An improved SUMO fusion protein system for effective production of native proteins (original) (raw)
Abstract
Expression of recombinant proteins as fusions with SUMO (small ubiquitin-related modifier) protein has significantly increased the yield of difficult-to-express proteins in Escherichia coli. The benefit of this technique is further enhanced by the availability of naturally occurring SUMO proteases, which remove SUMO from the fusion protein. Here we have improved the exiting SUMO fusion protein approach for effective production of native proteins. First, a sticky-end PCR strategy was applied to design a new SUMO fusion protein vector that allows directional cloning of any target gene using two universal cloning sites (Sfo1 at the 5′-end and XhoI at the 3′-end). No restriction digestion is required for the target gene PCR product, even the insert target gene contains a SfoI or XhoI restriction site. This vector produces a fusion protein (denoted as His6-Smt3-X) in which the protein of interest (X) is fused to a hexahistidine (His6)-tagged Smt3. Smt3 is the yeast SUMO protein. His6-Smt3-X was purified by Ni2+ resin. Removal of His6-Smt3 was performed on the Ni2+ resin by an engineered SUMO protease, His6-Ulp1403–621-His6. Because of its dual His6 tags, His6-Ulp1403–621-His6 exhibits a high affinity for Ni2 resin and associates with Ni2+ resin after cleavage reaction. One can carry out both fusion protein purification and SUMO protease cleavage using one Ni2+-resin column. The eluant contains only the native target protein. Such a one-column protocol is useful in developing a better high-throughput platform. Finally, this new system was shown to be effective for cloning, expression, and rapid purification of several difficult-to-produce authentic proteins.
Keywords: fusion protein, SUMO, Rad51, RecA, enterovirus, foot-and-mouth disease virus
In humans, many diseases, including cancer, aging, anemia, and developmental disorders arise because of gene mutations that result in aberrant proteins. Therefore, proteins are targets for therapeutic drugs, and protein production for structural and functional analysis is a major task in modern biology and medicine. Protein production is never a simple task because proteins are extremely diverse in their physio-chemical properties, and there is no generic protocol available for expression and purification. In addition, proteins are frequently expressed at low quantities or as insoluble aggregated folding intermediates, known as inclusion bodies. A fusion protein approach with expression tags has been widely used to overcome these problems (Wang and Wang 2004; Hu et al. 2007).
Because of concerns about the impact of expression tags on the structure and function of target proteins, removing expression tags by enzymatic cleavage is a goal. For therapeutic proteins, a prerequisite is that the final products contain only the authentic or native amino acid sequences; therefore, the fusion protein must be designed to contain at least one specific protease cleavage site between the target protein and the expression tags. A situation commonly arises in which fusion carriers cannot be processed effectively because of steric hindrance at the cleavage sites. Moreover, most proteases (e.g., factor Xa, tobacco etch virus protease, enterokinase, and thrombin) used in the fusion protein approach bring with them the challenge that cleavage reactions often occur at unexpected locations. This problem can be overcome by using a ubiquitin (Ub) or SUMO (small ubiquitin-related modifier) fusion protein system (Mossessova and Lima 2000). In this case, Ub or SUMO proteases are used for specific cleavage of Ub and SUMO from their fusion partner, respectively. Ub or SUMO serves not only as a solubility enhancer but also as a protease recognition site. Ub and SUMO proteases also have the advantage of recognizing the tertiary structures of Ub or SUMO, but not a linear amino acid sequence like other proteases. This characteristic prevents Ub and SUMO from erroneously cleaving within the target protein (Mossessova and Lima 2000; Catanzariti et al. 2004; Malakhov et al. 2004; Butt et al. 2005; Marblestone et al. 2006). The Escherichia coli protein ElaD has recently been identified as a Ub protease, which specifically cleaves the Ub conjugates, but not the SUMO conjugates (Catic et al. 2007). Therefore, SUMO may be a better expression tag than Ub for a fusion protein approach in E. coli.
The aim of this study was to establish a simpler and more efficient SUMO fusion protein expression system. Our new design allows cloning of any gene into this vector using two unique cloning sites (i.e., Sfo1 at the 5′-end and XhoI at the 3′-end) without restriction digestion of the target PCR products. In addition, we have developed a one-column production strategy to rapidly generate native or authentic proteins from the SUMO fusion proteins.
