Anaerobic Sulfatase-maturating Enzymes, First Dual Substrate Radical S-Adenosylmethionine Enzymes (original) (raw)
Abstract
Sulfatases are a major group of enzymes involved in many critical physiological processes as reflected by their broad distribution in all three domains of life. This class of hydrolases is unique in requiring an essential post-translational modification of a critical active-site cysteine or serine residue to Cα-formylglycine. This modification is catalyzed by at least three nonhomologous enzymatic systems in bacteria. Each enzymatic system is currently considered to be dedicated to the modification of either cysteine or serine residues encoded in the sulfatase-active site and has been accordingly categorized as Cys-type and Ser-type sulfatase-maturating enzymes. We report here the first detailed characterization of two bacterial anaerobic sulfatase-maturating enzymes (anSMEs) that are physiologically responsible for either Cys-type or Ser-type sulfatase maturation. The activity of both enzymes was investigated in vivo and in vitro using synthetic substrates and the successful purification of both enzymes facilitated the first biochemical and spectroscopic characterization of this class of enzyme. We demonstrate that reconstituted anSMEs are radical_S_-adenosyl-l-methionine enzymes containing a redox active [4Fe-4S]2+,+ cluster that initiates the radical reaction by binding and reductively cleaving _S_-adenosyl-l-methionine to yield 5 ′-deoxyadenosine and methionine. Surprisingly, our results show that anSMEs are dual substrate enzymes able to oxidize both cysteine and serine residues to Cα-formylglycine. Taken together, the results support a radical modification mechanism that is initiated by hydrogen abstraction from a serine or cysteine residue located in an appropriate target sequence.
Bacterial sulfatases are widely distributed among the major bacterial phyla. Nevertheless, they have attracted only few studies compared with their eukaryotic counterparts (1). One of the difficulties in studying sulfatases is the requirement for post-translational modification of a critical active-site cysteine or serine residue to generate an active enzyme. Indeed, despite high homologies in the sulfatase domain (PFAM domain PF00884), sulfatases are divided into two sub-classes, Cys-type and Ser-type, according to the genetically encoded active site residue. In eukaryotes, only Cys-type sulfatases have been identified so far, whereas in bacteria, both types of sulfatases exist. Nevertheless, eukaryotic and prokaryotic sulfatases undergo the same post-translational modification involving oxidation of cysteine or serine to oxoalanine, also called Cα-formylglycine (FGly).3 To date, two distinct enzymatic systems have been identified for catalyzing this reaction. One is the formylglycine generating enzyme system, an oxygen-dependent oxidoreductase that has been extensively studied in humans and is functional only on Cys-type sulfatases (2). The other is AtsB, a putative member of the _S_-adenosyl-l-methionine (AdoMet)-dependent family of radical enzymes, which is responsible for maturation of Ser-type sulfatases and was first identified in Klebsiella aeruginosa (3,4). Thus, the maturation of Cys-type sulfatases, the most abundant type, in anaerobic environments such as the human gut, has long remained unknown.
We recently identified an enzyme related to the AtsB enzyme from K. aeruginosa in the obligate anaerobe Clostridum perfringens, which is surprisingly involved in Cys-type sulfatase maturation (5). Although the K. aeruginosa enzyme, AtsB, appeared to be restricted to Ser-type sulfatases harboring a signal peptide (4,6), we grouped these enzymes into a family called anaerobic sulfatase-maturating enzymes (anSMEs) (5). These results raised the possibility that anSMEs could be divided into two subgroups that differ in specificity for sulfatase FGly generation via oxidation of an active-site cysteine or serine residue.
Subsequent purification and characterization of C. perfringens anSME showed it to be a member of the radical-AdoMet superfamily of iron-sulfur enzymes (7). All the members of this superfamily share a strictly conserved cysteine motif C_X_3C_X_2C, which coordinates an unconventional [4Fe-4S] cluster. The [4Fe-4S] cluster is directly involved in the reductive cleavage of AdoMet to generate a 5′-deoxyadenosyl radical, which then initiates various radical-based reactions according to the functional specificity of the enzyme. In addition to the C_X_3C_X_2C motif, all the anSMEs identified to date have two other strictly conserved cysteine clusters (Fig. 1), which could also be involved in the coordination of iron-sulfur centers. However, no experimental data have been produced to substantiate the existence of additional iron-sulfur clusters and, other than UV-visible absorption, no spectroscopic data have been reported for these enzymes.
FIGURE 1.
Sequence alignment of the three anSME putative clusters, AtsB (K. pneumoniae), anSMEcpe (CPF_0616, C. perfringens), and anSMEbt (BT_0238, B. thetaiotaomicron). Sequence positions in the proteins are in brackets. In black are the conserved cysteines and in gray are the other conserved residues.
To get insights into this group of enzymes, we have investigated the in vitro and in vivo activity of bacterial anSMEs that are responsible for modification of either Cys-type or Ser-type sulfatases: anSMEcpe cloned from C. perfringens, which is responsible for maturation of a Cys-type sulfatase and anSMEbt cloned from Bacteroides thetaiotaomicron, a prominent gut symbiont, which possesses only Ser-type sulfatases. We report here the first detailed characterization of purified anSMEs and demonstrate that both enzymes can maturate cysteine and serine residues in vitro and in vivo. Furthermore, the ability to purify these unstable enzymes allowed us to provide the first spectroscopic characterization of their iron-sulfur centers.
EXPERIMENTAL PROCEDURES
_Chemicals_—_p-_Nitrophenylsulfate was purchased from Sigma. Enzymes for molecular biology were obtained from New England Biolabs (Ipswich, MA). Oligonucleotides were purchased from Eurogenetec (Seraing, Belgium). Other chemicals and reagents were obtained from commercial sources and were of analytical grade.
Bacterial Strains, Plasmids, and DNA Manipulations_—The_B. thetaiotaomicron strain used in this study was the VPI-5482 strain. Escherichia coli DH5α was used for routine DNA manipulations. E. coli BL21(DE3) (Stratagene) was used for enzyme overexpression. The pET-28(a) and pRSF plasmids were used to express the various proteins (Novagen, Inc.). T4 DNA ligase was from Promega, Inc. The plasmid DNA purification kit and QIAprep spin were from Qiagen, Inc. DNA fragments were extracted from agarose gel and purified with the Wizard SV Gel and PCR clean up system kit (Promega, Inc.). DNA sequencing was performed by MWG.
Cloning and Construction of the pET-6His-anSMEcpe and pET-6His-anSMEbt Overexpressing Plasmids_—_B. thetaiotaomicron VPI-5482 was grown anaerobically in BHI medium, pH 7.0, and the cells were harvested to extract genomic DNA using the Wizard Genomic Kit (Promega). The BT_0238 gene encoding the putative anSME was amplified by a PCR-based method using genomic DNA as a template. The following primers were used: 5′-cat ATG aaa gca act act tat gca cct ttt gcc-3′ (NdeI site underlined, ATG codon in uppercase) hybridized to the non-coding strand at the 5′ terminus of the gene and 5′-ctc gag tta ata ttc tat ttt taa act tcc gtc_-3′ (XhoI site underlined) hybridized to the coding strand. PCR was run as follows: genomic DNA (1 μg) in the presence of the primers (0.5 μm each) was mixed with the Hot Start Kit (Promega) and 30 cycles of PCR were performed (1 min at 95 °C, 1 min at 50 °C, 2 min at 72 °C), followed by a final 10-min elongation step at 72 °C. The PCR product was digested with NdeI and XhoI and then ligated with T4 DNA ligase into pET28(a) plasmid previously digested with the same restriction enzymes. The entire sequence of the cloned gene was sequenced to ensure that no errors were introduced during PCR. The plasmid was designated pET-6His_-anSMEbt. The pET-6His-anSMEcpe construction containing the CPF_0616 gene from C. perfringens ATCC 13124 has been previously reported (5).
anSMEcpe and anSMEbt Protein Expression and Purification_—_E. coli BL21(DE3) were transformed with pET-6His-anSMEcpe or pET-6His-anSMEbt, then grown aerobically overnight at 37 °C in LB medium (100 ml) supplemented with ampicillin (100 μgml–1). An overnight culture was then used to inoculate fresh LB medium (12 liters) supplemented with the same antibiotic and bacterial growth proceeded at 37 °C until the _A_600 reached 0.6. The cells were induced by adding 500 μm isopropyl 1-thio-β-D-galactopyranoside and were collected after overnight growth at 25 °C. After resuspension in Tris buffer (50 mm Tris, 150 mm KCl, 10% glycerol, pH 7.5), the cells were disrupted by sonication and centrifuged at 220,000 × g at 4 °C for 1 h. The solution was then loaded onto a Ni-NTA Sepharose column equilibrated with Tris buffer, pH 7.5. The column was washed extensively with the same buffer. Some of the adsorbed proteins were eluted by a washing step with 25 and 100 mm imidazole and the overexpressed protein was eluted by applying 500 mm imidazole. Fractions containing the anSMEcpe or anSMEbt proteins were immediately concentrated in Ultrafree cells (Millipore) with a molecular cut-off of 10 kDa.
