Comparative Genome Analysis of Filamentous Fungi Reveals Gene Family Expansions Associated with Fungal Pathogenesis (original) (raw)
Abstract
Fungi and oomycetes are the causal agents of many of the most serious diseases of plants. Here we report a detailed comparative analysis of the genome sequences of thirty-six species of fungi and oomycetes, including seven plant pathogenic species, that aims to explore the common genetic features associated with plant disease-causing species. The predicted translational products of each genome have been clustered into groups of potential orthologues using Markov Chain Clustering and the data integrated into the e-Fungi object-oriented data warehouse (http://www.e-fungi.org.uk/). Analysis of the species distribution of members of these clusters has identified proteins that are specific to filamentous fungal species and a group of proteins found only in plant pathogens. By comparing the gene inventories of filamentous, ascomycetous phytopathogenic and free-living species of fungi, we have identified a set of gene families that appear to have expanded during the evolution of phytopathogens and may therefore serve important roles in plant disease. We have also characterised the predicted set of secreted proteins encoded by each genome and identified a set of protein families which are significantly over-represented in the secretomes of plant pathogenic fungi, including putative effector proteins that might perturb host cell biology during plant infection. The results demonstrate the potential of comparative genome analysis for exploring the evolution of eukaryotic microbial pathogenesis.
Introduction
Fungi and oomycetes are responsible for many of the world's most devastating plant diseases including late blight disease of potato, caused by the oomycete pathogen Phytophthora infestans and rice blast disease caused by the ascomycete fungus Magnaporthe grisea, both of which are responsible for very significant harvest losses each year. The enormous diversity of crop diseases caused by these eukaryotic micro-organisms poses a difficult challenge to the development of durable disease control strategies. Identifying common underlying molecular mechanisms necessary for pathogenesis in a wide range of pathogenic species is therefore a major goal of current research. Approximately 100,000 species of fungi have so far been described, but only a very small proportion of these are pathogenic [1]. Phylogenetic studies have, meanwhile, shown that disease-causing pathogens are not necessarily closely-related to each other, and in fact are spread throughout all taxonomic groups of fungi, often showing a close evolutionary relationship to non-pathogenic species [2], [3]. It therefore seems likely that phytopathogenicity has evolved as a trait many times during fungal and oomycete evolution [1] and in some groups may be ancestral to the more recent emergence of saprotrophic species. A significant effort has gone into the identification of pathogenicity determinants– individual genes that are essential for a pathogen to invade a host plant successfully, but which are dispensable for saprophytic growth [4], [5]. However, far from being novel proteins encoded only by the genomes of pathogenic fungi, many of the genes identified so far encode components of conserved signalling pathways that are found in all species of fungi, such as the mitogen activated protein (MAP) kinases [6], adenylate cyclase [7] and G-protein subunits [8]. The MAP kinase pathways, for example, have been studied extensively in the budding yeast Saccharomyces cerevisiae and trigger morphological and biochemical changes in response to external stimuli such as starvation stress or hyperosmotic conditions [9]. In pathogenic fungi, components of these pathways have evolved instead to regulate the morphological changes associated with plant infection. For example, appressorium formation in the rice blast fungus Magnaporthe grisea, stimulated by hard, hydrophobic surfaces is regulated by a MAP kinase cascade [10]. This pathway deploys novel classes of G-protein coupled receptors not found in the genome of S. cerevisiae [11], but the inductive signal is transmitted via a MAP kinase, Pmk1, that is a functional homologue of the yeast Fus3 MAP kinase where it serves a role in pheromone signalling [10]. Similarly, conserved metabolic pathways such as the glyoxylate cycle and amino acid biosynthesis are also important for pathogenesis [12]–[14]. This may in some cases reflect the nutritional environment the pathogen encounters when growing in the host plant tissue, and in others shows the importance of simple metabolites for pathogenic processes, such as the role of glycerol as a compatible solute for generating turgor pressure in the appressorium of M. grisea [15]. It is undoubtedly the case, however, that identification of such genes has also been a consequence of the manner in which these studies have been carried out, often using yeast as a model organism to test hypotheses concerning the developmental biology and biochemistry of plant pathogenic species.
Other pathogenicity factors identified to date have been shown to be involved in functions associated with host infection, such as plant cell wall degradation, toxin biosynthesis and protection against plant defences [reviewed in 5]. Identification of a pathogenicity factor generally involves making a mutant fungal strain with a non-functioning version of the gene by targeted gene deletion and assaying the ability of the mutant to cause disease. Therefore, most pathogenicity factors identified so far, have been validated in only a small number of genetically tractable pathogenic fungi, such as M. grisea and the corn smut Ustilago maydis and many of the advances in understanding the developmental biology of plant infection have occurred in these model pathogens [16], [17]. However, there are severe limitations to studying pathogenicity by mutating one gene at a time and working predominantly with a hypothesis-driven, reverse genetics approach. Many virulence-associated processes, for instance, such as the development of infection structures and haustoria, are likely to involve a large number of gene products and so there is likely to be redundancy in gene function. One example of this is cutinase, a type of methyl esterase that hydrolyses the protective cutin layer present on the outside of the plant epidermis. Cutinase was excluded as a pathogencity factor for M. grisea on the basis that a mutant strain containing a non-functional cutinase-encoding gene was still able to cause rice blast disease [18]. However, sequencing of the M. grisea genome has shown the presence of eight potential cutinase-encoding genes implicated in virulence [19]. Additionally, targeted gene deletion is not feasible in many important pathogens and the normal definition of fungal pathogenicity cannot be applied in the case of obligate biotrophs, such as the powdery mildew fungus Blumeria graminis, which cannot be cultured away from living host plants. Therefore, new approaches are needed to identify genes that are vital for the process of pathogenicity. These include high-throughput methods such as microarray analysis, serial analysis of gene expression (SAGE), insertional mutagenesis, proteomics and metabolomics [19], [20] and are dependent on the availability of genome sequence information.
After the initial release of the genome of the budding yeast S. cerevisiae in 1996 [21], the number of publicly available sequenced fungal genomes has recently risen very quickly. A large number of fungal genome sequences are now publicly available, including those from several phytopathogenic fungi, including M. grisea [22], Ustilago maydis [23], Gibberella zeae [24] (the causal agent of head blight of wheat and barley), Stagonospora nodorum [25] (the causal agent of glume blotch of wheat), the grey mould fungus Botrytis cinerea and the white mould fungus Sclerotinia sclerotiorum [reviewed in 19]. Comparison of gene inventories of pathogenic and non-pathogenic organisms offers the most direct means of providing new information concerning the mechanisms involved in fungal and oomycete pathogenicity. In this report, we have developed and utilized the e-Fungi object-oriented data warehouse [26], which contains data from 36 species of fungi and oomycetes and deploys a range of querying tools to allow interrogation of a significant amount of genome data in unparalleled detail. We report the identification of new gene families that are over represented in the genomes of filamentous ascomycete phytopathogens and define gene sets that are specific to diverse fungal pathogen species. We also report the putatively secreted protein sets which are produced by plant pathogenic fungi and which may play significant roles in plant infection.
Results
Identification of orthologous gene sets from fungal and oomcyete genomes
Genome sequences and sets of predicted proteins were analysed from 34 species of fungi and 2 species of oomycete (Table 1). In order to compare such a large number of genomes, an object-oriented data warehouse has been constructed known as e-Fungi [26] which integrates genomic data with a variety of functional data and has a powerful set of queries that enables sophisticated, whole-genome comparisons to be performed. To compare genome inventories, the entire set of predicted proteins from the 36 species (348,787 proteins) were clustered using Markov Chain Clustering [27] as described previously [28], [29]. A total of 282,061 predicted proteins were grouped into 23,724 clusters, each cluster representing a group of putative orthologues. The remaining 66,934 sequences were singletons, the products of unique genes. A total of 165 clusters contained proteins from all 36 species used in this study (Table S1). Not surprisingly, they included many proteins involved in basic cellular processes, such as ribosomal proteins, components of transcription, translation and DNA replication apparatus, cytoskeletal proteins, histones, proteins involved in the secretory pathway, protein folding, protein sorting and ubiquitin-mediated proteolysis and enzymes involved in primary metabolism. Only 16 clusters contained proteins that were found in all 34 species of fungi, but which were absent from the two species of oomycete (Table S2). This number of fungal-specific clusters is surprisingly low considering the phylogenetic distance between the oomycetes and fungi [30]. The list however, is consistent with the fundamental differences in biology between fungi and oomycetes and included proteins involved in fungal septation, glycosylation, transcriptional regulation, cell signalling, as well as two amino-acyl tRNA synthetases. The obligate mammalian pathogen Encephalitozoon cuniculi, a microsporidian fungus, has a reduced genome that codes only for 1,997 proteins and lacks genes encoding enzymes of many primary metabolic pathways such as the tricarboxylic acid cycle, fatty acid β-oxidation, biosynthetic enzymes of the vast majority of amino acids, fatty acids and nucleotides, as well as components of the respiratory electron transport chain and F1-F0 ATP synthase. It also lacks mitochondria and peroxisomes [31]. Therefore, we reasoned that the inclusion of this species in the analysis of MCL clusters is likely to result in underestimation of the number of groups of conserved proteins. By discarding E. cuniculi, there are 377 clusters that contained proteins from 35 species of fungi and oomycetes (Table S3). This relatively small number of fungal-conserved clusters reflects the large evolutionary distance between members of the fungal kingdom, as well as complex patterns of gene gains and losses during the evolution of fungi. Basidiomycetes and ascomycetes are thought to have diverged nearly 1,000 million years ago [32] and the Saccharomycotina alone are more evolutionarily diverged than the Chordate phylum of the animal kingdom [33]. Since the divergence of Saccharomycotina (hemiascomycetes) and Pezizomycotina (euascomycetes), the genomes of the latter have greatly increased in size, partly due to the appearance of novel genes related to the filamentous lifestyle. Lineage-specific gene losses have also been shown in a number of hemiascomycete species [34]. As well as the groups of proteins mentioned above (Table S1), the fungal-conserved clusters included those containing enzymes from primary metabolic pathways not present in E. cuniculi, such as the tricarboxylic acid cycle, amino acid metabolism, fatty acid biosynthesis, cholesterol biosynthesis and nucleotide metabolism, as well as components of the respiratory electron transport chain and F1-F0 ATP synthase. The conserved protein clusters also include a number of transporters (including mitochondrial transporters), enzymes involved in haem biosynthesis, autophagy-related proteins, those involved in protein targeting to the peroxisome and vacuole and additional groups of proteins involved in signal transduction that are not present in E. cuniculi (including those involved in inosine triphosphate and leukotriene metabolism). The analysis also showed there were 105 clusters that contained proteins from 33 species of fungi (excluding E. cuniculi), but not from the two species of oomycete (see Table S4). As well as those mentioned previously (Table S2), the group includes a number of clusters of transporters that are conserved in fungi but not found in oomycetes, as well as proteins involved in fungal cell wall synthesis, and lipid metabolism. It may be the case that the genomes of oomycete species do not possess orthologues of the fungal genes in these clusters, or alternatively, the large evolutionary distance between the oomycetes and fungi mean that the corresponding orthologues from each Kingdom cluster separately.
Table 1. Fungal species used in this study.
