Perturbation of the T cell repertoire in rheumatoid arthritis (original) (raw)
Abstract
Aberrations in the T cell repertoire with the emergence of oligoclonal populations have been described in patients with rheumatoid arthritis (RA). However, the extent of the repertoire perturbations as well as the underlying mechanisms are not known. We now have examined the diversity of the peripheral CD4 T cell repertoire by determining the frequencies of arbitrarily selected T cell receptor (TCR) β-chain sequences. Healthy individuals displayed a highly diverse repertoire, with a median frequency of individual TCR β-chain sequences of 1 in 2.4 × 107 CD4 T cells. In RA patients, the median TCR β-chain frequency was increased 10-fold, indicating marked contraction of the repertoire (P < 0.001). The loss in TCR diversity was not limited to CD4 memory T cells but also involved the compartment of naive T cells, suggesting that it reflected an abnormality in T cell repertoire formation and not a consequence of antigen recognition in the synovium. Also, control patients with chronic inflammatory disease such as hepatitis C expressed a diverse repertoire indistinguishable from that of normals. Telomere length studies indicated an increased replicative history of peripheral CD4 T cells in RA patients, suggesting an enhanced turnover within the CD4 compartment. Compared with age-matched controls, terminal restriction fragment sizes were 1.7 kilobases shorter (P < 0.001). These data demonstrate an altered CD4 T cell homeostasis in RA that may contribute to the autoimmune response as well as to the immunodeficiency in these patients.
T lymphocytes are the dominant cell population infiltrating the synovial membrane in rheumatoid arthritis (RA) (1). In some patients, tissue-infiltrating lymphocytes are organized into follicles that are similar in appearance to germinal centers (2). The formation of lymphoid tissue-like structures strongly supports the notion that rheumatoid synovitis is the sequela of an antigen-driven immune response. The model of RA as an antigen-induced inflammation also accommodates the major histocompatibility complex class II association of RA (3, 4). However, the synovial T cell response is highly diverse, and it remains an issue of controversy whether RA can be explained as an immune response toward an immunodominant arthritogenic antigen (5).
More recent studies have raised the possibility that the role of T cells in RA goes beyond the recognition of antigens in the joint. Aberrations in the global and in the synovial T cell receptor (TCR) repertoire of RA patients have been reported by several investigators (6–8). The most impressive difference in the T cell repertoire of RA patients has been associated with the emergence of clonal T cell populations. CD4 T cell clones, which expand to a considerable size, are uniformly present in the circulation and infiltrate into synovial lesions (9, 10). CD8 T cell clones are not limited to RA patients but are more frequent and larger. Besides the tendency for clonal expansion, the in vivo expanded CD4 T cell clones have additional unusual features. They are autoreactive to ubiquitous antigen (11), lack the expression of the CD28 molecule, and do not require costimulatory signals to secrete lymphokines (12). Because they are detectable in very early stages of disease and even in unaffected siblings of RA patients, they are unlikely to represent an epiphenomenon of the chronic inflammatory process (10). Clonal expansion of CD4 T cells is particularly evident in patients with extra-articular manifestations of RA, raising the possibility that they have a role in rheumatoid organ disease (13).
We were interested in whether clonality is a restricted phenomenon and only involves a limited number of CD4 T cells in the repertoire or whether the tendency for T cells to proliferate is widespread throughout the T cell compartment of RA patients. To estimate the diversity of the CD4 T cell repertoire, the frequency of individual TCR β-chains used by circulating CD4 T cells was determined, and the diversity of the TCR repertoire in healthy donors and RA patients was compared. RA patients differed from controls in that the majority of TCR β-chains were present at increased frequencies and that infrequent TCR β-chains were the exception. Restricted diversity of TCR β-chains was associated with an increased number of peripheral CD4 T cells undergoing proliferation and a loss of telomere length in the CD4 T cell compartment. In contrast to the findings in RA, chronic T cell responses to viral antigens in hepatitis C-infected patients did not shift the distribution of TCR β-chains. These data suggest that the entire TCR repertoire is altered in RA with a loss of diversity and multiclonal growth of a high proportion of T cells.
MATERIALS AND METHODS
Study Population.
Peripheral blood mononuclear cells (PBMC) were obtained from patients with RA (n = 26), patients with chronic hepatitis C infection (n = 3), and healthy unrelated controls (n = 24). RA patients were positive for rheumatoid factor and had disease durations between 1 and 12 years. None of the patients had received alkylating drugs or anti-T cell directed therapy. Hepatitis C patients had a minimal disease duration of 5 years and were positive for viral DNA by PCR. None of the patients had received interferon α therapy. RA patients and controls were characterized for their HLA-DRB1 alleles by PCR and oligonucleotide hybridization (Biotest Diagnostics, Danville, NJ).
Cell Purification.
