Genome-wide Analysis Identifies Interleukin-10 mRNA as Target of Tristetraprolin (original) (raw)
Abstract
Tristetraprolin (TTP) is an RNA-binding protein required for the rapid degradation of mRNAs containing AU-rich elements. Targets regulated by TTP include the mRNAs encoding tumor necrosis factor-α, granulocyte-macrophage colony-stimulating factor, interleukin-2 (IL-2), and immediate early response 3. To identify novel target mRNAs of TTP in macrophages, we used a genome-wide approach that combines RNA immunoprecipitation and microarray analysis. A list was compiled of 137 mRNAs that are associated with TTP with an estimated accuracy on the order of 90%. Sequence analysis revealed a highly significant enrichment of AU-rich element motifs, with AUUUA pentamers present in 96% and UUAUUUAUU nonamers present in 44% of TTP-associated mRNAs. We further show that IL-10 is a novel target regulated by TTP. IL-10 mRNA levels were found to be elevated because of a reduced decay rate in primary macrophages from TTP-/- mice. Our study demonstrates the importance of experimental approaches for identifying targets of RNA-binding proteins.
Tristetraprolin (TTP)2 is an RNA-binding protein that recognizes AU-rich elements (AREs), which are characteristic for a group of short-lived mRNAs (for review, see Ref.1). Importantly, binding of TTP causes the rapid degradation of its target mRNAs. The first physiological target was identified by the careful analysis of mutant mice lacking TTP. Nullizygous mice develop an inflammatory syndrome characterized by cachexia, spontaneous arthritis, dermatitis, and neutrophilia (2). The inflammatory features of this syndrome are similar to those observed in TNFα transgenic mice (3), and neutralizing antibodies reactive with TNFα prevent most of the inflammatory symptoms in TTP-/- mice (2). Indeed macrophages derived from TTP-/- mice were found to overexpress TNFα as a consequence of a prolonged mRNA half-life. TTP was shown to specifically bind to the ARE of TNFα mRNA and cause its rapid degradation (4). Subsequent analysis of primary cells from TTP-/- mice showed that TTP has additional targets including granulocyte-macrophage colony-stimulating-factor and interleukin-2 (IL-2) mRNA (5,6). Immediate early response 3 mRNA has recently been identified as a novel target in TTP-/- mouse embryonic fibroblasts (MEFs) (7). Other possible targets of TTP were suggested by alternative approaches; knock down of TTP via RNA-interference showed that the rapid degradation of 1,4-galactosyltransferase 1, c-myc, cyclin D1, and vasculo-endothelial growth factor mRNA is dependent on TTP (8-10).In vivo binding as well as in vitro decay studies provided evidence that TTP destabilizes Pitx2 mRNA (11), and overexpression studies indicated that IL-3, IL-6, cyclooxygenase-2, plasminogen activator inhibitor type 2, and TTP mRNA itself may also be destabilized by TTP (12-16). Finally, knock down of TTP in macrophages was shown to cause increased production of IL-6, IL-12, and macrophage inflammatory protein-2, although mRNA stability was not addressed in this study (17). Because data base analysis suggests that up to 5-8% of all mRNAs possess putative AREs in their 3′-UTR (18), TTP has the potential to coordinately dampen the expression of a large number of transcripts. Indeed, a recent genome-wide analysis found 250 mRNAs to be stabilized with statistical significance in TTP-/- MEFs (7).
The ARE is a loosely defined cis element located in the 3′-untranslated region of many short-lived mRNAs. It was first characterized as a destabilizing element that promotes the degradation of granulocyte-macrophage colony-stimulating factor transcripts (19). The study of additional short-lived mRNAs led to a classification scheme based on the number and spacing of a canonical AUUUA pentamer (20). Class I AREs (e.g. c-myc, c-fos) have a few scattered pentamers, class II AREs (e.g. granulocyte-macrophage colony-stimulating factor, TNFα) have a cluster of 4-7 partially overlapping pentamers within a U-rich context, and class III AREs (e.g. c-jun) lack pentamers. The pentamer is critical to the function of class II AREs as point mutations within the pentamer prevent rapid mRNA decay (21). Class II AREs typically have an extended UUAUUUAUU nonamer, and this motif is also the minimal sequence that can effectively induce mRNA decay in reporter systems (22). The same nonamer also binds to TTP with maximum affinity in vitro (23) and is, thus, considered a canonical TTP binding site. However, these sequence features are not sufficient to reliably identify short-lived transcripts or predict the binding of destabilizing factors. A global analysis of mRNA decay rates found that only 10-15% of all mRNAs containing typical ARE motifs have a half-life below 2 h (24).
This situation calls for alternative approaches to identify mRNAs that are regulated at the level of mRNA stability. As an unbiased approach, RNA can be isolated from affinity-purified RNA-binding proteins (RNA immunoprecipitation (RIP)) and subjected to genome-scale microarray analysis (RIP-Chip) (25,26). This strategy has revealed the spectrum of mRNAs associated with several RNA-binding proteins including HuB, eIF4E, PABP (27), FMRP (28), Puf (29), TIA-1 (30), and TIAR (31). In this report we have used RIP-Chip to identify transcripts that associate with TTP in macrophages activated by lipopolysaccharide (LPS). Our results show that the AUUUA pentamer and the UUAUUUAUU nonamer are highly enriched in 3′-UTRs of TTP-associated mRNAs. By analyzing macrophages from TTP-/- mice we confirmed that a subset of TTP-associated mRNAs including IL-10 mRNA is regulated by TTP. The ability of TTP to sequentially suppress the expression of TNFα and IL-10 suggests that its activity may regulate both the activation and the deactivation phase of the inflammatory response.
EXPERIMENTAL PROCEDURES
_RIP_—RAW264.7 cells were lysed for 10 min on ice in RIP buffer containing 50 mm Tris, pH 8.0, 150 mm NaCl, 1 mm MgCl2, 1.0% Nonidet P-40, and 10% glycerol and freshly supplemented with 100 μm dithiothreitol, 1 mm sodium vanadate, 50 mm NaF, 20 nm okadaic acid, and EDTA-free complete protease inhibitors (Roche Applied Science). The lysate was cleared from nuclei by centrifugation for 2 min at 2000 × g before the addition of 1 mg/ml heparin (Sigma) and 10 units/ml RNasin (Promega). 10% of the lysate was removed to isolate the input RNA using Trizol-LS (Invitrogen). The remaining lysate was precleared for 1 h at 4 °C with 5% (v/v) protein A/G beads (Pierce). Beads were removed by centrifugation, and lysates were incubated with either rabbit-TTP (CARP-3) or rabbit hemagglutinin (HA.11, Covance) antibody (5 μg/ml) for 2 h. 5% (v/v) protein A/G beads were added for another 2 h and washed 8 times with RIP buffer before extraction of RNA using Trizol-LS. RIP using polysome lysis buffer was performed as described earlier (25).
