Protein Kinase D1 Stimulates MEF2 Activity in Skeletal Muscle and Enhances Muscle Performance (original) (raw)
Abstract
Skeletal muscle consists of type I and type II myofibers, which exhibit different metabolic and contractile properties. Type I fibers display an oxidative metabolism and are resistant to fatigue, whereas type II fibers are primarily glycolytic and suited for rapid bursts of activity. These properties can be modified by changes in workload, activity, and hormonal stimuli, facilitating muscle adaptation to physiological demand. The MEF2 transcription factor promotes the formation of slow-twitch (type I) muscle fibers in response to activity. MEF2 activity is repressed by class II histone deacetylases (HDACs) and is enhanced by calcium-regulated protein kinases that promote the export of class II HDACs from the nucleus to the cytoplasm. However, the identities of skeletal muscle class II HDAC kinases are not well defined. Here we demonstrate that protein kinase D1 (PKD1), a highly effective class II HDAC kinase, is predominantly expressed in type I myofibers and, when misexpressed in type II myofibers, promotes transformation to a type I, slow-twitch, fatigue-resistant phenotype. Conversely, genetic deletion of PKD1 in type I myofibers increases susceptibility to fatigue. PKD1 cooperates with calcineurin to facilitate slow-twitch-fiber transformation. These findings identify PKD1 as a key regulator of skeletal muscle function and phenotype.
Skeletal muscles consist of a heterogeneous mixture of myofibers that display different metabolic, contractile, and endurance properties. Based on the expression of myosin heavy chain (MyHC) isoforms, myofibers can be classified as type I or type II fibers; type II fibers are further categorized into type IIa, type IId/x, and type IIb (3). Type I fibers are oxidative, slow-twitch fibers with high endurance, whereas type IIb fibers are glycolytic, fast-twitch fibers with low endurance. Type IIa fibers can use both aerobic and anaerobic metabolism but display fast-twitch contracting properties. Type I myofibers play a major role in regulating whole-body energy metabolism and insulin sensitivity. Slow-twitch oxidative skeletal muscle has greater insulin binding capacity, insulin receptor kinase activity, and a higher content of glucose transporter 4 than fast-twitch glycolytic skeletal muscle (3). In fact, type 2 diabetes mellitus is associated with defects in insulin signaling in skeletal muscle (17), and insulin resistance correlates with a reduced percentage of slow oxidative type I fibers and reduced oxidative enzyme capacity (29).
In response to various stimuli, including work load, hormonal influences, and innervation, myofibers modify their phenotype to maintain a balance between physiological demand and functional capacity. This muscle plasticity is achieved through activation of intracellular signaling pathways that modulate myofiber gene expression (38). Several calcium-dependent signaling pathways have been implicated in the modulation of myofiber phenotypes. In particular, the calcium/calmodulin-dependent protein phosphatase calcineurin, which responds selectively to sustained calcium waves (11), promotes the formation of slow-twitch, type I myofibers (6, 27, 31, 32), and blockade of calcineurin signaling promotes the formation of fast myofibers (31). Forced expression of calcium- and calmodulin-dependent protein kinase IV (CaMKIV) in skeletal muscle also stimulates the formation of slow-twitch myofibers (44). However, CaMKIV is not expressed in skeletal muscle (1), suggesting that other endogenous kinases may normally play this role.
The myocyte enhancer factor-2 (MEF2) transcription factor acts as a transcriptional regulator of skeletal muscle remodeling and fiber type specification (33, 34, 46). We previously showed that skeletal muscle deletion of MEF2C or MEF2D resulted in reduction in levels of slow-twitch fibers within the soleus (34). In adult skeletal muscle, the activity of MEF2 is tightly regulated through association with class II histone deacetylases (HDACs), which act as signal-dependent repressors of gene expression (24, 25). In response to differentiation signals or motor innervation, class II HDACs are phosphorylated, a process which provides docking sites for the 14-3-3 chaperone protein and leads to nuclear export of HDACs accompanied by derepression of MEF2 and activation of MEF2 target genes (24, 25, 42). Because phosphorylation of class II HDACs governs the activity of MEF2 and the downstream gene programs it regulates, identifying the kinases responsible for phosphorylation and nuclear export of class II HDACs in skeletal muscle may provide opportunities for manipulating skeletal muscle function.
Recently, we showed that protein kinase D1 (PKD1) in cardiomyocytes acts as an HDAC export kinase that controls hypertrophic growth in response to G-protein receptor-coupled agonists (12, 41). When excessively activated in the heart, PKD1 drives adverse cardiac remodeling, heart failure, and death (14). Conversely, genetic deletion of PKD1 in the heart diminishes hypertrophy and pathological remodeling in response to pressure overload and chronic adrenergic signaling (12).
Three PKD isoforms, PKD1, -2, and -3, encoded by different genes (36), share homology with the CaMK and protein kinase C (PKC) families. However, unlike CaMK and PKC, which are directly regulated by calcium, PKD activity is independent of calcium regulation and is instead activated through phosphorylation by PKC (36, 40, 49). PKD1 has been implicated in the regulation of a variety of biological processes, including membrane trafficking, cell survival, proliferation, differentiation, and migration (36).
The potential involvement of PKD in the control of skeletal muscle development, growth, and remodeling has not been investigated. Here we show that PKD1 is preferentially expressed in type I myofibers. Forced expression of constitutively active PKD1 (caPKD1) in type II fibers of transgenic mice potently stimulates the transcriptional activity of MEF2, as revealed by expression of an MEF2-dependent transgene, and in turn promotes their conversion toward a slow myofiber phenotype. Consistent with this fiber type switch, skeletal muscles derived from caPKD1 transgenic mice are resistant to fatigue during repetitive contractions. In contrast, skeletal-muscle-specific deletion of PKD1, by use of a conditional PKD1 null allele, increases the susceptibility to fatigue of type I soleus muscle, but not that of type II extensor digitorum longus (EDL) muscle, where PKD1 expression is low. Calcineurin enhances the ability of PKD1 to activate slow-twitch- and oxidative-myofiber-specific gene expression. We conclude that PKD1 signaling plays an important role in the control of skeletal muscle fiber type via its stimulatory activity with respect to MEF2 and that PKD1 and calcineurin act in a cooperative manner to modulate skeletal muscle function and phenotype.
MATERIALS AND METHODS
Generation of MCK-caPKD1 transgenic mice.
An MCK-caPKD1 transgene was generated by placing a myc-tagged human constitutively active PKD1 (S738E/S742E) cDNA (39) downstream from the 4.8-kb muscle creatine kinase (MCK) promoter (16). The construct contained a downstream human growth hormone poly(A) signal. DNA isolation and oocyte injections were performed as described previously (26). Genomic DNA was isolated from mouse tail snips and analyzed by PCR using a PKD1-specific primer (5′-GTGGTGGGTACCCCCGCTTAC-3′) and a primer specific for human growth hormone poly(A) (5′-CACTCCGCTTGGTTCCCGAATAGAC-3′). Nontransgenic littermates were used for comparison with MCK-caPKD1 mice in all experiments. Mice 8 to 11 weeks of age were used for all experiments. All experimental procedures involving animals in this study were reviewed and approved by the Institutional Animal Care and Research Advisory Committee at the University of Texas Southwestern Medical Center.
Skeletal-muscle-specific deletion of PKD1.
The generation of a conditional PKD1 (Prkcm) allele has been described previously (12). We generated a skeletal-myocyte-specific deletion of PKD1 by use of myogenin-Cre transgenic mice (18) that express Cre recombinase specifically in myocytes of skeletal muscle and heterozygous PKD1(loxP/KO) mice that were obtained using CAG-Cre transgenic mice which express Cre recombinase in the embryo at the zygote stage (37). The breeding scheme involved crossing PKD1(loxP/loxP), myogenin-Cre, and PKD1(loxP/KO) mice to obtain PKD1(loxP/KO), myogenin-Cre mice (referred to as PKD1 skKO) and PKD1(loxP/loxP) mice (referred to as wild type [WT]) as controls. For analysis, 8- to 10-week-old male mice were used. Genotyping was performed by PCR using specific primers as described previously (12).
