Tracking molecular evolution of photosynthesis by characterization of a major photosynthesis gene cluster from Heliobacillus mobilis (original) (raw)
Abstract
A DNA sequence has been obtained for a 35.6-kb genomic segment from Heliobacillus mobilis that contains a major cluster of photosynthesis genes. A total of 30 ORFs were identified, 20 of which encode enzymes for bacteriochlorophyll and carotenoid biosynthesis, reaction-center (RC) apoprotein, and cytochromes for cyclic electron transport. Donor side electron-transfer components to the RC include a putative RC-associated cytochrome _c_553 and a unique four-large-subunit cytochrome bc complex consisting of Rieske Fe-S protein (encoded by petC), cytochrome b6 (petB), subunit IV (petD), and a diheme cytochrome c (petX). Phylogenetic analysis of various photosynthesis gene products indicates a consistent grouping of oxygenic lineages that are distinct and descendent from anoxygenic lineages. In addition, H. mobilis was placed as the closest relative to cyanobacteria, which form a monophyletic origin to chloroplast-based photosynthetic lineages. The consensus of the photosynthesis gene trees also indicates that purple bacteria are the earliest emerging photosynthetic lineage. Our analysis also indicates that an ancient gene-duplication event giving rise to the paralogous bchI and bchD genes predates the divergence of all photosynthetic groups. In addition, our analysis of gene duplication of the photosystem I and photosystem II core polypeptides supports a “heterologous fusion model” for the origin and evolution of oxygenic photosynthesis.
Keywords: bacteriochlorophyll biosynthesis/cytochrome bc complex/phylogenetic analysis/photosystem origin
Bacterial photosynthesis is found in five eubacterial phyla: cyanobacteria, purple bacteria, heliobacteria, green sulfur bacteria, and green nonsulfur bacteria. Analysis of photosystems from purple and green nonsulfur bacteria shows that these organisms synthesize primitive photosystems that are ancestral to the more complex oxygen-evolving photosystem II (PSII) from cyanobacteria and chloroplasts (1). Additional studies of heliobacteria and green sulfur bacteria indicate that they synthesize photosystems that are ancestral to the photosystem I (PSI) complex from cyanobacteria and chloroplasts (2).
With the exception of the sequence analysis of PSI and PSII structural polypeptides, there is little information on the origin and evolution of photosynthesis. Complete sequence information on photosynthesis genes is only available for the purple nonsulfur bacterium, Rhodobacter capsulatus, which has a major clustering of photosynthesis genes (3), and for the cyanobacterium Synechocystis sp. PCC 6803, for which sequence analysis has been completed for the entire chromosome. Information on photosynthesis genes from the three other eubacterial photosynthetic phyla is limited mainly to the reaction-center (RC) core polypeptides and a few cytochromes.
With the aim of providing more substantive information on the evolution of photosynthesis, we have undertaken an extensive characterization of photosynthesis genes from Heliobacillus mobilis. A gene cluster was found to encode enzymes for bacteriochlorophyll and carotenoid biosynthesis, as well as the RC core polypeptide and cytochromes involved in photosynthetic electron transfer. Phylogenetic studies based on this new sequence information provide important clues for the unique evolutionary position of H. mobilis in the course of development of photosynthesis.
MATERIALS AND METHODS
DNA Sequencing.
A genomic library was constructed by partial digestion of H. mobilis (ATCC 43427) genomic DNA with _Sau_3A I, followed by ligating the DNA fragments into the _Bam_HI site of the cosmid vector pJRD215 (4). Cosmid containing transformants were pooled and used for complementation of bacteriochlorophyll-biosynthesis mutants of R. capsulatus through bipartite matings as described (5). One cosmid, pHM6, which complemented a bchB mutant (6), was chosen for sequencing analysis. Additional DNA sequences flanking the insert in pHM6 were obtained by the inverse PCR method as described (7). DNA sequencing was performed with a fluorescent dye-labeled dideoxynucleotide sequencing reaction kit (Amersham) with an Applied Biosystems DNA sequencer (model 377, Perkin—Elmer). Oligonucleotide primers were synthesized by Operon Technologies (Alameda, CA) and GIBCO/BRL. All of the DNA sequences reported in this study were determined completely on both strands.
Phylogenetic Analysis.
The nucleotide sequence was analyzed with sequencher software (version 3.0, Gene Codes, Ann Arbor, MI). Database searches were performed by using the Basic Local Alignment Search Tool (blast; ref. 8). Sequence compilation and analysis of translated ORFs were performed with the aid of the Genetics Computer Group’s sequence-analysis package (version 9.0, Madison, WI). Multiple sequence alignments of translated gene sequences were carried out with the computer program clustal w (version 1.7; ref. 9). Phylogenetic trees were generated by the neighbor-joining method with the clustal w program (with 100 bootstrap replicates), by the maximum-parsimony method (with 100 branch-bound replicates) with the paup program (version 3.1.1; ref. 10), and by the maximum-likelihood method (1,000 replicates) with the puzzle programs (version 3.1).
RESULTS AND DISCUSSION
Photosynthesis Gene Cluster.
