RelA Ser276 Phosphorylation Is Required for Activation of a Subset of NF-κB-Dependent Genes by Recruiting Cyclin-Dependent Kinase 9/Cyclin T1 Complexes (original) (raw)
Abstract
NF-κB plays a central role in cytokine-inducible inflammatory gene expression. Previously we empirically determined the identity of 92 members of the genetic network under direct NF-κB/RelA control that show marked heterogeneity in magnitude of transcriptional induction and kinetics of peak activation. To investigate this network further, we have applied a recently developed two-step chromatin immunoprecipitation assay that accurately reflects association and disassociation of RelA binding to its chromatin targets. Although inducible RelA binding occurs with similar kinetics on all NF-κB-dependent genes, serine 276 (Ser276)-phosphorylated RelA binding is seen primarily on a subset of genes that are rapidly induced by tumor necrosis factor (TNF), including Gro-β, interleukin-8 (IL-8), and IκBα. Previous work has shown that TNF-inducible RelA Ser276 phosphorylation is controlled by a reactive oxygen species (ROS)-protein kinase A signaling pathway. To further understand the role of phospho-Ser276 RelA in target gene expression, we inhibited its formation by ROS scavengers and antioxidants, treatments that disrupt phospho-Ser276 formation but not the translocation and DNA binding of nonphosphorylated RelA. Here we find that phospho-Ser276 RelA is required only for activation of IL-8 and Gro-β, with IκBα being unaffected. These data were confirmed in experiments using RelA−/− murine embryonic fibroblasts reconstituted with a RelA Ser276Ala mutation. In addition, we observe that phospho-Ser276 RelA binds the positive transcription elongation factor b (P-TEFb), a complex containing the cyclin-dependent kinase 9 (CDK-9) and cyclin T1 subunits. Inhibition of P-TEFb activity by short interfering RNA (siRNA)-mediated knockdown shows that the phospho-Ser276 RelA-P-TEFb complex is required for IL-8 and Gro-β gene activation but not for IκBα gene activation. These studies indicate that TNF induces target gene expression by heterogeneous mechanisms. One is mediated by phospho-Ser276 RelA formation and chromatin targeting of P-TEFb controlling polymerase II (Pol II) recruitment and carboxy-terminal domain phosphorylation on the IL-8 and Gro-β genes. The second involves a phospho-Ser276 RelA-independent activation of genes preloaded with Pol II, exemplified by the IκBα gene. Together, these data suggest that the binding kinetics, selection of genomic targets, and mechanisms of promoter induction by RelA are controlled by a phosphorylation code influencing its interactions with coactivators and transcriptional elongation factors.
Infectious and inflammatory stimuli induce expression of gene networks controlling proinflammatory and innate immune responses. One important arm of the inflammatory response is mediated by monocyte-derived tumor necrosis factor (TNF), a cytokine that activates gene expression programs in adjacent epithelial cells to propagate mucosal inflammation (42, 51). Here, the spectrum of functional activities and the magnitude and timing of TNF-induced gene expression are important determinants of tissue homeostasis. A central mediator of epithelial genomic response is nuclear factor-κB (NF-κB), an inducible transcription factor that controls the expression of proinflammatory genes and whose dysregulation has also been implicated in the pathogenesis of inflammatory, neoplastic, and autoimmune diseases (4, 6, 17).
In most non-B-lymphoid cells, NF-κB is sequestered as an inactive complex in the cytoplasm by the inhibitors of κB (IκBs). The mechanism by which NF-κB is activated by TNF, referred to as the “canonical” activation pathway, has been extensively investigated (22). The canonical pathway is activated by ligands of the TNF superfamily including TNF alpha, the prototypical macrophage-derived cytokine that binds to the ubiquitously expressed TNF receptor 1 (TNFRI) (50). TNF-induced TNFRI trimerization induces the recruitment of cytoplasmic signal adapters, including the TNF receptor-associated death domain (TRADD), TNF receptor-associated factor 2 (TRAF2) and TRAF6, receptor-interacting protein (RIP), mitogen/extracellular signal-regulated kinase kinase 3, and others (23-25). This activated submembranous TNFRI complex transiently recruits the IκB kinase (IKK) complex, resulting in the phosphorylation of the catalytic IKKα and -β kinases followed by its cytosolic release (15, 43). In the cytosol, activated IKK phosphorylates serine (Ser) residues 32 and 36 on the NH2 terminal regulatory domain of IκBα, targeting IκBα for proteolytic destruction by ubiquitination and calpain pathways (20, 29). IκBα degradation releases NF-κB to rapidly translocate into the nucleus.
In the nucleus, the activated NF-κB complex binds to highly conserved regulatory sequences corresponding to the 5′-GGGRNNYYCC-3′ consensus (30). Although NF-κB binding in vitro is quite stable, with a half-life of 45 min or longer (8), fluorescence recovery after photobleaching and fluorescence lifetime measurements have shown that NF-κB interacting with genomic targets within native chromatin is in a hyperdynamic exchange, with an observed half-life of seconds (8). Studies of highly inducible promoters have suggested that NF-κB interaction induces target gene activation through a mechanism that induces the formation of a multiprotein complex known as an “enhanceosome” (34). The enhanceosome contains non-DNA-binding chromatin-modifying proteins, including the CBP/p300 coactivator (49), a histone acetylase that modifies promoter-associated nucleosomes, as well as incompletely characterized methylases, kinases, and chromatin-organizing proteins. This activity results in enhanced transcriptional initiation.
Although nuclear translocation is required for the activation of NF-κB-dependent genes, we have recently shown a requirement for a TNF-inducible reactive oxygen species (ROS) pathway in licensing the transactivating potential of NF-κB (26, 58). In this pathway, TNF induces the formation of ROS, whose formation occurs at times well after the occurrence of IκBα proteolysis and NF-κB nuclear translocation (58). This second signal activates the catalytic subunit of protein kinase A (PKAc), resulting in the selective phosphorylation of the RelA-transactivating subunit at Ser residue 276, a modification that permits complex formation with incompletely characterized transcriptional activating proteins, including p300/CBP (60), and serves as a switch for additional posttranslational modifications, such as acetylation (12). We have recently found that the inhibition of the TNF-induced ROS prevents phospho-Ser276 RelA formation, resulting in the nuclear translocation of hypophosphorylated RelA and the formation of an unstable enhanceosome (26).
Despite intense study, a systematic definition of the NF-κB-dependent gene network has only recently been approached (53, 54, 56). High-density microarray analyses of cells expressing a regulated dominant-negative NF-κB inhibitor have elucidated the genes under control of the canonical NF-κB activation pathway in epithelial cells subjected to viral infections (40, 55) and cytokine stimulation (56) and those controlled by its distinct activation modes (54). These studies have shown that in response to TNF stimulation, NF-κB binding controls the expression of a noncontiguous group of genes whose products control a variety of biological processes, including leukocyte activation/chemotaxis, negative regulators of the TNF-IKK pathway, cellular metabolism, antigen processing, and others (54).
In this study, we examine the detailed mechanisms for highly inducible NF-κB-dependent genes by use of an efficient two-step chromatin immunoprecipitation (ChIP) assay (39). We observed indistinguishable patterns of inducible RelA binding to all promoters irrespective of their degree of induction or activation kinetics. However, distinct binding patterns of Ser276-phosphorylated RelA binding were observed, with rapid phospho-Ser276 RelA binding to the most highly inducible promoters, and this was at times coincident with their maximal expression. To establish the role of the phospho-Ser276 RelA modification, we blocked phospho-Ser276 RelA formation using ROS scavengers (dimethyl sulfoxide [DMSO]) and antioxidants (_N_-acetyl cysteine [NAC]), agents that blocked RelA Ser276 phosphorylation without affecting its translocation. We found that interleukin-8 (IL-8) and Gro-β expression were highly dependent on phospho-Ser276 RelA binding, whereas IκBα was not. Further study of this mechanism revealed that the phospho-Ser276 RelA associates with the cyclin-dependent kinase 9 (CDK-9)-cyclin T1 (Ccn T1) complex, positive transcription elongation factor b (P-TEFb). Short interfering RNA (siRNA)-mediated knockdown showed that RelA recruitment of P-TEFb activity is required only for a subset of NF-κB-dependent genes (the IL-8 and Gro-β genes), genes characterized by TNF-induced polymerase II (Pol II) recruitment. Together, these data show for the first time that phospho-Ser276 RelA formation is responsible for the activation of a subset of NF-κB-dependent genes via the P-TEFb transcriptional elongation complex.
MATERIALS AND METHODS
Cell culture and reagents.
HeLa S3 and A549 cells were from ATCC and maintained as previously described (54, 56). HeLa expressing the nondegradable IκBα Mut inhibitor in Tet-Off (TtA) cells were described earlier (55, 56). Clonal HeLa cells stably expressing the enhanced green fluorescent protein (EGFP)-RelA fusion protein (33) were selected in G418 (250 μg/ml). In all experiments, unless specifically noted, cells were serum starved in Dulbecco's modified Eagle's medium containing 0.5% bovine serum albumin for 24 h prior to stimulation. Stimulation was performed by addition of recombinant TNF alpha (30 ng/ml; Peprotech) to the culture medium. Flavopiridol (FP) was used at 500 nM concentrations for 1 h prior to TNF stimulation. For RelA site mutations, eukaryotic expression vector encoding FLAG epitope-tagged RelA was generated by ligating the monomeric strawberry cDNA (47) into a pcDNA3-FLAG backbone, generating the plasmid pcDNA-FLAG-Straw. Wild-type RelA (RelAWT) and a RelA (Ser276Ala) site mutation were generated by rolling-circle mutagenesis and cloned into pcDNA-FLAG-Straw. The transcription unit containing the cytomegalovirus promoter and FLAG-mStraw-RelA coding sequences was excised as a BglII/XbaI fragment and ligated into pEF6/V5 plasmid digested with BglII/XbaI, producing pXFS-RelA. The pXFS plasmids confer blasticidin resistance and were used to generate stable transfectants in a RelA−/− background. For this purpose, RelA−/− murine embryonic fibroblasts (MEFs) (a gift of D. Baltimore [7]) were transfected with 8 μg of pXFS-RelA and 24 μl of Targefect-MEF transfection reagent (Targeting Systems Inc.) in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. After 24 h, cells were subdivided and stable transfectants selected by the addition of blasticidin S (10 μg/ml) for 3 weeks. Pools of cells were confirmed by fluorescence microscopy to have similar levels of Straw fluorescence, and amounts of protein expression were confirmed by Western immunoblotting.
EMSA.
Sucrose cushion-purified nuclear extracts were prepared as described previously (19). Nuclear proteins (30 μg) were bound to a 40,000-cpm 32P-labeled duplex NF-κB site (27) in 1× binding buffer (10 mM HEPES, pH 7.9, 5 mM MgCl2, 1 mM EDTA, 0.5 mM dithiothreitol, 0.1 M KCl) with 1 μg poly(dA-dT) per sample. Binding reaction mixtures were incubated for 1 h at 4°C and fractionated by nondenaturing 6% Tris-borate-EDTA gels on ice. For supershift assays, the same conditions were used except that 2 μg of the indicated NF-κB isoform-specific antibody (Ab) (Santa Cruz) was preincubated for 1 h at 4°C prior to the addition of labeled probe. Supershift electromobility shift assay (EMSA) samples were fractionated on a 5% acrylamide 1× Tris-borate-EDTA gel.
Western immunoblots.
