Saccharomyces cerevisiae septins: Supramolecular organization of heterooligomers and the mechanism of filament assembly (original) (raw)
Abstract
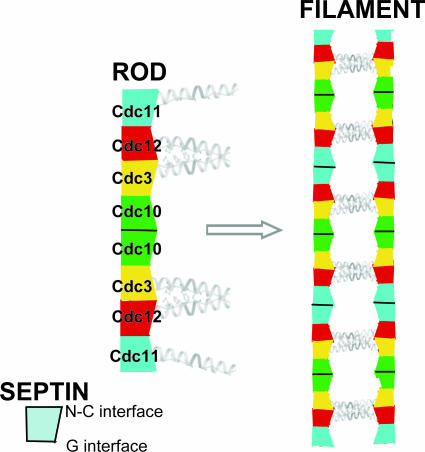
Mitotic yeast cells express five septins (Cdc3, Cdc10, Cdc11, Cdc12, and Shs1/Sep7). Only Shs1 is nonessential. The four essential septins form a complex containing two copies of each, but their arrangement was not known. Single-particle analysis by EM confirmed that the heterooligomer is octameric and revealed that the subunits are arrayed in a linear rod. Identity of each subunit was determined by examining complexes lacking a given septin, by antibody decoration, and by fusion to marker proteins (GFP or maltose binding protein). The rod has the order Cdc11–Cdc12–Cdc3–Cdc10–Cdc10–Cdc3–Cdc12–Cdc11 and, hence, lacks polarity. At low ionic strength, rods assemble end-to-end to form filaments but not when Cdc11 is absent or its N terminus is altered. Filaments invariably pair into long parallel “railroad tracks.” Lateral association seems to be mediated by heterotetrameric coiled coils between the paired C-terminal extensions of Cdc3 and Cdc12 projecting orthogonally from each filament. Shs1 may be able to replace Cdc11 at the end of the rod. Our findings provide insights into the molecular mechanisms underlying the function and regulation of cellular septin structures.
Keywords: electron microscopy, yeast, complexes, GTP
Septins comprise a discrete family of GTP-binding proteins conserved from fungi to humans (1). Budding yeast (Saccharomyces cerevisiae) encodes seven paralogous septins. Five (Cdc3, Cdc10, Cdc11, Cdc12, and Shs1) are expressed in mitosis, found in filaments at the mother-bud neck, and participate in cytokinesis (2). Cdc3, Cdc10, Cdc11, and Cdc12 (but not Shs1) are essential for viability and were first identified in the collection of temperature-sensitive cell division cycle (cdc) mutants isolated by Hartwell (3). All contain a ≈300-residue GTP-binding domain (G domain, 30–40% pairwise identity) but are distinguished by more variable N- and C-terminal extensions [supporting information (SI) Fig. S1]. Cdc3 has the longest N-terminal extension, whereas Shs1 has the longest C-terminal extension (CTE). At their distal end, the CTEs of Cdc3, Cdc11, Cdc12, and Shs1 (but not Cdc10) contain a segment of 40–50 residues with coiled coil-forming heptad repeats (4). The CTEs are essential for Cdc3 and Cdc12 function in vivo but not for Cdc11 (5). Available data indicate that guanine nucleotide binding promotes septin folding and stability, but mutations that block cycles of GTP binding and hydrolysis cause no overt phenotypes in vivo (6, 7). The collar of filaments at the bud neck is apposed to the plasma membrane, an interaction attributed to a polybasic segment situated proximal to the G domain (8, 9). The collar filaments impose a barrier to diffusion of integral membrane proteins between mother and bud (10, 11) and act as a scaffold to recruit proteins required for bud-site selection and a morphogenesis checkpoint (12).
Native yeast septin complexes purified by immunoaffinity contain near-equimolar Cdc3, Cdc10, Cdc11, and Cdc12 and have an apparent molecular mass compatible with 2:2:2:2 stoichiometry (13, 14). Likewise, Cdc3, Cdc10, Cdc11, and Cdc12 coexpressed in and purified from bacterial cells form octameric complexes (5, 15). The native and recombinant complexes resist dissociation at high ionic strength (0.5–1 M KCl) but polymerize into filaments when the salt concentration is reduced (≤50 mM KCl) (5, 13). Various strategies have been used to ascertain the interactions within the core septin complex. Two-hybrid (16) is fraught with potential problems because septin complexes do not normally assemble in the nucleus and because apparent associations might arise indirectly via “bridging” through other members of the complex or other septin-associated proteins (15). By using purified septins for in vitro binding to assess their capacity for pairwise interactions, apparent self-association could occur in the absence of a preferred partner, and ability to associate with another septin could be influenced by the presence of a third (5). The predicted coiled coil might account for such nonphysiological associations. An analogy is c-Jun, which by itself forms homodimers but yields c-Fos-c-Jun heterodimers preferentially when both are present (17).
Coexpression in bacteria of various combinations of yeast septins gave a more consistent picture of the organization of the complex (5). When Cdc10, Cdc11, and (His)6–Cdc12 were coexpressed, stoichiometric (His)6–Cdc12–Cdc11 binary complexes were recovered, but no significant amount of Cdc10 was incorporated, suggesting that Cdc11 interacts directly with Cdc12 and that Cdc3 is needed to recruit Cdc10. However, when (His)6–Cdc3, Cdc10, and Cdc11 were coexpressed, no protein other than (His)6–Cdc3 was recovered. This result suggested that, in the absence of Cdc12, Cdc3 is not competent to associate with Cdc10. Indeed, when coexpressed, Cdc3 and Cdc12 associate avidly, and Cdc3, Cdc10 and (His)6–Cdc12 form stoichiometric ternary complexes. Collectively, these findings indicated that assembly of the octameric complex has an intrinsic order, with Cdc12 serving as the linchpin because it associates directly with both Cdc11 and Cdc3. However, significant ambiguities remained. In binding studies in vitro, Cdc10 displayed separate interactions with both Cdc3 and Cdc12. Thus, incorporation of Cdc10 into ternary complexes with Cdc3 and Cdc12 could reflect simultaneous contact of Cdc10 with both Cdc3 and Cdc12, as initially proposed (5) Alternatively, association of Cdc12 with Cdc3 may induce a conformational change in Cdc3 that allows it to interact with Cdc10, as suggested above. Hence, the structural organization of the rod-like native (13) and recombinant (18) yeast septin complexes seen by negative staining in the EM was unknown.
