Characterization and Biological Function of the ISOCHORISMATE SYNTHASE2 Gene of Arabidopsis (original) (raw)
Abstract
Salicylic acid (SA) is an important mediator of plant defense response. In Arabidopsis (Arabidopsis thaliana), this compound was proposed to derive mainly from isochorismate, itself produced from chorismate through the activity of ISOCHORISMATE SYNTHASE1 (ICS1). Null ics1 mutants still accumulate some SA, suggesting the existence of an enzymatic activity redundant with ICS1 or of an alternative ICS-independent SA biosynthetic route. Here, we studied the role of ICS2, a second ICS gene of the Arabidopsis genome, in the production of SA. We have shown that ICS2 encodes a functional ICS enzyme and that, similar to ICS1, ICS2 is targeted to the plastids. Comparison of SA accumulation in the ics1, ics2, and ics1 ics2 mutants indicates that ICS2 participates in the synthesis of SA, but in limited amounts that become clearly detectable only when ICS1 is lacking. This unequal redundancy relationship was also observed for phylloquinone, another isochorismate-derived end product. Furthermore, detection of SA in the double ics1 ics2 double mutant that is completely devoid of phylloquinone provides genetic evidence of the existence of an ICS-independent SA biosynthetic pathway in Arabidopsis.
Salicylic acid (SA) has been linked in various species with diverse physiological aspects, like thermogenesis, stomatal closure, senescence, leaf abscission, or resistance to abiotic stresses (Raskin, 1992; Morris et al., 2000; Martinez et al., 2004). SA is also a well-established regulatory component of the induced defense response in many plant species (Sticher et al., 1997). An increase in endogenous concentration of SA after an infection has been observed in many plants and correlated to the activation of defense mechanisms. The importance of the involvement of SA in the induction of resistance to oomycetes, bacterial or viral pathogens was demonstrated with mutants and transgenic plants that exhibit altered levels of SA (Sticher et al., 1997; Métraux and Durner, 2004; Garcion and Métraux, 2006). The pathway for SA biosynthesis and its regulation during infection has therefore become a central question in the understanding of induced plant resistance mechanisms. The biosynthesis of SA was first proposed to proceed through the benzoate pathway, as shown by studies based on radiolabeled compounds (Garcion and Métraux, 2006). In Arabidopsis (Arabidopsis thaliana), a second pathway was proposed that is based on isochorismate, similar to the pathway described in some Pseudomonas species (Wildermuth et al., 2001). In this pathway, chorismate is converted into isochorismate through the action of an isochorismate synthase (ICS), and SA is generated from isochorismate by an isochorismate pyruvate lyase. This scheme gained strong support from studies with ics1 mutants that accumulate only low levels of SA, although the conversion from isochorismate to SA has not yet been demonstrated in Arabidopsis (Wildermuth et al., 2001). The ICS gene product was confirmed to possess ICS activity and to be targeted to the plastidic compartment (Strawn et al., 2007). Synthesis of SA following exposure to ozone in Arabidopsis was also suggested to proceed through the activity of ICS enzymes (Ogawa et al., 2005). The ICS pathway was recently shown to be active in tomato (Solanum lycopersicum; Uppalapati et al., 2007) and tobacco (Nicotiana benthamiana; Catinot et al., 2008). Furthermore, the isochorismate generated by ICS is a precursor of phylloquinone, known as vitamin K1, which is a component of PSI (Gross et al., 2006). The involvement of isochorismate in the synthesis of this compound was further confirmed in transgenic tobacco plants overexpressing an ICS of Catharanthus roseus and accumulating higher amounts of phylloquinone (Verberne et al., 2007). C. roseus or members of the Rubiaceae family also use isochorismate as a precursor for compounds such as anthraquinone or dihydroxybenzoates (Muljono et al., 2002; Mustafa and Verpoorte, 2005). However, these molecules are taxon specific and so far have not been investigated in Arabidopsis.
The Arabidopsis genome contains a second ICS gene, named ICS2 (Wildermuth et al., 2001), but its biochemical activity relative to isochorismate production and its contribution to SA synthesis has not yet been clarified. In this article, we have addressed this question by isolating a full-length clone of ICS2 and testing the activity and localization of its product relative to that of ICS1. We have also determined the amount of remaining SA in the ics1, ics2, and ics1 ics2 mutants.
RESULTS
Sequence Analysis of ICS1 and ICS2
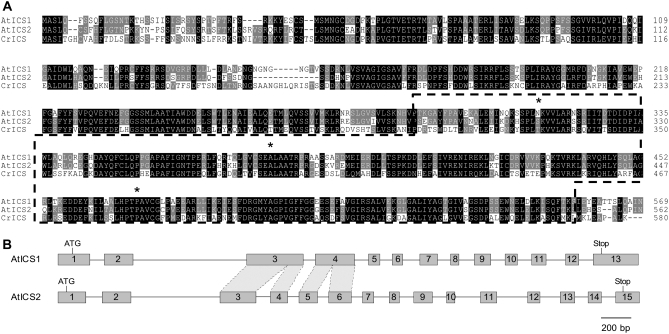
Two isochorismate synthase genes, ICS1 (At1g74710) and ICS2 (At1g18870), are present in the genome of Arabidopsis. These two genes belong to two blocks of approximately 3.5 Mb containing ordered fragments of similar sequence and therefore presumably originate from an ancient genomic duplication event (see http://wolfe.gen.tcd.i.e./athal/dup; Blanc et al., 2003). We first obtained a complete sequence of ICS2 to compare the nucleotide sequences of ICS1 and ICS2. No full-length ICS2 cDNA sequence was available from public databases and the current conceptual translation of the ICS2 coding sequence, relying on ESTs, predicted an ICS2 protein sequence lacking an N-terminal extension compared to ICS1. We have isolated the 5′-end of the ICS2 messenger by using the RACE-PCR technique and then its full-length coding sequence by reverse transcription-PCR (accession no. EU589462). We found that the most 5′-located EST (N96097) matched our ICS2 cDNA sequence but started at position 216 only, and so therefore did not include the first ATG start codon located at position 58 and suggested an erroneous start codon. The translation of the complete ICS2 coding sequence (562 amino acids) includes an N-terminal extension predicted to be a plastid-targeting peptide, unlike the current annotation for ICS2, but similar to ICS1. Overall, ICS1 and ICS2 share 78% identity and 88% similarity at the amino acid level and are close to the characterized C. roseus CrICS (72% similarity with both ICS1 and ICS2; Fig. 1A; van Tegelen et al., 1999). ICS1, ICS2, and CrICS contain the so-called chorismate-binding domain (Pfam accession no. PF00425) and conserved key residues consistent with an ICS catalytic activity (Kolappan et al., 2007).
