Mitochondrial nucleoids maintain genetic autonomy but allow for functional complementation (original) (raw)
Abstract
Mitochondrial DNA (mtDNA) is packaged into DNA-protein assemblies called nucleoids, but the mode of mtDNA propagation via the nucleoid remains controversial. Two mechanisms have been proposed: nucleoids may consistently maintain their mtDNA content faithfully, or nucleoids may exchange mtDNAs dynamically. To test these models directly, two cell lines were fused, each homoplasmic for a partially deleted mtDNA in which the deletions were nonoverlapping and each deficient in mitochondrial protein synthesis, thus allowing the first unequivocal visualization of two mtDNAs at the nucleoid level. The two mtDNAs transcomplemented to restore mitochondrial protein synthesis but were consistently maintained in discrete nucleoids that did not intermix stably. These results indicate that mitochondrial nucleoids tightly regulate their genetic content rather than freely exchanging mtDNAs. This genetic autonomy provides a molecular mechanism to explain patterns of mitochondrial genetic inheritance, in addition to facilitating therapeutic methods to eliminate deleterious mtDNA mutations.
Introduction
Unique among human organelles, mitochondria have a dual nuclear/mitochondrial genetic makeup: nuclear-encoded proteins combine with those encoded by mitochondrial DNA (mtDNA) to comprise the mitochondrial respiratory chain, the five multisubunit complexes of oxidative phosphorylation which produce the bulk of cellular ATP. mtDNA is a 16.6-kb double-stranded circular molecule encoding 22 transfer RNAs, two ribosomal RNAs, and 13 polypeptides (Fig. 1 A). mtDNA exists in hundreds of identical copies per cell in mammals (homoplasmy) but can also exist in multiple nonidentical species within an individual tissue or cell (heteroplasmy). Mutations of mtDNA, including point mutations and Δ-mtDNAs (partial deletions), are associated with a wide range of systemic and tissue-specific diseases. High levels of Δ-mtDNAs are found in the substantia nigra of individuals with Parkinson's disease and aged individuals (Kraytsberg et al., 2004). Δ-mtDNAs cause the mitochondrial diseases Kearns-Sayre syndrome and progressive external ophthalmoplegia. In these disorders, affected tissues typically carry both mutant and wild-type (WT) mtDNAs. The proportion of mutant versus WT mtDNA is critically important for pathology (for review see DiMauro and Schon, 2003). Despite this, it is unclear how mtDNA genotypes are propagated and maintained. Mitotic segregation caused by random genetic drift has been reported, particularly in mammalian oocytes (Jenuth et al., 1996; Brown et al., 2001). Conversely, directed segregation has been demonstrated both for (Yoneda et al., 1992) and against (Rajasimha et al., 2008) mutated mtDNAs. To understand the propagation and maintenance of mtDNA heteroplasmy, it is necessary to examine the mechanisms of mtDNA organization and inheritance within the mitochondrion itself.
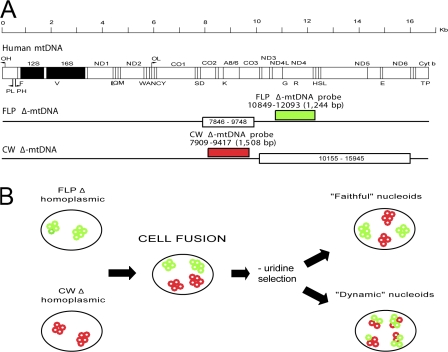
Figure 1.
mtDNAs and experimental design. (A) Linearized maps of mtDNAs discussed. The WT human mtDNA is shown at top. FLP and CW Δ-mtDNAs are shown below. White box denotes deleted region for each Δ-mtDNA. Red box denotes CW Δ-mtDNA FISH probe position and size. Green box denotes FLP Δ-mtDNA FISH probe position and size. (B) Schematic of cell fusion experiment. FLPΔ and CWΔ homoplasmic cell lines are fused in the presence of PEG, allowed to recover, and placed in uridine-minus medium to select for mitochondrial function. Homoplasmic FLPΔ (green) and CWΔ (red) nucleoids are shown with four to five copies of mtDNA per nucleoid. Two models for complementation are shown: faithful, in which each Δ-mtDNA (red only or green only) remains segregated from the other in homoplasmic nucleoids, and dynamic, in which the Δ-mtDNAs are exchanged among nucleoids, resulting in heteroplasmic nucleoids (red plus green).
Within the organelle, mtDNA is organized into assemblies called nucleoids. Each nucleoid is composed of 1–10 copies of mtDNA (Satoh and Kuroiwa, 1991; Iborra et al., 2004; Legros et al., 2004) and associated proteins, including TFAM (which acts as both a transcription factor and a DNA packaging protein), aconitase (in yeast), polymerase γ, mitochondrial single-strand binding protein, branched chain α-ketoacid dehydrogenase (in yeast), and the mitochondrial helicase Twinkle (Chen and Butow, 2005; Chen et al., 2005; Wang and Bogenhagen, 2006). Nucleoids are distributed throughout the mitochondrial network at regular spatial intervals (Capaldi et al., 2002) and are sufficiently mobile to repopulate cells lacking mtDNA when introduced via cell fusion. When mitochondria transition to a fragmented morphology, each individual organelle contains at least one nucleoid (Margineantu et al., 2002). This is similar to bacterial nucleoid organization, in which pathways exist to ensure the proper distribution of nucleoids during bacterial cell division (Rothfield et al., 2005). However, it is unclear how mtDNA heteroplasmy is organized at the organellar level. In particular, how are mtDNAs organized and propagated via the nucleoid? How does nucleoid organization explain the observed patterns of mtDNA segregation?
Two models have been proposed to explain how mitochondrial nucleoids mediate mtDNA inheritance. Jacobs et al. (2000) proposed that individual nucleoids replicate only their own exact genetic content (Fig. 1 B). This model maintains that mtDNA content within a nucleoid is generally static and that nucleoids do not exchange mtDNAs between each other (the “faithful nucleoid” model). Alternatively, D'Aurelio et al. (2004) proposed that nucleoids are subject to remodeling and have a dynamic organization allowing nucleoids to exchange mtDNAs freely (the “dynamic nucleoid” model; Fig. 1 B). To date, this fundamental question of mitochondrial cell biology and genetics has remained unanswered because of the experimentally intractable nature of the problem, as it demands both the isolation of two cell lines with the same nuclear background and with different mtDNAs and an experimental approach capable of clearly distinguishing between the two heterologous mtDNAs at the level of the nucleoid, a suborganellar macromolecular assembly.
To determine which model is more accurate, we observed the segregation of mtDNA nucleoids after the fusion of two homoplasmic cell lines carrying nonoverlapping partial deletions (FLP6a39.32Δ, hereafter referred to as FLPΔ [Santra et al., 2004] and CW420-115Δ, hereafter referred to as CWΔ [Mita et al., 1990; Pallotti et al., 2004]; Fig. 1 A). The two nucleoid populations were clearly distinguishable from each other when visualized by two-color red-green FISH, using probes specific to each Δ-mtDNA. The use of nonoverlapping deletions of mtDNA reveals the mode of mtDNA nucleoid inheritance through unequivocal subcellular tracking of the two heterologous mtDNAs.
Results
Validation of mtDNA genotype in parental cell lines with nonoverlapping Δ-mtDNAs
To visualize the two heterologous mtDNA nucleoid populations, we used two parental cybrid cell lines homoplasmic for nonoverlapping deletions (Fig. 1 A). By using FISH probes specific to the deleted regions, it is possible to distinguish between the two mtDNA genotypes at nucleoid resolution. We therefore characterized the mtDNA genotypes of the two parental cell lines by conventional and long-distance PCR to ensure the genetic suitability of these cell lines for use in cell fusion experiments.