_Microarray and Data Analysis_—RNA isolated from input as well as IP samples was checked for quality using a nanodrop spectrophotometer and an Agilent Bioanalyzer. 50 ng of isolated RNA was converted to labeled fragmented cRNA using the Affymetrix small sample protocol (Version II). Briefly, the RNA was converted to single-stranded cDNA using Superscript II reverse transcriptase and the GeneChip T7 promoter primer kit (Affymetrix). The single-stranded cDNA was converted to double-stranded cDNA using DNA polymerase I, DNA ligase, and RNase H from Escherichia coli. The double-stranded cDNA was subsequently purified using a cleanup kit from Affymetrix and converted to cRNA by in vitro transcription using the MEGAscript T7 polymerase kit (Ambion). The cRNA was further converted to double-stranded cDNA using a random primed first-strand synthesis and a T7-oligo(dT)-primed second step. This double-stranded cDNA was then cleaned up and in vitro transcribed to biotin-labeled cRNA. 35-200-base fragments were generated by metal-induced hydrolysis and hybridized to Affymetrix Mouse Genome 430 2.0 oligonucleotide arrays. After hybridization, the chip was washed, stained with streptavidin-phycoerythrin before being scanned. An antibody amplification staining protocol that uses biotinylated goat IgG followed by a second streptavidin-phycoerythrin staining increases the sensitivity of the assay. The chip was then scanned, and images were analyzed qualitatively using Affymetrix GeneChip Operating System software. RIP-Chip was repeated three times independently (biological repeats), and the signal intensities were background-corrected and summarized using two independent algorithms, MAS 5.0 (32) and gcRMA (33), as recommended by comparative analysis of microarray normalization methods (34-37). Algorithms were provided by the Affymetrix package (38) in Version 1.7 of the BioConductor suite (39). For each transcript, a retention coefficient (RC) was calculated by dividing the signal intensity from the TTP-IP RNA by the signal intensity from the input RNA. A false-positive likelihood was also calculated for each transcript by dividing the signal from the hemagglutinin-IP RNA sample by the signal from the input RNA. A list was assembled with all transcripts that had both RC(gcRMA) and RC(MAS5) values >4, and the average RC was calculated (R analysis script is available on request).
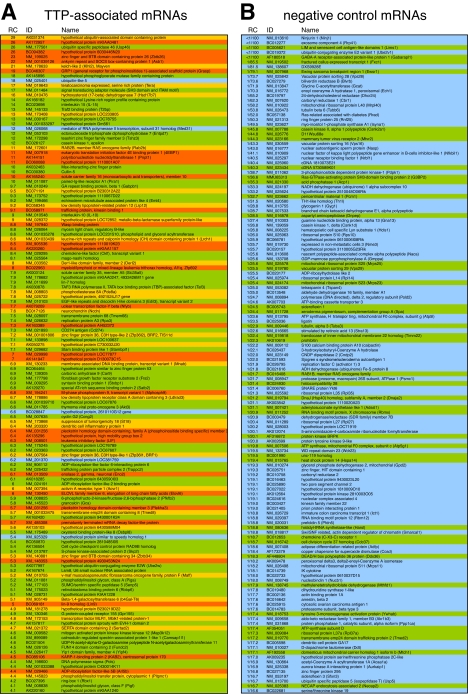
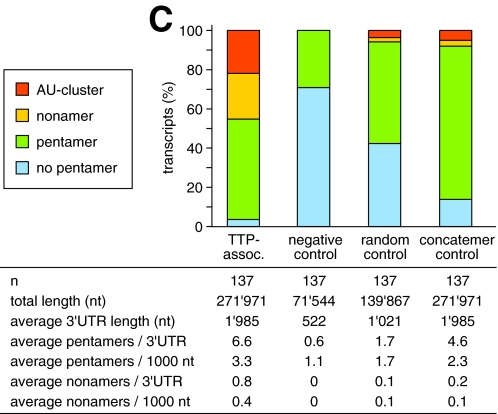
_3_′_-UTR Sequence Analysis_—Sequences retrieved from the Affymetrix array were blasted onwww.ncbi.nlm.nih.gov/BLAST to identify sense-matching full-length cDNAs. 3′-UTRs were defined at their upstream boundary by an annotated coding region and at their downstream boundary by the presence of a polyadenylation site within the last 50 nt of the cDNA, including non-canonical polyadenylation sites as described by Beaudoing et al. (40). Alternatively, the presence of ≥10 3′-terminal A residues was taken as evidence for a 3′-UTR downstream boundary. In some cases evidence for the 3′-UTR boundaries was obtained from alternative cDNA data base entries. AREs were assessed using an AU score as follows: AU score = 1 (blue inFig. 2 and supplemental Table S1) if no AUUUA pentamer is present in the 3′-UTR; AU score = 2 (green) if ≥1 AUUUA pentamer but no UUAUUUAUU nonamer is present; AU score = 3 (yellow) if ≥1 UUAUUUAUU nonamer but no AU cluster is present; AU score = 4 (orange) if ≥1 AU cluster is present, defined as a nonamer linked to a pentamer solely by U-residues (e.g. UUAUUUAUUUA or UUAUUUAUUAUUUA).
FIGURE 2.
Distribution of AU-rich elements in the 3′-UTRs of TTP-associated mRNAs. A, RNA was isolated from RAW264.7 cells stimulated for 2 h with LPS as in Fig. 1_B_, and TTP-associated mRNAs were identified by RIP-Chip. From 280 transcripts with RC values >4, a total of 137 full-length sense-matching mRNAs were identified by BLAST analysis, and their 3′-UTRs were analyzed for the presence of AU clusters (orange), UUAUUUAUU nonamers (yellow), AUUUA pentamers (green), or the absence of any of these motifs (blue). B, as a negative control, the 3′-UTRs of the 137 mRNAs with lowest RC values were analyzed in the same way. C, frequency of 3′-UTRs containing either AU clusters, UUAUUUAUU nonamers, AUUUA pentamers, or none of these motifs is plotted for the following groups; TTP-associated mRNAs as described in panel A, negative control mRNAs as described in panel B, mRNAs randomly chosen from the UCSC KnownGene murine mm8 data base (random control), and size-matched sequences of concatenated 3′-UTRs (concatemer control) from the UCSC KnowGene human hg17 assembly.
Cell Culture_—RAW264.7 and COS-7 cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal calf serum (Sigma) as well as 2 mml-glutamine, 100 units/ml penicillin (Sigma), and 100 μg/ml streptomycin (Sigma). COS-7 cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Bone marrow-derived macrophages (BMDM) were isolated from the femur and tibia of wt and TTP-/- C57BL/6 mice and cultured for 7-10 days in macrophage medium containing RPMI supplemented with 10% fetal calf serum (Sigma), 20 mm HEPES (Invitrogen), essential and non-essential amino acids (Invitrogen), 1 mm sodium pyruvate (Invitrogen), 55 μm 2-mercaptoethanol (Invitrogen), 10 mm sodium hydroxide, 100 units/ml penicillin (Sigma), and 100 μg/ml streptomycin (Sigma). For BMDM, 5% conditioned medium from L929 cells was added as a source of macrophage colony-stimulating-factor. LPS (L3755 from_E. coli 026:B6, Sigma) was used at a concentration of 10 ng/ml to stimulate RAW264.7 cells and at 100 ng/ml to stimulate BMDM. Blocking anti-mouse TNFα antibody (AF-410-NA, R&D Systems) was used at a concentration of 1 and 10 μg/ml. Freshly isolated spleen cells were stimulated with 100 nm 12-_O_-tetradecanoylphorbol-13-acetate (TPA) and 1 μm ionomycin in macrophage medium.
_Western Blot Analysis_—Cytoplasmic lysates were resolved by SDS-PAGE using 4-20% polyacrylamide gradient Tris-glycine gels (Invitrogen) as described previously (41). Goat α-TTP (sc-8458, Santa Cruz) and goat α-TIAR (sc-1749, Santa Cruz) were used as primary antibodies.
_Northern Blot Analysis_—Northern blots were prepared as described previously (42). Digoxigenin-labeled DNA probes were generated by PCR using custom reagents (Roche Applied Science), corresponding cDNAs, and the following primer pairs: 5′-glyceraldehyde phosphate dehydrogenase (GAPDH)/3′-GAPDH for GAPDH, G29/G30 for globin, G78/G79 for S7, G83/G84 for nucleolin (Ncl), G152/G153 for IL-10, G154/G155 for leukemia inhibitory factor (LIF), G198/G199 for RAN, and G254/G255 for IL-15. Digoxigenin-labeled RNA probes were generated by SP6 in vitro transcription using custom reagents (Roche Applied Science), and cDNA templates were generated with the following primer pairs: G314/G316 for TNFα, G366/G390 for ubiquitin specific peptidase 46 (Usp46), G370/G392 for Kelch-like protein 2 (Klhl2), G376/G395 for teratocarcinoma expressed protein (Tera), G380/G397 for AK166182, G382/G398 for TNF receptor-associated factor-2-binding protein (T2BP), G386/G400 for Gm561, and G388/G401 for eIF4E-binding protein 1 (4EBP1). All primer sequences are listed in supplemental Table S2.