Comparative analysis of yeasts and filamentous fungi
One striking difference in the morphology of species of fungi is between those that have a filamentous, multi-cellular growth habit and those that grow as single yeast cells. There is some overlap between these two groups; because some fungi are dimorphic or even pleiomorphic, switching between different growth forms depending on environmental conditions or the stage of their life cycle. For example, the corn-smut fungus Ustilago maydis can exist saprophytically as haploid yeast-like cells, but needs to form a dikaryotic filamentous growth form in order to infect the host plant [23]. Generally the genomes of the filamentous fungi contain more protein-encoding genes (9,000–17,000) than those from unicellular yeasts (5,000–7,000), perhaps reflecting their greater morphological complexity and secondary metabolic capacity. U. maydis, however, has 6,522 protein encoding genes, perhaps reflecting its lack of extensive secondary metabolic pathways and its potential usefulness in defining the minimal gene sets associated with biotrophic growth [23]. The increase in proteome size in filamentous ascomycetes may be due to the expansion of certain gene families or the presence of novel genes that are essential for the filamentous lifestyle.
For the purposes of this study, the filamentous fungi were defined as the filamentous ascomycetes (subphylum Pezizomycotina), basidiomycetes and zygomycetes and the unicellular fungi were defined as the budding yeasts (order Saccharomycetales), the archiascomycete Schizosaccharomyces pombe and the microsporidian fungus Encephalitozoon cuniculi. A total of 37 MCL clusters contained proteins from all species of filamentous fungi, but no species of unicellular fungi (Table 2). Interestingly, eight of these clusters also contained proteins from both species of oomycete represented in e-Fungi. The filamentous-fungal specific clusters included a number of proteins that are involved in cytoskeletal rearrangements (dedicator of cytokinesis protein, integrin beta-1-binding protein, dynactin p62 family, dynein light intermediate chain 2), it seems likely that these are required for the complex morphological changes that filamentous fungi undergo during their lifecycle and the production of differentiated cells, such as spores, fruiting bodies and infection structures. The results also suggest that filamentous fungal species make a greater use of lipids as signalling molecules than yeast species. For example, the occurrence of filamentous fungal-specific clusters representing two groups of lysophospholipases, as well as ceramidases that are involved in sphingolipid signalling [35] and linoleate diol synthases that can catalyse the formation of leukotrienes [36]. Interestingly, one of the products of linoleate diol synthase has been shown to be a sporulation hormone in Aspergillus nidulans [37]. There is also a cluster that represents homologues of a novel human gene (LRP16) that acts downstream of a steroid receptor and promotes cell proliferation [38]. Two clusters of filamentous fungal-specific proteins represent enzymes involved in molypterin biosynthesis (MCL2420, MCL2581). Molypterin is a molybdenum-containing co-factor for nitrate reductase, an enzyme that is known to be absent from the species of yeast used in this study [39]. Both these clusters are also found in oomycetes. There are other clusters representing proteins important for activities specific to filamentous fungi, such as homologues of Pro11 (striatin) which regulates fruiting body formation in Sordaria macrospora [40], the vegetatible incompatibility protein HET-E-1, which prevents the formation of heterokaryons between incompatible fungal strains in Podospora anserina [41], anucleate primary sterigmata protein A from Aspergillus nidulans, which is essential for nuclear migration and conidiophore development [42] and cytochrome P450 and polyketide synthase-encoding genes, both of which are involved in a number of secondary metabolic pathways including toxin biosynthesis [43].
Table 2. A list of MCL clusters that are conserved in and specific to filamentous fungi.
| Cluster ID1 | Predicted function of members of cluster2 |
|---|---|
| MCL94 | O-methylsterigmatocystin oxidoreductase (cytochrome P450) (O13345) |
| MCL147 | polyketide synthase (P37693) |
| MCL924 | linoleate diol synthase (Q9UUS2) |
| MCL1613 | acetoacetyl-coenzyme A synthetase (Q9Z3R3) |
| MCL1912 | neutral/alkaline non-lysosomal ceramidase (PF04734) |
| MCL2061 | homogentisate 1,2-dioxygenase (Q00667) |
| MCL2420 | molybdenum cofactor biosynthesis protein (Q9NZB8) |
| MCL2503 | metal tolerance protein (Q9M2P2) |
| MCL2515 | serine protease (Q9QXE5) |
| MCL2581 | gephyrin (Q9NQX3) |
| MCL2664 | similar to bacterial membrane protein (Q8YSU5) |
| MCL2812 | vegetatible incompatibility protein HET-E-1 (Q00808) |
| MCL2938 | 2-nitropropane dioxygenase (PF03060) |
| MCL3026 | saccharopine dehydrogenase (Q8R127) |
| MCL3203 | lysophospholipase (O88202) |
| MCL3466 | cAMP-regulated guanine nucleotide exchange factor II (Q9EQZ6) |
| MCL3490 | cytosolic phospholipase A2 (P50392) |
| MCL3518 | similar to human LRP16 (Q9BQ69) |
| MCL3545 | COP9 signalosome complex subunit 6 (O88545) |
| MCL3546 | anucleate primary sterigmata protein A (Q00083) |
| MCL3547 | dynein light intermediate chain 2, cytosolic (O43237) |
| MCL3573 | 3-oxoacyl-[acyl-carrier-protein] reductase (Q9X248) |
| MCL3665 | dedicator of cytokinesis protein 1 (Q14185) |
| MCL3670 | ketosamine-3-kinase (Q8K274) |
| MCL3770 | unknown |
| MCL3945 | integrin beta-1 binding protein 2 (Q9R000) |
| MCL4010 | dynactin p62 family (PF05502) |
| MCL4033 | citrate lyase beta chain (O53078) |
| MCL4036 | peroxisomal hydratase-dehydrogenase-epimerase (multifunctional beta-oxidation protein) (Q01373) |
| MCL4037 | striatin Pro11 (Q70M86) |
| MCL4054 | histone-lysine N-methyltransferase (Q04089) |
| MCL4055 | unknown |
| MCL4057 | protein of unknown function (PF06884) |
| MCL4058 | UV radiation resistance-associated gene protein (Q9P2Y5) |
| MCL4062 | intramembrane protease (P49049) |
| MCL4068 | unknown |
| MCL4082 | mitochondrial protein cyt-4 (P47950) |
Pathogenicity-associated gene functions in fungi
As the selected set of fungi includes both saprotrophic and pathogenic species, this allows us to compare the gene inventories of phytopathogenic and closely related non-pathogenic fungi to look for genes that are unique to phytopathogens. Analysis of MCL clusters showed that there were no clusters that contained proteins from all species of fungal phytopathogen in e-Fungi (namely B. cinerea, Eremothecium gossypii, G. zeae, M. grisea, S. sclerotiorum, S. nodorum and U. maydis) but did not contain proteins from non-pathogenic species. There were, however, four clusters that were exclusive to filamentous ascomycete phytopathogens (namely B. cinerea, G. zeae, M. grisea, S. sclerotiorum, S. nodorum as shown in Table 3). Significantly, none of the members of these clusters had homology to any known proteins or contained motifs from the Pfam database [44], so we were unable to predict their function, although two of the clusters (MCL4854 and MCL8229) consisted entirely of proteins that were predicted to be secreted. Taken together, the observations indicate that a battery of completely novel secreted proteins may be associated with ascomycete fungal pathogens.
Table 3. Ascomycete phytopathogen-specific MCL clusters.
| Cluster ID | B. cinerea | G. zeae | M. grisea | S. sclerotiorum | S. nodorum |
|---|---|---|---|---|---|
| MCL4854 | 1 | 1 | 6 | 6 | 1 |
| MCL8229 | 1 | 1 | 2 | 1 | 2 |
| MCL9641 | 1 | 1 | 1 | 1 | 1 |
| MCL9651 | 1 | 1 | 1 | 1 | 1 |
Pathogenicity factors have been defined as genes that are essential for successful completion of the pathogen lifecycle but dispensable for saprophytic growth [4]. This is an experimental definition based on whether null mutations of a given gene reduce the virulence of the pathogen on its host. We wished to ascertain whether homologues of previously characterised and experimentally-validated pathogenicity factors were limited to the genomes of pathogenic species. A search was therefore made for pathogenicity factors that have been identified experimentally for the species of phytopathogens represented in e-Fungi using PHI-base, the plant-host interaction database [45]. The matching locus was identified for each pathogenicity factor in the corresponding genome sequence by comparing a published protein sequence with sets of predicted proteins for each genome using BLASTP. This produced a list of 105 pathogenicity factors, although corresponding loci could not be found in genome sequences for all the published genes (see Table S5). MCL clusters containing these proteins were identified (76 unique clusters) and the species distribution of members of these clusters analysed. In total, 29 of the MCL clusters contained pathogenicity factors with members from at least 34 of the 36 species represented in e-Fungi (Table 4). Not surprisingly, many of these clusters contain conserved components of signalling pathways such as protein kinases, adenylate cyclases, G-proteins and cell cycle regulators. Cellular morphogenesis is known to be important for infection of the host plant by many phytopathogens, for example, in appressorium formation in Magnaporthe grisea [46] or the switch in the growth form of Ustilago maydis from yeast-like growth to filamentous invasive growth [47]. Links between successful plant infection and cell cycle control have also been demonstrated [48]. It seems likely that conserved signalling pathways that control activities, such as mating and morphogenesis in all fungi, have evolved to control processes essential for pathogencity in phytopathogens. Other conserved pathogenicity factors encode enzymes of metabolic pathways that are present in nearly all fungi, but seem to be important for the life cycle of particular pathogenic species, for example, enzymes involved in beta-oxidation of fatty acids, the glyoxylate shunt, amino acid metabolism and the utilisation of stored sugars. When considered together, this may indicate that nutritional conditions which fungi encounter when invading host plant tissue require mobilisation of stored lipids prior to nutrition being extracted from the host plant. Seventeen of the MCL clusters containing pathogenicity factors were specific to filamentous ascomycetes (Table 5). These include a number of enzymes involved in secondary metabolism, such as those involved in the synthesis of the fungal toxin trichothecene in G. zeae [43] and those involved in melanin biosynthesis [49], as well as structural proteins, some of which are components of differentiated cell types not seen in yeasts, for example, hydrophobins which are components of aerial structures such as fruiting bodies [50] but are also involved in pathogenicity [16]. There also seems to be a number of filamentous ascomycete specific receptor proteins (transducin beta-subunit, G-protein coupled receptor, tetraspanins) that have evolved in pathogens to be used in sensing environmental cues that are essential for successful infection of the host [51]. The Woronin body is a structure found only in filamentous ascomycetes, and has been shown to be essential for pathogenicity in M. grisea [52]. A major constituent of the woronin body, encoded by MVP1, is a pathogenicity factor for M. grisea, but also has homologues in nearly all species of filamentous ascomycetes. Two proteins that were initially discovered as being highly expressed in the appressoria of M. grisea and essential for pathogenicity (Mas1 and Mas3) [53] also have homologues in a number of species of filamentous fungi (Table 5). Thus, many innovations that have allowed filamentous ascomycetes to have a more complex morphology than unicellular yeasts have also evolved to be essential for plant infection by phytopathogenic species. Interestingly, none of the MCL clusters containing known pathogenicity factors contained members only from phytopathogenic fungi, apart from those that were restricted to just one species. These are therefore likely to represent highly-specialised proteins that have evolved for the specific lifecycle of just one species of phytopathogen, for example the Pwl proteins involved in determining host range of different strains of M. grisea [54]. Two of the proteins specific to M. grisea, the metallothionein Mmt1 [55] and the hydrophobin Mpg1 [56] are small polypeptides and are members of highly divergent gene families, other members of which do not cluster together using BLASTP.