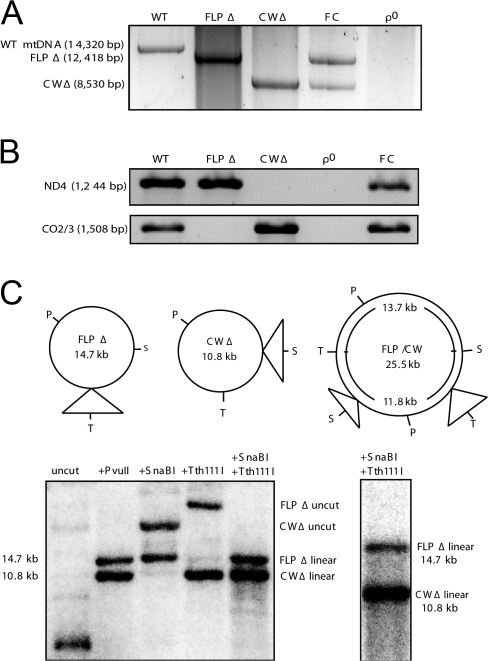
The homoplasmic FLPΔ (deleted for nt 7846–9748) and CWΔ (deleted for nt 10155–15945) cell lines have been reported previously (Pallotti et al., 2004; Santra et al., 2004). Long-distance PCR of mtDNA from FLP6a39.2 (100% WT mtDNA) yielded an amplification product of the expected size (14,320 bp), whereas FLPΔ and CWΔ total DNA templates each yielded a single product consistent with the expected FLP and CW Δ-mtDNA products (12,418 bp and 8,530 bp, respectively; Fig. 2 A). 143B206 ρ0 cells that are devoid of mtDNA (King and Attardi, 1989) gave no product, as expected (Fig. 2 A). Conventional PCR of total DNA isolated from WT cells gave an amplification product of the predicted sizes for a fragment containing a portion of MTND4 (1,244 bp; nt 10849-12093), hereafter termed ND4, and a fragment containing MTATP8 and MTATP6 and portions of the flanking MTCO2 and MTCO3 genes (1,508 bp; nt 7909–9417), hereafter termed CO2/3 (Fig. 2 B). FLPΔ cells showed an amplification product for ND4 but not for CO2/3, whereas CWΔ cells showed a band for CO2/3 but not ND4, which is consistent with the predicted Δ-mtDNAs present (Mita et al., 1990; Pallotti et al., 2004; Santra et al., 2004). As expected, WT cells gave both products, whereas 143B206 ρ0 cells gave no product in either amplification (Fig. 2 B).
Figure 2.
mtDNA analysis of cybrid cell lines. (A) Long-distance PCR of mtDNAs. Lanes denote long-distance amplification from 1 ng of template from total cellular DNA of the indicated cell line. Primers amplify 14,320 bp from nt 3066 (forward primer) to nt 812 (reverse primer) of the WT mtDNA molecule, encompassing both deleted regions. (B) Conventional PCR of ND4 and CO2/3 from isolated total DNA of cybrid cell lines. Lanes denote PCR reactions from 100 ng of template. (C) Southern blot analysis of FC II day-90 cells, including schematic of CW and FLP Δ-mtDNA restriction sites, as well as that of a potential FLP/CW recombinant molecule. Both Δ-mtDNAs carry a PvuII site. FLP Δ-mtDNA carries a SnaBI site but not a Tth111I site, whereas CW Δ-mtDNA carries a Tth111I site but not a SnaBI. 2 μg of total cellular DNA was digested with the enzymes listed, transferred to PVDF, and probed with DNA corresponding to nt 3778–6051 of human mtDNA. Single-lane panel shows a Southern blot of FC II cells subjected to ketogenic selection, digested with SnaBI and Tth111I, and electrophoresed for several days to achieve better separation of the FLP and CW Δ-mtDNA–linearized molecules to examine for the presence of FC recombinant restriction digest products (13.7 and 11.8 kb) intermediate to the two parental Δ-mtDNA restriction products (FLP Δ-mtDNA, 14.7 kb; CW Δ-mtDNA, 10.8 kb).
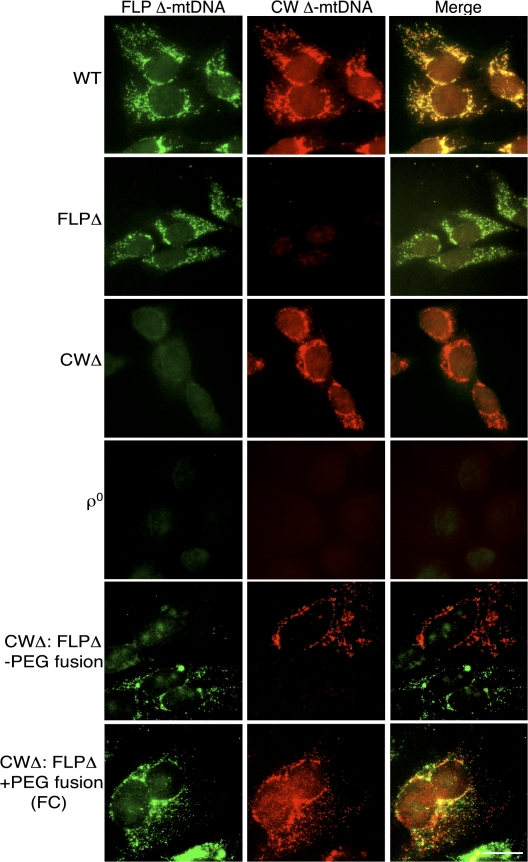
We examined the mtDNA species present in these cell lines using FISH, as described previously (Santra et al., 2004). Because the deletions in the FLP and CW Δ-mtDNAs are nonoverlapping (Fig. 1 A), we were able to design in situ probes unique to each deletion, allowing for sequence-specific visualization of each species of Δ-mtDNA. The 1,244-bp FLP Δ-mtDNA probe (Fig. 1 A, green) corresponded to mtDNA from nt 10849-12093, per the ND4 PCR in Fig. 2 B, and was predicted to hybridize to FLP Δ-mtDNA but not to CW Δ-mtDNA, whereas the 1,508-bp CW Δ-mtDNA probe (Fig. 1 A, red) corresponded to mtDNA from nt 7909–9417, per the CO2/3 PCR in Fig. 2 B, and was predicted to hybridize to CW Δ-mtDNA but not to FLP Δ-mtDNA (Fig. 1 A). FISH probes localized to the mitochondria (Fig. S1 a, available at http://www.jcb.org/cgi/content/full/jcb.200712101/DC1) and showed the punctate distribution typical of mtDNA in fluorescence microscopy (Garrido et al., 2003; Legros et al., 2004). When these probes were mixed to examine cells carrying solely WT mtDNA, cells hybridized with both red and green probes, as WT mtDNA carries both probe target sequences (colocalizing as yellow; Fig. 3, merge). In contrast, FLPΔ cells displayed strong green signal but no significant red signal, whereas CWΔ cells displayed strong red signal but no appreciable green signal. 143B206 ρ0 cells displayed no significant signal for either probe, as expected (Fig. 3). These results confirmed that the cells used had the expected mtDNA genotypes and validated our use of FISH in this system.
Figure 3.
Two-color FISH of cultured cells. Green (FLP Δ-mtDNA) and red (CW Δ-mtDNA) images were captured sequentially. Bar, 20 μm.
FC fusion cells carry both CW and FLP Δ-mtDNAs
To examine the segregation of the FLP and CW Δ-mtDNAs, we fused the two homoplasmic cell lines FLPΔ and CWΔ via polyethylene glycol (PEG)–mediated fusion, as previously described (Tang et al., 2000). Upon fusion, cells were plated in nonselective high-glucose medium for 48 h to recover, after which they were given medium lacking uridine to select for mitochondrial function (King and Attardi, 1989; Fig. 1 B). Long-distance PCR of FLPΔ/CWΔ-fused cells (hereafter referred to as FC cells) revealed two mtDNA species of the same sizes as the respective parental FLP and CW Δ-mtDNA amplification products (Fig. 2 A). Conventional PCR amplification of ND4 and CO2/3 from total DNA of FC cells revealed amplification products of the predicted sizes for both ND4 and CO2/3 (Fig. 2 B). Conventional PCR to detect WT-mtDNA molecules demonstrated that no WT mtDNAs were present in FC cells (unpublished data). Southern blot analysis of FC cells revealed the presence of both CW and FLP Δ-mtDNAs (Fig. 2 C). Although both mtDNAs possess a single PvuII restriction site, the FLP Δ-mtDNA harbors a SnaBI restriction site at position 10737 but lacks a Tth111I site at position 7851, whereas the CW Δ-mtDNA harbors the Tth111I site but not the SnaBI site. Single cuts with each restriction enzyme revealed the appropriate linearized Δ-mtDNA, whereas the SnaBI–Tth111I double digest revealed the presence of both linearized Δ-mtDNAs. The single-lane panel shows a SnaBI–Tth111I lane from a Southern blot of FC cells grown for 14 d under strong ketogenic selection for mitochondrial function (Santra et al., 2004), in which the gel was run for several days to better resolve the linearized FLP and CW Δ-mtDNAs. Importantly, no bands consistent with the expected FC recombinant restriction digest products (13.7 and 11.8 kb) were detected migrating on the gel between the FLP- (14.7 kb) and CW (10.8 kb)-linearized Δ-mtDNAs, even at long-term exposure (Fig. 2 C).