_Plasmid Constructs_—Plasmid pTet-7B and pTet-7B-TNF-ARE have been described previously (42). For pTet-7B-IL-10-UTR, the murine IL-10 3′-UTR was amplified by reverse transcription-PCR using primers G192 and G193 and ligated as a BamHI - BglII fragment into the BglII site of pTet-7B. For pTet-7B-RAN-UTR, the murine RAN 3′-UTR was amplified by reverse transcription-PCR using primers G228 and G229 and ligated as a NheI-BglII fragment into the NheI-BglII sites of pTet-7B-IL-10-UTR. For pTet-7B-IL15-UTR, the murine IL15 3′-UTR was first amplified by reverse transcription-PCR using primers G196 and G197 followed by secondary PCR using primers G226 and G227. The product was ligated as a NheI-BglII fragment into the NheI-BglII sites of pTet-7B-IL-10-UTR. For pTet-7B-LIF-UTR, the murine LIF 3′-UTR was amplified by reverse transcription-PCR using primers G245 and G249. After secondary PCR with primers G232 and G262, the LIF 3′-UTR was ligated as a XbaI-BglII fragment into the NheI-BglII sites of pTet-7B-IL-10-UTR. Primer sequences are listed in supplemental Table S2.
_MTT Assay_—L929 cells were grown over night in 96-well plates at a density of 3.5 × 104 cells per well, and medium containing recombinant mouse TNFα (554589, BD Pharmingen) or TNFα-containing supernatants were added together with 1 μg/ml actinomycin D for another 24 h. After the addition of MTT (25 μg/ml) for 4 h, cells were lysed in 4% SDS/10% dimethyl formamide over night. Spectrophotometric absorbance was measured at 570 nm on a microtiter plate reader.
RESULTS
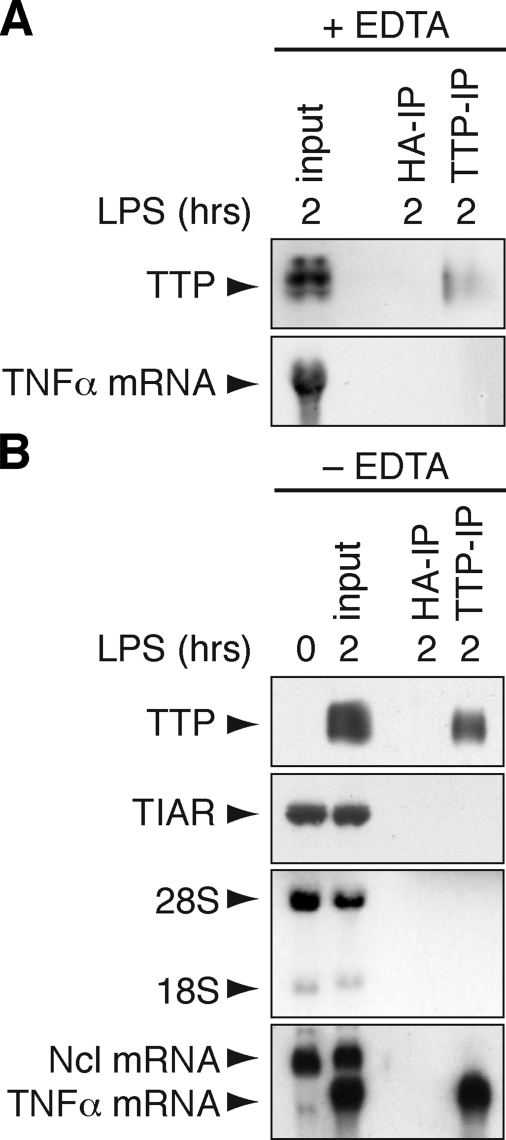
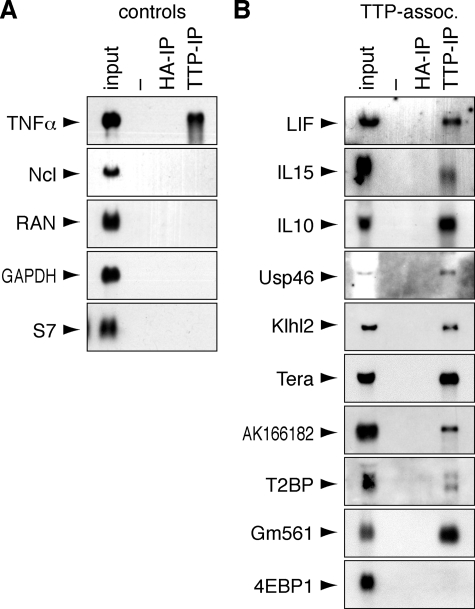
_RNA Immunoprecipitation of TTP-associated mRNA_—With the aim of isolating TTP along with its associated mRNAs, RAW264.7 macrophages were stimulated for 2 h with LPS to induce the expression of TTP. Importantly, stimulation by LPS also induces and stabilizes many ARE-containing mRNAs, which may be advantageous for their isolation. We first tested an EDTA-containing polysome lysis buffer that has been successfully used to isolate mRNAs associated with HuB, eIF4E, and PABP (27). TTP was immunoprecipitated using CARP-3 antibody (43) from the cytoplasmic lysate, yet recovery of TTP after IP was poor, and TNFα as a TTP-associated mRNA (4) could not be detected in the IP material (Fig. 1_A_). Because TTP is a zinc-finger protein and, therefore, requires zinc ions for structural integrity, we next tested a lysis buffer lacking EDTA (Fig. 1_B_). With this method, TTP was recovered efficiently after IP along with TNFα mRNA, whereas neither TTP nor TNFα mRNA was recovered with a control hemagglutinin antibody. Another ARE-binding protein, TIAR (44), was not immunoprecipitated with the TTP antibody. In contrast to TNFα mRNA, neither ribosomal 18 S/28 S RNA nor nucleolin mRNA (which does not contain an ARE) were detected in the IP material. We concluded that this method could efficiently and specifically isolate TTP-associated mRNAs. It is important to note, however, that this technique does not distinguish between mRNAs bound to TTP directly and mRNAs bound indirectly via other RNA-binding proteins that may interact with TTP.
FIGURE 1.
Immunoprecipitation of TTP-associated mRNA. A, RAW264.7 macrophages were stimulated for 2 h with LPS (10 μg/ml) before lysis in EDTA-containing polysome lysis buffer. IP was carried out with TTP antibody or hemagglutinin (HA) antibody as negative control. Recovery of TTP protein was monitored by Western blot analysis (upper panel), and TTP-associated TNFα mRNA was visualized by Northern blot analysis (lower panel). B, RNA immunoprecipitation was carried out as in panel A except that EDTA-free RIP buffer was used for lysis. Specificity of the IP was addressed by Western blot analysis of TTP and TIAR as an unrelated RNA-binding protein. 18 S and 28 S RNA were visualized using ethidium bromide staining, and TNFα mRNA was detected by Northern blot analysis together with Ncl mRNA that does not contain an ARE.