_Reconstitution of Iron-Sulfur Clusters on anSMEbt and anSMEcpe_—Reconstitution was carried out anaerobically in a glove box (Bactron IV). As purified anSMEs (200 μm monomer) were treated with 5 mm dithiothreitol and incubated overnight with a 7-fold molar excess of both Na2S (Fluka) and (NH4)2Fe(SO4)2 (Aldrich) at 12 °C. The protein was desalted using a Sephadex G-25 column (Amersham Biosciences) and the colored fractions were concentrated on Nanosep 10 (Amicon). Protein concentrations were determined by the DC protein assay (Bio-Rad), using bovine serum albumin as a standard. Iron concentrations were determined colorimetrically using bathophenanthroline under reducing conditions, after digestion of the protein in 0.8% KMnO4, 0.2m HCl.
Construction of Plasmid Systems Expressing C. perfringens Sulfatase Alone or with anSMEbt or anSMEcpe_—The CPF_0221 gene encoding C. perfringens Cys-type sulfatase was amplified by a PCR-based method using genomic DNA as a template. The following primers were used: 5′-gga tcc ATG aag cca aat att gtg tta atc atg gtt-3′ (BamHI site underlined and ATG codon in uppercase), which corresponded to the 5′ terminus of the gene, and 5′-ctg cag tta tta tct tat atg ttt taa agt gct tac-3′ (PstI site underlined), complementary to the 3′-end of the gene. PCR was run as follows. Genomic DNA (1 μg) in the presence of the primers (0.5 μm each) was mixed with the Hot Start kit (Promega), and 30 cycles of PCR were performed (1 min at 94 °C, 1 min at 52 °C, and 1.5 min at 72 °C), followed by a final 10-min elongation step at 72 °C. The PCR product was digested with BamHI and PstI and inserted into pRSF plasmid site one. The obtained plasmid was designated pRSF-Cys-Sulf. The anSMEbt or anSMEcpe genes previously cloned were inserted into pRSF-Cys-Sulf plasmid (site two) previously digested by the NdeI and XhoI enzymes. Each ligation step was performed with T4 DNA ligase. The entire sequences of the cloned genes were sequenced to ensure that no errors were introduced during the PCR. The plasmid obtained containing the_C. perfringens sulfatase and anSMEbt or anSMEcpe were designated pRSF-anSMEbt-Cys-Sulf and pRSF-anSMEcpe-Cys-Sulf.
Construction of the Serine Variant of C. perfringens Cys-type Sulfatase_—Site-directed mutagenesis was performed on plasmid pRSF-Cys-Sulf to mutate cysteine 67 into serine. The cysteine to serine mutation of the C. perfringens sulfatase was performed with the QuikChange™ site-directed mutagenesis kit (Stratagene) using the following primers and a two-step PCR method (8). Sulfatase Cys to Ser primers were: forward, 5′-aca gca gtt cca agt AGC att gca tct agg gca-3′; and reverse, 5′-tgc cct aga tgc aat GCT act tgg aac tgc tgt-3′ (mutated codon in uppercase). After verification of the correct mutation by sequencing, the plasmid obtained was designated pRSF-Ser-Sulf. The_anSMEbt or anSMEcpe genes previously cloned were inserted into pRSF-Ser-Sulf plasmid (site two) previously digested by the NdeI and XhoI enzymes and, after a ligation step, the following plasmids were obtained pRSF-anSMEbt-Ser-Sulf and pRSF-anSMEcpe-Ser-Sulf.
_Peptide Synthesis_—The following 23-mer peptides (with the critical residue in bold): Ac-FENAYTAVPSCIASRASILTGMS-NH2, Ac-FENAYTAVPSSIASRASILTGMS-NH2, and Ac-FENAYTAVPSAIASRASILTGMS-NH2, were synthesized (0.1 mmol scale) by the solid phase methodology on a Rink amide 4-methylbenzhydrylamine resin (Biochem, Meudon, France) using a 433A Applied Biosystems peptide synthesizer (Applera-France, Courtaboeuf, France) and the standard Fmoc (_N_-(9-fluorenyl)methoxycarbonyl) procedure from the manufacturer. The synthetic peptides were purified by reverse-phase HPLC on a 2.2 × 25-cm Vydac 218TP1022 C18 column (Alltech, Templemars, France) using a linear gradient (10–50% over 45 min) of acetonitrile/trifluoroacetic acid (99.9:0.1; v/v) at a flow rate of 10 ml/min. Analytical HPLC, performed on a 0.46 × 25-cm Vydac 218TP54 C18 column (Alltech), showed that the purity of the peptides was >99.1%. The purified peptides were characterized by MALDI-TOF mass spectrometry on a Voyager DE PRO (Applera, France) in the reflector mode with α-cyano-4-hydroxycinnamic acid as a matrix.
_Peptide Maturation_—Samples containing 6 mm dithiothreitol, 3 mm sodium dithionite, 20 μm reconstituted anSMEbt or anSMEcpe were added with 500 μm peptides and 1 mm AdoMet in Tris buffer, pH 7.5. The reactions were performed in an anaerobic glovebox (Bactron IV) and incubated at 25 °C for 60, 120, 180, 240, and 360 min. Samples were divided in half, and one part was used to test the maturation activity by mass spectrometry, whereas the other half was diluted in 10 volumes of H2O, 0.1% trifluoroacetic acid and then loaded onto the C18 column to assay the reductive cleavage of AdoMet. Control samples were prepared without enzyme to verify the peptides and AdoMet stability over time.
_HPLC Detection of AdoMet Cleavage_—Standards of 3 mm AdoMet and 3 mm 5′-deoxyadenosine were run over a C18 column (LicroSphere, 5-μm, 4.6 × 150-mm) at 1 ml/min with the following gradient: after a 1-ml step of Milli-Q H2O, 0.1% trifluoroacetic acid, a linear gradient from 0 to 30% acetonitrile with 0.1% trifluoroacetic acid was used to elute the samples. Detection was carried out at 260 nm.
_Electron Paramagnetic Resonance (EPR)_—X-band EPR spectra were recorded on a Bruker Instruments ESP 300D spectrometer equipped with an Oxford Instruments ESR 900 flow cryostat (4.2–300 K). Spectra were quantified under non-saturating conditions by double integration against a 1 mm CuEDTA standard.
_Resonance Raman_—Resonance Raman spectra were recorded using an Instruments SA U1000 spectrometer fitted with a cooled RCA 31034 photomultiplier tube with 90° scattering geometry. Spectra were recorded digitally using photon-counting electronics, and improvements in signal-to-noise were achieved by signal averaging multiple scans. Band positions were calibrated using the excitation frequency and CCl4 and are accurate to ±1 cm–1. Lines from a Coherent Sabre 10-W argon ion laser were used for excitation, and plasma lines were removed using a Pellin Broca prism premonochromator. For each sample, ∼140 milliwatts of incident laser power was used, and slit widths were adjusted to give 7.0 cm–1 spectral resolution. Scattering was collected from the surface of a frozen 18-μl droplet of sample using a custom designed anaerobic sample cell (9) attached to the cold finger of an Air Products Displex model CSA-202E closed cycle refrigerator. This enables samples to be cooled down to 17 K, which facilitates improved spectral resolution and prevents laser-induced sample degradation.
