TWEAK-FN14 signaling induces lysosomal degradation of a cIAP1–TRAF2 complex to sensitize tumor cells to TNFα (original) (raw)
Abstract
Synthetic inhibitor of apoptosis (IAP) antagonists induce degradation of IAP proteins such as cellular IAP1 (cIAP1), activate nuclear factor κB (NF-κB) signaling, and sensitize cells to tumor necrosis factor α (TNFα). The physiological relevance of these discoveries to cIAP1 function remains undetermined. We show that upon ligand binding, the TNF superfamily receptor FN14 recruits a cIAP1–Tnf receptor-associated factor 2 (TRAF2) complex. Unlike IAP antagonists that cause rapid proteasomal degradation of cIAP1, signaling by FN14 promotes the lysosomal degradation of cIAP1–TRAF2 in a cIAP1-dependent manner. TNF-like weak inducer of apoptosis (TWEAK)/FN14 signaling nevertheless promotes the same noncanonical NF-κB signaling elicited by IAP antagonists and, in sensitive cells, the same autocrine TNFα-induced death occurs. TWEAK-induced loss of the cIAP1–TRAF2 complex sensitizes immortalized and minimally passaged tumor cells to TNFα-induced death, whereas primary cells remain resistant. Conversely, cIAP1–TRAF2 complex overexpression limits FN14 signaling and protects tumor cells from TWEAK-induced TNFα sensitization. Lysosomal degradation of cIAP1–TRAF2 by TWEAK/FN14 therefore critically alters the balance of life/death signals emanating from TNF-R1 in immortalized cells.
Introduction
Upon binding of their cognate ligand, TNF receptor superfamily (TNFRSF) members transmit signals via their cytoplasmic domains. Several TNF receptors bear death domains (DD) that allow them to directly promote apoptotic cell death. Activation of the TNFRSF receptors, such Fas or TNF-related apoptosis-inducing ligand (TRAIL)–R2 (Tartaglia et al., 1993), allows the binding of FADD in a DD–DD interaction, which initiates apoptotic signaling by the recruitment and activation of caspase 8 or 10 by oligomerization. TNF-R1–induced activation of caspase 8 or 10 is less direct, involving recruitment of the DD-containing adaptor TRADD, followed by the formation of an internalized secondary complex which can bind FADD and caspase 8 to initiate the apoptotic program (Micheau and Tschopp, 2003).
Despite its name, most tumor cells do not die when exposed to TNFα but must also be treated with inhibitors of translation or transcription, such as actinomycin D or cycloheximide. These agents are thought to sensitize cells to TNFα by preventing production of survival proteins induced via NF-κB. Many of the TNFRSF members, including FN14, contain a consensus Tnf receptor-associated factor (TRAF) binding motif (Park et al., 1999; Ye et al., 1999) that recruits TRAFs to activate transcription factors including NF-κB and AP1 (Lee et al., 1997; Yeh et al., 1997).
TRAF1 and TRAF2 were initially identified in protein complexes that bound to the cytoplasmic domain of TNF-R2 (Rothe et al., 1994), together with cellular inhibitor of apoptosis 1 (cIAP1) and 2 (Rothe et al., 1995). However, another cellular IAP homologue, XIAP (Duckett et al., 1996; Listen et al., 1996; Uren et al., 1996), became the focus of attention because it was shown to directly inhibit activated downstream caspases (Deveraux et al., 1997) and the N-terminally processed form of the initiator caspase, caspase 9 (Srinivasula et al., 2001), whereas neither cIAP1 nor cIAP2 can inhibit caspase activity at concentrations that are reached in vivo (Tenev et al., 2005; Eckelman and Salvesen, 2006)
Although the function of cIAP1 has remained unclear for some time, recent studies have identified genetic abnormalities in cIAP1 from patients with multiple myeloma that correlate with reduced cIAP1 protein levels and enhanced noncanonical NF-κB activity (Annunziata et al., 2007; Keats et al., 2007). Consistent with this work, it has recently been demonstrated that genetic deletion of cIAP1 in immortalized mouse embryonic fibroblasts (MEFs) causes constitutive noncanonical NF-κB activity and sensitization to TNFα-induced apoptosis (Vince et al., 2007), and loss of cIAP1 sensitizes cells to TNFα (Gaither et al., 2007). Strikingly, synthetic IAP antagonists, or smac mimetics, which deplete both cIAP1 and 2 protein levels, also activate NF-κB signaling and enhance TNFα death signaling (Li et al., 2004; Gaither et al., 2007; Varfolomeev et al., 2007; Vince et al., 2007). Therefore, although the intended target of Smac mimetics was XIAP, it appears that their ability to effectively inhibit cIAP1 and or cIAP2 plays a central role in tumor cell killing and that cIAP1 is a central player in regulating the survival and death signals initiated from TNF-R1 in tumor cells.
cIAP1 and 2 were identified via their indirect binding to TNF-R2, but they are also present in the TNF-R1 complex (Micheau and Tschopp, 2003; Vince et al., 2007) and have the potential to regulate the signaling from ∼17 TNF superfamily receptors that contain TRAF2 consensus binding sites. Despite this, it is still unknown which receptors cIAP1 does bind and how it might be physiologically regulated to control signaling from these receptors.
TNF-like weak inducer of apoptosis (TWEAK) is a member of the TNF superfamily (TNFSF12) that engages a receptor termed FN14 (TNFRSF12A). FN14 has been shown to bind TRAF2 (and TRAFs 1, 3, and 5) in a yeast two-hybrid screen (Brown et al., 2003) and can initiate both canonical and noncanonical NF-κB activation (Saitoh et al., 2003). It is unknown how FN14 activates NF-κB, but physiological TWEAK/FN14 signaling can inhibit cellular differentiation, promote angiogenesis, cytokine production, and cellular proliferation, and has been suggested to play a role in the wound response because of its induction in wounded tissues and organs (Vince and Silke, 2006; Winkles et al., 2007). TWEAK also induces apoptosis in HT29 and KATO-III cells pretreated with IFNγ and has growth-suppressive effects on several cell types (Felli et al., 2005; Maecker et al., 2005).
In this paper, we show that binding of TWEAK to endogenous FN14 recruits a complex containing both cIAP1 and TRAF2. This complex is subsequently recruited to a lysosomal compartment where it is degraded. Consistent with lysosomal degradation, TWEAK-induced TRAF2 degradation is prevented by several different inhibitors of lysosomal function but, interestingly, still requires cIAP1 function. The loss of cIAP1 or TRAF2, but not cIAP2 or XIAP, preferentially sensitizes immortalized cells, but not primary cells, to killing by TNFα. In contrast, overexpression of both cIAP1 and TRAF2 correlates with tumor cell resistance to TWEAK/TNFα-induced cell death. These results provide mechanistic insights into how the cIAP1–TRAF2 complex functions in tumor cells to inhibit apoptosis and how this complex can be physiologically regulated.
Results
FN14 is expressed by most tumor cell lines
To understand how endogenous cIAPs regulate TNFRSF signaling, we used TNFSF ligands to screen for cell lines containing detectable levels of endogenous TNFSF receptors. To facilitate the screen, we generated TNFSF ligands as recombinant proteins tagged with the Fc portion of human IgG (Fig. S1A, available at http://www.jcb.org/cgi/content/full/jcb.200801010/DC1; Bossen et al., 2006). These molecules are cross-linked via the Fc portion that promotes higher order aggregation of the corresponding receptors, closely mimicking engagement by membrane-bound ligands (Holler et al., 2003). The Fc portion also facilitates reliable detection of these proteins by Western blot (Fig. S1 B) and allows for their simple purification with protein A (Fig. S1 B, right) and for immunoprecipitation of interacting protein complexes (see Fig. 1 D).
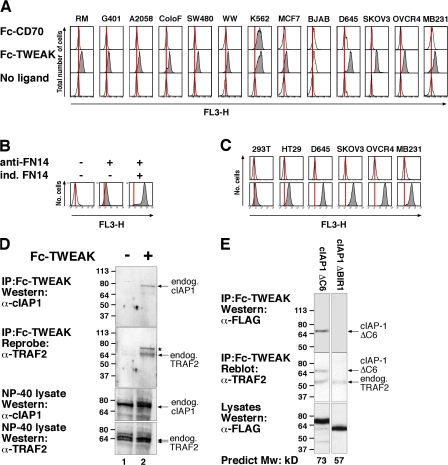
Figure 1.
TWEAK specifically binds to endogenous FN14 in many tumor cell lines and cIAP1 binds to FN14 via its TRAF2 binding domain. (A) Cells were harvested and incubated with Fc-CD70 or Fc-TWEAK, Tricolor-labeled anti-Fc, and analyzed by flow cytometry. (B) FN14 antibody specifically detects FN14. Stable FlpIn T-REx 293 cells inducible for FN14 were induced with or without doxycycline overnight, stained with anti-FN14, and analyzed as in A. Controls were stained with secondary antibody alone. (C) Cell lines that bind TWEAK also stain with antibodies to FN14. Cells were harvested and stained with the FN14 antibody as in B. (D) D645 cells were harvested and then treated for 15 min at 37°C with or without 2 μg Fc-TWEAK. Cells were lysed and Fc-TWEAK protein complexes precipitated with protein A beads and analyzed by Western blot. *, carryover signal from cIAP1 blot. (E) Binding of cIAP1 to FN14 is mediated by its association with TRAF2. D645 glioma cells were transfected with the indicated FLAG-cIAP1 constructs, harvested, and then treated with Fc-TWEAK for 20 min at 37°C. Cells were lysed and Fc-TWEAK complexes precipitated and analyzed as in D. Molecular mass is indicated in kD on the left of the autoradiograph.
We tested the purified Fc ligands for specific binding to their cognate receptor using FlpIn stable cell lines inducibly expressing different TNFSF receptors and only observed binding when the ligand was added to cells in which expression of the corresponding cognate receptor was induced, i.e., CD27/CD70 and FN14/TWEAK (Fig. S1 C). Satisfied with the specificity of the ligands, we used them to screen a panel of tumor cell lines (including those from kidney, brain, colon, melanoma, breast, and ovarian cancers). Only one of the ligands, TWEAK, bound to a high proportion of the tumor cell lines examined (Fig. 1 A, Fig. S1 D, and not depicted), suggesting that in culture, many tumor cells constitutively express the TWEAK receptor FN14.