For measurement of phospho-Ser276 RelA formation, whole-cell extracts (WCEs) were prepared by lysis in 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer. Protein from 3 × 106 cells was fractionated by SDS-PAGE. For measurement of NF-κB subunit nuclear translocation, 75 μg of sucrose cushion-purified nuclear protein was fractionated by SDS-PAGE. Gels were electrotransferred to polyvinylidene difluoride membranes (Millipore) overnight and blocked with 5% nonfat dried milk in Tris-buffered saline (TBS)-0.1% Tween 20. Anti-RelA, -RelB, and -C-Rel Abs were from Santa Cruz. Phospho-Ser276 RelA was detected using anti-phospho-Ser276 RelA (1:1,000; Santa Cruz) in 5% bovine serum albumin-TBS-Tween 0.1% and incubated for 72 h at 4°C. Secondary Abs (IRD800 anti-rabbit immunoglobulin G [IgG] or IRD680 anti-mouse IgG; Rockland) were diluted 1:5,000 in TBS-Tween 0.1% and incubated for 1 h at 22°C, and the immune complexes were visualized by near-infrared fluorescence (Odyssey imaging system; LiCor BioSciences).
Gene expression analysis.
Quantitative real-time PCR (Q-RT-PCR) was performed with SYBR green I by use of a MyiQ single-color RT-PCR detection system and iQ SYBR green supermix (Bio-Rad, Hercules, CA) according to the manufacturer's protocols. First-strand cDNA was prepared with an iScript cDNA synthesis kit (Bio-Rad). cDNA (1 μl) was added to a reaction master mix (20 μl) containing 2.5 mM MgCl2, HotStart Taq DNA polymerase, SYBR green I, deoxynucleoside triphosphates, fluorescein (10 nM), and gene-specific primers (200 nM each of forward and reverse primers). For each experimental sample, triplicate reactions were conducted in 96-well plates. PCR cycling conditions consisted of a hot-start activation of HotStart Taq DNA polymerase (94°C, 15 min) and 40 cycles of denaturation (95°C, 15 s), annealing (55 to 58°C, 30 s), and extension (72°C, 30 s). Formation of PCR product was monitored in real time by measuring SYBR green I fluorescence at 72°C.
Primer sets used for human transcripts were as follows: for IL-8, 5′-ACTGAGAGTGATTGAGAGTGGAC-3′ and 5′-AACCCTCTGCACCCAGTTTTC-3′; for Gro-β, 5′-AGAATGGGCAGAAAGCTTGTCT-3′ and 5′-CAGCATCTTTTCGATGATTTTCTTAA-3′; for the IκBα 3′ untranslated region, 5′-TGCCTAGCCCAAAACGTCTT-3′ and 5′-CGTCCCCTACAAAAAGTTCACAA-3′; for IκBα coding sequences, 5′-CTCCGAGACTTTCGAGGAAATAC-3′ and 5′-GCCATTGTAGTTGGTAGCCTTCA-3′; for TNFAIP3/A20, 5′-AAGCTGTGAAGATACGGGAGA-3′ and 5′-CGATGAGGGCTTTGTGGATGAT-3′; for Naf1, 5′-AGAGGACCGTACCGGATCTAC-3′ and 5′-CCTTCACTAGGCGCTCAAAAG-3′; and for GAPDH, 5′-ATGGGGAAGGTGAAGGTCG-3′ and 5′-GGGGTCATTGATGGCAACAATA-3′. Primers for mouse transcripts were as follows: for mGro-β, 5′-CACTCTCAAGGGCGGTCAA-3′ and 5′-TGGTTCTTCCGTTGAGGGAC-3′; and for mIκBα, 5′-TCCTGCACTTGGCAATCATC-3′ and 5′-AGCCAGCTCTCAGAAGTGCC-3′. Relative gene expression was determined using the 2−ΔΔ_CT_ method (56). The mean threshold cycle (CT) of triplicate measures was computed for each sample. The sample mean CT of cyclophilin (internal control) was subtracted from the sample mean CT of the respective gene of interest (Δ_CT_). The mean Δ_CT_ of the control (untreated) sample was selected as a calibrator and subtracted from the mean Δ_CT_ of each experimental sample (ΔΔ_CT_). The severalfold change in gene expression (normalized to the internal control gene and relative to the control sample) was expressed as 2−ΔΔ_CT_.
Two-step ChIP assay.
Two-step ChIP was performed as described previously (39). The Abs for RelA, RelB, c-Rel, NF-κB1, NF-κB2, total RNA Pol II, CDK-9, and Ccn T1 were those used in Western blotting. Mouse monoclonal Abs for phosphorylated RNA Pol II were from Covance Research Products. Monoclonal Ab-chromatin complexes were captured with a rabbit anti-mouse IgM Ab (Rockland) prior to binding to protein A magnetic beads (38). DNA binding was detected by qualitative PCR using primers for IL-8, Gro-β, IκBα, Naf1, NF-κB2, and TRAF1 as described previously (39, 56). The primers used for TRAF1 were 5′-GATGTGCCCAGCGAAGTGG-3′ and 5′-TGAGTCACAGCAGGGATGGAG-3′. For Q-RT-PCR, the primers used were as follows: for Gro-β, 5′-TCGCCTTCCTTCCGAACTC-3′ and 5′-CGAACCCCTTTTATGCATGGT-3′; and for Naf1, 5′-GGTCTAGGAAATCCCAGTCTGTTG-3′ and 5′-CGGGTGGGCAAATCCA-3′.
Intracellular ROS assay.
Changes in intracellular ROS were determined as described previously (26). Briefly, cells were suspended in phosphate-buffered saline (PBS) and loaded with 5 μM of 5 (and 6)-carboxy-2′,7′-dichlorodihydro-fluorescein diacetate (H2DCF-DA; Molecular Probes) for 15 min at 37°C. After removal of excess H2DCF-DA, cells were TNF treated, and changes in DCF fluorescence were determined by flow cytometry (FACScan; Becton Dickinson). The fluorescences for 12,000 cells from three or more independent experiments were analyzed and expressed as means ± standard errors of the means.
Changes in levels of carbonylated proteins were determined as described previously (31). Briefly, TNF-stimulated HeLa cells were lysed in radioimmunoprecipitation assay buffer containing 5 mM dithiothreitol and protease inhibitors. The carbonyl groups in the side chains of proteins (20 μg) were derivatized to 2,4-dinitrophenylhydrazone by reaction with 2,4-dinitrophenylhydrazine. The reaction was neutralized with neutralization buffer provided by the Chemicon OxyBlot assay kit. The 2,4-dinitrophenyl-derivatized proteins were fractionated on an SDS-10% polyacrylamide gel and proteins transferred to polyvinylidene difluoride membranes (Amersham, Inc.). After transfer, the membranes were blocked with 5% dry milk in PBS-Tween 20 (0.1%) for 3 h at room temperature than incubated with primary Ab to NAD (Chemicon, Inc.). After the washing, horseradish peroxidase-conjugated secondary Ab (Amersham, Inc.) was added for 1 h. Detection was performed by enhanced chemiluminescence (ECL; Amersham, Inc.).
RelA-CDK-9 colocalization analysis.
HeLa S3 cells stably expressing the EGFP-RelA fusion protein were plated on glass coverslips. Once the cultures reached 50 to 70% confluence, individual coverslips were exposed to TNF for 0 (control), 30, and 60 min in triplicate and fixed with 4% paraformaldehyde in PBS at room temperature for 20 min. Immunofluorescence staining was done using rabbit anti-CDK-9 Ab as the primary Ab (1:100) and the Alexa Fluor 568 F(ab′)2 fragment of goat anti-rabbit IgG (heavy plus light chains) (Invitrogen, Eugene, OR) as the secondary Ab (5 μg/ml). Then, the cells were counterstained with 500 nM TO-PRO-3 (Invitrogen) and mounted on glass slides by use of Vectashield mounting medium (Vector Laboratories, Burlingame, CA). Fluorescence imaging was performed with a Zeiss LSM510META confocal microscope with a Plan-Apochromat 63× 1.4-numerical-aperture oil immersion lens. The GFP, Alexa 568, and TO-PRO-3 image components were obtained using 488-nm (argon ion), 543-nm (green He/Ne), and 633-nm (red He/Ne) laser excitation lines; the corresponding emission collection ranges were 505 to 530 nm, 560 to 620 nm, and longer than 650 nm, respectively. At the image collection settings used (laser intensity and detection gain), no bleed-through was observed. The pinhole size and scan settings were adjusted for a section thickness of 0.7 μm and a pixel size of 0.1 μm, respectively. Images were acquired using a bit depth of 12- and 16-frame Kallman averaging. At least five fields were captured per coverslip.
For image processing and analysis, the .lsm files generated by the Zeiss confocal microscope were translated into TIFF files by use of Image J (http://rsb.info.nih.gov/ij/) and the LSM reader plug-in (http://rsb.info.nih.gov/ij/plugins/lsm-reader.html). Metamorph V6.1 (Molecular Devices, Downingtown, PA) was used for background correction and image segmentation and to quantify translocation. The TO-PRO-3 stain was used to create segmentation masks separating individual nuclei. Dilation of these masks was used to select perinuclear cytoplasmic rings for each cell. Average intensity measurements within each nuclear region and the corresponding cytoplasmic ring were used to quantify RelA translocation. To quantify colocalization, each nucleus was analyzed separately. First, an intensity threshold was used to exclude the nucleolus, an area which showed low levels of both RelA and CDK-9 stains. Each masked nuclear image was exported to Imaris (Bitplane, Zurich, Switzerland) and the degree of colocalization between RelA and CDK-9 was assessed using the “colocalization analysis module.” This module uses correlation analysis first to test the significance of true colocalization over random color overlap and then to quantify the degree of colocalization by use of an automatic threshold search algorithm to avoid the bias of visual interpretation. The algorithm used in this software was described previously (13). Results are expressed as percentages of EGFP colocalized with the Alexa-568 signal.
siRNA-mediated CDK-9 knockdown.
For CDK-9 knockdown, control and CDK-9 siRNAs were from Ambion, Inc. (Austin, TX). siRNAs were transfected at a 100 nM concentration into A549 cells by use of TransIT-TKO transfection reagent (Mirus Bio Corp., Madison, WI). For Western blotting, 72 h later, cells were lysed in modified radioimmunoprecipitation assay buffer; for RNA extraction, cells were TNF stimulated for the indicated times and lysed in TRI reagent (Sigma-Aldrich Inc., St. Louis, MO).
RESULTS
Kinetics of NF-κB-dependent gene transcription.
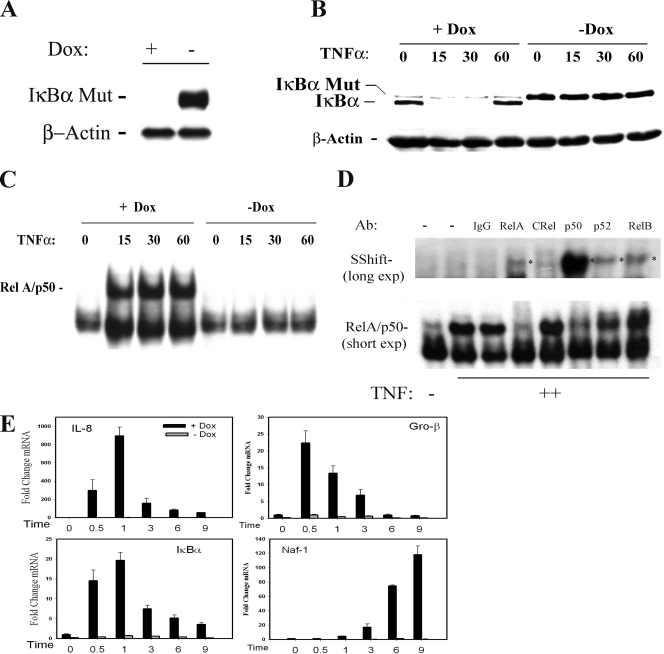
To explore the genetic networks under NF-κB control, we have developed a tetracycline (Tet)-regulated cell system (Tet-Off [18]) in which a nondegradable FLAG epitope-tagged IκBα (IκBα Ser32Ala/Ser36Ala, termed FLAG-IκBα Mut) under control of the Tet operator was stably introduced into cells expressing the Tet transactivator (tTA). FLAG-IκBα Mut contains site mutations in the serine phosphoacceptor sites of IKKβ that block its inducible degradation and functions as a potent dominant-negative inhibitor of the canonical NF-κB activation pathway (11, 55). In these cells, doxycycline (Dox) inhibits FLAG-IκBα Mut expression; upon Dox withdrawal, FLAG-IκBα Mut expression is strongly upregulated (Fig. 1A). To confirm that FLAG-IκBα Mut expression was inert to TNF-induced proteolysis and can inhibit NF-κB activation, a time course of TNF stimulation was performed. In cells cultured in the continuous presence of Dox, little detectable FLAG-IκBα Mut is detected (Fig. 1B). Within 15 min of TNF stimulation, endogenous IκBα is rapidly proteolyzed to undetectable levels, and by 60 min, endogenous IκBα is resynthesized. In contrast, upon Dox withdrawal, FLAG-IκBα Mut is strongly expressed at amounts similar to those for endogenous IκBα and does not degrade in response to TNF (Fig. 1B, right).