Figure 1.
A, Sequence alignment of Arabidopsis ICS1 (NP_565090) and ICS2 (ACC60228) and C. roseus ICS (CAA06837). The chorismate-binding domain is located in the dashed area and the position of residues conserved in ICS enzymes (Kolappan et al., 2007) is indicated with a star. B, Exon/intron structure of ICS1 and ICS2. Exons are represented by gray boxes and introns by thin lines. Sequence similarity between exon 3 of AtICS1 and 3 and 4 of ICS2, and 4 of AtICS1 and 5 and 6 of ICS2 is highlighted by dashes.
At the genomic level, the ICS2 gene contains 15 exons, compared to 13 for ICS1, but the overall exon/intron organization has been retained since the event that duplicated the ICS genes (Fig. 1B). Minor exceptions are the splitting of exons 3 and 4 of ICS1 into exons 3 to 6 of ICS2 or, alternatively, the fusion of exons 3 to 6 of ICS2 into exons 3 and 4 of ICS1.
Subcellular Localization of ICS1 and ICS2
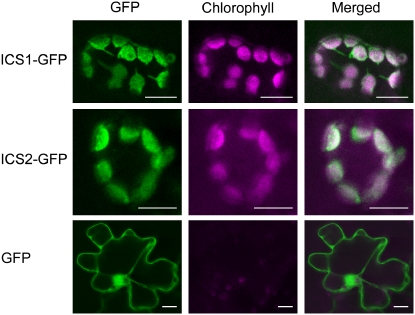
Analysis of the ICS1 and ICS2 sequences with TargetP (Emanuelsson et al., 2000) and Predotar (Small et al., 2004) software suggested that both proteins contained a plastid-targeting signal. The predicted localization in plastids is consistent with the production of the chorismate substrate in these organelles. Plastid localization of ICS1 and ICS2 was tested by fusing their coding sequence to GFP and transiently expressing these constructs in tobacco cells. Observations of the transformed cells with a laser confocal microscope clearly showed that the GFP fluorescence colocalized with chlorophyll fluorescence, indicating that both fusion proteins were located within the chloroplasts (Fig. 2). As a control, we expressed GFP alone following the same protocol and observed a cytosolic signal.
Figure 2.
Subcellular localization of ICS1 and ICS2. GFP fusion constructs were transiently expressed in tobacco cells by agroinfiltration. GFP signal (green) and chlorophyll autofluorescence (magenta) were observed using confocal microscopy. Magenta and green overlay is shown in white. Bars = 10 _μ_m.
Characterization of ICS1 and ICS2 Enzymatic Activity
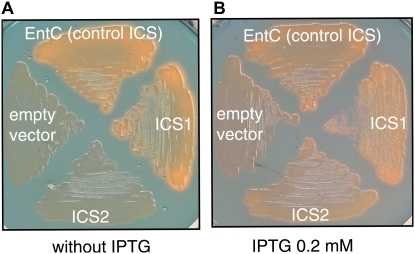
ICS1 was initially proposed to catalyze the conversion of chorismate into isochorismate based on sequence similarity with a characterized ICS from C. roseus (Wildermuth et al., 2001). To determine experimentally whether ICS1, but also ICS2, truly possess such an ICS enzymatic activity, we relied on a functional complementation assay. In Escherichia coli, an endogenous ICS activity is required for production of enterobactin, a high-affinity iron ligand secreted in the medium and internalized following iron chelation (Raymond et al., 2003). Iron scavenging by microorganisms can be monitored easily using the chrome azurol sulfonate (CAS) medium devised by Schwyn and Neilands (1987). This medium contains a dye that forms an intense blue complex with iron and turns orange when iron has been transferred to another iron-binding molecule. For our functional assay, we used PBB8, a mutant of E. coli that lacks endogenous ICS activity and is therefore unable to produce enterobactin (Muller et al., 1996; Fig. 3). As a positive control, we have used the E. coli EntC gene encoding an ICS that restored enterobactin production after introduction into the PBB8 strain, and entailed the formation of an orange halo around positive colonies (Fig. 3). PBB8 cells expressing ICS1 under the control of the strong isopropylthio-_β_-galactoside (IPTG)-inducible Ptac promoter also showed an orange halo even without induction by IPTG, indicating that ICS1 could complement the PBB8 ICS deficiency. No significant coloration was observed for cells expressing ICS2 under the same conditions. However, induction by IPTG at 0.2 mm led to the appearance of the orange signature for siderophore production (Fig. 3), thus demonstrating that ICS2 encodes functional ICS enzymes similarly to ICS1.
Figure 3.
Functional complementation of the _PBB8 ICS_-deficient strain of E. coli by expression of Arabidopsis ICS1 and ICS2. PBB8 cells were transformed with constructs expressing ICS and ICS-GFP fusions and spread onto CAS medium. Orange coloration indicate iron uptake from the medium and therefore restoration of siderophore production by functional ICS activity. Bacteria were plated either on CAS medium without IPTG (A) or CAS medium containing 0.2 mm of IPTG (B).
The apparent difference of response to IPTG between ICS1 and ICS2 in the CAS assay was not due to variations of the expression system because both were cloned in the same vector, under the control of the same promoter and 5′-untranslated region, after removal of the transit peptide-coding sequence. Expression problems due to codon bias were also prevented by using the pRARE plasmid throughout the experiment (Novy et al., 2001). Instead, we observed that the ICS2 protein accumulated at much lower levels than ICS1 in E. coli cells by using tagged versions of ICS1 and ICS2 (data not shown).