Materials and Methods
Protein expression and purification
Ulp1403-621, a fragment of Saccharomyces cerevisiae Ulp1 protein (amino acid residues 403–621), has been shown to cleave a C-terminal tagged yeast Smt3 in vitro, producing its mature forms (i.e., C-terminal “Gly-Gly”) (Mossessova and Lima 2000). The open reading frame (ORF) of Ulp1403–621 was cloned into the pET28a vector (Novagen) to express a His6-Ulp1403–621-His6 fusion protein, in which both the C and N termini were tagged with a hexahistidine tag (His6). This recombinant enzyme was soluble in E. coli and could be purified from crude extracts using Ni2+ resins. The final yield was ∼20 mg/L E. coli culture. The protein migrated as a single band on an SDS-PAGE gel stained with Coomassie blue, and with >99% purity as determined by densitometry (data not shown). Notably, because of its dual His6 tags, His6-Ulp1403–621-His6 exhibited a high affinity for Ni2+ resin. It could not be released from Ni2+ resins unless >300 mM imidazole or 100 mM EDTA was added to the elution buffer (data not shown).
The open reading frame of the E. coli RecA protein was cloned into a His6-Smt3 fusion protein expression vector modified from the pET32-Xa/LIC vector (Novagen). The His6-Smt3-RecA expression vectors were then transformed into _JM109(DE3)-competent cells. An overnight cell culture (15 mL) was grown at 37°C in the presence of 100 mg/L ampicillin. After transfer of the cell culture to 1 L of Luria-Bertani medium, the cell suspension was allowed to reach an OD600 of ∼0.5–0.6 before addition of IPTG (1 mM). Cells were then grown for 12 h at 20°C and then centrifuged at 9000_g for 30 min. Whole-cell lysates were prepared according to a protocol described previously (Wang et al. 1993), except that a different lysis buffer (50 mM Tris-HCl [pH 7.4], 300 mM NaCl, 0.2 mM EGTA [pH 8.0]) was used here to prevent nonspecific association of His6-Smt3-RecA with bacterial DNA. The soluble protein fraction was then mixed with 2 mL of Ni2+ resins (Amersham) to capture the His6-Smt3-RecA fusion proteins. The Ni2+ resins were then washed three times with 30 mL of wash buffer (50 mM Tris-HCl [pH 7.4], 300 mM NaCl, 0.2 mM EGTA [pH 8.0], 40 mM imidazole [pH 8.0]). Without elution, the Ni2+ resins bound with His6-Smt3-RecA fusion proteins were incubated with 0.1 mg of His6-Ulp1403–621-His6 for 10 h at 4°C to separate His6-Smt3 and RecA. The cleaved RecA proteins were then released from the Ni2+ resin. The flow-through was collected and dialyzed against buffer Q (50 mM Tris-HCl pH 8.0, 5% glycerol, 1 mM dithiothretol).
DNA substrates
The ΦX174 viral (+) strand DNA and the replicative form II DNA were purchased from New England Biolabs and GIBCO BRL, respectively. The P1656 single-stranded (ss) DNA primer (50 nucleotides) and plasmid DNA GW1 used in the D-loop formation assay have been described previously (Chen et al. 2004). The P1656 ssDNA was also used in the nuclease activity and ATPase activity assays. Exonuclease I (ExoI) was also purchased from New England Biolabs.
Enzymatic assays
Unless stated otherwise, all enzymatic reactions were carried out at 37°C. An electrophoretic mobility-shift assay was performed for DNA binding according to a previously described protocol (Chen et al. 2004), except that purified RecA protein was used here. A nuclease assay was carried out in 50 μL of buffer D (25 mM Tris-HCl [pH 7.4], 10 mM Mg-acetate, 1 mM ATPγS, 100 mM Na-acetate) with 3 μM P1656 ssDNA for 30 min. ExoI was used as a positive control for the nuclease assay. The ssDNA-stimulated ATPase activity assay was performed as described previously (Lee et al. 2004). The strand assimilation or D-loop formation assay was performed as follows. RecA proteins (1) were preincubated for 5 min at 37°C with 3 μM (in nucleotides) 5′ 32P-end-labeled P1656 ssDNA in the presence of 1 mM magnesium acetate and 2 mM AMP-PNP. A D-loop formation reaction was initiated by the addition of an equal volume (10 μL) of a solution containing a supercoiled double-stranded (ds) DNA plasmid GW1 (20 μM in base pairs). The reactions were allowed to proceed for 5 min. The reactions were then stopped by incubation with both 2 μL of SDS (5.5%) and proteinase K (6 mg/mL) for 5 min to remove the proteins. DNA from the reaction mixtures was resolved by electrophoresis for 2 h at 4 V/cm on a 0.8% agarose gel in Tris–acetate–EDTA buffer (40 mM Tris, 1 mM Na2–EDTA, and 20 mM acetic acid, pH 8.0). A phosphorimage of the agarose gel was taken to show the D-loop formation in the presence of RecA proteins (Lee et al. 2004).