FIG. 1.
Dox-regulated NF-κB-dependent gene expression. (A) Tet-regulated IκBα Mut expression. HeLa Tet-Off cells expressing FLAG epitope-tagged IκBα Mut (Ser32Ala/Ser36Ala) were cultured in the presence (+) or absence (−) of 2 μg/ml Dox. Cytoplasmic extracts were extracted and assayed by Western blotting. (Top) Anti-FLAG Ab; (bottom) β-actin used as a loading control. (B) IκBα Mut is TNF resistant. Tet-regulated FLAG-IκBα Mut cells cultured in the presence or absence of Dox were TNF stimulated for the indicated times (min). (Top) Cytoplasmic extracts were assayed for IκBα abundance using anti-IκBα Ab. The endogenous and IκBα Mut isoforms are shown. (Bottom) β-Actin staining. (C) NF-κB binding in EMSA. Nuclear extracts from the experiment shown in panel B were assayed for NF-κB binding using EMSA. Shown are the bound complexes. The RelA/NF-κB1 (p50)-containing complex is indicated. (D) Supershift assay in EMSA. EMSA was performed on control (−) or TNF-stimulated (+) nuclear extracts in the absence or presence of indicated Ab (top). (Top) Long exposure (exp) of the supershifted (SShift) bands (indicated by *); (bottom) short exposure to demonstrate depletion of the TNF-inducible complex. The TNF-inducible NF-κB complex is composed predominantly of RelA·p50 heterodimers. (E) mRNA expression is NF-κB translocation dependent. RNA from a time series of TNF-stimulated HeLa IκBα Mut cells was analyzed by Q-RT-PCR. (The IκBα primers were designed to hybridize selectively to the 3′ untranslated region of the endogenous gene and do not detect IκBα Mut expression.) Shown are mRNA expression profiles in the presence or absence of Dox. For each gene, change is determined relative to unstimulated values.
We sought to establish whether this level of IκBα Mut expression was sufficient to inhibit NF-κB activation by performing EMSA on nuclear extracts prepared from the same experiment. In the presence of TNF, a rapid and potent induction of the RelA-NF-κB1 (p50) heterodimer binding was observed, and its abundance peaked at 15 min (Fig. 1C). In the absence of Dox, no TNF-inducible changes in NF-κB binding above what were seen for the unstimulated control were observed by EMSA (Fig. 1C, right). The rapidly induced DNA-binding complex is composed primarily of RelA-NF-κB1(p50) subunits, because the complex is disrupted by anti-RelA and anti-NF-κB1 Abs (Fig. 1D). Although a strong supershift of NF-κB1 is seen, only a faint supershift of RelA is seen, a finding consistent with the destabilization of the Ab-RelA complex in EMSA (19).
The effects of FLAG-IκBα Mut on TNF-induced endogenous gene expression was measured for four NF-κB-dependent genes (the Gro-β, IL-8, IκBα, and Naf-1 genes) by use of Q-RT-PCR. In the presence of Dox, TNF stimulation rapidly induces IL-8, Gro-β, and IκBα gene expression, peaking within 30 min to 1 h after stimulation at 900-fold, 23-fold, and 22-fold, respectively, over the control level (Fig. 1E). In the absence of Dox, the TNF-induced mRNA expression is almost completely abrogated, indicating that these genes are NF-κB dependent. Although the expression of the Naf1 gene is kinetically distinct, peaking 6 to 9 h after TNF administration, its expression is also NF-κB dependent.
Kinetics of NF-κB subunit recruitment to the target genes in chromatin.
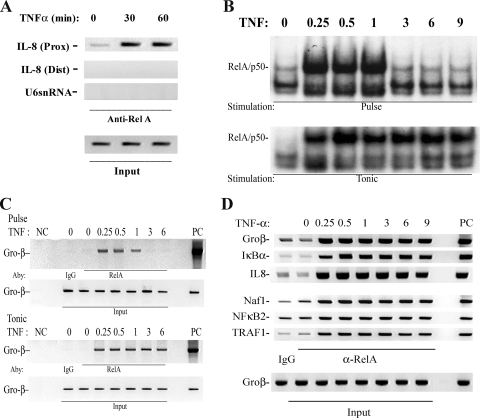
To better characterize the kinetics of NF-κB recruitment on individual promoters within their native chromatin context, we developed a highly efficient two-step ChIP assay that first immobilizes RelA by _N_-hydroxysuccinimide ester-mediated protein-protein cross-linking prior to a second formaldehyde-mediated protein-DNA cross-linking step (39). The protein cross-linking step was necessary because fluorescence photobleaching experiments of NF-κB interacting with its multimeric binding sites in vivo have shown that RelA is in rapid exchange with its target DNA, being completely replaced within 30 s (8). Although we earlier demonstrated that this assay was robust for distinct NF-κB isoforms and captured targets at levels greater than seen for IgG controls (39), the performance of this modified ChIP assay to enrich for RelA binding to target genes was further examined. First, to establish whether the two-step ChIP specifically measured RelA binding to target genes containing NF-κB sites, chromatin from a time series of TNF-stimulated HeLa cells was cross-linked, sonicated, and used as input for immunoprecipitation (IP) using anti-RelA Ab. After capture, washing, and reversal of the cross-links, the DNA was subjected to PCR using primers spanning the high-affinity RelA site located at nucleotides −99 to −61 in the human IL-8 promoter [IL-8 (Prox); Fig. 2A], primers to an upstream region of IL-8 not known to bind RelA spanning nucleotides −1042 to −826 [IL-8 (Dist) (9, 38)], and primers spanning an unrelated U6 snRNA spanning nucleotides −245 to +85, an RNA Pol III-driven gene. We observed specific PCR products only in the IL-8 (Prox) samples spanning the high-affinity RelA site, with a faint signal being detected at time zero and a strong induction 30 and 60 min after TNF administration relative to what was seen for input DNA used as a loading control (Fig. 2A). Specificity was indicated by a lack of signal produced by the IL-8 (Dist) and U6 primers (Fig. 2A).
FIG. 2.
Recruitment kinetics of NF-κB/RelA. (A) Specificity of ChIP assay. Two-step ChIP assay was performed on a time series of TNF-stimulated cells (39). Chromatin was subjected to IP using anti-RelA Ab. Eluted DNA was subjected to PCR using primers spanning the high-affinity RelA site located at nucleotides −99 to −61 in the human IL-8 promoter [IL-8 (Prox)] (top), primers to an upstream region of IL-8 not known to bind RelA and spanning nucleotides −1042 to −826 [(IL-8(Dist)] (middle), and primers spanning the Pol III-driven gene, U6 snRNA (bottom). Input DNA was used as a loading control. Shown is an ethidium bromide-stained agarose gel of the PCR products. Note that only the IL-8 (Prox) primers produce a TNF-inducible signal. (B) EMSA of TNF-stimulated cells. HeLa cells were exposed to either a 15-min pulse or continuous (tonic) stimulation. Nuclear extracts were subjected to EMSA using a high-affinity RelA-NF-κB1 binding site (10). Shown is an autoradiograph of the specifically bound species. (Top) Pulse stimulation; (bottom) tonic stimulation. (C) ChIP of tonic versus pulse TNF stimulation protocols. HeLa cells were stimulated for the indicated times after 15-min pulse (top) or tonic (bottom) TNF stimulation. Chromatin was prepared using two-step ChIP and immunoprecipitated with anti-RelA Ab. Shown is an ethidium bromide-stained gel of the PCR products. Input DNA was used as an IP control. (D) Time course of RelA recruitment to target genes. Cells were stained for indicated times and chromatin immunoprecipitated with anti-RelA Ab. Shown is an ethidium bromide-stained gel for PCR of the IL-8, IκBα, Gro-β, Naf1, NF-κB2, and TRAF1 genes by use of gene-specific primers flanking a high-affinity NF-κB binding site (56). Each PCR was run with 25 ng of genomic DNA as a positive control (PC). The negative control (NC) for the IP was IgG.
To determine whether the two-step ChIP assay could detect dynamic differences in the association and dissociation of RelA, we exploited the findings that distinct NF-κB activation kinetics can be produced by different TNF stimulation protocols. Previous work using dynamic fluorescence imaging has shown that RelA can be induced to translocate in separate modes, depending on the nature of the TNF stimulation protocol and the nucleus-cytoplasm volumetric ratio (8, 32, 36, 54). If the TNF is applied as a transient pulse (15 min), the initial NF-κB translocation is monophasic. After a rapid initial translocation, RelA induces the resynthesis of its IκBα inhibitor, resulting in its cytoplasmic reaccumulation (19, 52). Upon reaccumulation of IκBα, nuclear RelA is captured and relocated to the cytoplasm, resulting in a single spike in nuclear NF-κB binding. Conversely, if the TNF agonist is applied to the cells without removal (tonic stimulation), RelA is constitutively nuclear in cells with large nuclear volumes and oscillatory in cells with small nuclear volumes. The tonic exposure of agonist results in continuous IκBα degradation, in spite of its nuclear resynthesis. To illustrate, nuclear extracts prepared from TNF-stimulated HeLa cells for various times were bound to a radiolabeled NF-κB consensus site in EMSA (Fig. 2B). In response to pulsatile TNF stimulation, NF-κB DNA-binding activity was rapidly induced, peaking from 15 to 60 min and declining rapidly thereafter to control levels (32, 36, 54-56). By contrast, in response to tonic TNF stimulation, NF-κB binding activity was induced and persisted over 9 h of stimulation (Fig. 2B). Together, these data indicated that we could induce distinct RelA activation modes using different TNF stimulation protocols.
The two-step ChIP was applied to chromatin prepared from cells induced by the two different stimulation protocols. The cross-linked chromatin was immunoprecipitated using anti-RelA Ab, and Gro-β binding was detected by qualitative PCR. In response to pulsatile TNF stimulation, we observed that RelA binding was rapidly induced at 15 to 30 min, whereupon it dissociated from the Gro-β promoter coincidentally with its cytoplasmic recapture (Fig. 2C, top). By contrast, in response to tonic TNF stimulation, RelA binding to Gro-β was stable over the entire time course (Fig. 2C, bottom). These findings indicated that the two-step ChIP could detect the association and dissociation of RelA binding with its endogenous gene targets and that there was no detectable lag in RelA binding between RelA's appearance in the nucleoplasm (detected by EMSA) and its association with chromatin (detected by ChIP) on the Gro-β promoter. This experimental result is further compatible with previous findings that nuclear RelA is in rapid exchange with its target sites in chromatin (8). Finally, this result validated that the modified two-step ChIP was capable of measuring both the association and the dissociation of RelA on its endogenous binding sites.