Expression of ICS1 and ICS2 in E. coli also allowed us to test whether these two enzymes could generate SA directly from chorismate. No SA accumulated in cell cultures, unlike cells expressing the positive control PchB from Pseudomonas aeruginosa (Gaille et al., 2002), therefore suggesting that ICS1 and ICS2 have no isochorismate pyruvate lyase activity (data not shown).
Phylloquinone Accumulation in ics Mutants
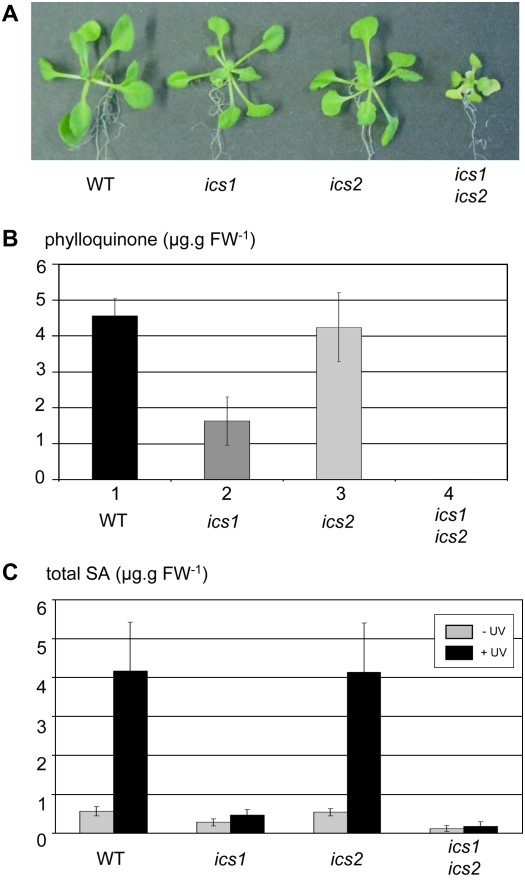
To determine the relative contribution of ICS1 and ICS2 to isochorismate production, we used the following mutants: ics1 (sid2-1 allele [Nawrath and Métraux, 1999]) and ics2 (carrying a T-DNA insertion in the ICS2 gene of Arabidopsis [SALK_084635]). Neither of these single mutants exhibited a striking visual phenotype compared to the wild type (Fig. 4A). We focused our study on phylloquinone and SA, presumably both derived from isochorismate in Arabidopsis. Phylloquinone was measured in the ics1 and ics2 mutant lines by fluorescence HPLC after reduction of phylloquinone to the phyllohydroquinone form according to Lohmann et al. (2006). The pylloquinone content in leaves of ics1 was reduced to approximately 35% of wild type, indicating that the ICS1 gene product provides the predominant amount of isochorismate required for phylloquinone synthesis (Fig. 4B). The amount of phylloquinone of the homozygous ics2 mutant was not different from wild type (Fig. 4B). Although the phylloquinone content in ics1 was strongly reduced, the total chlorophyll content was not affected (1,169 ± 62, 1,182 ± 61, and 1,208 ± 103 _μ_g g−1 fresh weight in wild type, ics1, and ics2, respectively).
Figure 4.
Functional roles of ICS1 and ICS2. A, Phenotype of wild type, ics1, ics2, and ics1 ics2 double mutant when grown in vitro. ics1 ics2 double mutants can be more affected and display a more yellowish pigmentation. B and C, Accumulation of isochorismate-derived compounds in the mutants: phylloquinone (B) and total SA accumulation following UV induction (C).
The functional overlap between ICS1 and ICS2 was investigated in double homozygous ics1 ics2 mutants. F2 plants derived from a cross of ics1 and ics2 were raised on Suc-containing medium and screened for double homozygous lines by phylloquinone measurements and by PCR using oligonucleotides designed for the ics2 locus. Double homozygous ics1 ics2 mutants remained smaller than the wild type and single mutants, and displayed a pale green to yellowish pigmentation (Fig. 4A). Besides this strong reduction in growth, they could only be maintained on Suc medium. The novel growth defects present in the double mutant compared to the single mutants indicated that both genes were dispensable, but not together, and that at least one was required and sufficient for normal growth under these conditions. This defines a symmetrical or equal redundancy between ICS1 and ICS2 with respect to the ability to grow under standard conditions.
Double homozygous mutant lines (ics1 ics2) were totally devoid of phylloquinone (Fig. 4B). Plants homozygous for the ics1 mutation, but heterozygous for ics2, contained significantly less phylloquinone (19% of wild type; data not shown) than ics1 single mutants (35% of wild type; see above; see also Gross et al., 2006), suggesting that the ICS2 enzyme becomes limiting for isochorismate production in the ics1 mutant background.
SA Accumulation in ics Mutants
SA is presumed to derive from isochorismate in Arabidopsis because SA accumulation was shown to be severely impaired in the ics1 mutant upon inducing conditions (Nawrath and Métraux, 1999; Wildermuth et al., 2001). We have analyzed the induction of SA accumulation in the ics1, ics2, and ics1 ics2 mutants after UV treatment, a known stimulus for SA accumulation (Nawrath et al., 2002). As expected, the ics1 mutant accumulated roughly 10% of total SA compared to the wild type (0.47 ± 0.14 versus 4.17 ± 1.26 _μ_g g−1 fresh weight, respectively; Fig. 4C). The mean value for the ics2 mutant was similar to the wild-type reference (4.13 ± 1.27 versus 4.17 ± 1.26 _μ_g g−1 fresh weight; Fig. 4C). Therefore, in the wild type, the ics2 mutation does not have a high impact on SA accumulation upon UV exposure or in a noninduced state (0.54 ± 0.09 versus 0.56 ± 0.12 _μ_g g−1 fresh weight). This result was expected following the simple reasoning that if (1) ICS1 and ICS2 were independently regulated, and (2) ICS1 accounted for approximately 90% of the total amount of isochorismate produced in these conditions (phenotype of the ics1 mutant), then ICS2, at best, would contribute to only 10% of isochorismate production in these conditions and consequently an ics2 mutant should not be strongly affected in SA accumulation.