RNA isolation and RT-PCR.
Total RNA was isolated from skeletal muscles by use of Trizol reagent (Invitrogen) following the manufacturer's instructions. Two micrograms of RNA was converted to cDNA by use of random primers and Superscript III reverse transcriptase (RT) (Invitrogen). Gene expression was analyzed using either semiquantitative PCR or real-time PCR. Real-time PCR was performed using an ABI PRISM 7000 sequence detection system with TaqMan primers (Applied Biosystems) or with SYBR green Master Mix reagent (Applied Biosystems) and the primer set listed below. Real-time PCR values were normalized with L7 and glyceraldehyde-3-phosphate dehydrogenase expression. Primer sequences were as follows: for Myh2 (type IIa) forward, 5′-CCAGCTGCACCTTCTCGTTTGCCAG-3′; for Myh2 reverse, 5′-CATGGGGAAGATCTGGTCTTCTTTCACGGTCAC-3′; for Myh1 (type IIx) forward, 5′-GCGCAACGTGGAAGCTATCAAGGGTCTG-3′; for Myh1 reverse, 5′-GATCTTCACATTTTGCTCATCTTTTGGTCACT-3′; for Myh4 (type IIb) forward, 5′-CCTGGAACAGACAGAGAGGAGCAGGAGAG-3′; for Myh4 reverse, 5′-GTGAGTTCCTTCACTCTGCGCTCGTGC-3′; for Troponin I (slow) forward, 5′-GTGCCTGGAACATCCCTAAT-3′; for Troponin I (slow) reverse, 5′-TGAGAGGCTGTTCTCTCTGC-3′; for Myoglobin forward, 5′-CATGGTTGCACCGTGCTCACAG-3′; for Myoglobin reverse, 5′-GAGCCCATGGCTCAGCCCTG-3′; for Cpt1 forward, 5′-ATCATGTATCGCCGCAAACT-3′; for Cpt1 reverse, 5′-ATCTGGTAGGAGCACATGGGC-3′; for COXIV forward, 5′-GTTCAGTTGTACCGCATCCA-3′; for COXIV reverse, 5′-TTGTCATAGTCCCACTTGGC-3′; for UCP3 forward, 5′-TTTCTGCGTCTGGGAGCTT-3′; for UCP3 reverse, 5′-GGCCCTCTTCAGTTGCTCAT-3′; for L7 forward, 5′-GGAGGAAGCTCATCTATGAGAAGGCA-3′; and for L7 reverse, 5′-AAGATCTGTCGAAGACGAAGGAGCT-3′.
β-Galactosidase staining of skeletal muscle.
Dissected muscles from MCK-caPKD1 transgenic mice and WT littermates were fixed in 2% formaldehyde-0.2% glutaraldehyde in phosphate-buffered saline for 45 to 60 min on ice. After fixation, muscles were washed twice with phosphate-buffered saline for 10 min on ice and stained in X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) solution containing 5 mM ferrocyanide, 5 mM ferricyanide, 2 mM MgCl2, 1 mg/ml X-gal, 0.01% sodium deoxycholate, and 0.02% NP-40 for 1 h at room temperature.
Fiber type analysis by metachromatic ATPase staining.
Individual muscles from wild-type and PKD1 transgenic mice were dissected and embedded in medium containing gum tagacanth (Sigma, St. Louis, MO) and tissue-freezing medium (Triangle Biomedical Sciences, Durham, NC). Muscles were quickly frozen in liquid nitrogen-cooled isopentane and cut at 200-μm intervals by use of a cryotome and put on glass slides. For fiber type analysis, metachromatic ATPase staining was performed as previously described (30).
Electrophoretic analysis of MyHC isoforms.
Dissected muscles were homogenized in extraction buffer (300 mM KCl, 100 mM KH2PO4, 50 mM K2HPO4, 10 mM EDTA, 1 tablet of protease inhibitor cocktail [Roche], pH 6.5). After 30 min of incubation on ice, homogenized tissues were subjected to centrifugation at 18,000 × g for 5 min at 4°C. Supernatant fractions were diluted in 2× Laemmli buffer, and 0.5 μg of protein was separated by use of 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis containing 30% glycerol for 40 h at 60 V and 4°C. Gels were stained using a silver staining kit (Bio-Rad) following the manufacturer's instructions. The relative expression levels of MyHC isoforms were calculated by densitometry using Image J (version 1.38X; National Institutes of Health).
Immunoblotting of muscle extracts.
Tissues were homogenized in extraction buffer and subjected to centrifugation at 12,000 × g for 10 min at 4°C. The soluble fractions were collected, and 20 μg of proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed by immunoblotting with antibodies against c-Myc (Santa Cruz Biotechnology Inc.) (1:1,000), PKD1 (Santa Cruz Biotechnology Inc.) (1:1,000), myoglobin (DACO) (1:3,000), actin (Sigma-Aldrich) (1:2,000), and β-tubulin (Sigma-Aldrich) (1:2,000). To detect HDAC phosphorylation, immunoprecipitation was performed using ExactaCruz reagent (Santa Cruz Biotechnology Inc.) with HDAC4 and HDAC5 antibodies (24). The relative levels of phospho-HDAC and total HDAC were calculated by densitometry.
Muscle performance assay.
Mice were euthanized, and EDL and soleus muscles were excised and mounted on a Grass FT03 force transducer and bathed in an oxygenized (95% oxygen and 5% CO2) physiological salt solution (120.5 mM NaCl, 4.8 mM KCl, 1.2 mM Na2HPO4, 20.4 mM NaHCO3, 1.5 mM CaCl2, 1.2 mM MgSO4) at 30°C. Muscles were adjusted to the optimal length showing maximal isometric twitch tension and rested for 30 min. To measure skeletal muscle performance, muscles were continuously stimulated at 100 Hz until the force output reached 10% of the initial force. The time taken to reach 30% of the initial force output is the fatigue index (13).
Transient transfection assays.
C2C12 myoblast cells were cotransfected with a luciferase reporter plasmid controlled by the 2-kb myoglobin promoter (6), an expression vector encoding constitutively active calcineurin (6), an expression vector encoding the inactive form of PKD1 (K614W) (39), an expression vector encoding the constitutively active form of PKD1 (S738E/S742E), an expression vector encoding the constitutively active form of PKCθ (R145I/R146W) (20), and an expression vector encoding the inactive form of PKCθ (K409W) (20) as indicated. At 24 h following transfection, cells were changed to differentiation medium containing 2% horse serum. Cells were harvested 48 h following transfection and lysed in passive lysis buffer (Promega). Luciferase activity was measured using the luciferase assay system (Promega) following the manufacturer's instructions. The values were normalized to levels of β-galactosidase expression by use of a FluoReporter lacZ quantitiation kit (Molecular Probes).
RESULTS
PKD1 is enriched in slow-twitch type I muscle fibers.