We next sought to determine RelA binding kinetics on target genes showing a rapid transcriptional spike. Using qualitative PCR, we could detect changes in target DNA over a concentration range of 0.2 to 20 ng, as shown for IL-8, Gro-β, and IκBα (see Fig. S1 in the supplemental material). For all genes examined, we found that the kinetics of TNF-induced RelA binding was rapid and was maximal within 15 to 30 min after stimulation (Fig. 2D). Interestingly, for genes that are strongly induced by TNF, such as the IL-8, IκBα, and Gro-β genes, as well as genes that are not, such as the Naf1, NF-κB1, and TRAF2 genes, RelA binding was rapidly inducible, peaking 15 min after stimulation. Moreover, the binding profiles were indistinguishable in nature for the duration of the stimulation. To confirm these findings, we measured RelA-associated target genes by use of Q-RT-PCR, where a three- to fourfold change in RelA binding for Gro-β and Naf1 was observed at identical times (see Fig. S2 in the supplemental material). These findings were surprising because previous work has suggested that RelA is occluded from accessing chromatin sites in late-response genes in monocytes (45).
Also of note is that RelA binding to the IL-8, IκBα, and Gro-β promoters was persistent over the 9-h time course, even though mRNA expression by these genes was terminating (cf. Fig. 1E and 2D). These findings suggested to us that the differential patterns of NF-κB-dependent gene expression were not due to different rates of RelA association.
Promoter-specific phospho-Ser276 RelA recruitment.
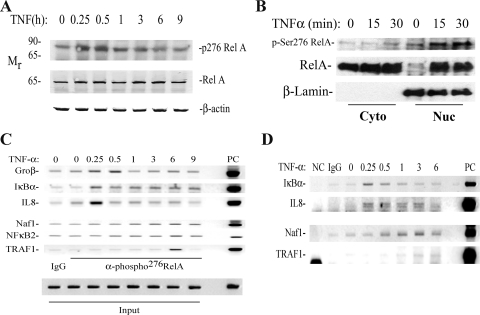
Because the binding of bulk RelA did not accurately track with the kinetics of transcriptional activation (e.g., Naf1, TRAF1, and NF-κB2) or termination (IL-8, IκBα and Gro-β), we next examined the kinetics of formation and chromatin binding of phospho-Ser276, as this phosphorylation results in a potent activating species required for IL-8 gene induction (26, 59). Western blots were performed on a time series of TNF-stimulated cells extracted by denaturing lysis using a phospho-Ser276 RelA-specific Ab (Fig. 3A). In unstimulated cells, low amounts of phospho-Ser276 RelA could be detected, a form which migrates aberrantly at 80 kDa, consistent with the findings of others (59, 60). TNF induced rapid formation of phospho-Ser276 RelA, peaking 15 to 30 min later at ∼3-fold induction, which rapidly returned to unstimulated levels 1 h after stimulation (Fig. 3A). To determine whether this isoform translocated into the nucleus, Western blots of cytoplasmic and sucrose cushion-purified nuclear extracts were performed. Phospho-Ser276 RelA was highly enriched in the nuclear fractions, with little detectable cytoplasmic staining (Fig. 3B). The kinetics of nuclear accumulation parallel those of the whole-cell lysate. Together, these data indicate that phospho-Ser276 RelA is rapidly translocated into the nucleus.
FIG. 3.
(A) Time course of phospho-Ser276 RelA formation. WCEs prepared from a time course of TNF-stimulated cells were fractionated by SDS-PAGE and blotted with anti-phospho-Ser276 RelA (top), anti-RelA (middle), and β-actin as an internal control (bottom). (B) Nucleus-cytoplasm distribution of phospho-Ser276 RelA. HeLa cells were stimulated with TNF for the times indicated at the top and fractionated into cytoplasmic and nuclear fractions. (Top) Western immunoblotting using anti-phospho-Ser276 RelA Ab. (Middle) Blot was reprobed for RelA as a loading control. Note the inducible accumulation of RelA in the nuclear fraction within 15 min of stimulation. (Bottom) β Lamin staining as a nuclear fraction control. The nuclear fractions stain strongly for β lamin. (C) Time course of phospho-Ser276 RelA recruitment to NF-κB-dependent genes. HeLa cells were subjected to a time course of TNF stimulation and chromatin was prepared. Shown is an ethidium bromide-stained gel of the PCR products. IgG, negative control for IP using nonimmune IgG. Each row represents a separate PCR assay. (D) Time course of phospho-Ser276 RelA binding in A549 cells. Experiment is as described for panel C. A549 cells show similar patterns of phospho-Ser276 RelA recruitment.
We next measured the binding kinetics of phospho-Ser276 RelA to NF-κB-dependent genes. Chromatin from a time course of TNF stimulation was prepared and two-step ChIP performed using anti-phospho-Ser276 RelA Ab. Here we found that in the absence of stimulation, phospho-Ser276 RelA binding was very low, but phospho-Ser276 RelA was rapidly recruited to the IL-8, IκBα, and Gro-β promoters, peaking 15 to 30 min after stimulation (Fig. 3C). Importantly, Ser276-phosphorylated RelA binding to IL-8, IκBα, and Gro-β gene promoters occurred at times corresponding to their peak transcriptional activations (compare Fig. 1A and 3C). In contrast, phospho-Ser276 RelA binding to Naf1 and NF-κB2 promoters was distinct from this pattern, with no detectable change 15 to 30 min after stimulation, but we did note a gradual increase in binding over 3 to 9 h that was significantly reduced in magnitude (Fig. 3C, bottom).
To determine whether distinct differential phospho-Ser276 RelA recruitment was cell type dependent, we performed the same experiment on TNF-stimulated type II alveolar epithelial cells (A549). As seen in Fig. 3D, similar results were obtained, with an apparent peak in phospho-Ser276 RelA binding to IL-8 and IκBα 15 to 30 min after stimulation and binding to Naf1 and TRAF1 ∼3 h later. The mechanisms controlling the expression of late genes will require further study. For the purposes of this work, we sought to evaluate the role of phospho-Ser276 RelA in the expression of the rapidly inducible genes because phospho-Ser276 RelA binding was coincident with gene expression.
Requirement of phospho-Ser276 binding in NF-κB-dependent gene expression.
Previous work in our laboratory has shown that a TNF-induced ROS signaling pathway mediates the phosphorylation of Ser276 on RelA in monocytic cells (26). Here, intracellular ROS activate IκBα-RelA-associated PKAc to phosphorylate RelA selectively on Ser276. We further showed that phospho-Ser276 RelA formation can be selectively disrupted by a variety of chemically unrelated antioxidants, including the ROS scavenger DMSO, an agent that inhibits PKAc but does not inhibit TNF-induced Jun N-terminal protein kinase activation or mitogen-stress related kinase or induce cellular toxicity (26).
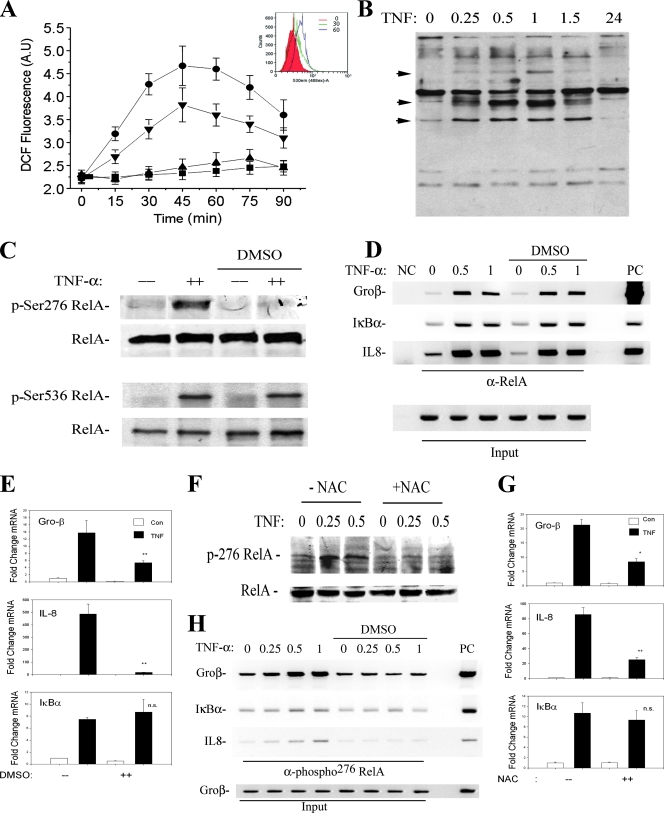
To confirm that DMSO interferes with TNF-induced ROS formation in HeLa cells, DCFDA-loaded HeLa cells were stimulated in the absence or presence of various concentrations of DMSO. TNF induces a detectable increase in ROS formation within 15 min and produces a broad peak from 30 to over 90 min after stimulation (Fig. 4A). These kinetics of ROS formation are consistent with previous studies (3, 21). Although a significant inhibition of ROS was observed with 0.5% DMSO, complete inhibition was observed using 2% DMSO (Fig. 4A).
FIG. 4.
Inhibition of phospho-Ser276 RelA formation by antioxidant. (A) Kinetics of TNF-induced ROS formation and inhibition by DMSO. HeLa cells were loaded with DCFDA and stimulated with TNF (20 ng/ml) in the absence or presence of DMSO (0.5%, 2%). Fluorescence measurements were performed using a fluorescence-activated cell sorter at indicated times. Symbols: ▪, TNF; ▾, TNF plus 2% DMSO; ▴, TNF plus 0.5% DMSO; •, untreated. TNF induces detectable ROS formation within 15 min of stimulation. (Inset) Histograms of fluorescence-activated cell sorter analysis at 0, 30, and 60 min of treatment. (B) Kinetics of TNF-induced protein carbonylation. HeLa cells were stimulated with TNF for the times indicated. Shown is a Western blot for carbonylated proteins. Inducible carbonylation is seen within 15 min of stimulation (indicated by arrow). (C) Selective inhibition of TNF-induced phospho-Ser276 RelA formation by DMSO. HeLa cells were stimulated with TNF for 30 min the absence or presence of nontoxic concentrations of DMSO (2%) (26, 58). (Top) WCEs were prepared and blotted with anti-phospho-Ser276 RelA or pan anti-RelA Abs; (bottom) WCEs were blotted with anti-phospho-Ser536 RelA or anti-RelA Abs. (D) Antioxidant does not affect total RelA recruitment to target promoters. HeLa cells were stimulated for indicated times in the absence or presence of DMSO. Shown is a two-step ChIP assay using pan-anti-RelA Ab in the IP. Input is the loading control for the IP. DMSO has no impact on inducible RelA binding. (E) Effect of DMSO on NF-κB-dependent gene expression. mRNA was prepared from TNF-stimulated HeLa cells in the absence or presence of DMSO. Q-RT-PCR analysis was performed for the indicated genes. Shown is change relative to level at time zero and normalized to 18S RNA as an internal control. **, P < 0.01, t test. (F) Inhibition of TNF-induced phospho-Ser276 RelA formation by NAC. HeLa cells were stimulated with TNF for 30 min in the absence or presence of 10 mM NAC (26, 58). WCEs were prepared and blotted with anti-phospho-Ser276 RelA or anti-RelA Abs. (G) Effect of NAC on NF-κB-dependent gene expression. mRNA was prepared from a time course of TNF-stimulated HeLa cells in the absence or presence of NAC (10 mM). Q-RT-PCR analysis was performed as described for panel E. *, P < 0.05; **, P < 0.01, t test. (H) Antioxidant blocks phospho-Ser276 RelA recruitment. HeLa cells were TNF stimulated for the indicated times in the absence or presence of DMSO. Shown is a two-step ChIP assay using anti-phospho-Ser276 RelA Ab as the immunoprecipitating Ab. DMSO completely blocks TNF-inducible phospho-Ser276 RelA binding.
