Phospholipase D Activity Regulates Integrin-mediated Cell Spreading and Migration by Inducing GTP-Rac Translocation to the Plasma Membrane (original) (raw)
Abstract
Small GTPase Rac is a crucial regulator of actin cytoskeletal rearrangement, and it plays an important role in cell spreading, migration, mitogenesis, phagocytosis, superoxide generation, and axonal growth. It is generally accepted that Rac activity is regulated by the guanosine triphosphate (GTP)/guanosine diphosphate (GDP) cycle. But, it is suggested that in addition to Rac-GTP loading, membrane localization is required for the initiation of downstream effector signaling. However, the molecular mechanisms that control the targeting of GTP-Rac to the plasma membrane remain largely unknown. Here, we have uncovered a signaling pathway linking phospholipase D (PLD) to the localized functions of Rac1. We show that PLD product phosphatidic acid (PA) acts as a membrane anchor of Rac1. The C-terminal polybasic motif of Rac1 is responsible for direct interaction with PA, and Rac1 mutated in this region is incapable of translocating to the plasma membrane and of activating downstream target p21-activated kinase upon integrin activation. Finally, we show that PA induces dissociation of Rho-guanine nucleotide dissociation inhibitor from Rac1 and that PA-mediated Rac1 localization is important for integrin-mediated lamellipodia formation, cell spreading, and migration. These results provide a novel molecular mechanism for the GTP-Rac1 localization through the elevating PLD activity, and they suggest a general mechanism for diverse cellular functions that is required localized Rac activation.
INTRODUCTION
Cell spreading and migration are essentially required for development and for the tissue repair, but they are also important during inflammatory conditions and cancer metastasis (Lauffenburger and Horwitz, 1996; Ridley et al., 2003). Cell spreading and migration are complex processes that include the formation of membrane protrusions called lamellipodia, and lamellae at the leading edges of cells, and membrane adhesive interactions with migratory substrates, in addition, these processes require cell polarization and coordinated cytoskeletal dynamics (Lauffenburger and Horwitz, 1996; Ridley et al., 2003). In many cases, these physiological processes are orchestrated by a combination of signals from the extracellular matrix (ECM) through integrins and growth factors (Giancotti and Tarone, 2003). The integrin family of cell adhesion receptors mediates cellular contacts with the ECM, and upon ECM engagement, a large number of signaling molecules are recruited and actin cytoskeletal rearrangement occurs. Moreover, these processes are essential for establishing cell spreading and migration. The key downstream signaling molecules of integrin-mediated actin cytoskeletal rearrangements include small GTPases of the Rho family, such as Rho, Cdc42, and Rac (Ridley et al., 2003). Among these molecules, Rac is crucial for generating actin-rich lamellipodial protrusions, a key component of cell movement and cell spreading (Etienne-Manneville and Hall, 2002).
To regulate accurately cell spreading and directional cell migration, spatial and temporal control of Rac activation is required. Rac activity is regulated by cycling between an active guanosine triphosphate (GTP)-bound state and an inactive guanosine diphosphate (GDP)-bound state, and this GTP/GDP cycling is tightly regulated by guanine nucleotide exchange factors (GEFs), which stimulate GTP loading; GTPase-activating proteins (GAPs) that catalyze GTP hydrolysis; and guanine nucleotide dissociation inhibitors (GDIs), which antagonize both GEFs and GAPs (Raftopoulou and Hall, 2004). Rac executes its biological functions by activating downstream effectors, and one of the best-characterized downstream effectors of Rac is p21-activated kinase (PAK). It is generally accepted that GTP-loaded Rac spontaneously activates PAK, and for this reason, Rac activity is usually measured by following changes in Rac-GTP loading. However, Rac also cycles between the membrane and cytosol and at the plasma membrane activated GTPases interact with downstream effectors. Recently, according to the accepted model of Rac activation, independently of the effects of Rac on GTP loading, Rac membrane translocation seems to be critical for the activation of its effector, PAK (del Pozo et al., 2004; Moissoglu et al., 2006). In its inactive status, Rac remains in the cytoplasm and becomes activated upon cell stimulation, when it is translocated to the membrane (Dinauer, 2003). A significant portion of GAP-insensitive, constitutively GTP-loaded form V12-Rac exists in the cytosol (Del Pozo et al., 2002). Moreover, it has been reported that integrins promote membrane targeting by regulating the availability of membrane binding sites so that, in nonadherent cells, despite GTP loading status, Rac is present in the cytosol (Del Pozo et al., 2002; del Pozo et al., 2004). Therefore, these observations suggest that the GTP loading and membrane targeting of Rac are separable events and that Rac activity is regulated by both the GDP/GTP exchange cycle and the membrane association/dissociation cycle. However, the dynamics of Rac membrane translocation and the signaling pathway involved have not been described previously. Moreover, regulators of Rac cycling between cytosol and the plasma membrane have not been elucidated previously; thus, it remains at issue as to how Rac dynamically translocates to its proper membrane target and activates downstream effector proteins.
Two mammalian phospholipase Ds (PLDs) have been cloned, PLD1 and PLD2, and these two species share similar regulatory domains and enzymatic characteristics. PLD hydrolyzes membrane phosphatidylcholine to generate phosphatidic acid (PA) and choline in response to a variety of signals, including hormones, neurotransmitters, and growth factors (Exton, 1999), and PA itself is an intracellular lipid second messenger that is involved in many physiological events such as mitogenesis, membrane vesicle trafficking, superoxide production, and cytoskeletal dynamics in a large number of cell types (Frohman et al., 1999). In particular, the functions of PA during actin cytoskeletal reorganization are well established. PLD activity has been found in the detergent-insoluble fraction of various cell types that contain a wide range of cytoskeletal proteins, such as, F-actin, α-actinin, vinculin, paxillin, and talin (Iyer and Kusner, 1999). Moreover, the stimulation of PLD with physiological and pharmacological agonists results in its association with actin filaments (Iyer and Kusner, 1999), and actin polymerization is tightly coupled to the activation of PLD (Ha and Exton, 1993). Furthermore, the formations of lamellipodia structures and membrane ruffles are blocked by PLD inhibition (Santy and Casanova, 2001; O'Luanaigh et al., 2002), and PLD activity is critical for leukocyte cell migration (Lehman et al., 2006). However, the downstream targets of PLD that signal actin reorganization are poorly understood, and it is not known what conditions and signaling pathways induce PLD-mediated actin cytoskeletal reorganization.
Here, we identify for the first time that Rac is a downstream target of PLD during PLD-mediated actin cytoskeleton rearrangement. In addition, we show that GTP-Rac1 membrane translocation occurs via a direct interaction between the C-terminal polybasic motif of Rac1 and PA. We also show that this interaction and Rac translocation occur via an integrin-mediated signaling pathway and that this is important for downstream PAK activation, lamellipodia formation, cell spreading, and migration. Rac membrane translocation by PA seems to be a novel regulatory mechanism for the localization of the functions of Rac. Previous studies have shown that lipid second messenger PA and small G protein Rac are involved in cell migration and actin cytoskeletal rearrangement. However, the link between PA and Rac with respect to these cellular functions is not known. Our results bridge these two important regulators of diverse cellular physiological functions, and they provide a novel molecular mechanism for the localized Rac function by PA-mediated Rac membrane translocation.
MATERIALS AND METHODS
Cell Culture and Transfection
OVCAR-3 cells were maintained in DMEM supplemented with 10% (vol/vol) fetal bovine serum at 37°C in a humidified, CO2-controlled (5%) incubator. For transfection and the transient expression of proteins, cells were transfected using Lipofectamine (Invitrogen, Carlsbad, CA), as described previously (Chae et al., 2005). To silence PLDs, cells were transfected with synthetic small interfering RNA (siRNA) specific for human PLDs, as described previously (Lee et al., 2006). To effect the restoration of the expressions of PLDs, cells were transiently expressed with PLDs that resist siRNA silencing, 1 d after being transfected with siRNA.
Cell Suspension and Replating Assays
Cell suspension and replating assays were performed as described previously (del Pozo et al., 2004). For fibronectin or polylysine coating, plastic Petri dishes were first rinsed with 0.25% gelatin solution, and then they were coated with 25 μg/ml fibronectin or 100 μg/ml polylysine. For replating assays, cells were detached with 0.5% trypsin and 5.4 mM EDTA. Cells were diluted into suspension media (DMEM, 10 mM HEPES, pH 7.4, 0.5 mg/ml fatty acid-free bovine serum albumin, and 0.5 mg/ml soybean trypsin inhibitor type II), collected by centrifugation at 1000 × g for 3 min, and resuspended in suspension media. Cells were then incubated for 3 h at 37°C and gently shake to prevent cell clumping. Suspended cells were replated on fibronectin- or polylysine-coated plastic Petri dishes.
In Vivo PLD Activity Assay
In vivo PLD activity was determined, as described previously (Chae et al., 2005). In brief, OVCAR-3 ovarian cancer cells were serum starved for 24 h, loaded with [3H]myristic acid (10 μCi/ml) for 4 h, and then the unincorporated [3H]myristic acid was removed by two washings with serum-free DMEM. Cells were detached from culture dishes and maintained in suspension for 3 h, and then they were replated on fibronectin- or polylysine-coated plastic Petri dishes in the presence of 0.4% 1-butanol. Reactions were stopped by removing medium and adding 0.4 ml of ice-cold methanol to each well. Cells were then scraped into tubes and 0.4 ml of 1 M NaCl and 0.4 ml of chloroform were added. After vortexing, tubes were centrifuged at 15,000 × g for 1 min, solvents were removed, and residual lipids were separated by thin layer chromatography on silica-gel 60 plates (developed with chloroform:methanol:acetic acid; 90:10:10, vol/vol). Amounts of labeled phosphatidylbutanol (PBt) and total lipids were determined using a Fuji BAS-2000 Image Analyzer (Fuji, Tokyo, Japan). The amount of PBt formed was expressed as a percentage of total 3H-lipid to account for cell labeling efficiency differences.