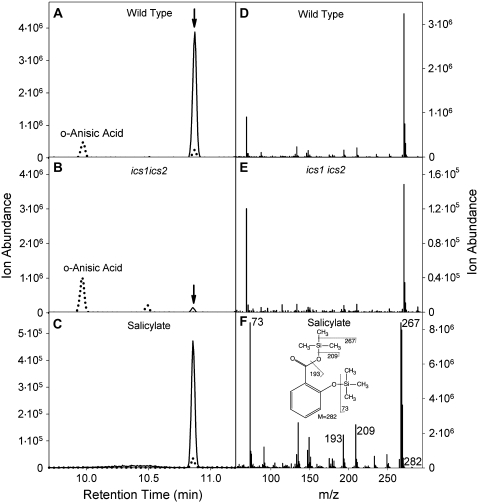
We then determined whether ICS2 was required for the production of the remaining SA in the ics1 mutant and evaluated the genetic relationship between ICS1 and ICS2 in the course of SA biosynthesis after UV induction using the ics1 ics2 double mutant. Without any induction, ics1 ics2 double mutants accumulated approximately 43% of total SA relative to the ics1 mutant (0.12 ± 0.08 versus 0.28 ± 0.09 _μ_g g−1 fresh weight; Fig. 4C). The difference in SA accumulation between the ics1 and ics1 ics2 mutants can be assigned to the activity of the ICS2 gene product. The same effect was observed after UV stimulation because the ics1 ics2 double mutant produced total SA in a similar range of about 36% relative to the ics1 mutant (0.17 ± 0.12 versus 0.47 ± 0.14 _μ_g g−1 fresh weight; Fig. 4C). The structure of SA in the double mutant was verified by gas chromatography-mass spectrometry (GC-MS; Fig. 5). These results imply that ICS2 can be involved in SA biosynthesis through isochorismate production; however, its contribution is marginal compared to ICS1. Because the values before and after UV induction are not statistically very different both for the ics1 and ics1 ics2 mutants, we can further conclude that, unlike ICS1, ICS2 does not display a strong inducible activity following UV irradiation. The detection of SA in the double ics1 ics2 mutant that is devoid of plastid ICS activity, as suggested from the absence of phylloquinone, indicates that an alternative SA biosynthetic pathway is active. Our data show that this pathway is not inducible by UV irradiation and accounts for minor amounts of total SA production (about 20% in uninduced state and 4% after UV induction).
Figure 5.
Identification of SA in ics1 ics2 by GC-MS. SA in leaf extracts was silylated and separated by GC-MS. _o_-Anisic acid was used as internal standard. A (wild type), B (ics1 ics2), and C (SA standard) show the ion traces for m/z 209 (dotted line, characteristic for silylated _o_-anisic acid) and 267 (solid line, silylated SA). D to F, Mass spectra for the peaks eluting at 10.8 min (arrows in A and B) for wild type, ics1 ics2, and the SA standard, respectively. The inset in F shows the fragmentation pattern of silylated SA.
DISCUSSION
SA plays a major role in a number of physiological responses and defense reactions; yet, some aspects of its biosynthesis still remain unknown. In particular, this concerns the implication of the two Arabidopsis ICS genes in the biosynthesis of SA. In this article, we have characterized the Arabidopsis ICS1 and ICS2 gene products and evaluated their respective contribution to isochorismate-derived compounds in this species, namely, SA and phylloquinone.
The existence of two ICS genes in Arabidopsis raises the issue of their specificity and the extent of their redundancy. However, not all higher plants have two distinct ICS genes. In rice (Oryza sativa ‘japonica’), there is only one ICS gene and this gene has not been studied so far. In this species, SA derives from a Phe ammonia lyase-dependent pathway (Silverman et al., 1995; Sawada et al., 2006). However, the regulation of SA synthesis and metabolism in rice might be different from other plant species because SA is present in very high levels as a free acid and might serve alternative functions (Yang et al., 2004). In poplar (Populus spp.), a single ICS gene has been detected (Tsai et al., 2006). For this species, the involvement of ICS in the synthesis of its numerous potential SA glucoside and of phylloquinone has not yet been explored. So far, no generalization can be drawn from these model organisms because each of them is known to have a specific metabolism for SA. The recent draft sequence of the grapevine genome also suggests that there is only one ICS gene in this species, although three ancestral genomes have been detected (Jaillon et al., 2007). The uniqueness of the ICS gene in poplar, rice, and grapevine may constitute a state of evolution where other duplicated ICS genes have been lost and a single gene takes over the production of the common precursor of phylloquinone and SA. The existence of two ICS genes in the Arabidopsis genome may thus appear as an exception arising from our consideration of this genome at this time instead of in the course of its evolution. The two ICS genes occur within duplicated blocks that originate from a large genomic duplication (Blanc et al., 2003). On the one hand, the less obvious role of ICS2 compared to ICS1 might indicate that ICS2 could be lost in the future without loss of fitness. On the other hand, this gene could also provide a benefit for the plant in the field or yet undiscovered conditions, which would justify why ICS2 has not already been lost. It is interesting to note that in C. roseus only one gene, but two different ICS isoforms, have been detected after elicitation of cell cultures (van Tegelen et al., 1999). It is possible that in this species the supplementary function of isochorismate as a precursor of 2,3-dihydroxybenzoic acid is associated with specific regulation of the ICS gene product.
It is not known whether only one of the two Arabidopsis ICS genes would contribute to a specific biological process. The data from the ics mutants (Fig. 4) indicate that both participate in the synthesis of SA and phylloquinone. This is consistent with the observation that both are targeted to the same subcellular compartment. Moreover, a recent study has suggested that the gene products involved in the phylloquinone biosynthesis may form enzymatic complexes in the stroma (Kim et al., 2008). However, the ubiquitous stromal localization of both of the ICS:GFP fusions (Fig. 3) indicates that neither ICS1 nor ICS2 is presumably restricted to these complexes. This localization suggests that the isochorismate produced by both of the enzymes then diffuses up to the subsequent active centers that catalyze phylloquinone formation.