Results
Using the SUMO fusion protein approach to express the E. coli RecA protein
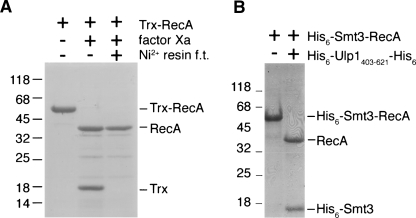
E. coli RecA was the first identified member of the RecA protein family, which also includes archaeal RadA and the eukaryotic proteins Dmc1 and Rad51. These proteins are DNA strand-exchange proteins that mediate homologous DNA recombination as well as the SOS response in bacteria. Because of their biological significance, RecA family proteins have been extensively studied during the past two decades. However, the molecular mechanisms of these proteins have not yet been fully elucidated (Cox 2007). Although RecA family proteins can be isolated directly from crude cell extracts (Cox et al. 1981; Sung 1994), this process requires significant effort in protein purification because nuclease contamination is a common problem. We recently reported that the thioredoxin (Trx) fusion protein was a good solution for production of the archaeal RadA protein (Chen et al. 2007a, 2007b). Specifically, we first expressed and purified the Trx-RadA fusion protein in E. coli using the pET32-Xa/LIC expression vector (Novagen). Native RadA was then obtained by proteolytic cleavage of the fusion protein with factor Xa. The resulting RadA protein is catalytically active, because it hydrolyzes more ATP in response to ssDNA binding and also promotes D-loop formation between a 5′ 32P-end-labeled ssDNA and a homologous dsDNA target (Lee et al. 2004; Chen et al. 2007a). When the same method was applied to the E. coli RecA protein, we found that a significant portion of Trx-RecA was cleaved by factor Xa at unexpected locations (Fig. 1A). To solve this problem, we replaced the ORF of the Trx protein with that of His6-Smt3 to generate the His6-Smt3-RecA expression vector, pSUMO-RecA. The His6-Smt3-RecA fusion protein was soluble in E. coli and could be specifically cleaved by an engineered SUMO protease, His6-Ulp1403–621-His6, to yield His6-Smt3 and RecA (see below) (Fig. 1B).
Figure 1.
Proteolytic cleavage of Trx-RecA and His6-Smt3-RecA fusion proteins. (A) An SDS-PAGE gel stained with Coomassie blue with purified Trx-RecA fusion proteins (first lane) before and (second lane) after digestion by factor Xa. After the factor Xa digestion, the protein solution was passed through a Ni2+-resin column. (Third lane) The flow-through (f.t.) was collected and analyzed by SDS-PAGE. Trx was retained in the Ni2+ resin, because it contained a His6 tag. (B) Purified His6-Smt3-RecA fusion proteins (first lane) before and (second lane) after digestion by an engineered SUMO protease, His6-Ulp1403–621-His6. The molecular weight standards are shown on the left (×1000).
On-resin proteolytic cleavage of the His6-Smt3-RecA fusion protein to yield authentic RecA protein
A one-column production protocol was developed here to produce authentic or native RecA proteins from the His6-Smt3-RecA fusion proteins. As described above, His6-Smt3-RecA was first affinity purified from E. coli cell lysates using Ni2+ resins. We used high-salt buffers (with 300 mM NaCl) throughout the cell disruption and protein purification processes to avoid nonspecific association of His6-Smt3-RecA fusion proteins with bacterial DNA. Without elution, the Ni2+ resins bound with His6-Smt3-RecA fusion proteins were then mixed with His6-Ulp1403–621-His6 to perform proteolytic cleavage. Because of its dual His6 tags, His6-Ulp1403–621-His6 exhibited a high affinity for Ni2+ resins. After the cleavage reaction, both His6-Smt3 and His6-Ulp1403–621-His6 were retained on Ni2+ resins, and only the RecA proteins were released from Ni2+ resins. The final yield was ∼10 mg of protein per liter of cell culture.
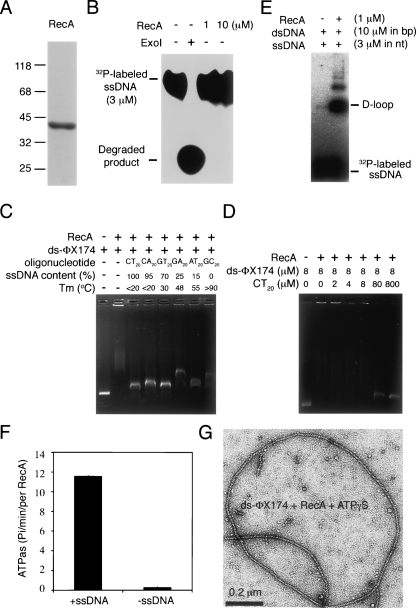
Native RecA protein has 352 amino acids and begins with alanine. The N terminus of the cleaved and purified RecA protein was sequenced by Edman degradation to confirm precise cleavage of His6-Ulp1403–621-His6, returning a sequence identical to the expected amino acid sequence. The molecular weight of purified RecA was determined by mass spectrometry to be 37,843, and the predicted molecular weight of native RecA protein is 37,842. Although the purified RecA protein looked reasonably pure (Fig. 2A), we still performed tests to determine if it was contaminated with nuclease using a 5′ 32P-end-labeled ssDNA substrate (P1656, 50 nt). ExoI was used here as a positive control for the nuclease assay. We found that all 5′ 32P-end-labeled ssDNA substrates were degraded after incubation with ExoI for 30 min. In contrast, the purified RecA proteins cleaved no 5′ 32P-end-labeled ssDNA substrate under the same conditions (Fig. 2B). Therefore, nuclease contamination is not a problem for the one-column production protocol.