Table 4. MCL clusters containing known pathogenicity factors that have members in at least 34 out of the 36 fungal and oomycete genomes found in e-Fungi.
| Cluster ID | Pathogenicity factor1 | Function | Number of species |
|---|---|---|---|
| MCL11 | MGG_06368.5 (CPKA), UM04456.1 (ADR1), UM04956.1 (UKC1), UM03315.1 (UKB1) | cAMP-dependent protein kinase catalytic subunit | 36 |
| MCL1121 | SNU09357.1 (ALS1) | delta-aminolevulinic acid synthase | 35 |
| MCL120 | UM01643.1 (RAS2) | guanyl nucleotide exchange factor | 35 |
| MCL122 | MGG_03860.5 (TPS1) | trehalose-6-phosphate synthase subunit 1 | 36 |
| MCL1224 | MGG_05201.5 (MGB1) | heterotrimeric G-protein beta subunit | 34 |
| MCL1495 | FG10825.1 (MSY1) | methionine synthase | 34 |
| MCL150 | BC1G_03430.1 (PIC5) | FKBP-type peptidyl-prolyl cis-trans isomerase | 35 |
| MCL1545 | MGG_07528.5 (PTH3) | imidazoleglycerol-phosphate dehydratase | 34 |
| MCL157 | MGG_12855.5 (MST11), UM04258.1 (KPP4) | MAP kinase kinase kinase | 35 |
| MCL175 | UM02588.1 (CLB2), UM04791.1 (CLN1) | cyclin | 35 |
| MCL179 | MGG_00800.5 (MST7), UM01514.1 (FUZ7) | MAP kinase kinase | 34 |
| MCL193 | BC1G_01681.1 (BCG1), SNU10086.1 (GNA1), MGG_00365.5 (MAGB), UM04474.1 (GPA3) | G alpha protein subunit | 35 |
| MCL196 | MGG_01721.5 (PTH2) | carnitine acetyl transferase | 34 |
| MCL24 | UM04218.1 (KIN2) | kinesin motor protein | 36 |
| MCL244 | FG01932.1 (CBL1) | cystathionine beta-lyase | 35 |
| MCL248 | UM01516.1 (SQL2) | guanyl nucleotide exchange factor | 34 |
| MCL295 | UM03917.1 (CRU1) | cell cycle regulatory protein | 36 |
| MCL42 | MGG_00529.5 (PEX6) | peroxin, peroxisome biogenesis | 36 |
| MCL421 | MGG_06148.5 (MFP1) | multifunctional beta-oxidation enzyme | 34 |
| MCL446 | MGG_04895.5 (ICL1) | isocitrate lyase | 35 |
| MCL46 | BC1G_13966.1 (BMP1), FG10313.1 (MGV1), FG06385.1 (MAP1), MGG_04943.5 (MPS1), MGG_09565.5 (PMK1), SNU03299.1 (MAK2), UM03305.1 (KPP2), UM02331.1 (KPP6), UM03305.1 (UBC3) | MAP kinase | 36 |
| MCL49 | BC1G_01740.1 (BCP1), MGG_10447.5 (CYP1) | cyclophillin | 36 |
| MCL54 | MGG_06320.5 (CHM1), UM04583.1 (SMU1), UM02406.1 (CLA4) | PAK kinase | 35 |
| MCL618 | MGG_07335.5 (SUM1), UM06450.1 (UBC1) | cAMP-dependent protein kinase regulatory subunit | 35 |
| MCL726 | SNU03643.1 (ODC) | ornithine decarboxylase | 34 |
| MCL761 | SNU07548.1 (MLS1) | malate synthase | 34 |
| MCL892 | UM04405.1 (GAS1) | alpha-glucosidase | 34 |
| MCL9 | BC1G_04420.1 (BcatrB), MGG_13624.5 (ABC1) | ABC transporter | 35 |
| MCL95 | MGG_00111.5 (PDE1), MGG_02767.5 (APT2) | P-type ATPase, aminophospholipid translocase | 35 |
Table 5. MCL clusters containing known pathogenicity factors that have members only in the genomes of filamentous ascomycetes.
| Cluster ID | Pathogenicity factor1 | Function | Number of species |
|---|---|---|---|
| MCL11972 | FG03537.1 (TRI5) | trichodiene synthase | 3 |
| MCL14401 | FG03536.1 (TRI6) | transcription factor, trichothecene biosynthesis pathway | 2 |
| MCL18766 | MGG_04301.5 (PWL1/2), MGG_13863.5 (PWL1/2) | host species-specificity protein | 1 |
| MCL2795 | FG01555.1 (ZIF1) | b-ZIP transcription factor | 13 |
| MCL29 | BC1G_13298.1 (BTP1), MGG_05871.5 (PTH11) | G-protein coupled receptor | 13 |
| MCL4777 | MGG_02696.5 (MVP1) | vacuolar ATPase, woronin body protein | 12 |
| MCL48738 | FG03543.1 (TRI14) | trichothecene biosynthesis gene | 1 |
| MCL52178 | MGG_06873.5 (ORP1) | essential for penetration of host leaves | 1 |
| MCL52784 | MGG_09730.5 (MMT1) | metallothionein | 1 |
| MCL52927 | MGG_10315.5 (MPG1) | class I hydrophobin | 1 |
| MCL6180 | MGG_04202.5 (MAS1) | highly expressed in appressoria | 9 |
| MCL6560 | MGG_05059.5 (RSY) | scytalone dehydratase | 9 |
| MCL7081 | MGG_01173.5 (MHP1) | class II hydrophobin | 7 |
| MCL7423 | BC1G_09439.1 9 (BcPLS1) | tetraspanin | 9 |
| MCL8295 | FG00332.1 (TBL1) | transducin beta-subunit | 7 |
| MCL8340 | MGG_12337.5 (MAS3) | highly expressed in appressoria | 6 |
| MCL8912 | MGG_00527.5 (EMP1) | extracellular matrix protein | 6 |
Comparative analysis of plant-pathogenic and saprotrophic filamentous ascomycetes
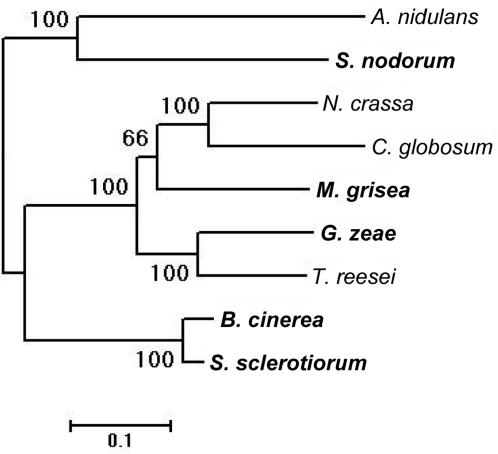
Based on the analysis reported, it is likely that in general there are a large number of differences in gene inventories between filamentous and yeast-like fungi. Therefore, in order to compare the genomes of phytopathogens and saprotrophs, we focused on filamentous ascomycetes in order to resolve in greater detail the distinct differences in gene sets between these two ecologically separate groups of fungi. In this way differences due to phylogeny between the species would be minimised. We compared the gene inventories of the phytopathogens B. cinerea, G. zeae, M. grisea, S. sclerotiorum, S. nodorum with the non-pathogens Aspergillus nidulans, Chaetomium globosum, Neurospora crassa and Trichoderma reesei. Phylogenetic analysis suggests that the phytopathogenic species do not form a separate clade from the pathogenic species (Figure 1), [3] and we assumed that differences in gene inventory should therefore reflect lifestyle rather than evolutionary distance. In order for such a comparison to be considered valid, the completeness and quality of the fungal genome sequences used should, however, also be comparable. Table S6 summarises the available data about genome sequence coverage, genome size and the number of predicted proteins for each species. This shows that the genome coverage is greater than 5x and the number of predicted proteins in the range of 10,000–16,000 for all genomes used, suggesting a high level of equivalence between species with regard to sequence quality. From our work it seems unlikely that there are pathogenicity factors conserved in, and specific to, all species of phytopathogen. It may, for instance, be the case that differences in the gene inventories are due to the expansion of certain gene families in the genomes of phytopathogenic species associated with functions necessary for pathogenesis. To define protein families, we used the Pfam database which contains protein family models based on Hidden Markov Models [44], [57]. Sets of predicted proteins for each fungal species in e-Fungi were analysed for the occurrence of Pfam motifs and the number of proteins containing each domain across fungal species ascertained. The sets of predicted protein sequences used in this study have been automatically predicted as part of each individual genome project and are likely to contain a number of artefactual sequences. The use of Pfam motifs to define gene families in this study reduces the likelihood of such sequences affecting the data, since Pfam motifs are based on multiple sequence alignments of well-studied proteins.
Figure 1. Species tree of filamentous ascomycetes used in this study based on concatenated sequences from 60 universal fungal protein families.
Support values shown for each branch (based on 100 bootstraps). Phytopathogenic species are highlighted in bold type. A more detailed methodology has been described previously [26].
A small number of Pfam motifs were not found in the proteomes of the filamentous ascomycete non-pathogens, but were found in the proteomes of at least three species of filamentous ascomycete phytopathogens (Table 6). These include the Cas1p-like motif (PF07779), found in 4 species of phytopathogen, including five copies in G. zeae, and the Yeast cell wall synthesis protein KRE9/KNH1 motif (PF05390), which was found in three species of phytopathogen. Cas1p is a membrane protein necessary for the O-acetylation of the capsular polysaccharide of the basidiomycete animal pathogen Cryptococcus neoformans [58]. KRE9 and KNH1 are involved in the synthesis of cell surface polysaccharides in S. cerevisiae [59]. Taken together this suggests that synthesis of cell surface polysaccharides is important for phytopathogens, perhaps helping to shroud the fungus from plant defences. The function of the YDG/SRA domain motif (PF02182) is unknown, but is found in a novel mouse cell proliferation protein Np95, in which the domain is important both for the interaction with histones and for chromatin binding in vivo [60]. As well as domains of unknown function, the list of phytopathogen-specific Pfam motifs includes Allophanate hydrolase (PF02682) which is found in an enzyme involved in the ATP-dependent urea degradation pathway [61], a peptidase motif, an opioid growth receptor motif (PF04664) and Mnd1 (PF03962), which is involved in recombination and meiotic nuclear division [62].
Table 6. Pfam motifs that are found in the proteomes from at least three species of phytopathogen, but in no species of filamentous ascomycete non-pathogen. Table shows the number of predicted proteins that contain each Pfam motif.