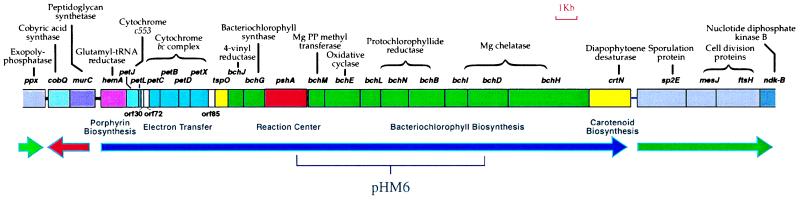
H. mobilis photosynthesis genes were cloned initially by complementing R. capsulatus bacteriochlorophyll-biosynthesis mutants with a H. mobilis genomic-cosmid library. Complete sequence analysis of cosmid pHM6, which complemented mutations in the protochlorophyllide reductase genes bchL, bchN, and bchB, indicated that it contained an 8,355-bp region coding for several genes involved in bacteriochlorophyll biosynthesis (bchM, bchE, bchL, bchN, bchB, bchI, and bchD), as well as part of the RC structural polypeptide coded by the previously sequenced pshA gene (Fig. 1). The DNA sequence flanking the region contained in pHM6 was determined by chromosome walking (7) until regions of nonphotosynthesis genes were obtained. This analysis generated a 35,618-bp contiguous sequence that contains 30 closely linked ORFs. Functions for all but three of the gene products were assigned with a relatively high degree of confidence based on sequence homology analysis (Table 1). We identified 20 clustered genes encoding enzymes and proteins involved in photosynthesis. Most photosynthesis genes are closely spaced, with several gene pairs (bchE–bchL, _bchL-_-bchN, bchN_–_bchB, and bchI_–_bchD) overlapping. With the exception of two genes, cobQ and murC, all of the genes are transcribed in the same orientation.
Figure 1.
The H. mobilis photosynthesis gene cluster. Directions of transcription are indicated by thick solid arrows with the plasmid pHM6 insert sequence delineated with a bracket.
Table 1.
The putative functions and similarity matches of the ORFs of Heliobacillus mobilis identified in this study
| Gene | Best database match, organism, function | Alignment score, _E_-value |
|---|---|---|
| hemA | Glutamyl-tRNA reductase, Bacillus subtilis, heme biosynthesis | 319, 2 × 10−86 |
| petJ | Cytochrome c553, Heliobacterium gestii, electron donor to the photosynthetic reaction center | 163, 4 × 10−40 |
| petC | Rieske iron sulfur protein, Thermus thermophilus, subunit of cytochrome bc complex | 63, 9 × 10−10 |
| petB | Cytochrome b6, liverwort, subunit of cytochrome b6f complex | 220, 4 × 10−57 |
| petD | Cytochrome b/subunit IV, Cairina moschata, subunit of cytochrome bc1 complex | 79, 1 × 10−14 |
| petX | Cytochrome c, Bradyrhizobium japonicum, subunit of cytochrome bc complex | 56, 2 × 10−7 |
| tspO | Peripheral-type benzodiazepine receptor, Archaeoglobus fulgidus, involved in transport of porphyrin | 105, 2 × 10−22 |
| bchJ | Bacteriochlorophyll synthase, Rhodobacter capsulatus, 4-vinyl reduction | 94, 4 × 10−19 |
| bchG | Chlorophyll synthase, Arabidopsis thaliana, esterification of chlorophyllide with geranylgeraniol-PP | 203, 1 × 10−51 |
| pshA | Reaction-center core polypeptide, Heliobacillus mobilis, primary photochemical reaction | 1182, 0 |
| bchM | Mg protoporphyrin IX methyl transferase, Synechocystis sp. 6803 | 142, 1 × 10−33 |
| bchE | Mg-protoporphyrin IX monomethyl ester oxidative cyclase, Rhodobacter capsulatus | 624, 1 × 10−178 |
| bchL | Subunit of light-independent protochlorophyllide reductase, Cyanophora paradoxa | 260, 6 × 10−69 |
| bchN | Subunit of light-independent protochlorophyllide reductase, Plectonema boryanum | 345, 2 × 10−94 |
| bchB | Subunit of light-independent protochlorophyllide reductase, Plectonema boryanum | 375, 1 × 10−103 |
| bchI | Subunit of Mg-chelatase, Synechocystis sp. 6803 | 393, 1 × 10−109 |
| bchD | Subunit of Mg-chelatase, Pisum sativum | 509, 1 × 10−143 |
| bchH | Subunit of Mg-chelatase, Synechocystis sp. 6803 | 1165, 0 |
| crtN | Diapophytoene dehydrogenase, Myxococcus xanthus, carotenoid biosynthesis | 351, 7 × 10−96 |
| ppx | Exopolyphosphatase, Synechocystis sp., response to signals of amino acid starvation | 129, 2 × 10−29 |
| cobQ | Cobyric acid synthase, Methanobacterium thermoautotrophicum, coenzyme B12 biosynthesis | 65, 4 × 10−10 |
| murC | UDP-_N_-acetylmuramyl tripeptide synthetase, Methanobacterium thermoautotrophicum, cell-wall synthesis | 130, 2 × 10−29 |
| sp2E | Stage II sporulation protein E, Bacillus megaterium, sporulation | 236, 4 × 10−61 |
| mesJ | Cell-cycle protein, Bacillus subtilis, cell division | 115, 7 × 10−25 |
| ftsH | ATP-dependent zinc metallopeptidase, Bacillus subtilis, cell division | 677, 0 |
| ndk-B | Nucleoside diphosphate kinase B, Flaveria bidentis, biosynthesis of nucleoside triphosphates | 159, 6 × 10−39 |
Pigment-Biosynthesis Genes.