Sulfatase Protein Expression and Purification_—_E. coli BL21(DE3) were transformed with pRSF-Cys-Sulf, pRSF-_anSMEbt_-Cys-Sulf, or pRSF-_anSMEcpe_-Cys-Sulf to produce the Cys-type sulfatase expressed alone or with the corresponding anSMEs but also with pRSF-Ser-Sulf, pRSF-_anSMEbt_-Ser-Sulf, or pRSF-_anSMEcpe_-Ser-Sulf to obtain the corresponding Ser-type sulfatases. The six strains were grown overnight anaerobically in stoppered flasks under an H2/CO2/N2 (5:5:90) atmosphere at 37 °C in LB medium (100 ml) supplemented with kanamycin (50 μgml–1). The overnight culture was then used to inoculate fresh LB medium (1 liter) supplemented with the same antibiotic and bacterial growth proceeded at 37 °C until the _A_600 reached 0.2. The cells were induced by adding 500 μm isopropyl 1-thio-β-d-galactopyranoside and were collected after overnight growth at 30 °C. After resuspension in Tris buffer, the cells were disrupted by sonication and centrifuged at 220,000 × g at 4 °C for 1 h. The solutions were then loaded onto Ni-NTA-Sepharose columns equilibrated with Tris buffer, pH 7.5. The columns were washed extensively with the same buffer. After two washing steps with 25 and 100 mm imidazole, the overexpressed proteins were eluted by applying 500 mm imidazole.
_Sulfatase Assay_—Sulfatase activity was assayed using the chromogenic substrate _p_-nitrophenylsulfate as substrate. Standard assays were performed at 30 °C using 50 mm _p_-nitrophenylsulfate in 100 mm Tris buffer containing 10 mm MgCl2, pH 7.15. Formation of _p_-nitrophenol was measured spectrophotometrically at 405 nm (ε = 9,000m–1 cm–1 at pH 7.15).
_Sulfatase Treatment for MALDI-TOF Analysis_—Purified sulfatases were dissolved (150 pmol μl–1) into 0.4m urea, then digested overnight with trypsin (20 ng μl–1) in ammonium carbonate buffer, pH 8.0, at 37 °C. 5 μl of digested sulfatase was further hydrolyzed with 5 μl of CNBr (20 mg ml–1 in 0.1 m HCl) in the dark overnight at room temperature, then lyophilized and purified on ZipTip (Millipore, Inc.) before MALDI-TOF mass spectrometry analysis.
_MALDI-TOF MS Analysis_—The α-cyano-4-hydroxycinnamic acid matrix was prepared at 4 mg ml–1 in 0.15% trifluoroacetic acid, 50% acetonitrile. The 2,4-dinitrophenylhydrazone acid matrix (DNPH) was prepared at 100 mg ml–1 in 0.15% trifluoroacetic acid, 50% acetonitrile. Equal volumes (1 μl) of matrix and sample were spotted onto the MALDI-TOF target plate. MALDI-TOF analysis was then performed on a Voyager DE STR Instrument (Applied Biosystems, Framingham, CA). Spectra were acquired in the reflector mode with: 20 kV accelerating voltage, 62% grid voltage, and a 120-ns delay.
RESULTS
_Identification of B. thetaiotaomicron anSME_—We previously reported the identification of the C. perfringens anaerobic sulfatase-maturating enzyme, anSMEcpe (5,7). Using its sequence as a probe, only one enzyme sharing 30% identity with anSMEcpe and possessing the three strictly conserved cysteine clusters was identified by Blast searching the B. thetaiotaomicron genome, which encodes only Ser-type sulfatases (10) (Fig. 1). This protein, encoded by the BT_0238 gene, annotated as a transcriptional regulator and named ChuR for chondroitin sulfate/heparin utilization regulator (11), was amplified by PCR and named anSMEbt.
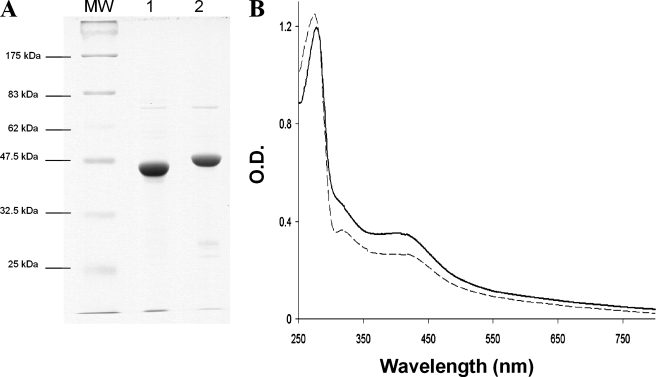
Overexpression and Purification of anSMEbt and anSMEcpe with an N-terminal Hexahistidine Tag_—Both anSMEcpe and_anSMEbt genes were subcloned into the pET-28(a) plasmid and the resulting expression vectors, pET-6His-anSMEbt and pET-6His-anSMEcpe, were used to transform E. coli BL21(DE3) for overproduction of histidine-tagged anSMEbt and anSMEcpe. Both enzymes were essentially expressed as inclusion bodies and induction under low temperature was necessary to obtain small amounts of the soluble proteins. Typically ∼5 mg of pure proteins were obtained from a 12-liter culture. Both proteins eluted as dark brown bands from Ni-NTA affinity chromatography, which was consistent with the presence of iron-sulfur clusters, and each protein migrated according to its predicted molecular mass (at ∼45 and 50 kDa for anSMEcpe and anSMEbt, respectively) on SDS-PAGE (Fig. 2_A_). Anaerobic reconstitution with Fe2+ and S2–, followed repurification, increased the iron-sulfur cluster content as evidenced by the increase in visible absorption at ∼420 nm relative to the protein band at 280 nm. As shown inFig. 2_B_, the UV-visible spectra of the reconstituted anSMEs were very similar and both exhibited the characteristic absorption bands associated with iron-sulfur clusters. The nature and stoichiometry of the iron-sulfur clusters in as isolated and reconstituted anSMEcpe were assessed by protein and iron determinations and the combination of UV-visible absorption, EPR, and resonance Raman spectroscopies.
FIGURE 2.
A, gel electrophoresis analysis of anSMEcpe (lane 1) and anSMEbt (lane 2)(MW, molecular weight markers). B, UV-visible absorption spectra of reconstituted anSMEbt (dotted line) and anSMEcpe (solid line).