As an independent confirmation of TNF-inducible oxidative stress, TNF-induced protein carbonylation was assayed. Carbonylation is an irreversible oxidation of Arg, Lys, and Pro side chains that targets proteins for proteasomal degradation (31). TNF stimulation resulted in rapid carbonylation within 15 min of treatment (Fig. 4B). These data confirm that TNF rapidly induces biologically significant oxidative stress and are consistent with our previous findings of enhanced 8-oxoguanosine adduct formation on DNA (16). To test whether the TNF-induced ROS pathway was required for phospho-Ser276 RelA formation in HeLa cells, a Western blot on TNF-stimulated cells in the absence or presence of antioxidant was performed. Here we found that the highly inducible phospho-Ser276 RelA formation after TNF stimulation was completely abrogated after DMSO treatment (Fig. 4C, top), whereas there was no effect on phospho-Ser536 RelA formation (Fig. 4C, bottom). DMSO, therefore, selectively interfered with the formation of Ser276-phosphorylated RelA without apparently inhibiting its other phosphorylation steps or nuclear translocation (26).
To exclude the possibility that DMSO interfered with bulk NF-κB binding to its sites in chromatin, ChIP assays were performed on a time course of TNF-stimulated cells in the presence or absence of DMSO by use of an anti-RelA Ab that binds both phosphorylated and nonphosphorylated (“bulk”) RelA (Fig. 4D). Again, TNF stimulation alone induced robust bulk RelA binding. Importantly, bulk RelA binding was not affected in the presence of DMSO (Fig. 4D, right). From these data, we concluded that inhibition of the ROS pathway has no effect on total RelA nuclear translocation or DNA binding to target genes, findings consistent with our earlier studies (26, 58).
Inhibition of the ROS pathway by DMSO, then, allows an examination of the role of phospho-Ser276 RelA in NF-κB-dependent gene expression. For this purpose, TNF-induced expression of Gro-β, IκBα, and IL-8 mRNA was determined by Q-RT-PCR in the absence or presence of DMSO. We found that DMSO completely inhibits TNF-inducible Gro-β and IL-8 expression but does not affect IκBα expression (Fig. 4E). These data suggest that Gro-β and IL-8 require phospho-Ser276 RelA for inducible gene expression but IκBα does not.
To confirm these observations using a chemically unrelated antioxidant, we performed TNF stimulations using NAC at concentrations that inhibited TNF-induced DCF oxidation (58). NAC blocked phospho-Ser276 formation (Fig. 4F) and significantly inhibited Gro-β and IL-8 gene expression without affecting IκBα (Fig. 4G).
To confirm that DMSO reduced phospho-Ser276 RelA binding, two-step ChIP was conducted using a phospho-Ser276 RelA Ab. In this experiment, we plated the cells at a twofold increase in cell density to maximize the detection of phospho-Ser276 RelA signal by the two-step ChIP assay. As a result, a basal signal of phospho-Ser276 RelA binding could be detected for Gro-β and, to a lesser extent, for IL-8 in unstimulated cells due to the increase in DNA input into the PCR (Fig. 4H). Moreover, for reasons not yet completely understood, the kinetics of phospho-RelA binding were slightly delayed by 15 min relative to the experimental results shown earlier in Fig. 3C, suggesting that the kinetics of ROS formation are cell density dependent. Nevertheless, and most importantly, inducible phospho-Ser276 RelA binding to Gro-β, IL-8, and IκBα was completely inhibited (Fig. 4H). These data indicated that the inhibition of the ROS pathway selectively ablates the phospho-Ser276 RelA formation and recruitment of phospho-Ser276 RelA, but not bulk RelA, to target genes. Taken together, this series of experiments shows that phospho-Ser276 RelA is rapidly formed after TNF stimulation in an oxidant-dependent manner and that its binding is necessary for the expression of a subset of NF-κB-dependent genes, including the Gro-β and IL-8 genes.
RelA Ser276 mediates differential patterns of gene expression.
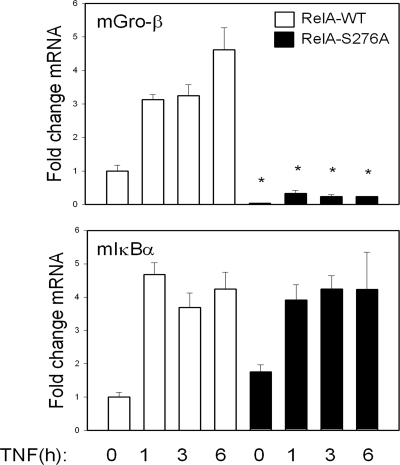
To further determine the requirement of RelA Ser276 phosphorylation in the activation of a subset of NF-κB-dependent genes, we examined the effect of expressing a RelA Ser-Ala site mutation at residue 276 (RelA Ser276Ala). RelA−/− MEFs were transfected with FLAG epitope-tagged strawberry-RelAWT or RelA Ser276Ala expression vectors and pools of stable transfectants isolated. Cells were then TNF stimulated for a 6-h time course, and Q-RT-PCR was performed. We selected for measurement the mouse homologs of the Gro-β and IκBα genes (note that mice do not have an IL-8 gene). Although RelAWT potently transactivated the mGro-β gene, the RelA Ser276Ala mutant did not (Fig. 5). Conversely, both proteins strongly transactivated the mIκBα gene by ∼5-fold. These data confirmed our earlier findings that phospho-Ser276 RelA controls a subset of NF-κB gene expression.
FIG. 5.
Effect of RelA Ser276Ala mutation on target gene expression. RelA−/− MEFs were transfected with expression vectors encoding a FLAG epitope-tagged monomeric strawberry fusion with RelAWT or RelA Ser276Ala, and pooled transfectants were isolated. Cells were stimulated for indicated times with TNF, and mRNA was extracted. Q-RT-PCR was performed to determine the activation of endogenous NF-κB-dependent genes.
The CDK-9-Ccn T1 complex, P-TEFb, inducibly binds phospho-Ser276 RelA.
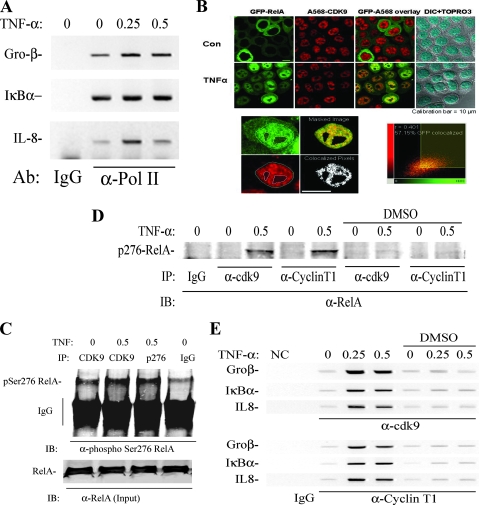
Our data suggested that inducible phospho-Ser276 RelA binding is important for the activation of the IL-8 and Gro-β genes but not for that of the IκBα gene. Of these, the IL-8 and Gro-β genes show inducible RNA Pol II recruitment, whereas the IκBα gene is preloaded with Pol II (Fig. 6A). This finding suggested to us that phospho-Ser276 RelA may regulate transcriptional elongation of the IκBα gene. In this process, inducible association of RelA with specific kinases that phosphorylate the Pol II carboxy-terminal domain (CTD) allows it to enter a processive phase of mRNA transcription. Previous work has shown that P-TEFb, a complex containing CDK-9-Ccn T1, is responsible for phosphorylating RNA Pol II on the Ser2 CTD, licensing Pol II to enter the active transcriptional elongation mode (37, 44).
FIG. 6.
TNF-inducible Pol II recruitment and RelA-P-TEFb complex formation. (A) Effect of TNF on Pol II loading. ChIP from TNF-stimulated HeLa cells, which were immunoprecipitated with anti-Pol II Ab. (B) Colocalization analysis. HeLa cells stably expressing EGFP-RelA were stimulated in the absence (control [Con]) or presence of TNF (30 min). Cells were fixed and stained with rabbit anti-CDK-9 Ab and Alexa Fluor 568-labeled goat anti-rabbit IgG. Nuclei were counterstained with TO-PRO-3 and are shown as overlays with differential interference contrast. (Bottom left) Sequential stages in image analysis with masking and colocalized pixels; (bottom right) corresponding two-dimensional histogram of EGFP-RelA and CDK-9 correlation. Shown is least-squares correlation analysis and the extent of colocalization for this image. After analysis of 39 individual cells, the Pearson's correlation coefficient was 0.428 ± 0.127, and the association was highly statistically significant (P < 0.001). (C) CDK-9 recruitment to RelA. FLAG-RelA-expressing HeLa cells were stimulated with TNF for 0 and 0.5 h in the absence or presence of DMSO as indicated. RelA-associated complexes were immunoprecipitated using anti-FLAG Ab, and the association of CDK-9 was detected by immunoblotting. (Bottom) IB of FLAG-RelA as a recovery control for the immunoprecipitation (IP). CDK-9-RelA complex association is TNF inducible and is reduced by DMSO treatment. (D) Coimmunoprecipitation assay for RelA-P-TEFb complex formation. HeLa cells were stimulated with TNF for 0 and 0.5 h in the absence or presence of antioxidant (DMSO). Nuclear extracts were prepared and immunoprecipitated with indicated anti-CDK-9 or Ccn T1 Abs. The presence of RelA in the immune complexes was then detected using anti-RelA Ab in the immunoblot (IB). A TNF-inducible complex was detected between Ser276-phosphorylated RelA, CDK-9, and Ccn T1. (Right) The inducible RelA-P-TEFb complex is quantitatively prevented by DMSO treatment. (E) Inducible P-TEFb binding. HeLa cells were stimulated with TNF for 0, 0.25, and 0.5 h in the absence or presence of DMSO as indicated. (Top) Two-step ChIP assay using anti-CDK-9 Ab as the immunoprecipitating Ab; (bottom) anti-Ccn T1 Ab was used. Shown is an ethidium bromide-stained gel of PCRs for IL-8, Gro-β, and IκBα as indicated (left). TNF-inducible recruitment of each member of the pTEF complex is rapid and indistinguishable. Inducible P-TEFb recruitment is significantly inhibited by DMSO.
We therefore investigated whether RelA spatially colocalized with CDK-9 by use of fluorescence confocal microscopy. For this purpose, cells stably expressing a TNF-regulated EGFP-RelA fusion protein were stimulated in the presence or absence of TNF. Staining with anti-CDK-9 Ab was then used to visualize endogenous CDK-9, tagging it with Alexa 568 (Fig. 6B). In the absence of stimulation, EGFP-RelA was strongly cytoplasmic; in response to TNF, EGFP-RelA was translocated into the nucleus in 100% of cells in a nonrandom distribution, excluding nucleoli (Fig. 6B). By contrast, CDK-9 was constitutively nuclear and did not change in response to TNF. To rigorously demonstrate colocalization, we analyzed their nuclear spatial distributions for correlation in excess of random overlap by use of a validated statistical algorithm (13), where the observed correlation between two color channels is compared against a probability distribution of random overlap generated by repetitive image scrambling (13). The extent of colocalization is calculated by using an automatic threshold search method that avoids the subjective bias that usually occurs with other methods that rely more in visual inspection. Using this approach, we observed that 56.65% ± 19.9% of EGFP-RelA was colocalized with CDK-9, an association that was highly statistically significant (P < 0.001). These data suggested that CDK-9 colocalized with activated nuclear RelA.
To examine whether phospho-Ser276 RelA associated with CDK-9, nondenaturing coimmunoprecipitation experiments were performed. Control or TNF-stimulated nuclear extracts were immunoprecipitated with anti CDK-9 Ab, and the presence of RelA isoforms was detected by Western blotting (Fig. 6C). We detected twofold-increased binding of phospho-Ser276 RelA in the CDK-9 complexes (Fig. 6C). RelA comigrated with the 80-kDa species that was produced by anti-phospho-Ser276-RelA Ab (Fig. 6C and reference 12). The binding of phospho-RelA to Ccn T1 and CDK-9 in the presence of TNF stimulation was significantly inhibited by the presence of DMSO (Fig. 6D). Together, these data suggest that the TNF-inducible Ser276 phosphorylation of RelA is required for association with the P-TEFb Pol II CTD kinase.