Figure 2.
Purified RecA protein is catalytically active and is not contaminated with nuclease. (A) Coomassie-blue-stained SDS-PAGE gel with purified RecA protein (5 μg). The molecular weight standards are shown on the left (×1000). (B) Nuclease activity assay. Purified RecA protein (1 or 10 μM) or exonuclease I (20 units; New England Biolabs) was incubated with 5′ 32P-end-labeled P1656 ssDNA (50 nucleotides, 3 μM), respectively. The reaction mixtures were then treated with proteinase K to remove proteins and then subjected to electrophoresis with 20% native acrylamide gel to separate the 5′ 32P-end-labeled P1656 ssDNA from its degraded products. The result was visualized by a PhosphorImager. (C,D) Purified RecA binds both dsDNA and ssDNA. DNA binding specificity of purified RecA proteins was determined by mixing ds-ΦX174 DNA with various dinucleotide repeated oligonucleotides (40 nt). The melting temperature (T m) and ssDNA contents of these dinucleotide repeat oligonucleotides have been indicated (Biet et al. 1999). (E) D-loop formation. The ability to form a D-loop between a 5′ 32P-end-labeled P1656 oligonucleotide and dsDNA plasmid GW1 was assayed in buffers containing AMP-PNP and Mg2+. The reaction mixtures were then deproteinated by addition of 0.5% SDS and 0.55 mg/mL proteinase K. The assimilated and free oligonucleotides were separated on a 0.8% agarose gel and visualized by a PhosphorImager. (F) ATPase assay. Purified RecA proteins (0.5 μM) were incubated in the presence of 1 mM Mg2+ either with or without ΦX174 ssDNA (1 mM nucleotides). ATP hydrolysis was initiated by adding 1 mM ATP (with 0.6 nM [γ-32P]ATP) at 37°C. At different time points, 0.3-mL aliquots were withdrawn and spotted on thin layer chromatography paper to separate [γ-32P]ATP from 32P-labled inorganic phosphate. Shown are RecA ATPase activities in the presence and the absence of ssDNA. (G) Purified RecA proteins formed a nucleoprotein helical filament with ssDNA. Shown is the negative-staining electron microscopy image of the purified RecA proteins with a ds-ΦX174 DNA substrate in the presence of ATPγS.
The purified RecA protein was catalytically active. First, we showed, by electrophoretic mobility-shift assay, that purified RecA could bind DNA. Incubation of 10 μM RecA with 8 μM (in base pairs) ds-ΦX174 DNA resulted in a substantial decrease in the electrophoretic mobility of ds-ΦX174. To further substantiate the preference of RecA for dsDNA or ssDNA, we examined the relative ability of six oligonucleotides (80 μM in each case) to compete with circular ds-ΦX174 (8 μM) for RecA binding. These oligonucleotides were previously designed to contain different fractions of single-stranded regions under the same reaction conditions and were used to demonstrate that RecA could bind both ssDNA and dsDNA. In earlier experiments, thermal denaturation profiles showed a relative thermal stability for the six oligonucleotides of (CT)20 > (CA)20 > (GT)20 > (GA)20 > (AT)20 > (CG)20. The fraction of each oligonucleotide that presents as ssDNA at 37°C was shown (Biet et al. 1999). We found that all six oligonucleotides could compete with ds-ΦX174 for RecA binding (Fig. 2C). The purified RecA protein exhibits no apparent preference for ssDNA than for dsDNA, because the addition of oligonucleotide (CT)20 at low concentrations (2, 4, or 8 μM) resulted in no significant increase in the electrophoretic mobility of ds-ΦX174 (Fig. 2D).
Second, the purified RecA protein could promote a homology-dependent strand-exchange reaction, as revealed by D-loop formation between a 5′ 32P-end-labeled P1656 ssDNA and a supercoiled dsDNA plasmid, GW1 (Fig. 2E; Chen et al. 2004, 2007a,b). Third, it also exhibited ssDNA-activated ATPase activity, as determined by release of 32P inorganic phosphate from [γ-32P]ATP in the presence or absence of ss-ΦX174 DNA (Fig. 2F). Finally, we confirmed by electron microscopy that purified RecA proteins formed helical filaments on a circular ds-ΦX174 substrate (Fig. 2G), indicating that the purified RecA proteins have no apparent polymerization defect. Taken together, we conclude that our new SUMO fusion protein system and one-column production protocol have successfully produced native RecA proteins.