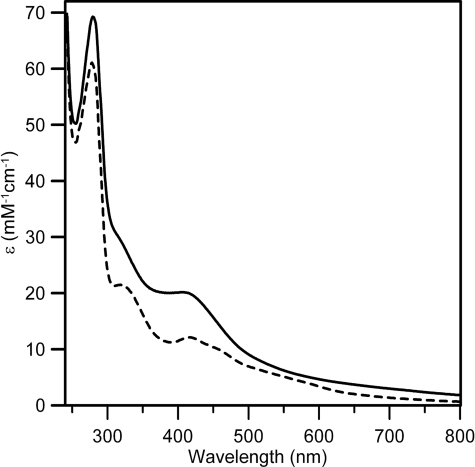
_Nature and Stoichiometry of Iron-Sulfur Centers_—In accord with previous studies (7), analytical data for the anSMEcpe samples used for spectroscopic analyses indicated 2.5 ± 0.3 iron/monomer for aerobically purified samples and 5.7 ± 0.5 iron/monomer for samples anaerobically reconstituted with Fe2+ and S2–. The form and visible extinction coefficients of corresponding UV-visible absorption spectra (seeFig. 3) are consistent with approximately one [2Fe-2S]2+ and one [4Fe-4S]2+ cluster in the as isolated and reconstituted samples, respectively. As isolated anSMEcpe has resolved bands at 320 and 420 nm (_A_420/_A_280 = 0.19 ± 0.02) and shoulders centered at ∼460 and ∼550 nm that are characteristic of [2Fe-2S]2+ clusters and extinction coefficient at 420 nm (ε420 = 11.8 ± 1.2 mm–1 cm–1) is just above the high end of the range generally associated with a single [2Fe-2S]2+ cluster (ε420 = 8–11 mm–1 cm–1) (12). Reconstituted anSMEcpe exhibits broad shoulders centered at ∼320 and ∼400 nm that are characteristic of a [4Fe-4S]2+ cluster (_A_420/_A_280 = 0.29 ± 0.03) and the extinction coefficient at 400 nm (ε400 = 20 ± 2 mm–1 cm–1) is just above the range generally associated with a single [4Fe-4S]2+ cluster (ε400 = 14–18 mm–1 cm–1) (13).
FIGURE 3.
UV-visible absorption spectra of as isolated (dashed line) and reconstituted (solid line) anSMEcpe.
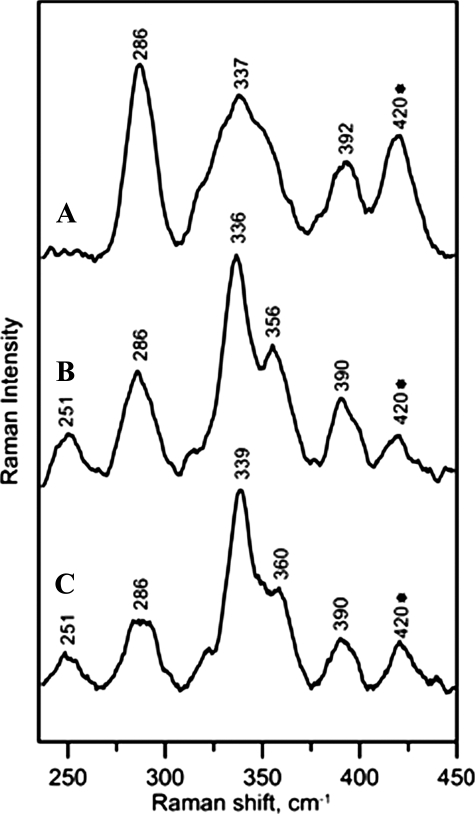
More definitive information concerning the nature of the iron-sulfur clusters in as isolated and reconstituted anSMEcpe and for AdoMet binding to the [4Fe-4S]2+ cluster in the reconstituted enzyme was provided by resonance Raman spectroscopy (Fig. 4). The resonance Raman spectrum of as isolated anSMEcpe comprises broad bands centered at 286, 337, and 392 cm–1 (with the possibility of an additional weak band underlying the glycerol band at 420 cm–1) (Fig. 4_A_) and is characteristic of a cluster [2Fe-2S]2+ that is derived from O2 degradation of a radical-AdoMet-type [4Fe-4S]2+ cluster. For example, very similar spectra have been observed in the as purified pyruvate formate lyase activating enzyme and in [4Fe-4S]2+ cluster-containing forms of biotin synthase after exposure to O2 (14,15). Hence we conclude that the [2Fe-2S]2+ cluster in the as isolated anSMEcpe is wholly or predominantly derived from O2-induced degradation of a radical-AdoMet type [4Fe-4S]2+ cluster during aerobic isolation. Reconstituted anSMEcpe has a resonance Raman spectrum that is dominated by bands at 251, 336, 356, and 390 cm–1 that are characteristic of a [4Fe-4S]2+ cluster (15) (Fig. 4_B_). Bands associated with the [2Fe-2S]2+ cluster that is present in the as isolated enzyme are still apparent, e.g. at 286 cm–1, but they tend to overestimate the [2Fe-2S]2+ cluster content as resonance enhancement of [2Fe-2S]2+ clusters are 5–10 times greater than that of [4Fe-4S]2+ clusters with 458 nm excitation (15). Based on parallel Mössbauer and resonance Raman studies of a range of radical-AdoMet enzymes with mixtures of [4Fe-4S]2+ clusters and their [2Fe-2S]2+ cluster O2 degradation products, we estimate the cluster composition of reconstituted anSMEcpe to be 10–20% [2Fe-2S]2+ clusters and 90–80% [4Fe-4S]2+ clusters. Hence, the majority of the [2Fe-2S]2+ clusters present in the as isolated anSMEcpe have been replaced by [4Fe-4S]2+ clusters during anaerobic reconstitution. Moreover, the [4Fe-4S]2+ clusters have been assembled in the radical-AdoMet cluster binding site, as evidenced by changes in resonance Raman spectrum of reconstituted anSMEcpe on addition of excess AdoMet (Fig. 4_C_). Crystallographic studies have demonstrated that AdoMet binds to the unique iron of the active site [4Fe-4S]2+ cluster in radical-AdoMet enzymes via the methionine amide and carboxylate groups (16) and this is manifest by 3–4 cm–1 upshifts in both dominant bands in the resonance Raman spectrum, i.e. the symmetric breathing mode of the [4Fe-4S] core and the asymmetric stretching mode of the terminal Fe-S(Cys) ligands (17). These bands are observed at 336 and 356 cm–1, respectively, in reconstituted anSMEcpe and upshift to 339 and 360 cm–1, respectively, on addition of excess AdoMet.
FIGURE 4.
Resonance Raman spectra of anSMEcpe as isolated (A), reconstituted (B), and reconstituted in the presence of a 20-fold excess of SAM (C). The spectra were recorded with 458-nm excitation, using samples that were ∼3 mm in anSMEcpe frozen at 20 K, with 140 milliwatt laser power at the sample. Each scan involved photon counting for 1 s at 1.0 cm–1 increments with 7 cm–1 spectral resolution, and each spectrum is the sum of ∼100 scans. Bands resulting from the lattice modes have been subtracted from each spectrum and the band at 420 cm–1 that is marked with as asterisk contains a contribution from glycerol.
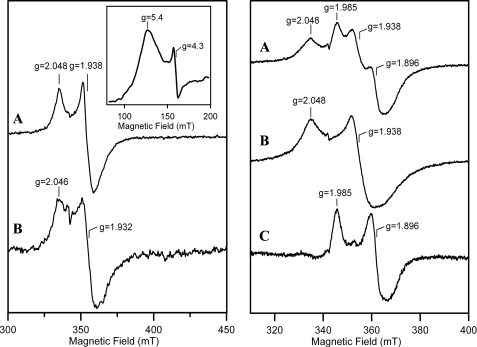
EPR was used to investigate the type and properties of the reduced forms of anSMEcpe that were generated by anaerobic reduction with dithionite (Fig. 5). The EPR properties of dithionite-reduced reconstituted anSMEcpe were characteristic of a [4Fe-4S]+ cluster in a radical-AdoMet enzyme (18,19) (Fig. 5_A_, left panel), i.e. mixed spin system with a rhombic S = 3/2 component (as evidenced by the broad low-field feature centered at g = 5.4, shown in the inset) and a near-axial, fast-relaxing S = 1/2 component (_g_∥ ∼ 2.05, _g_⊥ ∼ 1.94) accounting for ∼0.2 spins/mol. A very weak S = 1/2 resonance (∼0.03 spins/mol) with similar g values and relaxation behavior was observed on dithionite reduction of the as isolated enzyme (Fig. 5_B_, left panel). We attributed this to [4Fe-4S]+ clusters formed either by reduction of residual [4Fe-4S]2+ clusters in the as isolated sample or to [4Fe-4S] clusters that were reassembled in the radical-AdoMet cluster binding site on reductive degradation of the [2Fe-2S]2+ cluster. The [4Fe-4S]+ cluster in the reconstituted enzyme interacted with AdoMet as evidenced by partial conversion of the S = 1/2 EPR signal to a new species with significantly lower g values on addition of AdoMet (_g_∥ ∼ 1.99,_g_⊥ ∼ 1.90) (Fig. 5, right panel). Similar changes in the S = 1/2 [4Fe-4S]+ EPR signal on addition of AdoMet have been observed with many radical-AdoMet enzymes (20–22). Because the radical-AdoMet active site can turn over on addition of dithionite in the presence of AdoMet, it is often difficult to get the [4Fe-4S]+ cluster in a fully AdoMet-bound form.