| Pfam accession | Pfam description | B. cinerea | G. zeae | M. grisea | S. sclerotiorum | S. nodorum |
|---|---|---|---|---|---|---|
| PF07779 | Cas1p-like protein | 1 | 5 | 1 | 1 | 0 |
| PF02182 | YDG/SRA domain | 1 | 0 | 1 | 0 | 1 |
| PF02682 | Allophanate hydrolase subunit 1 | 0 | 1 | 1 | 0 | 1 |
| PF03577 | Peptidase family C69 | 0 | 1 | 1 | 0 | 1 |
| PF03962 | Mnd1 family | 1 | 0 | 0 | 1 | 1 |
| PF04664 | Opioid growth factor receptor (OGFr) conserved region | 1 | 0 | 0 | 1 | 1 |
| PF05390 | Yeast cell wall synthesis protein KRE9/KNH1 | 1 | 0 | 0 | 1 | 1 |
| PF05899 | Protein of unknown function (DUF861) | 1 | 3 | 0 | 1 | 0 |
| PF06916 | Protein of unknown function (DUF1279) | 1 | 0 | 1 | 1 | 0 |
| PF06993 | Protein of unknown function (DUF1304) | 0 | 1 | 1 | 0 | 1 |
To detect potential gene family expansion, we decided to identify Pfam motifs that were present in both phytopathogenic and non-pathogenic species of filamentous ascomycetes, but that were more common in the genomes of the former. The Pfam motifs were ranked on the ratio of the mean number of proteins containing each motif in phytopathogens, when compared to non-pathogens (Table 7). The tables only show ratios of greater than or equal to 2.5. Pfam motifs that were more common in the proteomes of pathogens, include some found in enzymes involved in secondary metabolic pathways. These include novel enzymes that have only previously been studied in non-fungal species, such as the chalcone synthases; type III polyketide synthases involved in the biosynthesis of flavonoids in plants [63] and lipoxygenases; components of metabolic pathways resulting in the synthesis of physiologically-active compounds such as eicosanoids in mammals [64] and jasmonic acid in plants [65] as well as antibiotic synthesis monooxygenases. It seems likely that secondary metabolism is essential in phytopathogenic species for the synthesis of mycotoxins, antibiotics, siderophores and pigments [66], but it may also offer fungal pathogens a distinct alternative means of perturbing host metabolism, cell signalling or plant defence, in contrast to bacterial pathogens that rely on protein secretion to achieve this. There also seems to be number of protease and peptidase domains that are more common in the genomes of phytopathogens as well as domains from two classes of cell-wall degrading enzymes: namely cutinase (PF01083) and Glycosyl hydrolase family 53 (PF07745) which is found in arabinogalactan endo-1,4-beta-galactosidases that hydrolyze the galactan side chains that form part of the complex carbohydrate structure of pectin [67]. Two other domains found in enzymes involved in pectin degradation, pectinesterase (PF01095) and Glycosyl hydrolases family 28 (PF00295) are both more than twice as common in the genomes of phytopathogens than saprotrophs. In contrast, domains found in cellulases have fairly equal distribution between the proteomes of phytopathogens and non-pathogens (data not shown). Therefore, for phytopathogens the most essential enzymes for pathogenesis may well be those that allow the fungus to penetrate the protective cutin layer of the plant epidermis and disrupt the pectin matrix of the plant cell wall in which cellulose fibrils are embedded. Pectin-degrading enzymes have already been shown to be pathogenicity factors in a number of fungi [68]. NPP1 motifs are characteristic of a group of proteins called NLPs (Nep1-like proteins) that trigger defence responses, necrosis and cell death in plants and may act as virulence factors [69]. The NLPs are more common in the genomes of phytopathogenic, when compared to non-pathogenic ascomycetes, but are even more numerous in the proteomes of the oomycetes (64 proteins in Phytophthora ramorum and 75 in Phytophthora sojae). Proteins containing the Chitin recognition protein domain (PF00187) are also very common in the proteomes of phytopathogens (18 in M. grisea and 16 in S. nodorum). A role for chitin-binding proteins has been proposed in protecting the fungal cell wall from chitinases produced by host plants [70]. There are also two other Pfam motifs, which are more common in the proteomes of phytopathogens, that are found in enzymes involved in the catabolism of toxic compounds, namely arylesterase (PF01731) and EthD protein (PF07110) which breakdown organophosphorus esters [71] and ethyl tert-butyl ether [72], respectively.
Table 7. Pfam motifs that are at least twice as common in the proteomes of filamentous ascomycete phytopathogens, compared to filamentous ascomycete non-pathogens.
| Accession | Pfam description | B. cin | G. zea | M. gri | S. scl | S. nod | A. nid | C. glo | N. cra | T. ree | path1 | non-path2 | Ratio3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PF00195 | Chalcone and stilbene synthases, N-terminal domain | 1 | 1 | 2 | 1 | 2 | 0 | 0 | 1 | 0 | 1.4 | 0.25 | 5.6 |
| PF01731 | Arylesterase | 1 | 3 | 0 | 2 | 1 | 0 | 0 | 0 | 1 | 1.4 | 0.25 | 5.6 |
| PF07110 | EthD protein | 1 | 2 | 0 | 2 | 2 | 0 | 0 | 0 | 1 | 1.4 | 0.25 | 5.6 |
| PF03935 | Beta-glucan synthesis-associated protein (SKN1) | 2 | 0 | 0 | 2 | 2 | 1 | 0 | 0 | 0 | 1.2 | 0.25 | 4.8 |
| PF00024 | PAN domain | 2 | 12 | 2 | 1 | 5 | 0 | 0 | 2 | 2 | 4.4 | 1 | 4.4 |
| PF02128 | Fungalysin metallopeptidase (M36) | 0 | 1 | 2 | 0 | 2 | 0 | 0 | 0 | 1 | 1 | 0.25 | 4.0 |
| PF02705 | K+ potassium transporter | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 0.25 | 4.0 |
| PF07504 | Fungalysin/Thermolysin Propeptide Motif | 0 | 1 | 2 | 0 | 2 | 0 | 0 | 0 | 1 | 1 | 0.25 | 4.0 |
| PF00754 | F5/8 type C domain | 2 | 4 | 1 | 1 | 1 | 0 | 0 | 2 | 0 | 1.8 | 0.5 | 3.6 |
| PF03992 | Antibiotic biosynthesis monooxygenase | 3 | 1 | 1 | 2 | 2 | 1 | 0 | 1 | 0 | 1.8 | 0.5 | 3.6 |
| PF03572 | Peptidase family S41 | 2 | 1 | 3 | 1 | 6 | 0 | 0 | 2 | 1 | 2.6 | 0.75 | 3.5 |
| PF00209 | Sodium:neurotransmitter symporter family | 0 | 3 | 3 | 0 | 2 | 1 | 0 | 0 | 1 | 1.6 | 0.5 | 3.2 |
| PF00659 | POLO box duplicated region | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0.8 | 0.25 | 3.2 |
| PF01400 | Astacin (Peptidase family M12A) | 0 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0.8 | 0.25 | 3.2 |
| PF02018 | Carbohydrate binding domain | 0 | 2 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0.8 | 0.25 | 3.2 |
| PF02116 | Fungal pheromone mating factor STE2 GPCR | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0.8 | 0.25 | 3.2 |
| PF02244 | Carboxypeptidase activation peptide | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0.8 | 0.25 | 3.2 |
| PF03928 | Domain of unknown function (DUF336) | 1 | 1 | 1 | 1 | 4 | 1 | 0 | 0 | 1 | 1.6 | 0.5 | 3.2 |
| PF05051 | Cytochrome C oxidase copper chaperone (COX17) | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0.8 | 0.25 | 3.2 |
| PF05493 | ATP synthase subunit H | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0.8 | 0.25 | 3.2 |
| PF05631 | Protein of unknown function (DUF791) | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0.8 | 0.25 | 3.2 |
| PF05783 | Dynein light intermediate chain (DLIC) | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0.8 | 0.25 | 3.2 |
| PF01083 | Cutinase | 11 | 12 | 17 | 8 | 11 | 4 | 5 | 3 | 4 | 11.8 | 4 | 3.0 |
| PF00187 | Chitin recognition protein | 6 | 8 | 18 | 8 | 16 | 8 | 7 | 0 | 1 | 11.2 | 4 | 2.8 |
| PF00305 | Lipoxygenase | 3 | 1 | 1 | 2 | 0 | 0 | 1 | 1 | 0 | 1.4 | 0.5 | 2.8 |
| PF00314 | Thaumatin family | 2 | 1 | 1 | 2 | 1 | 0 | 0 | 1 | 1 | 1.4 | 0.5 | 2.8 |
| PF02797 | Chalcone and stilbene synthases, C-terminal domain | 1 | 1 | 2 | 1 | 2 | 0 | 1 | 1 | 0 | 1.4 | 0.5 | 2.8 |
| PF05630 | Necrosis inducing protein (NPP1) | 2 | 4 | 4 | 2 | 2 | 2 | 1 | 1 | 0 | 2.8 | 1 | 2.8 |
| PF07745 | Glycosyl hydrolase family 53 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 0 | 1.4 | 0.5 | 2.8 |
| PF02129 | X-Pro dipeptidyl-peptidase (S15 family) | 2 | 4 | 3 | 1 | 7 | 3 | 1 | 1 | 0 | 3.4 | 1.25 | 2.7 |
Comparative secretome analysis of phytopathogenic and saprotrophic filamentous ascomycetes
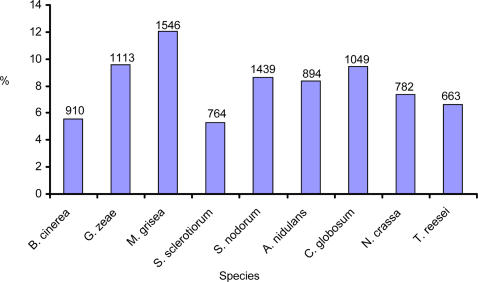
Studies in bacterial pathogens and oomycetes have shown that a range of secreted proteins known as effectors are important for establishing infection of the host plant [73], [74]. These secreted proteins may disable plant defences and subvert cellular processes to suit the needs of invading pathogens. Therefore, we decided also to compare gene family size in the secretomes of phytopathogens and non-pathogens. There are a number of programs available that predict whether a protein is likely to be secreted, although the predictions they give significantly differ from each other. Therefore we defined the secretome of each fungal species based on those proteins that are predicted to be secreted by two different programs: SignalP 3.0 [75] and WoLFPSORT [76]. The size of each secretome is summarised in Figure 2. Even when using two programs, the sizes of predicted secretomes can vary greatly. For example, a similar analysis for M. grisea using SignalP and ProtComp (www.Softberry.com) predicted only 739 secreted proteins (out of a proteome of 11,109) compared to our prediction of 1,546 secreted proteins (out of a proteome of 12,841) [22]. The size of the secretomes for each species varied from 5%–12% of the total proteome. Overall, the size of the secretomes from phytopathogens did not differ greatly from that of non-pathogens.
Figure 2. Bar chart showing the percentage of the total proteome that is predicted to be secreted in each fungal species.
The number of secreted proteins is indicated at the top of each bar.
Table 8 shows a list of Pfam motifs, not found in the secretomes of non-pathogenic filamentous ascomycetes, that were present in at least three phytopathogenic fungal species. The Isochorismatase motif (PF00857) was found in the secretomes of all five species of phytopathogen. Isochorismatase catalyses the conversion of isochorismate to 2,3-dihydroxybenzoate and pyruvate. It has been implicated in the synthesis of the anti-microbial compound phenazine by Pseudomonas aeruginosa [77] and the siderophore, enterobactin, by Escherichia coli [78]. The isochorismatase motif is also found in a number of hydrolases, such as nicotinamidase that converts nicotinamide to nicotinic acid [79]. Members of this family are found in all filamentous ascomycetes, but interestingly they are only secreted in phytopathogens. Salicylic acid is synthesised in plants in response to pathogen attack and mediates plant defences. As isochorismate is a precursor of salicyclic acid [80], it may be worth speculating that isochorismatases secreted by fungi could act to reduce salicylic acid accumulation in response to pathogen attack and thus inhibit plant defence responses. The secreted isochorismatases (apart from one of the proteins from S. nodorum) all show sequence similarity to ycaC from E. coli, an octameric hydrolase of unknown function [81]. Pfam motifs found in the secretomes of at least three species of phytopathogens, but not in any of the non-pathogens also include those found in enzymes potentially involved in detoxification, such as arylesterase and amidohydrolase, and also beta-ketoacyl synthase, which catalyses the condensation of malonyl-ACP with a growing fatty acid chain and is found as a component of a number of enzyme systems, including fatty acid synthases and polyketide synthases [82], [83].