When observed by FISH, FC cells displayed both FLP Δ-mtDNA (Fig. 3, green) and CW Δ-mtDNA (Fig. 3, red) signal, indicating that these cells carried both parental Δ-mtDNAs (Fig. 3), which is consistent with the results observed using long-distance (Fig. 2 A) and conventional (Fig. 2 B) PCR, as well as Southern blotting (Fig. 2 C). In contrast, in a control experiment in which FLPΔ and CWΔ cells were mixed together but not fused with PEG, the cells showed either only red or only green signal, indicating that these unfused control cells remained homoplasmic for their respective Δ-mtDNA (Fig. 3).
FC cells recover mitochondrial protein synthesis because of complementation of CW and FLP Δ-mtDNAs
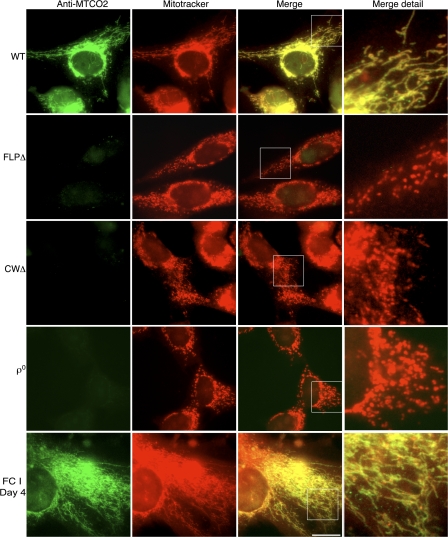
Homoplasmic cell lines carrying large-scale Δ-mtDNAs are incapable of synthesizing mtDNA-encoded proteins, including those protein-coding genes that are still intact (as is the case with CWΔ, in which MTCO2 is not deleted; Nakase et al., 1990; Tang et al., 2000; Santra et al., 2004) because of the loss of mtDNA-encoded tRNAs. Both FLPΔ and CWΔ parental cell lines are deficient in mitochondrial protein synthesis and respiratory function (Pallotti et al., 2004). We examined mitochondrial protein synthesis of all cell lines using immunofluorescence microscopy (Fig. 4). Cells containing 100% WT mtDNA expressed high levels of MTCO2 protein (mtDNA-encoded cytochrome c oxidase II; green), which colocalized clearly with MitoTracker (Fig. 4, red). In contrast, FLPΔ, CWΔ, and 143B206 ρ0 cell lines displayed no significant signal for MTCO2 in green (Fig. 4). When FC cells were examined by anti-MTCO2 immunolabeling, we observed a strong MTCO2 signal (Fig. 4, green) that colocalized with MitoTracker (Fig. 4, red), indicating a restoration of mitochondrial protein synthesis. Moreover, when viewed in detail, the respiratory-deficient FLPΔ, CWΔ, and 143B206 ρ0 cell lines had a fragmented mitochondrial morphology, whereas the complementing FC cells showed a restoration of reticular mitochondrial morphology similar to that in WT cells, concomitant with restoration of the MTCO2 signal (Fig. 4; Santra et al., 2004).
Figure 4.
Immunofluorescence microscopy of cybrid cell lines. Cells were immunolabeled for fluorescence microscopy with an antibody against mitochondrially encoded MTCO2 (green). Mitochondria were visualized with MitoTracker (red). Bar, 20 μm. Outlined boxes in merge are enlarged in merge detail.
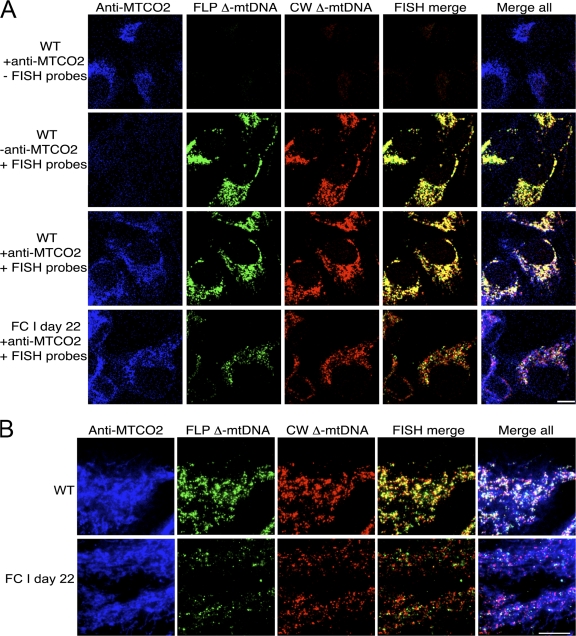
To determine whether the recovery of MTCO2 protein in FC cells was caused by complementation of the CW and FLP Δ-mtDNAs, we developed a method to simultaneously examine MTCO2 protein and the mtDNA species present using three different fluorescent probes. By first immunolabeling for MTCO2 using a blue Alexa Fluor 350–conjugated secondary antibody and then performing red/green FISH as before, we were able to localize MTCO2 protein and the two Δ-mtDNAs within individual cells. Control experiments (Fig. 5 A) demonstrated the specificity of the technique. When WT cells were processed for immunofluorescence/FISH labeling and the FISH probes were omitted during hybridization, MTCO2 immunolabeling (Fig. 5 A, blue) was the only signal observed. When WT cells were processed omitting the anti-MTCO2 primary antibody, only a yellow signal was observed, indicating colocalization of the red/green in situ signals. When both the anti-MTCO2 antibody and the FISH probes were included, we observed colocalization of all three signals (Fig. 5 A). Conversely, no signals were observed in ρ0 cells (unpublished data).
Figure 5.
A novel combined immunofluorescence/FISH method. (A) Cells were first immunolabeled for MTCO2 protein (blue) and subsequently labeled for mtDNA via FISH with probes against FLP Δ-mtDNA (green) and CW Δ-mtDNA (red). Indicated controls lacking primary antibody and/or FISH probes demonstrate signal specificity. Cells were visualized by confocal microscopy. Bar, 10 μm. (B) High-resolution immunofluorescence/FISH images of WT and FC cells. Cells were visualized by conventional microscopy. Bar, 10 μm.
When viewed at higher resolution, immunofluorescence/FISH of FC cells showed a strong signal for MTCO2 coinciding with the FLP Δ-mtDNA and CW Δ-mtDNA in situ signals within the same cells. The red and green FISH signals showed a heterogeneous distribution, unlike the near-complete overlay of these probes in WT cells. MTCO2 signal delineates the mitochondria, with red and green signal apparent as nucleoids distributed throughout (Fig. 5 B). These results demonstrate that FC cells that recovered mitochondrial protein synthesis, as assayed by MTCO2 immunolabeling, presumably did so via complementation of polypeptides encoded by the FLP and CW Δ-mtDNAs within the cell.