We developed reliable recombinant methods for producing and purifying yeast septin complexes. By using single-particle EM analysis, we confirm that the complex of the four essential septins is a linear octameric rod. We then applied various tactics to pinpoint the position of each septin within the rod. Our results show that the rod has two-fold rotational symmetry about an axis running orthogonally through its center. Recently, a septin complex isolated from the nematode Caenorhabditis elegans, which encodes only two septins (UNC-59 and UNC-61), was shown to form symmetric rods containing two copies of each subunit (19). Likewise, a crystal structure and EM analysis of a complex comprising three human septins (SEPT2, SEPT6, and SEPT7) revealed a symmetrical heterohexameric rod (18). These findings suggest that all eukaryotic septin complexes will be organized as linear symmetrical rods. The human heterohexamer forms filaments through the interface surrounding the guanine nucleotide-binding site in SEPT7 (18), whereas the yeast complex forms filaments through a salt-sensitive interface involving the N and C termini of Cdc11. Moreover, the yeast heterooctamer cooperatively assembles into parallel filaments paired through the predicted C-terminal coiled-coil domains of Cdc3 and Cdc12. Thus, septins in different organisms not only form complexes with different numbers of subunits but also can reversibly form filaments via different interfaces.
Results and Discussion
Yeast Core Septin Complex Is a Linear Rod.
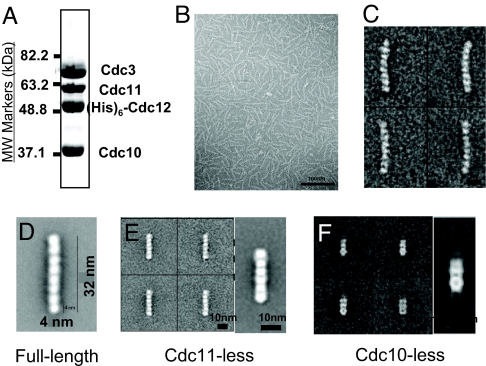
In negatively stained EM images, the purified recombinant Cdc3–Cdc10–Cdc11–(His)6–Cdc12 complex prepared at high ionic strength (Fig. 1A) was a beaded rod with a long dimension of 32–35 nm (Fig. 1B). Averaging images of large numbers of classes of individual rods (Fig. 1C) afforded enhanced resolution that revealed that each rod is composed of eight globular densities of roughly equivalent size (≈4–5-nm diameter) (Fig. 1D). This latter dimension corresponds well to other small GTP-binding proteins (20), suggesting that the globular densities reflect the conserved G domain common to Cdc3, Cdc10, Cdc11, and Cdc12 (Fig. S1). The calculated molecular mass for eight protein spheres of such dimension agrees well with that determined for native and recombinant septin complexes in which the apparent stoichiometry of the four septins is 2:2:2:2. Thus, by its appearance in the EM, each rod is a heterooctamer, presumably containing two molecules of each of the four different septins. Human hexameric septin rods often displayed a sharp kink in EM images, suggesting that they can flex at their center (18). Although the yeast octameric rod is longer, we did not observe any kinked rods, although some had a slight smooth curvature (Fig. 1 B and C).
Fig. 1.
Characterization of yeast heterooligomeric septin complexes. (A) A sample (50 μg) of the purified Cdc3–Cdc10–Cdc11–(His)6–Cdc12 heterooctamers used in most of these studies, prepared as described in Materials and Methods, was resolved by electrophoresis on a 10% SDS/PAGE gel and stained with Coomassie Brilliant blue. MW, molecular weight. (B) Low magnification view of the preparation shown in (A) suspended in high-salt (300 mM) buffer, adsorbed on a carbon grid, stained, and viewed in the EM as described in Materials and Methods. (C) Four representative class averages (≈150 particles each) derived from processing a total of 11,000 particles from micrographs of Cdc3–Cdc10–Cdc11–(His)6–Cdc12 complexes, as in (B). (D) Higher magnification view of an average of 1,986 particles of the Cdc3–Cdc10–Cdc11–(His)6–Cdc12 (full-length) complex. (E) Four representative class averages (100 particles each) (Left) and a higher magnification view of the average of the whole data set of 2,093 particles (Right) observed in a preparation of Cdc3–Cdc10–(His)6–Cdc12 (Cdc11-less) complexes, examined as in B. (F) Four representative class averages (10 particles each) (Left) and a higher magnification view of the average of the whole data set of 824 particles (Right) observed in a preparation of Cdc3–Cdc11–(His)6–Cdc12 (Cdc10-less) complexes, examined as in B.
Arrangement of Subunits Within the Octameric Rod.
We applied several strategies to delineate the position of each septin in the rod. First, we examined images of complexes prepared in the absence of a given septin and ascertained the effect on rod structure. Second, we decorated rods with an antibody directed against a septin or epitope-tagged septin and located the bound IgG. Third, we incorporated into complexes septins tagged at their N- or C-terminal end by fusion to a small monomeric marker protein, and located the extra nearby density.