PAK Kinase Assay
Endogenous PAK was immunoprecipitated from 400 μg of cell lysate using anti-PAK antibodies (Santa Cruz Biotechnology, Santa Cruz, CA). Immunoprecipitated PAK was incubated in kinase buffer (50 mM HEPES, pH 7.5, 10 mM MgCl2, 2 mM MnCl2, and 0, 2 mM dithiothreitol) containing 20 μM ATP and 5 μCi of [γ-32P]ATP for 30 min at 30°C, and 5 μg of myelin basic protein (MBP) was used as a substrate. Samples were separated on SDS-polyacrylamide gels, and then they were transferred to nitrocellulose for immunoblotting and/or autoradiography. Radiolabeled phosphate incorporation was measured using a Fuji BAS-2000 Image Analyzer.
Measurement of Rac-GTP Loading (p21 Binding Domain [PBD] Binding Assay)
Cells were washed with phosphate-buffered saline (PBS) and lysed in ice-cold buffer (50 mM Tris, pH 7.6, 0.5 M NaCl, 0.1% SDS, 0.5% deoxycholate, 1% Triton X-100, 0.5 mM MgCl2, 0.2 mM sodium vanadate, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 μg/ml aprotinin, and 1 μg/ml leupeptin). Lysates were centrifuged at 14,000 × g for 10 min at 4°C. Supernatants were incubated for 30 min at 4°C with 20 μg of glutathione-agarose beads coupled to glutathione transferase (GST)-PBD. Beads were sedimented and washed four times with 50 mM Tris-HCl, pH 7.6, containing 150 mM NaCl, 1% Triton X-100, and 0.5 mM MgCl2. Bound Rac proteins were detected by Western blotting, and whole cell lysates also were analyzed for Rac for normalization purposes.
Membrane Translocation Assays
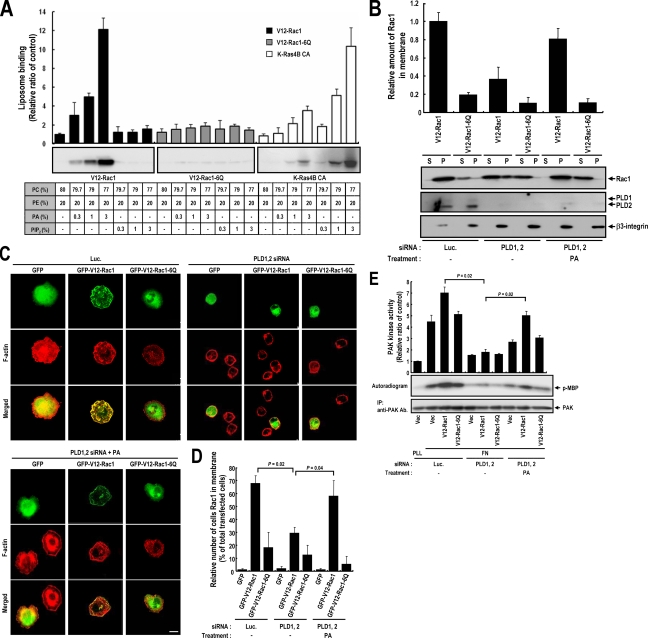
Rac membrane translocation assays were performed as described previously (Del Pozo et al., 2002). Cells were lysed with hypotonic lysis buffer (10 mM HEPES, pH 7.5, 1.5 mM MgCl2, 5 mM KCl, 1 mM dithiothreitol, 0.2 mM sodium vanadate, 1 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 mM PMSF) for 30 min, lysates were homogenized with a Dounce homogenizer, and centrifuged at 700 × g for 3 min to pellet nuclei and intact cells. Supernatants were then centrifuged at 40,000 × g for 30 min at 4°C. Cytosol-containing supernatants were removed, and membrane pellets were gently washed twice with hypotonic lysis buffer. Membrane and cytosol fractions were then assayed by Western blotting. Typically, 15 μg of proteins was analyzed, representing 1–2% of the cytosolic fraction and 10–20% of the membrane fraction. Membrane-translocated Rac was quantitated by scanning densitometry. Data were normalized to β3-integrin level, and the relative amount of Rac1 in membrane fractions was calculated.
Purification of Flag-Fusion Proteins
To purify isoprenylated V12-Rac1 and V12-Rac1-6Q and KRas4B CA proteins, COS-7 cells were transfected with Flag-V12-Rac1 or Flag-V12-Rac1-6Q or KRas4B-CA. Cells were washed with PBS and lysed in ice-cold buffer (50 mM Tris, pH 7.6, 0.5 M NaCl, 0.1% SDS, 0.5% deoxycholate, 1% Triton X-100, 0.5 mM MgCl2 0.2 mM sodium vanadate, 1 mM PMSF, 1 μg/ml aprotinin, and 1 μg/ml leupeptin). Lysates were centrifuged at 14,000 × g for 10 min at 4°C, and Flag-tagged proteins were isolated using Flag antibody-coupled beads (Sigma-Aldrich, St. Louis, MO). Samples were then washed five times with lysis buffer, and Flag-tagged proteins were eluted from the immune complex with 100 μg/ml Flag peptide (Sigma-Aldrich) in Tris-buffered saline buffer (50 mM Tris, pH 7.4, 150 mM NaCl).
Liposome Binding Assay
Liposome preparation and binding assays were performed as described previously (del Pozo et al., 2004). In brief, phosphatidylcholine (1,2-dimyristoyl-_sn_-glycero-3-phosphocholine), phosphatidyl ethanolamine (1,2-dimyristoyl-_sn_-glycero-3-phosphoethanolamine), and phosphatidic acid (1,2-dimyristoyl-_sn_-glycero-3-phosphate) (Avanti Polar Lipids, Alabaster, AL) were dissolved in chloroform at 10 mM and mixed in varying proportions. Chloroform was evaporated off under a stream of N2, the dried lipids were resuspended in buffer (10 mM HEPES, pH 7.5, and 140 mM NaCl), and then they were sonicated for 10 min at 25°C to form small unilamellar liposomes. Purified isoprenylated Rac proteins were incubated with liposomes for 30 min at 37°C, centrifuged at 40,000 × g for 15 min at 4°C, washed twice and resuspended in incubation buffer. Bound Rac proteins were detected by Western blotting and quantitated by scanning densitometry.
Immunocytochemistry
Immunocytochemistry was performed using a previously reported procedure (Chae et al., 2005). Cells were rinsed with ice-cold PBS four times, and fixed with 4% (wt/vol) paraformaldehyde 30 min at room temperature. After rinsing with PBS, cells were blocked with PBS containing 1% goat serum and 0.1% Triton X (TX)-100 for 30 min at room temperature. To visualize actin filaments, cells were incubated with rhodamine-labeled phalloidin for 40 min at room temperature, and to visualize Rac1 or β3-integrin, cells were incubated with 2 μg/ml mouse Rac1 or β3-integrin monoclonal antibodies (BD Biosciences Transduction Laboratories, Lexington, KY) for 2 h at room temperature. After washing six times with PBS, the cells were incubated with rhodamine-conjugated goat anti-mouse antibodies. After a further six washings with PBS, slides were examined under a confocal microscope (LSM-510 Meta; Carl Zeiss, Jena, Germany) driven by LSM5 software (Carl Zeiss). Pictures shown were taken with an Apochromat 40×/1.2 W korr objective (Carl Zeiss).
Quantification of Cell Adhesion and Spreading
To quantify cell adhesion and spreading, cells were washed three times with PBS and fixed in 4% formaldehyde. To visualize membrane ruffles, actin filaments were stained with tetramethylrhodamine B isothiocyanate (TRITC)-labeled phalloidin (0.1 μg/ml) for 30 min. Ratios of spread cells to adherent cells were determined by confocal microscopy. The mean number of adherent cells ± SD per 1.1 mm2 was determined from 10 different images from at least two separate experiments in three independent assays. And cells that exhibited flattening and lamellipodia were scored as spread cells. Percentages of spreading cells were defined as ((spreading cells/total number of adherent cells) × 100).
Generation of Pan-PLD Polyclonal Antibody
Polypeptide TKLVPMEVWT (homology sequence containing C-terminal amino acids of PLD) was used to immunize rabbits. The same polypeptide was used to prepare affinity column for antibody purification. Serum was loaded onto a washed (10 mM Tris-HCl, pH 7.4, followed by 0.5 N NaCl in equilibration buffer), pre-equilibrated column, and anti-Pan-PLD polyclonal antibody was eluted with 100 mM glycine, pH 2.5. Eluted antibody was immediately neutralized with 1 M Tris-HCl, pH 8.0, and concentrated by using ultrafiltration (Centricon; Millipore, Billerica, MA).
Statistical Analysis
The statistical significance was accessed by Student's t test.