Heliobacteria synthesize a unique photopigment, bacteriochlorophyll g, which has an interesting structural similarity to chlorophyll a from oxygenic phototrophs (11). Bacteriochlorophyll g contains an ethylidene side group at C8 of ring II that, in the presence of oxygen and light, isomerizes to produce a vinyl group, giving rise to a ring structure that is the same as in chlorophyll a. Inspection of ORFs in the photosynthesis gene cluster indicates that all of the genes involved in steps of the bacteriochlorophyll/chlorophyll biosynthesis pathway between Mg-chelation and formation of chlorophyllide (bchI, bchD, bchH, bchJ, bchM, bchE, bchL, bchN, and bchB) are present in the cluster. The cluster also contains bchG, which is involved in esterification of the tetrapyrrole with farnesol. No unique genes are present in the photosynthesis gene cluster that have an obvious role in synthesizing/stabilizing the ethylidene side group at C8. This indicates that either an unidentified gene(s) may be involved in the formation of the ethylidene side group or that it may be generated as a product by one of the homologs of the chlorophyll a biosynthesis genes. bchI, bchD, and bchH, which code for subunits of Mg-chelatase, are linked tightly, as are bchL, bchN, and bchB, which code for three subunits of light-independent protochlorophyllide reductase. The gene cluster also contains both hemA, which codes for glutamyl tRNA reductase, the enzyme involved in the first step of tetrapyrrole biosynthesis, and tspO (previously known as crtK), which codes for a homolog to a peripheral-type benzodiazepine receptor that has been implicated in the transport of porphyrins (12).
Heliobacteria are unique among photosynthetic organisms in that they synthesize a C30 carotenoid (4,4′-diaponeurosporene) rather than a C40 carotenoid (neurosporene; ref. 13). There are only a few other eubacterial species, such as Staphylococcus aureus, known to synthesize C30 carotenoids. Two genes in S. aureus carotenoid biosynthesis have been identified. One (CrtM) codes for dehydrosqualene synthase, and the other (CrtN) codes for diapophytoene desaturase (14). In the H. mobilis photosynthesis gene cluster, a homolog to crtN was found downstream of bchH.
Reaction Center and Cytochromes.
The gene cluster contains at least six genes involved in primary photochemistry (RC) and electron transport (cytochromes). The RC gene pshA is located among the major group of bch genes. Although this gene was previously sequenced from the same strain used in our study (15), our analysis indicated several variations from the published sequence, including four frame shifts that resulted in an alteration of 34 amino acid codons (5.5% of the entire sequence) near the center and the C terminus of the protein. There is also a putative rho-independent termination site located in the intervening sequence between pshA and bchM, indicating that pshA may not be cotranscribed with the large cluster of downstream bch genes.
The cytochrome gene petJ is found downstream of hemA. This gene was found to encode cytochrome _c_553, and a partial sequence of petJ was recently reported (16). It also shows a high degree of similarity to cytochrome _c_553 from Heliobacterium gestii (17). Cytochrome _c_553 is involved in electron transfer between the cytochrome bc complex and the RC in H. mobilis (18). PetJ has a single heme c binding motif, CNSCH (starting at amino acid residue 79), with Cys-79 and Cys-82 as covalent heme binding ligands and His-83 and Met-125 as axial heme ligands. The petJ coding sequence is followed immediately by a putative rho-independent transcription-termination site.
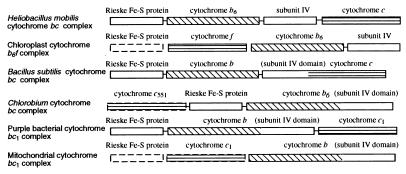
Located downstream of petJ is a cluster of pet genes that form an apparent operon that codes for a unique cytochrome bc complex. A comparison of the H. mobilis pet gene organization with the pet operons from several other species and organelles is shown in Fig. 2. Purple bacteria synthesize a three-subunit cytochrome _bc_1 complex consisting of cytochrome b, cytochrome _c_1, and the Rieske Fe-S protein (19). Cyanobacteria, algae, and plants synthesize a functionally similar cytochrome b_6_f complex that is composed of four major core subunits, the Rieske Fe-S protein, cytochrome f (equivalent to cytochrome _c_1), cytochrome _b_6, and subunit IV (20). Cytochrome _b_6 and subunit IV are considered equivalent to cytochrome b of purple bacteria, in which these two regions are fused into a single polypeptide.
Figure 2.
Comparison of organizations of pet genes coding for cytochrome _bc/bc_1/b6f complexes from various organisms. The nuclear-encoded genes are enclosed in dashed lines. Separated boxes indicate nonlinked genes on chromosome.
Inspection of pet genes of H. mobilis indicates that this species synthesizes an unconventional bc complex resembling both cytochrome _bc_1 of purple bacteria and cytochrome b_6_f of cyanobacteria. Specifically, this putative operon contains petC, which codes for a Rieske Fe-S protein that has a conserved 2Fe-2S site (Cys-87, His-89, Cys-108, and His-111) as observed in other species. The Rieske Fe-S protein of H. mobilis also shares high sequence similarity to a homolog from the green sulfur bacterium Chlorobium limicola, which coincides with a finding from the EPR spectral analysis that showed a close relationship between the Rieske Fe-S proteins of these two species (21).