FIGURE 5.
EPR spectra of anSMEcpe after anaerobic reduction with a 10-fold excess of sodium dithionite. Spectra were recorded for samples of 0.2 mm anSMEcpe at 10 K with a microwave frequency of 9.603 GHz, a modulation amplitude of 0.63 millitesla, and a microwave power of 10 milliwatt, unless otherwise indicated. Left panel, reconstituted anSMEcpe (A) and as isolated anSMEcpe (B). Inset shows the low field region of the spectrum for reconstituted anSMEcpe recorded at 4.3 K and 50 mm. Right panel, reconstituted anSMEcpe in the presence (A) or absence (B) of a 20-fold excess of SAM. The difference spectrum corresponding to: A minus 0.5 ×B is shown in C.
Taken together, the spectroscopic and analytical data are consistent with the anSMEcpe samples investigated in this work only having clusters assembled in the radical-AdoMet cluster binding site involving the rigorously conserved C_X_3C_X_2C motif. Reconstituted samples have a catalytically competent [4Fe-4S]2+,+ cluster assembled in this site that can interact with AdoMet in both the oxidized and reduced states. In aerobically isolated samples, the [4Fe-4S]2+,+ cluster degrades to an O2-stable [2Fe-2S]2+ cluster that is rapidly and irreversibly degraded on reduction. Nevertheless, further experiments would be required to definitively rule out the presence of a second iron-sulfur cluster.
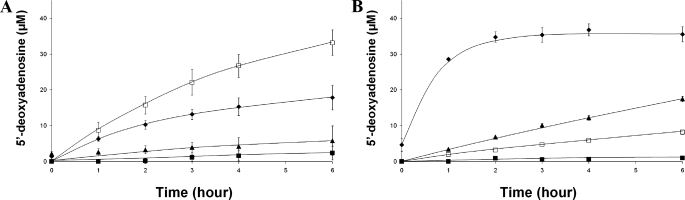
_In Vitro Activity of anSMEbt and anSMEcpe_—Purified and reconstituted anSMEs were tested in vitro under anaerobic conditions for their ability to cleave AdoMet and to mature two peptides mimicking the Cys-type and Ser-type sulfatases. Each reconstituted protein (20 μm) was incubated with 1 mm AdoMet in the presence or absence of peptides using dithionite as reductant and, in each case, 5′-deoxyadenosine production was followed by HPLC analysis. In the absence of peptides, anSMEcpe and anSMEbt exhibited different behaviors (Fig. 6, open square). After a 6-h incubation, anSMEcpe was able to produce only low amounts of 5′-deoxyadenosine (8 μm), whereas anSMEbt produced 4 times more 5′-deoxyadenosine (33 μm). In contrast, both enzymes behaved similarly in the presence of 23-mer peptides (500 μm). Both enzymes were sensitive to the nature of the first residue in the C_X_A_X_R motif, exhibiting substantially higher levels of 5′-deoxyadenosine production in the presence of the cysteine-containing peptide (23C) (35 μm for anSMEcpe and 17 μm for anSMEbt) (Fig. 6,diamonds) compared with the serine-containing peptide (23S) (17 and 6 μm, respectively) (Fig. 6, triangles). Furthermore, substitution of the cysteine residue by alanine (23A) resulted in inhibition of AdoMet cleavage for both enzymes and became almost undetectable for anSMEcpe.
FIGURE 6.
AdoMet reductive cleavage assayed by reverse phase HPLC. Incubation of 20 μm reconstituted anSMEbt (A) or anSMEcpe (B), with 1 mm AdoMet alone (□) or with 23-mer peptides (500 μm) containing a cysteine (♦), serine (▴), or alanine (▪) as target residue.
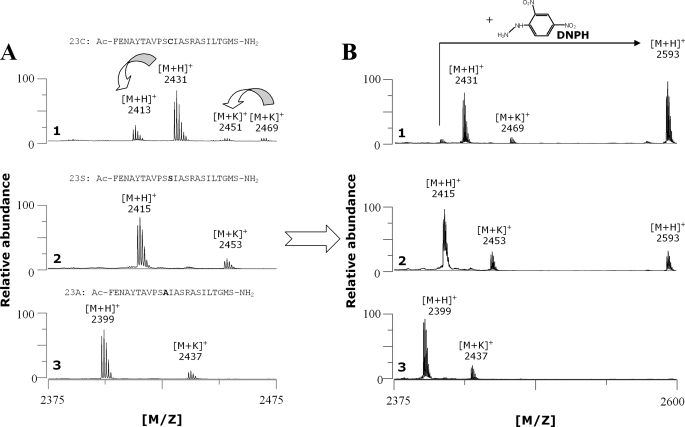
_In Vitro Peptide Maturation_—Using MALDI-TOF MS, peptide maturation was followed both by direct measurement in a α-cyano-4-hydroxycinnamic acid matrix and derivatization with DNPH (23). anSMEcpe efficiently converted the cysteine of peptide 23C as shown by the production of a peptide with an 18-Da mass shift (Fig. 7_A_, panel 1). The nature of the modification was assessed using DNPH, which specifically reacts with the FGly aldehyde function leading to a hydrazone derivative with a 180.13-Da mass increment (Fig. 7_B_, panel 1, and supplemental Fig. S1). For the incubation with the 23S peptide, we were unable to directly observe a FGly-containing peptide that should have a 2-Da shift from the 23S peptide (Fig. 7_A_, panel 2). Nevertheless, derivatization with DNPH allowed us to detect a product with the expected mass for the hydrazone derivative of the FGly-containing peptide (Fig. 7_B_, panel 2, and supplemental Fig. S1). Indeed DNPH not only gives a mass increment but also enhances peptide ionization that is then more easily detected.
FIGURE 7.
In vitro maturation of 23-mer peptides with reconstituted anSMEcpe. After 6 h of incubation under anaerobic conditions in the presence of AdoMet and a cysteine (panel 1), serine (panel 2), or alanine-containing peptide (panel 3), maturation was analyzed by MALDI-TOF MS either with a α-cyano-4-hydroxycinnamic acid (A) or a DNPH matrix (B).
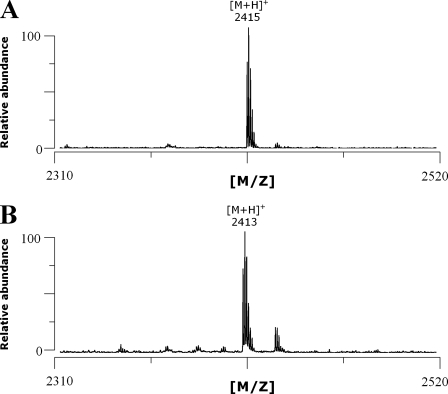
To definitively demonstrate the formation of an FGly-peptide from the 23S peptide, the peptides from the incubation mixture were purified using C18-HPLC and the fractions obtained were analyzed by MALDI-TOF MS. In our conditions, the 23S peptide elutes at 24 min as confirmed by MALDI-TOF MS analysis (molecular mass: 2415 Da, Fig. 8_A_), and in a preceding fraction we were able to isolate the FGly-containing peptide as assessed by its molecular mass (2413 Da) (Fig. 8_B_).
FIGURE 8.
MALDI-TOF MS analysis of the serine (A) and FGly-containing peptide (B) purified by HPLC.