RESULTS
PLD Activity Is Regulated by Integrin Signaling
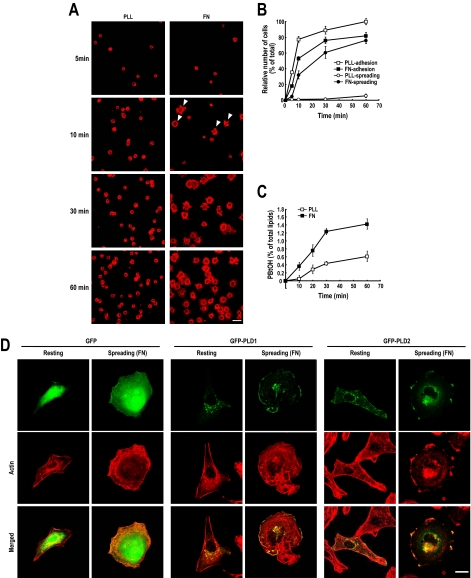
Integrin and ECM protein interactions induce cell adhesion and spreading, such that cells acquire a flattened morphology through complex dynamic rearrangements of the actin cytoskeleton (Price et al., 1998). As shown in Figure 1, A and B, when detached ovarian cancer cells OVCAR-3 were replated onto fibronectin-coated dishes, the cells rapidly made contact with the substrate. Moreover, these adherent cells rapidly spread and adopted a flat and elongated morphology with lamellipodia and filopodia over an hour or so. However, on polylysine, although cells adhered at the same rate, they did not spread and retained a round morphology at 1 h (Figure 1, A and B). These results suggest that in OVCAR-3 cells, fibronectin-mediated integrin activation specifically induces cell spreading. At the same time, we found that fibronectin, but not polylysine, up-regulated PLD activity in these cells (Figure 1C). Fibronectin induced PLD activation during cell spreading was also observed in NIH-3T3 fibroblasts, in COS-7 and in human embryonic kidney (HEK)-293T cells (data not shown). PLD1 is known to localize to diverse subcellular membrane structures, including the endoplasmic reticulum (ER), Golgi complex, endosomes, and lysosomes, and to the plasma membrane, whereas PLD2 is more prevalent at the plasma membrane (Brown et al., 1998; Jones et al., 1999; Kim et al., 1999; Roth et al., 1999; Toda et al., 1999). In resting OVCAR3 cells, PLD1 was predominantly localized to vesicular structure in the cytosol, whereas PLD2 was localized to plasma membranes. However, during integrin-mediated spreading cells, both PLD1 and PLD2 predominantly localized at the plasma membrane and distribution of PLD1 closely resembled that of PLD2 (Figure 1D). It is known that under stimulatory conditions, such as, those induced by phorbol 12-myristate 13-acetate or epidermal growth factor, PLD1 translocates to the plasma membrane (Emoto et al., 2000; Han et al., 2002; Du et al., 2003). Furthermore, it has been reported that the engagement of β3-integrin by fibronectin leads to an association between specific proteins involved in signaling and the actin cytoskeleton, and that this results in the recovery of Triton-insoluble caveolin-enriched fractions (Bodin et al., 2005). In the present study, fibronectin induced actin polymerization and the incorporation of β3-integrin and PLD in Triton-insoluble membrane fractions (Supplemental Figure S1). Taken, our data suggest that both PLD1 and PLD2, but especially PLD1, are recruited to the plasma membrane during the integrin-mediated cell spreading process and that PLD is specifically regulated at this time by integrin signaling.
Figure 1.
PLD activation during integrin-mediated cell spreading. (A) OVCAR-3 human ovarian cancer cells were serum starved for 24 h, detached from culture dishes, maintained in suspension for 3 h, and then replated on fibronectin (FN) or polylysine (PLL) for the indicated times (5, 10, 30, and 60 min). After removing unbounded cells, cells were fixed and stained with TRITC-labeled phalloidin. Arrowheads indicate spreading cells. Bars, 50 μm. (B) This graph shows adherent and spreading cell quantifications. Error bars represent means ± SD (SD; n = 3). (C) PLD activities were measured in cells replated on FN or PLL for the indicated times (0, 5, 10, 30, and 60 min). 0 represents the control and indicates PLD activity for 5 min in suspension cells. Error bars represent means ± SD (n = 3). (D) Cells were transfected with GFP control, GFP-PLD1, or GFP-PLD2. Two days after transfection, cells were serum starved for 18 h, and then they were fixed and stained with TRITC-labeled phalloidin (Resting). For cell spreading, the same serum-starved cells were detached from culture dishes and maintained in suspension for 3 h. Detached cells were replated on FN or PLL for 20 min, and cells were then fixed and stained with TRITC-labeled phalloidin (Spreading). Bars, 10 μm.
PLD Activity Is Required for Integrin-mediated Cell Spreading
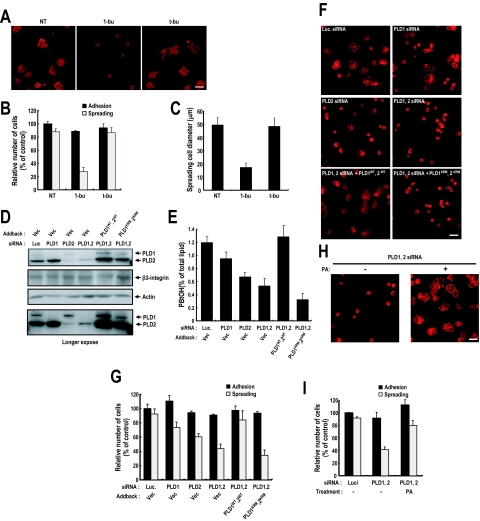
Specific PLD activation during the integrin-mediated cell spreading process implies that PLD activity is associated with integrin function. To determine whether PLD activity is required for integrin-mediated cell spreading, we prevented the formation of the PLD product, PA, by adding 1-butanol to cell medium. Although inhibiting PA formation did not affect fibronectin-induced cell adhesion, it significantly decreased cell spreading; most cells retained a round morphology like those that adhered to polylysine-coated dishes (Figure 2, A and B). PLD RNA interference (RNAi) experiments demonstrated that fibronectin-mediated cell spreading requires PLD activation. RNAi targeting PLD1 or PLD2, or both isoforms reduced integrin-mediated PA generation (Figure 2, D and E). In correlation with PA generation, PLD knockdown reduced cell spreading, but it had no effect on cell adhesion; adding wild-type PLDs, but not lipase-inactive mutants, restored cell spreading (Figure 2, F and G). Moreover, reduced cell spreading by PLD silencing was restored by direct PA treatment (Figure 2, H and I). From these results, we conclude that PA generated PLD is critical for integrin-mediated cell spreading. Furthermore, this role of PA in integrin-mediated cell spreading, and the roles of PLD isozymes in cell spreading were examined in other cells types expressing either PLD1 or PLD2 isoforms. We used COS-7 and HEK293T cells because in COS-7 cells PLD2 is the major isotype and in HEK293T cells, PLD1 is major isotype. Thus, in these cells, we could minimize the effects of other enzymes during enzyme silencing and addback. It was found that PLD1 knockdown reduced cell spreading, and adding wild-type PLD1 restored cell spreading in HEK293 cells. Similarly, in COS-7 cells PLD2 knockdown reduced cell spreading and adding wild-type PLD2 restored cell spreading. These observations suggest that both PLD1 and PLD2 contribute to integrin-mediated cell spreading (Supplemental Figure S6). Cell spreading is closely related with cell migration. Therefore, we examined whether PLD activity is also required for integrin-mediated cell migration, and it was found, in agreement with our cell spreading results, that PLD activity is critical for integrin-mediated cell migration (Supplemental Figure S4A). Together, these results indicate that PA generated by both PLD isozymes through integrin activation is critical for integrin-mediated cell spreading and migration, but not for cell adhesion.
Figure 2.
PLD activity regulates integrin-mediated cell spreading, but not cell adhesion. (A) Detached cells were replated on FN for 20 min with medium containing 1-butanol or _t_-butanol (0.2%, vol/vol). After removing unbound cells, cells were fixed and stained with TRITC-labeled phalloidin. Bars, 50 μm. (B) The graph shows numbers of adherent and spreading cells from A. Error bars represent means ± SD (n = 4). (C) The diameter of spreading cells 20 min after plating was calculated as the mean ± SD (n = 4). (D) Cells were transfected as indicated with siRNA PLD1 or PLD2, or luciferase as a control. One day after siRNA transfection, cells were transfected with empty vector or PLD 1 wild type (WT) plus 2 WT (PLD1WT, 2WT) or PLD 1 lipase inactive mutant plus 2 lipase inactive mutant (PLD1KRM, 2KRM), as indicated. Cells were then harvested, and PLD and β3-integrin expression levels were analyzed by Western blotting using antibodies directed against PLDs or β3-integrin. Actin was used as a loading control for Western blotting. The result shown is representative of three independent experiments. (E) Cells treated as in D were detached from culture dishes and maintained in suspension for 3 h. PLD activities were measured in cells replated on FN for 20 min. Error bars represent means ± SD (n = 3). (F) Cells treated as in D were replated on FN for 20 min. After removing unbounded cells, cells are fixed and stained with TRITC-labeled phalloidin (n = 3). Bar, 50 μm. (G) The graph shows numbers of adherent and spreading cells from F. Error bars represent means ± SD (n = 4). (H) PLD1 and PLD2 silenced cells were replated on FN for 20 min with/without PA (50 μM). After removing unbounded cells, cells were fixed and stained with TRITC-labeled phalloidin (n = 3). Bars, 50 μm. (I) The graph shows numbers of adherent and spreading cells from F. Error bars represent means ± SD (n = 3).