_Identification of TTP-associated mRNAs by RIP-Chip_—We next performed RIP-Chip analysis to identify TTP-associated mRNAs on a genome-wide scale. RAW264.7 macrophages were stimulated for 2 h with LPS, and RNA was isolated from both the cytoplasmic lysate (input) and the material immunoprecipitated with the TTP antibody (TTP-IP). To account for losses during the IP procedure, 10 times more lysate was used for the IP than for the input. The two RNA pools were converted into biotinylated cRNA and hybridized with Affymetrix Mouse Genome 430 2.0 oligonucleotide arrays.
RIP-Chip analysis of TTP was repeated three times, and the signal intensities were calculated using two independent algorithms (specified under “Experimental Procedures”). For each transcript, a RC was calculated by dividing the signal intensity from the TTP-IP by the signal intensity from the input RNA. From the 280 probe sets with average RC values >4, a total number of 137 sense-matching full-length mRNAs could be retrieved by BLAST analysis. These mRNAs were compiled in a final list of TTP-associated mRNAs (Fig. 2_A_ and supplemental Table S1).
_AREs Are Overrepresented among TTP-associated mRNAs_—Because the known target mRNAs of TTP contain recognizable AREs, we first examined the 3′-UTRs of the TTP-associated mRNAs for the presence of characteristic ARE motifs. An AU score was applied that distinguishes four categories (color-coded in Fig. 2_A_ and supplemental Table S1); presence of an AU cluster (orange), presence of ≥1 UUAUUUAUU nonamer (yellow), presence of ≥1 AUUUA pentamer (green), absence of AUUUA pentamers (blue). Among the TTP-associated mRNAs, the nonamer sequence, which is considered a canonical TTP binding site (23), occurs in 44% of the 3′-UTRs (Fig. 2_C_, AU cluster and nonamer-containing 3′-UTRs combined). 96% of the 3′-UTRs contain at least one pentamer, whereas only 4% do not contain the pentamer motif. As a negative control group we analyzed the 137 mRNAs with the lowest RC values (Fig. 2_B_). None of the negative control mRNAs contain an AU cluster or a nonamer in the 3′-UTR, 29% contain ≥1 pentamer, and 71% do not contain any pentamer in the 3′-UTR. This comparison shows that typical ARE motifs are vastly overrepresented among the TTP-associated mRNAs. It is also noteworthy that the 3′-UTRs of the TTP-associated mRNAs are on average 4 times longer than those of the non-associated mRNAs (1985 versus 522 nt).
To compensate for this difference, a second control group was generated with 137 murine mRNAs randomly selected from the UCSC KnownGene (45) data base (Fig. 2_C_, random control). In this group the average 3′-UTR (1021 nt) was still about half as long as in the group of TTP-associated mRNAs (1985 nt). A third control group with sequences identical in length to the group of TTP-associated 3′-UTRs was generated from the concatenated string of all KnownGene human 3′-UTRs (Fig. 2_C_, concatemer control). In the latter two control groups, the frequency of 3′-UTRs with ≥1 nonamer was 6 and 8%, respectively, compared with a frequency of 44% among the TTP-associated 3′-UTRs. Taken together, all three control groups show that AU clusters, nonamers, and pentamers are strongly enriched among the TTP-associated mRNAs identified by RIP-Chip.
Another way to address the distribution of ARE motifs among the TTP-associated mRNAs is to calculate the statistical likelihood of finding 44% nonamers and 96% pentamers in a random set of 137 3′-UTRs. Among the 35,728 3′-UTRs of all genes represented on the Mouse Genome 430 2.0 oligonucleotide array, the observed frequency of pentamers is 57%, and the likelihood of randomly choosing a set with a frequency of 96% is 8.4 × 10-26. Likewise, the observed frequency of nonamers is 5%, and the likelihood of randomly choosing a set with a frequency of 44% is 3.3 × 10-42. In other words the TTP-associated mRNAs represent a unique population specifically enriched for pentamers and nonamers. This finding is in good agreement with the notion that pentamers, and in particular nonamers, are core motifs within AREs and important for TTP binding.
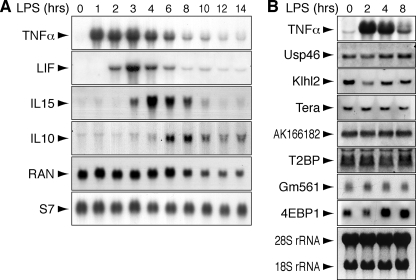
_Confirmation of TTP-associated mRNAs_—To further validate the list of TTP-associated mRNAs, we selected 10 candidate mRNAs (Table 1) and examined their expression by Northern blot analysis. Three cytokine mRNAs encoding LIF, IL-15, and IL-10 showed a transient pattern of expression in LPS-stimulated RAW264.7 macrophages (Fig. 3_A_). Whereas induction of TNFα mRNA was maximal within 1 h, LIF, IL-15, and IL-10 mRNAs reached peak levels after 3, 4, and 6 h, respectively. Seven candidate mRNAs, encoding Usp46, Klhl2, Tera, hypothetical protein AK166182, T2BP, hypothetical protein Gm561, and 4EBP1, showed a constitutive pattern of expression in RAW264.7 cells (Fig. 3_B_). We used RIP followed by Northern blot analysis to determine whether the candidate mRNAs bind to TTP. Macrophages were activated for 2 h with LPS, except in the case of IL-15 and IL-10, where we took later time points (4 and 8 h, respectively) to account for their delayed induction. Four non-associated control transcripts encoding Ncl, RAN, glyceraldehyde phosphate dehydrogenase (GAPDH), and ribosomal protein S7 were not retained by TTP-IP (Fig. 4_A_). Of the 10 candidate mRNAs, 9 were confirmed to bind to TTP in this assay, and only 4EBP1 mRNA was not retained by TTP-IP (Fig. 4_B_). Taken together, these results indicate that the true positive rate of TTP-associated mRNAs in supplemental Table S1 is on the order of 90%.
TABLE 1.
Selection of candidate mRNAs associated with TTP
FIGURE 3.
Expression profile of TTP-associated mRNAs in LPS-stimulated RAW264.7 macrophages. A, the induction of TTP-associated cytokine mRNAs encoding TNFα, LIF, IL-15, and IL-10 after LPS stimulation was determined by Northern blot analysis. Total RNA was extracted from RAW264.7 cells at different intervals after treatment with LPS (10 ng/ml). Non-associated mRNAs encoding RAN and S7 serve as controls. B, TTP-associated mRNAs encoding Usp46, Klhl2, Tera, AK166182, T2BP, Gm561, and 4EBP1 were analyzed as in panel A and show constitutive expression in LPS-stimulated RAW264.7 cells. TNFα mRNA induction is shown as control for LPS activity, and ribosomal 28 S and 18 S RNAs are shown as loading controls.
FIGURE 4.
Confirmation of TTP-mRNA binding by RIP and Northern blot analysis. A, binding of control mRNAs to TTP was analyzed by RIP from RAW264.7 cells stimulated for 2 h with 10 ng/ml LPS. The controls include TNFα mRNA known to bind to TTP as well as four non-associated mRNAs encoding Ncl, RAN, glyceraldehyde phosphate dehydrogenase (GAPDH), and S7. mRNAs were visualized by Northern blot analysis. HA, hemagglutinin.B, ten candidate transcripts (Table 1) were selected from the list of 137 TTP-associated mRNAs and tested for binding to TTP as in_panel A_. Of the 10 mRNAs tested, 9 are retained by RIP, whereas 4EBP1 mRNA is not retained. RNA was extracted from RAW264.7 cells stimulated for 2 h with 10 ng/ml LPS. In the case of IL-15 and IL-10, cells were stimulated for 4 and 8 h, respectively, to account for the delayed induction of these cytokines.