Table 8. Pfam motifs that are found in the secretomes from at least three species of phytopathogen but in no species of filamentous ascomycete non-pathogen.
| Pfam accession | Pfam description | B. cinerea | G. zeae | M. grisea | S. sclerotiorum | S. nodorum |
|---|---|---|---|---|---|---|
| PF00857 | Isochorismatase family | 1 | 1 | 1 | 1 | 2 |
| PF01731 | Arylesterase | 1 | 2 | 0 | 1 | 1 |
| PF04113 | Gpi16 subunit, GPI transamidase component | 1 | 1 | 1 | 1 | 0 |
| PF07969 | Amidohydrolase family | 1 | 2 | 0 | 1 | 1 |
| PF00109 | Beta-ketoacyl synthase, N-terminal domain | 1 | 0 | 1 | 1 | 0 |
| PF01156 | Inosine-uridine preferring nucleoside hydrolase | 1 | 0 | 1 | 1 | 0 |
| PF02801 | Beta-ketoacyl synthase, C-terminal domain | 1 | 0 | 1 | 1 | 0 |
| PF03134 | TB2/DP1, HVA22 family | 0 | 0 | 1 | 1 | 1 |
| PF04253 | Transferrin receptor-like dimerisation domain | 0 | 2 | 1 | 0 | 1 |
| PF05390 | Yeast cell wall synthesis protein KRE9/KNH1 | 1 | 0 | 0 | 1 | 1 |
Table 9 shows a list of Pfam motifs that are more common in the secretomes of phytopathogens as compared to saprotrophs. These include a number of secreted proteases, transcription factors and components of signal transduction pathways. The Kelch domain (PF01344) shows the most striking difference in distribution between phytopathogenic and non-pathogenic genomes. This 50-residue domain is found in a number of actin-binding proteins [84], as well as enzymes such as galactose oxidase and neuraminidase. The putative function of each secreted Kelch domain-containing protein was ascertained by performing a BLAST search against the NCBI non-redundant protein database (Table 10). A number of these seem to be galactose oxidases, enzymes which catalyse the oxidation of a range of primary alcohols, including galactose, to the corresponding aldehyde with the concomitant reduction of oxygen to hydrogen peroxide (H2O2) [85]. Galactose oxidase shares a copper radical oxidase motif with the hydrogen peroxide-generating glyoxal oxidases involved in lignin-degradation in Phanerochaete chrysosporium [86]. H2O2-producing copper oxidases have been shown to have roles in morphogenesis, in the corn-smut fungus Ustilago maydis for example, a glyoxal oxidase is required for filamentous growth and pathogenicity [87] and a galactose oxidase is involved in fruiting body formation in the gram-negative bacterium Stigmatella aurantiaca [88]. Interestingly, the list of Pfam motifs more common in the secretomes of phytopathogens also includes those found in copper amine oxidases, H2O2-generating enzymes that catalyse the oxidative deamination of primary amines to the corresponding aldehydes [89] and peroxidases, haem-containing enzymes that use hydrogen peroxide as the electron acceptor to catalyse a number of oxidative reactions. Secreted fungal peroxidases include enzymes involved in lignin breakdown by the white rot fungus Phanerochaete chrysosporium [90], but in plants they generate reactive oxygen species and are involved in defence responses and growth induction [91]. A number of other secreted Kelch domain-containing proteins have similarity to proteins of unknown function from species of the bacterial phytopathogen Xanthomonas. Many Kelch domain-containing proteins are involved in cytoskeletal rearrangement and cell morphology [92], [93]. It may be worth speculating that secreted Kelch domain-containing proteins could act as effectors, causing changes in the arrangement of the cytoskeleton of infected plants to aid the proliferation of fungal hyphae. It has recently been shown, for example, that M. grisea co-opts plasmodesmata to move from cell to cell in infected rice leaves [94] and would therefore need to peturb cytoskeletal organisation in rice epidermal cells. There are other Pfam domains that are more common in the secretomes of phytopathogens that may potentially be found in effectors such as the PAN domain (PF00024), that mediates protein-protein and protein-carbohydrate interactions [95] and the F5/8 type C domain (PF00754), found in the discoidin family of proteins involved in cell-adhesion or developmental processes [96].
Table 9. Pfam motifs that are at least twice as common in the secretomes of filamentous ascomycete phytopathogens as compared to filamentous ascomycete non-pathogens.
| Accession | Pfam description | B. cin | G. zea | M. gri | S. scl | S. nod | A. nid | C. glo | N. cra | T. ree | path1 | non-path2 | Ratio3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PF01344 | Kelch motif | 2 | 3 | 4 | 1 | 5 | 0 | 1 | 0 | 0 | 3 | 0.25 | 12.0 |
| PF00024 | PAN domain | 2 | 9 | 0 | 1 | 3 | 0 | 0 | 0 | 2 | 3 | 0.5 | 6.0 |
| PF04082 | Fungal specific transcription factor domain | 2 | 2 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 1.2 | 0.25 | 4.8 |
| PF00089 | Trypsin | 1 | 2 | 3 | 1 | 3 | 1 | 0 | 0 | 1 | 2 | 0.5 | 4.0 |
| PF00232 | Glycosyl hydrolase family 1 | 0 | 1 | 1 | 2 | 1 | 1 | 0 | 0 | 0 | 1 | 0.25 | 4.0 |
| PF01019 | Gamma-glutamyltranspeptidase | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0.25 | 4.0 |
| PF01161 | Phosphatidylethanolamine-binding protein | 1 | 2 | 4 | 0 | 3 | 1 | 1 | 0 | 0 | 2 | 0.5 | 4.0 |
| PF02128 | Fungalysin metallopeptidase (M36) | 0 | 1 | 2 | 0 | 2 | 0 | 0 | 0 | 1 | 1 | 0.25 | 4.0 |
| PF03403 | Platelet-activating factor acetylhydrolase, plasma/intracellular isoform II | 1 | 0 | 1 | 1 | 2 | 0 | 1 | 0 | 0 | 1 | 0.25 | 4.0 |
| PF04909 | Amidohydrolase | 1 | 1 | 0 | 1 | 2 | 0 | 0 | 1 | 0 | 1 | 0.25 | 4.0 |
| PF07504 | Fungalysin/Thermolysin Propeptide Motif | 0 | 1 | 2 | 0 | 2 | 0 | 0 | 0 | 1 | 1 | 0.25 | 4.0 |
| PF08244 | Glycosyl hydrolases family 32 C terminal | 0 | 1 | 1 | 1 | 2 | 0 | 0 | 1 | 0 | 1 | 0.25 | 4.0 |
| PF00246 | Zinc carboxypeptidase | 2 | 7 | 9 | 1 | 8 | 1 | 1 | 2 | 2 | 5.4 | 1.5 | 3.6 |
| PF00445 | Ribonuclease T2 family | 2 | 3 | 1 | 2 | 1 | 1 | 0 | 1 | 0 | 1.8 | 0.5 | 3.6 |
| PF03572 | Peptidase family S41 | 1 | 0 | 3 | 0 | 5 | 0 | 0 | 1 | 1 | 1.8 | 0.5 | 3.6 |
| PF07883 | Cupin domain | 2 | 2 | 1 | 2 | 2 | 1 | 0 | 0 | 1 | 1.8 | 0.5 | 3.6 |
| PF01083 | Cutinase | 9 | 10 | 12 | 7 | 9 | 2 | 5 | 2 | 2 | 9.4 | 2.75 | 3.4 |
| PF00710 | Asparaginase | 1 | 1 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0.8 | 0.25 | 3.2 |
| PF00753 | Metallo-beta-lactamase superfamily | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0.8 | 0.25 | 3.2 |
| PF00754 | F5/8 type C domain | 1 | 2 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0.8 | 0.25 | 3.2 |
| PF01179 | Copper amine oxidase, enzyme domain | 0 | 3 | 2 | 0 | 3 | 1 | 0 | 1 | 0 | 1.6 | 0.5 | 3.2 |
| PF01679 | Uncharacterized protein family UPF0057 | 0 | 1 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0.8 | 0.25 | 3.2 |
| PF02244 | Carboxypeptidase activation peptide | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0.8 | 0.25 | 3.2 |
| PF03694 | Erg28 like protein | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0.8 | 0.25 | 3.2 |
| PF00141 | Peroxidase | 1 | 3 | 4 | 1 | 5 | 0 | 1 | 1 | 2 | 2.8 | 1 | 2.8 |
| PF00194 | Eukaryotic-type carbonic anhydrase | 1 | 2 | 2 | 1 | 1 | 0 | 1 | 0 | 1 | 1.4 | 0.5 | 2.8 |
| PF00187 | Chitin recognition protein | 4 | 7 | 17 | 5 | 14 | 6 | 7 | 0 | 1 | 9.4 | 3.5 | 2.7 |
| PF00295 | Glycosyl hydrolases family 28 | 17 | 6 | 3 | 17 | 4 | 9 | 1 | 2 | 3 | 9.4 | 3.75 | 2.5 |
Table 10. Secreted Kelch-domain containing proteins.
| Gene locus | Species | Top non-hypothetical hit vs NCBI non-redundant protein database1 |
|---|---|---|
| BC1G_02702.1 | Botrytis cinerea | ring canal kelch-like protein (Xanthomonas campestris pv. campestris) (AAM43333.1) (8e-30) |
| BC1G_12145.1 | Botrytis cinerea | galactose oxidase (Gibberella zeae) (XP_391208.1) (1e-160) |
| FG00251.1 | Gibberella zeae | galactose oxidase (Gibberella zeae) (XP_391208.1) (0) |
| FG09093.1 | Gibberella zeae | galactose oxidase (Cladobotryum dendroides) (A38084) (1e-126) |
| FG09142.1 | Gibberella zeae | ring canal kelch-like protein (Xanthomonas campestris pv. campestris) (AAM43333.1) (5e-32) |
| MGG_02368.5 | Magnaporthe grisea | galactose oxidase (Cladobotryum dendroides) (A38084) (1e-117) |
| MGG_03826.5 | Magnaporthe grisea | ring canal kelch-like protein (Xanthomonas campestris pv. campestris) (AAM43333.1) (7e-22) |
| MGG_04086.5 | Magnaporthe grisea | ring canal kelch-like protein (Xanthomonas campestris pv. campestris) (AAM43333.1) (9e-24) |
| MGG_10013.5 | Magnaporthe grisea | ring canal kelch-like protein (Xanthomonas campestris pv. campestris) (AAM43333.1) (9e-34) |
| SS1G_03276.1 | Sclerotinia sclerotiorum | beta-scruin (Limulus polyphemus) (Q25386) (1e-07) |
| SNU05548.1 | Stagonospora nodorum | Kelch repeat (Herpetosiphon aurantiacus) (ZP_01426654) (2e-18) |
| SNU06096.1 | Stagonospora nodorum | ring canal kelch-like protein (Xanthomonas axonopodis pv. citri) (NP_644535.1) (9e-30) |
| SNU08346.1 | Stagonospora nodorum | epithiospecifier (Arabidopsis thaliana) (AAL14622.1) (3e-11) |
| SNU11576.1 | Stagonospora nodorum | galactose oxidase (Gibberella zeae) (XP_391208.1) (1e-120) |
| SNU15302.1 | Stagonospora nodorum | galactose oxidase (Gibberella zeae) (XP_391208.1) (0) |
| CHG08026.1 | Chaetomium globosum | Kelch (Herpetosiphon aurantiacus) (ZP_01423335.1) (1e-09) |
Discussion
One of the most fundamental aims in plant pathology research is to define precisely the difference between pathogenic and non-pathogenic microorganisms. The answer cannot be one of simple phylogeny, because phytopathogenic species are found in all taxonomic divisions of fungi and are often closely related to non-pathogenic species [3]. Before the availability of genomic sequences and high throughput approaches to study gene function [20], research was concentrated on the search for single pathogenicity factors; genes that are dispensable for saprophytic growth but essential for successful infection of the host plant [4], [97]. However, rather than encoding novel proteins found only in phytopathogens, the majority of pathogenicity factors discovered in this way have been found to be involved in signalling cascades and metabolic pathways and hence are conserved in most species of fungi [5]. Components of signalling cascades that in the budding yeast S. cerevisiae are responsible for responses to pheromones, nutritional starvation and osmotic stress [9] have in many cases evolved different roles in the life cycle of pathogens, such as controlling appressorium formation, dimorphism and growth [10]. Although the central components of signalling are conserved between phytopathogens and S. cerevisiae, the receptors are often different, reflecting the different environmental cues to which the pathogen needs to respond [11], [98].