Incubation with the 23A peptide did not lead to any new peptide despite the presence of four serine residues within the peptide sequence (Fig. 7, A panel 3 and_B, panel 3_). MALDI-TOF MS analysis performed on the peptides incubated with B. thetaiotaomicron enzyme (anSMEbt) gave similar results (supplemental Figs. S2 and S3).
Both, anSMEbt and anSMEcpe were thus active on cysteine and serine residues. Nevertheless, the MALDI-TOF MS experiment did not allow quantifying the efficiency of maturation. Notably, the 23S peptide ionizes much more efficiently than the 23C and the FGly-containing peptides. Attempts to quantify peptide maturation using HPLC failed mostly because the 23C peptide proved to be unstable. We thus devised an in vivo strategy to compare anSME maturation efficiency on both residues.
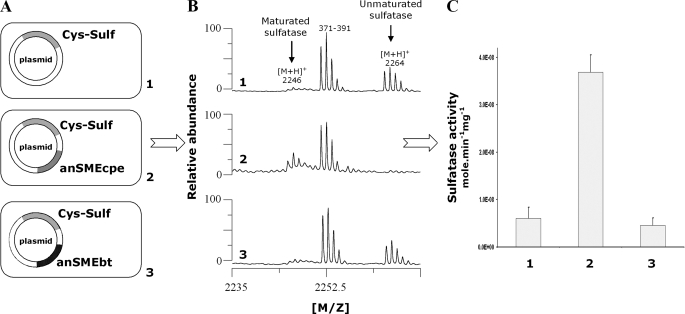
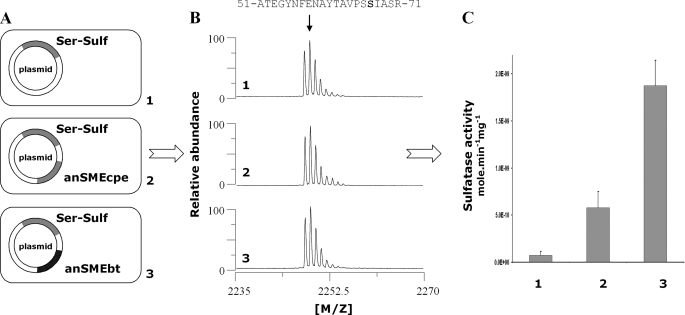
_In Vivo Maturation of C. perfringens Cys-type Sulfatase_—We used the Cys-type sulfatase from C. perfringens as a reporter gene and constructed three pRSF plasmids containing the Cys-type sulfatase into subsite 1 alone or with anSMEbt or anSMEcpe into subsite 2. These plasmids were named pRSF-Cys-Sulf, pRSF-anSMEbt-Cys-Sulf, and pRSF-anSMEcpe-Cys-Sulf. Using these plasmids, we obtained three strains expressing the Cys-type sulfatase alone as a His tag protein or co-expressed with anSMEbt or anSMEcpe. These strains were grown under anaerobic conditions as our previous investigations have demonstrated that E. coli constitutively matures Cys-type sulfatases but this maturation was partly repressed under anaerobic conditions (24). After overnight induction with isopropyl 1-thio-β-d-galactopyranoside, the expressed sulfatases were purified (see “Experimental Procedures”) and their maturation assessed by MALDI-TOF mass spectrometry. The conversion of cysteine into FGly is expected to result in a mass decrease of 18 Da, which could be evidenced in the peptides obtained after trypsin/CNBr treatment of the sulfatases (7). We also measured the specific activity of the purified sulfatases using the chromogenic substrate_p_-nitrophenylsulfate as the sulfatase activity is directly proportional to the maturation level. As shown inFig. 9_B_, panel 1, when the Cys-type sulfatase was expressed alone under anaerobic conditions, one relevant peptide was obtained containing the critical Cys-67 and encompassing residues from 51 to 71:51ATEGYNFENAYTAVPSCIASR71 (theoretical molecular mass 2264.03 Da). This peptide with an intact cysteine residue thus corresponds to unmaturated sulfatase. When the sulfatase was coexpressed with anSMEcpe (Fig. 9_B_,panel 2), the 2264-Da peptide disappeared and was replaced by a new peptide with a mass shift of 18 Da. This peptide (theoretical molecular mass 2246.03 Da) corresponded to maturated sulfatase. Surprisingly, when we expressed the Cys-type sulfatase with anSMEbt, MALDI-TOF MS indicated principally the presence of unmaturated sulfatase (Fig. 9_B_, panel 3).
FIGURE 9.
In vivo_maturation of_C. perfringens sulfatase expressed alone (panel 1) or in the presence of anSMEcpe (panel 2) or anSMEbt (panel 3). A, maps of the plasmids used for the anaerobic production of the C. perfringens sulfatase. B, MALDI-TOF MS analysis of purified sulfatase digested with trypsin and CNBr (numbers indicate the amino acid residues). C, specific activity of the purified sulfatases.
The specific sulfatase activity assay of each enzyme confirmed the results obtained by MALDI-TOF MS (Fig. 9_C_). No differences in the sulfatase activity was evidenced between the Cys-type sulfatase expressed alone or in the presence of anSMEbt, whereas anSMEcpe significantly increased the specific sulfatase activity from 6 to 36 nmol min–1 mg–1. The basal sulfatase activity in the absence of known maturation enzyme was probably due to an imperfect inhibition of the E. coli sulfatase maturation system as trace maturation could be seen in all the MALDI-TOF MS spectra.
_Construction of the Serine Variant of C. perfringens Cys-type Sulfatase_—To investigate the activity of both anSMEs toward the serine residue, we converted the C. perfringens Cys-type sulfatase into a Ser-type sulfatase by site-directed mutagenesis. We obtained three strains harboring the following plasmids named pRSF-Ser-Sulf, pRSF-anSMEbt-Ser-Sulf, and pRSF-anSMEcpe-Ser-Sulf, respectively. These three strains were also grown under anaerobic conditions (Fig. 10) although identical results were obtained under aerobic conditions (data not shown). The MALDI-TOF MS analysis of purified sulfatases, after trypsin/CNBr treatment, confirmed the mutation of the Cys-67 into Ser, as a 2248-Da peptide was observed consistent with the following sequence:51ATEGYNFENAYTAVPSSIASR71 (Fig. 10_B_). Like in the in vitro experiments, the ionization properties of this peptide differed considerably from those of the cysteine and FGly-containing peptide. As a result, this peptide dominated the whole spectrum and prevented the direct measurement of maturation by MALDI-TOF MS.
FIGURE 10.
In vivo maturation of the serine mutant of the C. perfringens sulfatase expressed alone (panel 1) or in the presence of anSMEcpe (panel 2) or anSMEbt (panel 3). A, maps of the plasmids used for the anaerobic production of the serine mutant of C. perfringens sulfatase. B, MALDI-TOF MS analysis of purified sulfatases digested with trypsin and CNBr (numbers indicate the amino acid residues). C, specific activity of the purified sulfatases.
The sulfatase activity of the serine mutant expressed alone in E. coli was almost undetectable (Fig. 10_C_, panel 1), consistent with previous reports on Ser-type sulfatases (3,25). This result confirms that the unknown E. coli sulfatase maturation system is restricted to the Cys-type sulfatases (24). Nevertheless, when the serine mutant was co-expressed with anSMEcpe or anSMEbt, we clearly measured sulfatase activity which was 25 times higher than in the control experiment. anSMEbt and anSMEcpe are thus able to activate Ser-type sulfatases but with an apparent lower efficiency than on Cys-type sulfatase.
DISCUSSION
Sulfatases are widely distributed in eukaryotes and prokaryotes (1) due to their biological significance and their high substrate versatility. Sulfatases are involved in the metabolism of various compounds ranging from small organic molecules to macromolecules such as mucins and glycosaminoglycans (1). They are usually considered to be restricted to sulfate supply but they probably also have other functions that remain to be explored. Consequently, sulfatase genes are identified in many bacterial genomes and metagenomes notably from those arising from marine and gut environments (10,26–28). Despite their functional importance, little is known about sulfatase maturation. Until recently, it was thought that an enzymatic system, formylglycine generating enzyme, was dedicated to the maturation of Cys-type sulfatases (29,30) and another, AtsB, to the Ser-type sulfatases (3).