Some studies have suggested that TWEAK binds other receptors in addition to FN14 (Polek et al., 2003; Bover et al., 2007). To confirm that the signal caused by binding of TWEAK correlated with expression of FN14, we used a commercial antibody against FN14. The specificity of this FN14 antibody was demonstrated by flow cytometry using cells inducible for FN14 expression (Fig. 1 B). Importantly, cell lines that bound TWEAK also stained strongly with the antibody to FN14 (Fig. 1 C). These results demonstrate that a large number of transformed cell lines of both human and mouse origin constitutively express the TWEAK receptor FN14.
TWEAK binding to FN14 recruits TRAF2 and cIAP1
Because yeast two-hybrid screens suggested a potential interaction between TRAF2 and FN14 (Brown et al., 2003), we tested whether TRAF2 could interact with FN14 in vivo. Recombinant Fc-TWEAK successfully immunoprecipitated endogenous TRAF2 and cIAP1 in D645 glioma cells, whereas in the absence of TWEAK, no TRAF2 or cIAP1 was detected (Fig. 1 D).
To test whether cIAP1 binding to FN14 was indirectly mediated through TRAF2, we transiently transfected cIAP1 ΔC6 (a stable mutant of cIAP1 lacking the last six residues) and cIAP1 ΔBIR1 constructs into D645 glioma cells and immunoprecipitated endogenous FN14 with Fc-TWEAK. Consistent with previous observations (Samuel et al., 2006; Varfolomeev et al., 2006), cIAP1 constructs that lacked a BIR1 domain were unable to bind endogenous TRAF2, whereas mutations to other regions of cIAP1 did not affect the TRAF2 interaction (Fig. S1 E and not depicted). As for endogenous cIAP1 (Fig. 1 D), transfected cIAP1 ΔC6 could be immunoprecipitated by Fc-TWEAK (Fig. 1 E), whereas ΔBIR1 cIAP1 could not be detected under the same conditions, even though endogenous TRAF2 was immunoprecipitated (Fig. 1 E). Similarly, when vesicular stomatitis virus (VSV)–tagged FN14 was induced and immunoprecipitated with anti-VSV in the presence of TWEAK, cIAP1 ΔC6 could be detected, whereas neither ΔBIR1 cIAP1 nor ΔBIR1 cIAP1 ΔC6 associated with FN14 (Fig. S1 F). These results strongly suggest that endogenous cIAP1 binds indirectly to FN14 through its association with TRAF2.
Signaling from FN14 induces the degradation of both TRAF2 and cIAP1
The role of TRAF2 and cIAP1 in TNFSF signaling is still unclear. Several studies suggest that TRAF2 is the ubiquitin E3 ligase that ubiquitylates RIP (Lee et al., 2004), thereby promoting activation of canonical NF-κB (Chen et al., 2006). Other studies suggest cIAP1 is the ubiquitin E3 ligase for RIP (Park et al., 2004) and for TRAF2 itself (Li et al., 2002). Indeed, the most widely accepted model for cIAP1 function, based largely on overexpression data, states that cIAP1 ubiquitylates TRAF2 causing its proteasomal degradation (Li et al., 2002). A followup study using primary B cells also showed that TRAF2 degradation after stimulation with agonistic TNF-R2 antibodies did not occur in cIAP1 knockout cells (Zhao et al., 2007), lending credence to this model. We therefore tested whether TWEAK induced TRAF2 degradation. OVCAR4, SKOV3, Kym1 (Fig. 2 A), and transformed MEF cells (Fig. 2 C and not depicted) were treated with TWEAK for 0–6 h and cellular levels of endogenous cIAP1 and TRAF2 analyzed by Western blot (Fig. 2, A and C). After 1–6 h of TWEAK addition, cellular levels of both TRAF2 and cIAP1 were reduced in all cell lines examined.
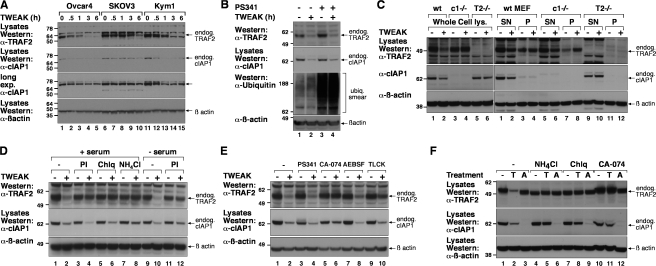
Figure 2.
FN14 signaling decreases cellular cIAP1 and TRAF2 levels by lysosomal degradation. (A) OVCAR4, SKOV3, or Kym1 cells were treated with 100 ng/ml Fc-TWEAK at 37°C for the indicated time, lysed, and analyzed by Western blot. (B) Proteasome inhibition does not block TWEAK-mediated cIAP1 or TRAF2 degradation. D645 cells were pretreated with or without 1 μM PS341 for 2 h and then incubated with 100 ng/ml Fc-TWEAK for 4 h. The cells were lysed and analyzed by Western blot. (C) TRAF2 accumulates in the insoluble fraction in cIAP1 knockout MEF cells after TWEAK stimulation. The indicated MEF cell lines were treated with or without TWEAK for 6 h, and whole cell lysates (left) or Triton X-100 supernatant (SN) or pellet (P) fractions were analyzed by Western blot (right). (D) D645 cells were pretreated with a protease inhibitor cocktail (PI), 200 μM chloroquine (Chlq), or 100 mM NH4Cl, with or without serum, and then stimulated with 100 ng/ml Fc-TWEAK for 4 h. Cells were lysed in 1% SDS and total cell lysate was analyzed by Western blot. (E) D645 cells were pretreated with 1 μM PS341, 20 μM CA-074Me, 40 μM AEBSF, or 100 μM TLCK for 2 h and then stimulated with 100 ng/ml Fc-TWEAK for 4 h. Cell lysates were analyzed as in D. (F) D645 cells were pretreated for 2 h with 200 μM chloroquine, 100 mM NH4Cl, or 20 μM CA-074Me before stimulation with Fc-TWEAK (T) or the IAP antagonist compound A (A) for a further 6 h. Cell lysates were analyzed as in D. Molecular mass is indicated in kD on the left of the autoradiograph.
TWEAK/FN14 promotes cathepsin dependent lysosomal degradation of the cIAP1–TRAF2 complex
Although we observed substantial degradation of TRAF2 and cIAP1 after TWEAK treatment, we were surprised that the degradation of TRAF2 and cIAP1 could not be blocked by preincubating cells with proteasome inhibitors such as MG132 (Fig. S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200801010/DC1) or PS341 (Fig. 2 B) before TWEAK stimulation, despite the fact that these inhibitors efficiently blocked proteasome function, as indicated by enhanced levels of total cellular ubiquitylated proteins (Fig. 2 B).
TNF-R2–induced TRAF2 degradation has been reported to occur by the E3 ubiquitin ligase activity of cIAP1 targeting it for proteasomal degradation (Li et al., 2002). To examine the requirement of cIAP1 for TWEAK-induced TRAF2 loss, and vice versa, we used gene knockout transformed MEF cell lines and stimulated endogenous FN14 with TWEAK. Although TWEAK stimulation resulted in decreased levels of cIAP1 in wild-type MEFs, it was not degraded in TRAF2−/− knockout MEFs (Fig. 2 C, left). TRAF2-mediated binding of cIAP1 to FN14 is therefore required for TWEAK-induced degradation of cIAP1. Consistent with previous studies, cIAP1 was required for the degradation of TRAF2 because TWEAK-stimulated TRAF2 depletion did not occur in cIAP1−/− MEFs (Fig. 2 C, left).
To examine the requirement for cIAP1 in TRAF2 degradation in greater detail, we performed further experiments where we lysed cells in Triton X-100 and examined the detergent soluble and insoluble membrane fractions. Remarkably, TRAF2 disappeared from the Triton X-100–soluble fraction in cIAP1 knockout cells as it did from wild-type cells (Fig. 2 C, right; and Fig. S2 B). These two results suggest that TRAF2 translocation to an insoluble compartment occurs in the absence of cIAP1 but its degradation requires the activity of cIAP1. This notion is consistent with previous reports demonstrating that TRAF2 relocalizes to a detergent-insoluble fraction and becomes degraded after signaling from other TNFSF receptors (Habelhah et al., 2004; Wu et al., 2005).
Because TWEAK did not induce proteasomal degradation of the cIAP1–TRAF2 complex, we tested other protease inhibitors. Cells preincubated with a protease inhibitor cocktail showed reduced TWEAK-mediated degradation of TRAF2 and a modest protecton of cIAP1 when serum was removed from the medium before addition of the inhibitor (Fig. 2 D). We therefore tested whether TWEAK-mediated TRAF2 and cIAP1 depletion was dependent upon lysosomal function. Consistent with this hypothesis, inhibitors of lysosomal function, such as chloroquine and ammonium chloride, prevented TWEAK-mediated TRAF2 degradation, whereas ammonium chloride also substantially blocked TWEAK-mediated cIAP1 degradation, although not to the same extent as it blocked TRAF2 depletion (Fig. 2, D and F).
To further test a role for lysosomal proteases, we used specific protease inhibitors. The serine protease inhibitor AEBSF failed to block TWEAK-mediated cIAP1–TRAF2 degradation, whereas TLCK, which can inhibit both serine and cysteine proteases, partially blocked TWEAK-mediated TRAF2 loss (Fig. 2 E). The cathepsin B inhibitor CA-074Me (Fig. 2, E and F) also provided protection against loss of both cIAP1 and TRAF2, implying that lysosomal cathepsins may be important for the degradation of this complex. Importantly, neither CA-074Me nor the inhibitors of lysosomal function perturbed the proteasomal degradation pathway because they did not prevent the loss of cIAP1 induced by IAP antagonist (compound A) treatment (Fig. 2 F), which we have previously shown is proteasomal dependent (Vince et al., 2007).
Although endogenous TRAF2 and cIAP1 was difficult to detect by confocal microscopy, analysis of D645 cells transiently transfected with FLAG-TRAF2 revealed that in unstimulated cells, TRAF2 was exclusively cytosolic and did not overlap with the acidotropic lysosome marker lysotracker (Fig. S2, C and D). However upon stimulation with TWEAK ligand for 3–6 h, TRAF2 showed a significant redistribution to punctate vesicles (Fig. S2, C and E). TRAF2-containing vesicles were juxtaposed with lysotracker-stained compartments and often directly overlapped (Fig. S2 E), suggesting that TRAF2 degradation occurs in the lysosome or in compartments that are in close association. It is probable that the Triton X-100–insoluble fraction contains MVB/lysosomal membranes because the inhibitors NH4Cl and CA-074Me significantly blocked degradation of TRAF2 and cIAP1 in the Triton X-100–insoluble fraction (unpublished data).