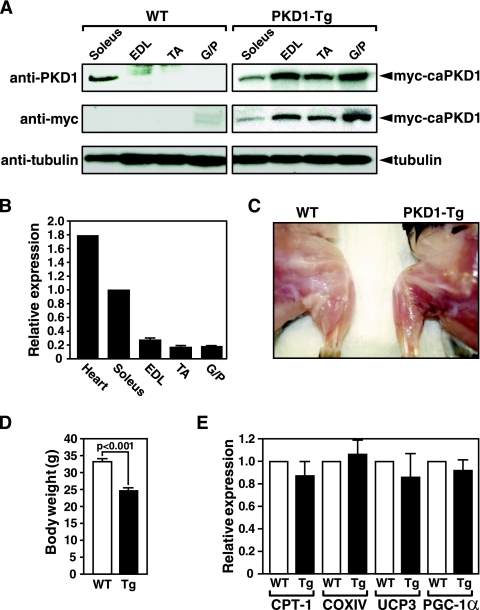
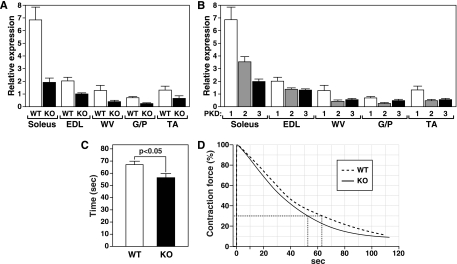
Soleus muscle is comprised exclusively of slow-twitch type I and fast-twitch, oxidative type IIa myofibers, whereas EDL muscle is predominantly comprised of fast-twitch, glycolytic type IIb myofibers. Tibialis anterior (TA) muscles and gastrocnemius and plantaris (G/P) muscles contain heterogenous myofiber types but are comprised primarily of type II myofibers. Prior to exploring the potential involvement of PKD1 in the modulation of muscle fiber type, we used Western blot analysis (Fig. 1A) and real-time PCR (Fig. 1B) to examine expression of PKD1 in each of the muscle categories named above. PKD1 protein and mRNA in soleus muscle were highly enriched. Despite detectable PKD1 mRNA expression, only a trace of PKD1 protein expression was detectable in EDL muscle, and no PKD1 protein was detected in TA and G/P muscle, suggesting a posttranscriptional mechanism for limiting PKD1 protein expression. Overall, the level of PKD1 expression correlated closely with the oxidative, slow-twitch-fiber phenotype.
FIG. 1.
Expression of PKD1 in skeletal muscle. (A) Immunoblots of skeletal muscle lysates isolated from MCK-myc-caPKD1 transgenic mice (PKD1-Tg) and WT mice prepared using anti-PKD1, anti-Myc, and antitubulin antibodies. Tubulin was used as a loading control. (B) Real-time PCR showing expression of endogenous PKD1 in striated muscles of 8-week-old male mice. Muscles from three mice were pooled for each sample. (C) Gross morphology of hindlimb from 2-month-old WT and PKD1-Tg mice showing a lean phenotype of the transgenic mouse. (D) Measurement of body weight of WT and PKD1-Tg mice shows a 25% weight reduction in Tg mice (n = 4 for each group). (E) Real-time PCR showing no change in the expression of carnitine palmitoyl transferase (CPT-1), subunit IV of cytochrome c oxidase (COX IV), uncoupling protein 3 (UCP3), and PGC-1α, proteins involved in fatty acid synthesis and oxidative metabolism (n = 3 for each group). Error bars indicate standard errors of the means.
Forced expression of constitutively active PKD1 in skeletal muscle.
To determine whether PKD1 signaling was sufficient to modulate skeletal muscle fiber types, we generated transgenic mice overexpressing caPKD1 under the control of the MCK promoter, which is preferentially active in type IIb and type IIa fast-twitch myofibers (6), which express endogenous PKD1 at diminished levels relative to soleus results. Examination of transgene expression by immunoblotting with anti-Myc antibody, which recognizes the epitope-tagged caPKD1 protein, showed, as expected, the highest expression of caPKD1 in EDL, TA, and G/P muscles, which are primarily composed of type II fibers, and less expression of the transgene in the soleus muscle (Fig. 1A).
MCK-caPKD1 mice were readily distinguishable from WT littermates by their unusually lean appearance and weighed approximately 25% less than normal (Fig. 1C and D). Given the importance of skeletal muscle in the regulation of whole-body glucose and fatty acid metabolism, we examined expression of enzymes involved in fatty acid metabolism to determine whether the lean phenotype of the MCK-caPKD1 transgenic mice might reflect enhanced metabolic activity. RNA analyses showed no increase in mRNAs encoding CPT-1, COXIV, UCP3, or peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1α) (Fig. 1E). Furthermore, studies using metabolic cages to measure food and liquid intake, ambulation, and oxygen and carbon dioxide consumption showed no alteration in the respiratory exchange ratios, indicating that there was no overall change in the metabolic properties of MCK-caPKD1 mice (data not shown). These findings suggest that the lean phenotype of MCK-caPKD1 transgenic mice does not result from altered metabolism.
MCK-caPKD1 transgenic mice display fiber type conversion.
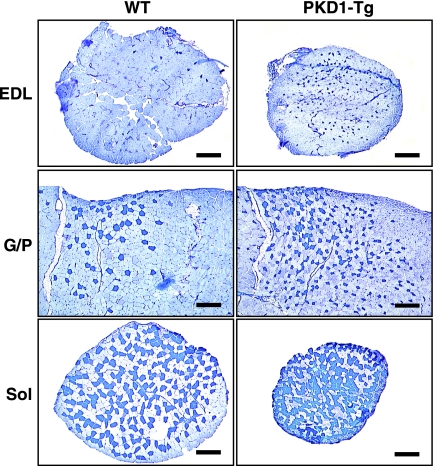
Type I fibers can be distinguished from type II fibers by their red color, which primarily reflects the high content of myoglobin. Muscles of MCK-caPKD1 transgenic mice appeared redder than those of wild-type mice, suggesting an increase in type I fiber numbers (Fig. 1C). We analyzed fiber type composition of MCK-caPKD1 transgenic mice by use of a metachromatic ATPase staining method, which allows identification of individual fiber types. When the metachromatic ATPase staining method is used, type I fibers stain dark blue and type II fibers stain different shades of lighter blue. Type I fibers were concentrated in specific regions of the G/P muscles of wild-type mice. However, G/P muscles from MCK-caPKD1 transgenic mice displayed an increase in type I fibers throughout the muscle (Fig. 2). In addition, both the EDL and soleus muscles displayed an increased number of type I and type IIa fibers in MCK-caPKD1 transgenic mice (Fig. 2).
FIG. 2.
Skeletal muscles of MCK-caPKD1 transgenic mice exhibit an increase in type I fiber numbers. EDL, G/P, and soleus (Sol) muscles were isolated from WT and MCK-myc-caPKD1 transgenic (PKD1-Tg) mice. Metachromatic ATPase fiber type analysis of skeletal muscle cross sections shows an increase in type I fibers (dark blue) compared to type II fibers (light blue) in PKD1-Tg mice. Scale bar, 300 μm.
In wild-type mice, the soleus muscle is comprised of approximately 45% type I fibers and the remaining fibers are type IIa (Fig. 2). In contrast, up to 60% of the fibers from the soleus muscles of MCK-caPKD1 transgenic mice were type I fibers. Notably, the individual sizes of fibers, as well as the overall size of the muscle, were reduced in MCK-caPKD1 transgenic mice regardless of fiber type. Cross-sectional measurements of the soleus showed that the total number of fibers was decreased by only approximately 10% in MCK-caPKD1 transgenic mice. Therefore, a decrease in the overall muscle size primarily reflects a reduction in the size of individual myofibers. These findings show that overexpression of caPKD1 in myofibers promotes slow-twitch-fiber formation in adult skeletal muscle and suggest that the lean phenotype might be explained, at least in part, by a reduction in myofiber size.
MCK-caPKD1 mice show an increase in levels of specific contractile proteins.