FC cells can rapidly sort FLP and CW Δ-mtDNAs to near homoplasmy
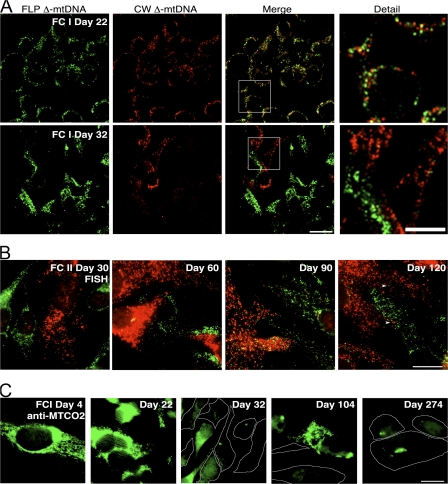
Two independent fusion experiments were performed, yielding essentially the same results. Fig. 6 A shows confocal FISH micrographs of FC cells from the first fusion experiment (FC I) after 22 and 32 d of selection in medium lacking uridine. At day 22, all cells observed (100%) were found to have signal for both FLP and CW Δ-mtDNAs (Fig. 6 A and Table I), indicating that all the homoplasmic parental cells had died, which is consistent with the parental cells' inability to grow in uridine-minus selection for mitochondrial function. Each surviving cell displayed signal for both probes and appeared to contain high levels of both red and green nucleoids (Fig. 6 A, detail). At day 32, however, we observed that the distribution of the two Δ-mtDNAs was dramatically skewed: most cells observed were essentially homoplasmic for either red (55%) or green (42%) nucleoids, having segregated back to near homoplasmy for one genotype or the other (Fig. 6 A and Table I). Only 3% of cells were still obviously heteroplasmic. A similar shift occurred in the second fusion experiment (FC II), in which the percentage of heteroplasmic cells dropped from 78% at day 22 to 1% at day 32. In addition, FISH analysis performed at intermediate time points (days 25, 28, and 30) revealed a steady decrease in heteroplasmic cells over this period of time (Table I).
Figure 6.
Time course of FC cell fusion. (A) Confocal microscopy images of FC I cells viewed by green/red in situ hybridization at days 22 and 32 of selection in medium lacking uridine. Bar, 20 μm. Outlined boxes in merge are enlarged in detail. Bar, 10 μm. (B) Long-term culture of FC II cells visualized by FISH using conventional microscopy. Arrowheads (day 120) denote minority genotype nucleoids in essentially homoplasmic cells (Table I). Bar, 20 μm. (C) Anti-MTCO2 immunofluorescence microscopy of FC I cells at days 4, 22, 32, 104, and 274 of selection. The boundaries of cells with little or no MTCO2 signal were added freehand in Photoshop. Days 22 and 32 were detected with an Alexa 350 (blue) goat anti–rabbit secondary as in Fig. 5, and images were pseudocolored green via ImageJ. Bar, 20 μm.
Table I. FC cell heteroplasmy as scored by FISH.
| Homoplasmic CWΔ | Heteroplasmic CWΔ:FLPΔ | Homoplasmic FLPΔ | Number of cells examined | |
|---|---|---|---|---|
| % | % | % | ||
| FC I | ||||
| Day 22 | 0 | 100 | 0 | 93 |
| Day 32 | 55 | 3 | 42 | 109 |
| Day 51 | 46 | 4 | 50 | 172 |
| Day 59 | 12 | 41 | 47 | 116 |
| Day 104 | 42 | 55 | 3 | 121 |
| FC II | ||||
| Day 22 | 3 | 78 | 18 | 132 |
| Day 25 | 19 | 50 | 31 | 74 |
| Day 28 | 42 | 16 | 41 | 93 |
| Day 30 | 54 | 6 | 40 | 110 |
| Day 32 | 49 | 1 | 50 | 156 |
| Day 60 | 47 | 9 | 44 | 115 |
| Day 90 | 22 | 46 | 32 | 122 |
| Day 120 | 40 | 12 | 48 | 130 |
Long-term culture reveals cycling of complementation and Δ-mtDNA heteroplasmy
When examined by FISH, long-term culture revealed fluctuating heteroplasmy of the two mtDNAs. Fig. 6 B shows FISH images from the second fusion experiment (FC II) at days 30, 60, 90, and 120. At days 30 and 60, a relatively high proportion of cells were essentially homoplasmic for one mtDNA or the other, whereas at day 90, a higher proportion of cells carried both mtDNAs, as demonstrated in the day-90 image. At day 120, there was again a net segregation toward homoplasmy (Fig. 6 B). This periodic variation in heteroplasmy was borne out in the FISH cell scoring of heteroplasmy in both fusion experiments (Table I). Very long-term culture revealed a loss of most CW Δ-mtDNA, as FC I day-274 cells had high levels of FLP Δ-mtDNA but very little CW Δ-mtDNA as demonstrated by FISH (Fig. S2 b, available at http://www.jcb.org/cgi/content/full/jcb.200712101/DC1) and PCR (not depicted).
When examined by MTCO2 immunolabeling, mitochondrial protein synthesis was restored in a subset of FC I cells as early as day 4 of selection (Fig. 6 C). At day 22, all surviving FC I cells carried MTCO2 signal, which is consistent with widespread complementation of the two mtDNAs after intercellular selection. However, at day 32, most cells had either lost MTCO2 labeling or displayed only small patches of signal, indicating a loss of complementation (cells with little or no MTCO2 signal have been outlined to demonstrate the loss of MTCO2 signal). This gain-and-loss of MTCO2 signal was also evident at later time points, as a subset of cells at day 104 displayed high levels of MTCO2 signal, whereas cells at day 274 displayed either no signal or small subcellular pockets of MTCO2 signal (Fig. 6 C). In a separate third fusion experiment, FC clones displayed a periodic gain and loss of MTCO2 immunolabeling similar to that shown here (not depicted), mirroring the cyclical segregation of FLP and CW Δ-mtDNAs observed (Fig. 6 B and Table I). Further, long-term culture of these clones resulted in loss of most copies of the CW Δ-mtDNA, whereas the FLP Δ-mtDNA was maintained stably, as assayed by Southern blot (unpublished data).
CW and FLP Δ-mtDNAs do not show frequent stable colocalization within nucleoids
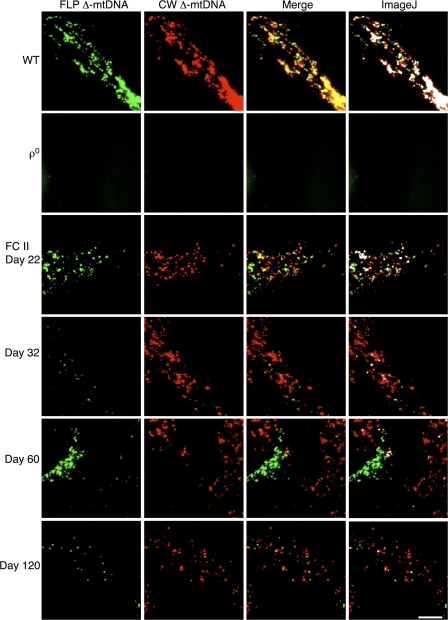
To examine the colocalization of the FLP and CW Δ-mtDNA nucleoids, we conducted image analysis of FC cells in both fusion experiments at several time points. Image analysis using ImageJ allowed us to set a threshold for significant FISH signal. 143B206 ρ0 cells lacking mtDNA were processed for FISH with the same probes used for the other samples. When the threshold was set to eliminate nonspecific background, we observed no meaningful signal, as expected for cells carrying no mtDNA (Fig. 7, ρ0). This technique was applied to numerous images of FC cells at each time point visualized by FISH. The percentage of green, red, and white (colocalized) pixels were calculated as the mean of all micrographs for each time point (Table II). WT cells displayed a 58% colocalization of red and green probes (Table II), a value similar to that obtained when these cells were double labeled with both a red and a green CW Δ-mtDNA probe (unpublished data). This seems to indicate that the 58% colocalization observed for the two probes represents the maximal colocalization of FISH probes using this protocol.
Figure 7.
Nucleoid segregation in FC cells. Representative FISH images, visualized with conventional microscopy, used to generate the colocalization data in Table II. FLP Δ-mtDNA is labeled in green, whereas the CW Δ-mtDNA is labeled in red, with colocalization appearing as yellow in the merge. ImageJ colocalization is shown in white in the far right, illustrating the pixels having both significant red and green signal. WT and ρ0 controls are also shown. Bar, 0.5 μm.