TWEAK activates noncanonical NF-κB by depleting cIAP1 and TRAF2
TWEAK/FN14 signaling has previously been shown to activate both canonical and noncanonical NF-κB (Saitoh et al., 2003). Because TRAF2 knockout B cells and either immortalized cIAP1 or TRAF2 knockout MEFs show constitutive activation of noncanonical NF-κB (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200801010/DC1; Grech et al., 2004; Vince et al., 2007), we hypothesized that TWEAK-mediated NF-κB signaling may be a direct result of the depletion of cIAP1 and TRAF2 and noncanonical in nature, despite the fact that degradation of cIAP1 after TWEAK signaling is never complete.
To measure TWEAK-induced NF-κB activation, we created stable cell lines containing an NF-κB reporter, where expression of EGFP is driven by a promoter containing four NF-κB binding elements. As expected, NIH 3T3 cells bearing the NF-κB reporter showed strong NF-κB induction when stimulated with TNFα (Fig. 3 A). TWEAK also induced a significant NF-κB response, although this was slower and not as large as the TNFα response (Fig. 3 A). TWEAK-induced NF-κB was not dependent upon autocrine-produced TNFα because induction of NF-κB could not be blocked by anti-TNFα (unpublished data). To investigate whether NF-κB was noncanonical, we examined processing of the NF-κB2 subunit from the p100 form to the activated, processed p52 form. In both OVCAR4 and KYM1 cell lines, processing of p100 to p52 became visible after 1 h of TWEAK stimulation (Fig. 3, B and C) and correlated well with the TWEAK-induced loss of cIAP1–TRAF2 (Fig. 2 A). Also consistent with noncanonical activation of NF-κB, we observed that TWEAK treatment caused a remarkable stabilization of NIK (Fig. 3, C and E), which correlated with processing of p100 to p52, but observed no change in NF-κB1 p105 processing (Fig. 3, C and E).
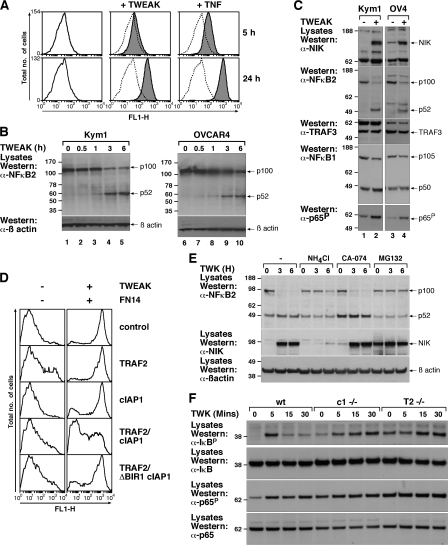
Figure 3.
TWEAK-induced cIAP1–TRAF2 loss activates noncanonical NF-κB. (A) An NIH 3T3 cell clone stably transformed with a lentiviral NF-κB reporter vector was stimulated with Fc-TNF or Fc-TWEAK for the indicated times. (B) Kym1 or OVCAR4 cells were stimulated with 100 ng/ml Fc-TWEAK for the indicated times and analyzed by Western blot for p100 processing to p52. (C) Kym1 or OVCAR4 cells were treated with or without Fc-TWEAK for 6 h and lysates were analyzed by Western blot. (D) FN14-inducible FlpIn 293 cells infected with the lentiviral NF-κB reporter were transiently transfected with the indicated constructs for 24 h. FN14 was induced overnight, the cells were treated with 100 ng/ml Fc-TWEAK for a further 24 h, and NF-κB activity was measured by flow cytometry. Histograms are representative of three independent experiments. (E) NH4Cl blocks TWEAK-induced noncanonical NF-κB. Cells were pretreated with NH4Cl, CA-074Me, or MG132 for 2 h and then treated with 100 ng/ml TWEAK for the indicated times, lysed, and analyzed by Western blot. (F) Wild-type and knockout MEFs were treated with TWEAK for the indicated times, and lysates were analyzed as in E. Molecular mass is indicated in kD on the left of the autoradiograph.
If TWEAK-induced loss of the cIAP1–TRAF2 complex is required to activate the noncanonical pathway, then genetic loss of either cIAP1 or TRAF2 might also result in constitutive activation of this pathway. Consistent with this model and our previous observations (Vince et al., 2007), MEFs deleted for either cIAP1 or TRAF2 showed elevated p52 levels and an increase in p52 localization to a nucleus-containing fraction (Fig. S3 A). In contrast, p50 localization was unaffected by loss of these genes and was predominantly present in the unprocessed p105 form in the cytoplasm (Fig. S3 A).
If depletion of the cIAP1–TRAF2 complex is sufficient to activate NF-κB, then overexpression of these two proteins should inhibit TWEAK/FN14-induced NF-κB activity. To test this hypothesis, we used FN14-inducible NF-κB EGFP reporter cells in which maximal NF-κB activity was detected in cells that were simultaneously induced for FN14 expression and stimulated with TWEAK ligand and tested the effect of transiently transfecting cIAP1, TRAF2, or both (Fig. 3 D) in this system. Individual expression of either TRAF2 or cIAP1 failed to block TWEAK/FN14-induced NF-κB activation (Fig. 3 D). However, the overexpression of both proteins together significantly reduced the amount of NF-κB activation (Fig. 3 D). Importantly, this was dependent on cIAP1 binding to TRAF2, because coexpression of TRAF2 with the ΔBIR1 cIAP1 mutant that is unable to bind TRAF2 (Fig. S1 E), was unable to inhibit FN14/TWEAK-induced activation of NF-κB (Fig. 3 D). NIK stabilization and p100 processing to p52 could be blocked by pretreatment of cells with NH4Cl but not by pretreatment with CA-074Me (Fig. 3 E). This suggests that relocalization to the lysosomal compartment is sufficient to trigger stabilization of NIK and subsequent processing of p100 rather than degradation in the lysosome per se.
Because TWEAK has been reported to activate the canonical pathway, we also examined the effects of TWEAK and cIAP1 or TRAF2 loss on canonical signaling markers. Consistent with previous observations (Saitoh et al., 2003), we observed TWEAK-induced rapid phosphorylation of IκB and p65. Loss of either cIAP1 or TRAF2 resulted in almost identical responses, with higher basal phosphorylation of IκB and p65 and a significantly delayed TWEAK-induced increase (Fig. 3 F). This highlights that the cIAP1–TRAF2 complex plays an important role in both NF-κB pathways induced by TWEAK.
TWEAK induces cell death through NF-κB–dependent induction of TNFα
Tumor cell lines sensitive to synthetic IAP antagonists are killed through NF-κB–dependent autocrine production of TNFα (Vince et al., 2007). Moreover, it has been described that TWEAK can kill Kym1 cells in a TNFα-dependent manner (Schneider et al., 1999), although how TWEAK stimulated TNFα in Kym1 cells remains unknown. We therefore asked whether TWEAK acted in a similar manner to synthetic IAP antagonists by causing an increase in the abundance of TNFα driven through the activation of NF-κB.
We observed that the levels of TNFα in the cell lysate of TWEAK-treated cell lines increased significantly in all three cell types that are killed by TWEAK treatment alone (Fig. 4, A and B; and Fig 5 A) with a concomitant increase of TNFα released into the media supernatant (Fig. 4 C). In contrast, cell lines that are not killed by TWEAK treatment alone did not produce TNFα when TWEAK was added (unpublished data), suggesting that induction of TNFα is necessary for TWEAK to cause apoptosis.
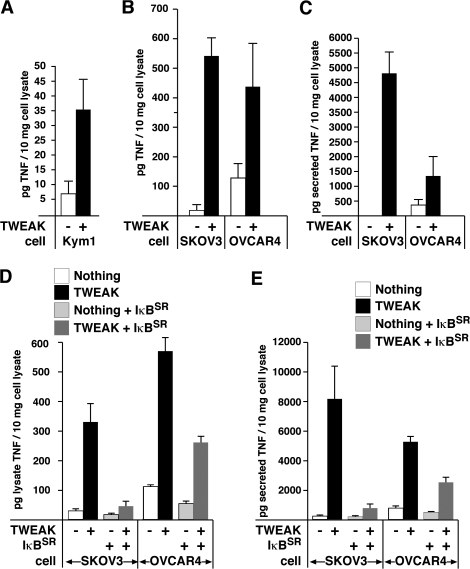
Figure 4.
TWEAK-mediated NF-κB activation induces TNFα in TWEAK-sensitive lines. Kym1 (A) or SKOV3 and OVCAR4 (B) cells were treated with Fc-TWEAK for 8 or 24 h, respectively and the amount of TNFα in cell lysates was measured by ELISA. (C) Supernatant from TWEAK-treated SKOV3 and OVCAR4 cells was collected and filtered and TNFα was measured by ELISA. (D and E) SKOV3 and OVCAR4 cell lines containing inducible IκBSR were induced or not before Fc-TWEAK treatment for 24 h. The levels of TNFα in the cell lysate (D) or cell supernatant (E) was measured by ELISA. Error bars represent SEM from three to five independent experiments.
To test whether activation of NF-κB by TWEAK/FN14 was required for the enhanced TNFα production observed in TWEAK-sensitive cell lines, we created stable inducible nondegradable IκBSR (IκB superrepressor) SKOV3 and OVCAR4 cell lines. Induction of IκBSR inhibited TWEAK-induced NF-κB activity (Fig. S3 A) and significantly reduced the TWEAK-dependent increase in levels of cellular and secreted TNFα in both SKOV3 and OVCAR4 cells (Fig. 4, D and E).
Inhibition of TNFα signaling or caspase 8 blocks TWEAK/FN14 cell death
Previous work with synthetic IAP antagonists (Gaither et al., 2007; Varfolomeev et al., 2007; Vince et al., 2007) and the data presented here with TWEAK demonstrate that either treatment results in an increase in TNFα, which is driven by NF-κB. Remarkably, tumor cell lines that are killed by treatment with a synthetic IAP antagonist alone, such as OVCAR4, SKOV3, and Kym1 cells, (Vince et al., 2007) are also killed by TWEAK.