Construction of a new SUMO fusion protein expression vector, pHD
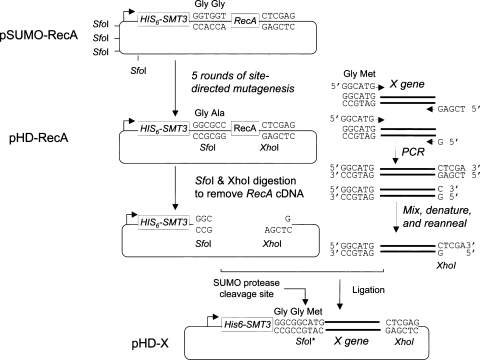
To develop a simpler and better system for high-throughput molecular cloning and screening of soluble SUMO fusion proteins, we engineered the pSUMO-RecA vector into pHD-RecA by five rounds of site-directed mutagenesis reactions. First, the four Sfo1 (5′-GGCGCC-3′) restriction sites in the backbone of pET32-Xa/LIC were mutated into 5′-GGCTCC-3′ or 5′-GGCACC-3′, respectively. Second, a new Sfo1 restriction site was generated at the SUMO protease cleavage site, leading to a point mutation from “GlyGly” to “GlyAla.” This design allows application of the sticky-end PCR cloning method (Shih et al. 2002, 2005) to insert any target gene (denoted X) into the pHD vector using two universal cloning sites, that is, SfoI at the 5′-end and XhoI at the 3′-end.
As illustrated in the right panel of Figure 3, sticky-end PCR cloning requires two PCR reactions in two separate tubes. Both PCR products are purified and mixed equally. After denaturation and renaturation, 50% of the final products carry one Sfo1 blunt end and one XhoI cohesive end and are ready for ligation even without restriction digestion of the PCR products. Moreover, a new “GlyGly” SUMO cleavage site is generated right before the first amino acid codon of the X gene (Fig. 2). Therefore, the resulting expression constructs allow production of His6-Smt3-X fusion proteins. Native or authentic protein X can then be obtained via digestion of His6-Smt3-X with His6-Ulp1403–621-His6 protease.
Figure 3.
Design of the new SUMO fusion protein expression vector, pHD. The pSUMO-RecA expression vector was subjected to five rounds of site-directed mutagenesis to generate the pHD-RecA vector. The latter was then digested with Sfo1 and XhoI, and ready to use for cloning a new target gene X (left panel). A sticky-end PCR cloning method was applied here to prepare the PCR insert of X gene. Two separate PCR reactions were carried out using one forward primer and two reverse primers. The two PCR products were mixed and were 5′-end phosphorylated by T4 polynucleotide kinase. After denaturation at 95°C and annealing at 65°C, ∼50% of the products carried 5′ Sfo1 blunt ends and 3′ XhoI ends, and were ready for ligation into the pHD vectors. After ligation, the SfoI was mutated (SfoI*). The final expression plasmid pHD-X can be used to produce the His6-Smt3-X fused protein, which can be cleaved by His6-Ulp1403–621-His6 protease to release the native X protein with any amino acid residue (e.g., Met) at its NH2 end next to the Gly-Gly end of Smt3.
Uses of the pHD vector to produce proteins previously known to be poorly expressed
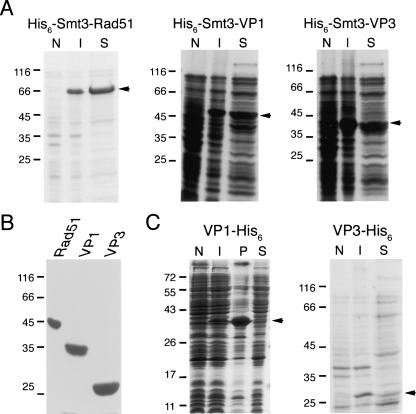
We have successfully applied the pHD vector to clone and produce three other authentic proteins. The first one is the yeast Rad51. Rad51, the eukaryotic homolog of E. coli RecA, plays important roles in mitotic and meiotic homologous DNA recombination. Like RecA, Rad51 is difficult to overexpress in E. coli. Direct purification of Rad51 protein from yeast cell extracts also requires laborious effort (Van Komen et al. 2006). Here, we showed that His6-Smt3-Rad51 fusion protein is soluble in E. coli (Fig. 4A). Using the same protocols described above for RecA protein, we have successfully produced native yeast Rad51 with a final yield of ∼10 mg/L of E. coli culture. The purified yeast Rad51 protein displays as a single band on an SDS-PAGE gel stained with Coomassie blue (Fig. 4B). The molecular weight of Rad51 was determined by mass spectrometry to be 42,964, and the predicted molecular weight is 42,963. Edman degradation was also performed to confirm that the N terminus of purified yeast Rad51 was identical to the expected amino acid sequence.