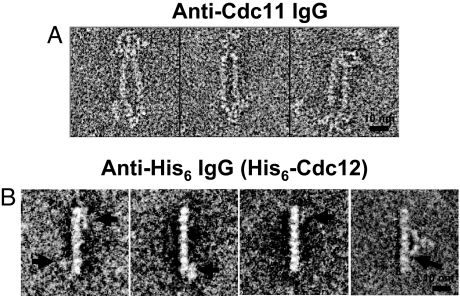
When coexpressed in bacteria in the absence of Cdc11, stoichiometric complexes of Cdc3, Cdc10, and (His)6–Cdc12 were obtained, as before (5, 15). These complexes appeared as rods with six clear-cut globular densities (Fig. 1E). Thus, Cdc3, Cdc10, and Cdc12 formed a stable hexameric rod when no Cdc11 was present, suggesting that Cdc11 occupies a terminal position in the rod, with a Cdc11–Cdc11 dimer at one end or one Cdc11 at each end. When Cdc3–Cdc10–Cdc11–(His)6–Cdc12 octamers were incubated with purified rabbit polyclonal anti-Cdc11 antibodies, the IgG bound to both termini of an individual octameric rod (Fig. 2A). Moreover, given their bivalent Y-shaped nature, the anti-Cdc11 IgG often was found holding two octameric rods together side-by-side (Fig. 2A). These results established that Cdc11 occupies the outermost position at both termini of the octameric rod.
Fig. 2.
Location of Cdc11 and (His)6–Cdc12 determined by antibody decoration. (A) A sample of the preparation of purified Cdc3–Cdc10–Cdc11–(His)6–Cdc12 heterooctamers shown in Fig. 1 A–D was incubated on ice in high-salt buffer with rabbit polyclonal anti-Cdc11 antibodies. Three representative raw (unaveraged) images of the resulting complexes observed are shown. (B) As in A, except incubation was with a mouse monoclonal anti-(His)6 antibody. Although binding of this antibody was substoichiometric under the high-salt conditions, image processing readily identified particles with antibody bound. Left, three representative raw (unaveraged) images of the resulting complexes observed are shown, wherein the extra density (horizontal arrows) is most often positioned closest to the second subunit in from one or both ends of the rod. Right, one of the class averages obtained after multiple rounds of multireference alignment and classification, in which the antibody is seen bound symmetrically around the center of the rod, with at least one of its Fab arms in contact with the second subunit from the end (most clearly seen for the contact indicated by the arrow).
The only direct interaction with another core septin detected for Cdc11 is with Cdc12, and Cdc12 (but not Cdc11) interacts with Cdc3, and Cdc3, in turn, interacts with Cdc10 (even when Cdc11 is absent, as long as Cdc12 is present) (5, 15). These data suggested that the arrangement in the rod must include the order Cdc11–Cdc12–Cdc3–Cdc10. Because we established that Cdc11 is at each end, Cdc12 was likely the second subunit in from each end. To test whether Cdc12 occupies this position, we first tried polyclonal anti-Cdc12 IgG directed against its G domain. However, the Cdc3–Cdc10–Cdc11–(His)6–Cdc12 complexes were only inefficiently decorated, presumably because the main antigenic determinants are buried in the rod [consistent with rod formation requiring contacts made by Cdc12 via its globular G domain (18)]. As an alternative, we incubated the Cdc3–Cdc10–Cdc11–(His)6–Cdc12 complexes with a commercial anti-(His)6 mAb. To examine particles only at their periphery, a mask was used (see Materials and Methods), and a total of 49 particles displayed density with an antibody-like profile in this region. As shown in the class average from those 49 particles (Fig. 2B), the antibody density was juxtaposed to the second subunit from both ends. These findings confirmed that Cdc12 is the septin that occupies the penultimate position at each end of the octameric rod.
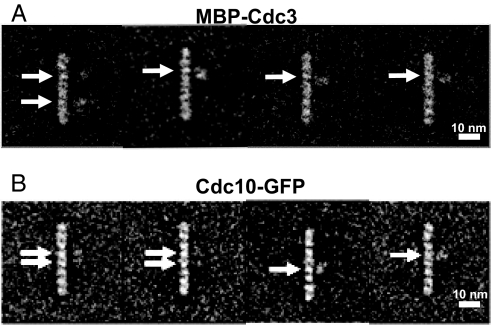
Based on biochemical data, the subunit adjacent to Cdc12 on the side opposite that contacting Cdc11 was likely Cdc3. To establish the position of Cdc3, we first tried a polyclonal antibody directed against its 14 C-terminal residues. Approximately 14% of the treated rods displayed a bound IgG-like profile (data not shown); however, the extra density was found in diverse positions with respect to the rod (36% close to the middle, 36% near the ends, and 28% at intermediate positions). If in α-helical conformation, the Cdc3 CTE (113 residues) could be more than 160 Å long and, if flexible, capable of sampling from the center to the ends of the rod, explaining our observation. As an alternative means to locate Cdc3, we prepared complexes containing an MBP–Cdc3 fusion as the sole source of this septin. The maltose binding protein (MBP) sequence was fused in-frame to the N terminus of Cdc3, which was expected to be more structured than the CTE and hold the 42-kDa MBP appendage more rigidly. Indeed, MBP–Cdc3–Cdc10–Cdc11–(His)6–Cdc12 complexes yielded stable octameric rods with the same overall structure observed for complexes with untagged Cdc3, except that extra density was often juxtaposed to the third subunit from one or both ends (Fig. 3A).
Fig. 3.