A tight regulation of duplicated genes is observed in prokaryotes and correlates with their genomic organization in operons. In E. coli, two ICS genes have been described and each of them is linked to a distinct pathway. The EntC gene is part of an operon associated with the synthesis of enterobactin, a dihydroxybenzoate-derived siderophore, and is regulated following iron requirement (Kwon et al., 1996). The MenF gene is involved in the synthesis of menaquinone (also known as vitamin K2), an electron carrier in the respiratory chain during anaerobic growth (Jiang et al., 2007). MenF is induced only during anoxygenic conditions. Due to this tight regulation, the two ICS genes show low redundancy (Dahm et al., 1998; Buss et al., 2001). A somewhat different situation has been described for Bacillus subtilis, where one of the two ICS genes can provide isochorismate for dihydroxybenzoate and menaquinone production, but the other one is active only in menaquinone synthesis (Rowland and Taber, 1996). In Arabidopsis, our study established that, although ICS1 and ICS2 are involved in both SA and phylloquinone production, the absence of ICS2 produced a less dramatic effect than the absence of ICS1 on SA and phylloquinone accumulation. That is, the activity of ICS1 could largely compensate for the absence of ICS2, but the ICS2 activity could not make up for a genetic lesion in ICS1. Therefore, ICS1 and ICS2 constitute an example of unequal redundancy as defined by Briggs et al. (2006).
Transcriptional regulation might provide part of a mechanistic interpretation of these results. Microarray data available through Genevestigator (Zimmermann et al., 2005) indicate that both ICS1 and ICS2 are expressed at low levels in noninducing conditions. The resulting low-level constitutive ICS activity correlates with the continuous requirement of phylloquinone from the plant to sustain continuous growth and development. Phylloquinone is an essential component of PSI, where it functions as an electron acceptor (Gross et al., 2006). Visual inspection of the single ics1 and ics2 mutant would suggest that ICS1 and ICS2 are equally redundant; however, phylloquinone measurement revealed that ICS1 played a greater role in phylloquinone accumulation than ICS2. A threshold effect for phylloquinone requirement for growth and development is likely responsible for this discrepancy. The different requirements for the respective ICS gene products suggest that in these noninducing conditions ICS1 is more accumulated or active than ICS2, maybe because of differences in the transcript accumulation rate, translation efficiency, other posttranslational control or catalytic activity or regulation. A less straightforward possibility to interpret the phenotype of phylloquinone accumulation of the single mutants would be that the absence of one ICS gene product is compensated by increased activity of the other remaining functional ICS gene; differences in the efficiency of this negative feedback control loop for regulation of ICS1 or ICS2 would also lead to apparent unequal importance of the two genes.
Specific induction of ICS1 mRNA accumulation, but not ICS2 mRNA, under some SA-inducing conditions such as biotic (Wildermuth et al., 2001) or abiotic stresses, for example, UVC and ozone (data not shown; Zimmermann et al., 2005), provides an obvious explanation to the stronger requirement for ICS1 rather than ICS2 for SA biosynthesis, and therefore explains the phenotype of the mutants relative to their SA accumulation. Alternatively, posttranscriptional mechanisms or enzymatic properties could also be postulated to explain the major role of ICS1 compared to ICS2. Interestingly, specific stimuli can also induce the ICS2 gene either in conjunction with ICS1 (i.e. senescence), or without induction of ICS1 (i.e. abscisic acid treatment; data not shown; Zimmermann et al., 2005), suggesting that ICS2 could play a role yet to be discovered. Careful inspection of the ics2 mutant subjected to such stimuli did not reveal obvious phenotypical changes (data not shown). So far, these data suggest that a major difference between ICS1 and ICS2 relies on transcriptional regulation.
SA biosynthesis in Arabidopsis has been proposed to proceed for a large extent through the catalytic activity of the ICS genes (Wildermuth et al., 2001). However, the ics1 mutant still exhibits 5% to 10% of SA compared to the wild type after adequate stimulation. The presence of SA in this mutant indicates either redundant ICS activity or an alternative biosynthetic pathway, such as the phenylpropanoid pathway as proposed by previous studies in Arabidopsis (Mauch-Mani and Slusarenko, 1996; Ferrari et al., 2003). Here, we have assessed the contribution of ICS2 in the production of SA in the ics1 mutant using a genetic approach. Our data indicate a low, but detectable, residual level of SA in the double mutant ics1 ics2, supporting the existence of an ICS-independent biosynthetic pathway for SA. The susceptibility of the ics1 mutant suggests that SA produced through ICS1 is biologically active and important for defense reactions (Wildermuth et al., 2001). Other studies in Arabidopsis have proposed that biologically active SA is mainly made from a pathway derived from phenylpropanoids. However, these results might be questioned for the following reasons. In the first study, SA levels were determined using HPLC separation and detected by absorption at 280 nm (Mauch-Mani and Slusarenko, 1996). At this wavelength, SA is hardly detectable for the amounts present in leaf extracts of Arabidopsis and fluorescence detection is required for a reasonable readout. This leaves some uncertainty over the nature of the metabolite measured as SA in this report. In another study, the production of biologically relevant SA from an ICS-independent pathway was mainly inferred from biological effects on resistance against B. cinerea using treatments with 2-aminoindane 2-phosphonic acid, an inhibitor of Phe ammonia lyase (Ferrari et al., 2003). The effect of these treatments on SA accumulation in the tissue was, however, not determined.
In conclusion, we have demonstrated the function and localization of ICS2 involved in SA biosynthesis. Using ics1, ics2, and ics1 ics2 mutants we have demonstrated that ICS2 contributes to SA, but in limited amounts, detectable only when ICS1 is lacking. This unequal redundancy was also observed for phylloquinone production. Furthermore, detection of SA in ics1 ics2 that is completely devoid of phylloquinone provided genetic evidence of the existence of an ICS-independent SA biosynthetic pathway in Arabidopsis.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Plants were grown on a pasteurized soil mix of humus:perlite (3:1) under a 12-h-light/12-h-dark cycle, with a night temperature of 16°C and a day temperature of 20°C to 22°C. Arabidopsis (Arabidopsis thaliana) accession Columbia-0 was obtained from the Arabidopsis Biological Resource Center (Columbus, OH). The T-DNA insertion mutant ics2 (SALK_084635; Alonso et al., 2003) was obtained from the Nottingham Arabidopsis Stock Centre.