The next gene is petB, the product of which exhibits a significant sequence similarity to cytochrome _b_6 from plants and cyanobacteria. This cytochrome _c_ontains two conserved histidine pairs (His-84 and His-185 pair and His-98 and His-200 pair), which noncovalently bind two heme groups, located in the transmembrane helices II and IV. The presence of cytochrome _b_6 in H. mobilis is supported by recent electron-transfer studies that indicated that flash-induced kinetics of cytochrome b oxidation of the H. mobilis bc complex were more similar to those of algal and plant b_6_f than those of purple bacterial _bc_1 (22). Interestingly, the C. limicola bc complex contains a cytochrome b composed of fused _b_6 and subunit IV domains (Fig. 2). This bc complex also has biochemical characteristics between those of the _bc_1 and b_6_f complexes with key features resembling the b_6_f type (23).
Downstream of petB is petD, the product of which exhibits significant homology (30% identity and 61% similarity) to subunit IV from plants and cyanobacteria. Like its counterparts, the H. mobilis subunit IV contains three transmembrane domains (determined by hydropathy analysis; data not shown). Study with subunit IV of the b_6_f complex has indicated a role for the chloroplast b_6_f complex in quinone binding, which is essential for the catalytic function (24).
Immediately downstream of petD is an ORF that codes for a protein exhibiting similarity to a membrane-bound diheme cytochrome c from Bradyrhizobium japonicum (25). This cytochrome contains two putative heme c binding sites, CASCH and CAACH, starting at amino acid residues 35 and 133, respectively, with His-39, Met-81, His-137, and Met-176 forming potential axial ligands. In the absence of biochemical data, we tentatively designated this putative cytochrome c subunit of the bc complex as PetX. This ORF also shows homology to a monoheme cytochrome (CytM) of cyanobacteria (with a slightly lower score), which apparently is involved in electron transfer between plastocyanin/cytochrome _c_553 and cytochrome oxidase (26). As heliobacteria are obligate anaerobes, it is possible that this protein performs a dual function, first in its association with other subunits of the bc complex and second as an alternative electron donor to the RC, as was suggested for cytochrome _c_551 in C. limicola (23).
In addition to genes coding for the major cytochrome bc subunits, there are three additional small ORFs located upstream of petC (orf29, orf30, and orf72) and one downstream of petX (orf85). Biochemical analysis indicates that cytochrome b_6_f has three small subunits (subunits V, VI, and VII, or PetG, PetL, and PetM, respectively) in addition to the four major subunits discussed above. These additional subunits have a common feature of being of low molecular mass (≈4 kDa) and having one single transmembrane helix (27). Direct sequence comparison of the H. mobilis small ORFs with the b_6_f small subunits indicated considerable sequence similarity between PetL of Cyanophora paradoxa and orf29 (28% identity and 38% similarity). This ORF was thus designated as petL. No obvious similarity is, however, observed between orf30, which does not seem to contain transmembrane segments, and the small b_6_f subunits or any known sequences in GenBank. In addition, there is limited sequence similarity of orf72 and orf85 with subunits 10 and 11, respectively, of the bovine mitochondrial cytochrome _bc_1 complex (19.4% identity and 51.6% similarity relative to subunit 10; 17.9% identity and 58.9% similarity for subunit 11). These additional H. mobilis ORFs may thus code for components of this unique cytochrome bc complex.
Evolutionary Analyses.
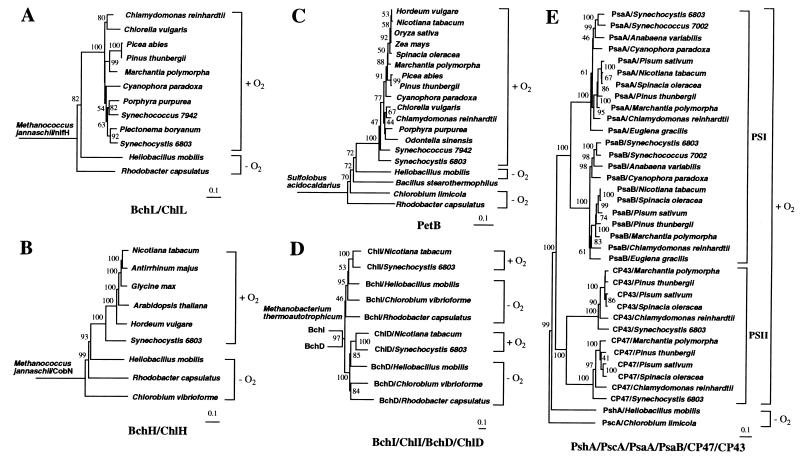
H. mobilis photosynthesis genes were compared with homologous sequences in the GenBank database to study evolutionary relationships. Phylogenetic trees were generated from multiple aligned sequences by using the maximum-parsimony, neighbor-joining, and maximum-likelihood methods. As the results from the three methods are highly congruent, we shall mainly discuss results obtained from the neighbor-joining tree analysis. A BchL/ChlL phylogenetic tree was constructed by using the nitrogenase iron protein NifH from an archaeon species Methanococcus jannaschii as an outgroup (Fig. 3A). Fujita et al. (28) and Burke et al. (29) found remarkable similarity between NifH and BchL. NifH was also used previously as an outgroup for studying phylogenetic relationships among BchL homologs from bacteria and plants (30). Its choice as an outgroup is based on the premise that nitrogen fixation occurs in both archaea and bacteria, whereas tetrapyrrole-based photosynthesis occurs only in bacteria and bacterially derived chloroplasts. Thus, unlike nitrogen fixation, photosynthesis may not predate the divergence of archaea and bacteria. A notable feature of the highly supported BchL tree is that oxygenic phototrophs together form a monophyletic group distinct from the anoxygenic phototrophs R. capsulatus and H. mobilis (Fig. 3A). The paraphyletic R. capsulatus and H. mobilis lineages are rooted closer to the base of the tree and form an outgroup to the oxygenic clade, suggesting that anoxygenic phototrophs are ancestral to the oxygenic phototrophs. This result is in agreement with the previous phylogenetic analysis by Burke et al. (30), who concluded that bacteriochlorophyll biosynthesis evolved earlier than chlorophyllous biosynthesis. However, this conclusion was later challenged by Lockhart et al. (31) who argued for an alternative hypothesis. Another notable feature of the tree is that R. capsulatus is a deeper branching lineage than H. mobilis, suggesting its more primitive evolutionary status.