Table II. CW Δ-mtDNA/FLP Δ-mtDNA colocalization in FC cells.
| ImageJ colocalization | Normalizedcolocalizationa | |
|---|---|---|
| % ± SD | % | |
| WT | 58 ± 10 (n = 14) | 100 |
| FC I | ||
| Day 22 | 18 ± 7 (n = 10) | 31 |
| Day 32 | 3 ± 3 (n = 10) | 5 |
| Day 51 | 7 ± 9 (n = 10) | 12 |
| Day 59 | 11 ± 7 (n = 9) | 19 |
| Day 104 | 4 ± 3 (n = 10) | 7 |
| Day 274 | 2 ± 2 (n = 11) | 3 |
| FC II | ||
| Day 22 | 16 ± 9 (n = 11) | 28 |
| Day 32 | 2 ± 4 (n = 10) | 4 |
| Day 60 | 2 ± 2 (n = 10) | 4 |
| Day 90 | 7 ± 4 (n = 9) | 12 |
| Day 120 | 7 ± 6 (n = 8) | 12 |
| Day 150 | 1 ± 2 (n = 10) | 2 |
Fig. 7 shows examples of the FISH images used to quantitate colocalization, demonstrating the segregation of the CW and FLP Δ-mtDNAs in FC II cells (second fusion experiment). Colocalization was demonstrated in Fig. 7 (right) using the ImageJ analysis described in the previous paragraph (appears as white). As in FC I, each cell had an intermixed population of red, green, and red/green colocalized (i.e., white) nucleoids at day 22, whereas at day 32, most nucleoids had segregated away from each other, such that only a handful of colocalization sites were found. Colocalization sites indicated on the micrograph frequently appeared as partial overlap between red and green nucleoids (Fig. S2 b, day 22). At day 22, 16% of pixels were determined to have colocalization of red and green signal. At day 32, this value fell to 2%, demonstrating a significant decrease in the incidence of colocalization (Table II). Even cells having roughly equal proportions of the two Δ-mtDNAs had a low incidence of colocalization at day 32 (unpublished data). This quantitation confirmed that colocalization of FLP and CW Δ-mtDNAs underwent a dramatic decrease between days 22 and 32 and implies that the observed colocalization at day 22 was transient in nature, including that observed at subsequent time points. Moreover, although colocalization fluctuates, at no point did we observe a high occurrence of red/green colocalization (Fig. 7). The maximum colocalization observed was only 31% of WT (Table II, FC I, day 22), and most time points were well below even this level.
Discussion
The experiments shown here provide the first suborganellar visualization of the segregation of two heterologous mtDNAs. When cells carrying mtDNA nucleoids from two different genetic backgrounds were fused, the two heterologous nucleoid populations complemented each other functionally to restore mitochondrial protein synthesis. During this process, the two nucleoid populations faithfully maintained their original genetic composition rather than frequently exchanging mtDNAs, which is consistent with the faithful nucleoid model (Jacobs et al., 2000).
Complementary mtDNAs restore mitochondrial protein synthesis
Upon fusion of two cell lines, each homoplasmic for one of two nonoverlapping deletions, the presence of both complementary mtDNAs led to a restoration of mitochondrial protein synthesis in a subset of cells at the first time point examined (day 4 of selection for mitochondrial function). Intercellular selection expanded this to all remaining cells by day 22, as nonfused parental cells died by this time point. Using a novel immunofluorescence/FISH method, both Δ-mtDNAs colocalized with MTCO2 protein within FC cells, demonstrating that the observed restoration of mitochondrial protein synthesis resulted from transcomplementation of the two mtDNAs. FC cells showed restoration of mitochondrial protein synthesis, as assayed by 35S methionine labeling (Fig. S3 a, available at http://www.jcb.org/cgi/content/full/jcb.200712101/DC1), as well as recovery of WT-like reticular mitochondrial morphology (Fig. 4), correlating with WT mitochondrial function (Gilkerson et al., 2000) and cytochrome c oxidase activity (Fig. S3 b), indicating that FC cells recover mitochondrial function because of transcomplementation of the two mtDNAs. The recovery of mitochondrial protein synthesis, cytochrome c oxidase activity, and reticular mitochondrial morphology are in agreement with Ono et al. (2001), who found similar results in a fusion of two cell lines carrying complementary patient-derived point mutations of mtDNA. This suggests that heterologous mtDNAs need not be in the same nucleoid to complement each other. It is likely that the complementing mRNAs and/or proteins need only be within diffusible distance of each other within the mitochondrial matrix to synthesize mtDNA-encoded proteins and restore normal mitochondrial function. This result further implies that the mitochondrial compartment maintains the ability to fuse and exchange heterologous mitochondrial components, even in respiratory-deficient backgrounds. mtDNA transcomplementation has been demonstrated previously, both between two complementing point mutations (Ono et al., 2001) and between a point mutation and a large-scale deletion (Takai et al., 1999), but has never been shown at nucleoid resolution.
mtDNA nucleoids distribute within cells but remain genetically independent
Strikingly, at no time point in either fusion experiment did we observe frequent colocalization of the two mtDNAs, as visualized by deletion-specific FISH probes. Even in cells carrying both mtDNAs at roughly equal levels, the maximal colocalization observed was only 31% of WT colocalization (FC I, day 22). Most other time points were well below this value (2–19%; Table II). Further, the majority of colocalization we observed typically involved a partial overlap of adjacent red and green nucleoids (Fig. S2 b, day 22, arrowheads). These findings suggest that the majority of colocalization is not equivalent to a bona fide stable fusion of two nucleoids but rather is indicative of transient proximity of two heterologous nucleoids. Our results demonstrate that although nucleoids migrate relatively freely within the cell, which is in agreement with previous studies (Okamoto et al., 1998; Iborra et al., 2004; Legros et al., 2004), heterologous nucleoids do not exchange genomes with each other frequently, which is consistent with the faithful nucleoid model (Jacobs et al., 2000).
The faithful nucleoid model provides a mechanism to explain both stable persistent heteroplasmy and rapid changes in heteroplasmy. In both cases, the mtDNA content of the individual nucleoids is the key determinant of cellular heteroplasmy as a whole. In a cell fusion with two heterologous nucleoid populations, such as ours, the faithful nucleoid model predicts that the two nucleoid populations will swiftly undergo mitotic segregation toward homoplasmy (Jacobs et al., 2000). We observed such segregation experimentally in both fusions (for example, Fig. 6 A and Table I, days 22 and 32). Our results mirror examples of rapid mitotic segregation caused by random genetic drift, particularly as has been reported to occur in mammalian oocytes (Jenuth et al., 1996; Brown et al., 2001). This mitotic segregation allows the cell to undergo a purification of deleterious mtDNA mutations. Conversely, this stochastic segregation can also allow mutations to reach pathogenically high levels in some oocytes before mtDNA amplification during development (Cree et al., 2008). Although the faithful nucleoid model predicts that mitotic segregation occurs in the absence of selection for mitochondrial function, we interpret the restoration of mitochondrial protein synthesis and function in FC cells (as at day 22) as a temporary suspension of selective pressure within the cell, allowing random genetic drift to result in the mitotic segregation we observe. Accordingly, FC cells removed from selection and grown in uridine-plus media displayed similar rapid mitotic segregation (unpublished data). As functional respiratory chain complexes are turned over and are not replaced because of segregation via random genetic drift, we believe that selection is reinforced, resulting in another round of intercellular selection for cells carrying both mtDNAs, which is consistent with the outgrowth of colonies heteroplasmic for the two mtDNAs (Fig. S2 a).
In the absence of selective pressure, as in the oocyte examples described in the previous paragraph, random segregation events can result in shifts toward homoplasmy when the two mtDNAs present occur in heterologous nucleoid populations. In the presence of selective pressure or replicative advantage, faithful nucleoid inheritance provides a mechanism allowing for more directed changes in heteroplasmy, both for (Yoneda et al., 1992; Blok et al., 1997) and against (Fan et al., 2008; Rajasimha et al., 2008) mtDNA mutations. Any mtDNA with a selective or replicative advantage can thus be freely and preferentially propagated. Nuclear genetic control of mtDNA segregation has been observed (Lehtinen et al., 1999; Battersby et al., 2003); however, it remains to be determined how nucleoid organization, selective pressure, and nuclear-encoded factors combine to effect mtDNA segregation. Remodeling of bacterial nucleoids by DNA binding proteins has been shown to result in widespread transcriptional and metabolic changes (Kar et al., 2005). Mitochondrial nucleoid organization may play a central role in regulating mitochondrial metabolism by modulating mtDNA transcription and assembly of respiratory chain complexes.
mtDNA exchange between nucleoids and mtDNA recombination can occur but are low-frequency events
If heterologous nucleoids did undergo frequent exchange of mtDNAs, we would have observed a relatively stable appreciable frequency of red/green colocalization at all time points and a concomitant stable restoration of MTCO2 immunolabeling. Instead, we observed a low incidence of colocalization, which fluctuated greatly as the two genotypes underwent mitotic segregation. The rare stable exchange of mtDNAs between nucleoids is most likely represented by the small number of nucleoids displaying red/green colocalization at day 274. At this time point, very few nucleoids have a red CW signal that neatly overlays with green FLP nucleoids, which is consistent with a truly fused nucleoid carrying both mtDNAs (Fig. S2 b, day 274, arrowhead). Further, frequent exchange of Δ-mtDNAs between heterologous nucleoids would likely have led to some appreciable level of intermolecular recombination, but we observed no evidence of recombination by Southern blotting (Fig. 2 C), by long-distance PCR, or even by Southern blotting of long-distance PCR products (not depicted). No heteroduplexes or reconstituted WT molecules were detected.