Analysis of all available genome sequences from a wider range of fungal species has for the first time allowed us to address the differences between phytopathogens and non-pathogens at a whole genome level. For this purpose, the e-Fungi data warehouse provides a means to interrogate the vast amounts of genomic and functional data available in a simple integrated manner [26]. Previous research, in which EST datasets were compared with genomic sequences, suggested that the expressed gene inventories of phytopathogenic species were not significantly more similar to one another than to those of saprotrophic filamentous fungi [99]. We clustered sets of predicted proteins from 36 different species of fungi and oomycetes into groups of potential orthologues and the species distribution of members of each cluster was ascertained. There were no clusters that were completely specific to phytopathogenic species across both fungi and oomycetes, suggesting that the presence of novel, universal pathogenicity factors in the genomes of phytopathogens is unlikely. This was confirmed by looking at clusters containing empirically defined pathogenicity factors, where homologues of many of these were found in all species studied and none were conserved in the genomes only of phytopathogens. A small number were only found in a single species of fungus and probably represented proteins that are highly specialised for a particular role in a specific pathogenic species, for example in host-plant recognition [54]. Previous research also suggested that the gene inventories of filamentous fungi were more similar to each other than to those of unicellular yeasts [99]. Analysis of the clusters of similar proteins show some clusters that are found in all species of filamentous fungi (including ascomycetes, basidiomycetes and zygomycetes) but are not present in the genomes of yeasts, consistent with the original conclusion. These contain a number of proteins that are likely to be involved in morphological changes associated with the more complex filamentous lifestyle, as well those involved in secondary metabolism and signalling cascades that are not found in yeasts. In particular, our results suggest that filamentous fungi use a wider variety of lipid molecules for the purpose of signalling. Some of these may act as pheromones, or hormones– chemical messengers diffusing from one cell to another to elicit a physiological or developmental response [37]. A number of these innovations to the filamentous lifestyle may serve important roles in pathogenesis as well, because homologues of a number of pathogenicity factors are found only in filamentous ascomycetes. The distribution of filamentous fungi-specific proteins, such as involved in those cytoskeletal rearrangements and fruiting body formation, throughout the fungal kingdom (and in some cases in oomycetes as well), suggests that the last common ancestral fungus may well have been multi-cellular and the evolution of uni-cellular fungi was likely associated with massive gene loss. For example, it has been shown that early in ascomycete evolution there was a proliferation of subtilase-type protease-encoding genes that have been retained in some filamentous ascomycete lineages, but lost in the yeast lineage [100].
It has previously been speculated that the evolution of phytopathogenesis was associated with the expansion of certain gene families [1]. Duplication of an ancestral gene, followed by mutation allows members of the family to take on new functions [101]. For example, genomes of the filamentous ascomycetes studied here have between 40 and 140 cytochrome P450-encoding genes (data not shown) that are involved in toxin biosynthesis, lipid metabolism, alkane assimilation and detoxification [102] and which probably arose via gene duplication and functional diversification. In contrast, the genome of the budding yeast S. cerevisiae has only three cytochrome P450-encoding enzymes. We have shown here that there are likely to be large differences in the gene inventories of filamentous fungi compared to unicellular yeasts.
To study the differences between phytopathogenic and saprophytic fungi, we concentrated on the filamentous ascomycetes where there are a number of phytopathogenic species genomes have been sequenced along with closely related non-pathogens. Protein families were defined using Pfam motifs [57] and the predicted protein sets for each species analysed in order to identify domains that were specific to or more common in the genomes of phytopathogens. Not surprisingly, many of the protein families we identified are likely to be associated with pathogenic processes such as plant cell wall degradation, toxin biosynthesis, formation of reactive oxygen species and detoxification [5]. Studies of bacterial phytopathogens have shown the importance of effectors, secreted proteins that disable plant defences and subvert metabolic and morphological processes for the benefit of the invading pathogen and which require delivery via a type III secretion system that are often deployed during pathogenesis [73]. Bacterial type III secreted effectors (T3SEs) have been shown to target salicyclic acid and abscisic acid-dependent defences, host vesicle trafficking, transcription and RNA metabolism, and several components of the plant defence signalling networks [103]. Very recently, potential effector-encoding genes have been identified in the genomes of several species of oomycete pathogens and are defined by the presence of a conserved RXLR-EER motif downstream of the signal peptide sequence [74]. The RXLR-EER motif is necessary for delivery of effector proteins into host plant cells and is therefore critical to their biological activity [74].
To identify potential fungal effectors, we compared Pfam motif frequency between the secretomes of phytopathogens and non-pathogens. This analysis identified potential effector-encoding genes, including secreted proteases, transcription factors and proteins that may be involved in cytoskeletal rearrangements (such as Kelch-domain containing proteins) and protein-protein interactions, as well as a group of pathogen-specific secreted isochorisimatases that potentially could suppress salicyclic acid-dependent host plant defences. Bacterial T3SEs are injected directly into the host cytoplasm via the type III secretion injection apparatus [73]. In contrast, the potential fungal effectors identified in this study appear to be secreted by the normal cellular secretory pathway via the endoplasmic reticulum and the mechanism by which fungal effectors might be taken up by plant cells and enter into the host cytoplasm is currently unknown.
Although the evolution of phytopathogenicity is likely to have happened several times and the lifestyles of these fungi are diverse, a comparison of gene inventories of a number of species using a powerful resource, such as e-Fungi, has allowed us to pinpoint new gene families that may serve important roles in the virulence of phytopathogens, allowing their selection for gene functional studies, that are currently in progress. The analyses deployed here may also offer a blueprint for the types of larger, more comprehensive studies that will be necessary to interpret the large flow of genetic data that will result from next generation DNA sequence analysis utilizing both a much wider variety of fungal pathogen species and also large sets of individual isolates of existing species.
Materials and Methods
Clustering of sequences
Sets of predicted proteins were downloaded for each of the 36 genomes from respective sequencing project websites (Table 1). Proteins less than 40 amino acids in length were not included in this analysis. Proteins were clustered using “all against all” BLASTP [104] followed by Markov Chain Clustering (MCL) [27] with 2.5 as a moderate inflation value and 10−10 as an E-value cut-off, as described previously [28], [29]. Clusters were annotated based on best hit against Swiss-Prot protein database [105] of members of that cluster (e-value <10−20 using BLASTP), or Pfam motifs contained in proteins from the cluster in the absence of Swiss-Prot hits.
Identification of Pfam motifs
The Pfam-A library from release 18.0 of the Pfam database was downloaded from the Pfam website (http://www.sanger.ac.uk/Software/Pfam/). This library contains 7973 protein models constructed from manually curated multiple alignments and covers 75% of proteins in UniProt [44], [57]. This library was used to analyse the sequences of predicted proteins for all 36 fungal genomes to identify the Pfam motifs that each protein contains. The analysis was performed using the “pfam_scan” perl script (version 0.5) downloaded from the Pfam website and HMMER software (downloaded from http://hmmer.wustl.edu/). Default thresholds were used, which are hand-curated for every family and designed to minimise false positives [44].
Identification of secreted proteins
The N-terminal sequence of each predicted protein from the 36 fungal genomes used in this study was analysed for the presence of a signal peptide using SignalP 3.0 [75] and sub-cellular localisation was predicted using WoLF PSORT [76]. Both these programs were installed locally. SignalP 3.0 uses two different algorithms to identify signal sequences. The secretome for each fungal species was defined as containing those proteins that were predicted have a signal peptide by both prediction algorithms from SignalP 3.0 and also predicted to be extracellular by WoLF PSORT.
Data analysis
All the data produced, as described above, was stored in the e-Fungi data warehouse [26] from which it can be accessed via a web-interface (http://www.e-fungi.org.uk/). Analyses described in this study were performed using the e-Fungi database.
Supporting Information
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The authors would like to acknowledge the financial support of the Biotechnology and Biological Sciences Research Council (BBSRC). The development of e-Fungi has been funded by the BBSRC Bioinformatics and e-Science programme II.