Figure 4.
Uses of pHD expression vector to produce authentic yeast Rad51, HFDV-VP1, and FMD-VP3 proteins. (A) Solubility analysis of the His6-Smt3-Rad51, His6-Smt3-VP1, and His6-Smt3-VP3 fusion proteins. To express the fusion proteins, each SUMO fusion protein expression vector was transformed into E. coli host cells, respectively. The expression of SUMO fusion proteins was induced with 1 mM isopropyl β-D-thiogalactoside at OD600 = 0.4 and incubated for 6 h at 20°C. A solubility test was performed as described previously (Shih et al. 2002). Samples of total proteins and soluble protein fractions were separated on a 12% SDS-PAGE under reducing conditions and stained with Coomassie blue. (N) Whole-cell lysates of uninduced cells; (I) whole-cell lysates of induced cells; (S) soluble proteins with induction. The position of each fusion protein in the SDS-PAGE gel is marked by an arrow on the right, respectively. (B) Production of yeast Rad51, HFMDV-VP1, and FMD-VP3 proteins using the one-column production protocol described in this study. Shown is a Coomassie blue stained SDS-PAGE gel with each purified protein (5 μg). (C) Solubility analysis of the VP1-His6 and VP3-His6 proteins. The band of each protein in the SDS-PAGE gel is marked by an arrow on the right, respectively. (P) Insoluble proteins with induction. The molecular weight standards are shown on the left (×1000).
The same approach also was successfully used to express and purify the capsid protein VP1 of enterovirus EV71 (HFMDV-VP1) and the capsid protein VP3 of foot-and-mouth disease virus (FMDV-VP3). In 1998, an epidemic of enterovirus infection caused hand-foot-and-mouth disease (HFMD) and herpangina in thousands of people in Taiwan. Although several enteroviruses were circulating during this 1998 epidemic, enterovirus EV71 was associated with most of the serious clinical manifestations and with nearly all of the deaths (Ho 2000). Foot-and-mouth disease is one of the most contagious animal diseases, often causing extensive epidemic in domestic cattle and swine (Davies 2002). Producing soluble capsid proteins for developing subunit vaccines or detection reagents is important. However, nearly all capsid proteins in both viruses, including HFMDV-VP1 and FMDV-VP3, are insoluble when overexpressed in E. coli (Fig. 4C). We showed here that both His6-Smt3-VP1 and His6-Smt3-VP3 proteins were soluble (Fig. 4A) in E. coli and yield authentic VP1 and VP3 proteins after proteolytic cleavage with His6-Ulp1403–621-His6 (Fig. 4B). Edman degradation was performed to confirm that the N termini of purified HFDV-VP1 and FMDV-VP3 were identical to the expected amino acid sequences, respectively. Mass spectrometry analysis revealed that the molecular weights of HFMDV-VP1 and FMDV-VP3 were 32,829 and 23,816, respectively. The predicted molecular weights of these two proteins are 32,744 and 23,817, respectively. Because HFMDV-VP1 and FMDV-VP3 are still soluble after the cleavage/removal of Smt3, it suggests an important advantage of the SUMO expression system. SUMO is not simply promoting the solubility through its attachment to these proteins, but it actually may promote the folding of VP1/VP3 in a particular conformation that enhances solvent interactions.
Discussion
SUMO fusion technology has advanced beyond the traditional gene fusion system for difficult-to-express proteins (Mossessova and Lima 2000; Malakhov et al. 2004; Butt et al. 2005; Marblestone et al. 2006). Here, we have designed a new SUMO fusion protein expression vector that allows rapid cloning of any gene with two unique cloning sites using the sticky-end PCR method. No restriction digestion to insert PCR products is required. In addition, a one-column production protocol was developed here to carry out protein purification as well as the SUMO cleavage reaction. We have tested this protocol on four different difficult-to-produce proteins with excellent results.
Our results indicate that the SUMO fusion protein approach is a better solution than the traditional purification approaches for production of authentic or native RecA family proteins. We showed that RecA was cleaved by factor Xa at unexpected locations. In contrast, His6-Ulp1403–621-His6 specifically cleaved the SUMO fusion proteins to yield native RecA and Rad51 proteins. Moreover, the expression level of the Trx-Rad51 fusion protein in E. coli was relatively low (data not shown). Therefore, SUMO is superior to other fusion tags (e.g., Trx) not only for expression of RecA and Rad51 proteins but also for proteolytic removal of RecA and Rad51 from their fusion proteins. More importantly, we have overcome the nuclease contamination problem for production of RecA family proteins, because His6-Smt3 allows for affinity purification with Ni2+ resin. We showed that the purified RecA proteins exhibited no detectable nuclease activity. We are currently applying the same approach to rapidly produce mutant RecA and Rad51 proteins for structural and functional studies.