Although these experiments were conducted in selective pressure for mitochondrial function, uridine-minus selection in glucose-containing medium is the gentlest such selection available, as cells can still synthesize ATP via glycolysis. We placed FC cells in glucose-minus media, using ketones as a carbon source to force mitochondrial ATP production (Santra et al., 2004), resulting in a much harsher selective environment. FC cells grown in ketogenic media contained no recombinant molecules as assayed by Southern blot (Fig. 2 C, bottom right). Thus, even a harsh selective environment did not induce or select for recombination of the two parental mtDNAs. Although D'Aurelio et al. (2004) did observe intermolecular recombination at low levels in a similar cell fusion, it was necessary to deplete mtDNA with ethidium bromide to enrich for recombinants. Collectively, these results suggest that mtDNA nucleoids typically maintain a consistent genetic composition and are not generally inclined to fuse and exchange mtDNAs.
Collectively, these results suggest that mitochondrial nucleoid organization typically results in very little free mtDNA, instead remaining tightly complexed by mtDNA binding proteins (such as TFAM [Kaufman et al., 2007]) into nucleoids. It is possible that the tightly constrained nature of nucleoid maintenance does not permit a high degree of physical interaction of the two mtDNAs, as is required for intramolecular recombination. Thus, two heterologous mtDNAs may be spatially and temporally adjacent yet still have little opportunity to recombine.
Although the faithful nucleoid model of Jacobs et al. (2000) assumes that an individual nucleoid carries multiple (approximately five) mtDNAs, the number of mtDNAs carried within a nucleoid remains somewhat controversial. Using quantitative PCR (to determine copy number per cell) in conjunction with confocal Z-stack reconstruction (to quantitate nucleoid number per cell), we found that nucleoids in FC cells contain a mean of 5.1 ± 2.4 mtDNAs per nucleoid (Fig. S1, b and c), which is similar to the values determined by others (Iborra et al., 2004; Legros et al., 2004). It remains a formal possibility that nucleoids may carry only one or two mtDNAs in some cell types and contexts, which is in accordance with the original estimates of Satoh and Kuroiwa (1991); however, our results are consistent with the faithful nucleoid model of multigenomic nucleoids that maintain a consistent genetic composition. Additional experiments may reveal differences in nucleoid organization between rapidly dividing and postmitotic cell types.
It remains to be determined whether our findings are a general property of mtDNA propagation. Different cell and tissue types in other conditions may yield different results. For example, it has long been known that mtDNA duplications are particularly abundant in human heart (Fromenty et al., 1997; Kajander et al., 2001) and that mtDNA isolated from human heart appears as multigenomic tangles of monomeric and dimeric mtDNA species (J. Pohjoismaki and H. Jacobs, personal communication), implying that there may be many mtDNAs per nucleoid in this tissue. Our experiments involved the use of a transformed cybrid cell culture system, in which neither Δ-mtDNA is typical of the region commonly deleted in mtDNA (Schon et al., 1989; Mita et al., 1990).
Despite these caveats, the experiments presented here are the first unequivocal subcellular tracking of the segregation of two heterologous mtDNAs, demonstrating that mitochondrial nucleoids are faithfully inherited as independent genetic entities. Moreover, the genetic autonomy of mitochondrial nucleoids provides a mechanism by which to therapeutically eliminate deleterious mtDNA mutations (Santra et al., 2004; Bayona-Bafaluy et al., 2005). A better understanding of the mechanisms of mtDNA propagation and inheritance will allow the development of mechanism-based therapeutic methods to rescue mitochondrial function in patients carrying deleterious mtDNA mutations.
Materials and methods
Cell lines, cell culture, and cell fusion
We studied cybrid cell lines containing mtDNA derived from patients reported previously. The FLP Δ-mtDNA is derived from a heteroplasmic patient with Kearns-Sayre syndrome (Patient 4 [Zeviani et al., 1988] and Patient K11 [Mita et al., 1990]). This deletion removes 1,902 bp from nt 7846 (notation of Anderson et al. [1981]) in the MTCO2 gene to nt 9748 in the MTCO3 gene (Fig. 1 A). The CW Δ-mtDNA is derived from Patient K13 (Mita et al., 1990). This deletion removes 5,790 bp from nt 10155 in MTND3 to nt 15945 in MTTT (Fig. 1 A). Cybrid cell lines FLP6a39.2WT (100% WT), FLP6a39.32Δ (100% Δ), CW420-115Δ (100% Δ), and 143B206 ρ0 (lacking mtDNA) have been previously described (Pallotti et al., 2004; Santra et al., 2004). All cells were grown in high glucose DME with 10% FBS and 50 μg/ml uridine before selection in the same medium lacking uridine after fusion. CW420-115Δ and FLP6a39.32Δ cell lines were fused in the presence of PEG (as per King and Attardi [1989]) and allowed to recover in DME with 10% FBS with uridine. After 48 h of recovery, fusion medium was replaced with DME with 10% FBS minus uridine to select for mitochondrial function. For ketogenic selection, cells were grown in DME lacking glucose and uridine, supplemented with 5 mM dl-β-hydroxybutyrate (Santra et al., 2004). FBS was dialyzed with eight changes of buffer before use in cell culture.
Immunocytochemistry
Cultured cells were seeded to 22 × 22-mm glass coverslips before processing. Coverslips were incubated with MitoTracker Red CMXRos (Invitrogen) before fixation in 4% PFA in PBS for 30 min, permeabilized with 0.1% Triton X-100 in PBS for 10 min, and blocked in 10% normal goat serum for 30 min. Coverslips were incubated with a rabbit polyclonal antibody to MTCO2 (gift from R. Doolittle, University of California, San Diego, La Jolla, CA) at 1:100 dilution in PBS for 1 h, followed by three washes of 5 min each in PBS. Coverslips were then incubated with Alexa 488–conjugated goat anti–rabbit secondary antibody (Invitrogen) at 1:100 dilution in PBS for 1 h, again followed by three washes of 5 min each in PBS. Coverslips were washed with PBS and mounted with 50% glycerol in PBS.
PCR from isolated total cellular DNA
Total cellular DNA was isolated as previously described (Zeviani et al., 1988), using proteinase K digestion followed by organic extraction and isopropanol/ethanol precipitation. Long-distance PCR was conducted using the LA Taq system (Takara Bio Inc.) with a forward primer corresponding to mtDNA nt 3068–3100 and reverse primer corresponding to mtDNA nt 817–782. Reactions were run on a 0.8% agarose gel. Conventional PCR amplifications were as follows: the CO2/3 fragment, corresponding to mtDNA from nt 7909–9417, used forward primer ACGAGTACACCGACTACGGC (nt 7909–7928) and reverse primer GTGGCCTTGGTATGTGCTTT (nt 9397–9417); the ND4 fragment, corresponding to mtDNA from nt 10849–12093, used forward primer CCACCCACAGCCTAATTATTAGC (nt 10849–10865) and reverse primer GAATGGGGGATAGGTGTATGAAC (nt 12093–12071). Reactions were electrophoresed through a 1% agarose gel and photographed using an AlphaImager2200 gel imaging system (Imgen Technologies).
Southern blotting
2.5 μg FC total cellular DNA was digested with PvuII, SnaBI, Tth111I, or both SnaBI and Tth111I, run on a 0.8% agarose gel, and transferred to PVDF. A PCR fragment corresponding to nt 3778–6051 of human mtDNA was radiolabeled with 32P-labeled deoxy-CTP. Membranes were prehybridized in 0.5% sodium dodecyl sulfate in 0.25 M disodium hydrogen phosphate and hybridized with radiolabeled probe DNA overnight at 65°. Membranes were washed, exposed to a screen (Bio-Rad Laboratories), and visualized using a Molecular Imager FX phosphorimager (Bio-Rad Laboratories).