CD4+, CD4+ CD45RO+, and CD45RO− T cells were purified from PBMC on a FACSVantage (Becton Dickinson) to obtain highly purified subsets for TCR sequencing and probe design. For limiting dilution analysis, PBMC were incubated with anti-CD8 antibody conjugated to magnetic beads (Miltenyi Biotec, Auburn, CA) at 4°C for 30 minutes and then were separated on columns in a magnetic cell separator (VarioMACS, Miltenyi Biotec). The purity of CD8 T cells in the positively selected fraction and of CD4 T cells in the negatively selected fraction were >95% as determined by flow cytometry. Negatively selected CD4 T cells further were separated into CD45RO+ and CD45RO− cells by using an anti-CD45RO antibody coupled to magnetobeads.
Amplification of TCR β-Chain Sequences.
cDNA was amplified in a seminested reverse transcription–PCR with TCR BV8 (ATTTACTTTAACAACAACGTTCCG), BV18 (GATGAGTCAGGAATG CCAAAGGAA), and BC-specific primers (GTGGGAGATCTCTGCTTCTG). Products were reamplified with the appropriate BV-specific primer and either a BJ1S4 (AGACAGAGAGCTGGGTTCCAC)- or BJ2S5 (ACCAGGAGCCGCGTGCCT)-specific primer. TCR BV8-BJ1S4 and BV18-BJ2S5 PCR products were cloned by using a TA cloning kit (Invitrogen) and were sequenced. For each BV-BJ combination, five 20-bp biotinylated probes (Biotin-On Phosphoramidite, CLONTECH) spanning the N-D-N region were synthesized. In three RA patients, but none of the normal controls, one TCR sequence was found repeatedly in the sample sequenced. These sequences were excluded to avoid a skewing of the study toward clonally expanded TCR in RA.
Liquid Phase Hybridization Assay.
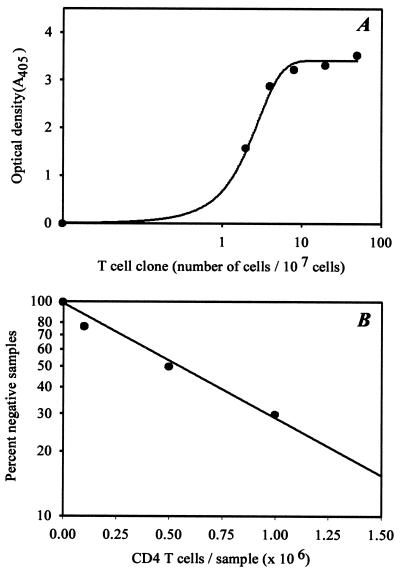
Digoxigenin-labeled TCR BV-BJ PCR products were hybridized with biotinylated TCR N-D-N-specific probes (50 ng/ml) in streptavidin-coated microtiter plates at 55°C by using a commercially available PCR ELISA kit (Boehringer Mannheim). Hybridized probes were detected by using a horseradish peroxidase-conjugated anti-digoxigenin antibody and a colorimetric reaction. To assess assay sensitivity, serial dilutions of T cell clones with known TCR β-chain sequences were mixed with 1 × 107 polyclonal unrelated T cells. The assay system was sensitive to detect a single T cell in a population of 1 × 107 cells (Fig. 1).
Figure 1.
Estimation of TCR β-chain frequencies. (A) Serial dilutions of a T cell clone with known TCR β-chain sequence were mixed with 1 × 107 unrelated T cells. The mixtures were analyzed by nested BV- and BC/BJ-specific PCR followed by hybridization with the TCR N-D-N-specific probe. (B) TCR β-chain frequencies were determined in a limiting dilution system. A TCR β-chain sequence was chosen arbitrarily from peripheral blood CD4 T cells, and a TCR N-D-N specific probe was designed. Multiple serially diluted CD4 T cell samples were probed for the presence or absence of the particular TCR β-chain sequence by TCR BV-BC- and BV-BJ-specific PCR and subsequent hybridization. The frequency of the TCR β-chain was determined from the slope of the curve to be 1 in 8 × 105 CD4 T cells.
Limiting Dilution Analysis.
The frequencies of individual TCR sequences were determined by limiting dilution analysis. CD4 T cells were divided into multiple aliquots of 1 × 105, 5 × 105, 1 × 106, 2 × 106, and 5 × 106 cells (8–10 aliquots/dilution except the highest concentration). cDNA from serially diluted CD4 T cells were amplified with BV- and BJ-specific primers in a seminested PCR, and the amplified product was hybridized with the appropriate TCR N-D-N probes. The samples from which the sequences for the probes originally were derived were used as positive controls. Samples from CD8 T cells from the same patient served as negative controls. Hybridization signals of >10% of the positive control were considered positive. Frequencies were determined by assuming a Poisson distribution. A representative experiment is shown in Fig. 1B. To estimate the specificity of the assay, 15 probes were analyzed in detail. For each probe, all wells yielding a positive hybridization reaction were eluted, and the material was reamplified with the appropriate primers and was sequenced directly. A crossreactive TCR sequence was found only with two probes, suggesting a high specificity of the assay.
Cell Cycle Analysis of Peripheral CD4 T Cells.
PBMC were stained at 4°C with fluorescein isothiocyanate-labeled mouse anti-CD4 antibody (Becton Dickinson), were fixed with PBS/1% paraformaldehyde, and were permeabilized in PBS containing 0.05% Tween 20 (Sigma). Cells then were stained with 25 mg/ml 7-amino-actinomycin D (Calbiochem, San Diego, CA). The percentage of CD4 T cells in the S-G2/M phase was determined on a FACScan flow cytometer (Becton Dickinson).