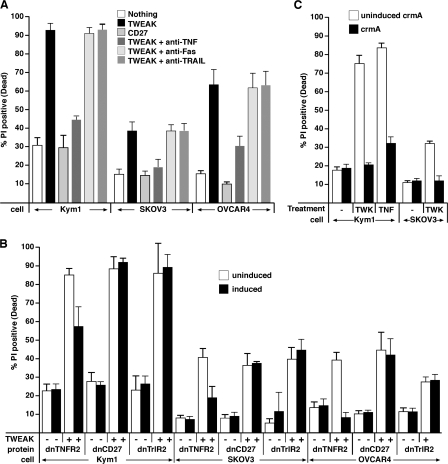
TWEAK killing of sensitive cell lines was prevented by TNFα-blocking antibodies but not by TRAIL- or Fas ligand–neutralizing antibodies in both short-term (Fig. 5 A), and long-term clonogenic survival assays (Fig. S4 A, available at http://www.jcb.org/cgi/content/full/jcb.200801010/DC1), which is consistent with a conserved mechanism of cell death between synthetic IAP antagonist compounds and TWEAK. In addition, expression of the extracellular domain of TNF-R2 fused to a GPI-anchor (dnTNFR2), which is able to sequester and hence neutralize TNFα (Vince et al., 2007), significantly inhibited cell death caused by TWEAK (Fig. 5 B). In contrast, neither dnCD27 nor dnTRAIL-R2 had any protective effect (Fig. 5 B).
Figure 5.
TWEAK-induced cell death is mediated by TNFα. (A) TWEAK-induced death is blocked by neutralizing TNFα antibodies. Kym1, SKOV3, and OVCAR4 cells were incubated with Fc-TWEAK or Fc-CD27 (control) for 24 (Kym1) or 48 (SKOV3 and OVCAR4) h in the absence or presence of 10 μg/ml of neutralizing antibodies against TNFα, FasL, or TRAIL. Cell death was measured by propidium iodide staining and flow cytometry. (B) Dominant-negative (dn) TNF receptor blocks TWEAK-induced cell death. Cells containing inducible dominant-negative GPI-anchored TNF-R2, CD27, or TRAILR2 receptors were induced for 24 h before Fc-TWEAK treatment. Cell death was measured as in A. (C) TWEAK-induced death is blocked by crmA. CrmA-inducible Kym1 or SKOV3 cells were induced for 24 h before Fc-TWEAK or Fc-TNFα treatment for 24 (Kym1) or 48 (SKOV3) h. Cell death was measured as in A. All errors bars represent SEM of at least three independent experiments.
Caspase 8 activity was necessary for TWEAK to induce apoptosis because Kym1 and SKOV3 cell lines inducibly expressing the caspase 8 inhibitor crmA were significantly resistant to TWEAK killing in both short-term (Fig. 5 C) and long-term clonogenic survival assays with Kym1 cells (Fig. S4 B).
To provide a nongenetic test that TWEAK-driven NF-κB was sufficient to kill cells, we used Geldanamycin because it completely blocked TWEAK-induced NF-κB (Fig. S4 C). As has been shown before (Wang et al., 2006), inhibiting the IKK1/2 complex with Geldanamycin is sufficient to sensitize OVCAR4 and wild-type MEFs to TNFα (Fig. S4 E), presumably by blocking NF-κB–induced transcription of prosurvival genes (Wang et al., 2006). Remarkably, however, Geldanamycin was able to block TWEAK-induced NF-κB (Fig. S4, C and D) and TWEAK-induced cell death of Kym1 and OVCAR4 cells (Fig. S4 E). Moreover, although Geldanamycin-treated Kym1 cells showed reduced survival in long-term clonogenic growth assays, cells treated with TWEAK and Geldanamycin still showed clonogenic protection when compared with TWEAK treatment alone (Fig. S4 A).
TWEAK/FN14 signaling sensitizes cells to exogenously supplied TNFα
Although TWEAK kills OVCAR4 and SKOV3 cells through induction of autocrine TNFα, it is known that, like most other cell types, these cells are resistant to TNFα treatment alone (Fig. 6 A). It has been recently shown that removal of cIAP1 by either synthetic IAP antagonists or in gene knockout MEFs sensitizes these cells to TNFα killing (Li et al., 2004; Gaither et al., 2007; Vince et al., 2007). Therefore, we hypothesized that TWEAK not only induces TNFα but also sensitizes cells to TNFα-induced cell death through degradation of the cIAP1–TRAF2 complex in a similar manner to synthetic IAP antagonists, which sensitize tumor cells to TNFα killing by depleting cIAP1, albeit in a mechanistically distinct fashion.
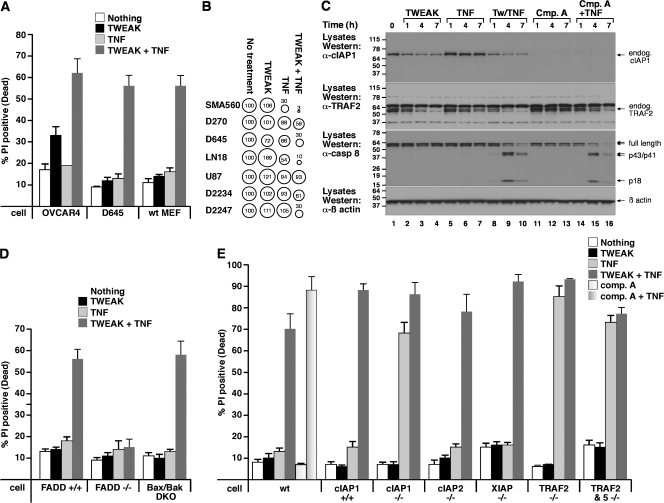
Figure 6.
TWEAK-induced cIAP1–TRAF2 loss sensitizes transformed cells to TNFα-induced death. (A) The indicated cell lines were treated with 100 ng/ml Fc-TWEAK and/or 100 ng/ml Fc-TNFα for 24 h, followed by propidium iodide staining and flow cytometry to measure cell death. (B) TWEAK sensitizes primary human tumor lines D2234 and D2247 to TNFα. Cells from the indicated primary or established glioma cell lines were incubated in vitro with media and 100 ng/ml TNFα, 100 ng/ml TWEAK, or both for 72 h. Survival was assayed with CellTitre Glo and depicted by “bubble” graphs. The areas of the circles and accompanying numbers denote net survival after each treatment, relative to untreated cells (set at 100%). (C) D645 cells were treated with the indicated combinations of Fc-TWEAK, 100 ng/ml Fc-TNFα, or 500 nM compound A for the indicated times and analyzed by Western blot. (D) TWEAK sensitization to TNFα killing requires FADD and is independent of Bax/Bak. The indicated cell lines were treated with 100 ng/ml Fc-TWEAK and/or 100 ng/ml Fc-TNFα. (E) Loss of cIAP1 or TRAF2 sensitizes immortalized MEFs to TNFα-induced death. MEFs were treated for 24 h with 100 ng/ml Fc-TWEAK and/or 100 ng/ml Fc-TNFα. Wild-type MEFs were also treated with 500 nM compound A with or without 100 ng/ml TNFα. Cell death was measured as in A. All errors bars represent SEM of at least three independent experiments. Molecular mass is indicated in kD on the left of the autoradiograph.
To test this hypothesis, exogenous TNFα was applied to TWEAK-sensitive (OVCAR4) and -resistant (D645 and MEF) cell lines alone or in combination with TWEAK for 24 h. Consistent with an additional sensitizing role for TWEAK, OVCAR4 cells were killed by TWEAK/TNFα treatment far more efficiently and rapidly than with TWEAK alone. Even more significantly, D645 and MEF cells (among many other cell types; not depicted) were resistant to treatment with TWEAK or TNFα alone but were extremely sensitive to combined TWEAK/TNFα treatment (Fig. 6, A and B). Even a subset (2/12) of primary human tumor lines was significantly sensitized to TNFα by TWEAK treatment (Fig. 6 B).
TWEAK sensitization to TNFα killing was examined further by Western blot on the TWEAK (and TNFα)-resistant D645 glioma cell line. As in TWEAK-sensitive cell lines, TWEAK treatment reduced cIAP1 and TRAF2 levels, whereas TNFα treatment alone had no effect (Fig. 6 C). Consistent with the lack of cell death (Fig. 6 A), the individual treatments of TWEAK or TNFα did not alter caspase 8 cleavage (Fig. 6 C). In contrast, upon cotreatment of TWEAK with TNFα, processing of caspase 8 into the p43/p41 forms and the active p18 subunit was observed within 3 h (Fig. 6 C) and correlated with the loss of cIAP1–TRAF2 and the rapid death of these cells (Fig. 6 A). To allow a direct comparison with our synthetic IAP antagonist, we also incubated D645 cells with compound A alone or compound A and TNFα. Treatment with compound A alone resulted in the rapid loss of cIAP1 but did not affect either TRAF2 or caspase 8 levels (Fig. 6 C). Significantly, cIAP1 loss alone was sufficient to sensitize D645 cells to TNF to a similar level as that of TWEAK-induced depletion of the cIAP1–TRAF2 complex (Fig. 6 C).
Further evidence supporting the observation that TWEAK/TNFα kill in a death receptor–dependent pathway was obtained using FADD−/− MEFs, as these were completely resistant to TWEAK/TNFα-induced death (Fig. 6 D). In contrast, TWEAK/TNFα killing was independent of the Bax/Bak-dependent apoptotic pathway, as Bax/Bak double knockout MEFs showed a similar TWEAK/TNFα sensitivity to wild-type MEFs (Fig. 6 D).
As expected, cIAP1−/− (Vince et al., 2007), TRAF2−/−, and TRAF2/TRAF5−/− double knockout or compound A–treated MEFs were all extremely sensitive to killing by TNFα alone (Fig. 6 E; Tada et al., 2001), supporting the hypothesis that TWEAK-induced loss of the cIAP1–TRAF2 complex is sufficient to sensitize MEFs to TNFα killing. Surprisingly, cIAP2−/− MEFs were not sensitive to TNFα-mediated cell death (Fig. 6 E), making it unlikely that cIAP2 has a role in TWEAK-mediated sensitization to TNFα.
Pretreating wild-type MEFs with TWEAK for 8 h before addition of TNFα caused a reduction in the total canonical response. However simultaneous treatment with TWEAK/TNFα resulted in an augmented canonical response (Fig. S5 A, available at http://www.jcb.org/cgi/content/full/jcb.200801010/DC1), making it unlikely that a reduction in prosurvival NF-kB signal from TNFα is the reason for TWEAK-induced sensitization to TNFα when the two cytokines are added simultaneously. Consistent with this data, pretreating wild-type MEFs with either TNFα or TWEAK alone for 24 h before cotreatment with TWEAK/TNFα or compound A/TNFα did not change the amount of cell death observed when cells were cotreated for the same time period (Fig. S5 B). This suggests that the prosurvival signals elicited by TNFα, such as NF-κB–induced gene transcription, are not sufficient to counteract TWEAK/TNFα killing.