Location of MBP–Cdc3 and Cdc10–GFP. (A) Four representative class averages of purified MBP–Cdc3–Cdc10–Cdc11–(His)6–Cdc12 heterooctamers resulting from three rounds of multireference alignment and classification (2,620 particles total). The extra density (horizontal arrows) is most often positioned closest to the third subunit in from one or both ends of the rod. (B) Four representative class averages of purified Cdc3–Cdc10–GFP–Cdc11–(His)6–Cdc12 heterooctamers resulting from three rounds of multireference alignment and classification (3,377 particles total). The extra density (horizontal arrows) is most often positioned closest to one or both of the two central subunits in the rod. In A and B, the majority of the averages yielded only one extra density per octamer, most likely because of flexibility in the joint between the marker protein and septin, making detection of the second marker improbable.
We used similar strategies to confirm that the subunit adjacent to Cdc3 at the center of the rod is Cdc10. First, when Cdc3, Cdc11, and (His)6–Cdc12 were coexpressed, stable ternary complexes were formed, as seen before (5, 15). These Cdc10-less complexes appeared as short rods composed of three distinct densities (Fig. 1F), consistent with the idea that a central Cdc10 dimer is needed to join two outer trimeric arms (Cdc11–Cdc12–Cdc3) together to form the octameric rod. The fact that two of the densities in the Cdc11–Cdc12–Cdc3 trimers seem distinctly less globular than the subunits in octameric rods suggests that, in the absence of Cdc10, the remaining subunits undergo conformational transitions that affect their packing and/or relative orientation. Second, we prepared complexes containing a Cdc10–GFP fusion as the sole source of this septin. This same chimera (21) fully complements a _cdc10_Δ mutation, indicating that the 27-kDa GFP appendage does not compromise Cdc10 folding or function. The Cdc3–Cdc10–GFP–Cdc11–(His)6–Cdc12 complexes yielded stable octameric rods in which extra density was juxtaposed to the center of the rod, projecting from the fourth subunit from one or both ends (Fig. 3B), thus demonstrating that a pair of Cdc10 subunits is at the center of the rod. It has been reported that haploid cells lacking Cdc10 can be isolated and appear to lack neck filaments (13, 22). Although these cells are viable at 20–25°C, they manifest gross morphological abnormalities at 30° and are inviable at 37° (5, 13, 22). Thus, the inability to form the full heterooctameric complex (and presumably the absence of normal filaments) is not well tolerated.
Based on the molecular organization (UNC-59–UNC-61–UNC61–UNC59) of the minimal tetrameric septin rod from C. elegans (and the presumption that UNC-59 and UNC-61 are likely orthologs of Cdc12 and Cdc3, respectively), it was proposed by others (19) that the more elaborate yeast septin complex would be symmetrically organized around a central Cdc12–Cdc3–Cdc3–Cdc12 core. Our data show that this prediction was incorrect. In addition, a model based on prior findings (5) suggesting that yeast septin complexes might form polar filaments also requires revision. Our results show that the yeast septin complex has the order Cdc11–Cdc12–Cdc3–Cdc10–Cdc10–Cdc3–Cdc12–Cdc11. Thus, like the worm (19) and human (18) septin rods, the yeast rod lacks polarity because it has an axis of two-fold rotational symmetry situated between and perpendicular to the two central Cdc10 subunits.
Filament Formation by Salt-Dependent End-to-End Rod Polymerization.
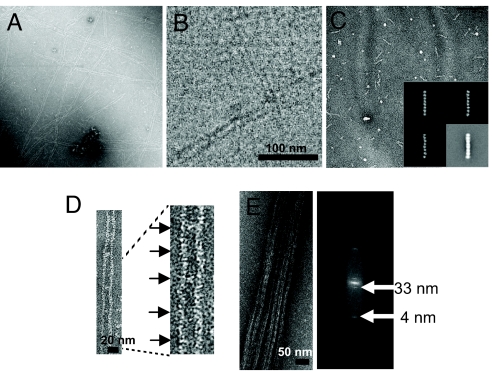
At ≥250 mM salt, Cdc3–Cdc10–Cdc11–(His)6–Cdc12 complexes are octameric rods (Fig. 1B). When ionic strength was lowered (≤50 mM salt), we observed very long paired filaments (Fig. 4A), each with the same diameter as that of the rod (5, 15), suggesting that electrostatic interactions drive filament assembly. When complexes were deposited onto EM grids immediately after lowering the salt, we saw very occasionally, in addition to filaments, possible intermediates that are twice the length of the rod, or even less frequently, three or four times the rod length (Fig. S2).
Fig. 4.
End-on-end polymerization of the rods forms paired filaments. (A) A sample of the purified Cdc3–Cdc10–Cdc11–(His)6–Cdc12 heterooctamers shown in Fig. 1 A–D were diluted from high salt (300 mM) to low salt (50 mM), incubated for 2 h, and then examined in the EM after negative staining. (B) As in A, except that the sample was viewed via cryo-EM after vitrification in liquid ethane, as described in Materials and Methods. (C) Purified Cdc3–Cdc10–Cdc11(Δα0)–(His)6–Cdc12 heterooctamers were examined after dilution and incubation in low salt, as in A. (Inset) Three representative class averages and the average of the most common class (Bottom Right) of the same sample of purified Cdc3–Cdc10–Cdc11(Δα0)–(His)6–Cdc12 heterooctamers examined in high salt. (D) Paired filaments displaying periodic densities between the filaments (Left) and at higher magnification (Right). (E) Example of a bundle of paired filaments formed after prolonged incubation under low-salt conditions at relatively high protein concentration (>1 μg ml−1) and above neutral pH (Left) and a representative power spectrum (Right) derived from the Fourier transform of the image of such a bundle of paired filaments.
Filament formation seems a highly cooperative process, for two reasons. First, filaments are very long and invariably align pairwise (like railroad tracks), separated by a 15- to 25-nm gap. Paired filaments are observed whether visualized by negative staining (Fig. 4A) or viewed in vitreous ice by cryo-EM (Fig. 4B). Thus, pairing is not an artifact of staining or the support used. Second, paired filaments appear to be in register because the ends of each filament are flush with each other, not staggered (Fig. 4A). These observations suggest further that filament elongation and filament pairing are coupled. Moreover, the filaments have no intrinsic polarity because the rod of which they are composed is symmetric.