Telomere Length Analysis.
DNA (5 μg/20 μl) from purified CD4 T cells was digested with _Hin_fI and RSA I (Boehringer Mannheim) overnight at 37°C and was separated on 0.5% agarose gels. Gels were dried at 60°C for 25 minutes, were denatured, were prehybridized for 2 h, and then were hybridized in 20× standard saline citrate (SSC) buffer, 10× Denhardt’s solution, and 0.1 M dibasic sodiumphosphate with a 32P end-labeled (CCCTAA)3-probe at 37°C overnight. After five washings with 0.5× SSC buffer at room temperature, gels were scanned by using a PhosphorImager (Bio-Rad).
Statistical Analysis.
The distributions of TCR β-chain frequencies and of telomere sizes were compared by using the nonparametric Mann–Whitney test (sigma stat, SPSS, Chicago).
RESULTS
The Diversity of the TCR β-Chain Repertoire of Circulating CD4 T Cells in Normal Individuals and RA Patients.
Diversity is one of the fundamental principles in the immune system and is achieved by the formation of an enormously high number of structurally different TCR molecules (14). It has, therefore, been assumed that each TCR is present at a very low frequency under nonstimulated conditions. To examine the diversities of the CD4 T cell compartment, the frequencies of arbitrarily chosen TCR β-chain sequences in CD4 T cells purified from PBMC of seven healthy individuals were determined. Results from experiments analyzing the BV8-BJ1S4 combination are summarized in Fig. 2. Of 36 TCR β-chain sequences examined in the control individuals, 44% could be detected only in the positive control and were not present in any of the serially diluted samples, suggesting that their frequencies were <1 in 5 × 107 cells. Half of the receptor sequences were present at frequencies between 1 in 106 and 1 in 5 × 107 CD4 T cells. TCR β-chains expressed at frequencies >1 in 106 cells were the exception, and only two such sequences were found in one of the seven donors. In summary, the frequency analysis of individual TCR β-chains showed that the TCR repertoire was highly diverse. Unique TCR β-chains were found with a median frequency of 1 in 2.1 × 107 CD4 T cells.
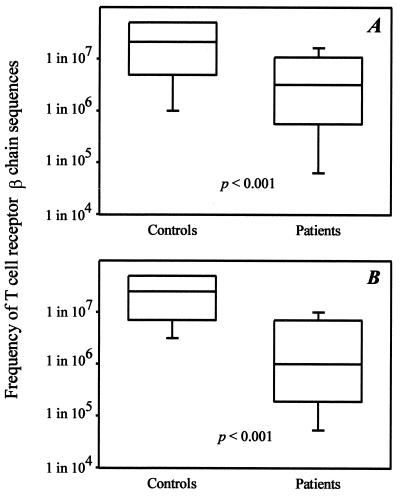
Figure 2.
Restricted TCR β-chain diversity in RA. (A) The frequencies of 36 arbitrarily chosen TCR BV8-BJ1S4 sequences from seven controls and 27 sequences from six RA patients were determined by limiting dilution analysis as shown in Fig. 1. Results are shown as box plots displaying medians and 25th and 75th percentiles. (B) TCR BV18-BJ2S5 sequences were analyzed from an additional five RA patients and five age-matched controls. Again, results are shown as box plots.
A different picture emerged for patients with RA. The median frequency of 27 BV8-BJ1S4 TCR β-chain sequences derived from six patients was significantly higher compared with the controls (1 in 3.2 × 106, P < 0.001). Only 4% of the TCR β-chains in the RA patients were present at frequencies too low to be detected, indicating that most T cells were expanded clonally. TCR β-chains expressed at frequencies of >1 in 106 cells accounted for 25% of all sequences. The TCR BV8-BJ1S4 combination was chosen arbitrarily for analysis. To test whether the repertoire contraction was unique for this combination or whether these results could be generalized, a second TCR BV-BJ combination, BV18-BJ2S5, was examined in five additional RA patients and in five age-matched normal controls. The results again demonstrated a reduced diversity in RA patients (Fig. 2). Only 1 of 22 TCR sequences in the RA patients was infrequent (<1 in 5 × 107) compared with 8 of 22 sequences in normal controls. The median frequency (1 in 1 × 106) in the RA patients was again significantly different from the median frequency of 1 in 2.5 × 107 in the controls (P < 0.001). Taking together TCR sequences from both BV-BJ combinations, a total number of 58 and 49 TCR β-chains in controls and patients, respectively, were analyzed. The median frequency was increased 10-fold in RA patients (1 in 2.5 × 106 compared with 1 in 2.4 × 107 CD4 T cells; P < 0.001).
The Repertoire of CD4 TCR Sequences Is Stable Over Time.