TWEAK/TNFα treatment distinguishes between normal and transformed cells
Genetic knockout cIAP1 mice display no obvious phenotypic defects in apoptotic signaling (Conze et al., 2005; unpublished data), raising the possibility that primary cells may be less sensitive to TWEAK/TNFα-induced death. Consistent with this possibility, primary MEFs showed only a twofold increase in death after TWEAK/TNFα stimulation, whereas a 14-fold increase was observed in SV40 large T immortalized MEFs (Fig. 7 A). TWEAK-induced loss of cIAP1–TRAF2 was observed in both primary MEFs and transformed MEFs (Fig. 7 B), as was activation of noncanonical NF-κB (Fig. 7 B). Although similar levels of FN14 were initially present in both MEF lines, these increased dramatically after TWEAK stimulation (Fig. 7 B), implying that FN14 expression is regulated by TWEAK.
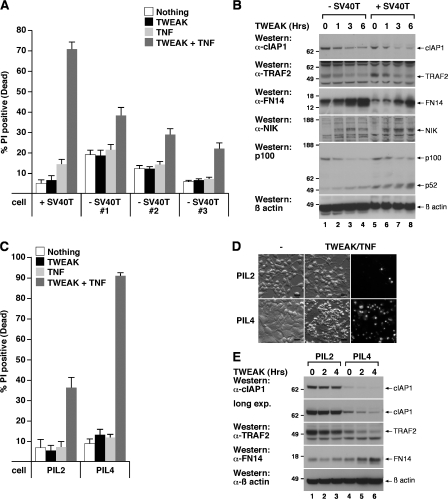
Figure 7.
TWEAK/TNFα treatment distinguishes between normal and transformed cells, and TWEAK/TNFα killing is suppressed by elevated cIAP1–TRAF2 levels. (A) Primary MEFs are resistant to TWEAK/TNFα-induced death. Three primary MEF cell lines derived from three separate embryos were treated with 100 ng/ml Fc-TWEAK and/or 100 ng/ml Fc-TNFα for 24 h, and the amount of cell death was measured by propidium iodide staining and flow cytometry. Error bars are SEM of three to five independent experiments. (B) TWEAK increases FN14 expression and induces cIAP1–TRAF2 degradation in primary and transformed MEFs. Primary MEFs or SV40T transformed MEFs were treated with 100 ng/ml Fc-TWEAK for the indicated times, and DISC lysates were analyzed by Western blot. (C) PIL2 cells or PIL4 cells were treated with 100 ng/ml Fc-TWEAK and/or 100 ng/ml Fc-TNFα for 24 h and cell death was analyzed as in A. Error bars are SEM of three independent experiments. (D) Fluorescence microscopy of propidium iodide–stained cells shown in C. Bars, 20 μm. (E) Enhanced cIAP1 and TRAF2 levels in PIL2 cells are resistant to TWEAK-induced degradation. PIL2 and PIL4 cells were treated with 100 ng/ml Fc-TWEAK for the indicated times and lysates were analyzed by Western blot. Molecular mass is indicated in kD on the left of the autoradiograph.
Because TWEAK-mediated loss of the cIAP1–TRAF2 complex is sufficient to sensitize tumor cells to TNFα-induced death, we tested whether a liver progenitor tumor cell line, PIL2, which expresses high levels of the cIAP1–TRAF2 complex (Fig. 7 E), was resistant to TWEAK/TNFα killing relative to a liver progenitor cell line, PIL4, with lower levels (Fig. 7 E). Treatment of these cells with TWEAK/TNFα killed >90% of PIL4 cells, whereas only 35% of PIL2 (cIAP1 high) cells were killed (Fig. 7, C and D). Western blot analysis showed that PIL2 and PIL4 cells expressed equal levels of FN14 (Fig. 7 E). Significantly, TWEAK-induced degradation of cIAP1–TRAF2, and increased FN14 levels, were attenuated in the PIL2 cells, implying that enhanced expression of cIAP1–TRAF2 inhibits FN14 signaling and counters TWEAK-induced sensitivity to TNFα-induced death.
Discussion
Recent work using synthetic IAP antagonists has shown that cIAPs play a pivotal role in regulating NF-κB signaling from TNF-R1 (Gaither et al., 2007; Varfolomeev et al., 2007; Vince et al., 2007). However, the binding of cIAP1 to TNF receptors other than TNF-R1 or TNF-R2, or the physiological regulation of cIAP1 by TNF receptor signaling, remains poorly characterized. In this paper, we show that when ligated with TWEAK, endogenous FN14 recruits a cIAP1–TRAF2 complex that is subsequently degraded by a cathepsin-mediated lysosomal pathway. TWEAK/FN14 signaling results in both canonical and noncanonical NF-κB activity. Noncanonical NF-κB activity is most probably the result of relocalization of the cIAP1–TRAF2 complex to lysosomes and subsequent degradation. In cell lines that can be killed by TWEAK, NF-κB induces production of TNFα and simultaneously sensitizes tumor cells to TNF-R1–induced death. Transformation of MEFs renders these cells significantly more sensitive to killing by TWEAK/TNFα than their nontransformed progenitors, but tumor cells that express high levels of cIAP1–TRAF2 are comparatively resistant to TWEAK/TNFα killing (Fig. 8).
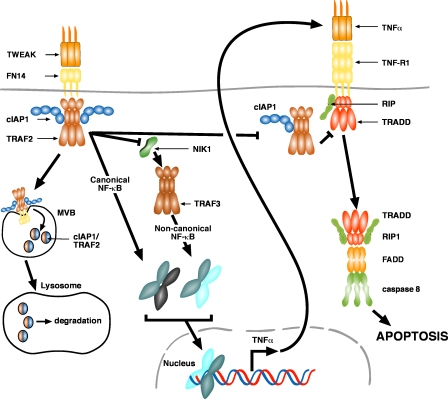
Figure 8.
Model for TWEAK killing. TWEAK binding to FN14 induces the recruitment of a cIAP1–TRAF2 complex. This complex is then targeted for lysosomal degradation by the MVB pathway. The loss of cIAP1 or TRAF2 induces the stabilization of NIK and subsequent activation of noncanonical NF-κB. The canonical NF-κB pathway is also activated in response to TWEAK. Activation of NF-κB drives autocrine TNFα production in TWEAK-sensitive cell lines. The loss of cellular cIAP1 and TRAF2 by FN14 signaling prevents their recruitment to TNF-R1 upon autocrine TNFα binding, and this promotes the formation of a death-inducing complex and apoptotic signaling.
Our data demonstrate that tumor cell sensitivity to TWEAK correlates with their sensitivity to synthetic IAP antagonists and that the mechanism of tumor cell killing between this class of chemical compounds and naturally occurring ligand is remarkably similar. Synthetic IAP antagonists target cIAP1 and cIAP2 for complete proteasomal degradation (Gaither et al., 2007; Varfolomeev et al., 2007; Vince et al., 2007), whereas TWEAK targets a proportion of the cIAP1–TRAF2 complex (presumably the proportion that can be recruited to FN14) for degradation in a lysosomal cathepsin-dependent manner. The end result is, however, the same, with activation of NF-κB. In sensitive tumor cell lines, activated NF-κB drives TNFα production that, in the absence of the cIAP1–TRAF2 complex, kills them. Likewise, both TWEAK and synthetic IAP antagonists sensitize tumor cells to exogenously added TNFα by depleting the cIAP1–TRAF2 complex.
TRAF2-mediated loss by CD40 stimulation requires an intact ubiquitylation pathway (Brown et al., 2001, 2002). Previous work has implicated cIAP1 in the ubiquitylation and proteasomal degradation of TRAF2 by TNF-R2 (Li et al., 2002; Conze et al., 2005; Zhao et al., 2007), and cIAP2 can ubiquitylate and degrade TRAF1 (Lee et al., 2004). In this context, it was completely unexpected that inhibition of the proteasome did not prevent TWEAK/FN14-mediated loss of cIAP1–TRAF2, although TRAF2 depletion required cIAP1 function. However, TWEAK/FN14-induced cIAP1–TRAF2 degradation was prevented by several different classes of inhibitors of lysosome proteases or function and could be specifically blocked by an inhibitor of the lysosome cysteine protease cathepsin B. Consistent with this observation, TWEAK mediated relocalization of TRAF2 to punctate vesicles that often overlapped with, and were in contact with, lysotracker-stained vesicles.
Given previous reports implicating the proteasome in TRAF2 degradation, it remains unclear whether other TNFSF ligands can also stimulate cIAP1–TRAF2 loss through lysosomal mechanisms. However, it has been demonstrated that CD30-induced TRAF2 degradation was blocked by nonproteasomal inhibitors (Duckett and Thompson, 1997). It is likely that lysosomal degradation of the cIAP1–TRAF2 complex occurs by the well documented multivesicular body (MVB) pathway, whereby endocytosed cell surface material is further internalized to form endosomal MVBs, which subsequently fuse with lysosomes (Fig. 8; Williams and Urbé, 2007). The finding that cIAP1 was required for TRAF2 degradation raises the possibility that cIAP1-mediated ubiquitylation of TRAF2/FN14 targets the FN14 complex to the MVB pathway, as has previously been demonstrated for ubiquitin-dependent MVB targeting of several cell surface receptors. Depletion of cIAP1–TRAF2 by TWEAK/FN14 is the most likely cause of noncanonical NF-κB activation. Both cIAP1 and TRAF2 are required to inhibit noncanonical NF-κB activation from TNF receptors, and genetic knockout of either component alone results in spontaneous noncanonical NF-κB activity. Underlining the requirement for their concerted action, overexpression of either cIAP1 or TRAF2 alone does not affect FN14 signaling, but overexpression of both components is able to block TWEAK-induced NF-κB.
The NF-κB activity observed after FN14 signaling resulted in an increase in production of TNFα in TWEAK-sensitive cells but not in TWEAK-resistant cells. TWEAK-resistant cell lines were nevertheless sensitized to exogenously added TNFα, emphasizing that it is the production of autocrine TNFα that determines sensitivity of cells to TWEAK. This finding is consistent with previous observations that activation of TNF-R2, CD30, or CD40 can also induce TNFα and enhance TNF-R1 apoptotic signaling (Grell et al., 1999).
We demonstrated TWEAK binding and FN14 expression on both primary and tumor cells in culture. Surprisingly, however, primary nontransformed MEFs were insensitive to TWEAK/TNFα-induced death, whereas the same MEFs transformed by SV40 Large T were efficiently killed by this treatment. It is not clear why tumor cells show heightened sensitivity to TWEAK/TNFα-induced apoptosis, as TWEAK stimulation of primary MEFs still resulted in the loss of cIAP1–TRAF2 and NF-κB activation.