PA Can Target Rac1 to the Plasma Membrane
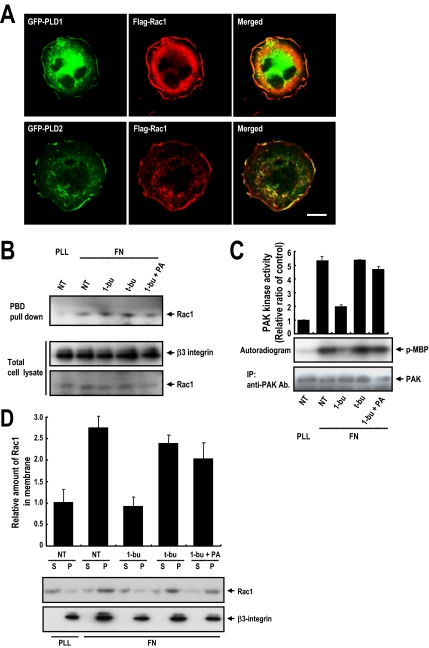
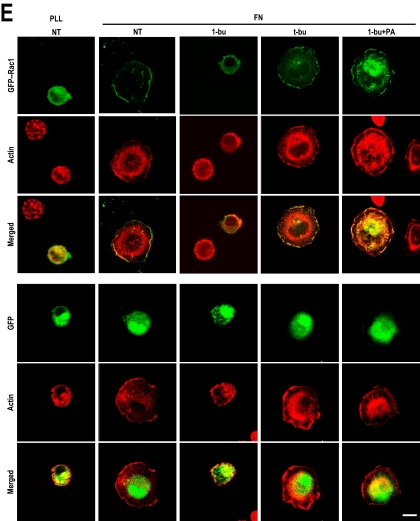
One of the earliest key steps in the transduction of extracellular cues through integrins to the cytoskeleton is the activation of tyrosine kinases Src and focal adhesion kinase (FAK), and others (Schwartz et al., 1995; Giancotti and Tarone, 2003), but the blocking of PLD activity did not affect the integrin-mediated total tyrosine phosphorylation level in whole cell lysates or the kinase activities of FAK and Src (Supplemental Figure S2, A and B). These data suggest that PLD activity does not affect the integrin–fibronectin interaction or the kinase activities of key downstream regulators. This suggestion is supported by the observation that PLD activity did not affect integrin-mediated cell adhesion. When detached cells adhere, integrin-induced Rac activation leads to lamellipodial extension and cell spreading (Price et al., 1998; Etienne-Manneville and Hall, 2002), indicating that lamellipodia formation and cell spreading are closely related Rac1 functions. To explore the relationship between PLD and Rac1, we investigated the intracellular localizations of PLD and Rac1 during integrin-mediated cell spreading. In spreading OVCAR-3 cells, Rac1 localized at cell peripheries in lamellipodia, and PLD also localized at peripheries where polymerized actin was associated with membrane ruffles (Figure 3A), suggesting that PLD activity is involved in Rac functionality. However, PLD activity did not affect integrin-induced Rac1-GTP loading (Figure 3B), although interestingly, Rac1 downstream PAK kinase activity was reduced by 1-butanol treatment and restored by PA treatment (Figure 3C and Supplemental Figure S5). Thus, our results indicate that PLD activity may be additionally required for GTP-Rac1 functions such as downstream PAK activation. Previously, it was generally believed that GTP-loaded Rac spontaneously activates downstream effectors. However, recently it was suggested that independently of their effects on GTP loading, integrins control the translocation of Rac1 to the plasma membrane and that this membrane targeting of Rac1 is critical for the activation of its effector, PAK (del Pozo et al., 2000, 2002). Because 1-buthanol and PA affect PAK without perturbing Rac1-GTP loading, we hypothesize that PLD regulates GTP-Rac1 localization to the plasma membrane after integrin activation. To address this hypothesis, we performed a fractionation experiment. As shown in Figure 3D, fibronectin prompted Rac1 partitioning to the particulate fraction, which contained β3-integrin resides. Treatment with 1-butanol, but not _t_-butanol, inhibited Rac1 partitioning to this particulate fraction, but this was restored by PA treatment. Correspondingly, treatment of cells with 1-buthaol, but not _t_-butanol, inhibited the plasma membrane localization of green fluorescent protein (GFP)-Rac1 on fibronectin plates (Figure 3E). In addition, the dispersed cytosolic localization of GFP-Rac1 by 1-butanol was recovered by adding PA. Together, these results suggest that PA generation by PLD activation is required for GTP-Rac1 membrane translocation.
Figure 3.
PLD activity regulates Rac1 membrane targeting. (A) Cells were transfected with GFP control vector or GFP-PLD1 or GFP-PLD2, as indicated, and then they were cotransfected with Flag-Rac1 and either GFP-PLD1 or GFP-PLD2, serum-starved for 24 h, detached from culture dishes, maintained in suspension for 3 h, and replated on FN. The cells were then fixed and stained with TRITC-labeled phalloidin or anti-Flag monoclonal antibody to detect either actin or Rac1 expression. Images are single confocal sections (n = 3). Bars, 20 μm. (B) After serum starvation for 24 h, cells were detached from culture dishes, maintained in suspension for 3 h, and replated on FN or PLL with medium containing 1-butanol (0.2%, vol/vol), _t_-butanol (0.2% vol/vol), or 1-butanol plus PA (50 μM) for 20 min. Cells were then harvested, and Rac-GTP loadings were quantified by PBD pull-down assays as described in Materials and Methods. The result shown is representative of three independent experiments. (C) PAK proteins were immunoprecipitated and kinase activities were assayed in cells treated as described in B. PAK kinase activities were assayed densitometrically, and precipitated PAK was visualized by Western blotting using antibodies directed against PAK. Densitometric PAK kinase activities were normalized versus precipitated PAK protein levels. Error bars represent means ± SD (n = 3). (D) Cells treated as in B were separated into particulate (P) and cytosolic (S) fractions. Samples were analyzed by Western blotting using antibodies directed against Rac1 or β3-integrin. β3-integrin was used as a marker for the membrane fraction. Quantitative data concerning the membrane localization of Rac1 were normalized to β3-integrin levels. Error bars represent means ± SD (n = 3). (E) Cells were transfected with GFP control vector or GFP-Rac1 as indicated and treated as described in B, and then they were fixed and stained with TRITC-labeled phalloidin. Images are of single confocal sections (n = 4). Bars, 20 μm.
PA-mediated Membrane Translocation Is Dependent on the Polybasic Motif of Rac1
Rac1 contains a polybasic motif (KKRKRK) near its C terminus that is located in the anchor region immediately upstream of an isoprenylated CAAX box. It has been suggested that this polybasic motif facilitates the plasma membrane targeting of Rac1 and that mutation of this polybasic residue to glutamine reduces translocation, even when mutants have an additional mutation that makes them constitutively GTP loaded (V12-Rac1-6Q) (Hancock et al., 1990; Del Pozo et al., 2002). Because PA is an acidic lipid that may be a direct target of the Rac1 polybasic motif, we examined whether Rac1 can directly bind PA in vitro. As shown in Figure 4A, purified isoprenylated V12-Rac1 was found to bind PA containing liposomes in a concentration-dependent manner. However, phosphatidylinositol 4, 5-bisphosphate (PIP2), another acidic phospholipid enriched in the plasma membrane, could not recruit V12-Rac1 to liposomes. Furthermore, mutation of the polybasic motif in Rac1 (V12-Rac1-6Q) showed failure in binding to PA containing liposomes. Small GTPase KRas4B also has a polybasic motif (KKKKKK) and a prenyl targeting motif, and it is suggested that this sequence is necessary for membrane association and it interaction with negative charged membrane phospholipids (Hancock, 2003). To determine whether the each polybasic motif has different lipid binding properties, we performed a liposome binding experiment with purified isoprenylated Kras4B. The data obtained shows that Kras4B has a higher affinity for PIP2 than PA. These results suggest that the polybasic motifs, such as, Rac1 and KRas4B, can interact with membrane lipid in a manner that depends on specific anion phospholipids. Our liposome binding results strongly suggest that membrane PA might be a direct plasma membrane receptor for GTP-loaded Rac1. In-line with the effect of PLD knockdown on cellular spreading (Figure 2), the knockdown of PLDs reduced V12-Rac1 translocation during integrin-mediated cell spreading (Figure 4, B–D). Moreover, the addition of PA to PLD-silenced cells restored V12-Rac1 membrane targeting. However, V12-Rac1-6Q mutant did not translocate to the plasma membrane under any condition (Figure 4, B–D). These results show that PLD is the enzyme responsible for the generation of PA that is required for GTP-Rac1 translocation. Also, the membrane targeting of Rac1 mutants was found to be correlated with downstream PAK kinase activity (Figure 4E). Because purified V12-Rac1-6Q mutant can interact with PAK in a GTP-dependent manner, like V12-Rac1 in vitro (Supplemental Figure S3), differences in PAK kinase activities by V12-Racl and 6Q mutants might be caused by differences in the membrane targeting of Rac1, which is mediated by interaction between PA and the polybasic motif of Rac1.
Figure 4.
Rac1 directly interacts with PA via a polybasic sequence in its C-terminal region. (A) Purified isoprenylated Flag-V12-Rac1, Flag-V12-Rac1-6Q, and Flag-Kras-4B-CA mutants were incubated with the indicated liposomes. Liposomes were sedimented, and the pellets obtained were analyzed by Western blotting using anti-Flag antibody. Liposome binding assay results were quantified by densitometry. Error bars represent means ± SD (n = 3). (B) Cells were transfected with the indicated control luciferase or siRNA PLD1 and PLD2. One day after siRNA transfection, cells were transfected with Flag-V12-Rac1 or Flag-V12-Rac1-6Q. After serum starvation for 24 h, cells were detached from culture dishes, maintained in suspension for 3 h, and replated on fibronectin with or without 50 μM PA for 20 min. Adherent cells were harvested and particulate (P) and soluble (S) fractions were isolated. Samples were analyzed by Western blotting using anti-Flag antibody. β3-integrin was used as a membrane fraction marker. Quantitative data concerning the membrane localization of Rac1 were normalized to β3-integrin levels. Error bars represent means ± SD (n = 3). (C) Cells were transfected with the indicated control luciferase or siRNA PLD1 and PLD2. One day after siRNA transfection, cells were transfected with GFP control vector or with GFP-V12-Rac1 or GFP-V12-Rac1-6Q, as indicated. After serum-starvation for 24 h, cells were detached from culture dishes, maintained in suspension for 3 h, and replated on fibronectin with or without 50 μM PA for 20 min. Cells were then fixed and stained with TRITC-labeled phalloidin. Images are single confocal sections (n = 4). Bars, 20 μm. (D) The graph shows quantification of the cells expressing Rac1 in membrane from C. Error bars represent means ± SD (n = 4), 100 cells per group were assessed in each experiment. (E) PAK proteins were immunoprecipitated, and kinase activities in cells treated as described in B were assayed. PAK kinase activity assay results were quantified by densitometry and normalized versus PAK protein levels, and precipitated PAK was visualized by Western blotting using antibodies directed against PAK. Error bars represent means ± SD (n = 3).