Clonal expansion of T cells is a consequence of antigen-specific stimulation. To explore whether high frequencies of particular TCR sequences are a transient phenomenon reflecting acute immune responses, the stability of TCR β-chain frequencies was investigated in blood samples collected 2 years apart. Results for nine TCR β-chains derived from two individuals, a normal donor (GH) and a RA patient (PN), are given in Table 1. Three of the nine sequences were too infrequent at both timepoints to be detectable in any of the serially diluted T cell samples. For the remaining TCR β-chains, the frequencies at the two timepoints were very similar. Thus, TCR β-chains are characterized by a stable representation within the CD4 repertoire over a timespan of at least 2 years.
Table 1.
Stability of TCR β-chain frequencies over time
| Frequency | ||
|---|---|---|
| Probe | Sample I* | Sample II |
| GH1 | n.d. | n.d. |
| GH2 | 1 in 1.2 × 107 | 1 in 1.6 × 107 |
| GH3 | 1 in 1 × 106 | 1 in 3.3 × 106 |
| GH4 | 1 in 5 × 106 | 1 in 6.6 × 106 |
| GH5 | n.d. | n.d. |
| PN1 | 1 in 1 × 107 | 1 in 2.6 × 106 |
| PN2 | 1 in 1 × 107 | 1 in 6.8 × 106 |
| PN3 | n.d. | n.d. |
| PN5 | 1 in 6 × 104 | 1 in 6 × 104 |
Chronic Antigenic Stimulation and the Diversity of the TCR Repertoire.
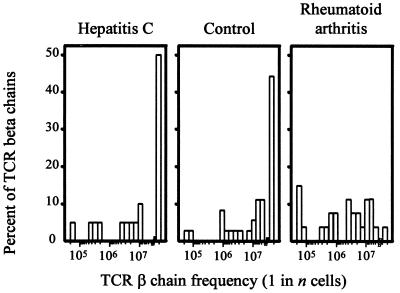
A likely explanation for the increased frequencies of TCR β-chains in RA patients is clonal proliferation of CD4 T cells leading to a significant loss in diversity. To explore whether chronic stimulation with antigen can induce the global changes in the composition of the TCR β-chain repertoire seen in the RA patients, the repertoire in three patients with chronic hepatitis C infection was analyzed. The frequency distribution in the hepatitis C-infected donors was indistinguishable from that found in healthy individuals and different from RA patients, demonstrating that a chronic T cell response to viral antigens does not induce a multiclonal expansion resulting in a loss of diversity (Fig. 3). The median frequency was 1 in 1.9 × 107 in the hepatitis patients, compared with 1 in 2.1 × 107 in the controls. Of the TCR β-chain sequences isolated from hepatitis C-infected hosts, 50% were too infrequent to be detectable, compared with 42% in controls and 4% in RA patients.
Figure 3.
The influence of chronic stimulation with viral antigen on the diversity of the CD4 TCR repertoire. Frequencies of 17 BV8-BJ1S4 TCR β-chains from three patients with chronic hepatitis C infection were determined by limiting dilution analysis. Distributions of TCR β-chain frequencies in CD4 T cells are shown in comparison to normal donors and RA patients. The chronic antiviral response did not alter the diversity of the CD4 T cell repertoire.
Contraction of the TCR Repertoire in RA Involves Naive as well as Memory CD4 T Cells.
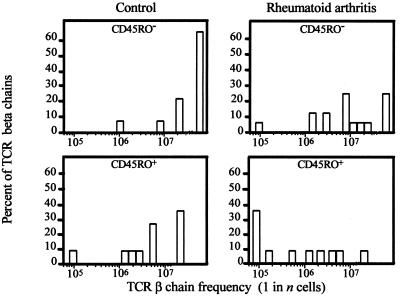
Clonal expansion of peripheral CD4 T cells in RA patients theoretically could result from a spillover of T cell activation in the synovium. If that were the case, repertoire contraction would be limited to memory CD4 T cells. To address this question, CD45RO+ memory and CD45RO− naive CD4 T cells were examined separately in three additional RA patients and three age-matched controls. The size of the CD4+ CD45RO+ T cell subset was not increased in the patients, as compared with the naive T cell subset (data not shown). As shown in Fig. 4, TCR sequences isolated from naive T cells of the control individuals were highly infrequent. More than 60% of the naive TCR β-chains were below the threshold of the assay system that detected sequences expressed at >1 in 5 × 107 T cells. In contrast, only 25% of the naive TCR β-chains from RA patients fell into the category of low frequency. Of the naive T cells in RA patients, 75% were represented in increased frequencies (P = 0.008).
Figure 4.
Repertoire contraction in RA includes memory as well as naive CD4 T cells. TCR BV8-BJ1S4 sequences were obtained from purified CD4+ CD45RO+ memory (n = 22) and CD4+ CD45RO− naive T cells (n = 36) from three RA patients and three controls, and the frequencies in the respective T cell subsets were determined. Compared with controls, TCR β-chain frequencies in RA patients were increased for the memory (P = 0.04) as well as the naive (P = 0.008) T cell subset.
Compared with CD45RO− naive T cells, TCR β-chains derived from the CD45RO+ population were enriched by a factor of 10 in both patients and controls. However, the frequencies of individual memory TCR sequences were still significantly higher in the patients, as compared with the controls (P = 0.04). These results indicated that perturbations in the T cell repertoire of RA patients were not limited to antigen-exposed memory T cells and suggested increased proliferation of naive T cells.