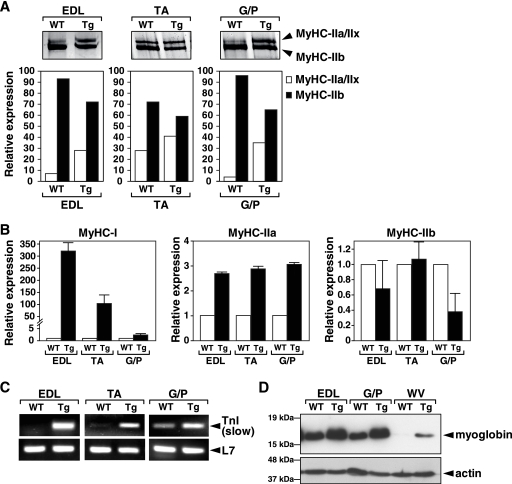
To further examine the fiber type switch in the skeletal muscle of MCK-caPKD1 transgenic mice, we examined the composition of MyHC isoforms in EDL, TA, and G/P muscles by use of silver staining-high-resolution glycerol gels. Two bands were present in muscle extracts of EDL, TA, and G/P muscles isolated from wild-type mice that represented two major MyHC proteins: the faster migrating MyHC IIb protein and the slower migrating MyHC IIa/IIx protein. In WT mice, approximately 90% of the MyHC protein in the G/P muscles consists of the MyHC IIb isoform, whereas in the MCK-caPKD1 transgenic mice, the G/P muscles consist of 65% MyHC IIb and 35% MyHC IIa/IIx protein (Fig. 3A).
FIG. 3.
Type I and type IIa fiber numbers are increased in MCK-myc-caPKD1 transgenic (Tg) mice. (A) Silver-stained, high-resolution glycerol gel resolving MyHC isoforms MyHC-IIa, MyHC-IIx, and MyHC-IIb in protein lysates from EDL, TA, and G/P muscles from MCK-myc-caPKD1 Tg mice and WT mice. The graph shows relative expression levels of MyHC isoforms in Tg and WT muscles. (B) Real-time PCR showing expression profiles of MyHC isoforms in EDL, TA, and G/P muscles isolated from PKD1-Tg and WT mice (n = 3). (C) Semiquantitative RT-PCR showed an increase in expression of troponin I (TnI) slow, a type I fiber-specific isoform of TnI, in EDL, TA, and G/P skeletal muscle of PKD1-Tg compared to WT mice. (D) Immunoblot results from EDL, G/P, and WV lysates isolated from WT and PKD1-Tg mice analyzed by use of antibodies to myoglobin and actin revealed increased expression of myoglobin in PKD1-Tg mice compared to WT mice. Actin was used as a loading control. Error bars indicate standard errors of the means.
The MyHC IIa and IIx proteins were not separable on glycerol gels, and so we examined their relative levels of abundance by real-time PCR, which revealed increased expression of MyHC I and IIa RNA and unaltered or decreased expression of MyHC IIb in the EDL, TA, and G/P muscle of MCK-caPKD1 transgenic mice relative to wild-type littermate controls (Fig. 3B). The expression of MyHC IIx was not significantly different from normal in EDL, TA, and G/P muscle isolated from MCK-caPKD1 transgenic mice (data not shown). The expression of troponin I (slow twitch), another type-I-fiber-specific contractile protein, was up-regulated in MCK-caPKD1 transgenic muscles (Fig. 3C). Myoglobin, a protein highly expressed in the soleus muscle and absent from the white vastus (WV) of wild-type mice, was expressed in the WV, as well as in the EDL and G/P muscles, of MCK-caPKD1 transgenic mice, as revealed by immunoblot and real-time PCR analysis (Fig. 3D and data not shown). These findings show that overexpression of caPKD1 in skeletal muscle induces a switch from fast- to slow-twitch myofibers.
Skeletal muscles of MCK-caPKD1 transgenic mice display fatigue resistance.
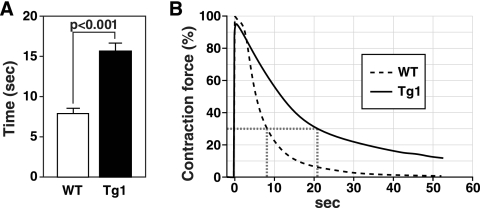
Fatigue resistance during repetitive contraction is a hallmark of type I fibers. To assess whether fiber type conversion was accompanied by changes in the physiological properties of skeletal muscle in MCK-caPKD1 transgenic mice, we measured susceptibility to fatigue during repetitive contractions. EDL and soleus muscles were isolated from wild-type and MCK-caPKD1 transgenic mice and subjected to continuous electrical stimulation, which mimics muscle contraction during exercise. We measured the time required to lose 70% of the initial contractile force as an index of muscle fatigue. EDL and soleus muscles derived from MCK-caPKD1 transgenic mice fatigued slower than those from wild-type mice (Fig. 4 and data not shown).
FIG. 4.
MCK-myc-caPKD1 skeletal muscle is more resistant to fatigue during repetitive contraction. EDL muscle isolated from WT mice and PKD1 transgenic (Tg1) mice was subjected to continuous electrical stimulation. (A) Time in seconds required to reach 30% of initial contraction is defined as the fatigue index (n = five WT and n = six Tg mice) (P < 0.001 by unpaired two-tailed Student's t test). (B) Overlay of representative traces showing the percentages of contraction force over time in PKD1-Tg and WT EDL muscles. Error bars indicate standard errors of the means.
Deletion of PKD1 in skeletal muscle enhances susceptibility to fatigue.
To further investigate the potential involvement of PKD1 in muscle fatigue, we measured the susceptibility to fatigue during repetitive contractions in the soleus and EDL muscles of mice in which PKD1 was deleted from skeletal muscle. Skeletal-muscle-specific deletion of PKD1 (referred to as PKD1 skKO) was achieved by breeding mice harboring a conditional PKD1 null allele (12) with transgenic mice expressing Cre recombinase specifically in skeletal muscle (18). Mice with PKD1 skKO were grossly indistinguishable from their WT littermates. Quantification of PKD1 mRNA by real-time RT-PCR revealed a fivefold reduction in levels of PKD1 mRNA in PKD1 skKO skeletal muscle (Fig. 5A). The observed residual expression of PKD1 mRNA most likely reflects PKD1 expression in fibroblasts and endothelial, smooth muscle, and immune cells within the skeletal muscle. Prkcm2 (encoding PKD2) and Prkcn (encoding PKD3) are expressed in skeletal muscle (Fig. 5B) but were not up-regulated in PKD1 skKO mice (data not shown).
FIG. 5.
Deletion of PKD1 in soleus muscle leads to an increase in fatigue during repetitive contraction. (A) Expression of PKD1 transcripts detected by quantitative PCR. Total RNA isolated from soleus, EDL, G/P, and TA skeletal muscles of 8-week-old male mice was used for cDNA synthesis and subsequent quantitative RT-PCR of WT mice and PKD1 skKO (KO) mice (n = 5). (B) PKD1, -2, and -3 levels in the muscle groups of WT mice (n = 5) were quantified using quantitative RT-PCR. (C) Soleus muscles were isolated and subjected to continuous electrical stimulation. Time to reach 30% of initial contraction was measured as an index of fatigue. PKD1 skKO mice (n = 9) showed a significant increase in susceptibility to fatigue compared to WT littermate controls (n = 5) (P < 0.05 by two-way analysis of variance). (D) Overlay of representative traces showing the percentages of contraction force over time in PKD1 skKO and WT soleus muscles. Error bars indicate standard errors of the means.