References
- 1.Tunlid A, Talbot NJ. Genomics of parasitic and symbiotic fungi. Curr Opin Microbiol. 2002;5:513–519. doi: 10.1016/s1369-5274(02)00355-7. [DOI] [PubMed] [Google Scholar]
- 2.James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, et al. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature. 2006;443:818–822. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- 3.Fitzpatrick DA, Logue ME, Stajich JE, Butler G. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol Biol. 2006;6:99. doi: 10.1186/1471-2148-6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oliver R, Osbourn A. Molecular dissection of fungal phytopathogenicity. Microbiology. 1995;141:1–9. doi: 10.1099/00221287-141-1-1. [DOI] [PubMed] [Google Scholar]
- 5.Idnurm A, Howlett BJ. Pathogenicity genes of phytopathogenic fungi. Mol Plant Path. 2001;2:241–255. doi: 10.1046/j.1464-6722.2001.00070.x. [DOI] [PubMed] [Google Scholar]
- 6.Xu JR. Map kinases in fungal pathogens. Fungal Genet Biol. 2000;31:137–152. doi: 10.1006/fgbi.2000.1237. [DOI] [PubMed] [Google Scholar]
- 7.Choi W, Dean RA. The adenylate cyclase gene MAC1 of Magnaporthe grisea controls appressorium formation and other aspects of growth and development. Plant Cell. 1997;9:1973–1983. doi: 10.1105/tpc.9.11.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu S, Dean RA. G protein alpha subunit genes control growth, development, and pathogenicity of Magnaporthe grisea. Mol Plant Microbe Interact. 1997;10:1075–1086. doi: 10.1094/MPMI.1997.10.9.1075. [DOI] [PubMed] [Google Scholar]
- 9.Gustin MC, Albertyn J, Alexander M, Davenport K. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1998;62:1264–1300. doi: 10.1128/mmbr.62.4.1264-1300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu JR, Hamer JE. MAP kinase and cAMP signalling regulate infection structure formation and growth in the rice blast fungus Magnaporthe grisea. Genes Dev. 1996;10:2696–2706. doi: 10.1101/gad.10.21.2696. [DOI] [PubMed] [Google Scholar]
- 11.Kulkarni RD, Thon MR, Pan H, Dean RA. Novel G-protein-coupled receptor-like proteins in the plant pathogenic fungus Magnaporthe grisea. Genome Biol. 2005;6:R24. doi: 10.1186/gb-2005-6-3-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang ZY, Thornton CR, Kershaw MJ, Debao L, Talbot NJ. The glyoxylate cycle is required for temporal regulation of virulence by the plant pathogenic fungus Magnaporthe grisea. Mol Microbiol. 2003;47:1601–1612. doi: 10.1046/j.1365-2958.2003.03412.x. [DOI] [PubMed] [Google Scholar]
- 13.Solomon PS, Lee RC, Wilson TJ, Oliver RP. Pathogenicity of Stagonospora nodorum requires malate synthase. Mol Microbiol. 2004;53:1065–1073. doi: 10.1111/j.1365-2958.2004.04178.x. [DOI] [PubMed] [Google Scholar]
- 14.Seong K, Hou Z, Tracy M, Kistler HC, Xu JR. Random Insertional Mutagenesis Identifies Genes Associated with Virulence in the Wheat Scab Fungus Fusarium graminearum. Phytopathology. 2005;95:744–750. doi: 10.1094/PHYTO-95-0744. [DOI] [PubMed] [Google Scholar]
- 15.de Jong J, McCormack BJ, Smirnoff N, Talbot NJ. Glycerol generates turgor in rice blast. Nature. 1997;389:244–245. [Google Scholar]
- 16.Talbot NJ. On the trail of a cereal killer: Exploring the biology of Magnaporthe grisea. Annu Rev Microbiol. 2003;57:177–202. doi: 10.1146/annurev.micro.57.030502.090957. [DOI] [PubMed] [Google Scholar]
- 17.Bolker M. Ustilago maydis-a valuable model system for the study of fungal dimorphism and virulence. Microbiology. 2001;147:1395–1401. doi: 10.1099/00221287-147-6-1395. [DOI] [PubMed] [Google Scholar]
- 18.Sweigard JA, Chumley FG, Valent B. Disruption of a Magnaporthe grisea cutinase gene. Mol Gen Genet. 1992;232:183–190. [PubMed] [Google Scholar]
- 19.Xu JR, Peng YL, Dickman MB, Sharon A. The dawn of fungal pathogen genomics. Annu Rev Phytopathol. 2006;44:337–366. doi: 10.1146/annurev.phyto.44.070505.143412. [DOI] [PubMed] [Google Scholar]
- 20.Jeon J, Park SY, Chi MH, Choi J, Park J, et al. Genome-wide functional analysis of pathogenicity genes in the rice blast fungus. Nat Genet. 2007;39:561–565. doi: 10.1038/ng2002. [DOI] [PubMed] [Google Scholar]
- 21.Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, et al. Life with 6000 genes. Science. 1996;275:1051–1052. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 22.Dean RA, Talbot NJ, Ebbole DJ, Farman ML, Mitchell TK, et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature. 2005;434:980–986. doi: 10.1038/nature03449. [DOI] [PubMed] [Google Scholar]
- 23.Kamper J, Kahmann R, Bolker M, Ma LJ, Brefort T, et al. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature. 2006;444:97–101. doi: 10.1038/nature05248. [DOI] [PubMed] [Google Scholar]
- 24.Cuomo CA, Güldener U, Xu JR, Trail F, Turgeon BG, et al. The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science. 2007;317:1400–1402. doi: 10.1126/science.1143708. [DOI] [PubMed] [Google Scholar]
- 25.Hane JK, Lowe RG, Solomon PS, Tan KC, Schoch CL, et al. Dothideomycete plant interactions illuminated by genome sequencing and EST analysis of the wheat pathogen Stagonospora nodorum. Plant Cell. 2007;19:3347–3368. doi: 10.1105/tpc.107.052829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cornell M, Alam I, Soanes DM, Wong HM, Hedeler C, et al. Comparative genome analysis across a kingdom of eukaryotic organisms: Specialization and diversification in the Fungi. Genome Res. 2007;17:1809–1822. doi: 10.1101/gr.6531807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alam I, Cornell MJ, Soanes DM, Hedeler C, Wong HM, et al. A methodology for comparative functional genomics. Journal of Integrative Bioinformatics. 2007;4:69. [Google Scholar]
- 29.Hedeler C, Wong HM, Cornell MJ, Alam I, Soanes DM, et al. e-Fungi: a data resource for comparative analysis of fungal genomes. BMC Genomics. 2007;8:426. doi: 10.1186/1471-2164-8-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science. 2000;290:972–977. doi: 10.1126/science.290.5493.972. [DOI] [PubMed] [Google Scholar]
- 31.Katinka MD, Duprat S, Cornillot E, Metenier G, Thomarat F, et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414:450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- 32.Hedges SB, Blair JE, Venturi ML, Shoe JL. A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol Biol. 2004;4:2. doi: 10.1186/1471-2148-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, et al. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- 34.Wapinski I, Pfeffer A, Friedman N, Regev A. Natural history and evolutionary principles of gene duplication in fungi. Nature. 2007;449:54–61. doi: 10.1038/nature06107. [DOI] [PubMed] [Google Scholar]
- 35.Tani M, Okino N, Mori K, Tanigawa T, Izu H, et al. Molecular cloning of the full-length cDNA encoding mouse neutral ceramidase. A novel but highly conserved gene family of neutral/alkaline ceramidases. J Biol Chem. 2000;275:11229–11234. doi: 10.1074/jbc.275.15.11229. [DOI] [PubMed] [Google Scholar]
- 36.Hornsten L, Su C, Osbourn AE, Garosi P, Hellman U, et al. Cloning of linoleate diol synthase reveals homology with prostaglandin H synthases. J Biol Chem. 1999;274:28219–28224. doi: 10.1074/jbc.274.40.28219. [DOI] [PubMed] [Google Scholar]
- 37.Champe SP, el-Zayat AA. Isolation of a sexual sporulation hormone from Aspergillus nidulans. J Bacteriol. 1989;171:3982–3988. doi: 10.1128/jb.171.7.3982-3988.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han WD, Mu YM, Lu XC, Xu ZM, Li XJ, et al. Up-regulation of LRP16 mRNA by 17beta-estradiol through activation of estrogen receptor alpha (ERalpha), but not ERbeta, and promotion of human breast cancer MCF-7 cell proliferation: a preliminary report. Endocr Relat Cancer. 2003;10:217–224. doi: 10.1677/erc.0.0100217. [DOI] [PubMed] [Google Scholar]
- 39.Siverio JM. Assimilation of nitrate by yeasts. FEMS Microbiol Rev. 2002;26:277–284. doi: 10.1111/j.1574-6976.2002.tb00615.x. [DOI] [PubMed] [Google Scholar]
- 40.Poggeler S, Kuck U. WD40 repeat protein regulates fungal cell differentiation and can be replaced functionally by the mammalian homologue striatin. Eukaryot Cel. 2004;3:232–240. doi: 10.1128/EC.3.1.232-240.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saupe S, Turcq B, Begueret J. A gene responsible for vegetative incompatibility in the fungus Podospora anserina encodes a protein with a GTP-binding motif and G beta homologous domain. Gene. 1995;162:135–139. doi: 10.1016/0378-1119(95)00272-8. [DOI] [PubMed] [Google Scholar]
- 42.Fischer R, Timberlake WE. Aspergillus nidulans apsA (anucleate primary sterigmata) encodes a coiled-coil protein required for nuclear positioning and completion of asexual development. J Cell Biol. 1995;128:485–498. doi: 10.1083/jcb.128.4.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sweeney MJ, Dobson AD. Molecular biology of mycotoxin biosynthesis. FEMS Microbiol Lett. 1999;175:149–163. doi: 10.1111/j.1574-6968.1999.tb13614.x. [DOI] [PubMed] [Google Scholar]
- 44.Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34 (Database issue):D247–251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winnenburg R, Baldwin TK, Urban M, Rawlings C, Kohler J, et al. PHI-base: a new database for pathogen host interactions. Nucleic Acids Res. 2006;34:D459–464. doi: 10.1093/nar/gkj047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tucker SL, Talbot NJ. Surface attachment and pre-penetration stage development by plant pathogenic fungi. Annu Rev Phytopathol. 2001;39:385–417. doi: 10.1146/annurev.phyto.39.1.385. [DOI] [PubMed] [Google Scholar]
- 47.Perez-Martin J, Castillo-Lluva S, Sgarlata C, Flor-Parra I, Mielnichuk N, et al. Pathocycles: Ustilago maydis as a model to study the relationships between cell cycle and virulence in pathogenic fungi. Mol Genet Genomics. 2006;276:211–229. doi: 10.1007/s00438-006-0152-6. [DOI] [PubMed] [Google Scholar]
- 48.Veneault-Fourrey C, Barooah M, Egan M, Wakley G, Talbot NJ. Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science. 2006;312:580–583. doi: 10.1126/science.1124550. [DOI] [PubMed] [Google Scholar]
- 49.Langfelder K, Streibel M, Jahn B, Haase G, Brakhage AA. Biosynthesis of fungal melanins and their importance for human pathogenic fungi. Fungal Genet Biol. 2003;382:143–158. doi: 10.1016/s1087-1845(02)00526-1. [DOI] [PubMed] [Google Scholar]
- 50.Kershaw MJ, Talbot NJ. Hydrophobins and repellents: proteins with fundamental roles in fungal morphogenesis. Fungal Genet Biol. 1998;23:18–33. doi: 10.1006/fgbi.1997.1022. [DOI] [PubMed] [Google Scholar]
- 51.Clergeot PH, Gourgues M, Cots J, Laurans F, Latorse MP, et al. PLS1, a gene encoding a tetraspanin-like protein, is required for penetration of rice leaf by the fungal pathogen Magnaporthe grisea. Proc Natl Acad Sci U S A. 2001;98:6963–6968. doi: 10.1073/pnas.111132998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soundararajan S, Jedd G, Li X, Ramos-Pamplona M, Chua NH. Woronin body function in Magnaporthe grisea is essential for efficient pathogenesis and for survival during nitrogen starvation stress. Plant Cell. 2004;16:1564–1574. doi: 10.1105/tpc.020677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xue C, Park G, Choi W, Zheng L, Dean RA, et al. Two novel fungal virulence genes specifically expressed in appressoria of the rice blast fungus. Plant Cell. 2002;14:2107–2119. doi: 10.1105/tpc.003426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kang S, Sweigard JA, Valent B. The PWL host specificity gene family in the blast fungus Magnaporthe grisea. Mol Plant Microbe Interact. 1995;8:939–948. doi: 10.1094/mpmi-8-0939. [DOI] [PubMed] [Google Scholar]
- 55.Tucker SL, Thornton CR, Tasker K, Jacob C, Giles G, et al. A fungal metallothionein is required for pathogenicity of Magnaporthe grisea. Plant Cell. 2004;16:1575–1588. doi: 10.1105/tpc.021279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Talbot NJ, Ebbole DJ, Hamer JE. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell. 1993;5:1575–1590. doi: 10.1105/tpc.5.11.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sonnhammer EL, Eddy SR, Durbin R. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins. 1997;28:405–420. doi: 10.1002/(sici)1097-0134(199707)28:3<405::aid-prot10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 58.Janbon G, Himmelreich U, Moyrand F, Improvisi L, Dromer F. Cas1p is a membrane protein necessary for the O-acetylation of the Cryptococcus neoformans capsular polysaccharide. Mol Microbiol. 2001;42:453–467. doi: 10.1046/j.1365-2958.2001.02651.x. [DOI] [PubMed] [Google Scholar]
