, and (ii) growth is a nonlinear function of resource abundance. Both conditions are fulfilled in our experiment, the first obviously so and the second because the equilibrium population density in chemostats reaches a plateau of 6 × 107 cells per ml at 0.2 mM arginine (data not shown). However, selection is sufficiently weak that the range of environments where coexistence is possible will be tiny, if it exists at all. Not surprisingly, we find no evidence that mean fitness depends on allele frequency with seasons four generations long (data not shown). Frequency-dependent selection generated by environmental seasonality is unlikely to operate at argR.
Inherently unstable in local habitats (whether constant or temporally variable), the argR polymorphism may nevertheless persist in a spatially variable habitat. We envision a kind of serial monomorphism across temporally variable patches in the species range, with argRK12 dominating wherever 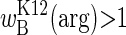 and argRB dominating wherever
and argRB dominating wherever 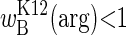 . The scarcity of auxotrophs among natural isolates suggests that patches where arginine is never limiting, or where seasons are so short that fitness converges on
. The scarcity of auxotrophs among natural isolates suggests that patches where arginine is never limiting, or where seasons are so short that fitness converges on 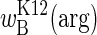 , are rare and short lived: when arginine disappears, the auxotrophs are rapidly purged by intense selection.
, are rare and short lived: when arginine disappears, the auxotrophs are rapidly purged by intense selection.
Demonstrating that deregulated gene expression is a preferred adaptive strategy in certain variable environments suggests a mechanism to maintain a fully functional regulatory system (argRK12) along side a deregulated system (argRB). How commonly this occurs is unknown. Surveys of regulatory polymorphisms are sparse (39, 40), and their functional consequences (41, 42) in relation to seasonal variability are largely unexplored. On the other hand, there are many instances where fitness is anticipated to be a concave function of resource availability, favoring one and then another allele at all manner of genes: regulatory, developmental, structural, and catalytic. For these, the outcome of competition will depend not only on the spatial structure of habitats, but equally on the pattern of environmental seasonality and how such variability transmogrifies fitness.
Acknowledgments
We thank Ben Kerr, Steve Miller, Lin Chao, Dan Dykhuizen, Dan Stroebel, Mike Feldgarden, and Dusty Brisson for their thoughtful and constructive criticisms and Lauren Merlo for sequencing the argR alleles. This work was supported by National Institutes of Health grants (to A.M.D.).
Abbreviation: Tmp, trimethoprim.
References
- 1.Buckling, A., Kassen, R., Bell, G. & Rainey, P. B. (2000) Nature 408 961–965. [DOI] [PubMed] [Google Scholar]
- 2.Oleksiak, M. F., Churchill, G. A. & Crawford, D. L. (2002) Nat. Genet. 32 261–265. [DOI] [PubMed] [Google Scholar]
- 3.Kerr, B., Riley, M. A., Feldman, M. W. & Bohannan, B. J. M. (2002) Nature 418 171–174. [DOI] [PubMed] [Google Scholar]
- 4.Meyers, L. A. & Bull, J. J. (2002) Trends Ecol. Evol. 17 551–557. [Google Scholar]
- 5.Savageau, M. A. (1998) Pacific. Symp. Biocomput. 1998 54–65. [PubMed] [Google Scholar]
- 6.Hedrick, P. W. (1986) Ann. Rev. Ecol. Syst. 17 535–566. [Google Scholar]
- 7.Watt, W. B. & Dean, A. M. (2002) Annu. Rev. Genet. 34 593–622. [DOI] [PubMed] [Google Scholar]
- 8.Ennis, H. L. & Gorini, L. (1961) J. Mol. Biol. 3 439–446. [DOI] [PubMed] [Google Scholar]
- 9.Gorini, L. & Gundersen, W. (1961) Proc. Natl. Acad. Sci. USA 47 961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tian, G., Lim, D. J., Oppenheim, D. & Maas, W. K. (1994) J. Mol. Biol. 235 221–230. [DOI] [PubMed] [Google Scholar]
- 11.Jacoby, G. A. & Gorini, L. (1967) J. Mol. Biol. 24 41–50. [Google Scholar]
- 12.Lim, D., Oppenheim, J., Eckhardt, T. & Maas, W. K. (1988) in Gene Expression and Regulation: The Legacy of Luigi Gorini, eds. Bissell, M., Deho, G., Sironi, G. & Torriani, A. (Excerpta Medica, New York), pp. 55–63.
- 13.Van Duyne, G. D., Ghosh, G., Maas, W. K. & Sigler, P. B. (1996) J. Mol. Biol. 256 377–391. [DOI] [PubMed] [Google Scholar]
- 14.Sunnerhagen, M., Nilges, M., Otting, G. & Carey, J. (1997) Nat. Struct. Biol. 4 819–826. [DOI] [PubMed] [Google Scholar]
- 15.Szwajkazer, D., Dai, L., Fukayama, J. W., Abramczyk, B., Fairman, R. & Carey, J. (2001) J. Mol. Biol. 312 949–962. [DOI] [PubMed] [Google Scholar]
- 16.Jacob, F., Ullmann, A. & Monod, J. (1964) C. R. Acad. Sci. 258 3125–3128. [PubMed] [Google Scholar]
- 17.Lunzer, M., Natarajan, A., Dykhuizen, D. E. & Dean, A. M. (2002) Genetics 162 485–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delbruck, M. & Luria, S. (1942) Arch. Biochem. 1 111–141. [Google Scholar]
- 19.Miller, J. (1992) A Short Course in Bacterial Genetics (Cold Spring Harbor Lab. Press, Plainview, NY).