As was consistent with the PKD1 overexpression experiment results, indicating a role for PKD1 in muscle endurance, the soleus muscles from PKD1 skKO mice fatigued faster than those from WT littermate controls (Fig. 5C), whereas no differences between PKD1 skKO and WT mice in fatigue response in EDL muscle were found (data not shown). To assess whether this phenotype was due to an effect of skeletal-muscle-specific deletion of PKD1 on fiber type composition, we used metachromatic ATPase staining, silver staining-high-resolution glycerol gels, and RT-PCR for MyHC I, IIa, IIb, and IIx. Surprisingly, no differences in fiber type composition of slow- or fast-twitch skeletal muscle were found between PKD1 skKO and WT littermates (data not shown), suggesting that fiber type switching does not account for the difference in susceptibility to fatigue seen in response to PKD1 deletion.
Activation of MEF2 in skeletal muscle of MCK-caPKD1 transgenic mice.
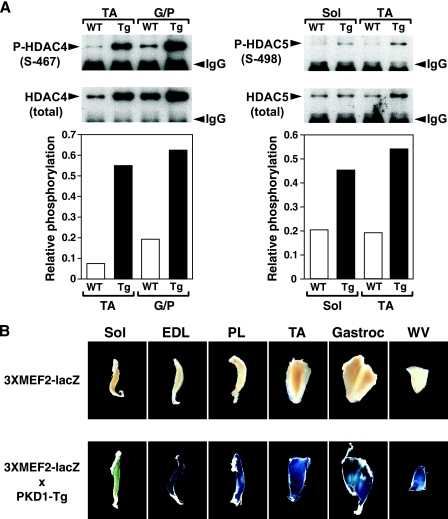
Previous studies have shown that PKD1 signaling stimulates MEF2 activity by promoting the phosphorylation and export of class II HDACs from the nucleus (14, 41). To determine whether overexpression of caPKD1 in skeletal muscle is sufficient to phosphorylate class II HDACs in vivo, we compared the phosphorylation statuses of HDAC4 and HDAC5 in skeletal muscle of wild-type and MCK-caPKD1 transgenic mice by immunoprecipitation and Western blot analysis with antibodies against phosphoserine-467 in HDAC4 and phosphoserine-498 in HDAC5, which mediate nuclear export (24). We observed increased phosphorylation of HDAC4 (S467) and HDAC5 (S498) in soleus, TA, and G/P muscles of the transgenic mice (Fig. 6A).
FIG. 6.
caPKD1 activates MEF2 in skeletal muscle through phosphorylation of class II HDACs. (A) Western blot analysis of immunoprecipitated endogenous HDAC4 or HDAC5 from TA, G/P, or soleus (Sol) muscles isolated from PKD1 transgenic (Tg) and WT mice. Blots were probed using an antibody that recognizes the phosphorylated form of HDAC4 (P-HDAC4 S-467) and HDAC5 (P-HDAC5 S-498). Blots were reprobed using commercial HDAC4 or HDAC5 antibodies. Graphs show the relative phosphorylation levels of HDAC4 and HDAC5 in WT and MCK-caPKD1 (Tg) muscles. Relative phosphorylation levels were calculated as the ratio of phospho-HDAC to total HDAC densitometric signals. IgG, immunoglobulin G. (B) β-Galactosidase staining of soleus (Sol), EDL, plantaris (PL), TA, gastrocnemius (Gastroc), and WV muscles isolated from 2-month-old MEF2 indicator mice (3XMEF2-lacZ) or PKD1-Tg mice crossed with MEF2 indicator mice (3XMEF2-lacZ × PKD1-Tg). Expression of the lacZ transgene depended on MEF2 activity.
To determine whether increased phosphorylation of class II HDACs in MCK-caPKD1 transgenic mice leads to MEF2 activation in vivo, we bred MCK-caPKD1 transgenic mice with MEF2 indicator transgenic mice harboring a lacZ transgene linked to three copies of the MEF2 consensus sequence from the desmin promoter (3XMEF2-lacZ) (28). In 3XMEF2-lacZ mice, expression of lacZ depends on the activity of MEF2 (46). We examined the expression of the lacZ transgene in wild-type and MCK-caPKD1 transgenic muscles by β-galactosidase staining (Fig. 6B). In WT mice, MEF2 activity is normally detected preferentially in the soleus (45). In order to compare the levels of β-galactosidase expression in muscle of 3XMEF2-lacZ transgenic mice and 3XMEF2-lacZ × PKD1-Tg mice, we allowed the β-galactosidase staining reaction to proceed for a relatively limited period of time such that β-galactosidase activity was not detectable in the soleus of 3XMEF2-lacZ mice. We previously reported that lacZ activity was detectable only within soleus muscle of a subset (10 to 15%) of animals and was uniformly undetectable in EDL, plantaris, and WV muscles (45).
As is consistent with previous studies (45), only a basal level of MEF2 activity was detected in most adult skeletal muscle from wild-type mice. However, skeletal muscle derived from MCK-caPKD1 transgenic mice displayed intense lacZ expression indicative of the activation of MEF2 by caPKD1. The expression level of the lacZ transgene was higher in muscles comprised of type II fibers than in soleus muscle in which the caPKD1 transgene was expressed at a lower level (Fig. 1B).
PKD1 synergizes with calcineurin in skeletal muscle.
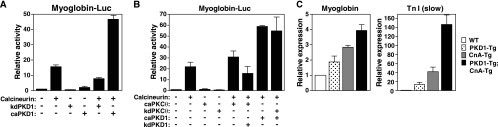
MEF2 is stimulated by calcineurin, which promotes the formation of slow-twitch myofibers (6, 27, 32, 46). To examine whether PKD1 cooperates with the calcineurin signaling pathway to drive the slow-twitch-fiber phenotype, we transfected C2C12 myogenic cells with a luciferase reporter gene controlled by the myoglobin promoter, which is activated by MEF2 (6), and a combination of expression plasmids encoding caPKD1 and/or constitutively active calcineurin. Cells were transferred to differentiation media 24 h following transfection, and luciferase activity was measured 48 h following transfection. We observed that caPKD1 and calcineurin increased activation of the myoglobin-luciferase reporter gene by 3- and 15-fold, respectively. When coexpressed in C2C12 cells, the combination of caPKD1 and calcineurin increased activation of the myoglobin promoter approximately 50-fold, whereas no synergy was seen between calcineurin and a kinase-dead mutant of PKD1 (kdPKD1) (Fig. 7A).
FIG. 7.
PKD1 acts synergistically with calcineurin. (A) C2C12 cells were cotransfected with a reporter plasmid consisting of the myoglobin promoter driving luciferase (Myoglobin-Luc) and expression plasmids encoding the constitutively active form of calcineurin, the dominant-negative form of PKD1 (kdPKD1), and/or the constitutively active form of PKD1 (caPKD1). Cytomegalovirus lacZ (CMV-lacZ) was included in all transfections, and the relative levels of expression of luciferase activity were normalized to β-galactosidase expression. The values represent the means ± standard errors of the means from three independent experiments. (B) C2C12 cells were cotransfected with myoglobin-luc and a combination of the expression plasmids encoding the constitutively active form of calcineruin, the constitutively active form of PKCθ (caPKCθ), the dominant-negative form of PKCθ (kdPKCθ), caPKD1, and/or kdPKD1. CMV-lacZ was included in all transfections, and the relative levels of expression of luciferase activity were normalized to β-galactosidase expression. Values represent the means ± standard errors of the means (n = 3). (C) Real-time PCR showed expression of myoglobin or troponin I slow-twitch fiber [TnI (slow)] in EDL muscle isolated from WT mice, MCK-myc-caPKD1 transgenic mice (PKD1-Tg), MCK-calcineurin transgenic mice (CnA-Tg), and MCK-myc-caPKD1 transgenic mice crossed with MCK-calcineurin transgenic mice (PKD1-Tg;CnA-Tg). The expression of myoglobin and TnI slow was up-regulated synergistically in the PKD1-Tg;CnA-Tg double transgenic mice (n = 3 for each group).