In this study, we have also successfully produced two soluble virus capsid proteins, HFMDV-VP1 and FMDV-VP3. The capability of Smt3 protein to enhance the solubility of virus capsid proteins may be physiologically relevant. Studies have revealed that the capsid and envelope proteins of several viruses could either interact with SUMO or Ubc9 (the SUMO E2 ligase, enzymes) or were SUMO modified during virus infection; these viruses including the Tula hantavirus, Epstein-Barr virus, cyto-Megalo virus, Dengue virus, herpes virus, and Molony murine leukemia virus (Wilson and Rangasamy 2001; Kaukinen et al. 2003; Lee et al. 2003; Yueh et al. 2006; Chiu et al. 2007). Moreover, quantitative SUMO modification of a vaccinia virus protein, A40R, prevents A40R proteins from self-polymerization and aggregation in vivo (Palacios et al. 2005). SUMO modification may be a common mechanism for virus proteins to retain their solubility or to prevent improper self-segregation before virus assembly or during targeting to proper cellular locations. It is of interest to address if the SUMO fusion approach can be applied for analyzing the mechanisms of virus assembly.
Acknowledgments
This work was supported by a Research Investigator Award from Academia Sinica (AS-97-FP-M02 to T.F.W.). We thank Yuan-Chih Chang (Institute of Physics, Academia Sinica) for EM image analysis.
Footnotes
Reprint requests to: Ting-Fang Wang, Institute of Biochemical Sciences, National Taiwan University, Taipei 106, Taiwan; e-mail: tfwang@gate.sinica.edu.tw; fax: 886-2-27889759; or Chih-Hsiang Leng, Vaccine Research and Development Center, National Health Research Center, Miaoli 350, Taiwan; e-mail: leoleng@nhri.org.tw; fax: 886-37-583009.
References
- Biet, E., Sun, J., Dutreix, M. Conserved sequence preference in DNA binding among recombination proteins: An effect of ssDNA secondary structure. Nucleic Acids Res. 1999;27:596–600. doi: 10.1093/nar/27.2.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt, T.R., Edavettal, S.C., Hall, J.P., Mattern, M.R. SUMO fusion technology for difficult-to-express proteins. Protein Expr. Purif. 2005;43:1–9. doi: 10.1016/j.pep.2005.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanzariti, A.M., Soboleva, T.A., Jans, D.A., Board, P.G., Baker, R.T. An efficient system for high-level expression and easy purification of authentic recombinant proteins. Protein Sci. 2004;13:1331–1339. doi: 10.1110/ps.04618904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catic, A., Misaghi, S., Korbel, G.A., Ploegh, H.L. ElaD, a deubiquitinating protease expressed by E. coli . PLoS ONE. 2007;2:e381. doi: 10.1371/journal.pone.0000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y.K., Leng, C.H., Olivares, H., Lee, M.H., Chang, Y.C., Kung, W.M., Ti, S.C., Lo, Y.H., Wang, A.H., Chang, C.S., et al. Heterodimeric complexes of Hop2 and Mnd1 function with Dmc1 to promote meiotic homolog juxtaposition and strand assimilation. Proc. Natl. Acad. Sci. 2004;101:10572–10577. doi: 10.1073/pnas.0404195101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L.T., Ko, T.P., Chang, Y.C., Lin, K.A., Chang, C.S., Wang, A.H., Wang, T.F. Crystal structure of the left-handed archaeal RadA helical filament: Identification of a functional motif for controlling quaternary structures and enzymatic functions of RecA family proteins. Nucleic Acids Res. 2007a;35:1787–1801. doi: 10.1093/nar/gkl1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L.T., Ko, T.P., Chang, Y.W., Lin, K.A., Wang, A.H., Wang, T.F. Structural and functional analyses of five conserved positively charged residues in the L1 and N-terminal DNA binding motifs of archaeal RADA protein. PLoS ONE. 2007b;2:e858. doi: 10.1371/journal.pone.0000858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, M.W., Shih, H.M., Yang, T.H., Yang, Y.L. The type 2 dengue virus envelope protein interacts with small ubiquitin-like modifier-1 (SUMO-1) conjugating enzyme 9 (Ubc9) J. Biomed. Sci. 2007;14:429–444. doi: 10.1007/s11373-007-9151-9. [DOI] [PubMed] [Google Scholar]
- Cox, M.M. Motoring along with the bacterial RecA protein. Nat. Rev. Mol. Cell Biol. 2007;8:127–138. doi: 10.1038/nrm2099. [DOI] [PubMed] [Google Scholar]