- 59.Dijkgraaf GJ, Brown JL, Bussey H. The KNH1 gene of Saccharomyces cerevisiae is a functional homolog of KRE9. Yeast. 1996;12:683–692. doi: 10.1002/(SICI)1097-0061(19960615)12:7%3C683::AID-YEA959%3E3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 60.Citterio E, Papait R, Nicassio F, Vecchi M, Gomiero P, Mantovani R, Di Fiore PP, Bonapace IM. Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol Cell Biol. 2004;24:2526–2535. doi: 10.1128/MCB.24.6.2526-2535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kanamori T, Kanou N, Kusakabe S, Atomi H, Imanaka T. Allophanate hydrolase of Oleomonas sagaranensis involved in an ATP-dependent degradation pathway specific to urea. FEMS Microbiol Lett. 2005;245:61–65. doi: 10.1016/j.femsle.2005.02.023. [DOI] [PubMed] [Google Scholar]
- 62.Tsubouchi H, Roeder GS. The Mnd1 protein forms a complex with hop2 to promote homologous chromosome pairing and meiotic double-strand break repair. Mol Cell Biol. 2002;22:3078–3088. doi: 10.1128/MCB.22.9.3078-3088.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ferrer JL, Jez JM, Bowman ME, Dixon RA, Noel JP. Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nat Struct Biol. 1999;6:775–784. doi: 10.1038/11553. [DOI] [PubMed] [Google Scholar]
- 64.Samuelsson B, Dahlen SE, Lindgren JA, Rouzer CA, Serhan CN. Leukotrienes and lipoxins: structures, biosynthesis, and biological effects. Science. 1987;237:1171–1176. doi: 10.1126/science.2820055. [DOI] [PubMed] [Google Scholar]
- 65.Baker A, Graham IA, Holdsworth M, Smith SM, Theodoulou FL. Chewing the fat: beta-oxidation in signalling and development. Trends Plant Sci. 2006;11:124–132. doi: 10.1016/j.tplants.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 66.Keller NP, Turner G, Bennett JW. Fungal secondary metabolism-from biochemistry to genomics. Nat Rev Microbiol. 2005;3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- 67.Le Nours J, Ryttersgaard C, Lo Leggio L, Ostergaard PR, Borchert TV, et al. Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. Protein Sci. 2003;12:1195–1204. doi: 10.1110/ps.0300103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.D'Ovidio R, Mattei B, Roberti S, Bellincampi D. Polygalacturonases, polygalacturonase-inhibiting proteins and pectic oligomers in plant-pathogen interactions. Biochim Biophys Acta. 2004;1696:237–244. doi: 10.1016/j.bbapap.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 69.Gijzen M, Nurnberger T. Nep1-like proteins from plant pathogens: recruitment and diversification of the NPP1 domain across taxa. Phytochemistry. 2006;67:1800–1807. doi: 10.1016/j.phytochem.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 70.van den Burg HA, Harrison SJ, Joosten MH, Vervoort J, de Wit PJ. Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol Plant Microbe Interact. 2006;19:1420–1430. doi: 10.1094/MPMI-19-1420. [DOI] [PubMed] [Google Scholar]
- 71.Primo-Parmo SL, Sorenson RC, Teiber J, La Du BN. The human serum paraoxonase/arylesterase gene (PON1) is one member of a multigene family. Genomics. 1996;33:498–507. doi: 10.1006/geno.1996.0225. [DOI] [PubMed] [Google Scholar]
- 72.Chauvaux S, Chevalier F, Le Dantec C, Fayolle F, Miras I, et al. Cloning of a genetically unstable cytochrome P-450 gene cluster involved in degradation of the pollutant ethyl tert-butyl ether by Rhodococcus ruber. J Bacteriol 183. 2001:6551–6557. doi: 10.1128/JB.183.22.6551-6557.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Alfano JR, Collmer A. Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu Rev Phytopathol. 2004;42:385–414. doi: 10.1146/annurev.phyto.42.040103.110731. [DOI] [PubMed] [Google Scholar]
- 74.Birch PR, Rehmany AP, Pritchard L, Kamoun S, Beynon JL. Trafficking arms: oomycete effectors enter host plant cells. Trends Microbiol. 2006;14:8–11. doi: 10.1016/j.tim.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 75.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 76.Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35 (Web Server issue):W585–587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Parsons JF, Calabrese K, Eisenstein E, Ladner JE. Structure and mechanism of Pseudomonas aeruginosa PhzD, an isochorismatase from the phenazine biosynthetic pathway. Biochemistry. 2003;42:5684–5693. doi: 10.1021/bi027385d. [DOI] [PubMed] [Google Scholar]
- 78.Gehring AM, Bradley KA, Walsh CT. Enterobactin biosynthesis in Escherichia coli: isochorismate lyase (EntB) is a bifunctional enzyme that is phosphopantetheinylated by EntD and then acylated by EntE using ATP and 2,3-dihydroxybenzoate. Biochemistry. 1997;36:8495–5803. doi: 10.1021/bi970453p. [DOI] [PubMed] [Google Scholar]
- 79.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature. 2003;423:181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wildermuth MC, Dewdney J, Wu G, Ausubel FM. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature. 2001;414:562–565. doi: 10.1038/35107108. [DOI] [PubMed] [Google Scholar]
- 81.Colovos C, Cascio D, Yeates TO. The 1.8 A crystal structure of the ycaC gene product from Escherichia coli reveals an octameric hydrolase of unknown specificity. Structure. 1998;6:1329–1337. doi: 10.1016/s0969-2126(98)00132-4. [DOI] [PubMed] [Google Scholar]
- 82.Chirala SS, Wakil SJ. Structure and function of animal fatty acid synthase. Lipids. 2004;39:1045–1053. doi: 10.1007/s11745-004-1329-9. [DOI] [PubMed] [Google Scholar]
- 83.Mayorga ME, Timberlake WE. The developmentally regulated Aspergillus nidulans wA gene encodes a polypeptide homologous to polyketide and fatty acid synthases. Mol Gen Genet. 1992;235:205–212. doi: 10.1007/BF00279362. [DOI] [PubMed] [Google Scholar]
- 84.Way M, Sanders M, Chafel M, Tu YH, Knight A, et al. beta-Scruin, a homologue of the actin crosslinking protein scruin, is localized to the acrosomal vesicle of Limulus sperm. J. Cell Sci. 1995;108:3155–3162. doi: 10.1242/jcs.108.10.3155. [DOI] [PubMed] [Google Scholar]
- 85.McPherson MJ, Ogel ZB, Stevens C, Yadav KD, Keen JN, et al. Galactose oxidase of Dactylium dendroides. Gene cloning and sequence analysis. J Biol Chem. 1992;267:8146–8152. [PubMed] [Google Scholar]
- 86.Vanden Wymelenberg A, Sabat G, Mozuch M, Kersten PJ, Cullen D, et al. Structure, organization, and transcriptional regulation of a family of copper radical oxidase genes in the lignin-degrading basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol. 2006;72:4871–4877. doi: 10.1128/AEM.00375-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Leuthner B, Aichinger C, Oehmen E, Koopmann E, Muller O, et al. A H2O2-producing glyoxal oxidase is required for filamentous growth and pathogenicity in Ustilago maydis. Mol Genet Genomics. 2005;272:639–650. doi: 10.1007/s00438-004-1085-6. [DOI] [PubMed] [Google Scholar]
- 88.Silakowski B, Ehret H, Schairer HU. fbfB, a gene encoding a putative galactose oxidase, is involved in Stigmatella aurantiaca fruiting body formation. J Bacteriol. 1998;180:1241–1247. doi: 10.1128/jb.180.5.1241-1247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Parsons MR, Convery MA, Wilmot CM, Yadav KD, Blakeley V, et al. Crystal structure of a quinoenzyme: copper amine oxidase of Escherichia coli at 2 A resolution. Structure. 1995;3:1171–1184. doi: 10.1016/s0969-2126(01)00253-2. [DOI] [PubMed] [Google Scholar]
- 90.Reddy CA, D'Souza TM. Physiology and molecular biology of the lignin peroxidases of Phanerochaete chrysosporium. FEMS Microbiol Rev. 1994;13:137–152. doi: 10.1111/j.1574-6976.1994.tb00040.x. [DOI] [PubMed] [Google Scholar]
- 91.Kawano T. Roles of the reactive oxygen species-generating peroxidase reactions in plant defense and growth induction. Plant Cell Rep. 2003;21:829–837. doi: 10.1007/s00299-003-0591-z. [DOI] [PubMed] [Google Scholar]
- 92.Robinson DN, Cooley L. Drosophila kelch is an oligomeric ring canal actin organizer. J Cell Biol. 1997;138:799–810. doi: 10.1083/jcb.138.4.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Philips J, Herskowitz I. Identification of Kel1p, a kelch domain-containing protein involved in cell fusion and morphology in Saccharomyces cerevisiae. J Cell Biol. 1998;143:375–389. doi: 10.1083/jcb.143.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kankanala P, Czymmek K, Valent B. Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell. 2007;19:706–724. doi: 10.1105/tpc.106.046300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tordai H, Banyai L, Patthy L. The PAN module: the N-terminal domains of plasminogen and hepatocyte growth factor are homologous with the apple domains of the prekallikrein family and with a novel domain found in numerous nematode proteins. FEBS Lett. 1999;461:63–67. doi: 10.1016/s0014-5793(99)01416-7. [DOI] [PubMed] [Google Scholar]
- 96.Vogel WF, Abdulhussein R, Ford CE. Sensing extracellular matrix: an update on discoidin domain receptor function. Cell Signal. 2006;18:1108–1116. doi: 10.1016/j.cellsig.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 97.Baldwin TK, Winnenburg R, Urban M, Rawlings C, Koehler J, Hammond-Kosack KE. The pathogen-host interactions database (PHI-base) provides insights into generic and novel themes of pathogenicity. Mol Plant Microbe Interact. 2006;19:1451–1462. doi: 10.1094/MPMI-19-1451. [DOI] [PubMed] [Google Scholar]
- 98.DeZwaan TM, Carroll AM, Valent B, Sweigard JA. Magnaporthe grisea pth11p is a novel plasma membrane protein that mediates appressorium differentiation in response to inductive substrate cues. Plant Cell. 1999;11:2013–2030. doi: 10.1105/tpc.11.10.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Soanes DM, Talbot NJ. Comparative genomic analysis of phytopathogenic fungi using expressed sequence tag (EST) collections. Molecular Plant Pathology. 2006;7:61–70. doi: 10.1111/j.1364-3703.2005.00317.x. [DOI] [PubMed] [Google Scholar]
- 100.Hu G, Leger RJ. A phylogenomic approach to reconstructing the diversification of serine proteases in fungi. J Evol Biol. 2004;17:1204–1214. doi: 10.1111/j.1420-9101.2004.00786.x. [DOI] [PubMed] [Google Scholar]
- 101.Ohno S. New York: Springer; 1970. Evolution by Gene Duplication. [Google Scholar]
- 102.van den Brink HM, van Gorcom RF, van den Hondel CA, Punt PJ. Cytochrome P450 enzyme systems in fungi. Fungal Genet Biol. 1998;23:1–17. doi: 10.1006/fgbi.1997.1021. [DOI] [PubMed] [Google Scholar]
- 103.Stavrinides J, McCann HC, Guttman DS. Host-pathogen interplay and the evolution of bacterial effectors. Cell Microbiol. 2007;10:285–292. doi: 10.1111/j.1462-5822.2007.01078.x. [DOI] [PubMed] [Google Scholar]
- 104.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 105.Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438:1092–1093. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- 107.Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- 108.Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, et al. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol. 2007;25:221–231. doi: 10.1038/nbt1282. [DOI] [PubMed] [Google Scholar]
- 109.Machida M, Asai K, Sano M, Tanaka T, Kumagai T, et al. Genome sequencing and analysis of Aspergillus oryzae. Nature. 2005;438:1157–1161. doi: 10.1038/nature04300. [DOI] [PubMed] [Google Scholar]
- 110.Jones T, Federspiel NA, Chibana H, Dungan J, Kalman S, et al. The diploid genome sequence of Candida albicans. Proc Natl Acad Sci USA. 2004;101:7329–7334. doi: 10.1073/pnas.0401648101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dietrich FS, Voegeli S, Brachat S, Lerch A, Gates K, et al. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science. 2004;304:304–307. doi: 10.1126/science.1095781. [DOI] [PubMed] [Google Scholar]
- 112.Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422:859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- 113.Martinez D, Larrondo LF, Putnam N, Gelpke MD, Huang K, et al. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat Biotechnol. 2004;22:695–700. doi: 10.1038/nbt967. [DOI] [PubMed] [Google Scholar]
- 114.Tyler BM, Tripathy S, Zhang X, Dehal P, Jiang RH, et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313:1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- 115.Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 116.Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.