PKCθ has also been shown to cooperate with calcineurin in the activation of slow skeletal muscle genes in cultured muscle cells (8). Since PKD1 is a downstream target of PKCθ (48), we examined whether the effect of PKCθ on slow skeletal muscle gene expression is mediated by PKD1. C2C12 myogenic cells were cotransfected with the myoglobin-luciferase reporter and a combination of expression plasmids encoding calcineurin, constitutively active PKCθ (caPKCθ), kinase-dead PKCθ (kdPKCθ), caPKD1, and/or kdPKD1. Activation of the myoglobin promoter by calcineurin and caPKCθ was abolished by coexpression of kinase-inactive PKD1 (Fig. 7B). In contrast, the synergistic effect of PKD1 and calcineurin on the myoglobin promoter was not inhibited by coexpression of kdPKCθ. These findings suggest that PKCθ-PKD1 signaling activates slow muscle genes through cooperation with calcineurin. To confirm this cooperation in vivo, we bred MCK-calcineurin transgenic mice with MCK-caPKD1 transgenic mice. Analysis of myoglobin and troponin I (slow) expression showed similar levels of enhancement in the EDL muscle of the doubly transgenic mice (Fig. 7C).
DISCUSSION
The results of this study demonstrate that PKD1 signaling promotes the phenotype of slow-twitch skeletal-muscle fibers and diminishes muscle susceptibility to fatigue in vivo. Activation of expression of slow-twitch-fiber genes by PKD1 is accompanied by the stimulation of MEF2, a transcription factor whose presence has been shown to be sufficient to promote expression of slow-fiber genes and muscle endurance (34).
Signaling pathways involved in slow-twitch-fiber gene expression.
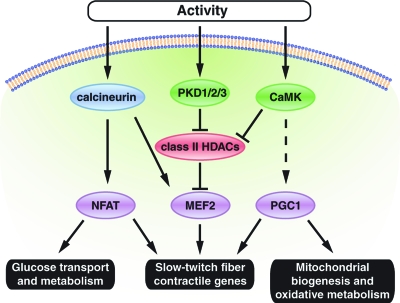
Prior studies showed that calcineurin and CaMKIV are sufficient to promote the formation of slow-twitch myofibers (6, 9, 27, 44). Similarly, PGC-1, peroxisome proliferator-activated receptor-delta (PPARδ) (19, 43), and MEF2 activate the expression of slow-twitch myofiber genes and serve as transcriptional targets of the upstream signaling pathways involved in specification of the slow myofiber phenotype (7). The ability of PKD1 to activate MEF2 and increase the abundance of type I fibers resembles the action of CaMKIV (47). The PKD1 and CaMK signaling pathways also cooperate with calcineurin. However, PKD1 appears to differ from these other regulators of the slow-twitch myofiber phenotype in that it selectively promotes the expression of slow-twitch contractile protein genes and myoglobin but not that of genes encoding mitochondrial enzymes involved in fatty acid metabolism and electron transport. A model to account for these observations is shown in Fig. 8.
FIG. 8.
Model for the PKD1/class II HDAC/MEF2 signaling pathway in skeletal muscle. Activation of PKD1 in skeletal muscle induces expression of the slow-twitch-fiber-specific contractile gene program and a subset of oxidative genes via phosphorylation of class II HDACs, which blocks their inhibitory influence on MEF2 as a consequence of their nuclear export. The calcineurin/NFAT pathway promotes the slow-twitch-fiber gene program and up-regulates a subset of oxidative metabolism genes (6, 9, 22, 31, 35). The CaMK signaling pathway activates slow-twitch-fiber contractile genes and the oxidative metabolism program via activation of MEF2 and coactivator PGC-1 (19, 44).
Mice overexpressing a constitutively active form of the lipid-activated nuclear receptor PPARδ in skeletal muscle show increased formation of type I fibers, mitochondrial biogenesis, exercise endurance, resistance to high-fat-induced obesity, and an improved metabolic profile (43). We therefore investigated whether transformed fibers in caPKD1 mice also confer improved metabolic properties. Although the expression of myoglobin was up-regulated, the MCK-caPKD1 mice displayed no overall metabolic changes. Furthermore, expression of PGC-1α, a transcriptional coactivator responsible for the regulation of mitochondrial biogenesis and oxidative metabolism (19), was not changed in MCK-caPKD1 mice. These data, together with the fact that caPKD1 and PGC-1α showed no cooperation on the myoglobin promoter in C2C12 muscle cells (data not shown), suggest that the PKD1-MEF2 signaling pathway is dedicated to the control of slow contractile protein gene expression rather than metabolic gene expression. It is interesting that PKD1 itself is preferentially expressed in slow-twitch myofibers, suggesting that the PKD1 gene may be regulated by PKD1 signaling through a positive amplification loop.
Another class II HDAC kinase, salt-inducible kinase-1 (SIK1), was recently reported (4). SIK1, a direct target of CREB, was shown to disrupt class II HDAC repression of MEF2, promote survival of skeletal myocytes, and partially rescue a dystrophic phenotype in mice. Whether SIK1 plays a role in myofiber remodeling remains to be determined.
Recently, several kinases were shown to mediate activity-dependent and -independent translocation of HDAC4 from the nucleus to the cytoplasm of adult skeletal muscle (21). Repetitive electrical stimulation of slow-twitch myofibers caused nuclear export of HDAC4 in cultured myofibers, and HDAC4 translocation was blocked by KN-62, the CaMK inhibitor. In unstimulated fibers, nucleocytoplasmic shuttling of HDAC4 was unaffected by KN-62 but was blocked by the general serine- and threonine-kinase inhibitor staurosporine. These findings suggest that two different pathways mediate nuclear export of HDAC4: an activity-dependent CaMK signaling pathway and an activity- and CaMK-independent pathway. PKD1 may play a role in mediating nuclear efflux of HDAC4 in unstimulated resting fibers.
Control by PKD1 of susceptibility of muscle to fatigue.
Muscle fatigue is manifested by a decline of muscle performance with activity. Several factors contribute to muscle fatigue, such as failure of excitation-contraction coupling, alteration in metabolism, and reactive oxygen species (2). A major determinant of fatigue resistance is the density of mitochondria and the capacity of the muscle to use oxidative metabolism. Since type I myofibers are metabolically oxidative, with high mitochondrial content, they are more resistant to fatigue than type II myofibers. Although MCK-caPKD1 transgenic mice showed no overall metabolic changes at the whole-body level, the expression of a subset of oxidative genes, including myoglobin, was up-regulated in skeletal muscle of these mice, which likely accounts for their fatigue resistance.
Skeletal-muscle-specific deletion of PKD1 led to an increase in susceptibility to fatigue in soleus but not in EDL muscles, consistent with the higher protein expression of PKD1 in soleus compared to EDL muscle. However, despite highly efficient deletion of PKD1 in skeletal muscles, no changes in fiber type composition were observed, suggesting that the increased muscle fatigue in PKD1 skKO soleus muscle was not due to an altered expression of contractile proteins. This phenotype might reflect the absence of phosphorylation of myofilament proteins by PKD, as shown for cardiomyocytes (15). Functional redundancy between different PKD isoforms might also have prevented changes in fiber type composition in PKD1 skKO mice. In this regard, PKD1 and -3 function redundantly to phosphorylate and export class II HDACs in B lymphocytes (23).
A striking phenotype of PKD1 transgenic mice is their reduction in muscle mass and myofiber size. The cross-sectional area of the individual fibers and the weight of the muscles of MCK-caPKD1 transgenic mice were decreased by 60% relative to the muscles of with WT mice. The reduced fiber size is unlikely to have been due to atrophy for several reasons. First, functional properties, such as fatigue resistance, are improved in MCK-caPKD1 transgenic muscle. Second, the expression profile of muscle-specific E3 ubiqutin ligases such as Muscle RING-finger 1 (Murf1), which mediates muscular atrophy (5), was not changed (data not shown). Finally, histological cross-sections of the muscles of MCK-caPKD1 transgenic mice revealed no increase in the number of central nuclei, an indicator of muscle injury (data not shown).