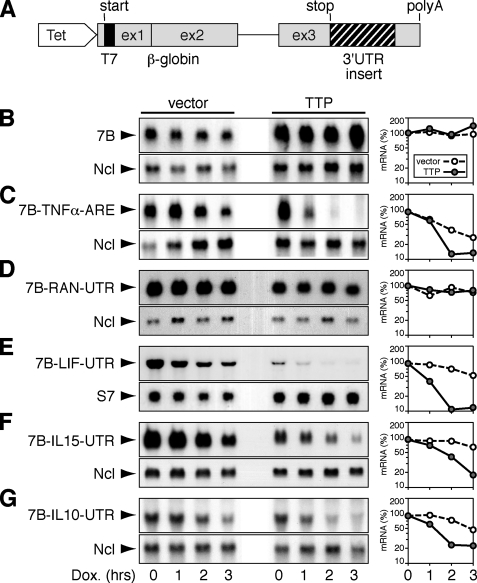
_TTP-mediated Decay of Reporter mRNAs_—As a first test of whether newly identified TTP-associated mRNAs would be targeted by TTP for decay, we generated β-globin reporter constructs containing the 3′-UTRs of either IL-10, IL-15, or LIF (Fig. 5_A_). The three sequences differ strongly in the number of ARE motifs. Whereas the IL-15 3′-UTR contains only one AUUUA pentamer, the IL-10 3′-UTR contains 6 pentamers, and the LIF 3′-UTR contains an AU cluster in addition to 5 pentamers (Table 1). Reporter constructs were transfected into COS-7 cells together with vector alone or with TTP. Because the reporter is attached to a tetracycline-sensitive promoter, its transcription can be blocked without affecting global transcription. The basic reporter mRNA termed 7B, which contains the coding sequence and 3′-UTR of β-globin, was stable both in the absence and presence of TTP (Fig. 5_B_). As a positive control, we tested the same reporter containing the ARE of TNFα. 7B-TNFα-ARE mRNA was labile, and co-expression of TTP strongly enhanced its decay (Fig. 5_C_). As a negative control, we inserted the RAN 3′-UTR, which was not found to associate with TTP. The 7B-RAN-UTR mRNA was stable both in the absence and presence of co-transfected TTP (Fig. 5_D_). We then tested the 3′-UTRs of LIF (Fig. 5_E_), IL-15 (Fig. 5_F_), and IL-10 (Fig. 5_G_). In all three cases, the reporter mRNA degraded weakly in the absence and strongly in the presence of co-transfected TTP. This result shows that TTP can promote the decay of the three transcripts in a heterologous system, indicating that the 3′-UTRs of LIF, IL-15, and IL-10 directly bind to TTP.
FIGURE 5.
Decay analysis of reporter mRNAs containing TTP-associated 3′-UTRs. A, schematic representation of the β-globin reporter gene pTet-7B. β-Globin sequences are shown in_gray_, the N-terminal T7 tag is in black. TTP-associated 3′-UTRs (hatched) were inserted between the stop codon and the β-globin 3′-UTR. A tetracycline-sensitive promoter allows specific inhibition of transcription of the reporter gene using a Tet-Off transactivator and doxycycline (Dox). B, COS-7 cells were transiently transfected with pTet-7B, Tet-Off, and either vector alone or a TTP-expressing plasmid. After 48 h, total RNA was isolated at 1-h intervals upon treatment with doxycycline (1 μg/ml). 7B reporter mRNA was visualized by Northern blot analysis using a globin probe; Ncl served as loading control. Signal intensities of 7B relative to Ncl were plotted against time in the graph. The same analysis was conducted for the 7B reporter mRNA containing the ARE of TNFα (C), the 3′-UTR of RAN (D), the 3′-UTR of LIF (E), the 3′-UTR of IL-15 (F), and the 3′-UTR of IL-10 (G).
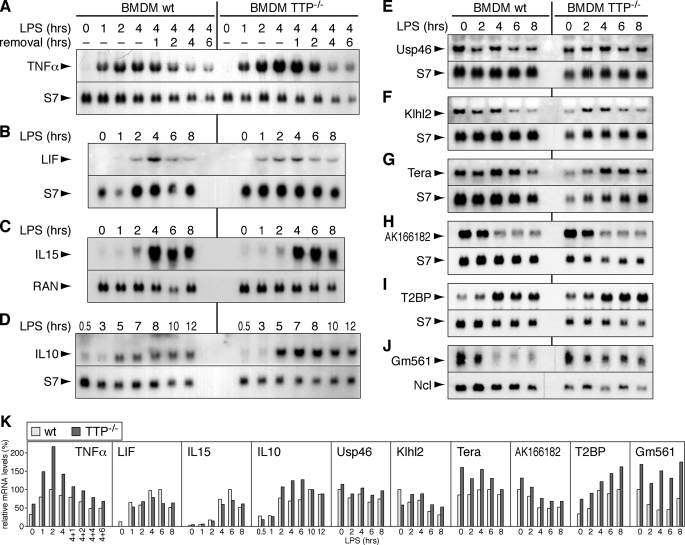
Elevated Levels of Some TTP-associated mRNAs in TTP_-/_- _Cells_—We next wanted to examine whether the endogenous expression levels of TTP-associated mRNAs would be altered in cells lacking TTP. To this end, we analyzed the nine mRNAs for which association with TTP was confirmed (Fig. 4_B_) in primary BMDM from wt and TTP-/- mice (Fig. 6). As a positive control, TNFα mRNA was examined in LPS-treated BMDM (Fig. 6_A_). Its expression was confirmed to be elevated in TTP-/- BMDM compared with wt BMDM, consistent with the established role of TTP in degrading TNFα mRNA (4). Quantification of the Northern blot signals is shown inFig. 6_K_. The expression levels of LIF and IL-15 mRNA were essentially the same in wt and TTP-/- BMDM (Fig. 6, B and C). In contrast, IL-10 mRNA levels were higher in TTP-/- BMDM, indicating that its expression is under control of TTP (Fig. 6_D_). Whereas the constitutively expressed Usp46 and Klhl2 mRNAs were expressed at similar levels in wt and TTP-/- BMDM, Tera, AK166182, T2BP, and Gm561 mRNAs showed a tendency toward higher expression levels in TTP-/- BMDM (Fig. 6, E-J).
FIGURE 6.
Expression profiles of TTP-associated mRNAs in wt and TTP-**/**- BMDM. BMDM were isolated from wt and TTP-/- C57BL/6 mice and cultured for 7-10 days, and total RNA was extracted after stimulation with 100 ng/ml LPS for the indicated time intervals. Northern blot analysis shows the expression of the following mRNAs: A, TNFα; B, LIF; C, IL-15;D, IL-10; E, Usp46; F, Klhl2; G, Tera;H, AK166182; I, T2BP; J, Gm561. The mRNAs encoding S7, RAN, or Ncl serve as loading controls. K, signal intensities in_panels A-J_ were quantified, normalized to the loading control signals, and plotted relative to the maximum level (100%) in wt BMDM.
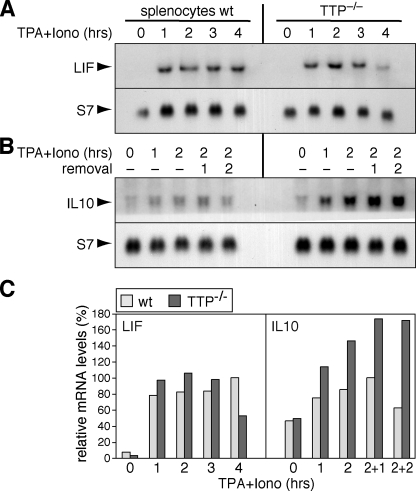
To verify the difference we observed between the two cytokines LIF and IL-10, we also analyzed freshly isolated spleen cells. After stimulation with TPA and ionomycin, LIF mRNA levels were again found to be similar in wt and TTP-/- splenocytes (Fig. 7_A_), whereas IL-10 mRNA levels were clearly elevated in TTP-/- splenocytes (Fig. 7_B_, quantification inFig. 7_C_).