Not all tumor cell lines were sensitive to TWEAK/TNFα treatment. In particular, PIL2 liver progenitor oval cells that express much higher levels of cIAP1 and TRAF2 than PIL4 cells were resistant to TWEAK/TNFα-induced death. TWEAK stimulation of PIL4 cells resulted in the loss of cIAP1–TRAF2, and these cells were highly sensitive to TWEAK/TNFα killing. In contrast, the increased levels of cIAP1–TRAF2 in the tumorigenic PIL2 cells appeared to block FN14 signaling, as the cIAP1–TRAF2 complex was resistant to TWEAK-induced degradation and the cells were more resistant to TWEAK/TNFα-induced death. These results are also consistent with our findings that the overexpression of cIAP1 and TRAF2 is sufficient to block FN14-induced NF-κB activity.
The fact that the cIAP1-cIAP2 locus is amplified in some human tumors and a mouse model of liver cancer (Zender et al., 2006) and that high levels of expression of cIAP1 have been observed in several cancers argues that increased cIAP1 can contribute to oncogenesis. Our results suggest that one possible mechanism by which it does this is by regulating the balance between life and death signaling from TNFSF receptors. Our findings provide further support for using IAP antagonists as tumor therapy and extend options by highlighting the fact that TWEAK is a physiological regulator of TNFSF signaling that targets the cIAP1–TRAF2 complex rather than the IAPs alone.
Materials and methods
Cell lines, transient transfections, antibodies, and protease inhibitor
SW480, K562, MCF7, BJAB, G401, ColoF, and NIH 3T3 cell lines were a gift from L. O'Reilly (The Walter and Eliza Hall Institute, Melbourne, Australia; O'Reilly et al., 2000, 2002), RM, WW, and A2058 were a gift from P. Hersey (Calvary Mater Newcastle Hospital, Newcastle, Australia; Zhang et al., 1999), and HT29, SKOV3, OVCAR4, and MDAMB231 were purchased from American Type Culture Collection. Kym1 cells were a gift from M. Grell (Institute of Cell Biology and Immunology, Stuttgart, Germany; Grell et al., 1999). D2234 and D2247 early passage lines were derived from specimens obtained from patients who had undergone tumor resection at Duke University Hospital (Durham, NC; Ashley et al., 2008). Transient transfections (typically using 1 μg of plasmid DNA per 10-cm plate of cells) were performed with effectene as described by the manufacturer (QIAGEN). Antibodies used in this study for flow cytometry were anti-FN14 (Abcam), Goat anti–mouse RPE (Millipore), and Goat anti–human RPE (SouthernBiotech). Antibodies used for Western blots were Goat anti–human IgG HRP (Jackson ImmunoResearch Laboratories), anti-cIAP1 (in house), anti-TRAF2 (Santa Cruz Biotechnology, Inc.), anti-FN14 (Cell Signaling Technology), anti-NIK (Cell Signaling Technology), anti–phospho–NF-κB (Ser536) p65 (Cell Signaling Technology), anti-IκB, (Cell Signaling Technology), anti–phospho-IκB (Ser32/36; Cell Signaling Technology), anti–NF-κB p65 (Santa Cruz), anti–NF-κB2 (Cell Signaling Technology), anti–NF-κB p50 (Santa Cruz), anti–β-actin (Sigma-Aldrich), anti-ubiquitin (Cell Signaling Technology), anti-TRAF3 (BD Biosciences), anti–FLAG M2 (Sigma-Aldrich), anti–IgG biotin (Jackson ImmunoResearch Laboratories), and anti-VSV (MBL International). Protease inhibitor cocktail final concentrations were the following: AEBSF, 1.3 mM; aprotinin 1.1 μM; bestatin, 66 μM; E-64, 20 μM; leupeptin, 27 μM; and pepstatin A, 13 μM.
Cell culture and lentivirus production
All cell lines were maintained in DME supplemented with 10% FCS, 2 mM l-glutamine, and penicillin/streptomycin and grown at 37°C in 10% CO2. PIL2, PIL4, and BMOL liver progenitor cells were maintained in Williams Media E supplemented with 2 mM l-glutamine, penicillin/streptomycin, 20 ng/ml mouse EGF, 30 ng/ml human IGF II, and 0.25 U/ml human insulin and grown at 37°C in 5% CO2.
To generate lentiviral particles, 293T cells were transfected with packaging constructs pCMV ðR8.2, VSVg, and the relevant lentiviral plasmid in the ratio of 1:0.4:0.6. After 24–48 h, the virus-containing supernatants were harvested and filtered. 12 μg/ml Polybrene was added and target cells were infected with virus supernatant for 24–48 h. The media was subsequently changed and successful infection selected for with 2–5 μg/ml puromycin (pF 5xUAS selection) or 100–500 μg/ml hygromycin B (GEV16 selection) or by screening for GFP fluorescence (pTRH). pF 5xUAS-inducible constructs were induced with 100 nM 4-hydroxy tamoxifen for 16 h before harvesting lysates for Western blotting or before death assays. Flp In T-Rex 293 cells (Invitrogen) containing doxycycline-inducible VSV-tagged FN14 were generated according to the manufacturer's instructions.
Constructs
The NF-κB lentiviral reporter vector pTRH1 mCMV NF-κB dscGFP was purchased from System Biosciences. Cre recombinase and SV40 Large T antigen were cloned into the lentiviral vector pFU. In the inducible lentiviral system, the inducible transcriptional activator Gal4 1–147 ERT2 VP16 (GEV16) was cloned into pFU PGK Hygro, and the genes dnTNF-R2, dnTRAIL-R2 or dnCD27 (Bossen et al., 2006), IκBSR super repressor (Van Antwerp et al., 1996), and N-Flag crmA, were cloned into a pF 5xUAS SV40 Puro vector. D. Baltimore (California Institute of Technology, Pasadena, CA) provided us with pFU and lentiviral packaging constructs and T. Mantamadiotis (Monash University, Melbourne, Australia) provided the ERT2 construct. Fc-TNFSF ligand DNA was provided by J. Tschopp (University of Lausanne, Lausanne, Switzerland). Complete sequence of all constructs can be obtained upon request.
Generation of MEFs
Knockout MEFs were generated from embryonic day–15 embryos from wild-type and XIAP−/− mice using standard procedures and infected with SV40 Large T antigen-expressing lentivirus. cIAP1, cIAP2, or TRAF2 conditional knockout MEFs were similarly generated from cIAP1 LoxP/LoxP or TRAF2 LoxP/LoxP embryonic day–15 embryos. To delete cIAP1 or TRAF2, the transformed MEFs were infected with a cre-expressing lentivirus (pFU cre SV40 puro), and deletion was confirmed by PCR and Western blotting. To delete cIAP2, the transformed MEFs were infected with a FlpE-expressing lentivirus (pFU FlpE PGK Hygro). W.-C. Yeh (University of Toronto, Toronto, Canada) provided FADD knockout MEFs, H. Nakano (Juntendo University, Tokyo, Japan) provided TRAF2/TRAF5 double knockout MEFs, and D. Huang (The Walter and Eliza Hall Institute, Melbourne, Australia) provided Bax/Bak double knockout MEFs.
Immunoflow cytometry
Approximately 3 × 106 cells were harvested, washed in PBS, and resuspended in buffer (KDS/BSS containing 3% FCS) on ice. 500 ng Fc-TWEAK or 1 μg FN14 antibody was incubated with cells on ice for 20 min, and then the cells were washed and incubated with anti–human IgG biotin for 20 min on ice, followed by Streptavidin Tri-color biotin (for Fc-TWEAK staining) or anti–mouse IgG-RPE (for anti-FN14 staining). Cells were washed and then analyzed by flow cytometry.
Death assays
Cells were seeded on 12-well tissue culture plates at ∼40% confluency and were allowed to adhere for 16–20 h. Compound A (5 nM Kym1 cells or 500 nM of all other cell types), 70 ng/ml of human Fc-TNFα, or 100 ng/ml of human Fc-TWEAK were added to cells for 24 or 48 h, and cell death was measured by propidium iodide staining and flow cytometry. In each sample, 10,000 events were measured and the cell death (percentage of propidium iodide-positive cells) was quantified.
Western blotting and immunoprecipitations
For immunoprecipitation of endogenous FN14, cells were grown on 15-cm tissue culture plates and, when approaching confluency, were harvested and resuspended in 800 μl of ice-cold DME. 1.6 μg Fc-TWEAK was added for 30 min on ice (or indicated times) at 37°C. Cells were subsequently washed in ice-cold PBS and lysed in DISC buffer (1% Triton X-100, 10% glycerol, 150 mM NaCl, 20 mM Tris, pH 7.5, 2 mM EDTA, and complete protease inhibitor cocktail [Roche]) on ice. Cell lysate was spun at 14,000 g for 10 min, and the soluble material was precleared with Sepharose 6B beads at 4°C for 1 h. Fc-TWEAK–bound material was immunoprecipitated by adding the precleared lysate to EZI view Protein A–agarose (Sigma-Aldrich) for 1–2 h at 4°C. Samples were washed four times with DISC buffer and then eluted with 1% SDS and β-mercaptoethanol at 95°C for 3 min. Samples were separated on 4–20 (Bio-Rad Laboratories) or 4–12% (Invitrogen) polyacrylamide gels and transferred to nitrocellulose membranes for antibody detection. All membrane-blocking steps and antibody dilutions were performed with 5% skim milk in PTBS (PBS containing 0.1% Tween 20), and washing steps were performed with PTBS. Proteins on Western blots were visualized by ECL (GE Healthcare) after incubation of membranes with HRP-coupled secondary antibodies.
Immunoprecipitation of FLAG-tagged cIAP1 constructs or VSV-tagged FN14 were performed similarly, except one 10-cm plate of cells was used per immunoprecipitation, and anti–FLAG M2–conjugated agarose (Sigma-Aldrich) or anti–VSV-conjugated agarose (Sigma-Aldrich) were used for immunoprecipitation of the relevant proteins.
Nuclear fractionation
Cells were harvested, washed in PBS, and resuspended in lysis buffer (10 mM Hepes-KOH, pH 7.9, 10 mM KCl, 1.5 mM MgCl, 0.5 mM DTT, and protease inhibitor cocktail) and incubated on ice for 15 min. The lysis buffer was adjusted to 0.6% NP-40 and immediately vortexed for 10 s, and the pellet (membrane) and supernatant (cytosol) fractions were separated by centrifugation at 14,500 g for 5 min. Equal amounts of membrane (nuclear) and cytosolic fractions were analyzed by SDS-PAGE and Western blotting.