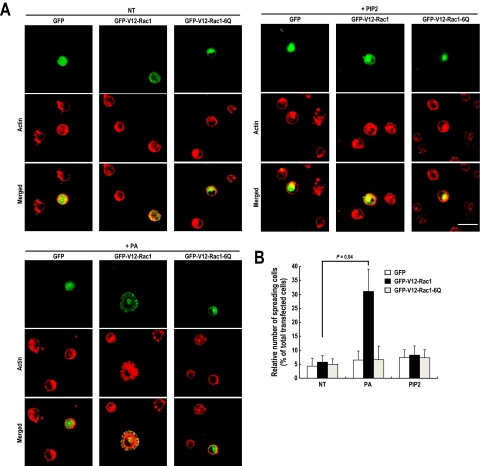
GTP Loading of Rac1 and PA Induces Cell Spreading
Integrin-mediated cell spreading and lamellipodia formation require Rac-GTP loading and PA-mediated GTP-Rac translocation. To test whether GTP-Rac and PA are sufficient for cell spreading and lamellipodia formation, we transfected cells with V12-Rac1 and treated them with PA before they were replated on polylysine-coated dishes. On polylysine-coated surfaces, integrin cannot be activated, and thus it would be expected that signaling leading to cell spreading would not occur. However, interestingly, we observed cell spreading and the formation of lamellipodia-like structures in cells treated with PA, not PIP2, and expressing V12-Rac1 (Figure 5, A and B). Moreover, cells expressing PA binding deficient mutant (V12-Rac1-6Q) did not show cell spreading even when treated with PA. These results suggest that both GTP loading of Rac and its translocation to the plasma membrane by PA are required and sufficient for Rac functions during the integrin-mediated cell spreading process.
Figure 5.
GTP-Rac1 and PA induced cell spreading with a lamellipodia-like structure. (A) Cells were transfected with GFP vector or GFP-V12-Rac1 or GFP-V12-Rac1-6Q. After being serum-starved for 24 h, cells were detached from culture dishes, maintained in suspension for 3 h, and replated on PLL with or without 50 μM PA or 50 μM PIP2 for 1 h. They were then fixed and stained with TRITC-labeled phalloidin (n = 4). Bars, 50 μm. (B) Graph illustrating the mean number of spreading cells among 230–270 transfected cells per group. Error bars represent means ± SD (n = 3).
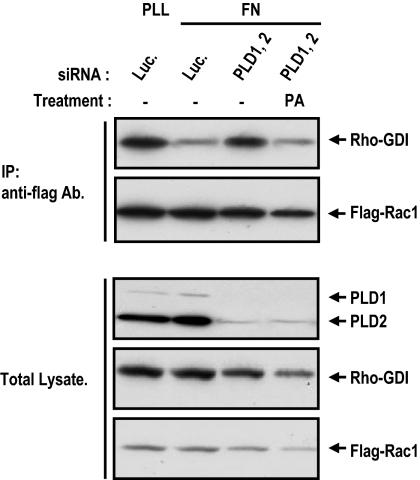
PA Blocks the Rho-GDI Binding of Rac
Rho-GDIs are thought to regulate the membrane association/dissociation cycle of Rho GTPases. Rho-GDI forms complexes with Rac in the cytoplasm and blocks the translocation of Rac to the membrane. In the membrane, Rac exists in a form dissociated from GDI (Lian et al., 2000; Scheffzek et al., 2000; Michaelson et al., 2001). If PA regulates GTP-Rac membrane targeting, it should modulate the association of Rac with Rho-GDI. To test whether PA blocks Rho-GDI binding for Rac membrane targeting, we examined whether PA can dissociate Rho-GDI from Rac. PLD knockdown increased the interaction of Rac-Rho-GDI complex, but in this status, PA treatment dissociated Rho-GDI from Rac. This result suggests that PA mediated GTP-Rac membrane targeting is accompanied by dissociation from Rho-GDI.
DISCUSSION
In addition to the GTP/GDP cycle, small G proteins also cycle between the membrane and the cytosol (Del Pozo et al., 2002). As well as GTP loading, this membrane targeting is required for the initiation of downstream effector signaling (del Pozo et al., 2004). It has been shown that the C termini of small GTPases mediate differential intracellular localization (Michaelson et al., 2001), and it has been demonstrated that the C termini of Rac1 and Rac2 dictate both subcellular localization and differential signaling (Filippi et al., 2004). These results suggest that the localizations of small G proteins to their proper targets are key step for the regulation of small G proteins functions and in this process, C terminus of small GTPase mediates their intracellular targeting. However, little is known about the mechanisms that determine the specificity of the Rac membrane binding site or how the C terminus of Rac is involved in this membrane targeting. The present study demonstrates that the polybasic motif in the C terminus of Rac1 mediates its interaction with PA, which induces Rac1 membrane targeting. In the absence of PLD activity, Rac1 does not translocate to the plasma membrane, and even in the GTP-loaded state, Rac1 does not activate the down stream effector PAK. The direct interaction between PA and the polybasic motif of Rac1 along and our mutant studies suggest that Rac1 membrane targeting is mediated by PA through an interaction with the polybasic motif in the Rac1 C terminus. And we showed that PA can dissociate Rho-GDI from Rac (Figure 6). Rho-GDI forms complexes with Rac in the cytoplasm, keeping it soluble by shielding the C-terminal hydrophobic isoprenoid moiety of Rac (Lian et al., 2000; Scheffzek et al., 2000). It has also been reported that Rho-GDI overexpression reduced and Rho-GDI down-regulation increased Rac1 membrane targeting (Michaelson et al., 2001). Rho-GDI also blocked the downstream effector binding (Del Pozo et al., 2002). Thus, the release of Rac from Rho-GDI is an important step that allows Rac1 to associate with the membrane and to activate downstream signaling. PA-mediated release of Rho-GDI from Rac can be caused by direct interaction with the C-terminal polybasic region. GDI interacts with the C-terminal isoprenyl region and the PA-binding polybasic region of Rac1 is located beside this isoprenyl region, so this PA–polybasic region interaction may inhibit GDI–isoprenyl group interaction. This notion is supported by the ability of negatively charged lipid to dissociate Rac1 from RhoGDI in vitro (Chuang et al., 1993; Ugolev et al., 2006). And it is reported that prenylated Rac1 mutant, lacking the polybasic region, was found defective in its interaction with Rho-GDI (Di-Poi et al., 2001). These results suggest that these Rac membrane targeting and downstream PAK activation effects may result from the Rho-GDI displacement activity of PA through the direct interaction with polybasic region of Rac. Rac membrane binding sites are within low-density, cholesterol-rich lipid rafts in the plasma membrane (del Pozo et al., 2004), and it has been previously reported that PLD is localized and that its activity is much enhanced in low-density lipid raft regions where Rac1 membrane binding sites are located (Czarny et al., 1999). In fact, during the integrin-mediated cell spreading process, PLD and Rac1 were found to colocalize at cortical regions where polymerized actin is associated with membrane ruffles (Figure 3A). These results suggest that Rac translocates to PA containing lipid rafts regions in the plasma membrane. Furthermore, this PA mediated Rac translocation can form a positive feedback loop. It is reported that Rac directly interacts with and activates PLD in a GTPγS-dependent manner (Ohguchi et al., 1997; Powner et al., 2002), leading to an increase in the levels of local PA synthesis. Together with the data from this study, recruitment of the GTP-Rac to the PA containing lipid raft where PLD resides would lead to enhanced synthesis of PA.
Figure 6.
PA induces dissociation of Rho-GDI from Rac1. Cells were transfected with the indicated control luciferase or siRNA PLD1 and PLD2. One day after siRNA transfection, cells were transfected with Flag-Rac1. After serum starvation for 24 h, cells were detached from culture dishes, maintained in suspension for 3 h, and then replated on fibronectin with or without 50 μM PA for 20 min. The cells were then harvested and immunoprecipitated using anti-Flag antibody-coupled agarose beads. Immunoprecipitates and total lysate were analyzed by SDS-PAGE and visualized by Western blotting using antibodies directed against the Flag tag, GDI, or PLDs. The result shown is representative of three independent experiments.