FIGURE 7.
Expression profiles of TTP-associated mRNAs in wt and TTP-**/**- splenocytes. Spleen cells were isolated from wt and TTP-/- mice and immediately stimulated with 100 nm TPA and 1 μm ionomycin (iono). Total RNA was extracted and subjected to Northern blot analysis using S7 mRNA as loading control. A, the expression of LIF mRNA was analyzed after stimulation with TPA and ionomycin for 0, 1, 2, 3, and 4 h. B, the expression of IL-10 mRNA was analyzed after stimulation with TPA and ionomycin for 0, 1, and 2 h followed by removal of the drugs for 1 and 2 h. C, signal intensities in panels A and B were quantified, normalized to the loading control signals, and plotted relative to the maximum level (100%) in wt splenocytes.
Another caveat may arise from the fact that in LPS-stimulated RAW264.7 macrophages and BMDM, IL-10 mRNA expression is induced 3-5 h after the induction of TNFα mRNA (see Figs.3 and6). Because TNFα itself can activate the expression of IL-10 in monocytes (46), it is conceivable that the elevated expression of IL-10 mRNA is due to the increased amounts of TNFα secreted by TTP-/- BMDM. To test this possibility, BMDM were stimulated for 5 h with LPS in the absence or presence of blocking TNFα antibody. Supplemental Fig. S1_A_ shows that the addition of blocking TNFα antibody at two different concentrations did not reduce the expression of IL-10 mRNA in either wt or TTP-/- BMDM. The antibody was effective at both concentrations as it fully blocked TNFα activity in an apoptosis assay (supplemental Fig. S1_B_). We concluded that elevated IL-10 mRNA expression in TTP-/- BMDM is not a secondary effect of increased TNFα secretion.
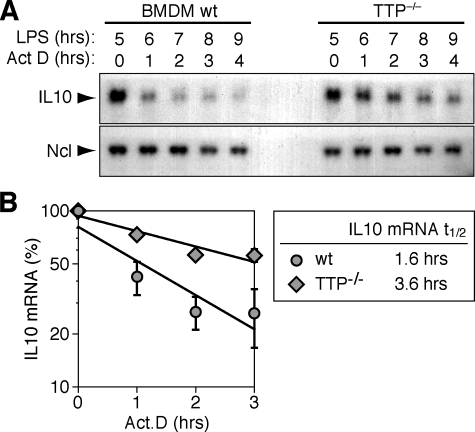
_TTP Accelerates IL-10 mRNA Decay_—To test whether IL-10 mRNA expression levels in TTP-/- BMDM were elevated due to a difference in half-life, we measured its decay rate in BMDM stimulated for 5 h with LPS. Whereas IL-10 mRNA showed a rapid decay in wt BMDM, it was clearly more stable in TTP-/- BMDM (Fig. 8_A_). Quantification of 3 repeat experiments showed that the half-life increased from 1.6 h in wt BMDM to 3.6 h in TTP-/- BMDM (Fig. 8_B_), a difference that is statistically significant (p = 0.016, n = 3). Therefore, stabilization of the mRNA is responsible for the accumulation of IL-10 mRNA in TTP-/- BMDM, indicating that TTP acts directly on this target by accelerating its degradation.
FIGURE 8.
IL-10 mRNA is stabilized in TTP-**/**- BMDM. A, BMDM isolated from wt and TTP-/- mice were stimulated for 5 h with LPS (100 ng/ml) prior to blocking transcription with actinomycin D (5 μg/ml). Total RNA was extracted at one h intervals, and IL-10 mRNA expression was monitored by Northern blot analysis. Ncl mRNA serves as loading control. B, Quantification of IL-10 mRNA decay in wt and TTP-/- BMDM stimulated for 5 h with LPS. Northern blot signal intensities were measured on autoradiograms using a digital camera and Chemiimager software. IL-10 mRNA signals were normalized to Ncl, and the value at 0 h actinomycin D was set 100%. Average percentages ± S.E. of 3 independent experiments were plotted against time, and exponential decay curves were calculated by best fit criteria.
DISCUSSION
In this study we have chosen a genome-wide approach to identify targets of TTP. Using RIP-Chip analysis, we found 280 transcripts that are retained by IP of TTP in LPS-stimulated macrophages. BLAST analysis of these transcripts allowed us to compile a list of 137 full-length mRNAs with defined coding and 3′-untranslated regions (Fig. 2_A_ and supplemental Table S1). Confirmation of binding by RIP and Northern blot analysis (Fig. 4) indicated that this list of TTP-associated mRNAs has an accuracy on the order of 90%.
A prominent absentee from this list is TNFα mRNA despite the fact that this mRNA is a known target of TTP (4) and efficiently co-immunoprecipitates with TTP (Fig. 1_B_). Most likely, this is due to the large variability in the quality and specificity of the Affymetrix probe sets. Also, we have chosen to analyze the data with a high specificity but low sensitivity approach by using very stringent criteria. This allows us to extract from these data a confident yet incomplete set of targets.
A second observation is that TTP-associated mRNAs encode a broad spectrum of proteins engaged in a variety of cellular processes. Supplemental Table S3 shows the most frequent functional groups as defined by gene ontology terms. To address whether certain groups are overrepresented, we compared the frequency among the TTP-associated mRNAs to the frequency among total mRNAs expressed in RAW264.7 macrophages stimulated for 2 h with LPS (supplemental Table S4). As an example, this comparison reveals that mRNAs encoding for proteins involved in development are about 2.5 times more abundant among the TTP-associated mRNAs compared with the total expressed mRNAs. We further noticed that the mRNAs encoding BRF1 and BRF2, two homologues of TTP, are in the list of TTP-associated mRNAs. BRF1 and BRF2 share highest homology with TTP in their RNA binding zinc finger domains; both proteins interact with AREs in a fashion similar to TTP, and both induce rapid mRNA degradation (47,48). This indicates that TTP may be part of a negative feedback mechanism, by which the induction of TTP dampens the expression of its related proteins BRF1 and BRF2. Via a similar autoregulatory loop, TTP has been proposed to down-regulate its own mRNA expression (16), although this notion could not be confirmed in TTP-/- MEFs (7).
A search for motifs that are characteristic for AREs revealed that AUUUA pentamers, UUAUUUAUU nonamers, and AU clusters are highly overrepresented among TTP-associated mRNAs (Fig. 2). The difference is most dramatic when compared with a group of 137 mRNAs with lowest RC values. Among this group of negative control mRNAs, not a single one contains a nonamer (or AU cluster, which by our definition contains a nonamer), whereas 44% of TTP-associated mRNAs contain at least one nonamer. In two additional control groups, the nonamer occurs in 6 and 8% of randomly selected or size-matched concatenated 3′-UTRs, respectively. Given their random nature, the latter two control groups may also contain TTP-associated 3′-UTRs yet only at a basal frequency. The presence of a pentamer alone does not appear to be a good predictor of whether an mRNA associates with TTP, since the average number of pentamers/3′-UTR is 6.6 for TTP-associated mRNAs and 4.6 for the size-matched concatemer control (Fig. 2_C_). In contrast, the nonamer seems to have a stronger predictive power, since the average number of nonamers/3′-UTR is 4 times higher for TTP-associated mRNAs (0.8) compared with the concatamer control (0.2).
Overall, our analysis of TTP-associated mRNAs confirms the known criteria of functional AREs, with pentamers being important but alone not sufficient, whereas nonamers and more extended AU clusters are indeed core motifs of many AREs. Yet our analysis also indicates that nonamers and AU clusters are not sufficient to predict the entire spectrum of TTP-associated mRNAs, of which more than half (56%) do not contain the nonamer. Among the 10 mRNAs that were tested by Northern blot analysis (summarized inTable 1), the 5 mRNAs lacking a nonamer all bound to TTP. This includes IL-10 mRNA whose stability was further shown to be directly regulated by TTP (Fig. 8). The IL-10 3′-UTR contains six AUUUA pentamers, four of which are surrounded by U residues forming octameric motifs that are very close to the nonamer (supplemental Fig. S2). These octameric motifs resemble TTP binding sites that were identified in the 3′-UTRs of immediate early response 3 and TTP itself (7,16).