Therapeutic implications.
The realization that PKD1 modulates the susceptibility of skeletal muscle to fatigue suggests potential opportunities for therapeutically manipulating PKD1 activity as a means of enhancing skeletal muscle function in the setting of human disease (3). For example, a reduced slow myofiber population in skeletal muscle could contribute to insulin resistance, resulting in type 2 diabetes, based on the fact that insulin-stimulated glucose transport in skeletal muscle directly correlates with the percentage of slow-twitch myofibers. However, a challenge to the manipulation of PKD1 activity in vivo is that, while stimulation of PKD1 activity in skeletal muscle may improve muscle function, PKD1 activation in the heart causes dilated cardiomyopathy and heart failure (14). Thus, an important challenge for the future will be to develop tissue-specific approaches to manipulate PKD activity.
Acknowledgments
We thank Jeffery Ryder, Cheryl Nolen, and Chanhee Kang for technical assistance and Jose Cabrera for assistance with graphics.
E.N.O. is supported by grants from the National Institutes of Health, the Donald W. Reynolds Cardiovascular Clinical Research Center, and the Robert A. Welch Foundation. J.F. was supported by a fellowship from the Muscular Dystrophy Association.
Footnotes
▿
Published ahead of print on 31 March 2008.
REFERENCES
- 1.Akimoto, T., T. J. Ribar, R. S. Williams, and Z. Yan. 2004. Skeletal muscle adaptation in response to voluntary running in Ca2+/calmodulin-dependent protein kinase IV-deficient mice. Am. J. Physiol. Cell Physiol. 287C1311-C1319. [DOI] [PubMed] [Google Scholar]
- 2.Allen, D. G., G. D. Lamb, and H. Westerblad. 2008. Skeletal muscle fatigue: cellular mechanisms. Physiol. Rev. 88287-332. [DOI] [PubMed] [Google Scholar]
- 3.Bassel-Duby, R., and E. N. Olson. 2006. Signaling pathways in skeletal muscle remodeling. Annu. Rev. Biochem. 7519-37. [DOI] [PubMed] [Google Scholar]
- 4.Berdeaux, R., N. Goebel, L. Banaszynski, H. Takemori, T. Wandless, G. D. Shelton, and M. Montminy. 2007. SIK1 is a class II HDAC kinase that promotes survival of skeletal myocytes. Nat. Med. 13597-603. [DOI] [PubMed] [Google Scholar]
- 5.Bodine, S. C., E. Latres, S. Baumhueter, V. K. Lai, L. Nunez, B. A. Clarke, W. T. Poueymirou, F. J. Panaro, E. Na, K. Dharmarajan, Z. Q. Pan, D. M. Valenzuela, T. M. DeChiara, T. N. Stitt, G. D. Yancopoulos, and D. J. Glass. 2001. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 2941704-1708. [DOI] [PubMed] [Google Scholar]
- 6.Chin, E. R., E. N. Olson, J. A. Richardson, Q. Yang, C. Humphries, J. M. Shelton, H. Wu, W. Zhu, R. Bassel-Duby, and R. S. Williams. 1998. A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Genes Dev. 122499-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Czubryt, M. P., J. McAnally, G. I. Fishman, and E. N. Olson. 2003. Regulation of peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC-1 alpha) and mitochondrial function by MEF2 and HDAC5. Proc. Natl. Acad. Sci. USA 1001711-1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D'Andrea, M., A. Pisaniello, C. Serra, M. I. Senni, L. Castaldi, M. Molinaro, and M. Bouche. 2006. Protein kinase C theta co-operates with calcineurin in the activation of slow muscle genes in cultured myogenic cells. J. Cell Physiol. 207379-388. [DOI] [PubMed] [Google Scholar]
- 9.Delling, U., J. Tureckova, H. W. Lim, L. J. De Windt, P. Rotwein, and J. D. Molkentin. 2000. A calcineurin-NFATc3-dependent pathway regulates skeletal muscle differentiation and slow myosin heavy-chain expression. Mol. Cell. Biol. 206600-6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dequiedt, F., J. Van Lint, E. Lecomte, V. Van Duppen, T. Seufferlein, J. R. Vandenheede, R. Wattiez, and R. Kettmann. 2005. Phosphorylation of histone deacetylase 7 by protein kinase D mediates T cell receptor-induced Nur77 expression and apoptosis. J. Exp. Med. 201793-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dolmetsch, R. E., R. S. Lewis, C. C. Goodnow, and J. I. Healy. 1997. Differential activation of transcription factors induced by Ca2+ response amplitude and duration. Nature 386855-858. [DOI] [PubMed] [Google Scholar]
- 12.Fielitz, J., M. S. Kim, J. M. Shelton, X. Qi, J. A. Hill, J. A. Richardson, R. Bassel-Duby, and E. N. Olson. 2008. Requirement of protein kinase D1 for pathological cardiac remodeling. Proc. Natl. Acad. Sci. USA 1053059-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grange, R. W., A. Meeson, E. Chin, K. S. Lau, J. T. Stull, J. M. Shelton, R. S. Williams, and D. J. Garry. 2001. Functional and molecular adaptations in skeletal muscle of myoglobin-mutant mice. Am. J. Physiol. Cell Physiol. 281C1487-C1494. [DOI] [PubMed] [Google Scholar]
- 14.Harrison, B. C., M.-S. Kim, E. van Rooij, C. F. Plato, P. J. Papst, R. B. Vega, J. A. McAnally, J. A. Richardson, R. Bassel-Duby, E. N. Olson, and T. A. McKinsey. 2006. Regulation of cardiac stress signaling by protein kinase D1. Mol. Cell. Biol. 263875-3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haworth, R. S., F. Cuello, T. J. Herron, G. Franzen, J. C. Kentish, M. Gautel, and M. Avkiran. 2004. Protein kinase D is a novel mediator of cardiac troponin I phosphorylation and regulates myofilament function. Circ. Res. 951091-1099. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, J. E., B. J. Wold, and S. D. Hauschka. 1989. Muscle creatine kinase sequence elements regulating skeletal and cardiac muscle expression in transgenic mice. Mol. Cell. Biol. 93393-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krook, A., M. Bjornholm, D. Galuska, X. J. Jiang, R. Fahlman, M. G. Myers, Jr., H. Wallberg-Henriksson, and J. R. Zierath. 2000. Characterization of signal transduction and glucose transport in skeletal muscle from type 2 diabetic patients. Diabetes 49284-292. [DOI] [PubMed] [Google Scholar]
- 18.Li, S., M. P. Czubryt, J. McAnally, R. Bassel-Duby, J. A. Richardson, F. F. Wiebel, A. Nordheim, and E. N. Olson. 2005. Requirement for serum response factor for skeletal muscle growth and maturation revealed by tissue-specific gene deletion in mice. Proc. Natl. Acad. Sci. USA 1021082-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin, J., H. Wu, P. T. Tarr, C. Y. Zhang, Z. Wu, O. Boss, L. F. Michael, P. Puigserver, E. Isotani, E. N. Olson, B. B. Lowell, R. Bassel-Duby, and B. M. Spiegelman. 2002. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature 418797-801. [DOI] [PubMed] [Google Scholar]
- 20.Liu, Y., C. Graham, V. Parravicini, M. J. Brown, J. Rivera, and S. Shaw. 2001. Protein kinase C theta is expressed in mast cells and is functionally involved in Fcepsilon receptor I signaling. J. Leukoc. Biol. 69831-840. [PubMed] [Google Scholar]