TNF-induced P-TEFb recruitment to target genes is dependent on phospho-Ser276 RelA formation.
To determine whether TNF induced P-TEFb recruitment on NF-κB-dependent genes, and if so, whether this recruitment required Ser276-phosphorylated RelA, a time course experiment using a ChIP assay of TNF stimulation in the presence or absence of DMSO was conducted. As seen in Fig. 6E, TNF-inducible CDK-9 and Ccn T1 binding was observed for IL-8, Gro-β, and IκBα. Here, P-TEFb binding occurred rapidly, within 15 min, at times coincident with target gene expression. Importantly, TNF-inducible CDK-9 and Ccn T1 binding was significantly inhibited in cells TNF stimulated in the presence of DMSO. Together, these data indicate that the P-TEFb complex is inducibly recruited to target promoters in a phospho-Ser276 RelA-dependent manner.
TNF-induced NF-κB-dependent gene expression requires CDK activity.
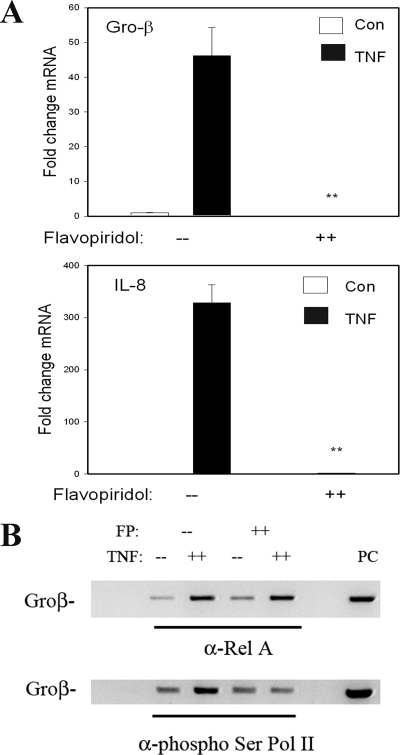
To further establish the functional role of CDK in NF-κB-dependent gene expression, we examined the effect of the CDK inhibitor FP (48). FP is a highly selective CDK inhibitor that potently inhibits the kinase activity of CDK-9 (37). In this experiment, vehicle- or FP (500 nM)-pretreated cells were stimulated with TNF, and the induction of NF-κB-dependent genes was measured by Q-RT-PCR. FP completely blocked the expression of IL-8 and Gro-β (Fig. 7A).
FIG. 7.
CDK inhibitor FP inhibits NF-κB-dependent gene expression by disrupting Pol II phospho-Ser2 CTD formation. (A) Gene expression HeLa cells were pretreated with vehicle or FP (500 nM) for 1 h prior to 30 min of TNF stimulation. mRNA expression was measured by Q-RT-PCR. Shown is change in mRNA normalized by 18S relative to unstimulated values. (B) FP inhibits P-TEFb-induced phospho-Ser Pol II CTD formation. HeLa cells were pretreated with vehicle or FP (500 nM) for 1 h prior to 30 min of TNF stimulation. (Top) Two-step ChIP assay using anti-RelA Ab as the immunoprecipitating Ab; (bottom) anti-phospho-Ser Pol II Ab was used. Although FP has no effect on inducible RelA binding, it significantly inhibits phospho-Ser Pol II recruitment.
We next investigated whether CDK kinase activity was required for TNF-induced Ser2 Pol II CTD phosphorylation. Chromatin prepared from control or TNF-stimulated cells in the absence or presence of FP was assayed for RelA and phospho-Pol II recruitment. Consistent with its selective effect on CDK-9 activity, FP had no effect on inducible RelA binding (Fig. 7B). By contrast, TNF-inducible phospho-Ser2 Pol II CTD binding was significantly inhibited, indicating that CDK activity was required to mediate this key posttranslational step in transforming Pol II to its transcriptional elongation mode. These data suggested that phospho-Ser276 RelA-mediated complex formation with P-TEFb mediates transcriptional elongation, a process important in NF-κB-dependent Gro-β expression.
CDK-9 subunit is required for optimal expression of Gro-β/IL-8 genes.
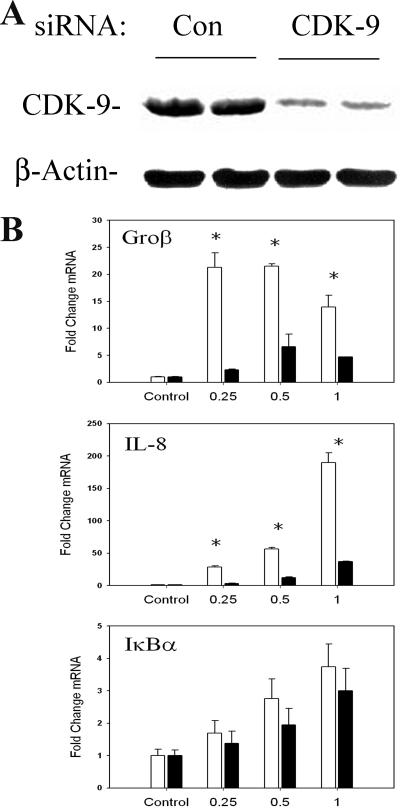
To more specifically implicate the role of CDK-9 in NF-κB-dependent gene expression, we conducted a series of experiments using siRNA knockdown. Relative to those transfected with control, cells transfected with CDK-9 siRNA showed a reduction of ∼50% in normalized CDK-9 steady-state levels (Fig. 8A). Under these conditions of CDK-9 knockdown, a time course of TNF stimulation was performed and gene expression measured by Q-RT-PCR. In contrast to what was seen for control transfectants, we observed a significant reduction in the peak expression of Gro-β and IL-8 mRNA but not in that of IκBα (Fig. 8B). We interpret this result to mean that CDK-9 is an important mediator of phospho-Ser276 RelA-dependent gene expression.
FIG. 8.
Effect of siRNA-mediated knockdown of CDK-9 on NF-κB-dependent gene expression. (A) Magnitude of siRNA-mediated knockdown. Cells were transfected with control (Con) or anti-CDK-9 siRNA prior to lysis and analysis of CDK-9 expression by Western blotting. Duplicate plates are shown for reproducibility. (Top) Staining with anti-CDK-9 Ab; the specific 42-kDa band is shown. (Bottom) Blot was reprobed with anti-β-actin Ab as a loading control. When normalized to β-actin, CDK-9 abundance is downregulated by ∼50%. (B) Effect of CDK-9 knockdown on inducible gene expression. Cells were transfected with control or CDK-9-targeted siRNA for 72 h prior to TNF stimulation for the indicated times. mRNA expression of early genes was measured by Q-RT-PCR. Shown is change in mRNA normalized by 18S relative to unstimulated values. *, P < 0.01, two-tailed t test.
DISCUSSION
TNF is a pleiotropic cytokine that mediates the pulmonary cytokine cascade, the hepatic acute-phase response, and the regulator of leukocyte activation and apoptosis. Upon its ligation to cell surface receptors, TNF induces protein recruitment to cytoplasmic death domains, assembling a signaling complex composed of TRADD, FADD, and TRAF2 (and others) that activates divergent intracellular signals, including the Jun N-terminal protein kinase-AP-1 and the IKK-NF-κB pathways to induce genomic responses in the target cell (23, 24). Although the canonical IKK-NF-κB pathway is critical for inducing tissue inflammation and preventing TNF-induced programmed cell death, surprisingly little is known about its mechanism for activating downstream gene targets. We have used high-density DNA microarrays of Tet-regulated dominant-negative cell lines to systematically identify the network of genes downstream of the NF-κB transcription factor (54-56). Surprisingly, these studies indicate that genes under direct NF-κB control are expressed in temporally distinct waves, with each group affecting a different biological process (54). In this study, we have exploited our recent developments in a quantitative two-step ChIP assay (39), an assay sensitive to binding and dissociation of NF-κB, to probe the diverse mechanisms governing NF-κB-inducible gene expression.
NF-κB subunits are nuclear phosphoproteins responsive to diverse signaling pathways (35). Specifically, the potent RelA-transactivating subunit can be phosphorylated at multiple sites, including Ser276 by PKAc (26, 60) and mitogen and stress kinase 1 (57) and Ser536 by IKKβ, NK-κB-inducing kinase, and c-Src (28, 46). These phosphorylation events lead to an increase in RelA nuclear translocation, DNA binding, and/or transcriptional activity, through the mechanisms not completely understood. Of relevance to this study is the fact that RelA Ser276 phosphorylation occurs upon the activation of the canonical NF-κB pathway and cytoplasmic release from IκBα. Although the relative roles of PKAc and mitogen and stress kinase 1 have yet to be fully understood, we have recently observed that PKAc mediates TNF-induced phospho-Ser276 RelA formation via an intracellular ROS signal (26). This second ROS-PKAc pathway is completely independent of that controlling RelA translocation and is necessary for the acquisition of full transcriptional activity (26). The consequence of Ser276 phosphorylation is thought to be a destabilization of the intermolecular association of the NH2- and COOH-terminal ends of RelA (60), producing a conformational change that may produce a more stable association with CBP/p300 and other coactivators (60). As a result, phospho-Ser276 RelA becomes a substrate for acetylation, resulting in enhanced transcriptional activity (12).
Our findings extend the range of protein interactions involving activated RelA, as we have observed that Ser276 phosphorylation is also required for interaction with the P-TEFb complex. This finding is important because it suggests that phospho-Ser276 RelA binding can mediate transcriptional elongation by recruiting P-TEFb to target genes as an additional mechanism for inducible gene expression over that mediated by p300/CBP recruitment. Importantly, the requirement for P-TEFb recruitment is not a uniform feature of NF-κB-dependent gene activation. Rather, our data suggest that phospho-Ser276 RelA plays only a restricted role in the activation of two NF-κB-dependent targets, the IL-8 and Gro-β genes (but not the IκBα gene). Our ChIP studies indicate that the IL-8 and Gro-β genes are highly inducible through a mechanism involving Pol II recruitment, whereas the IκBα gene is one whose magnitude of expression is not as highly inducible and is stably engaged with Pol II in the absence of TNF stimulation (Fig. 6A). Previously, using genomic footprinting of the IL-8 gene by ligation-mediated PCR, we found that NF-κB activation produced dramatic modifications of the proteins binding the TATA box and transcriptional initiation site, suggesting that NF-κB activated this gene by a promoter recruitment mechanism (9). Here, we extend this mechanism to include both Pol II and P-TEFb recruitment. In response to CDK-9-mediated CTD phosphorylation, recruited Pol II is able to produce full-length transcripts. In the case of the IκBα gene, others have shown that the gene is preloaded with RNA polymerase, making it able to rapidly respond to NF-κB binding without needing time-dependent formation of a preinitiation complex (1). Together, these studies indicate that NF-κB activates its downstream gene targets through pleiotropic mechanisms.
By comparing the kinetics of bulk RelA binding and those of phospho-Ser276 RelA binding to genomic targets, this study represents the first demonstration that there is a dissociation in the timing between phospho- and non-phospho-RelA recruitment. Specifically, our data show that phospho-Ser276 RelA binding is the functionally important complex in Gro-β and IL-8 gene expression because (i) phospho-Ser276 RelA binding temporally coincides with peak transcriptional activation and peak steady-state mRNA abundance, (ii) inhibition of phospho-Ser276 formation by antagonism of the ROS pathway selectively blocks phospho-RelA (but not bulk RelA) binding and transcriptional activation of IL-8/Gro-β, and (iii) a phosphorylation-deficient Ser-to-Ala site mutation at residue 276 is unable to efficiently transactivate Gro-β in a RelA-deficient MEFs. Others have that shown Ser276 phosphorylation was necessary for a strong response to the NF-κB-inducing stimuli TNF and IL-1 (2, 12, 60). It is possible that the size or shape of this multiprotein complex or the nature of chromatin where the NF-κB binding site is found may restrict the access of the phospho-Ser276 RelA complex to certain genes. It will be interesting to compare protein complexes formed by phospho-Ser276 RelA and nonphosphorylated RelA.