These assumptions were based on several observations. The specificity of formylglycine generating enzyme is based on experiments showing that eukaryotic cells are unable to maturate a serine sulfatase mutant (31). In contrast, the data are more complex for AtsB. Co-expression experiments performed with a serine sulfatase mutant obtained from the Cys-type sulfatase of Pseudomonas aeruginosa failed to produce an active enzyme. Nevertheless, an active sulfatase was obtained if this serine mutant carried a signal peptide in addition of the critical serine residue (6). This leads to the assumption that AtsB recognizes only Ser-type sulfatases with the help of a signal peptide (4,6).
We recently demonstrated that the C. perfringens AtsB ortholog is able to convert cysteine residue into FGly. In light of the homologies between the enzyme from K. pneumoniae and C. perfringens, these enzymes were gathered under the collective name of anSME (5). This raised the possibility of enzymes with different substrate specificities within the anSME group, some enzymes being dedicated to maturation of Ser-type sulfatases and others to the Cys-type sulfatases. To test this hypothesis, we decided to investigate the specificity of the anSMEs from C. perfringens (anSMEcpe) and B. thetaiotaomicron, a prominent gut symbiont (10), which possesses only Ser-type sulfatases and only one enzyme related to the anSME group. The B. thetaiotaomicron anSME is currently called ChuR (chondroitin heparin utilization regulator) and has been annotated as a transcriptional regulator as its inactivation prevents the bacterium to degrade various substrates (11). We cloned this enzyme and named it anSMEbt.
We successfully obtained both enzymes (anSMEbt and anSMEcpe) in soluble form after induction under low temperature and extraction in the presence of glycerol. Even though the yields were very low, sufficient amounts of anSMEcpe were obtained to enable characterization of the iron-sulfur centers in aerobically isolated and anaerobically reconstituted samples using the combination of UV-visible absorption, resonance Raman, and EPR spectroscopies. The results indicate that anaerobic reconstitution generates a [4Fe-4S]2+,+ cluster in the radical-AdoMet cluster binding site (C_X_3C_X_2C motif) that is able to bind AdoMet in both oxidation states. The aerobically isolated enzyme contains stoichiometric amounts of a reductively labile [2Fe-2S]2+ cluster with resonance Raman properties analogous to those established for [2Fe-2S]2+ clusters that form as a result of O2-induced degradation of radical-AdoMet active-site [4Fe-4S]2+ clusters. Hence it seems likely that the active-site [4Fe-4S]2+ cluster in anSME is not stable in air and degrades to yield an O2-stable [2Fe-2S]2+ cluster in the radical-AdoMet cluster binding site during aerobic isolation. Although the spectroscopic and analytical results presented in this work can be satisfactorily rationalized in terms of a single [4Fe-4S]2+ or [2Fe-2S]2+ cluster ligated by the radical-AdoMet C_X_3C_X_2C motif, it is not yet possible to definitively rule out the possibility that one or both of the two other cysteine clusters in anSMEs (Fig. 1) are involved in iron-sulfur cluster coordination as previously hypothesized (4). Notably, the third cysteine cluster is also strictly conserved in the AdoMet radical enzyme involved in quinohemoprotein amine dehydrogenase biosynthesis (32). Further mutagenesis studies coupled with spectroscopic analysis should be conducted to determine the definitive role of these cysteine clusters.
The in vitro assays performed with both enzymes revealed that anSMEs were able to produce 5′-deoxyadenosine from AdoMet. Furthermore, in the presence of 23-mer peptides, the activity of both enzymes was modulated and, surprisingly, greatly influenced by the first amino acid present in the target sulfatase motif (C_X_A_X_R). When a cysteine or serine residue was present, both enzymes produced 5′-deoxyadenosine from AdoMet although at different levels, according to the target amino acid residue. However, when this residue was substituted with alanine, the 5′-deoxyadenosine production was strongly inhibited, indicating a tight control of the anSME activity.
Maturation analysis of the different peptides showed that the serine residue of the 23S peptide and the cysteine residue of the 23C peptide were both converted into FGly by anSMEbt and anSMEcpe. This result indicated that both enzymes were able to oxidize serine or cysteine residues to FGly. Furthermore, our results also definitively established that the enzyme encoded by the chuR gene is not a transcriptional regulator but a radical-AdoMet enzyme involved in sulfatase maturation. We also showed that an alanine-containing peptide (23A), which contains four serines in its sequence, was not modified by either enzyme demonstrating the specificity of anSMEs for the sulfatase target sequence. Nevertheless, all critical peptide determinants that direct the selectivity of these enzymes are not precisely known and are currently under investigation.
With these in vitro experiments we were not able to compare the maturation efficiency of both enzymes toward the cysteine and serine residues. We thus devised an in vivo strategy to compare their activity on cysteine and serine residues. Using the C. perfringens sulfatase and its serine mutant we measured the in vivo activation catalyzed by anSMEbt and anSMEcpe. Both enzymes were able to activate the Ser-type sulfatase with sulfatase activity increased up to 25 times. Nevertheless, only anSMEcpe significantly increased the Cys-type sulfatase activity. This result suggests that anSMEs, despite being dual substrate enzymes, may exhibit substrate preferences. These in vivo experiments also demonstrated that anSMEs, contrary to previous reports (4,6), were able to maturate Ser-type sulfatases without the necessity of a signal peptide.
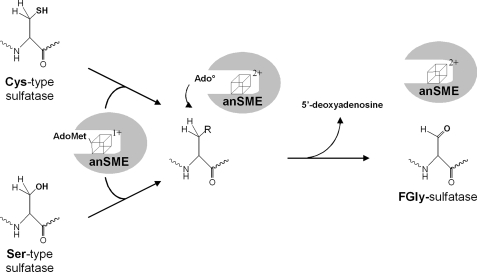
In conclusion, our data support the view that anSMEs are dual substrate radical-AdoMet enzymes rather than a class of enzymes divided into two subgroups dedicated to the maturation of either Ser-type or Cys-type sulfatases. Consistent with our experimental data, we propose herein the first mechanism for this fascinating class of enzymes (Fig. 11). The reduced [4Fe-4S]+ cluster in anSME interacts with AdoMet and reductively cleaves AdoMet to yield methionine and a 5′-deoxyadenosyl radical (Ado•), which abstracts a proton from either a cysteine or serine residue leading to the production of 5′-deoxyadenosine and to the formation of FGly. In this scheme, it is not yet possible to determine whether AdoMet is a co-substrate as is the case for the majority of radical-AdoMet enzymes, or a cofactor as demonstrated in the case of lysine 2,3-aminomutase or spore photoproduct lyase (33–35). Further investigations will thus be required to fully elucidate the mechanism of this class of radical-AdoMet enzymes.
FIGURE 11.
Proposed mechanism for the maturation reaction catalyzed by anSMEs.
Supplementary Material
[Supplemental Data]
Acknowledgments
We thank Dr. E. Mulliez (DRDC/CB, Grenoble) for providing S-adenosylmethionine. Mass spectrometry experiments were performed at the Plateau d'Analyse Protéomique par Séquençage et Spectrométrie de masse (PAPSS, INRA, Jouy-en-Josas).
*
This work was supported, in whole or in part, by National Institutes of Health Grant GM62524 (to M. K. J.). This work was also supported by a grant from the Région Ile-de-France (to A. B.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “_advertisement_” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
S⃞
The on-line version of this article (available athttp://www.jbc.org) contains supplemental Figs. S1–S3.
Footnotes
3
The abbreviations used are: FGly, Cα-formylglycine; AdoMet, _S_-adenosylmethionine; anSME, anaerobic sulfatase-maturating enzyme; DNPH, 2,4-dinitrophenylhydrazine; Ni-NTA, nickel-nitrilotriacetic acid; HPLC, high pressure liquid chromatography; MALDI-TOF, matrix-assisted laser desorption ionization time-of-flight; MS, mass spectrometry.