Under some conditions, still not well defined (but influenced by pH and protein concentration), the filament doublets coalesced into larger bundles, which are likely nonphysiological aggregates (Fig. 4E Left). These bundles appeared to be organized in register (Fig. 4E Left) and diffracted reasonably well. The power spectrum of the bundles (Fig. 4E Right) showed both a 4-nm axial repeat corresponding to each globular subunit and a ≈33-nm repeat corresponding to the length of the octameric rod. No clear repeat was found in the direction perpendicular to the bundle, suggesting lack of crystalline order in that direction. In low salt, anti-Cdc11 antibodies decorated these bundles with a periodicity compatible with the length of an octamer and with a banding pattern indicating that homotypic lateral interactions align like subunits across the bundle (Fig. S3). Furthermore, in agreement with our findings that Cdc11 is the terminal subunit at each end of a rod and that filaments form by end-to-end polymerization of the rods, when decorated with anti-Cdc11 IgG, filaments in bundles always had antibody bound at their free ends (Fig. S3).
Cdc11–Cdc11 Interaction Is Critical for Rod Polymerization.
Several lines of evidence demonstrated that direct Cdc11–Cdc11 interaction is the mechanism of filament assembly. First, Cdc11-less hexameric rods (Fig. 1E) never formed filaments detectable in the EM when the salt concentration was lowered (data not shown), in agreement with our prior finding that Cdc3–Cdc10–(His)6–Cdc12 complexes did not polymerize (5). Theoretically, Cdc10-less trimeric rods might be able to form “inside-out” Cdc3–Cdc12–Cdc11–Cdc11–Cdc12–Cdc3 hexamers. However, we have not detected such forms in our EM studies, suggesting that the central Cdc10–Cdc10 doublet may induce conformational effects propagated to the end of the rod or, more likely, the presence of Cdc10 may affect the orientation or conformation of the Cdc3 CTE, thereby promoting optimal filament pairing, which is coupled to filament elongation.
To define the interfaces that mediate rod and filament formation, we made mutations in contacts predicted from the crystal structures of human SEPT2 and the SEPT2–SEPT6–SEPT7 heterohexamer (18). In a linear array, a septin must contact each of its neighboring subunits asymmetrically. At its so-called G interface, a septin interacts via its G domain with the G domain in the adjacent septin, an association involving contacts around the guanine nucleotide-binding site in each monomer (18). On its other side, a septin binds its neighbor via a so-called N–C interface (18), which involves the N-terminal α-helix (α0) and the more distal α-helix (α6) (Fig. S1); the latter was at the C-terminal end of the visible electron density (18). These α0 and α6 helices distinguish septins from Ras-like GTPases. At an N–C interface, the paired septin CTEs (which were not visible in the human heterohexamer electron density map) project from the same face of the rod and are juxtaposed (18), permitting interaction of complementary sequences.
To confirm that Cdc11–Cdc11 association mediates filament formation and delineate what interface mediates this interaction, we generated two Cdc11 mutants, coexpressed them with Cdc3, Cdc10, and (His)6–Cdc12, and examined the efficiency of their incorporation into the resulting complexes and the ability of those complexes to form filaments. We found, first, that Cdc11(Δ357-415), in which the CTE (but not α6) was removed, was efficiently incorporated into octameric rods, which were indistinguishable in the EM from the rods containing full-length Cdc11, and that these rods polymerized efficiently into filaments in vitro at low salt (data not shown). These findings are consistent with our previous observations that CTE-less Cdc11 is capable of homotypic interaction in vitro and that yeast cells expressing Cdc11(Δ357-415) as the sole source of this septin, although displaying far from normal morphology, are nonetheless viable, whereas those expressing CTE-less Cdc3 or Cdc12 are inviable (5). Thus, EM observation confirmed that the Cdc11 CTE does not have an essential role in either octamer or filament assembly, and rather, may help recruit other proteins to rods or filaments. By contrast, deletion of the predicted Cdc11 α0 element [Cdc11(Δ2-18) or Cdc11(Δα0)] also produced octameric rods (Fig. 5A) indistinguishable in the EM from the rods containing full-length Cdc11 (Fig. 4C Inset), but these were unable to polymerize into filaments (Fig. 4C). These data corroborate that Cdc11–Cdc11 association is required for rod polymerization and indicate that this interaction is an N–C interface.
Fig. 5.
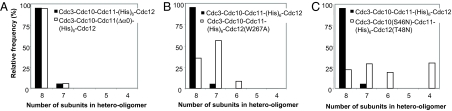
Effect of site-directed mutations on the efficiency of stable heterooctameric rod assembly. Purified preparations of the wild-type Cdc3–Cdc10–Cdc11–(His)6–Cdc12 complex (black bars) and the three mutant complexes indicated in each panel (white bars), Cdc3–Cdc10–Cdc11(Δα0)–(His)6–Cdc12 (A), Cdc3–Cdc10–Cdc11–(His)6–Cdc12(W267A) (B), and Cdc3–Cdc10(S46N)–Cdc11–(His)6–Cdc12(T48N) (C), were examined under high-salt conditions in the EM after negative staining, and the number of subunits in the rods formed were scored; 3,043 particles of the Cdc3–Cdc10–Cdc11(Δα0)–(His)6–Cdc12 complexes, 2,130 total particles for the Cdc3–Cdc10–Cdc11–(His)6–Cdc12(W267A) complexes, and 1,267 particles for the Cdc3–Cdc10(S46N)–Cdc11–(His)6–Cdc12(T48N) complexes were selected, aligned, and classified, first without any reference before a second round of alignment and classification by using references resulting from the first round of alignment, and classification was performed.