- Cox, M.M., McEntee, K., Lehman, I.R. A simple and rapid procedure for the large scale purification of the recA protein of Escherichia coli . J. Biol. Chem. 1981;256:4676–4678. [PubMed] [Google Scholar]
- Davies, G. Foot and mouth disease. Res. Vet. Sci. 2002;73:195–199. doi: 10.1016/s0034-5288(02)00105-4. [DOI] [PubMed] [Google Scholar]
- Ho, M. Enterovirus 71: The virus, its infections and outbreaks. J. Microbiol. Immunol. Infect. 2000;33:205–216. [PubMed] [Google Scholar]
- Hu, S.-M., Wang, A.H.-J., Wang, T.-F. Encyclopedia of life sciences. John Wiley; Chichester: 2007. Expression tags for protein production. [DOI] [Google Scholar]
- Kaukinen, P., Vaheri, A., Plyusnin, A. Non-covalent interaction between nucleocapsid protein of Tula hantavirus and small ubiquitin-related modifier-1, SUMO-1. Virus Res. 2003;92:37–45. doi: 10.1016/s0168-1702(02)00312-x. [DOI] [PubMed] [Google Scholar]
- Lee, B.H., Yoshimatsu, K., Maeda, A., Ochiai, K., Morimatsu, M., Araki, K., Ogino, M., Morikawa, S., Arikawa, J. Association of the nucleocapsid protein of the Seoul and Hantaan hantaviruses with small ubiquitin-like modifier-1-related molecules. Virus Res. 2003;98:83–91. doi: 10.1016/j.virusres.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Lee, M.H., Leng, C.H., Chang, Y.C., Chou, C.C., Chen, Y.K., Hsu, F.F., Chang, C.S., Wang, A.H., Wang, T.F. Self-polymerization of archaeal RadA protein into long and fine helical filaments. Biochem. Biophys. Res. Commun. 2004;323:845–851. doi: 10.1016/j.bbrc.2004.08.163. [DOI] [PubMed] [Google Scholar]
- Malakhov, M.P., Mattern, M.R., Malakhova, O.A., Drinker, M., Weeks, S.D., Butt, T.R. SUMO fusions and SUMO-specific protease for efficient expression and purification of proteins. J. Struct. Funct. Genomics. 2004;5:75–86. doi: 10.1023/B:JSFG.0000029237.70316.52. [DOI] [PubMed] [Google Scholar]
- Marblestone, J.G., Edavettal, S.C., Lim, Y., Lim, P., Zuo, X., Butt, T.R. Comparison of SUMO fusion technology with traditional gene fusion systems: Enhanced expression and solubility with SUMO. Protein Sci. 2006;15:182–189. doi: 10.1110/ps.051812706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossessova, E., Lima, C.D. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- Palacios, S., Perez, L.H., Welsch, S., Schleich, S., Chmielarska, K., Melchior, F., Locker, J.K. Quantitative SUMO-1 modification of a vaccinia virus protein is required for its specific localization and prevents its self-association. Mol. Biol. Cell. 2005;16:2822–2835. doi: 10.1091/mbc.E04-11-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih, Y.P., Kung, W.M., Chen, J.C., Yeh, C.H., Wang, A.H., Wang, T.F. High-throughput screening of soluble recombinant proteins. Protein Sci. 2002;11:1714–1719. doi: 10.1110/ps.0205202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih, Y.P., Wu, H.C., Hu, S.M., Wang, T.F., Wang, A.H. Self-cleavage of fusion protein in vivo using TEV protease to yield native protein. Protein Sci. 2005;14:936–941. doi: 10.1110/ps.041129605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- Van Komen, S., Macris, M., Sehorn, M.G., Sung, P. Purification and assays of Saccharomyces cerevisiae homologous recombination proteins. Methods Enzymol. 2006;408:445–462. doi: 10.1016/S0076-6879(06)08028-1. [DOI] [PubMed] [Google Scholar]
- Wang, T.-F., Wang, A.H.-J. High-throughput screening of soluble recombinant proteins. In: Simpson R.J., editor. Purifying proteins for proteomics: A laboratory manual. Cold Spring Harbor Laboratory Press; New York: 2004. Chapter 5. [Google Scholar]
- Wang, T.F., Chang, J.H., Wang, C. Identification of the peptide binding domain of hsc70. 18-Kilodalton fragment located immediately after ATPase domain is sufficient for high-affinity binding. J. Biol. Chem. 1993;268:26049–26051. [PubMed] [Google Scholar]
- Wilson, V.G., Rangasamy, D. Viral interaction with the host cell sumoylation system. Virus Res. 2001;81:17–27. doi: 10.1016/s0168-1702(01)00365-3. [DOI] [PubMed] [Google Scholar]
- Yueh, A., Leung, J., Bhattacharyya, S., Perrone, L.A., de los Santos, K., Pu, S.Y., Goff, S.P. Interaction of moloney murine leukemia virus capsid with Ubc9 and PIASy mediates SUMO-1 addition required early in infection. J. Virol. 2006;80:342–352. doi: 10.1128/JVI.80.1.342-352.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]