Figure 3.
(A) This phylogenetic tree of BchL/ChlL was created with the neighbor-joining method. NifH from M. jannaschii is used as an outgroup. Photosynthetic taxa are delineated by brackets and identified as oxygenic (+O2) or anoxygenic (−O2). Horizontal branch lengths represent relative evolutionary distances with the scale bar corresponding to 0.1 amino acid substitutions per site. Bootstrap values above 40% are indicated at each node, except basal nodes. (B) This phylogenetic tree of BchH/ChlH used CobN from M. jannaschii as an outgroup. (C) This phylogenetic tree of PetB used PetB from Sulfolobus acidocaldarius as an outgroup. (D) This composite phylogenetic tree of paralogous gene products Mg-chelatase subunits BchI/ChlI/BchD/ChlD used BchI and BchD homolog from Methanobacterium thermoautotrophicum as an outgroup. (C) This composite phylogenetic tree of PSI-like RC polypeptides (PshA/PscA/PsaA/PsaB) together with two PSII core antenna polypeptides (CP47 and CP43) used PscA of C. limicola as an outgroup.
A similar ancestral relationship between anoxygenic and oxygenic phototrophs is observed with the rooted BchH/ChlH tree with CobN from M. jannaschii as an outgroup (Fig. 3B). CobN is a subunit of Co-chelatase responsible for insertion of Co into tetrapyrrole during biosynthesis of coenzyme B12. Rooting with CobN is based on the extensive sequence similarity of CobN to the Mg-chelatase subunit BChH/ChlH (32) and on the argument that the biosynthesis of coenzyme B12, which is involved in a wide variety of cellular biochemical processes including the biosynthesis of nucleotides, may have preceded photosynthesis. A significant feature of this rooted tree is that it shows a better resolution of phylogenetic relationships among the phototrophic lineages with high bootstrap support. In addition to separation of anoxygenic and oxygenic lineages, with the former being an exclusive common ancestor to the latter, this tree places H. mobilis nearest to Synechocystis sp. PCC 6803, which is consistent with the previous 16S rRNA analysis (33). C. vibrioforme and R. capsulatus are deeper branching lineages relative to H. mobilis, although the placement of C. vibrioforme in a more basal position than R. capsulatus is considered tentative, because this placement is supported neither by the phylogenetic trees discussed below nor by the 16S rRNA tree (33). A monophyletic cyanobacterial origin of green plants is also evident in this tree with the support of an extremely high bootstrap value (100%), which confirms many lines of evidence that cyanobacteria are the endosymbiotic origin of plastids in photosynthetic eukaryotes (e.g., ref. 34).
A cytochrome b/_b_6 (PetB) parsimony tree was constructed that was rooted with PetB of an archeaon Sulfolobus acidocaldarius (Fig. 3C). This rooted tree corroborates the grouping of oxygenic lineages distinct from that of anoxygenic ones as was observed with the Bch/Chl trees. In this tree, H. mobilis is shown again as the closest relative to cyanobacteria. The neighbor-joining tree also places R. capsulatus as the deepest lineage and Bacillus stearothermophilus intermediately in between C. limicola and H. mobilis, which may indicate the possibility of lateral gene transfer for the cytochrome _b_6 gene from a green sulfur bacteria-like common ancestor to the low-G+C Gram-positive bacteria. The more basal placement of R. capsulatus relative to C. limicola in this tree is in congruence with the 16S rRNA tree without the inclusion of green nonsulfur bacteria (33). Another notable feature of this tree is significantly less sequence diversity among oxygenic photosynthetic lineages as shown by relatively short branch lengths, which reflects a highly conserved nature of cytochrome _b_6 in these organisms, indicating that PetB may also be a good phylogenetic marker for oxygenic photosynthesis.