PA plays a role as a lipid second messenger for recruitment to the membrane of some signaling molecules, such as, Raf-1, p47phox, and cAMP-PDE4A1. The PA binding sequences in these proteins are composed of multiple basic sequences (Andresen et al., 2002), and the multiple basic sequence in the Rac1 C-terminal region also mediates direct interaction between PA and Rac1 (Figure 4A). Previously, the polybasic motif of small GTPase was observed to nonspecifically associate with negatively charged lipid in vitro; this association does not seem to be anion specific and it depends only on the net negative charge (Leventis and Silvius, 1998; Ugolev et al., 2006). Polybasic peptide of Kras-4B interacts with negatively charged phospholipids such as phosphatidylserine (PS), phosphatidylinositol 4-phosphate [PI(4)P], phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2], and PA (Leventis and Silvius, 1998). The Rac1 polybasic motif was also reported to interact with acidic phospholipids, including phosphatidylinositol 3-phosphate, PI(4)P, phosphatidylinositol 5-phosphate, phosphatidylinositol-(3,4,5)-trisphosphate [PI(3,4,5)P3], and PA (Ueyama et al., 2005). However, we observed that Rac1 interacts with PA-containing liposome with higher affinity than with PI(4,5)P2, although PI(4,5)P2 has a lower net charge. And KRas4B interacts with PI(4,5)P2, with higher affinity than with PA (Figure 4A). PA-, PS-, and PI(4,5)P2-containing negatively charged lipid bilayers also reportedly have different binding affinity for polybasic peptide (Yeung et al., 2006). These results suggest that the polybasic motif of Rac1 can interact with membrane lipid depending not only on the net negative charge but also on specific anion phospholipids. In cells, the change in negatively charged lipid content of membrane differs, and it is localized according to cellular physiological events that are regulated by kinase, inositide lipase, phospholipase, and phosphatase activation. Therefore, the consequence of altered negatively charged membrane lipids in small GTPase membrane translocation events should be demonstrated in cells. It was shown that in cells, several negatively charged phospholipids, in particular, PS, PI(4,5)P2, and PI(3,4,5)P3, have important roles in membrane translocation of Rac. PS involve the Rac1 detachment from local phagosome membrane surface through the potential of inner leaflet decrease by depletion or flipping of PS during the phagocytosis (Yeung et al., 2006). Previously, we reported that PI(4,5)P2 and PI(3,4,5)P3 in combination are important for small GTPase membrane targeting (Heo et al., 2006). In this article, PIP2 and phosphatidylinositol 3,4,5-triphosphate depletion dissociate most small GTPases such as K-Ras, Rab35, and RhoE, which have a polybasic region and prenylation modification motif similar to Rac1. However, in Rac1, depletion of both PI(4,5)P2 and PI(3,4,5)P3 triggered only a minor reduction in plasma membrane localization. This suggests that another negatively charged lipid contributes to Rac1 membrane targeting. It was known that PI(4,5)P2 recruited vinculin, α-actinin, and syndecan-4 and that it induced actin polymerization and was found to be essential for the formation of focal adhesions (Fukami et al., 1992; Gilmore and Burridge, 1996). Moreover, PA activates phosphatidylinositol-4-phosphate 5-kinase and can generate PI(4,5)P2 (Jenkins et al., 1994). However, in the present study, in vitro liposome binding analysis showed that Rac1 did not interact with PI(4,5)P2-containing vesicles (Figure 4A), and PI(4,5)P2 treatment did not induce cell spreading (Figure 5). These data suggest that PI(4,5)P2 is not involved in Rac1 membrane translocation in the integrin signaling pathway. An investigation of the role of PI(3,4,5)P3 in Rac activity showed that phosphatidylinositol 3-kinase (PI3K) inhibitors and dominant-negative PI3K mutants reduce Rac-GTP loading. PI(3,4,5)P3 is involved in Rac-GEF activation such as Vav, Sos, Tiam, PIX, SWAP-70, and P-Rex families (Welch et al., 2003). And PI(3,4,5)P3 can also bind directly to Rac in vitro, and it can facilitate GTP loading through the dissociation of GDP and Rac (Missy et al., 1998). However, PI(3,4,5)P3 preferably binds to and stabilizes the nucleotide-free form of Rac rather than its GTP-loaded form (Missy et al., 1998). It was previously shown that platelet-derived growth factor-induced Rac-dependent lamellipodia formation is inhibited by PI3K inhibition but that V12-Rac1 can override this inhibitory effect (Hawkins et al., 1995). This suggests that PI(3,4,5)P3 is important for Rac GTP loading but that it is not involved in the targeting of Rac. However, PA did not affect Rac-GTP loading, and it induced Rac translocation, and the V12-Rac1 mutant did not restore the inhibition of lamellipodia formation caused by blocking PLD activity (data not shown). These findings suggest that PI(3,4,5)P3 is involved in Rac-GTP loading and that PA is involved in Rac targeting for downstream effector activation. The spatial and temporal control of Rac activation is important in many cellular functions, such as cell migration, phagocytosis, cytokinesis, and superoxide production (Bishop and Hall, 2000). Coincidentally, PLD and PI3K also are activated during these cellular process, and they are critical for the cellular functions required for the localization of Rac function (Santy and Casanova, 2001; Iyer et al., 2004). These findings suggest that the lipid second messengers PA and PI(3,4,5)P3 can cooperatively regulate localized Rac functions in a sophisticated manner by regulating GDP/GTP exchange and membrane targeting.
In the present study, we found a new signaling pathway that bridges lipid second messenger PA and small GTPase Rac, and we show that this pathway is part of the regulatory mechanism that underlies integrin-mediated cell spreading and migration involving actin cytoskeletal rearrangement (Figure 5 and Supplemental Figure S4). Integrin-mediated cell spreading and migration are essential for many diverse physiological processes, in particular, when inappropriate these cellular functions can cause cancer metastasis and tumor invasion; thus, they are the subjects of intense research scrutiny. αvβ3 integrins are up-regulated in invasive cancer cells and in angiogenic endothelial cells, and metastatic potential has been correlated with their expression. Moreover, integrins are known to guide matrix degradation and the activities of proteolytic enzymes such as matrix metalloproteinase (MMP)-2 and MMP-9 (Hood and Cheresh, 2002). Previously, it was suggested that PLD is also involved in cancer progression and metastasis, and elevated PLD expression and PLD activity have been reported in breast cancer, gastric, and prostate cancer tissues (Reich et al., 1995; Uchida et al., 1999; Noh et al., 2000). Moreover, PLD was found to stimulate cell protrusions in v-Src–transformed cells (Shen et al., 2002); interestingly, highly migrating and invading MDA-MB-231 human breast cancer cells have exceptionally high PLD activities, whereas MCF-7 breast cancer cells have relatively little (Sliva et al., 2002; Zhong et al., 2003). Although it has been suggested that PLD is involved in cancer motility and invasion, its relation with integrin receptor and the mode of action of PLD activity in cancer metastasis are still poorly understood. In the present study, we found that PLD activity is elevated by integrin receptor signaling pathway in OVCAR-3 cells (Figure 1). Furthermore, PLD block was found to inhibit integrin-mediated Rac translocation in and the spreading and migration of OVCAR-3 cells (Figures 2 and 4). This suggests that the PLD–PA–Rac pathway plays an important role in the metastasis of cancer cells, and it might provide a means for integrin and PLD-mediated cancer metastasis. In addition to integrin signaling, PLD and Rac have been found to be involved in diverse receptor signaling pathways. PA is known to regulate diverse physiological processes, such as, morphological change, migration, mitogenesis, phagocytosis, superoxide production, and axonal growth. (Cockcroft, 2001; Iyer et al., 2004; Watanabe et al., 2004; Lehman et al., 2006). Coincidentally with PA, Rac has also been found to be critical required for these cellular functions (Etienne-Manneville and Hall, 2002). Actin cytoskeletal rearrangements are centrally involved in many cellular functions, and in particular, cell migration, phagocytosis, and axonal growth require a sophisticated form of localized actin cytoskeletal regulation. Although the perhaps obvious relationship between PLD and Rac functions in these cellular functions has not been proven, it has been reported that PLD and Rac are commonly activated during phagocytosis and that PLD and Rac are localized to actin enriched phagosome surfaces (Ueyama et al., 2005; Lehman et al., 2006). Moreover, during axonal growth, PLD and Rac have been commonly found to positively regulate axonal growth. In addition, PLD2 and F-actin are colocalized in growth cones and Rac is localized in lamellipodia-rich areas of growth cones during axonal growth (Threadgill et al., 1997; Nikolic et al., 1998; Watanabe et al., 2004). These observations suggest that the PLD/PA/Rac pathway might be essential and general mechanism that underlies diverse cellular functions that involve Rac.
In summary, our findings show that PLD activity is regulated during integrin signaling and that this activation is important for integrin-mediated cell spreading and migration. We describe for the first time an endogenous regulatory pathway that controls GTP-Rac membrane translocation via direct interaction between Rac and the PLD product PA, and we demonstrate that this interaction is important for GTP-Rac1 translocation and Rho-GDI dissociation, which triggers downstream PAK activation, lamellipodia formation, cell spreading, and migration. Our results may explain the mechanisms underlying the functions of polarized cells in many systems, where precise spatiotemporal control of Rac is required. Moreover, our identification of the PLD–PA–Rac translocation pathway opens the way for further studies on the role of PA in Rac signaling in diverse cellular environments.
Supplementary Material
[Supplemental Materials]
ACKNOWLEDGMENTS
We thank J. C. Norman (University of Leicester, United Kingdom) for the gift of the β3-integrin plasmid and A. Salmeen and P. Vitorino (School of Medicine, Stanford University) for critical reading of the manuscript. This work was supported in part by a grant M102KM010001-03K1301-00710 from the 21st Frontier Functional Proteomics Research and National Core Research Center grant R15-2004-033-05001-0) in the Republic of Korea.
Abbreviations used:
MMP
matrix metalloproteinase
PA
phosphatidic acid
PAK
p21-activated kinase
PBD
p21-binding domain
PIP2
phosphatidylinositol 4,5-bisphosphate
PIP3
phosphatidylinositol 3,4,5-triphosphate
PI3K
phosphoinositide 3-kinase
PLD
phospholipase D
TX
Triton X.