FISH
mtDNA probes were prepared by PCR amplification of the mtDNA CO2/3 and ND4 fragments as in PCR from isolated total cellular DNA. Probe fragments were labeled by nick translation to incorporate an aminoallyl deoxy-UTP, followed by subsequent Alexa Fluor labeling using the ARES DNA labeling kit (Invitrogen). The FLP Δ-mtDNA probe was labeled with Alexa Fluor 488 (green). The CW Δ-mtDNA probe was labeled with Alexa Fluor 594 (red; Fig. 1 A). FISH was conducted essentially as described previously (Margineantu et al., 2002; Santra et al., 2004). In brief, cells were fixed in 4% PFA in PBS for 30 min, followed by serial dehydration/rehydration in graded ethanol solutions. Coverslips were treated with DNase-free RNase at 0.1 mg/ml in PBS for 1 h at 37°C. Coverslips were prehybridized in 2× SSC for 30–40 min at 37°C, dehydrated, air dried, and denatured in 70% formamide in 2× SSC for 5 min at 72°C. FLP Δ-mtDNA (green) and CW Δ-mtDNA (red) probes were combined to a final concentration of 2 ng/μl (each) in hybridization buffer (50% formamide/20% dextran sulfate in 2× SSC) and denatured at 72°C for 5 min. Probes were applied to coverslips and hybridized overnight at 37°C in a humid chamber. Coverslips were then washed in 0.3% Tween in 0.4× SSC for 5 min at 72°C, followed by washing in 0.1% Tween in 2× SSC for 1 min and 2× SSC for 2 min. Coverslips were mounted with 50% glycerol in PBS.
Combined immunofluorescence/FISH labeling
To examine mitochondrial protein synthesis and mtDNA distribution simultaneously, we developed a combined immunofluorescence/FISH method. To maintain adequate hydration, we first immunolabeled cells for MTCO2, followed by the harsher FISH procedure. In brief, cells were fixed and processed for immunocytochemistry as described in Immunocytochemistry. After blocking, cells were incubated with anti-MTCO2 primary as described, followed by incubation with Alexa Fluor 350–conjugated goat anti–rabbit antibody (blue; Invitrogen). After incubation with secondary antibody, cells were fixed in 4% PFA in PBS and processed for FISH exactly as described in the previous section. The immunolabeling did not work if FISH was performed first.
Microscopy
Cells were imaged using an inverted microscope (IX70; Olympus) with a UplanFl 100×/1.3 numerical aperture oil immersion objective (Olympus). Red, green, and blue images were captured sequentially using a SPOT RT digital camera and merged using SPOT RT software (Olympus). Images were processed equally after capture for HSV histogram and contrast adjustment in SPOT RT. Confocal images were obtained using an LSM 510 NLO multiphoton confocal microscope (Carl Zeiss, Inc.) with a Plan Neofluar 100×/1.3 numerical aperture objective (Olympus). Red, green, and blue images were acquired with LSM 510 NLO Standard software (Carl Zeiss, Inc.) sequentially and merged using ImageJ (National Institutes of Health). Confocal images were adjusted for brightness/contrast in ImageJ. Images in each figure were processed equally. Fluorochromes for FISH and immunofluorescence were Alexa 488, Alexa 594, and Alexa 350, or MitoTracker RedCMXRos (Invitrogen). All imaging was performed at room temperature. All samples were mounted in 50% glycerol in PBS.
Image analysis
Quantification of colocalization in FISH images was performed using ImageJ. By converting individual red and green TIFF images to binary look up table images and setting a threshold level that was empirically determined to eliminate background fluorescence (pixel intensity: green,29–255; red, 36–255 [arbitrary units]), we were able to obtain the number of green, red, and colocalized green/red pixels for each micrograph. By applying this method identically to all images, we compiled a comprehensive assessment of the level of green/red FISH colocalization for the cell lines quantitated in Table II. Colocalization mask images in Fig. 7 (ImageJ) were generated using colocalization finder in ImageJ, in which pixels with meaningful colocalization appear in the image as white (Fig. 7). The region of interest was generated by eliminating the 38 lowest pixel intensities in both the red and green axis.
Online supplemental material
Fig. S1 shows mtDNA nucleoid localization and quantification. Fig. S2 shows mitochondrial protein synthesis and function. Fig. S3 shows FC culture growth and nucleoid localization. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200712101/DC1.
Supplementary Material
[Supplemental Material Index]
Acknowledgments
We thank Adrianus DeGroof, Edina Torgyekes, Jorida Coku, Ali Naini, Eduardo Bonilla, Winsome Walker, Liza Pon, Theresa Swayne, and Sudhi Swamy for their assistance and suggestions. We also thank the Eric Schon, Michio Hirano, and Salvatore DiMauro laboratories for helpful discussions.
This research was supported by grants from the National Institutes of Health (HD83062, NS11766, and AG08702 to E.A. Schon), the Marriott Foundation, and a Muscular Dystrophy Association Development grant (MDA3869 to R.W. Gilkerson). We thank the Confocal and Specialized Microscopy Shared Resource of the Herbert Irving Comprehensive Cancer Center (supported by National Institutes of Health grant P30 CA13696 and National Institutes of Health Shared Instrumentation Grants S10 RR10506, S10 RR14701, and S10 RR017885).
The authors declare that they have no competing financial interests.
Abbreviations used in this paper: mtDNA, mitochondrial DNA; PEG, polyethylene glycol; WT, wild type.
References
- Anderson, S., A.T. Bankier, B.G. Barrell, M.H.L. de Bruijn, A.R. Coulson, J. Drouin, I.C. Eperon, D.P. Nierlich, B.A. Roe, F. Sanger, et al. 1981. Sequence and organization of the human mitochondrial genome. Nature. 290:457–465. [DOI] [PubMed] [Google Scholar]
- Battersby, B.J., J.C. Loredo-Osti, and E.A. Shoubridge. 2003. Nuclear genetic control of mitochondrial DNA segregation. Nat. Genet. 33:183–186. [DOI] [PubMed] [Google Scholar]
- Bayona-Bafaluy, M.P., B. Blits, B.J. Battersby, E.A. Shoubridge, and C.T. Moraes. 2005. Rapid directional shift of mitochondrial DNA heteroplasmy in animal tissues by a mitochondrially targeted restriction endonuclease. Proc. Natl. Acad. Sci. USA. 102:14392–14397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blok, R.B., D.A. Gook, D.R. Thorburn, and H.H. Dahl. 1997. Skewed segregation of the mtDNA nt 8993 (T→G) mutation in human oocytes. Am. J. Hum. Genet. 60:1495–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, D.T., D.C. Samuels, E.M. Michael, D.M. Turnbull, and P.F. Chinnery. 2001. Random genetic drift determines the level of mutant mtDNA in human primary oocytes. Am. J. Hum. Genet. 68:533–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capaldi, R.A., R. Aggeler, R. Gilkerson, G. Hanson, M. Knowles, A. Marcus, D. Margineantu, M. Marusich, J. Murray, D. Oglesbee, et al. 2002. A replicating module as the unit of mitochondrial structure and functioning. Biochim. Biophys. Acta. 1555:192–195. [DOI] [PubMed] [Google Scholar]
- Chen, X.J., and R.A. Butow. 2005. The organization and inheritance of the mitochondrial genome. Nat. Rev. Genet. 6:815–825. [DOI] [PubMed] [Google Scholar]
- Chen, X.J., X. Wang, B.A. Kaufman, and R.A. Butow. 2005. Aconitase couples metabolic regulation to mitochondrial DNA maintenance. Science. 307:714–717. [DOI] [PubMed] [Google Scholar]