Phylogenetic analyses of other photosynthesis gene products (BchI, BchD, BchB, BchN, and BchM) are in agreement with the three representative trees shown in Fig. 3 A–C (data not shown). Thus, one can conclude that photosynthesis genes from anoxygenic photosynthetic organisms are ancestral to those found in oxygenic photosynthetic organisms. Comparing phylogenetic trees of photosynthesis genes vs. nonphotosynthesis genes, however, has been complicated by the various versions of trees available in the literature (e.g., 16S rRNA; refs. 33, 35), which further justifies the use of photosynthesis genes for tracking the molecular evolution of photosynthesis.
In terms of phylogenetic position, it is the current belief, based on the common RC type, that heliobacteria share the closest relationship with green sulfur bacteria. However, our quantitative analyses of evolutionary distances for various other photosynthesis genes from cyanobacteria, heliobacteria, and green sulfur bacteria do not support this idea. The analysis of pairwise adjusted mean distances among the three bacterial groups calculated for 15 genes (data not shown) indicates that heliobacteria are closer to cyanobacteria for 12 genes and that all three groups are of equal distance for 2 genes (including the RC polypeptides), whereas only 1 gene supports the idea that heliobacteria are closer to green sulfur bacteria.
Gene-Duplication Events.
Analysis of several photosynthesis genes also indicates that some may have undergone a different developmental pathway, such as gene duplication. For example, bchI (or chlI) and bchD (or chlD), which encode two Mg-chelatase subunits, share significant similarity at the amino acid level (for H. mobilis, BchI and BchD polypeptides exhibit 31.0% identity and 67.9% similarity). These genes, which are nearly always linked in eubacterial genomes, are considered paralogous genes with a common evolutionary ancestry (36). To determine whether a gene-duplication event occurred before or after the divergence of current photosynthetic organisms, we conducted phylogenetic analysis of all available BchI and BchD pairs in the GenBank database. A joint phylogenetic tree with high bootstrap support was generated with a BchI and BchD homolog pair from Methanobacterium thermoautotrophicum as root. (The function of BchI and BchD homologs in the nonphotosynthetic archean is not yet known. However, we speculate that they may be subunits of a Ni-chelatase for the biosynthesis of the Ni-containing coenzyme F430, which is essential for the production of methane in methanogens.) As indicated in Fig. 3D, all of the BchI/ChlI sequences are clustered in a clade distinct from the BchD/ChlD clade, suggesting that an ancient gene duplication may have occurred well before photosynthetic speciation events. In addition, the topology in each subtree corroborates the result from the PetB tree (Fig. 3C), indicating that (i) H. mobilis is closest to the last common ancestor of all oxygenic lineages, (ii) C. vibrioforme and R. capsulatus are ancestral to H. mobilis, and (iii) in the absence of green nonsulfur bacteria, R. capsulatus is the earliest emerging photosynthetic lineage.
Another striking example of gene duplication is found with the gene for the PSI-like RC core polypeptide. H. mobilis and C. limicola use a single RC core polypeptide (PshA and PscA, respectively) to form a homodimeric PSI-type RC complex. In contrast, oxygenic species synthesize a heterodimeric PSI RC with two highly related polypeptides, PsaA and PsaB. A composite phylogenetic tree (Fig. 3E) of the PSI RC core polypeptides from a broad representation of oxygenic lineages with PscA of C. limicola as root indicates that PsaA and PsaB are divided into two distinct clades and are derived from an ancient gene duplication from a PscA/PshA-like common ancestor. Thus, the RC gene duplication seems to have occurred well before diversification of all oxygenic lineages. Interestingly, PSI RC polypeptides also share significant similarity to CP47 (PsbB) and CP43 (PsbC; ref. 2), both of which are core antenna polypeptides of PSII. Our phylogenetic analysis (Fig. 3E) indicates that these genes have undergone gene duplication from a _pshA_-like common ancestor that preceded the divergence of all oxygenic lineages.
Origin of Oxygenic Photosynthesis.
Several models have been put forward to account for the origin and evolution of PSI and PSII in oxygenic organisms. Many of these models (e.g., ref. 37) are based on subunit composition and functionality of photosystems rather than on rigorous phylogenetic reconstruction based on multiple sequence comparisons. Currently, there are three models that have obtained the most attention: the selective-loss model (38), a model proposed by Vermaas (2), and the fusion model (1, 39).
Olson and Pierson (38) propose a selective-loss model in which two linked photosystems analogous to PSI and PSII in cyanobacteria developed very early in evolution. Subsequent loss of one of the photosystems would give rise to the existing anoxygenic photosynthetic bacteria that have only a single PSI- or PSII-type photosystem. However, this model is not supported by our phylogenetic analysis (Fig. 3), which clearly indicates that photosynthesis genes from anoxygenic photosynthetic organisms are ancestral to those from oxygenic photosynthetic organisms.