Footnotes
REFERENCES
- Andresen B. T., Rizzo M. A., Shome K., Romero G. The role of phosphatidic acid in the regulation of the Ras/MEK/Erk signaling cascade. FEBS Lett. 2002;531:65–68. doi: 10.1016/s0014-5793(02)03483-x. [DOI] [PubMed] [Google Scholar]
- Bishop A. L., Hall A. Rho GTPases and their effector proteins. Biochem. J. 2000;348:241–255. [PMC free article] [PubMed] [Google Scholar]
- Bodin S., Soulet C., Tronchere H., Sie P., Gachet C., Plantavid M., Payrastre B. Integrin-dependent interaction of lipid rafts with the actin cytoskeleton in activated human platelets. J. Cell Sci. 2005;118:759–769. doi: 10.1242/jcs.01648. [DOI] [PubMed] [Google Scholar]
- Brown F. D., Thompson N., Saqib K. M., Clark J. M., Powner D., Thompson N. T., Solari R., Wakelam M. J. Phospholipase D1 localises to secretory granules and lysosomes and is plasma-membrane translocated on cellular stimulation. Curr. Biol. 1998;8:835–838. doi: 10.1016/s0960-9822(98)70326-4. [DOI] [PubMed] [Google Scholar]
- Chae Y. C., Lee S., Lee H. Y., Heo K., Kim J. H., Kim J. H., Suh P. G., Ryu S. H. Inhibition of muscarinic receptor-linked phospholipase D activation by association with tubulin. J. Biol. Chem. 2005;280:3723–3730. doi: 10.1074/jbc.M406987200. [DOI] [PubMed] [Google Scholar]
- Chuang T. H., Bohl B. P., Bokoch G. M. Biologically active lipids are regulators of Rac.GDI complexation. J. Biol. Chem. 1993;268:26206–26211. [PubMed] [Google Scholar]
- Cockcroft S. Signalling roles of mammalian phospholipase D1 and D2. Cell Mol. Life Sci. 2001;58:1674–1687. doi: 10.1007/PL00000805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarny M., Lavie Y., Fiucci G., Liscovitch M. Localization of phospholipase D in detergent-insoluble, caveolin-rich membrane domains. Modulation by caveolin-1 expression and caveolin-182–101. J. Biol. Chem. 1999;274:2717–2724. doi: 10.1074/jbc.274.5.2717. [DOI] [PubMed] [Google Scholar]
- del Pozo M. A., Alderson N. B., Kiosses W. B., Chiang H. H., Anderson R. G., Schwartz M. A. Integrins regulate Rac targeting by internalization of membrane domains. Science. 2004;303:839–842. doi: 10.1126/science.1092571. [DOI] [PubMed] [Google Scholar]
- Del Pozo M. A., Kiosses W. B., Alderson N. B., Meller N., Hahn K. M., Schwartz M. A. Integrins regulate GTP-Rac localized effector interactions through dissociation of Rho-GDI. Nat. Cell Biol. 2002;4:232–239. doi: 10.1038/ncb759. [DOI] [PubMed] [Google Scholar]
- del Pozo M. A., Price L. S., Alderson N. B., Ren X. D., Schwartz M. A. Adhesion to the extracellular matrix regulates the coupling of the small GTPase Rac to its effector PAK. EMBO J. 2000;19:2008–2014. doi: 10.1093/emboj/19.9.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di-Poi N., Faure J., Grizot S., Molnar G., Pick E., Dagher M. C. Mechanism of NADPH oxidase activation by the Rac/Rho-GDI complex. Biochemistry. 2001;40:10014–10022. doi: 10.1021/bi010289c. [DOI] [PubMed] [Google Scholar]
- Dinauer M. C. Regulation of neutrophil function by Rac GTPases. Curr. Opin. Hematol. 2003;10:8–15. doi: 10.1097/00062752-200301000-00003. [DOI] [PubMed] [Google Scholar]
- Du G., Altshuller Y. M., Vitale N., Huang P., Chasserot-Golaz S., Morris A. J., Bader M. F., Frohman M. A. Regulation of phospholipase D1 subcellular cycling through coordination of multiple membrane association motifs. J. Cell Biol. 2003;162:305–315. doi: 10.1083/jcb.200302033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emoto M., Klarlund J. K., Waters S. B., Hu V., Buxton J. M., Chawla A., Czech M. P. A role for phospholipase D in GLUT4 glucose transporter translocation. J. Biol. Chem. 2000;275:7144–7151. doi: 10.1074/jbc.275.10.7144. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S., Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- Exton J. H. Regulation of phospholipase D. Biochim. Biophys. Acta. 1999;1439:121–133. doi: 10.1016/s1388-1981(99)00089-x. [DOI] [PubMed] [Google Scholar]
- Filippi M. D., Harris C. E., Meller J., Gu Y., Zheng Y., Williams D. A. Localization of Rac2 via the C terminus and aspartic acid 150 specifies superoxide generation, actin polarity and chemotaxis in neutrophils. Nat. Immunol. 2004;5:744–751. doi: 10.1038/ni1081. [DOI] [PubMed] [Google Scholar]
- Frohman M. A., Sung T. C., Morris A. J. Mammalian phospholipase D structure and regulation. Biochim. Biophys. Acta. 1999;1439:175–186. doi: 10.1016/s1388-1981(99)00093-1. [DOI] [PubMed] [Google Scholar]
- Fukami K., Furuhashi K., Inagaki M., Endo T., Hatano S., Takenawa T. Requirement of phosphatidylinositol 4,5-bisphosphate for alpha-actinin function. Nature. 1992;359:150–152. doi: 10.1038/359150a0. [DOI] [PubMed] [Google Scholar]
- Giancotti F. G., Tarone G. Positional control of cell fate through joint integrin/receptor protein kinase signaling. Annu. Rev. Cell Dev. Biol. 2003;19:173–206. doi: 10.1146/annurev.cellbio.19.031103.133334. [DOI] [PubMed] [Google Scholar]
- Gilmore A. P., Burridge K. Regulation of vinculin binding to talin and actin by phosphatidyl-inositol-4–5-bisphosphate. Nature. 1996;381:531–535. doi: 10.1038/381531a0. [DOI] [PubMed] [Google Scholar]
- Ha K. S., Exton J. H. Activation of actin polymerization by phosphatidic acid derived from phosphatidylcholine in IIC9 fibroblasts. J. Cell Biol. 1993;123:1789–1796. doi: 10.1083/jcb.123.6.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J. M., Kim Y., Lee J. S., Lee C. S., Lee B. D., Ohba M., Kuroki T., Suh P. G., Ryu S. H. Localization of phospholipase D1 to caveolin-enriched membrane via palmitoylation: implications for epidermal growth factor signaling. Mol. Biol. Cell. 2002;13:3976–3988. doi: 10.1091/mbc.E02-02-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock J. F. Ras proteins: different signals from different locations. Nat. Rev. Mol. Cell Biol. 2003;4:373–384. doi: 10.1038/nrm1105. [DOI] [PubMed] [Google Scholar]
- Hancock J. F., Paterson H., Marshall C. J. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell. 1990;63:133–139. doi: 10.1016/0092-8674(90)90294-o. [DOI] [PubMed] [Google Scholar]
- Hawkins P. T., Eguinoa A., Qiu R. G., Stokoe D., Cooke F. T., Walters R., Wennstrom S., Claesson-Welsh L., Evans T., Symons M., et al. PDGF stimulates an increase in GTP-Rac via activation of phosphoinositide 3-kinase. Curr. Biol. 1995;5:393–403. doi: 10.1016/s0960-9822(95)00080-7. [DOI] [PubMed] [Google Scholar]
- Heo W. D., Inoue T., Park W. S., Kim M. L., Park B. O., Wandless T. J., Meyer T. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science. 2006;314:1458–1461. doi: 10.1126/science.1134389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood J. D., Cheresh D. A. Role of integrins in cell invasion and migration. Nat. Rev. 2002;2:91–100. doi: 10.1038/nrc727. [DOI] [PubMed] [Google Scholar]
- Iyer S. S., Barton J. A., Bourgoin S., Kusner D. J. Phospholipases D1 and D2 coordinately regulate macrophage phagocytosis. J. Immunol. 2004;173:2615–2623. doi: 10.4049/jimmunol.173.4.2615. [DOI] [PubMed] [Google Scholar]
- Iyer S. S., Kusner D. J. Association of phospholipase D activity with the detergent-insoluble cytoskeleton of U937 promonocytic leukocytes. J. Biol. Chem. 1999;274:2350–2359. doi: 10.1074/jbc.274.4.2350. [DOI] [PubMed] [Google Scholar]
- Jenkins G. H., Fisette P. L., Anderson R. A. Type I phosphatidylinositol 4-phosphate 5-kinase isoforms are specifically stimulated by phosphatidic acid. J. Biol. Chem. 1994;269:11547–11554. [PubMed] [Google Scholar]
- Jones D., Morgan C., Cockcroft S. Phospholipase D and membrane traffic. Potential roles in regulated exocytosis, membrane delivery and vesicle budding. Biochim. Biophys. Acta. 1999;1439:229–244. doi: 10.1016/s1388-1981(99)00097-9. [DOI] [PubMed] [Google Scholar]
- Kim J. H., Han J. M., Lee S., Kim Y., Lee T. G., Park J. B., Lee S. D., Suh P. G., Ryu S. H. Phospholipase D1 in caveolae: regulation by protein kinase Calpha and caveolin-1. Biochemistry. 1999;38:3763–3769. doi: 10.1021/bi982478+. [DOI] [PubMed] [Google Scholar]