Although the Ser276 RelA complex plays a key functional role in IL-8 and Gro-β gene expression, it is not required for IκBα expression, even though our ChIP assays show that phospho-Ser276 RelA rapidly and inducibly binds to this promoter. This conclusion is made based on the ability of hypo-Ser276-phosphorylated RelA to transactivate IκBα after the inhibition of the ROS-PKAc pathway, the ability of the RelA Ser276Ala mutant to transactivate IκBα in RelA−/− MEFs, and the lack of effect of CDK-9 downregulation on IκBα expression. Therefore, although we observed an induction of phospho-Ser276 RelA binding to IκBα in the ChIP assay, this RelA modification and P-TEFb recruitment were not required to activate productive transcription. In this regard, we note earlier studies that have shown phospho-defective RelA at residues Ser205, Ser276, and Ser281 are unable to mediate a subset of NF-κB-dependent genes (the ICAM-1, VCAM-1, and MIP-2 genes) in response to lipopolysaccharide and gamma interferon, whereas induction of two other NF-κB-dependent genes, the major histocompatibility complex class I and Mn superoxide genes, are essentially unaffected (2). These data suggest to us that a phosphorylation code controls NF-κB's ability to activate subnetworks of target genes, where distinct groups are induced by particular forms of phospho-RelA and the protein complexes that they form. Elucidation of the complete spectrum of genes under phospho-Ser276 RelA-dependent control will require further investigation.
CDK-9, along with Ccn T1, are core constituents of P-TEFb, a complex that functions as a transcriptional elongation factor mediating human immunodeficiency virus TAT-dependent transactivation (5) and the activation of heat shock genes in Drosophila melanogaster (37). P-TEFb mediates transcriptional elongation by its ability to phosphorylate several targets in the paused Pol II-dependent promoter, including serine 2 in the heptad repeat in the Pol II CTD, as well as inhibitory factors such as negative elongation factor (NELF) and DRB sensitivity-inducing factor (DRIF) (41). Our studies extend the understanding of inducible phospho-RelA protein interactions and suggest for the first time that phospho-Ser276 RelA is required for complex formation with the P-TEFb kinase. Our findings using a highly potent and specific CDK inhibitor and siRNA-mediated knockdown both suggest that IL-8 and Gro-β are activated by a mechanism involving P-TEFb recruitment. That CDK kinase activity mediates Pol II CTD phosphorylation is indicated by the reduction in CTD phosphorylation in response to FP (Fig. 6B). Further studies are needed to more precisely identify the spectrum of proteins modified by CDK-9 on NF-κB-dependent promoters, particularly NELF and DRIF.
Our fluorescence colocalization studies suggest that a significant fraction of RelA colocalizes with CDK-9 upon nuclear entry. Although spatial colocalization is subject to random error, especially in this case, where the concentration of one interacting protein is increasing in the cellular compartment of interest, we have analyzed these data using a formal statistical randomization algorithm which distinguishes random color overlap due to compartmentalization from that arising due to specific colocalization (13). Using this technique, we find that a significant fraction of RelA associated with CDK-9 in the nucleus. Because much of the nuclear fraction of RelA is not associated with chromatin, we believe that RelA-CDK-9 interaction occurs prior to promoter binding. To this end, we can coimmunoprecipitate NF-κB/RelA-CDK-9 from the nuclei of TNF-stimulated cells.
Recent work using fluorescence microscopy (photobleaching and fluorescence lifetime measurements) has shown that NF-κB transiently binds its chromatin targets with an exchange rate of seconds (8), resulting in the appreciation that chromatin-bound NF-κB is in dynamic exchange with its non-DNA-associated nucleoplasmic pool. This finding suggests that concentration changes in phospho-NF-κB isoforms entering the nuclear compartment can rapidly exchange with promoter-associated NF-κB. This phenomenon has been experimentally verified by us using Ang II, a ligand that activates phospho-Ser536 RelA formation without affecting total RelA nuclear abundance. In this case, an increase in the fraction of nuclear phospho-Ser536 RelA can rapidly exchange with promoter-bound hypophosphorylated RelA, producing a transition in gene expression from an inactive state to an activated one (14). Our ChIP analysis of phospho-Ser276 RelA binding, however, indicates that this isoform does not rapidly exchange with the hypophosphorylated RelA bound to the Naf1 gene. We suspect that phospho-Ser276 RelA complexed with p300/CBP and P-TEFb is a macromolecular complex whose promoter accessibility is dependent on the architecture and topology of the target promoters.
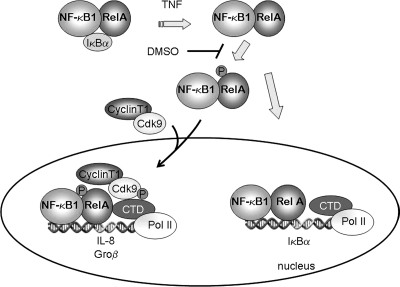
The findings of this study indicate that NF-κB controls downstream genes through at least two distinct mechanisms, schematically illustrated in Fig. 9. The first mechanism, exemplified by IL-8 and Gro-β, controls genes that are not significantly engaged with RNA Pol II in the absence of stimulation and require phospho-Ser276 RelA binding and P-TEFb recruitment for inducible Pol II recruitment and activation of transcriptional elongation. The second mechanism, exemplified by IκBα, binds with RNA Pol II in the absence of stimulation. Although IκBα inducibly binds phospho-Ser276 RelA and P-TEFb, this complex is not required for promoter activation.
FIG. 9.
Model of NF-κB-dependent initiation of transcription. This schematic represents the steps involved in the two mechanistically distinct pathways involved in the transcriptional initiation of NF-κB-dependent gene expression mediated by inducible Pol II loading. In the cytoplasm, TNF stimulation releases RelA from sequestered cytoplasmic sites by inducing IκBα proteolysis. A delayed ROS signal mediates phospho-Ser276 RelA formation on a subset of proteins. Phospho-Ser276 RelA binds the CDK-9-Ccn T1 P-TEFb complex, recruiting it to IL-8 and Gro-β gene promoters. Here, P-TEFb is involved in phosphorylation of Ser2 in the Pol II CTD, NELF, and DSIF, resulting in productive transcriptional elongation. Transcriptional activation of the IL-8 and Gro-β genes is sensitive to antioxidant DMSO by its ability to disrupt RelA phosphorylation at Ser276 and thereby disrupt association with P-TEFb complexes. Transcriptional activation of the IL-8 and Gro-β genes is disrupted by FP and CDK-9 siRNA by blocking the serine phosphorylation of RNA Pol II CTD. Although phospho-Ser276 RelA binds IκBα, it is not functionally required for gene expression.
Epithelial cells play important roles in the initiation and maintenance of innate immunity. For example, in the airway, epithelial cells respond to a variety of viral, environmental, and hormonal (cytokine) stimuli to activate NF-κB-dependent cytokine cascades important in the genesis and maintenance of airway inflammation (42, 51, 55, 56). Our delineation of distinct mechanisms by which NF-κB activates target genes has important implications for targeted anti-inflammatory therapy at epithelial surfaces, where distinct biological responses could be modified. For example, inhibition of the ROS-dependent phospho-Ser276 RelA-P-TEFb pathway may be useful to control the cytokine cascade without influencing other NF-κB-mediated antiapoptotic responses.
Supplementary Material
[Supplemental material]
Acknowledgments
We thank the NCI Developmental Therapeutics Program and Serono-Aventis for the gift of FP.
This project was supported by NIAID grants R01 AI40218 and P01 AI062885 (to A.R.B.) and a J. W. McLaughlin predoctoral fellowship (to D.E.N.). Core laboratory support was from NIEHS grant P30 ES06676 (to J. Halpert, UTMB).
Footnotes
▿
Published ahead of print on 24 March 2008.
REFERENCES
- 1.Ainbinder, E., M. Revach, O. Wolstein, S. Moshonov, N. Diamant, and R. Dikstein. 2002. Mechanism of rapid transcriptional induction of tumor necrosis factor alpha-responsive genes by NF-κB. Mol. Cell. Biol. 226354-6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anrather, J., G. Racchumi, and C. Iadecola. 2005. _cis_-acting element-specific transcriptional activity of differentially phosphorylated nuclear factor-{kappa}B. J. Biol. Chem. 280244-252. [DOI] [PubMed] [Google Scholar]
- 3.Babbar, N., and R. Casero, Jr. 2006. Tumor necrosis factor-A increases reactive oxygen species by inducing spermine oxidase in human lung epithelial cells: a potential mechanism for inflammation-induced carcinogenesis. Cancer Res. 6611125-11130. [DOI] [PubMed] [Google Scholar]
- 4.Baldwin, A. S. J. 2001. The transcription factor NF-kB and human disease. J. Clin. Investig. 1073-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barboric, M., R. M. Nissen, S. Kanazawa, N. Jabrane-Ferrat, and B. M. Peterlin. 2001. NF-kB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell 3327-337. [DOI] [PubMed] [Google Scholar]
- 6.Barnes, P. J., and M. Karin. 1997. Nuclear factor-kappaB: a pivotal transcription factor in chronic inflammatory diseases. N. Engl. J. Med. 3361066-1071. [DOI] [PubMed] [Google Scholar]
- 7.Beg, A. A., W. Sha, R. T. Bronson, S. Ghosh, and D. Baltimore. 1995. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-kB. Nature 376167-170. [DOI] [PubMed] [Google Scholar]
- 8.Bosisio, D., I. Marazzi, A. Agresti, N. Shimizu, M. E. Bianchi, and G. Natoli. 2006. A hyper-dynamic equilibrium between promoter-bound and nucleoplasmic dimers controls NF-kappaB-dependent gene activity. EMBO J. 25798-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brasier, A. R., M. Jamaluddin, A. Casola, W. Duan, Q. Shen, and R. Garofalo. 1998. A promoter recruitment mechanism for TNFα-induced IL-8 transcription in type II pulmonary epithelial cells: dependence on nuclear abundance of Rel A, NF-kB1 and c-Rel transcription factors. J. Biol. Chem. 2733551-3561. [DOI] [PubMed] [Google Scholar]
- 10.Brasier, A. R., D. Ron, J. E. Tate, and J. F. Habener. 1990. A family of constitutive C/EBP-like DNA binding proteins attenuate the IL-1 alpha induced, NF kappa B mediated trans-activation of the angiotensinogen gene acute-phase response element. EMBO J. 93933-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown, K., S. Gerstberger, L. Carlson, G. Franzoso, and U. Siebenlist. 1995. Control of IkB-alpha proteolysis by site-specific, signal-induced phosphorylation. Science 2671485-1488. [DOI] [PubMed] [Google Scholar]
- 12.Chen, L. F., S. A. Williams, Y. Mu, H. Nakano, J. M. Duerr, L. Buckbinder, and W. C. Greene. 2005. NF-κB RelA phosphorylation regulates RelA acetylation. Mol. Cell. Biol. 257966-7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costes, S. V., D. Daelemans, E. H. Cho, Z. Dobbin, G. Pavlakis, and S. Lockett. 2004. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys. J. 863993-4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui, R., B. Tieu, A. I. Recinos, R. G. Tilton, and A. R. Brasier. 2006. Rho A mediates angiotensin II-induced phospho-Ser536 NF-kB/RelA subunit exchange on the IL-6 promoter in VSMC. Circ. Res. 99723-730. [DOI] [PubMed] [Google Scholar]