A model described by Vermaas (2) involves the assumption that an ancestral RC resembling that of heliobacteria or Chlorobium evolved into both PSI-and PSII-type RCs. Part of the model suggests that PSI in oxygenic organisms was evolved from gene duplication and divergence from an anoxygenic homodimeric RC, a suggestion supported by our phylogenetic data (Fig. 3E). The model also proposes that the PSII-type RC in both anoxygenic and oxygenic organisms is derived from an anoxygenic PSI-type photosystem through gene splitting, operon duplication, and loss of the Fe-S center. The implicit assumptions are that heliobacteria and green sulfur bacteria preceded both oxygenic and purple bacteria and that purple bacteria were the latest emerging photosynthetic lineage. This part of the model is clearly not supported by our phylogenetic analysis, which indicates that purple bacteria are ancestral to heliobacteria and cyanobacteria (Fig. 3). Recent x-ray crystal-structure determination of PSI (37) has provided additional information and added complexity to the complete picture regarding the origin and evolution of oxygenic photosynthesis. The structure shows significant similarity between the purple bacterial RC and PSI core in cofactor arrangements and transmembrane helix topology; this similarity was taken to support the hypothesis that the two types of photosystems share a common ancestry and that the progenitor of all photosystems may be a PSI-like RC (37). However, the conclusion was drawn in the absence of identifiable sequence similarity between the two types of RC polypeptides. The structural similarity may thus indicate that the two types of RCs are either highly remote homologs or results from convergent evolution from two distinct origins. Our sequence comparisons support the convergent-evolution scenario.
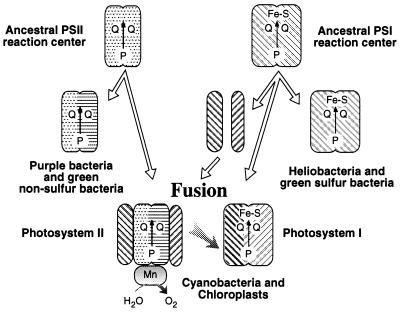
Mathis (39) and Blankenship (1) suggest a fusion model in which a primordial photosynthetic organism containing a single homodimeric photosystem gave rise to the existing lineages of anoxygenic bacteria that contain only a single PSI- or PSII-type photosystem. Subsequent fusion of these two types of anoxygenic photosystems and development of water-oxidizing capability gave rise to oxygenic photosynthesis. This model is supported by our rooted-parsimony analysis of various photosynthesis genes (Fig. 3), which clearly indicates that purple bacterial and heliobacterial photosynthesis genes are ancestral to cyanobacterial, algal, and plant photosynthesis genes. However, this model must be modified in light of the observation of Vermaas (2), who showed that the CP47 and CP43 PSII core antenna polypeptides exhibit sequence similarity with the light-harvesting domains of the anoxygenic PSI-type RC polypeptide, which is corroborated by our parsimony-tree analysis (Fig. 3E). This evidence indicates that the oxygenic PSII RC contains antenna components derived from an ancient anoxygenic PSI RC. Consequently, we propose a “heterologous fusion model” (Fig. 4) that indicates that PSII from oxygenic organisms are derived from a fusion of the ancestral form of anoxygenic PSI- and PSII-type RCs. In this model, the PSII D1 and D2 core polypeptides are obtained from an anoxygenic PSII-type ancestor, presumably a homodimer form that is most similar to the purple bacterial L/M polypeptides (1). In addition, the PSII CP47 and CP43 core polypeptides are derived from gene fragmentation and subsequent duplication from a common ancestor to the anoxygenic PSI-type RC polypeptide.
Figure 4.
A heterologous fusion model for the evolution of oxygenic photosynthesis based on phylogenetic analysis. Details of the model are provided in the text.
Acknowledgments
We thank Drs. Yuichi Fujita and Chris Parkinson for valuable suggestions. This work was supported by National Institutes of Health Grants GM53940 and GM00618 (to C.E.B.) and by a grant from the US–Japan Cooperative Program on Photoconversion and Photosynthesis Research (to K.I.).
ABBREVIATIONS
PSI
photosystem I
PSII
photosystem II
RC
reaction center
Footnotes
Data deposition: The nucleotide sequence reported in this paper has been deposited in the GenBank database (accession no. AF080002).
References
- 1.Blankenship R E. Photosynth Res. 1992;33:91–111. [PubMed] [Google Scholar]
- 2.Vermaas W F J. Photosynth Res. 1994;41:285–294. [PubMed] [Google Scholar]
- 3.Alberti M, Burke D H, Hearst J E. In: Anoxygenic Photosynthetic Bacteria. Blankenship R E, Madigan M T, Bauer C E, editors. Dordrecht, the Netherlands: Kluwer; 1995. pp. 1083–1106. [Google Scholar]
- 4.Davison J, Heusterspreute M, Chevalier N, Ha-Thi V, Brunel F. Gene. 1987;51:275–280. doi: 10.1016/0378-1119(87)90316-7. [DOI] [PubMed] [Google Scholar]