- Cree, L.M., D.C. Samuels, S.C. de Sousa Lopes, H.K. Rajasimha, P. Wonnapinij, J.R. Mann, H.H. Dahl, and P.F. Chinnery. 2008. A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes. Nat. Genet. 40:249–254. [DOI] [PubMed] [Google Scholar]
- D'Aurelio, M., C.D. Gajewski, M.T. Lin, W.M. Mauck, L.Z. Shao, G. Lenaz, C.T. Moraes, and G. Manfredi. 2004. Heterologous mitochondrial DNA recombination in human cells. Hum. Mol. Genet. 13:3171–3179. [DOI] [PubMed] [Google Scholar]
- DiMauro, S., and E.A. Schon. 2003. Mitochondrial respiratory-chain diseases. N. Engl. J. Med. 348:2656–2668. [DOI] [PubMed] [Google Scholar]
- Fan, W., K.G. Waymire, N. Narula, P. Li, C. Rocher, P.E. Coskun, M.A. Vannan, J. Narula, G.R. Macgregor, and D.C. Wallace. 2008. A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science. 319:958–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromenty, B., R. Carrozzo, S. Shanske, and E.A. Schon. 1997. High proportions of mtDNA duplications in patients with Kearns-Sayre syndrome occur in the heart. Am. J. Med. Genet. 71:443–452. [DOI] [PubMed] [Google Scholar]
- Garrido, N., L. Griparic, E. Jokitalo, J. Wartiovaara, A.M. van der Bliek, and J.N. Spelbrink. 2003. Composition and dynamics of human mitochondrial nucleoids. Mol. Biol. Cell. 14:1583–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkerson, R.W., D.H. Margineantu, R.A. Capaldi, and J.M. Selker. 2000. Mitochondrial DNA depletion causes morphological changes in the mitochondrial reticulum of cultured human cells. FEBS Lett. 474:1–4. [DOI] [PubMed] [Google Scholar]
- Iborra, F.J., H. Kimura, and P.R. Cook. 2004. The functional organization of mitochondrial genomes in human cells. BMC Biol. 2:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, H.T., S.K. Lehtinen, and J.N. Spelbrink. 2000. No sex please, we're mitochondria: a hypothesis on the somatic unit of inheritance of mammalian mtDNA. Bioessays. 22:564–572. [DOI] [PubMed] [Google Scholar]
- Jenuth, J.P., A.C. Peterson, K. Fu, and E.A. Shoubridge. 1996. Random genetic drift in the female germline explains the rapid segregation of mammalian mitochondrial DNA. Nat. Genet. 14:146–151. [DOI] [PubMed] [Google Scholar]
- Kajander, O.A., P.J. Karhunen, I.J. Holt, and H.T. Jacobs. 2001. Prominent mitochondrial DNA recombination intermediates in human heart muscle. EMBO Rep. 2:1007–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar, S., R. Edgar, and S. Adhya. 2005. Nucleoid remodeling by an altered HU protein: reorganization of the transcription program. Proc. Natl. Acad. Sci. USA. 102:16397–16402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman, B.A., N. Durisic, J.M. Mativetsky, S. Costantino, M.A. Hancock, P. Grutter, and E.A. Shoubridge. 2007. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol. Biol. Cell. 18:3225–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, M.P., and G. Attardi. 1989. Human cells lacking mtDNA: Repopulation with exogenous mitochondria by complementation. Science. 246:500–503. [DOI] [PubMed] [Google Scholar]
- Kraytsberg, Y., M. Schwartz, T.A. Brown, K. Ebralidse, W.S. Kunz, D.A. Clayton, J. Vissing, and K. Khrapko. 2004. Recombination of human mitochondrial DNA. Science. 304:981. [DOI] [PubMed] [Google Scholar]
- Legros, F., F. Malka, P. Frachon, A. Lombes, and M. Rojo. 2004. Organization and dynamics of human mitochondrial DNA. J. Cell Sci. 117:2653–2662. [DOI] [PubMed] [Google Scholar]
- Lehtinen, S.K., J.N. Spelbrink, and H.T. Jacobs. 1999. Heteroplasmic segregation associated with trisomy-9 in cultured human cells. Somat. Cell Mol. Genet. 25:263–274. [DOI] [PubMed] [Google Scholar]
- Margineantu, D.H., W.G. Cox, L. Sundell, S.W. Sherwood, J.M. Beechem, and R.A. Capaldi. 2002. Cell cycle dependent morphology changes and associated mitochondrial DNA. Mitochondrion. 1:425–435. [DOI] [PubMed] [Google Scholar]
- Mita, S., R. Rizzuto, C.T. Moraes, S. Shanske, E. Arnaudo, G.M. Fabrizi, Y. Koga, S. DiMauro, and E.A. Schon. 1990. Recombination via flanking direct repeats is a major cause of large-scale deletions of human mitochondrial DNA. Nucleic Acids Res. 18:561–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakase, H., C.T. Moraes, R. Rizzuto, A. Lombes, S. DiMauro, and E.A. Schon. 1990. Transcription and translation of deleted mitochondrial genomes in Kearns-Sayre syndrome: implications for pathogenesis. Am. J. Hum. Genet. 46:418–427. [PMC free article] [PubMed] [Google Scholar]
- Okamoto, K., P.S. Perlman, and R.A. Butow. 1998. The sorting of mitochondrial DNA and mitochondrial proteins in zygotes: preferential transmission of mitochondrial DNA to the medial bud. J. Cell Biol. 142:613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono, T., K. Isobe, K. Nakada, and J.I. Hayashi. 2001. Human cells are protected from mitochondrial dysfunction by complementation of DNA products in fused mitochondria. Nat. Genet. 28:272–275. [DOI] [PubMed] [Google Scholar]
- Pallotti, F., A. Baracca, E. Hernandez-Rosa, W.F. Walker, G. Solaini, G. Lenaz, G.V. Melzi D'Eril, S. Dimauro, E.A. Schon, and M.M. Davidson. 2004. Biochemical analysis of respiratory function in cybrid cell lines harbouring mitochondrial DNA mutations. Biochem. J. 384:287–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasimha, H.K., P.F. Chinnery, and D.C. Samuels. 2008. Selection against pathogenic mtDNA mutations in a stem cell population leads to the loss of the 3243A→G mutation in blood. Am. J. Hum. Genet. 82:333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothfield, L., A. Taghbalout, and Y.L. Shih. 2005. Spatial control of bacterial division-site placement. Nat. Rev. Microbiol. 3:959–968. [DOI] [PubMed] [Google Scholar]
- Santra, S., R. Gilkerson, M. Davidson, and E.A. Schon. 2004. Ketogenic treatment reduces the proportion of mutated mitochondrial DNAs in cells harboring mtDNA deletions. Ann. Neurol. 56:662–669. [DOI] [PubMed] [Google Scholar]
- Satoh, M., and T. Kuroiwa. 1991. Organization of multiple nucleoids and DNA molecules in mitochondria of a human cell. Exp. Cell Res. 196:137–140. [DOI] [PubMed] [Google Scholar]
- Schon, E.A., R. Rizzuto, C.T. Moraes, H. Nakase, M. Zeviani, and S. DiMauro. 1989. A direct repeat is a hotspot for large-scale deletions of human mitochondrial DNA. Science. 244:346–349. [DOI] [PubMed] [Google Scholar]
- Takai, D., K. Isobe, and J. Hayashi. 1999. Transcomplementation between different types of respiration-deficient mitochondria with different pathogenic mutant mitochondrial DNAs. J. Biol. Chem. 274:11199–11202. [DOI] [PubMed] [Google Scholar]
- Tang, Y., E.A. Schon, E. Wilichowski, M.E. Vazquez-Memije, E. Davidson, and M.P. King. 2000. Rearrangements of human mitochondrial DNA (mtDNA): new insights into the regulation of mtDNA copy number and gene expression. Mol. Biol. Cell. 11:1471–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., and D.F. Bogenhagen. 2006. Human mitochondrial DNA nucleoids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane. J. Biol. Chem. 281:25791–25802. [DOI] [PubMed] [Google Scholar]
- Yoneda, M., A. Chomyn, A. Martinuzzi, O. Hurko, and G. Attardi. 1992. Marked replicative advantage of human mtDNA carrying a point mutation that causes the MELAS encephalomyopathy. Proc. Natl. Acad. Sci. USA. 89:11164–11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeviani, M., C.T. Moraes, S. DiMauro, H. Nakase, E. Bonilla, E.A. Schon, and L.P. Rowland. 1988. Deletions of mitochondrial DNA in Kearns-Sayre syndrome. Neurology. 38:1339–1346. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Material Index]