Proliferative Activity Among Peripheral CD4 T Cells.
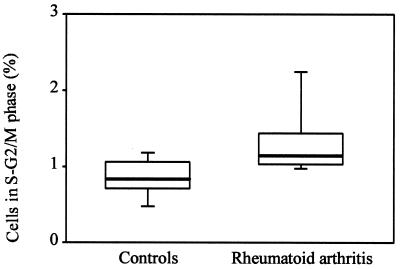
To analyze whether RA is associated with an increased turnover of peripheral CD4 T cells, the fraction of CD4 T cells in the S-G2/M phase of freshly drawn blood from 10 patients and 10 normal controls was determined by flow cytometric analysis. Results are shown in Fig. 5. In the normal individuals, a mean of 0.6% of all circulating CD4 T cells were in the S-G2/M phase. This number was increased significantly to 1.2% in the RA patients (P = 0.01). These data are consistent with the interpretation that the CD4 T cell compartment in RA patients is characterized by increased turnover.
Figure 5.
Proliferative activity in CD4 T cells. Peripheral T cells were isolated freshly from normal controls and RA patients, and the percentages of CD4 T cells in the S-G2/M phase were determined. Results are shown as box plots. Median, 25th, and 75th percentiles (box), and 10th and 90th percentiles (whiskers) are displayed. The fraction of CD4 T cells in the cell cycle was increased significantly in RA patients (P = 0.01).
Telomere Length of CD4 T Cells in RA Patients.
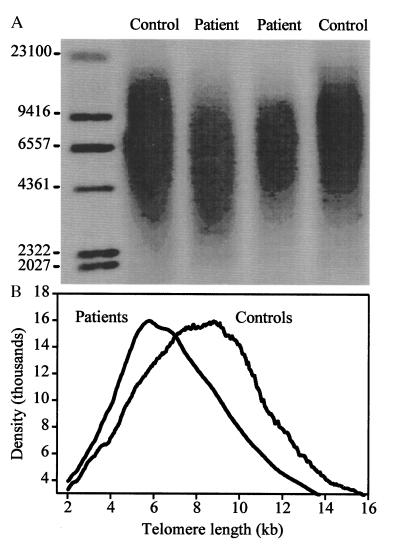
The reduction in diversity in the repertoire of RA patients suggested that a large proportion of peripheral CD4 T cells was involved in clonal expansion. To further support this hypothesis, the replicative history of CD4 cells was assessed by determining the telomere length. Because of the known age dependence of telomere length in naive as well as memory T cells, only individuals younger than 45 years were included (15). The results of the telomere length analysis of six RA patients and six age-matched controls are summarized in Fig. 6. The controls showed a Gaussian distribution of terminal restriction fragment sizes with a median length of 8.9 kilobases, which was significantly longer than the median length of 7.2 kilobases in the patients (P < 0.001). In addition, the Gaussian distribution was skewed toward shorter fragment sizes in RA patients, consistent with an extensive replicative history of a large fraction of peripheral CD4 T cells.
Figure 6.
Telomeric shortening of CD4 T cells from RA patients. (A) A representative telomeric restriction blot from peripheral blood CD4 T cells of two RA patients (lanes 2 and 3, ages 42 and 43 years) and two controls (lanes 1 and 4, both age 40 years) is shown. (B) Summary of PhosphorImage analysis of terminal restriction fragment sizes from CD4 T cells of six RA patients and six age-matched normal controls ranging in age from 29 to 43 years.
DISCUSSION
Studies of T cells in patients with RA have so far focused on their potential role in recognizing arthritogenic antigens in synovial lesions (16, 17). Aberrations in the peripheral T cell compartment have been described but have been interpreted as a consequence of ongoing synovial inflammation. Fluorescence-activated cell sorter studies have documented increased frequencies of peripheral T cells expressing activation markers or lacking CD28 and CD7 (11, 13). The analysis of the repertoire of peripheral CD4 and CD8 T cells has provided evidence of frequent oligoclonality (10, 18–20). By virtue of the applied technologies, these studies could only identify expanded clonotypes of a considerable size and expressed at frequencies larger than 1 in 1,000. The current study was designed to explore whether these clonal expansions are isolated events or whether the tendency to form clonal T cell populations is not limited to a small subset of T cells. The experimental design for the estimation of TCR frequencies used in this study was ≈1,000-fold more sensitive than previous techniques and therefore allowed for a better assessment of global T cell diversity (21). Our data suggest that RA is associated with a generalized defect in maintaining diversity of the TCR repertoire, resulting in clonal expansion of most peripheral T cells and marked repertoire contraction. Of importance, these aberrations equally involved memory as well as naive T cells, suggesting that they reflect intrinsic abnormalities of T cell homeostasis and not a consequence of antigen recognition in the synovium.