- 15.Devin, A., Y. Lin, S. Yamaoka, Z. Li, M. Karin, and Z.-G. Liu. 2001. The α and β subunits of IκB kinase (IKK) mediate TRAF2-dependent IKK recruitment to tumor necrosis factor (TNF) receptor 1 in response to TNF. Mol. Cell. Biol. 213986-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garofalo, R., M. Sabry, M. Jamaluddin, R. K. Yu, A. Casola, P. L. Ogra, and A. R. Brasier. 1996. Transcriptional activation of the interleukin-8 gene by respiratory syncytial virus infection in alveolar epithelial cells: nuclear translocation of the Rel A transcription factor as a mechanism producing airway mucosal inflammation. J. Virol. 708773-8781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghosh, S., M. J. May, and E. B. Kopp. 1998. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 60225-260. [DOI] [PubMed] [Google Scholar]
- 18.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 895547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han, Y., and A. R. Brasier. 1997. Mechanism for biphasic Rel A:NF-kB1 nuclear translocation in tumor necrosis factor α-stimulated hepatocytes. J. Biol. Chem. 2729823-9830. [DOI] [PubMed] [Google Scholar]
- 20.Han, Y., S. Weinman, I. Boldogh, R. K. Walker, and A. R. Brasier. 1999. Tumor necrosis factor-alpha-inducible IkappaBalpha proteolysis mediated by cytosolic m-calpain. A mechanism parallel to the ubiquitin-proteasome pathway for nuclear factor-kappab activation. J. Biol. Chem. 274787-794. [DOI] [PubMed] [Google Scholar]
- 21.Hansen, J. M., H. Zhang, and D. P. Jones. 2006. Mitochondrial thioredoxin-2 has a key role in determining tumor necrosis factor-a-induced reactive oxygen species generation, NF-kB activation, and apoptosis. Toxicol. Sci. 91643-650. [DOI] [PubMed] [Google Scholar]
- 22.Hayden, M. S., and S. Ghosh. 2004. Signaling to NF-kappa B. Genes Dev. 182195-2224. [DOI] [PubMed] [Google Scholar]
- 23.Hsu, H., J. Huang, H. B. Shu, V. Baichwal, and D. V. Goeddel. 1996. TNF-dependent recruitment of the protein kinase RIP to the TNF receptor-1 signaling complex. Immunity 4387-396. [DOI] [PubMed] [Google Scholar]
- 24.Hsu, H., H.-B. Shu, M.-G. Pan, and D. V. Goeddel. 1996. TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell 84299-308. [DOI] [PubMed] [Google Scholar]
- 25.Hsu, H., J. Xiong, and D. V. Goeddel. 1995. The TNF receptor1-associated protein TRADD signals cell death and NF-kB activation. Cell 81495-504. [DOI] [PubMed] [Google Scholar]
- 26.Jamaluddin, M., S. Wang, I. Boldogh, B. Tian, and A. R. Brasier. 2007. TNF-α-induced NF-κB/Rel A Ser 276 phosphorylation and enhanceosome formation on the IL-8 promoter is mediated by a reactive oxygen species (ROS)-dependent pathway. Cell. Signal. 91419-1433. [DOI] [PubMed] [Google Scholar]
- 27.Jamaluddin, M., T. Meng, J. Sun, I. Boldogh, Y. Han, and A. R. Brasier. 2000. Angiotensin II induces nuclear factor (NF)-kappaB1 isoforms to bind the angiotensinogen gene acute-phase response element: a stimulus-specific pathway for NF-kappaB activation. Mol. Endocrinol. 1499-113. [DOI] [PubMed] [Google Scholar]
- 28.Jiang, X., N. Takahashi, N. Matsui, T. Tetsuka, and T. Okamoto. 2003. The NF-kappa B activation in lymphotoxin beta receptor signaling depends on the phosphorylation of p65 at serine 536. J. Biol. Chem. 278919-926. [DOI] [PubMed] [Google Scholar]
- 29.Karin, M., and Y. Ben Neriah. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18621-663. [DOI] [PubMed] [Google Scholar]
- 30.Kunsch, C., S. M. Ruben, and C. A. Rosen. 1992. Selection of optimal kB/Rel DNA-binding motifs: interaction of both subunits of NF-κB with DNA is required for transcriptional activation. Mol. Cell. Biol. 124412-4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levine, R. L., D. Garland, C. N. Oliver, A. Amici, I. Climent, G. Lenz, W. Ahn, S. Shaltiel, and E. R. Stadman. 1990. Determination of carbonyl content in oxidatively modified proteins. Methods Enzymol. 186464-478. [DOI] [PubMed] [Google Scholar]
- 32.Lipniacki, T., P. Paszek, A. R. Brasier, B. Luxon, and M. Kimmel. 2004. Mathematical model of NF-κB regulatory module. J. Theor. Biol. 228195-215. [DOI] [PubMed] [Google Scholar]
- 33.Lu, Y., L. Jamieson, A. R. Brasier, and A. P. Fields. 2001. NF-kB/RelA transactivation is required for atypical protein kinase Ciota-mediated cell survival. Oncogene 204777-4792. [DOI] [PubMed] [Google Scholar]
- 34.Maniatis, T., J. V. Falvo, T. H. Kim, T. K. Kim, C. H. Lin, B. S. Parekh, and M. G. Wathelet. 1998. Structure and function of the interferon-beta enhanceosome. Cold Spring Harb. Symp. Quant. Biol. 63609-620. [DOI] [PubMed] [Google Scholar]
- 35.Naumann, M., and C. Scheidereit. 1994. Activation of NF-kB in vivo is regulated by multiple phosphorylations. EMBO J. 134597-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nelson, D. E., A. E. C. Ihekwaba, M. Elliott, J. R. Johnson, C. A. Gibney, B. E. Foreman, G. Nelson, V. See, C. A. Horton, D. G. Spiller, S. W. Edwards, H. P. McDowell, J. F. Unitt, E. Sullivan, R. Grimley, N. Benson, D. Broomhead, D. B. Kell, and M. R. H. White. 2004. Oscillations in NF-{kappa}B signaling control the dynamics of gene expression. Science 306704-708. [DOI] [PubMed] [Google Scholar]
- 37.Ni, Z., B. E. Schwartz, J. Werner, J. R. Suarez, and J. T. Lis. 2004. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol. Cell 1355-65. [DOI] [PubMed] [Google Scholar]
- 38.Nissen, R. M., and K. R. Yamamoto. 2000. The glucocorticoid receptor inhibits NFkappa B by interfering with serine-2 phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 142314-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nowak, D. E., B. Tian, and A. R. Brasier. 2005. Two-step cross-linking method for identification of NF-κB gene network by chromatin immunoprecipitation. BioTechniques 39715-725. [DOI] [PubMed] [Google Scholar]
- 40.O'Donnell, S. M., G. Holm, J. M. Pierce, B. Tian, R. S. Chari, D. W. Ballard, A. R. Brasier, and T. S. Dermody. 2006. Identification of an NF-κB-dependent gene network in cells infected by mammalian reovirus. J. Virol. 801077-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palangat, M., D. B. Renner, D. H. Price, and R. Landick. 2005. A negative elongation factor for human RNA polymerase II inhibits the anti-arrest transcript-cleavage factor TFIIS. Proc. Natl. Acad. Sci. USA 10215036-15041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Polito, A. J., and D. Proud. 1998. Epithelial cells as regulators of airway inflammation. J. Allergy Clin. Immunol. 102714-718. [DOI] [PubMed] [Google Scholar]
- 43.Poyet, J.-L., S. M. Srinivasula, J.-H. Lin, T. Fernandes-Alnemri, S. Yamaoka, P. N. Tsichlis, and E. S. Alnemri. 2000. Activation of the IkB kinases by RIP via IKKg/NEMO-mediated oligomerization. J. Biol. Chem. 27537966-37977. [DOI] [PubMed] [Google Scholar]
- 44.Price, D. H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 202629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saccani, S., S. Pantano, and G. Natoli. 2001. Two waves of nuclear factor kB recruitment to target promoters. J. Exp. Med. 1931351-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sakurai, H., H. Chiba, H. Miyoshi, T. Sugita, and W. Toriumi. 1999. IkappaB kinases phosphorylate NF-kappaB p65 subunit on serine 536 in the transactivation domain. J. Biol. Chem. 27430353-30366. [DOI] [PubMed] [Google Scholar]
- 47.Shaner, N. C., R. E. Campbell, P. A. Steinbach, B. N. Giepmans, A. E. Palmer, and R. Y. Tsien. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived by Discosoma sp. red fluorescent protein. Nat. Biotechnol. 221567-1572. [DOI] [PubMed] [Google Scholar]
- 48.Shapiro, G. I. 2004. Preclinical and clinical development of the cyclin-dependent kinase inhibitor flavopiridol. Clin. Cancer Res. 104270s-4275s. [DOI] [PubMed] [Google Scholar]
- 49.Shiama, N. 1997. The p300/CBP family: integrating signals with transcription factors and chromatin. Trends Cell Biol. 7230-236. [DOI] [PubMed] [Google Scholar]
- 50.Smith, C. A., T. Farrah, and R. G. Goodwin. 1994. The TNF receptor superfamily of cellular and viral proteins: activation, costimulation, and death. Cell 76959-962. [DOI] [PubMed] [Google Scholar]
- 51.Standiford, T. J., S. L. Kunkel, M. A. Basha, S. W. Chensue, J. P. I. Lynch, G. B. Toews, J. Westwick, and R. M. Strieter. 1990. Interleukin-8 gene expression by a pulmonary epithelial cell line: a model for cytokine networks in the lung. J. Clin. Investig. 861945-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun, S. C., P. A. Ganchi, D. W. Ballard, and W. C. Greene. 1993. NF-kappa B controls expression of inhibitor I kappa B alpha: evidence for an inducible autoregulatory pathway. Science 2591912-1915. [DOI] [PubMed] [Google Scholar]
- 53.Tian, B., and A. R. Brasier. 2005. The nuclear factor-κB (NF-κB) gene regulatory network. In M. F. Shannon and S. Rao (ed.), Microarrays and transcription factor networks. Landes Bioscience, Austin, TX.
- 54.Tian, B., D. Nowak, and A. R. Brasier. 2005. A TNF-induced gene expression program under oscillatory NF-κB control. BMC Genomics 6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tian, B., Y. Zhang, B. A. Luxon, R. P. Garofalo, A. Casola, M. Sinha, and A. R. Brasier. 2002. Identification of NF-κB-dependent gene networks in respiratory syncytial virus-infected cells. J. Virol. 766800-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tian, B., D. E. Nowak, M. Jamaluddin, S. Wang, and A. R. Brasier. 2005. Identification of direct genomic targets downstream of the NF-kappa B transcription factor mediating TNF signaling. J. Biol. Chem. 28017435-17448. [DOI] [PubMed] [Google Scholar]
- 57.Vermeulen, L., G. De Wilde, P. Van Damme, W. Vanden Berghe, and G. Haegeman. 2003. Transcriptional activation of the NF-kappaB p65 subunit by mitogen- and stress-activated protein kinase-1 (MSK1). EMBO J. 221313-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vlahopoulos, S., I. Boldogh, and A. R. Brasier. 1999. NF-κB dependent induction of interleukin-8 gene expression by tumor necrosis factor α: evidence for an antioxidant sensitive activating pathway distinct from nuclear translocation. Blood 941878-1889. [PubMed] [Google Scholar]
- 59.Zhong, H., H. SuYahng, H. Erdjument-Bromage, P. Tempst, and S. Ghosh. 1997. The transcriptional activity of NF-kB is regulated by the IkB-associated PKAc subunit through a cyclic AMP-independent mechanism. Cell 89413-424. [DOI] [PubMed] [Google Scholar]
- 60.Zhong, H., R. E. Voll, and S. Ghosh. 1998. Phosphorylation of NF-kB p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol. Cell 1661-671. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]