ELISA assays
Cells were grown on 10-cm plates, harvested, washed thoroughly with ice-cold PBS, and lysed in 300 μl DISC buffer for 20 min on ice. Cell lysate was spun for 10 min at 14000 g, and the soluble material was collected. Alternatively, the cell supernatant from the same plates was collected and filtered to remove cellular debris. Soluble cell lysate or the filtered cell supernatant was used for human or mouse TNF-α ELISA assays (R&D Systems) according to the manufacturer's protocol. Protein from the cell lysate was quantified using the BCA assay (Thermo Fisher Scientific).
Immunofluorescence, image acquisition, and processing
D645 cells grown on glass coverslips were fixed with 3.2% PFA for 20 min, washed in PBS, and permeabilized with 0.5% Triton X-100 for 5 min. Cells were blocked, incubated with primary antibody, washed four times with PBS, and then incubated with anti–rat or anti–mouse Alexa Fluor 488–conjugated secondary antibody (Invitrogen) and washed four times again. All blocking steps and antibody incubations were performed with PBS containing 1% BSA for 30 min. The primary antibodies used were anti-FLAG (Amrad) or anti-TRAF2 (BD Biosciences). Cells were viewed on an inverted confocal microscope (TCS-SP2; Leica) using a 63× 1.4 NA oil immersion objective at room temperature. Images were collected and analyzed with SP2 imaging software (Leica) or ImageJ software (National Institutes of Health; http://rsb.info.nih.gov/ij). All images were in TIF format and imported into Freehand MX (Macromedia) for the compilation of figures.
Online supplemental material
Fig. S1 shows that Fc-TWEAK binds to a large selection of adherent transformed cell lines. Fig. S2 shows analysis of TWEAK-induced cIAP1–TRAF2 degradation. Fig. S3 shows that oss of cIAP1 or TRAF2 results in constitutive activation of noncanonical NF-κB pathway and that an IκB superrepressor blocks TWEAK-induced NF-κB activity. Fig. S4 shows that inhibition of TNFα signaling or NF-κB activity provides clonogenic protection to TWEAK-treated cells. Fig. S5 shows the response of wild-type and knockout MEFs to TWEAK and TNFα treatment. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200801010/DC1.
Supplementary Material
[Supplemental Material Index]
Acknowledgments
We thank Jurg Tschopp for generous provision of materials and advice, Wen-Chen Yeh for FADD knockout MEFs, Hiroyasu Nakano for TRAF2/TRAF5 double knockout MEFs, David Baltimore for pFU and lentiviral packaging constructs, Theo Mantamadiotis for the ERT2 construct, Robert Gerl for early work with the GAL4 ERT2 VP16 system, David Huang for Bax/Bak double knockout MEFs, and an anonymous reviewer for their suggestions.
J. Silke is supported by NHMRC grants (433013 and 356256). D.L. Vaux is an Australian Fellow, funded by the Leukemia and Lymphoma Society and an NHMRC grant (461221). M. McKinlay, C.A. Benetatos, S.M. Condon, and SK. Chunduru are employees of TetraLogic Pharmaceuticals, J. Silke is a consultant, and D.L. Vaux is on the scientific advisory board.
Abbreviations used in this paper: cIAP1, cellular inhibitor of apoptosis 1; DD, death domain; MEF, mouse embryonic fibroblast; MVB, multivesicular body; TNFRSF, TNF receptor superfamily; TRAF, Tnf receptor-associated factor; TRAIL, TNF-related apoptosis-inducing ligand; TWEAK, TNF-like weak inducer of apoptosis; VSV, vesicular stomatitis virus.
References
- Annunziata, C.M., R.E. Davis, Y. Demchenko, W. Bellamy, A. Gabrea, F. Zhan, G. Lenz, I. Hanamura, G. Wright, W. Xiao, et al. 2007. Frequent engagement of the classical and alternative NF-κB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 12:115–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley, D.M., C.D. Riffkin, M.M. Lovric, A. Dobrovic, J.A. Maxwell, H.S. Friedman, T. Mikeska, K.J. Drummond, A.H. Kaye, H.K. Gan, et al. 2008. In vitro sensitivity testing of minimally passaged and uncultured gliomas with TRAIL and/or chemotherapy drugs. Br. J. Cancer. In press. [DOI] [PMC free article] [PubMed]
- Bossen, C., K. Ingold, A. Tardivel, J.L. Bodmer, O. Gaide, S. Hertig, C. Ambrose, J. Tschopp, and P. Schneider. 2006. Interactions of tumor necrosis factor (TNF) and TNF receptor family members in the mouse and human. J. Biol. Chem. 281:13964–13971. [DOI] [PubMed] [Google Scholar]
- Bover, L.C., M. Cardó-Vila, A. Kuniyasu, J. Sun, R. Rangel, M. Takeya, B.B. Aggarwal, W. Arap, and R. Pasqualini. 2007. A previously unrecognized protein-protein interaction between TWEAK and CD163: potential biological implications. J. Immunol. 178:8183–8194. [DOI] [PubMed] [Google Scholar]
- Brown, K.D., B.S. Hostager, and G.A. Bishop. 2001. Differential signaling and tumor necrosis factor receptor-associated factor (TRAF) degradation mediated by CD40 and the Epstein-Barr virus oncoprotein latent membrane protein 1 (LMP1). J. Exp. Med. 193:943–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, K.D., B.S. Hostager, and G.A. Bishop. 2002. Regulation of TRAF2 signaling by self-induced degradation. J. Biol. Chem. 277:19433–19438. [DOI] [PubMed] [Google Scholar]
- Brown, S.A., C.M. Richards, H.N. Hanscom, S.L. Feng, and J.A. Winkles. 2003. The Fn14 cytoplasmic tail binds TRAFs 1, 2, 3 and 5 and mediates NF-κB activation. Biochem. J. 371:395–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, F., D. Bhatia, Q. Chang, and V. Castranova. 2006. Finding NEMO by K63-linked polyubiquitin chain. Cell Death Differ. 13:1835–1838. [DOI] [PubMed] [Google Scholar]
- Conze, D.B., L. Albert, D.A. Ferrick, D.V. Goeddel, W.C. Yeh, T. Mak, and J.D. Ashwell. 2005. Posttranscriptional downregulation of c-IAP2 by the ubiquitin protein ligase c-IAP1 in vivo. Mol. Cell. Biol. 25:3348–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveraux, Q.L., R. Takahashi, G.S. Salvesen, and J.C. Reed. 1997. X-linked IAP is a direct inhibitor of cell-death proteases. Nature. 388:300–304. [DOI] [PubMed] [Google Scholar]
- Duckett, C.S., and C.B. Thompson. 1997. CD30-dependent degradation of TRAF2 - implications for negative regulation of TRAF signaling and the control of cell survival. Genes Dev. 11:2810–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckett, C.S., V.E. Nava, R.W. Gedrich, R.J. Clem, J.L. Vandongen, M.C. Gilfillan, H. Shiels, J.M. Hardwick, and C.B. Thompson. 1996. A conserved family of cellular genes related to the baculovirus iap gene and encoding apoptosis inhibitors. EMBO J. 15:2685–2694. [PMC free article] [PubMed] [Google Scholar]
- Eckelman, B.P., and G.S. Salvesen. 2006. The human anti-apoptotic proteins cIAP1 and cIAP2 bind but do not inhibit caspases. J. Biol. Chem. 281:3254–3260. [DOI] [PubMed] [Google Scholar]
- Felli, N., F. Pedini, A. Zeuner, E. Petrucci, U. Testa, C. Conticello, M. Biffoni, A. Di Cataldo, J.A. Winkles, C. Peschle, and R. De Maria. 2005. Multiple members of the TNF superfamily contribute to IFN-gamma-mediated inhibition of erythropoiesis. J. Immunol. 175:1464–1472. [DOI] [PubMed] [Google Scholar]
- Gaither, A., D. Porter, Y. Yao, J. Borawski, G. Yang, J. Donovan, D. Sage, J. Slisz, M. Tran, C. Straub, et al. 2007. A Smac mimetic rescue screen reveals roles for inhibitor of apoptosis proteins in tumor necrosis factor-alpha signaling. Cancer Res. 67:11493–11498. [DOI] [PubMed] [Google Scholar]
- Grech, A.P., M. Amesbury, T. Chan, S. Gardam, A. Basten, and R. Brink. 2004. TRAF2 differentially regulates the canonical and noncanonical pathways of NF-κB activation in mature B cells. Immunity. 21:629–642. [DOI] [PubMed] [Google Scholar]
- Grell, M., G. Zimmermann, E. Gottfried, C.M. Chen, U. Grunwald, D.C. Huang, Y.H. Wu Lee, H. Durkop, H. Engelmann, P. Scheurich, et al. 1999. Induction of cell death by tumour necrosis factor (TNF) receptor 2, CD40 and CD30: a role for TNF-R1 activation by endogenous membrane-anchored TNF. EMBO J. 18:3034–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habelhah, H., S. Takahashi, S.G. Cho, T. Kadoya, T. Watanabe, and Z. Ronai. 2004. Ubiquitination and translocation of TRAF2 is required for activation of JNK but not of p38 or NF-κB. EMBO J. 23:322–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holler, N., A. Tardivel, M. Kovacsovics-Bankowski, S. Hertig, O. Gaide, F. Martinon, A. Tinel, D. Deperthes, S. Calderara, T. Schulthess, et al. 2003. Two adjacent trimeric Fas ligands are required for Fas signaling and formation of a death-inducing signaling complex. Mol. Cell. Biol. 23:1428–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keats, J.J., R. Fonseca, M. Chesi, R. Schop, A. Baker, W.J. Chng, S. Van Wier, R. Tiedemann, C.X. Shi, M. Sebag, et al. 2007. Promiscuous mutations activate the noncanonical NF-κB pathway in multiple myeloma. Cancer Cell. 12:131–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.S., U.S. Hong, T.H. Lee, S.K. Yoon, and J.B. Yoon. 2004. Mass spectrometric analysis of TRAF1 ubiquitination mediated by cIAP2. Proteomics. 4:3376–3382. [DOI] [PubMed] [Google Scholar]