On the other hand, the single mRNA in our sample that did not bind TTP (encoding 4EBP1) does in fact have a AU cluster (which by our definition includes a nonamer). A similar observation was made in TTP-/- MEFs, where only 33 of 250 stabilized mRNAs contained two or more heptameric UAUUUAU core TTP binding sites (7). At this point we can only speculate that besides nonamers (or heptamers) and AU clusters, more loosely defined sequences or secondary structures may also support TTP binding. In addition, some transcripts may also interact with TTP indirectly via other RNA-binding proteins.
An unexpected result of our study was that the expression levels of some TTP-associated mRNAs were not altered in TTP-/- macrophages. Among the cytokine mRNAs tested, IL-10 mRNA was found to be elevated in TTP-/- macrophages and spleen cells, whereas IL-15 and LIF mRNAs were not (Figs. 6 and7). As a trivial explanation, TTP could bind to mRNAs that are not physiological targets after cell lysis, as has been observed for HuR-bound transcripts (49). Although we cannot formally rule out this possibility, the fact that TTP promotes decay of reporter mRNAs containing the 3′-UTRs of IL-15 and LIF (Fig. 5) indicates that TTP does bind to these 3′-UTRs within intact cells.
A second explanation for IL-15 and LIF mRNAs not being elevated in TTP-/- BMDM could be functional redundancy among the TTP/BRF family of proteins. Because BRF1 and BRF2 also recognize AREs and promote mRNA decay (47,48), they may compensate for the lack of TTP. In turn, one would have to postulate that mRNAs whose degradation is uniquely dependent on TTP (e.g. TNFα, granulocyte-macrophage colony-stimulating factor, IL-2, and IL-10) are not bound by BRF1/2. A recent study provides an example for such differential specificity; immediate early response 3 mRNA is stabilized in TTP-/- MEFs but not in BRF1-/- MEFs (7).
A third possibility is that some of the TTP-associated mRNAs may bind to TTP indirectly via other RNA-binding proteins. Inducible nitric-oxide synthase mRNA is such an example, as it interacts with TTP indirectly via the RNA-binding protein KSRP (50). In this case TTP does not induce degradation of inducible nitric-oxide synthase mRNA but, rather, seems to have a stabilizing function by antagonizing KSRP. However, our data with the reporter mRNAs (Fig. 5) indicate that the 3′-UTRs of IL-15 and LIF are directly bound by TTP.
Finally, it is possible that IL-15 and LIF mRNAs interact with additional RNA-binding proteins that are antagonistic to the destabilizing function of TTP. The fact that overexpression of TTP in a heterologous system accelerates the degradation of the LIF and IL-15 reporter mRNAs (Fig. 5) supports the notion that TTP-associated mRNAs are sensitive to the balance of different ARE-binding proteins. Such a combinatorial scenario would suggest that under physiological conditions, the composition of the entire RNA-protein complex rather than the binding of an individual protein determines the fate of an mRNA. This concept is consistent with post-transcriptional operon networks in eukaryotic systems that are regulated by groups of RNA-binding proteins, whose binding in addition may be modulated by regulatory RNAs (51,52). Given the complexity of this regulatory network and our limited capacity to predict targets for RNA-binding proteins, experimental approaches such as RIP-Chip together with genetic knock-out or RNA-interference-mediated knockdown models remain indispensable for deciphering the code of RNA-protein complexes.
Acknowledgments
We thank William F. Rigby (Dartmouth Medical School, Lebanon, NH) for generously providing the CARP-3 antibody and Jochen Kreth (German Cancer Research Center, Heidelberg) for technical assistance.
*
This work was supported by National Institutes of Health Grants AI-33600 and AI-50167 (to P. A.), National Human Genome Research Institute, National Institutes of Health Grants U01HG004571 and R21HG003679 (to S. A. T.), and a Young Investigators Grant from the Helmholtz Association of German Research Centres (to G. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “_advertisement_” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available athttp://www.jbc.org) contains supplemental Tables S1-S4 and Figs. S1 and S2.
Footnotes
2
The abbreviations used are: TTP, tristetraprolin; 3′-UTR, 3′-untranslated region; ARE, adenosine-uridine-rich element; BMDM, bone marrow-derived macrophages; IL, interleukin; IP, immunoprecipitation; Klhl2, Kelch-like protein 2; LIF, leukemia inhibitory factor; LPS, lipopolysaccharide; MEF, mouse embryonic fibroblast; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide; Ncl, nucleolin; RC, retention coefficient; RIP, RNA immunoprecipitation; T2BP, TNF receptor-associated factor-2 binding protein; Tera, teratocarcinoma expressed protein; TNFα, tumor necrosis factor-α; TPA, 12-_O_-tetradecanoylphorbol-13-acetate; wt, wild type; Usp46, ubiquitin-specific peptidase 46; 4EBP1, eIF4E-binding protein 1.
References
- 1.Blackshear, P. J. (2002) Biochem. Soc. Trans. 30 945-952 [DOI] [PubMed] [Google Scholar]
- 2.Taylor, G. A., Carballo, E., Lee, D. M., Lai, W. S., Thompson, M. J., Patel, D. D., Schenkman, D. I., Gilkeson, G. S., Broxmeyer, H. E., Haynes, B. F., and Blackshear, P. J. (1996) Immunity 4 445-454 [DOI] [PubMed] [Google Scholar]
- 3.Keffer, J., Probert, L., Cazlaris, H., Georgopoulos, S., Kaslaris, E., Kioussis, D., and Kollias, G. (1991) EMBO J. 10 4025-4031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carballo, E., Lai, W. S., and Blackshear, P. J. (1998) Science 281 1001-1005 [DOI] [PubMed] [Google Scholar]
- 5.Carballo, E., Lai, W. S., and Blackshear, P. J. (2000) Blood 95 1891-1899 [PubMed] [Google Scholar]
- 6.Ogilvie, R. L., Abelson, M., Hau, H. H., Vlasova, I., Blackshear, P. J., and Bohjanen, P. R. (2005) J. Immunol. 174 953-961 [DOI] [PubMed] [Google Scholar]
- 7.Lai, W. S., Parker, J. S., Grissom, S. F., Stumpo, D. J., and Blackshear, P. J. (2006) Mol. Cell. Biol. 26 9196-9208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gringhuis, S. I., Garcia-Vallejo, J. J., van Het Hof, B., and van Dijk, W. (2005) Mol. Cell. Biol. 25 6454-6463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marderosian, M., Sharma, A., Funk, A. P., Vartanian, R., Masri, J., Jo, O. D., and Gera, J. F. (2006) Oncogene 25 6277-6290 [DOI] [PubMed] [Google Scholar]