Theoretically, the mechanisms underlying TCR formation allow the generation of a repertoire of enormous diversity (14). It is estimated that the human body has 5 × 1011 T lymphocytes, the diversity of which is unknown. Our data demonstrate that the CD4 TCR β-chain repertoire in a normal individual is extremely large. In contrast, TCR β-chain sequences analyzed in RA patients were found at increased frequencies. Highly frequent TCR β-chain sequences were structurally diverse, were unique for each individual, and were not detected in unrelated individuals. This finding suggests that high frequency clonotypes do not represent a distinct subset of T cells characterized by the usage of less diverse TCR molecules as described by Dellabona et al. (22).
Different mechanisms could create the reduced TCR β-chain diversity in the RA patients. Identical TCR β-chains could be generated in independent rearrangements. This model would imply that mechanisms involved in generating TCR β-chain diversity would be disturbed in RA. It is also possible that the same TCR β-chain combines with a spectrum of different α-chains because thymocytes undergo proliferation between β-chain and α-chain rearrangement (23, 24). A defect in thymic selection would explain that the repertoire contraction already is seen for CD45RO− naive and not only for memory cells. The shortened telomere length in CD4 T cells from RA patients, however, suggests that the increased TCR β-chain frequencies do not result from thymic selection but from peripheral clonal proliferation of T cells. Assuming that each TCR β-chain sequence represents a single TCR α-β pair and taking into account the differences in clonal size, it can be estimated how many T cells have undergone expansion. From the data shown in Fig. 2, it can be concluded that 23% of all CD4 T cell specificities have replicated in normal donors whereas that fraction is increased to 77% in RA patients. This estimate suggests that the majority of CD4 T cells in RA patients contribute to proliferation and diversity loss. The realization that repertoire aberrations in RA are not limited to a small subset of memory T cells but rather involve a large fraction of the global T cell compartment is important in considering mechanisms underlying RA pathology.
It is now recognized that the size of the T cell compartment is controlled strictly at the level of total T cell number (25–27). Therefore, loss in diversity could result from insufficient influx of novel T cells. To maintain the size of the compartment, peripheral T cells would replicate to compensate for the lack of new production (28). There is evidence that de novo T cell generation is deficient in RA. T cell depletion with antibodies has been explored as a therapeutic modality in RA patients. One of the most intriguing observations from these treatment trials was the finding that patients developed long-standing CD4 T cell lymphopenia (29–31). Studies of the post-treatment repertoire have demonstrated extreme restriction in diversity with the emergence of oligoclonal T cell populations (32). RA may, therefore, be associated with a defect in T cell repopulation, and the aberrations described here may be sequel of this underlying dysfunction.
The initial abnormality also could stem from chronic antigenic stimulation in the synovium that either directly or indirectly influences T cell repertoire diversity. The experiments in patients with chronic active hepatitis C, however, documented that antiviral responses ongoing over >5 years did not bias the TCR repertoire to the extent that diversity was reduced. It is likely that hepatitis C-infected patients have expanded T cell clones to respond to viral antigens, but the spectrum of these T cells is obviously too small to alter the global TCR repertoire. Apparently, antigen-specific proliferation only then could explain the repertoire shifts if a large spectrum of antigens were involved. Chronic antigenic stimulation also would not explain the repertoire contraction in CD45RO− T cells. Based on previous studies in rats (33), it cannot be excluded completely that memory T cells lose CD45RO expression in the absence of antigenic stimulation and revert to the naive phenotype. However, this is unlikely to occur in a chronic inflammatory disease such as RA. Also, we have examined whether TCR β-chain sequences derived from cells with a naive phenotype frequently were represented within the CD45RO+ T cell compartment, which was not the case. Taken together, our data argue against the interpretation that the multiclonal T cell proliferation and the associated repertoire contraction in RA reflects the T cell response in the synovium.
Finally, our findings also could be explained by an increased peripheral T cell replication with competitive exclusion of nonproliferating clonotypes (34). The end result would again be a CD4 T cell repertoire with compromised diversity. Increased proliferative activity throughout the CD4 T cell repertoire could be maintained by growth-promoting cytokines. Production of interleukin 15, which has general T cell growth promoting activities, has been demonstrated in the synovial tissue of RA patients (35, 36). Interferon β has been implicated in driving the expansion of a wide spectrum of CD8 T cells during viral infection regardless of their antigen specificity (37). However, this mechanism exclusively targeted memory T cells and did not include naive T cells. Also, an increased turnover mediated by cytokines would not explain easily the skewing of the repertoire with the oligoclonal expansion of some and the competitive exclusion of other T cells. Besides cytokines, the recognition of self–major histocompatibility complex has been found to be pivotal for T cell homeostasis (38). Naive T cells have been estimated to have a very low self-replication rate compared to memory T cells (39), but their survival strictly depends on the continuous interaction with self–major histocompatibility complex (38). Individuals developing RA may have a heightened ability to proliferate to self-determinants, compatible with the model that they have a global defect in tolerance. Although the antigen specificities of TCR β-chain sequences described here are unknown, CD4 T cell clones derived from peripheral blood of RA patients that had expanded in vivo to a large clonal size have recently been isolated and characterized (11). These T cell clones proliferated when stimulated with autologous adherent cells, suggesting they recognize a ubiquitously distributed autoantigen in RA patients (11). A recent study by Kouskoff et al. has provided evidence for similar mechanisms in an animal model of RA (40). These investigators have described that a TCR transgenic mouse expressing a TCR recognizing self–major histocompatibility complex molecules develops arthritis resembling RA but no other autoimmune manifestations.