If so, the Cdc11–Cdc12 interaction is a G interface. To test this prediction, Cdc3, Cdc10, and Cdc11 were coexpressed with (His)6–Cdc12(W267A), which carries a point mutation in a conserved residue equivalent to a side chain that mediates an important contact for G dimer association of human SEPT2 (18). This mutation in Cdc12 greatly reduced Cdc11 incorporation onto the ends of the rod and greatly diminished the frequency of formation of full-length octamers (Fig. 5B), consistent with the conclusion that the Cdc11–Cdc12 interaction surface is a G interface. In agreement with the general need for guanine nucleotide binding for octamer formation and stability, coexpression of Cdc3 and Cdc11 with two mutants, Cdc10(S46N) and (His)6–Cdc12(T48N), previously demonstrated to be GTP binding-defective (7), markedly reduced formation of octameric rods (Fig. 5C).
The next interface in the rod, that between Cdc12 and Cdc3, should be an N–C interface given the prior finding that the Cdc3 and Cdc12 CTEs are critical for their association and for rod and filament formation both in vivo and in vitro (5). In some EM images, cross-bridges between the paired filaments were seen (Fig. 4D Left). At higher magnification (Fig. 4D Right), these structures had a periodicity consistent with their arising from interaction of the paired parallel CTEs of Cdc3 and Cdc12 from one filament associating with the paired parallel CTEs of Cdc3 and Cdc12 from another filament, presumably forming an antiparallel four-helix bundle (Fig. 6). The distance between filaments in a pair (15–25 nm) is compatible with the calculated lengths of the CTEs of these two proteins, assuming that they are α-helical and associate primarily via overlap of their distal coiled coil-forming elements. These results provide further evidence that filaments pair in register via homotypic contacts.
Fig. 6.
Schematic diagram of the yeast septin rod, filament, and paired filament assemblies. The yeast heterooctameric septin complex is a linear rod with the subunits arrayed in the order and with the interfaces indicated. Each septin forms associations with its neighbors through either a G interface or an N–C interface. The rod is nonpolar because it has a two-fold axis of rotational symmetry running left-to-right between and orthogonal to the central pair of Cdc10 septins. The CTEs of the two Cdc3 and Cdc12 pairs project from the same face of the rod and presumably associate to form parallel coiled coils that are essential for rod stability because no rod assembly or filament formation occurs in their absence. Filaments form via end-on-end assembly of the rods mediated by an ionic strength-dependent Cdc11–Cdc11 interaction through an N–C interface. The CTE of Cdc11 is not required either for rod assembly or for filament formation. Filament formation and pairing in register are coupled, with the pairing presumably mediated by lateral association between the parallel Cdc3–Cdc12 coiled coil on one filament with a corresponding parallel Cdc3–Cdc12 coiled coil on the neighboring filament, thereby forming an antiparallel four-helix bundle.
Given this model, a possible explanation for the observed coupling between rod polymerization and filament pairing is that polymerization of octamers places the CTEs of Cdc3 and Cdc12 in a position to promote interaction between filaments and vice versa. A more speculative possibility is that, at high salt, contact of the Cdc12 CTE with the surface of Cdc11 may be favored over its pairing with the Cdc3 CTE. If so, the Cdc12 CTE may mask the interface of Cdc11 required for its homotypic interaction. If this intraoctamer contact is alleviated at low salt, the surface of Cdc11 necessary for rod polymerization would be freed simultaneously with release and pairing of the Cdc12 CTE with the Cdc3 CTE, which then can engage in lateral contacts between elongating filaments. This simple masking idea predicts that at low salt, the Cdc3 and Cdc12 CTEs in Cdc11-less hexamers (which do not polymerize into filaments) should be paired and available to mediate side-by-side association of the hexamers, which we do not observe. Maybe binding of the Cdc11 G domain to Cdc12 influences pairing of its CTE with that of Cdc3 or the orientation or conformation of the resulting coiled coil. Whether the Cdc3 and Cdc12 CTEs are essential for filament pairing or elongation cannot be tested easily because no stable octamers form in their absence (5).
Shs1 May Serve as an Alternative Terminal Subunit.
Shs1 (23, 24) is often considered an accessory septin because null alleles (_shs1_Δ) have a mild phenotype (22, 24) and because it is recovered in substoichiometric amounts in native septin complexes purified under certain conditions (14). However, Shs1 is the mitotic septin that seems the most heavily modified by both phosphorylation and SUMOylation (reviewed in ref. 6), and its state of modification is correlated with the dynamic behavior of the septin filaments at the mother-bud neck (25). Cdc11 and Shs1 occupy a distinct phylogenetic subfamily (1) and are, among the five mitotic septins, the most closely related in primary structure both within their G domains and over the first half of their CTEs. Moreover, various assays to assess Shs1 interaction with other septins demonstrated binding to Cdc12 (15, 24). Thus, Shs1 may be able to replace Cdc11 at the end of the rod. If so, the resulting Shs1-containing octamers may generate septin filaments with special features or the ability to recruit unique proteins, thus fine-tuning yeast cell morphogenesis (22), although Shs1 is not essential for viability. Hence, it will be interesting to examine whether these predictions about Shs1 are correct.