RACE-PCR Reactions
We used the First-Choice RLM-RACE kit (Ambion) to determine the 5′-end of the ICS2 messenger, following instructions of the manufacturer.
Subcellular Localization
GFP fusions were realized by using Gateway technology. ICS1 and ICS2 cDNAs were amplified from reverse transcription products with Pfu polymerase (Stratagene) using primers ICS1-6 (CACCATGGCTTCACTTCAATTTTCT) and ICS1-8 (ATTAATCGCCTGTAGAGATG), and ICS2-5 (CACCATGGCGTCGCTTCAGTGTTCA) and ICS2-7 (GTTGATTGGTTGCAAAGCTGA), respectively, and then cloned into the entry vector pENTR-D-Topo (Invitrogen). Entry clones were then recombined with the pB7FWG2 vector (Karimi et al., 2005), generating cDNA-GFP fusions driven by the cauliflower mosaic virus (CaMV) 35S promoter. The pB7F2 control vector was generated by digestion of pB7FWG2 with _Eco_RV followed by self-ligation, thus removing the ccdB cassette and placing GFP directly under the control of the CaMV 35S promoter. The vectors were then transformed into Agrobacterium tumefaciens strain GV3101. The Agrobacterium infiltration procedure was realized as described (Burch-Smith et al., 2006), except that the 50-mL culture step was omitted and that acetosyringone was used at 100 _μ_m. Infiltrated patches of leaves were observed after 3 to 4 d with a Bio-Rad MRC 1024 laser confocal microscope. For imaging GFP and chlorophyll, excitation was at 488 and 647 nm, respectively, and emissions were collected with a 506- to 538-nm band-pass filter (referred as 522DF32) and a 664- to 696-nm band-pass filter (referred as 680DF32), respectively.
Functional Complementation of the PBB8 Strain of Escherichia coli
Empty vector pJF119EH1 and EntC-overexpressing construct pDF2 (Franke et al., 2003) were a kind gift from G. Sprenger (Stuttgart, Germany). ICS1 coding sequence without the first 45 amino acids was flanked with _Nco_I and _Bam_HI restriction sites by amplification with primers ICS1-1 (AACTTTAAGAAGGAGATATACCATGGCATATGCTATGTCTATGAATGGTTGTGAT) and ICS1-2 (GGATCCTCAATTAATCGCCTGTAGAGA) and was subcloned into the pGEM-T-Easy vector (Promega). An _Eco_RI-_Bam_HI fragment from this construct was then inserted into pJF119EH1 to give pJF202. The ICS2 coding sequence without the first 50 amino acids was amplified with primers ICS2-8 (CTTTAAGAAGGAGATATACCATGGCAAACGGATGTGAGGCTGACCAC) and ICS2-9 (GGATCCTTAGTTGATTGGTTGCAAAGC) and then subcloned into pJF202 as a _Nco_I-_Bam_HI fragment, giving pJF608. These constructs were introduced into the _entC_− _MenF_− PBB8 E. coli strain (Muller et al., 1996), previously transformed with the pRARE plasmid (Novy et al., 2001). The PBB8 strain was kindly provided by E. Leistner (Bonn). Individual colonies were allowed to grow for 6 h in 400 _μ_L of Luria-Bertani medium containing ampicillin and chloramphenicol; bacterial cells were then rinsed in MgSO4 (10 mm) and plated onto CAS agar (Schwyn and Neilands, 1987) supplemented with 10% casamino acids (BD Biosciences).
Determination of the Genotype of the ICS2 Locus
The primers ICS2-1 (GTCTTCAAAGTCTCCTCTGAT) and ICS2-2 (TGAATCACCTCTAGGCCTTGT) were used to detect a wild-type copy of the ICS2 gene by PCR. The T-DNA disrupted allele (Alonso et al., 2003) was detected by using primers ICS2-2 and Lba1 (TGGTTCACGTAGTGGGCCATCG). PCR reactions for investigating the presence of wild-type and mutant alleles were run separately.
Phylloquinone Determination
Phylloquinone was measured in the ics1 and ics2 mutants by fluorescence HPLC after reduction of phylloquinone to the phyllohydroquinone form (Lohmann et al., 2006).
SA Extraction and Quantification by HPLC
Samples were taken from leaves of plants grown on Murashige and Skoog medium supplemented with 1% Suc 18 h after exposure to UVC light for stimulation of SA accumulation (Nawrath and Métraux, 1999). SA was extracted and total SA (free and conjugated) quantified using a modification of the method previously described (Nawrath and Métraux, 1999). Leaf tissue (0.2–0.3 g with 1.5 _μ_g _o_-anisic acid as internal standard) was extracted twice with 2 mL of 100% methanol each. After evaporation of methanol from the combined extracts, acid hydrolysis was performed with 4 n HCl at 80°C for 1 h, and SA was extracted twice with 2 mL of ethyl acetate:hexane (1:1). The combined organic extracts were evaporated, redissolved in 50 _μ_L acetonitrile of which 20 _μ_L were injected on a reverse-phase HPLC column (ABZ+, 250 mm × 4.6 mm; Supelco) and separated at a flow rate of 1 mL min−1 (Agilent 1100 HPLC system; Böblingen). Elution began with an isocratic flow of 15% acetonitrile in water (pH 2.6 adjusted with phosphoric acid) for 1 min, followed by a linear increase to 20% acetonitrile over 5 min, isocratic elution at 20% for 10 min, a linear increase from 20% to 55% acetonitrile over 9.5 min, and to 90% in 5 min. For column regeneration, acetonitrile was decreased linearly from 90% to 0%, followed by a linear increase to 15% acetonitrile and an isocratic flow at 15% for 4 min. Fluorescence was recorded with excitation/emission wavelengths of 305/365 nm and 305/407 nm for _o_-anisic acid and SA, respectively (Dewdney et al., 2000).