Regardless of the precise mechanism for the loss of T cell diversity, these aberrations have important implications for the disease process and the way it is treated and studied. The fact that large proportions of the TCR repertoire are altered cannot remain without consequences for immunoresponsiveness. It is possible that the repertoire contraction will generate holes in the repertoire and therefore will lead to defective immune responses to selected antigens. The design of therapeutic approaches should consider that the RA repertoire already has lost diversity. So far, it has been assumed that it would be beneficial to deplete T cells. If these patients have difficulties repopulating the T cell compartment and have to generate new T cells through self-replication, T cell-directed therapies will compromise further their ability to maintain diversity. It is, therefore, not surprising that treatment trials using T cell depletion were not successful and had substantial side effects (31, 41). Very different therapeutic approaches will have to be taken to correct repertoire aberrations in an attempt to control the disease process and its complications.
Acknowledgments
This work was supported by grants from the National Institutes of Health (RO1 AR41974 and RO1 AR42527), by a Biomedical Science grant (AF 16) from the National Arthritis Foundation, and by the Mayo Foundation. U.G.W. was the recipient of a fellowship from the Deutscher Akademischer Austauschdienst and of a fellowship from the Stiftung Familie Klee.
ABBREVIATIONS
RA
rheumatoid arthritis
TCR
T cell receptor
PBMC
peripheral blood mononuclear cells
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Van Boxel J A, Paget S A. N Engl J Med. 1975;293:517–520. doi: 10.1056/NEJM197509112931101. [DOI] [PubMed] [Google Scholar]
- 2.Kurosaka M, Ziff M. J Exp Med. 1983;158:1191–1210. doi: 10.1084/jem.158.4.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winchester R. Adv Immunol. 1994;56:389–466. doi: 10.1016/s0065-2776(08)60456-3. [DOI] [PubMed] [Google Scholar]
- 4.Nepom G T, Erlich H. Annu Rev Immunol. 1991;9:493–525. doi: 10.1146/annurev.iy.09.040191.002425. [DOI] [PubMed] [Google Scholar]
- 5.Goronzy J J, Weyand C M. Rheum Dis Clin North Am. 1995;21:655–674. [PubMed] [Google Scholar]
- 6.Albani S, Carson D A. Immunol Today. 1996;17:466–470. doi: 10.1016/0167-5699(96)20029-g. [DOI] [PubMed] [Google Scholar]
- 7.Albani S, Keystone E C, Nelson J L, Ollier W E, La Cava A, Montemayor A C, Weber D A, Montecucco C, Martini A, Carson D A. Nat Med. 1995;1:448–452. doi: 10.1038/nm0595-448. [DOI] [PubMed] [Google Scholar]
- 8.Walser-Kuntz D R, Weyand C M, Weaver A J, O’Fallon W M, Goronzy J J. Immunity. 1995;2:597–605. doi: 10.1016/1074-7613(95)90004-7. [DOI] [PubMed] [Google Scholar]
- 9.Goronzy J J, Bartz-Bazzanella P, Hu W, Jendro M C, Walser-Kuntz D R, Weyand C M. J Clin Invest. 1994;94:2068–2076. doi: 10.1172/JCI117561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waase I, Kayser C, Carlson P J, Goronzy J J, Weyand C M. Arthritis Rheum. 1996;39:904–913. doi: 10.1002/art.1780390606. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt D, Goronzy J J, Weyand C M. J Clin Invest. 1996;97:2027–2037. doi: 10.1172/JCI118638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park W, Weyand C M, Schmidt D, Goronzy J J. Eur J Immunol. 1997;27:1082–1090. doi: 10.1002/eji.1830270507. [DOI] [PubMed] [Google Scholar]
- 13.Martens P B, Goronzy J J, Schaid D, Weyand C M. Arthritis Rheum. 1997;40:1106–1114. doi: 10.1002/art.1780400615. [DOI] [PubMed] [Google Scholar]
- 14.Davis M M, Bjorkman P J. Nature (London) 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- 15.Weng N P, Levine B L, June C H, Hodes R J. Proc Natl Acad Sci USA. 1995;92:11091–11094. doi: 10.1073/pnas.92.24.11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verheijden G F, Rijnders A W, Bos E, Coenen-de Roo C J, van Staveren C J, Miltenburg A M, Meijerink J H, Elewaut D, de Keyser F, Veys E, et al. Arthritis Rheum. 1997;40:1115–1125. doi: 10.1002/art.1780400616. [DOI] [PubMed] [Google Scholar]