In conclusion, our results suggest that the subunit that mediates end-on-end interactions between heterooligomers may exert control over septin polymerization into filaments. In contrast to the human SEPT7–SEPT6–SEPT2–SEPT2–SEPT6–SEPT7 rod, which can form filaments through the G interface of SEPT7, the yeast Cdc11–Cdc12–Cdc3–Cdc10–Cdc10–Cdc3–Cdc12–Cdc11 rod achieves filament formation through the N–C interface of Cdc11 (and perhaps, as argued above, Shs1 may be able to mediate similar end-on-end contacts). Thus, the yeast heterooctamer displays an assembly interface distinct from that of the mammalian septin heterohexamers (18, 26). This difference suggests that although assembled filament structures share alternating G- and N–C interfaces (Fig. 6), discrete mechanisms may regulate assembly of different septin filaments in the same or different organisms. Of course, the SEPT7–SEPT6–SEPT2–SEPT2–SEPT6–SEPT7 complex used for structural studies may not be the native form in any mammalian cell type. If mammalian heterohexamers are capped in vivo at each end by an additional Cdc11-like subunit, they would closely resemble the yeast heterooctamer in overall organization and in polymerizing via an N–C mode of interaction.
Materials and Methods
Septin Complex Expression and Purification.
Complexes containing Cdc3, Cdc10, Cdc11, and (His)6-tagged Cdc12 were expressed in Escherichia coli as described before (5) and then purified by immobilized metal affinity, size exclusion, and ion exchange chromatography (see SI Methods). Cdc3–Cdc11–(His)6–Cdc12 (Cdc10-less), Cdc3–Cdc10-(His)6–Cdc12 (Cdc11-less), _MBP–Cdc3_–Cdc10–Cdc11–Cdc12, and Cdc10–GFP_–Cdc3–Cdc11–(His)6–_Cdc12 complexes were prepared the same way by using the appropriate vectors to express the indicated combinations of proteins.
EM and Image Processing.
Purified septin complexes were dialyzed against buffer [2 mM MgCl2, 50 mM Tris·HCl (pH 8)] containing either 300 mM or 10 mM KCl (or NaCl) by using an 10- to 50-μl capacity Spindialyzer (Harvard Apparatus). For the high-salt samples, the septin complexes were diluted to 0.01 mg/ml, adsorbed onto carbon-coated grids, and stained with 2% uranyl formate. See SI Methods for details on antibodies used for EM studies.
Samples were examined by using a Tecnai T12 microscope (FEI) operated at 120 kV, and images ertr collected by using film (cat. no. S0163; Kodak) at a magnification of ×30,000 and ≈1-μm underfocus. Micrographs were digitized on a Nikon Coolscan 8000 scanner with a pixel size of 4.23 Å at the sample level. Particles were classified and aligned to generate class averages by using SPIDER (27). Particles were windowed out into 135 × 135 pixels images by using the Boxer interface of EMAN (28) and appended into a single SPIDER file, then normalized against the background. One round of reference-free alignment and classification was performed before references were selected from the first class averages. Several rounds of multireference alignment and classification were then performed, and new references were selected from the class averages until no further improvement was obtained. The following numbers of particles were analyzed for each of the indicated complexes [tagged subunit(s) in italics]. Cdc3–Cdc10–Cdc11–(_His)6–_Cdc12 complexes: 4,929 particles were selected and processed into 202 classes. Cdc3–Cdc10(S46N)–Cdc11–(His)6–_Cdc12(T48N) complexes: 1,267 particles were selected and classified into 10 classes. Cdc3–Cdc11–(His)6–_Cdc12 (Cdc10-less) complexes: 824 particles were selected and classified into 10 classes. Cdc3–Cdc10–(His)6–_Cdc12 (Cdc11-less) complexes: 2,093 particles were selected and processed into 20 classes. Cdc3–_Cdc10–GFP_–Cdc11–(His)6–_Cdc12 (Cdc10-tagged) complexes: 3,377 particles were windowed out and classified into 108 classes. MBP–Cdc3_–Cdc10–Cdc11–(His)6–_Cdc12 (Cdc3-tagged) complexes: 2,630 particles were windowed out and classified into 117 classes. Cdc3–Cdc10–Cdc11–(His)6–_Cdc12 complexes stained with anti-(His)6 mAb: 2,796 particles were selected and classified into 32 classes, corresponding to the main shapes and orientations of the septin complex in the data set. An additional subclassification of each of these classes was obtained by using a mask selecting only for the data at the periphery of the particles. The resulting data were then segregated into 150 final classes, five of which (total of 49 particles; 2% of the total data set) showed a clear density with an antibody-like profile. Cdc3–Cdc10–Cdc11–(His)6–_Cdc12 complexes stained with polyclonal anti-Cdc12 antibodies: 3,969 particles were selected and classified into 150 classes. Cdc3–Cdc10–Cdc11–(His)6–_Cdc12 complexes stained with polyclonal anti-Cdc3 C-terminal peptide antibodies: 11,575 particles were selected and classified into 773 classes. For Cdc3–Cdc10–Cdc11–(His)6–_Cdc12 complexes stained with polyclonal anti-Cdc11 antibodies, the IgG was sufficiently well immobilized that it was readily visualized in raw images.
Filaments formed under low-salt conditions were also vitrified in liquid ethane by using a Vitrobot (FEI) in a 100% humidity environment on a Quantifoil grid (Structure Probes, Inc.). The specimen was transferred to a cryo-specimen holder (Gatan) and observed on a Tecnai F20 (FEI) by using 200 kV. Micrographs were recorded under low-dose conditions at a magnification of ×30,000 and underfocus of ≈3 μm.
Supplementary Material
Supporting Information
Acknowledgments.