References
- 1.Hanson, S. R., Best, M. D., and Wong, C. H. (2004) Angew. Chem. Int. Ed. Engl. 43 5736–5763 [DOI] [PubMed] [Google Scholar]
- 2.Roeser, D., Preusser-Kunze, A., Schmidt, B., Gasow, K., Wittmann, J. G., Dierks, T., von Figura, K., and Rudolph, M. G. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 81–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szameit, C., Miech, C., Balleininger, M., Schmidt, B., von Figura, K., and Dierks, T. (1999) J. Biol. Chem. 274 15375–15381 [DOI] [PubMed] [Google Scholar]
- 4.Fang, Q., Peng, J., and Dierks, T. (2004) J. Biol. Chem. 279 14570–14578 [DOI] [PubMed] [Google Scholar]
- 5.Berteau, O., Guillot, A., Benjdia, A., and Rabot, S. (2006) J. Biol. Chem. 281 22464–22470 [DOI] [PubMed] [Google Scholar]
- 6.Marquordt, C., Fang, Q., Will, E., Peng, J., von Figura, K., and Dierks, T. (2003) J. Biol. Chem. 278 2212–2218 [DOI] [PubMed] [Google Scholar]
- 7.Benjdia, A., Leprince, J., Guillot, A., Vaudry, H., Rabot, S., and Berteau, O. (2007) J. Am. Chem. Soc. 129 3462–3463 [DOI] [PubMed] [Google Scholar]
- 8.Wang, W., and Malcolm, B. A. (1999) BioTechniques 26 680–682 [DOI] [PubMed] [Google Scholar]
- 9.Drozdzewski, P. M., and Johnson, M. K. (1988) Appl. Spectrosc. 42 1575–1577 [Google Scholar]
- 10.Xu, J., Bjursell, M. K., Himrod, J., Deng, S., Carmichael, L. K., Chiang, H. C., Hooper, L. V., and Gordon, J. I. (2003) Science 299 2074–2076 [DOI] [PubMed] [Google Scholar]
- 11.Cheng, Q., Hwa, V., and Salyers, A. A. (1992) J. Bacteriol. 174 7185–7193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dailey, H. A., Finnegan, M. G., and Johnson, M. K. (1994) Biochemistry 33 403–407 [DOI] [PubMed] [Google Scholar]
- 13.Agar, J. N., Krebs, C., Frazzon, J., Huynh, B. H., Dean, D. R., and Johnson, M. K. (2000) Biochemistry 39 7856–7862 [DOI] [PubMed] [Google Scholar]
- 14.Broderick, J. B., Duderstadt, R. E., Fernandez, D. C., Wojtuszewski, K., Henshaw, T. F., and Johnson, M. K. (1997) J. Am. Chem. Soc. 119 7396–7397 [Google Scholar]
- 15.Cosper, M. M., Jameson, G. N., Hernandez, H. L., Krebs, C., Huynh, B. H., and Johnson, M. K. (2004) Biochemistry 43 2007–2021 [DOI] [PubMed] [Google Scholar]
- 16.Layer, G., Kervio, E., Morlock, G., Heinz, D. W., Jahn, D., Retey, J., and Schubert, W. D. (2005) J. Biol. Chem. 386 971–980 [DOI] [PubMed] [Google Scholar]
- 17.Cosper, M. M., Jameson, G. N., Davydov, R., Eidsness, M. K., Hoffman, B. M., Huynh, B. H., and Johnson, M. K. (2002) J. Am. Chem. Soc. 124 14006–14007 [DOI] [PubMed] [Google Scholar]
- 18.Duin, E. C., Lafferty, M. E., Crouse, B. R., Allen, R. M., Sanyal, I., Flint, D. H., and Johnson, M. K. (1997) Biochemistry 36 11811–11820 [DOI] [PubMed] [Google Scholar]
- 19.Ollagnier, S., Meier, C., Mulliez, E., Gaillard, J., Schuenemann, V., Trautwein, A., Mattioli, T., Lutz, M., and Fontecave, M. (1999) J. Am. Chem. Soc. 121 6344–6350 [Google Scholar]
- 20.Ollagnier, S., Mulliez, E., Schmidt, P. P., Eliasson, R., Gaillard, J., Deronzier, C., Bergman, T., Graslund, A., Reichard, P., and Fontecave, M. (1997) J. Biol. Chem. 272 24216–24223 [DOI] [PubMed] [Google Scholar]
- 21.Walsby, C. J., Hong, W., Broderick, W. E., Cheek, J., Ortillo, D., Broderick, J. B., and Hoffman, B. M. (2002) J. Am. Chem. Soc. 124 3143–3151 [DOI] [PubMed] [Google Scholar]
- 22.Kriek, M., Martins, F., Leonardi, R., Fairhurst, S. A., Lowe, D. J., and Roach, P. L. (2007) J. Biol. Chem. 282 17413–17423 [DOI] [PubMed] [Google Scholar]
- 23.Peng, J., Schmidt, B., von Figura, K., and Dierks, T. (2003) J. Mass Spectrom. 38 80–86 [DOI] [PubMed] [Google Scholar]
- 24.Benjdia, A., Deho, G., Rabot, S., and Berteau, O. (2007) FEBS Lett. 581 1009–1014 [DOI] [PubMed] [Google Scholar]
- 25.Wright, D. P., Knight, C. G., Parkar, S. G., Christie, D. L., and Roberton, A. M. (2000) J. Bacteriol. 182 3002–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gill, S. R., Pop, M., Deboy, R. T., Eckburg, P. B., Turnbaugh, P. J., Samuel, B. S., Gordon, J. I., Relman, D. A., Fraser-Liggett, C. M., and Nelson, K. E. (2006) Science 312 1355–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glockner, F. O., Kube, M., Bauer, M., Teeling, H., Lombardot, T., Ludwig, W., Gade, D., Beck, A., Borzym, K., Heitmann, K., Rabus, R., Schlesner, H., Amann, R., and Reinhardt, R. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 8298–8303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu, J., Mahowald, M. A., Ley, R. E., Lozupone, C. A., Hamady, M., Martens, E. C., Henrissat, B., Coutinho, P. M., Minx, P., Latreille, P., Cordum, H., Van Brunt, A., Kim, K., Fulton, R. S., Fulton, L. A., Clifton, S. W., Wilson, R. K., Knight, R. D., and Gordon, J. I. (2007) PLoS Biol. 5 e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dierks, T., Schmidt, B., Borissenko, L. V., Peng, J. H., Preusser, A., Mariappan, M., and von Figura, K. (2003) Cell 113 435–444 [DOI] [PubMed] [Google Scholar]
- 30.Cosma, M. P., Pepe, S., Annunziata, I., Newbold, R. F., Grompe, M., Parenti, G., and Ballabio, A. (2003) Cell 113 445–456 [DOI] [PubMed] [Google Scholar]
- 31.Recksiek, M., Selmer, T., Dierks, T., Schmidt, B., and von Figura, K. (1998) J. Biol. Chem. 273 6096–6103 [DOI] [PubMed] [Google Scholar]
- 32.Ono, K., Okajima, T., Tani, M., Kuroda, S., Sun, D., Davidson, V. L., and Tanizawa, K. (2006) J. Biol. Chem. 281 13672–13684 [DOI] [PubMed] [Google Scholar]
- 33.Wang, S. C., and Frey, P. A. (2007) Trends Biochem. Sci. 32 101–110 [DOI] [PubMed] [Google Scholar]
- 34.Cheek, J., and Broderick, J. B. (2002) J. Am. Chem. Soc. 124 2860–2861 [DOI] [PubMed] [Google Scholar]
- 35.Fontecave, M., Atta, M., and Mulliez, E. (2004) Trends Biochem. Sci. 29 243–249 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Data]