- 17.Harris E D J. Rheumatoid Arthritis. Philadelphia: Saunders; 1997. p. 417. [Google Scholar]
- 18.Hingorani R, Monteiro J, Furie R, Chartash E, Navarrete C, Pergolizzi R, Gregersen P K. J Immunol. 1996;156:852–858. [PubMed] [Google Scholar]
- 19.Fitzgerald J E, Ricalton N S, Meyer A C, West S G, Kaplan H, Behrendt C, Kotzin B L. J Immunol. 1995;154:3538–3547. [PubMed] [Google Scholar]
- 20.DerSimonian H, Sugita M, Glass D N, Maier A L, Weinblatt M E, Reme T, Brenner M B. J Exp Med. 1993;177:1623–1631. doi: 10.1084/jem.177.6.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pannetier C, Cochet M, Darche S, Casrough A, Zoller M, Kourilsky P. Proc Natl Acad Sci USA. 1993;90:4319–4323. doi: 10.1073/pnas.90.9.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dellabona P, Padovan E, Casorati G, Brockhaus M, Lanzavecchia A. J Exp Med. 1994;180:1171–1176. doi: 10.1084/jem.180.3.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.von Boehmer H. Cell. 1994;76:219–228. doi: 10.1016/0092-8674(94)90330-1. [DOI] [PubMed] [Google Scholar]
- 24.Robey E, Fowlkes B J. Annu Rev Immunol. 1994;12:675–705. doi: 10.1146/annurev.iy.12.040194.003331. [DOI] [PubMed] [Google Scholar]
- 25.Freitas A A, Rocha B B. Immunol Today. 1993;14:25–29. doi: 10.1016/0167-5699(93)90320-K. [DOI] [PubMed] [Google Scholar]
- 26.Zijlstra M, Bix M, Simister N E, Loring J M, Raulet D H, Jaenisch R. Nature (London) 1990;344:742–746. doi: 10.1038/344742a0. [DOI] [PubMed] [Google Scholar]
- 27.Gosgrove D, Gray D, Dierich A, Kaufman J, Lemeur M, Benoist C, Mathis D. Cell. 1991;66:1051–1066. doi: 10.1016/0092-8674(91)90448-8. [DOI] [PubMed] [Google Scholar]
- 28.Mackall C L, Hakim F T, Gress R E. Immunol Today. 1997;18:245–251. doi: 10.1016/s0167-5699(97)81664-7. [DOI] [PubMed] [Google Scholar]
- 29.Horneff G, Burmester G R, Emmrich F, Kalden J R. Arthritis Rheum. 1991;34:129–140. doi: 10.1002/art.1780340202. [DOI] [PubMed] [Google Scholar]
- 30.Matteson E L, Yocum D E, St. Clair E W, Achkar A A, Thakor M S, Jacobs M R, Hays A E, Heitman C K, Johnston J M. Arthritis Rheum. 1995;38:1187–1193. doi: 10.1002/art.1780380903. [DOI] [PubMed] [Google Scholar]
- 31.Moreland L W, Pratt P W, Mayes M D, Postlethwaite A, Weisman M H, Schnitzer T, Lightfoot R, Calabrese L, Zelinger D J, Woody J N, et al. Arthritis Rheum. 1995;38:1581–1588. doi: 10.1002/art.1780381109. [DOI] [PubMed] [Google Scholar]
- 32.Jendro M C, Ganten T, Matteson E L, Weyand C M, Goronzy J J. Arthritis Rheum. 1995;38:1242–1251. doi: 10.1002/art.1780380912. [DOI] [PubMed] [Google Scholar]
- 33.Bell E B, Sparshot S M, Bunce C. Immunol Today. 1998;19:60–64. doi: 10.1016/s0167-5699(97)01211-5. [DOI] [PubMed] [Google Scholar]
- 34.De Boer R J, Perelson A S. J Theor Biol. 1994;169:375–390. doi: 10.1006/jtbi.1994.1160. [DOI] [PubMed] [Google Scholar]
- 35.McInnes I B, al-Mughales J, Field M, Leung B P, Huang F P, Dixon R, Sturrock R D, Wilkinson P C, Liew F Y. Nat Med. 1996;2:175–182. doi: 10.1038/nm0296-175. [DOI] [PubMed] [Google Scholar]
- 36.McInnes I B, Leung B P, Sturrock R D, Field M, Liew F Y. Nat Med. 1997;3:189–195. doi: 10.1038/nm0297-189. [DOI] [PubMed] [Google Scholar]
- 37.Tough D F, Borrow P, Sprent J. Science. 1996;272:1947–1950. doi: 10.1126/science.272.5270.1947. [DOI] [PubMed] [Google Scholar]
- 38.Tanchot C, Rosado M M, Agenes F, Freitas A A, Rocha B. Semin Immunol. 1997;9:331–337. doi: 10.1006/smim.1997.0090. [DOI] [PubMed] [Google Scholar]
- 39.McLean A R, Michie C A. Proc Natl Acad Sci USA. 1995;92:3707–3711. doi: 10.1073/pnas.92.9.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kouskoff V, Korganow A S, Duchatelle V, Degott C, Benoist C, Mathis D. Cell. 1996;87:811–822. doi: 10.1016/s0092-8674(00)81989-3. [DOI] [PubMed] [Google Scholar]
- 41.Matteson E L. Curr Opin Rheumatol. 1997;9:95–101. [PubMed] [Google Scholar]