- 10.Essafi-Benkhadir, K., Onesto, C., Stebe, E., Moroni, C., and Pages, G. (2007) Mol. Biol. Cell. 18 4648-4658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Briata, P., Ilengo, C., Corte, G., Moroni, C., Rosenfeld, M. G., Chen, C. Y., and Gherzi, R. (2003) Mol. Cell 12 1201-1211 [DOI] [PubMed] [Google Scholar]
- 12.Stoecklin, G., Ming, X. F., Looser, R., and Moroni, C. (2000) Mol. Cell. Biol. 20 3753-3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stoecklin, G., Stoeckle, P., Lu, M., Muehlemann, O., and Moroni, C. (2001) RNA 7 1578-1588 [PMC free article] [PubMed] [Google Scholar]
- 14.Yu, H., Stasinopoulos, S., Leedman, P., and Medcalf, R. L. (2003) J. Biol. Chem. 278 13912-13918 [DOI] [PubMed] [Google Scholar]
- 15.Sawaoka, H., Dixon, D. A., Oates, J. A., and Boutaud, O. (2003) J. Biol. Chem. 278 13928-13935 [DOI] [PubMed] [Google Scholar]
- 16.Brooks, S. A., Connolly, J. E., and Rigby, W. F. (2004) J. Immunol. 172 7263-7271 [DOI] [PubMed] [Google Scholar]
- 17.Jalonen, U., Nieminen, R., Vuolteenaho, K., Kankaanranta, H., and Moilanen, E. (2006) Mediators Inflamm. 2006 1-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bakheet, T., Williams, B. R., and Khabar, K. S. (2003) Nucleic Acids Res. 31 421-423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaw, G., and Kamen, R. (1986) Cell 46 659-667 [DOI] [PubMed] [Google Scholar]
- 20.Chen, C.-Y. A., and Shyu, A.-B. (1995) Trends Biochem. Sci. 20 465-470 [DOI] [PubMed] [Google Scholar]
- 21.Stoecklin, G., Hahn, S., and Moroni, C. (1994) J. Biol. Chem. 269 28591-28597 [PubMed] [Google Scholar]
- 22.Zubiaga, A. M., Belasco, J. G., and Greenberg, M. E. (1995) Mol. Cell. Biol. 15 2219-2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brewer, B. Y., Malicka, J., Blackshear, P. J., and Wilson, G. M. (2004) J. Biol. Chem. 279 27870-27877 [DOI] [PubMed] [Google Scholar]
- 24.Yang, E., van Nimwegen, E., Zavolan, M., Rajewsky, N., Schroeder, M., Magnasco, M., and Darnell, J. E., Jr. (2003) Genome Res. 13 1863-1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tenenbaum, S. A., Lager, P. J., Carson, C. C., and Keene, J. D. (2002) Methods 26 191-198 [DOI] [PubMed] [Google Scholar]
- 26.Keene, J. D., Komisarow, J. M., and Friedersdorf, M. B. (2006) Nat. Protoc. 1 302-307 [DOI] [PubMed] [Google Scholar]
- 27.Tenenbaum, S. A., Carson, C. C., Lager, P. J., and Keene, J. D. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 14085-14090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown, V., Jin, P., Ceman, S., Darnell, J. C., O'Donnell, W. T., Tenenbaum, S. A., Jin, X., Feng, Y., Wilkinson, K. D., Keene, J. D., Darnell, R. B., and Warren, S. T. (2001) Cell 107 477-487 [DOI] [PubMed] [Google Scholar]
- 29.Gerber, A. P., Herschlag, D., and Brown, P. O. (2004) PLoS Biol. 2 E79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopez de Silanes, I., Galban, S., Martindale, J. L., Yang, X., Mazan-Mamczarz, K., Indig, F. E., Falco, G., Zhan, M., and Gorospe, M. (2005) Mol. Cell. Biol. 25 9520-9531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mazan-Mamczarz, K., Lal, A., Martindale, J. L., Kawai, T., and Gorospe, M. (2006) Mol. Cell. Biol. 26 2716-2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, W. M., Mei, R., Di, X., Ryder, T. B., Hubbell, E., Dee, S., Webster, T. A., Harrington, C. A., Ho, M. H., Baid, J., and Smeekens, S. P. (2002) Bioinformatics 18 1593-1599 [DOI] [PubMed] [Google Scholar]
- 33.Wu, Z., Irizarry, R., Gentleman, R. C., Martinez-Murillo, F., and Spencer, F. (2004) J. Am. Stat. Assoc. 99 909-918 [Google Scholar]
- 34.Cope, L. M., Irizarry, R. A., Jaffee, H. A., Wu, Z., and Speed, T. P. (2004) Bioinformatics 20 323-331 [DOI] [PubMed] [Google Scholar]
- 35.Gentleman, R. C. (2005) Bioinformatics and Computational Biology Solutions Using R and Bioconductor, Springer-Verlag New York Inc., New York
- 36.Irizarry, R. A., Bolstad, B. M., Collin, F., Cope, L. M., Hobbs, B., and Speed, T. P. (2003) Nucleic Acids Res. 31 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Irizarry, R. A., Hobbs, B., Collin, F., Beazer-Barclay, Y. D., Antonellis, K. J., Scherf, U., and Speed, T. P. (2003) Biostatistics 4 249-264 [DOI] [PubMed] [Google Scholar]
- 38.Gautier, L., Cope, L., Bolstad, B. M., and Irizarry, R. A. (2004) Bioinformatics 20 307-315 [DOI] [PubMed] [Google Scholar]
- 39.Gentleman, R. C., Carey, V. J., Bates, D. M., Bolstad, B., Dettling, M., Dudoit, S., Ellis, B., Gautier, L., Ge, Y., Gentry, J., Hornik, K., Hothorn, T., Huber, W., Iacus, S., Irizarry, R., Leisch, F., Li, C., Maechler, M., Rossini, A. J., Sawitzki, G., Smith, C., Smyth, G., Tierney, L., Yang, J. Y., and Zhang, J. (2004) Genome Biology 5 R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beaudoing, E., Freier, S., Wyatt, J. R., Claverie, J. M., and Gautheret, D. (2000) Genome Res. 10 1001-1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kedersha, N., Cho, M. R., Li, W., Yacono, P. W., Chen, S., Gilks, N., Golan, D. E., and Anderson, P. (2000) J. Cell Biol. 151 1257-1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stoecklin, G., Stubbs, T., Kedersha, N., Wax, S., Rigby, W. F., Blackwell, T. K., and Anderson, P. (2004) EMBO J. 23 1313-1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brooks, S. A., Connolly, J. E., Diegel, R. J., Fava, R. A., and Rigby, W. F. (2002) Arthritis Rheum. 46 1362-1370 [DOI] [PubMed] [Google Scholar]
- 44.Gueydan, C., Droogmans, L., Chalon, P., Huez, G., Caput, D., and Kruys, V. (1999) J. Biol. Chem. 274 2322-2326 [DOI] [PubMed] [Google Scholar]
- 45.Kent, W. J., Sugnet, C. W., Furey, T. S., Roskin, K. M., Pringle, T. H., Zahler, A. M., and Haussler, D. (2002) Genome Res. 12 996-1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Platzer, C., Meisel, C., Vogt, K., Platzer, M., and Volk, H. D. (1995) Int. Immunol. 7 517-523 [DOI] [PubMed] [Google Scholar]
- 47.Lai, W. S., Carballo, E., Thorn, J. M., Kennington, E. A., and Blackshear, P. J. (2000) J. Biol. Chem. 275 17827-17837 [DOI] [PubMed] [Google Scholar]
- 48.Stoecklin, G., Colombi, M., Raineri, I., Leuenberger, S., Mallaun, M., Schmidlin, M., Gross, B., Lu, M., Kitamura, T., and Moroni, C. (2002) EMBO J. 21 4709-4718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mili, S., and Steitz, J. A. (2004) RNA 10 1692-1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fechir, M., Linker, K., Pautz, A., Hubrich, T., Forstermann, U., Rodriguez-Pascual, F., and Kleinert, H. (2005) Mol. Pharmacol. 67 2148-2161 [DOI] [PubMed] [Google Scholar]
- 51.Keene, J. D., and Tenenbaum, S. A. (2002) Mol. Cell 9 1161-1167 [DOI] [PubMed] [Google Scholar]
- 52.George, A. D., and Tenenbaum, S. A. (2006) RNA Biol. 3 57-59 [DOI] [PubMed] [Google Scholar]