- Lauffenburger D. A., Horwitz A. F. Cell migration: a physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- Lee C. S., Kim I. S., Park J. B., Lee M. N., Lee H. Y., Suh P. G., Ryu S. H. The phox homology domain of phospholipase D activates dynamin GTPase activity and accelerates EGFR endocytosis. Nat. Cell Biol. 2006;8:477–484. doi: 10.1038/ncb1401. [DOI] [PubMed] [Google Scholar]
- Lehman N., Di Fulvio M., McCray N., Campos I., Tabatabaian F., Gomez-Cambronero J. Phagocyte cell migration is mediated by phospholipases PLD1 and PLD2. Blood. 2006;108:3564–3572. doi: 10.1182/blood-2006-02-005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leventis R., Silvius J. R. Lipid-binding characteristics of the polybasic carboxy-terminal sequence of K-ras4B. Biochemistry. 1998;37:7640–7648. doi: 10.1021/bi973077h. [DOI] [PubMed] [Google Scholar]
- Lian L. Y., Barsukov I., Golovanov A. P., Hawkins D. I., Badii R., Sze K. H., Keep N. H., Bokoch G. M., Roberts G. C. Mapping the binding site for the GTP-binding protein Rac-1 on its inhibitor RhoGDI-1. Structure. 2000;8:47–55. doi: 10.1016/s0969-2126(00)00080-0. [DOI] [PubMed] [Google Scholar]
- Michaelson D., Silletti J., Murphy G., D'Eustachio P., Rush M., Philips M. R. Differential localization of Rho GTPases in live cells: regulation by hypervariable regions and RhoGDI binding. J. Cell Biol. 2001;152:111–126. doi: 10.1083/jcb.152.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missy K., Van Poucke V., Raynal P., Viala C., Mauco G., Plantavid M., Chap H., Payrastre B. Lipid products of phosphoinositide 3-kinase interact with Rac1 GTPase and stimulate GDP dissociation. J. Biol. Chem. 1998;273:30279–30286. doi: 10.1074/jbc.273.46.30279. [DOI] [PubMed] [Google Scholar]
- Moissoglu K., Slepchenko B. M., Meller N., Horwitz A. F., Schwartz M. A. In vivo dynamics of Rac-membrane interactions. Mol. Biol. Cell. 2006;17:2770–2779. doi: 10.1091/mbc.E06-01-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolic M., Chou M. M., Lu W., Mayer B. J., Tsai L. H. The p35/Cdk5 kinase is a neuron-specific Rac effector that inhibits Pak1 activity. Nature. 1998;395:194–198. doi: 10.1038/26034. [DOI] [PubMed] [Google Scholar]
- Noh D. Y., Ahn S. J., Lee R. A., Park I. A., Kim J. H., Suh P. G., Ryu S. H., Lee K. H., Han J. S. Overexpression of phospholipase D1 in human breast cancer tissues. Cancer Lett. 2000;161:207–214. doi: 10.1016/s0304-3835(00)00612-1. [DOI] [PubMed] [Google Scholar]
- O'Luanaigh N., Pardo R., Fensome A., Allen-Baume V., Jones D., Holt M. R., Cockcroft S. Continual production of phosphatidic acid by phospholipase D is essential for antigen-stimulated membrane ruffling in cultured mast cells. Mol. Biol. Cell. 2002;13:3730–3746. doi: 10.1091/mbc.E02-04-0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohguchi K., Nakashima S., Tan Z., Banno Y., Dohi S., Nozawa Y. Increased activity of small GTP-binding protein-dependent phospholipase D during differentiation in human promyelocytic leukemic HL60 cells. J. Biol. Chem. 1997;272:1990–1996. doi: 10.1074/jbc.272.3.1990. [DOI] [PubMed] [Google Scholar]
- Powner D. J., Hodgkin M. N., Wakelam M. J. Antigen-stimulated activation of phospholipase D1b by Rac1, ARF6, and PKCalpha in RBL-2H3 cells. Mol. Biol. Cell. 2002;13:1252–1262. doi: 10.1091/mbc.01-05-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price L. S., Leng J., Schwartz M. A., Bokoch G. M. Activation of Rac and Cdc42 by integrins mediates cell spreading. Mol. Biol. Cell. 1998;9:1863–1871. doi: 10.1091/mbc.9.7.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raftopoulou M., Hall A. Cell migration: Rho GTPases lead the way. Dev. Biol. 2004;265:23–32. doi: 10.1016/j.ydbio.2003.06.003. [DOI] [PubMed] [Google Scholar]
- Reich R., Blumenthal M., Liscovitch M. Role of phospholipase D in laminin-induced production of gelatinase A (MMP-2) in metastatic cells. Clin. Exp. Metastasis. 1995;13:134–140. doi: 10.1007/BF00133618. [DOI] [PubMed] [Google Scholar]
- Ridley A. J., Schwartz M. A., Burridge K., Firtel R. A., Ginsberg M. H., Borisy G., Parsons J. T., Horwitz A. R. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- Roth M. G., Bi K., Ktistakis N. T., Yu S. Phospholipase D as an effector for ADP-ribosylation factor in the regulation of vesicular traffic. Chem. Phys. Lipids. 1999;98:141–152. doi: 10.1016/s0009-3084(99)00026-2. [DOI] [PubMed] [Google Scholar]
- Santy L. C., Casanova J. E. Activation of ARF6 by ARNO stimulates epithelial cell migration through downstream activation of both Rac1 and phospholipase D. J. Cell Biol. 2001;154:599–610. doi: 10.1083/jcb.200104019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffzek K., Stephan I., Jensen O. N., Illenberger D., Gierschik P. The Rac-RhoGDI complex and the structural basis for the regulation of Rho proteins by RhoGDI. Nat. Struct. Biol. 2000;7:122–126. doi: 10.1038/72392. [DOI] [PubMed] [Google Scholar]
- Schwartz M. A., Schaller M. D., Ginsberg M. H. Integrins: emerging paradigms of signal transduction. Annu. Rev. Cell Dev. Biol. 1995;11:549–599. doi: 10.1146/annurev.cb.11.110195.003001. [DOI] [PubMed] [Google Scholar]
- Shen Y., Zheng Y., Foster D. A. Phospholipase D2 stimulates cell protrusion in v-Src-transformed cells. Biochem. Biophys. Res. Commun. 2002;293:201–206. doi: 10.1016/S0006-291X(02)00204-8. [DOI] [PubMed] [Google Scholar]
- Sliva D., Rizzo M. T., English D. Phosphatidylinositol 3-kinase and NF-kappaB regulate motility of invasive MDA-MB-231 human breast cancer cells by the secretion of urokinase-type plasminogen activator. J. Biol. Chem. 2002;277:3150–3157. doi: 10.1074/jbc.M109579200. [DOI] [PubMed] [Google Scholar]
- Threadgill R., Bobb K., Ghosh A. Regulation of dendritic growth and remodeling by Rho, Rac, and Cdc42. Neuron. 1997;19:625–634. doi: 10.1016/s0896-6273(00)80376-1. [DOI] [PubMed] [Google Scholar]
- Toda K., Nogami M., Murakami K., Kanaho Y., Nakayama K. Colocalization of phospholipase D1 and GTP-binding-defective mutant of ADP-ribosylation factor 6 to endosomes and lysosomes. FEBS Lett. 1999;442:221–225. doi: 10.1016/s0014-5793(98)01646-9. [DOI] [PubMed] [Google Scholar]
- Uchida N., Okamura S., Kuwano H. Phospholipase D activity in human gastric carcinoma. Anticancer Res. 1999;19:671–675. [PubMed] [Google Scholar]
- Ueyama T., Eto M., Kami K., Tatsuno T., Kobayashi T., Shirai Y., Lennartz M. R., Takeya R., Sumimoto H., Saito N. Isoform-specific membrane targeting mechanism of Rac during Fc gammaR-mediated phagocytosis: positive charge-dependent and independent targeting mechanism of Rac to the phagosome. J. Immunol. 2005;175:2381–2390. doi: 10.4049/jimmunol.175.4.2381. [DOI] [PubMed] [Google Scholar]
- Ugolev Y., Molshanski-Mor S., Weinbaum C., Pick E. Liposomes comprising anionic but not neutral phospholipids cause dissociation of Rac(1 or 2) x RhoGDI complexes and support amphiphile-independent NADPH oxidase activation by such complexes. J. Biol. Chem. 2006;281:19204–19219. doi: 10.1074/jbc.M600042200. [DOI] [PubMed] [Google Scholar]
- Watanabe H., Yokozeki T., Yamazaki M., Miyazaki H., Sasaki T., Maehama T., Itoh K., Frohman M. A., Kanaho Y. Essential role for phospholipase D2 activation downstream of ERK MAP kinase in nerve growth factor-stimulated neurite outgrowth from PC12 cells. J. Biol. Chem. 2004;279:37870–37877. doi: 10.1074/jbc.M402610200. [DOI] [PubMed] [Google Scholar]
- Welch H. C., Coadwell W. J., Stephens L. R., Hawkins P. T. Phosphoinositide 3-kinase-dependent activation of Rac. FEBS Lett. 2003;546:93–97. doi: 10.1016/s0014-5793(03)00454-x. [DOI] [PubMed] [Google Scholar]
- Yeung T., Terebiznik M., Yu L., Silvius J., Abidi W. M., Philips M., Levine T., Kapus A., Grinstein S. Receptor activation alters inner surface potential during phagocytosis. Science. 2006;313:347–351. doi: 10.1126/science.1129551. [DOI] [PubMed] [Google Scholar]
- Zhong M., Shen Y., Zheng Y., Joseph T., Jackson D., Foster D. A. Phospholipase D prevents apoptosis in v-Src-transformed rat fibroblasts and MDA-MB-231 breast cancer cells. Biochem. Biophys. Res. Commun. 2003;302:615–619. doi: 10.1016/s0006-291x(03)00229-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Materials]