GC-MS Analysis of SA
For GC-MS analysis, the SA samples were extracted as described above. After evaporation of solvent, 50 _μ_L of _N_-methyl-_N_-trimethylsilyl-trifluoracetamide was added, and the samples were incubated for 1 h at 80°C for silylation. After the evaporation of _N_-methyl-_N_-trimethylsilyl-trifluoracetamide, the extracts were redissolved in 40 _μ_L of hexane. The trimethylsilyl derivatives of SA were analyzed by GC using an Agilent 6890 system (Agilent) equipped with an Agilent 5973 quadrupole mass detector. The inlet was maintained at 260°C and operated in the splitless mode (injection volume 1 _μ_L). A 30 m × 530 _μ_m and 0.2-_μ_m film thickness SP-2380 column (Supelco) was used for analyses with a helium flow of 5.1 mL min−1. The initial oven temperature was 80°C and was then ramped to 180°C at a rate of 1°C min−1. The oven temperature was maintained at 180°C for 1 min and decreased to 80°C with a rate of 20°C min−1. Silylated SA and _o_-anisic acid were detected at m/z 267 and 209, respectively.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NP_565090, ACC60228, and CAA06837.
Acknowledgments
We acknowledge Linda Grainger for technical assistance, Martine Schorderet for confocal microscope assistance, and Eliane Abou-Mansour for help with SA analysis. We thank Professor G. Sprenger (Stuttgart, Germany) for sharing plasmids PJF119EH1 and pDF2, and Professor E. Leistner (Bonn) for providing the PBB8 E. coli strain.
1
This work was supported by the Swiss National Science Foundation (grant no. 3100A0–104224 to J.-P.M.) and by the German Science Foundation (grant no. DFG Do520/8 to P.D.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Jean-Pierre Métraux (jean-pierre.metraux@unifr.ch).
[OA]
Open Access articles can be viewed online without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen HM, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Blanc G, Hokamp K, Wolfe KH (2003) A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res 13 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs GC, Osmont KS, Shindo C, Sibout R, Hardtke CS (2006) Unequal genetic redundancies in Arabidopsis—a neglected phenomenon? Trends Plant Sci 11 492–498 [DOI] [PubMed] [Google Scholar]
- Burch-Smith TM, Schiff M, Liu YL, Dinesh-Kumar SP (2006) Efficient virus-induced gene silencing in Arabidopsis. Plant Physiol 142 21–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss K, Muller R, Dahm C, Gaitatzis N, Skrzypczak-Pietraszek E, Lohmann S, Gassen M, Leistner E (2001) Clustering of isochorismate synthase genes menF and entC and channeling of isochorismate in Escherichia coli. Biochim Biophys Acta 1522 151–157 [DOI] [PubMed] [Google Scholar]
- Catinot J, Buchala A, Abou-Mansour E, Métraux JP (2008) Salicylic acid production in response to biotic and abiotic stress depends on isochorismate in Nicotiana benthamiana. FEBS Lett 582 473–478 [DOI] [PubMed] [Google Scholar]
- Dahm C, Muller R, Schulte G, Schmidt K, Leistner E (1998) The role of isochorismate hydroxymutase genes entC and menF in enterobactin and menaquinone biosynthesis in Escherichia coli. Biochim Biophys Acta 1425 377–386 [DOI] [PubMed] [Google Scholar]
- Dewdney J, Reuber TL, Wildermuth MC, Devoto A, Cui JP, Stutius LM, Drummond EP, Ausubel FM (2000) Three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. Plant J 24 205–218 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300 1005–1016 [DOI] [PubMed] [Google Scholar]
- Ferrari S, Plotnikova JM, De Lorenzo G, Ausubel FM (2003) Arabidopsis local resistance to Botrytis cinerea involves salicylic acid and camalexin and requires EDS4 and PAD2, but not SID2, EDS5 or PAD4. Plant J 35 193–205 [DOI] [PubMed] [Google Scholar]
- Franke D, Lorbach V, Esser S, Dose C, Sprenger GA, Halfar M, Thommes J, Muller R, Takors R, Muller M (2003) (S,S)-2,3-Dihydroxy-2,3-dihydrobenzoic acid: microbial access with engineered cells of Escherichia coli and application as starting material in natural-product synthesis. Chemistry (Easton) 9 4188–4196 [DOI] [PubMed] [Google Scholar]
- Gaille C, Kast P, Haas D (2002) Salicylate biosynthesis in Pseudomonas aeruginosa. Purification and characterization of PchB, a novel bifunctional enzyme displaying isochorismate pyruvate-lyase and chorismate mutase activities. J Biol Chem 277 21768–21775 [DOI] [PubMed] [Google Scholar]
- Garcion C, Métraux JP (2006) Salicylic acid. In P Hedden, SG Thomas, eds, Plant Hormone Signaling. Annual Plant Reviews, Vol 24. Blackwell Press, Oxford, pp 229–257
- Gross J, Cho WK, Lezhneva L, Falk J, Krupinska K, Shinozaki K, Seki M, Herrmann RG, Meurer J (2006) A plant locus essential for phylloquinone (vitamin K1) biosynthesis originated from a fusion of four eubacterial genes. J Biol Chem 281 17189–17196 [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449 463–465 [DOI] [PubMed] [Google Scholar]
- Jiang M, Cao Y, Guo ZF, Chen MJ, Chen XL, Guo ZH (2007) Menaquinone biosynthesis in Escherichia coli: identification of 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-l-carboxylate as a novel intermediate and re-evaluation of MenD activity. Biochemistry 46 10979–10989 [DOI] [PubMed] [Google Scholar]
- Karimi M, De Meyer B, Hilson P (2005) Modular cloning in plant cells. Trends Plant Sci 10 103–105 [DOI] [PubMed] [Google Scholar]
- Kim HU, van Oostende C, Basset GJC, Browse J (2008) The AAE14 gene encodes the Arabidopsis _o_-succinylbenzoyl-CoA ligase that is essential for phylloquinone synthesis and photosystem-I function. Plant J 54 272–283 [DOI] [PubMed] [Google Scholar]
- Kolappan S, Zwahlen J, Zhou R, Truglio JJ, Tonge PJ, Kisker C (2007) Lysine 190 is the catalytic base in MenF, the menaquinone-specific isochorismate synthase from Escherichia coli: implications for an enzyme family. Biochemistry 46 946–953 [DOI] [PubMed] [Google Scholar]