- Lee, S.Y., A. Reichlin, A. Santana, K.A. Sokol, M.C. Nussenzweig, and Y. Choi. 1997. Traf2 is essential for jnk but not NF-κB activation and regulates lymphocyte proliferation and survival. Immunity. 7:703–713. [DOI] [PubMed] [Google Scholar]
- Li, L., R.M. Thomas, H. Suzuki, J.K. De Brabander, X. Wang, and P.G. Harran. 2004. A small molecule Smac mimic potentiates TRAIL- and TNFα-mediated cell death. Science. 305:1471–1474. [DOI] [PubMed] [Google Scholar]
- Li, X., Y. Yang, and J.D. Ashwell. 2002. TNF-RII and c-IAP1 mediate ubiquitination and degradation of TRAF2. Nature. 416:345–347. [DOI] [PubMed] [Google Scholar]
- Listen, P., N. Roy, K. Tamai, C. Lefebvre, S. Baird, G. Chertonhorvat, R. Farahani, M. Mclean, J.E. Ikeda, A. Mackenzie, and R.G. Korneluk. 1996. Suppression of apoptosis in mammalian cells by naip and a related family of iap genes. Nature. 379:349–353. [DOI] [PubMed] [Google Scholar]
- Maecker, H., E. Varfolomeev, F. Kischkel, D. Lawrence, H. LeBlanc, W. Lee, S. Hurst, D. Danilenko, J. Li, E. Filvaroff, et al. 2005. TWEAK attenuates the transition from innate to adaptive immunity. Cell. 123:931–944. [DOI] [PubMed] [Google Scholar]
- Micheau, O., and J. Tschopp. 2003. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 114:181–190. [DOI] [PubMed] [Google Scholar]
- O'Reilly, L.A., L. Cullen, J. Visvader, G.J. Lindeman, C. Print, M.L. Bath, D.C. Huang, and A. Strasser. 2000. The proapoptotic BH3-only protein bim is expressed in hematopoietic, epithelial, neuronal, and germ cells. Am. J. Pathol. 157:449–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly, L.A., P. Ekert, N. Harvey, V. Marsden, L. Cullen, D.L. Vaux, G. Hacker, C. Magnusson, M. Pakusch, F. Cecconi, et al. 2002. Caspase-2 is not required for thymocyte or neuronal apoptosis even though cleavage of caspase-2 is dependent on both Apaf-1 and caspase-9. Cell Death Differ. 9:832–841. [DOI] [PubMed] [Google Scholar]
- Park, S.M., J.B. Yoon, and T.H. Lee. 2004. Receptor interacting protein is ubiquitinated by cellular inhibitor of apoptosis proteins (c-IAP1 and c-IAP2) in vitro. FEBS Lett. 566:151–156. [DOI] [PubMed] [Google Scholar]
- Park, Y.C., V. Burkitt, A.R. Villa, L. Tong, and H. Wu. 1999. Structural basis for self-association and receptor recognition of human TRAF2. Nature. 398:533–538. [DOI] [PubMed] [Google Scholar]
- Polek, T.C., M. Talpaz, B.G. Darnay, and T. Spivak-Kroizman. 2003. TWEAK mediates signal transduction and differentiation of RAW264.7 cells in the absence of Fn14/TweakR. Evidence for a second TWEAK receptor. J. Biol. Chem. 278:32317–32323. [DOI] [PubMed] [Google Scholar]
- Rothe, M., S.C. Wong, W.J. Henzel, and D.V. Goeddel. 1994. A novel family of putative signal transducers associated with the cytoplasmic domain of the 75 kda tumor necrosis factor receptor. Cell. 78:681–692. [DOI] [PubMed] [Google Scholar]
- Rothe, M., M.G. Pan, W.J. Henzel, T.M. Ayres, and D.V. Goeddel. 1995. The TNF-R2-TRAF signaling complex contains two novel proteins related to baculoviral-inhibitor of apoptosis proteins. Cell. 83:1243–1252. [DOI] [PubMed] [Google Scholar]
- Saitoh, T., M. Nakayama, H. Nakano, H. Yagita, N. Yamamoto, and S. Yamaoka. 2003. TWEAK induces NF-κB2 p100 processing and long lasting NF-κB activation. J. Biol. Chem. 278:36005–36012. [DOI] [PubMed] [Google Scholar]
- Samuel, T., K. Welsh, T. Lober, S.H. Togo, J.M. Zapata, and J.C. Reed. 2006. Distinct BIR domains of cIAP1 mediate binding to and ubiquitination of TRAF2 and Smac. J. Biol. Chem. 281:1080–1090. [DOI] [PubMed] [Google Scholar]
- Schneider, P., R. Schwenzer, E. Haas, F. Muhlenbeck, G. Schubert, P. Scheurich, J. Tschopp, and H. Wajant. 1999. TWEAK can induce cell death via endogenous TNF and TNF receptor 1. Eur. J. Immunol. 29:1785–1792. [DOI] [PubMed] [Google Scholar]
- Srinivasula, S.M., R. Hegde, A. Saleh, P. Datta, E. Shiozaki, J. Chai, R.A. Lee, P.D. Robbins, T. Fernandes-Alnemri, Y. Shi, and E.S. Alnemri. 2001. A conserved XIAP-interaction motif in caspase-9 and Smac/DIABLO regulates caspase activity and apoptosis. Nature. 410:112–116. [DOI] [PubMed] [Google Scholar]
- Tada, K., T. Okazaki, S. Sakon, T. Kobarai, K. Kurosawa, S. Yamaoka, H. Hashimoto, T.W. Mak, H. Yagita, K. Okumura, et al. 2001. Critical roles of TRAF2 and TRAF5 in tumor necrosis factor-induced NF-κB activation and protection from cell death. J. Biol. Chem. 276:36530–36534. [DOI] [PubMed] [Google Scholar]
- Tartaglia, L.A., T.M. Ayres, G.H. Wong, and D.V. Goeddel. 1993. A novel domain within the 55 kd TNF receptor signals cell death. Cell. 74:845–853. [DOI] [PubMed] [Google Scholar]
- Tenev, T., A. Zachariou, R. Wilson, M. Ditzel, and P. Meier. 2005. IAPs are functionally non-equivalent and regulate effector caspases through distinct mechanisms. Nat. Cell Biol. 7:70–77. [DOI] [PubMed] [Google Scholar]
- Uren, A.G., M. Pakusch, C.J. Hawkins, K.L. Puls, and D.L. Vaux. 1996. Cloning and expression of apoptosis inhibitory protein homologs that function to inhibit apoptosis and/or bind TRAFs. Proc. Natl. Acad. Sci. USA. 93:4974–4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Antwerp, D.J., S.J. Martin, T. Kafri, D.R. Green, and I.M. Verma. 1996. Suppression of TNFα-induced apoptosis by NF-κB. Science. 274:787–789. [DOI] [PubMed] [Google Scholar]
- Varfolomeev, E., S.M. Wayson, V.M. Dixit, W.J. Fairbrother, and D. Vucic. 2006. The inhibitor of apoptosis protein fusion c-IAP2.MALT1 stimulates NF-κB activation independently of TRAF1 AND TRAF2. J. Biol. Chem. 281:29022–29029. [DOI] [PubMed] [Google Scholar]
- Varfolomeev, E., J.W. Blankenship, S.M. Wayson, A.V. Fedorova, N. Kayagaki, P. Garg, K. Zobel, J.N. Dynek, L.O. Elliott, H.J. Wallweber, et al. 2007. IAP antagonists induce autoubiquitination of c-IAPs, NF-κB activation, and TNFalpha-dependent apoptosis. Cell. 131:669–681. [DOI] [PubMed] [Google Scholar]
- Vince, J.E., and J. Silke. 2006. TWEAK shall inherit the earth. Cell Death Differ. 13:1842–1844. [DOI] [PubMed] [Google Scholar]
- Vince, J.E., W.W. Wong, N. Khan, R. Feltham, D. Chau, A.U. Ahmed, C.A. Benetatos, S.K. Chunduru, S.M. Condon, M. McKinlay, et al. 2007. IAP antagonists target cIAP1 to induce TNFα-dependent apoptosis. Cell. 131:682–693. [DOI] [PubMed] [Google Scholar]
- Wang, X., W. Ju, J. Renouard, J. Aden, S.A. Belinsky, and Y. Lin. 2006. 17-allylamino-17-demethoxygeldanamycin synergistically potentiates tumor necrosis factor-induced lung cancer cell death by blocking the NF-κB pathway. Cancer Res. 66:1089–1095. [DOI] [PubMed] [Google Scholar]
- Williams, R.L., and S. Urbé. 2007. The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell Biol. 8:355–368. [DOI] [PubMed] [Google Scholar]
- Winkles, J.A., N.L. Tran, S.A. Brown, N. Stains, H.E. Cunliffe, and M.E. Berens. 2007. Role of TWEAK and Fn14 in tumor biology. Front. Biosci. 12:2761–2771. [DOI] [PubMed] [Google Scholar]
- Wu, C.J., D.B. Conze, X. Li, S.X. Ying, J.A. Hanover, and J.D. Ashwell. 2005. TNF-alpha induced c-IAP1/TRAF2 complex translocation to a Ubc6-containing compartment and TRAF2 ubiquitination. EMBO J. 24:1886–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, H., Y.C. Park, M. Kreishman, E. Kieff, and H. Wu. 1999. The structural basis for the recognition of diverse receptor sequences by TRAF2. Mol. Cell. 4:321–330. [DOI] [PubMed] [Google Scholar]
- Yeh, W.C., A. Shahinian, D. Speiser, J. Kraunus, F. Billia, A. Wakeham, J.L. Delapompa, D. Ferrick, B. Hum, N. Iscove, et al. 1997. Early lethality, functional NF-κB activation, and increased sensitivity to TNF-induced cell death in TRAF2-deficient mice. Immunity. 7:715–725. [DOI] [PubMed] [Google Scholar]
- Zender, L., M.S. Spector, W. Xue, P. Flemming, C. Cordon-Cardo, J. Silke, S.T. Fan, J.M. Luk, M. Wigler, G.J. Hannon, et al. 2006. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 125:1253–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X.D., A. Franco, K. Myers, C. Gray, T. Nguyen, and P. Hersey. 1999. Relation of TNF-related apoptosis-inducing ligand (TRAIL) receptor and FLICE-inhibitory protein expression to TRAIL-induced apoptosis of melanoma. Cancer Res. 59:2747–2753. [PubMed] [Google Scholar]
- Zhao, Y., D.B. Conze, J.A. Hanover, and J.D. Ashwell. 2007. Tumor necrosis factor receptor 2 signaling induces selective c-IAP1-dependent ASK1 ubiquitination and terminates mitogen-activated protein kinase signaling. J. Biol. Chem. 282:7777–7782. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Material Index]