We thank Mark Longtine, Douglas Kellogg, and Michael Knop for the communication of unpublished information, useful conversations, and/or the gift of reagents. This work was supported by Postdoctoral Research Fellowships 61-1357 (to A.B.) and 61-1295 (to M.A.M.) from the Jane Coffin Childs Memorial Fund for Medical Research, National Institutes of Health Predoctoral Traineeship GM07232 (to G.G.), National Institutes of Health–National Research Service Award Kirschstein Postdoctoral Fellowship GM69165 (to H.-l.N.), National Institutes of Health Research Grant GM48958 (to T.A.), National Institutes of Health Research Grant GM21841 (to J.T.), and the Agouron Foundation, Department of Energy Office of Biological and Environmental Research, and the Howard Hughes Medical Institute (to E.N.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Pan F, et al. Analysis of septins across kingdoms reveals orthology and new motifs. BMC Evol Biol. 2007;7:103.1–103.17. doi: 10.1186/1471-2148-7-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Versele M, Thorner J. Some assembly required: Yeast septins provide the instruction manual. Trends Cell Biol. 2005;15:414–424. doi: 10.1016/j.tcb.2005.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartwell LH. Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp Cell Res. 1971;69:265–276. doi: 10.1016/0014-4827(71)90223-0. [DOI] [PubMed] [Google Scholar]
- 4.Harbury PB, et al. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 5.Versele M, et al. Protein-protein interactions governing septin heteropentamer assembly and septin filament organization in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:4568–4583. doi: 10.1091/mbc.E04-04-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McMurray MA, Thorner J. In: The Septins. Hall PA, Russell SHE, Pringle JR, editors. Chicester, West Sussex, UK: John Wiley & Sons, Ltd.; 2008. in press. [Google Scholar]
- 7.Versele M, Thorner J. Septin collar formation in budding yeast requires GTP binding and direct phosphorylation by the PAK, Cla4. J Cell Biol. 2004;164:701–715. doi: 10.1083/jcb.200312070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casamayor A, Snyder M. Molecular dissection of a yeast septin: Distinct domains are required for septin interaction, localization, and function. Mol Cell Biol. 2003;23:2762–2777. doi: 10.1128/MCB.23.8.2762-2777.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J, et al. Phosphatidylinositol polyphosphate binding to the mammalian septin H5 is modulated by GTP. Curr Biol. 1999;9:1458–1467. doi: 10.1016/s0960-9822(00)80115-3. [DOI] [PubMed] [Google Scholar]
- 10.Dobbelaere J, Barral Y. Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science. 2004;305:393–396. doi: 10.1126/science.1099892. [DOI] [PubMed] [Google Scholar]
- 11.Takizawa PA, et al. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science. 2000;290:341–344. doi: 10.1126/science.290.5490.341. [DOI] [PubMed] [Google Scholar]
- 12.Gladfelter AS, et al. The septin cortex at the yeast mother-bud neck. Curr Opin Microbiol. 2001;4:681–689. doi: 10.1016/s1369-5274(01)00269-7. [DOI] [PubMed] [Google Scholar]
- 13.Frazier JA, et al. Polymerization of purified yeast septins: Evidence that organized filament arrays may not be required for septin function. J Cell Biol. 1998;143:737–749. doi: 10.1083/jcb.143.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mortensen EM, et al. Cell cycle-dependent assembly of a Gin4-septin complex. Mol Biol Cell. 2002;13:2091–2105. doi: 10.1091/mbc.01-10-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farkasovsky M, et al. Nucleotide binding and filament assembly of recombinant yeast septin complexes. Biol Chem. 2005;386:643–656. doi: 10.1515/BC.2005.075. [DOI] [PubMed] [Google Scholar]
- 16.Fields S, Sternglanz R. The two-hybrid system: An assay for protein-protein interactions. Trends Genet. 1994;10:286–292. doi: 10.1016/0168-9525(90)90012-u. [DOI] [PubMed] [Google Scholar]
- 17.O'Shea EK, et al. Preferential heterodimer formation by isolated leucine zippers from fos and jun. Science. 1989;245:646–648. doi: 10.1126/science.2503872. [DOI] [PubMed] [Google Scholar]
- 18.Sirajuddin M, et al. Structural insight into filament formation by mammalian septins. Nature. 2007;449:311–315. doi: 10.1038/nature06052. [DOI] [PubMed] [Google Scholar]
- 19.John CM, et al. The Caenorhabditis elegans septin complex is nonpolar. EMBO J. 2007;26:3296–3307. doi: 10.1038/sj.emboj.7601775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stewart M, et al. The structure of the Q69L mutant of GDP-Ran shows a major conformational change in the switch II loop that accounts for its failure to bind nuclear transport factor 2 (NTF2) J Mol Biol. 1998;284:1517–1527. doi: 10.1006/jmbi.1998.2204. [DOI] [PubMed] [Google Scholar]
- 21.Cid VJ, et al. Cell integrity and morphogenesis in a budding yeast septin mutant. Microbiology (Reading) 1998;144:3463–3474. doi: 10.1099/00221287-144-12-3463. [DOI] [PubMed] [Google Scholar]
- 22.Iwase M, et al. Shs1 plays separable roles in septin organization and cytokinesis in Saccharomyces cerevisiae. Genetics. 2007;177:215–229. doi: 10.1534/genetics.107.073007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carroll CW, et al. The septins are required for the mitosis-specific activation of the Gin4 kinase. J Cell Biol. 1998;143:709–717. doi: 10.1083/jcb.143.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mino A, et al. Shs1p: A novel member of septin that interacts with spa2p, involved in polarized growth in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1998;251:732–736. doi: 10.1006/bbrc.1998.9541. [DOI] [PubMed] [Google Scholar]
- 25.Dobbelaere J, et al. Phosphorylation-dependent regulation of septin dynamics during the cell cycle. Dev Cell. 2003;4:345–357. doi: 10.1016/s1534-5807(03)00061-3. [DOI] [PubMed] [Google Scholar]
- 26.Lukoyanova N, et al. 3D reconstruction of mammalian septin filaments. J Mol Biol. 2007;376:1–7. doi: 10.1016/j.jmb.2007.11.029. [DOI] [PubMed] [Google Scholar]
- 27.Frank J, et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 28.Ludtke SJ, et al. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information