- 5.Young D A, Bauer C E, Williams J C, Marrs B L. Mol Gen Genet. 1989;218:1–12. doi: 10.1007/BF00330558. [DOI] [PubMed] [Google Scholar]
- 6.Bollivar D W, Suzuki J Y, Beatty J T, Dobrowski J M, Bauer C E. J Mol Biol. 1994;237:622–640. doi: 10.1006/jmbi.1994.1260. [DOI] [PubMed] [Google Scholar]
- 7.Ochman H, Gerber A S, Hartl D L. Genetics. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swofford D L. paup, Phylogenetic Analysis Using Parsimony. Champaign, IL: Illinois Natural History Survey; 1993. , Version 3.1.1. [Google Scholar]
- 11.Brockmann H, Jr, Lipiniski A. Arch Microbiol. 1983;136:17–19. [Google Scholar]
- 12.Yeliseev A A, Kaplan S. J Biol Chem. 1995;270:21167–21175. doi: 10.1074/jbc.270.36.21167. [DOI] [PubMed] [Google Scholar]
- 13.Takaichi S, Inoue K, Akaike M, Kobayashi M, Oh-oka H, Madigan M T. Arch Microbiol. 1997;168:277–281. doi: 10.1007/s002030050499. [DOI] [PubMed] [Google Scholar]
- 14.Marshall J, Wilmoth G. J Bacteriol. 1981;147:900–913. doi: 10.1128/jb.147.3.900-913.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liebl U, Mockensturm-Wilson M, Trost J T, Brune D C, Blankenship R E, Vermaas W. Proc Natl Acad Sci USA. 1993;90:7124–7128. doi: 10.1073/pnas.90.15.7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee W-Y, Blankenship R E, Kim S H. J Microbiol. 1997;35:206–212. [Google Scholar]
- 17.Albert I, Rutherford A W, Grav H, Kellermann J, Michel H. Biochemistry. 1998;37:9001–9008. doi: 10.1021/bi9731347. [DOI] [PubMed] [Google Scholar]
- 18.Nitschke W, Liebl U, Matsuura K, Kramer D M. Biochemistry. 1995;34:11831–11839. doi: 10.1021/bi00037a022. [DOI] [PubMed] [Google Scholar]
- 19.Trumpower B L. Microbiol Rev. 1990;54:101–129. doi: 10.1128/mr.54.2.101-129.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gray K A, Daldal F. In: Anoxygenic Photosynthetic Bacteria. Blankenship R E, Madigan M T, Bauer C E, editors. Dordrecht, the Netherlands: Kluwer; 1995. pp. 747–774. [Google Scholar]
- 21.Liebl U, Rutherford A W, Nitschke W. FEBS Lett. 1990;261:427–430. [Google Scholar]
- 22.Kramer D M, Schoepp B, Liebl U, Nitschke W. Biochemistry. 1997;36:4203–4211. doi: 10.1021/bi962241i. [DOI] [PubMed] [Google Scholar]
- 23.Schütz M, Zirngibl S, le Coutre J, Büttner M, Xie D-L, Nelson N, Deutzmann R, Hauska G. Photosynth Res. 1994;39:163–174. doi: 10.1007/BF00029383. [DOI] [PubMed] [Google Scholar]
- 24.Li L B, Zou Y P, Yu L, Yu C A. Biochim Biophys Acta. 1991;1057:215–222. doi: 10.1016/s0005-2728(05)80104-5. [DOI] [PubMed] [Google Scholar]
- 25.Preisig O, Anthamatten D, Hennecke H. Proc Natl Acad Sci USA. 1993;90:3309–3313. doi: 10.1073/pnas.90.8.3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manna P, Vermaas W. Plant Mol Biol. 1997;35:407–416. doi: 10.1023/a:1005875124387. [DOI] [PubMed] [Google Scholar]
- 27.de Vitry C, Breyton C, Pierre Y, Popot J-L. J Biol Chem. 1996;271:10667–10671. doi: 10.1074/jbc.271.18.10667. [DOI] [PubMed] [Google Scholar]
- 28.Fujita Y, Takahashi Y, Chuganji M, Matsubara H. Plant Cell Physiol. 1992;33:81–92. [Google Scholar]
- 29.Burke D H, Alberti M, Hearst J E. J Bacteriol. 1993;175:2407–2413. doi: 10.1128/jb.175.8.2407-2413.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burke D H, Hearst J E, Sidow A. Proc Natl Acad Sci USA. 1993;90:7134–7138. doi: 10.1073/pnas.90.15.7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lockhart P J, Larkum A W D, Steel M A, Waddell P J, Penny D. Proc Natl Acad Sci USA. 1996;93:1930–1934. doi: 10.1073/pnas.93.5.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hudson A, Carpenter R, Doyle S, Coen E S. EMBO J. 1993;12:3711–3719. doi: 10.1002/j.1460-2075.1993.tb06048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woese C R. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gray M W. Trends Genet. 1989;5:294–299. doi: 10.1016/0168-9525(89)90111-x. [DOI] [PubMed] [Google Scholar]
- 35.Neefs J-M, Van der Peer Y, De Rijk P, Chapelle S, De Wachter R. Nucleic Acids Res. 1993;21:3025–3049. doi: 10.1093/nar/21.13.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jensen P E, Gibson L C D, Hennigsen K W, Hunter C N. J Biol Chem. 1996;271:16662–16667. doi: 10.1074/jbc.271.28.16662. [DOI] [PubMed] [Google Scholar]
- 37.Schubert W-D, Klukas O, Saenger W, Witt H T, Fromme P, Krauss N. J Mol Biol. 1998;280:297–314. doi: 10.1006/jmbi.1998.1824. [DOI] [PubMed] [Google Scholar]
- 38.Olson J M, Pierson B K. Int Rev Cytol. 1987;108:209–248. doi: 10.1016/s0074-7696(08)61439-4. [DOI] [PubMed] [Google Scholar]
- 39.Mathis P. Biochim Biophys Acta. 1990;1018:163–167. doi: 10.1016/0005-2728(77)90167-0. [DOI] [PubMed] [Google Scholar]