- Kwon O, Hudspeth MES, Meganathan R (1996) Anaerobic biosynthesis of enterobactin in Escherichia coli: regulation of entC gene expression and evidence against its involvement in menaquinone (vitamin K-2) biosynthesis. J Bacteriol 178 3252–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann A, Schottler MA, Brehelin C, Kessler F, Bock R, Cahoon EB, Dormann P (2006) Deficiency in phylloquinone (vitamin K1) methylation affects prenyl quinone distribution, photosystem 1 abundance, and anthocyanin accumulation in the Arabidopsis atmeng mutant. J Biol Chem 281 40461–40472 [DOI] [PubMed] [Google Scholar]
- Martinez C, Pons E, Prats G, Leon J (2004) Salicylic acid regulates flowering time and links defence responses and reproductive development. Plant J 37 209–17 [DOI] [PubMed] [Google Scholar]
- Mauch-Mani B, Slusarenko AJ (1996) Production of salicylic acid precursors is a major function of phenylalanine ammonia-lyase in the resistance of Arabidopsis to Peronospora parasitica. Plant Cell 8 203–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Métraux JP, Durner J (2004) The role of salicylic acid and nitric oxide in programmed cell death and induced resistance. In H Sandermann, ed, Ecological Studies, Vol 170. Springer-Verlag, Berlin, pp 111–150
- Morris K, Mackerness SAH, Page T, John CF, Murphy AM, Carr JP, Buchanan-Wollaston V (2000) Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J 23 677–685 [DOI] [PubMed] [Google Scholar]
- Muljono RAB, Scheffer JC, Verpoorte R (2002) Isochorismate is an intermediate in 2,3-dihydroxybenzoic acid biosynthesis in Catharanthus roseus cell cultures. Plant Physiol Biochem 40 231–234 [Google Scholar]
- Muller R, Dahm C, Schulte G, Leistner E (1996) An isochorismate hydroxymutase isogene in Escherichia coli. FEBS Lett 378 131–134 [DOI] [PubMed] [Google Scholar]
- Mustafa NR, Verpoorte R (2005) Chorismate derived C6C1 compounds in plants. Planta 222 1–5 [DOI] [PubMed] [Google Scholar]
- Nawrath C, Heck S, Parinthawong N, Métraux JP (2002) EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C, Métraux JP (1999) Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novy R, Drott D, Yaeger K, Mierendorf R (2001) Overcoming the codon bias of E. coli for enhanced protein expression. Innovations 12 1–3 [Google Scholar]
- Ogawa D, Nakajima N, Sano T, Tamaoki M, Aono M, Kubo A, Kanna M, Ioki M, Kamada H, Saji H (2005) Salicylic acid accumulation under O-3 exposure is regulated by ethylene in tobacco plants. Plant Cell Physiol 46 1062–1072 [DOI] [PubMed] [Google Scholar]
- Raskin I (1992) Salicylate, a new plant hormone. Plant Physiol 99 799–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond KN, Dertz EA, Kim SS (2003) Enterobactin: an archetype for microbial iron transport. Proc Natl Acad Sci USA 100 3584–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland BM, Taber HW (1996) Duplicate isochorismate synthase genes of Bacillus subtilis: regulation and involvement in the biosyntheses of menaquinone and 2,3-dihydroxybenzoate. J Bacteriol 178 854–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada H, Shim IS, Usui K (2006) Induction of benzoic acid 2-hydroxylase and salicylic acid biosynthesis—modulation by salt stress in rice seedlings. Plant Sci 171 263–270 [Google Scholar]
- Schwyn B, Neilands JB (1987) Universal chemical-assay for the detection and determination of siderophores. Anal Biochem 160 47–56 [DOI] [PubMed] [Google Scholar]
- Silverman P, Seskar M, Kanter D, Schweizer P, Métraux JP, Raskin I (1995) Salicylic-acid in rice—biosynthesis, conjugation, and possible role. Plant Physiol 108 633–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small I, Peeters N, Legeai F, Lurin C (2004) Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4 1581–1590 [DOI] [PubMed] [Google Scholar]
- Sticher L, Mauch-Mani B, Métraux JP (1997) Systemic acquired resistance. Annu Rev Phytopathol 35 235–270 [DOI] [PubMed] [Google Scholar]
- Strawn MA, Marr SK, Inoue K, Inada N, Zubieta C, Wildermuth MC (2007) Arabidopsis isochorismate synthase functional in pathogen-induced salicylate biosynthesis exhibits properties consistent with a role in diverse stress responses. J Biol Chem 282 5919–5933 [DOI] [PubMed] [Google Scholar]
- Tsai CJ, Harding SA, Tschaplinski TJ, Lindroth RL, Yuan YN (2006) Genome-wide analysis of the structural genes regulating defense phenylpropanoid metabolism in Populus. New Phytol 172 47–62 [DOI] [PubMed] [Google Scholar]
- Uppalapati SR, Ishiga Y, Wangdi T, Kunkel BN, Anand A, Mysore KS, Bender CL (2007) The phytotoxin coronatine contributes to pathogen fitness and is required for suppression of salicylic acid accumulation in tomato inoculated with Pseudomonas syringae pv. tomato DC3000. Mol Plant Microbe Interact 20 955–965 [DOI] [PubMed] [Google Scholar]
- van Tegelen LJP, Moreno PRH, Croes AF, Verpoorte R, Wullems GJ (1999) Purification and cDNA cloning of isochorismate synthase from elicited cell cultures of Catharanthus roseus. Plant Physiol 119 705–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verberne MC, Sansuk K, Bol JF, Linthorst HJM, Verpoorte R (2007) Vitamin K-1 accumulation in tobacco plants overexpressing bacterial genes involved in the biosynthesis of salicylic acid. J Biotechnol 128 72–79 [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 417 562–565 [DOI] [PubMed] [Google Scholar]
- Yang YN, Qi M, Mei CS (2004) Endogenous salicylic acid protects rice plants from oxidative damage caused by aging as well as biotic and abiotic stress. Plant J 40 909–919 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hennig L, Gruissem W (2005) Gene-expression analysis and network discovery using Genevestigator. Trends Plant Sci 10 407–409 [DOI] [PubMed] [Google Scholar]