- 21.Liu, Y., W. R. Randall, and M. F. Schneider. 2005. Activity-dependent and -independent nuclear fluxes of HDAC4 mediated by different kinases in adult skeletal muscle. J. Cell Biol. 168887-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Long, Y. C., S. Glund, P. M. Garcia-Roves, and J. R. Zierath. 2007. Calcineurin regulates skeletal muscle metabolism via coordinated changes in gene expression. J. Biol. Chem. 2821607-1614. [DOI] [PubMed] [Google Scholar]
- 23.Matthews, S. A., P. Liu, M. Spitaler, E. N. Olson, T. A. McKinsey, D. A. Cantrell, and A. M. Scharenberg. 2006. Essential role for protein kinase D family kinases in the regulation of class II histone deacetylases in B lymphocytes. Mol. Cell. Biol. 261569-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKinsey, T. A., C. L. Zhang, J. Lu, and E. N. Olson. 2000. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 408106-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2002. Signaling chromatin to make muscle. Curr. Opin. Cell Biol. 14763-772. [DOI] [PubMed] [Google Scholar]
- 26.Molkentin, J. D., J. R. Lu, C. L. Antos, B. Markham, J. Richardson, J. Robbins, S. R. Grant, and E. N. Olson. 1998. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell 93215-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naya, F. J., B. Mercer, J. Shelton, J. A. Richardson, R. S. Williams, and E. N. Olson. 2000. Stimulation of slow skeletal muscle fiber gene expression by calcineurin in vivo. J. Biol. Chem. 2754545-4548. [DOI] [PubMed] [Google Scholar]
- 28.Naya, F. J., C. Wu, J. A. Richardson, P. Overbeek, and E. N. Olson. 1999. Transcriptional activity of MEF2 during mouse embryogenesis monitored with a MEF2-dependent transgene. Development 1262045-2052. [DOI] [PubMed] [Google Scholar]
- 29.Oberbach, A., Y. Bossenz, S. Lehmann, J. Niebauer, V. Adams, R. Paschke, M. R. Schon, M. Bluher, and K. Punkt. 2006. Altered fiber distribution and fiber-specific glycolytic and oxidative enzyme activity in skeletal muscle of patients with type 2 diabetes. Diabetes Care 29895-900. [DOI] [PubMed] [Google Scholar]
- 30.Ogilvie, R. W., and D. L. Feeback. 1990. A metachromatic dye-ATPase method for the simultaneous identification of skeletal muscle fiber types I, IIA, IIB and IIC. Stain Technol. 65231-241. [DOI] [PubMed] [Google Scholar]
- 31.Oh, M., Rybkin, I. I., V. Copeland, M. P. Czubryt, J. M. Shelton, E. van Rooij, J. A. Richardson, J. A. Hill, L. J. De Windt, R. Bassel-Duby, E. N. Olson, and B. A. Rothermel. 2005. Calcineurin is necessary for the maintenance but not embryonic development of slow muscle fibers. Mol. Cell. Biol. 256629-6638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parsons, S. A., B. J. Wilkins, O. F. Bueno, and J. D. Molkentin. 2003. Altered skeletal muscle phenotypes in calcineurin Aalpha and Abeta gene-targeted mice. Mol. Cell. Biol. 234331-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Potthoff, M. J., and E. N. Olson. 2007. MEF2: a central regulator of diverse developmental programs. Development 1344131-4140. [DOI] [PubMed] [Google Scholar]
- 34.Potthoff, M. J., H. Wu, M. A. Arnold, J. M. Shelton, J. Backs, J. McAnally, J. A. Richardson, R. Bassel-Duby, and E. N. Olson. 2007. Histone deacetylase degradation and MEF2 activation promote the formation of slow-twitch myofibers. J. Clin. Invest. 1172459-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ryder, J. W., R. Bassel-Duby, E. N. Olson, and J. R. Zierath. 2003. Skeletal muscle reprogramming by activation of calcineurin improves insulin action on metabolic pathways. J. Biol. Chem. 27844298-44304. [DOI] [PubMed] [Google Scholar]
- 36.Rykx, A., L. De Kimpe, S. Mikhalap, T. Vantus, T. Seufferlein, J. R. Vandenheede, and J. Van Lint. 2003. Protein kinase D: a family affair. FEBS Lett. 54681-86. [DOI] [PubMed] [Google Scholar]
- 37.Sakai, K., and J. Miyazaki. 1997. A transgenic mouse line that retains Cre recombinase activity in mature oocytes irrespective of the cre transgene transmission. Biochem. Biophys. Res. Commun. 237318-324. [DOI] [PubMed] [Google Scholar]
- 38.Schiaffino, S., M. Sandri, and M. Murgia. 2007. Activity-dependent signaling pathways controlling muscle diversity and plasticity. Physiology (Bethesda). 22269-278. [DOI] [PubMed] [Google Scholar]
- 39.Storz, P., and A. Toker. 2003. Protein kinase D mediates a stress-induced NF-kappaB activation and survival pathway. EMBO J. 22109-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Lint, J. V., J. Sinnett-Smith, and E. Rozengurt. 1995. Expression and characterization of PKD, a phorbol ester and diacylglycerol-stimulated serine protein kinase. J. Biol. Chem. 2701455-1461. [DOI] [PubMed] [Google Scholar]
- 41.Vega, R. B., B. C. Harrison, E. Meadows, C. R. Roberts, P. J. Papst, E. N. Olson, and T. A. McKinsey. 2004. Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol. Cell. Biol. 248374-8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verdin, E., F. Dequiedt, and H. G. Kasler. 2003. Class II histone deacetylases: versatile regulators. Trends Genet. 19286-293. [DOI] [PubMed] [Google Scholar]
- 43.Wang, Y. X., C. L. Zhang, R. T. Yu, H. K. Cho, M. C. Nelson, C. R. Bayuga-Ocampo, J. Ham, H. Kang, and R. M. Evans. 2004. Regulation of muscle fiber type and running endurance by PPARdelta. PLoS Biol. 2e294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu, H., S. B. Kanatous, F. A. Thurmond, T. Gallardo, E. Isotani, R. Bassel-Duby, and R. S. Williams. 2002. Regulation of mitochondrial biogenesis in skeletal muscle by CaMK. Science 296349-352. [DOI] [PubMed] [Google Scholar]
- 45.Wu, H., F. J. Naya, T. A. McKinsey, B. Mercer, J. M. Shelton, E. R. Chin, A. R. Simard, R. N. Michel, R. Bassel-Duby, E. N. Olson, and R. S. Williams. 2000. MEF2 responds to multiple calcium-regulated signals in the control of skeletal muscle fiber type. EMBO J. 191963-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu, H., B. Rothermel, S. Kanatous, P. Rosenberg, F. J. Naya, J. M. Shelton, K. A. Hutcheson, J. M. DiMaio, E. N. Olson, R. Bassel-Duby, and R. S. Williams. 2001. Activation of MEF2 by muscle activity is mediated through a calcineurin-dependent pathway. EMBO J. 206414-6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu, Y., R. J. Colbran, and M. E. Anderson. 2001. Calmodulin kinase is a molecular switch for cardiac excitation-contraction coupling. Proc. Natl. Acad. Sci. USA 982877-2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuan, J., D. Bae, D. Cantrell, A. E. Nel, and E. Rozengurt. 2002. Protein kinase D is a downstream target of protein kinase Ctheta. Biochem. Biophys. Res. Commun. 291444-452. [DOI] [PubMed] [Google Scholar]
- 49.Zugaza, J. L., J. Sinnett-Smith, J. Van Lint, and E. Rozengurt. 1996. Protein kinase D (PKD) activation in intact cells through a protein kinase C-dependent signal transduction pathway. EMBO J. 156220-6230. [PMC free article] [PubMed] [Google Scholar]