- 20.Rajagopal, B. S., DePonte, J., III, Tuchmann, M. & Malamy, M. (1998) Appl. Environ. Microbiol. 64 1805–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andresson, O. S., Magnusdottir, R. A. & Eggertssoon, G. (1976) Mol. Gen. Genet. 144 127–130. [DOI] [PubMed] [Google Scholar]
- 22.Nichols, B. P., Shafiq, O. & Meiners, V. (1998) J. Bacteriol. 180 6408–6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baumberg, S. (1970) Mol. Gen. Genet. 106 162–173. [DOI] [PubMed] [Google Scholar]
- 24.Sokal, R. R. & Rohlf, F. J. (1995) Biometry (Freeman, San Francisco), 3rd Ed.
- 25.Tian, G. & Maas, W. K. (1994) Mol. Microbiol. 13 599–608. [DOI] [PubMed] [Google Scholar]
- 26.Bradford, M. (1976) Anal. Biochem. 72 248–254. [DOI] [PubMed] [Google Scholar]
- 27.Glandsorff, N. (1996) in Escherichia coli and Salmonella: Cellular And Molecular Biology, eds. Neidhardt, F. C., Curtiss, R., III, Ingraham, J. L., Lin, E. C. C., Low, K. B., Magasanik, B., Reznikoff, W. S., Riley, M., Schaechter, M. & Umbarger, H. E. (Am. Soc. Microbiol. Press, Washington, DC), 2nd Ed., pp. 408–433.
- 28.Bachmann, B. (1996) in Escherichia coli and Salmonella: Cellular And Molecular Biology, eds. Neidhardt, F. C., Curtiss, R., III, Ingraham, J. L., Lin, E. C. C., Low, K. B., Magasanik, B., Reznikoff, W. S., Riley, M., Schaechter, M. & Umbarger, H. E. (Am. Soc. Microbiol. Press, Washington, DC), 2nd Ed., pp. 2460–2497.
- 29.Blattner, F. R., Plunkett, G., III, Bloch, C. A., Perna, N. T., Burland, V., Riley, M., Collado-Vides, J., Glasner, J. D., Rode, C. K., Mayhew, G. F., et al. (1997) Science 277 1453–1474. [DOI] [PubMed] [Google Scholar]
- 30.Welch, R. A., Burland, V., Plunkett, G., III, Redford, P., Roesch, P., Rasko, D., Buckles, E. L., Liou, S.-R., Boutin, A., Hackett, J., et al. (2002) Proc. Natl. Acad. Sci. USA 99 17020–17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayashi, T., Makino, K., Ohnishi, M., Kurokawa, K., Ishii, K., Yokoyama, K., Han, C.-G., Ohtsubo, E., Nakayama, K., Murata, T., et al. (2001) DNA Res. 8 11–22. [DOI] [PubMed] [Google Scholar]
- 32.Stewart, F. M. & Levin, B. R. (1973) Am. Nat. 107 171–198. [Google Scholar]
- 33.Armstrong, R. A. & McGehee, R. (1976) Theor. Popul. Biol. 9 317–328. [DOI] [PubMed] [Google Scholar]
- 34.Tilman, D. (1982) Resource Competition and Community Structure (Princeton Univ. Press, Princeton). [PubMed]
- 35.Lee, I. H., Fredricksen, A. G. & Tsuchiya, H. M. (1976) Biotechnol. Bioeng. 18 513–526. [DOI] [PubMed] [Google Scholar]
- 36.MeGee, R. D., III, Drake, J. F., Fredricksen, A. G. & Tsuchiya, H. M. (1972) Can. J. Microbiol. 18 1733–1742. [DOI] [PubMed] [Google Scholar]
- 37.Levins, R. (1979) Am. Nat. 114 765–783. [Google Scholar]
- 38.Armstrong, R. A. & McGehee, R. (1980) Am. Nat. 115 151–170. [Google Scholar]
- 39.Gasperini, R. & Gibson, G. (1999) J. Mol. Evol. 49 583–590. [DOI] [PubMed] [Google Scholar]
- 40.Purugganan, M. D. (2000) Mol. Ecol. 9 1451–1461. [DOI] [PubMed] [Google Scholar]
- 41.Hirano, H. Y., Eiguichi, M. & Sano, Y. (1998) Mol. Biol. Evol. 15 978–987. [DOI] [PubMed] [Google Scholar]
- 42.Schulte, P. M., Glemet, H. C., Fiebig, A. A. & Powers, D. A. (2000) Proc. Natl. Acad. Sci. USA 97 6597–6602. [DOI] [PMC free article] [PubMed] [Google Scholar]