Hierarchy of nonhomologous end-joining, single-strand annealing and gene conversion at site-directed DNA double-strand breaks (original) (raw)
Abstract
In mammalian cells, DNA double-strand breaks (DSBs) are repaired by three pathways, nonhomologous end-joining (NHEJ), gene conversion (GC) and single-strand annealing (SSA). These pathways are distinct with regard to repair efficiency and mutagenic potential and must be tightly controlled to preserve viability and genomic stability. Here, we employed chromosomal reporter constructs to characterize the hierarchy of NHEJ, GC and SSA at a single I-SceI-induced DSB in Chinese hamster ovary cells. We discovered that the use of GC and SSA was increased by 6- to 8-fold upon loss of Ku80 function, suggesting that NHEJ is dominant over the other two pathways. However, NHEJ efficiency was not altered if GC was impaired by Rad51 knockdown. Interestingly, when SSA was made available as an alternative mode for DSB repair, loss of Rad51 function led to an increase in SSA activity at the expense of NHEJ, implying that Rad51 may indirectly promote NHEJ by limiting SSA. We conclude that a repair hierarchy exists to limit the access of the most mutagenic mechanism, SSA, to the break site. Furthermore, the cellular choice of repair pathways is reversible and can be influenced at the level of effector proteins such as Ku80 or Rad51.
INTRODUCTION
DNA double-strand breaks (DSBs) are the most deleterious type of DNA damage that may lead to cell death or genomic instability. In mammalian cells, DSB repair is widely executed by two mechanistically distinct processes, nonhomologous end-joining (NHEJ) and homology-dependent recombination, which can be either conservative, i.e. gene conversion (GC), or nonconservative, i.e. single-strand annealing (SSA) (1–3).
GC is generally considered an error-free repair pathway, while NHEJ may introduce minor sequence alterations at the DNA ends and SSA is always associated with deletion of sequence. Additionally, all three pathways bear a risk of repair errors that may result in potentially oncogenic chromosomal aberrations. Failure of the NHEJ machinery to hold legitimate ends together allows promiscuous end-joining leading to deletions or translocations (4–7). GC is usually error-free if the homologous repair template is provided by the nearby sister chromatid in the S- or G2-phase of the cell cycle. In contrast, GC initiated in the G1-phase carries a high risk of chromosomal rearrangements because the homologous template can only be found on a distant chromosomal locus, i.e. the second allele, a pseudo gene or a repeat sequence (8,9). Crossover events coupled to GC will result in deletions, inversions, loss of heterozygosity (LOH) or gene amplification (10,11), all of which potentially promote carcinogenesis (12). Hence, GC should be generally suppressed in G1-phase. SSA is well characterized in yeast (3,13,14) but not in mammalian cells. Abundant repetitive elements in higher eukaryotes (15) should render SSA a suitable repair option but it is not known whether it actually contributes to overall DSB repair. Recently, however, SSA has also been identified as a significant pathway leading to translocations frequently inflicted in human cancers (16–18). It is important that cells control the choice of DSB pathways in order to optimize repair efficiency and to minimize the risk of genetic alterations. However, the relationship between the pathways and the mechanisms of regulation is poorly understood.
NHEJ is guided by the Ku70/80 heterodimer, which initially binds to free DNA ends. Ku then recruits and activates other components of the end-joining process (19), namely the DNA-PKcs, the polymerases μ and λ and the LigIV/XRCC4/XLF complex. Although Ku supports nearly all NHEJ functions and is essential for cellular radioresistance (20), we and others revealed a limited role of Ku80 for the repair of enzyme-induced chromosomal breaks (21–24). In contrast to XRCC4−/− mouse embryonic fibroblasts (MEFs), Ku80 knock-out cells were almost as competent to rejoin I-SceI-induced DSB as the wild-type cells (24). Analysis of repair products in Ku80-deficient cells showed that Ku-independent end-joining is mechanistically distinct from repair in wild-type cells, suggesting a switch to another pathway (23,24). One possible alternative is the PARP1/XRCC1/LigIII-dependent end-joining pathway, which was recently shown to operate in the absence of Ku (25,26). In addition, lack of Ku might favor the initiation of recombination processes such as GC (27–29) as well as Rad52-dependent SSA. Rad52 has been suggested to compete with Ku for end binding (30,31). However, the functional relationship between NHEJ and SSA has not been addressed in mammalian cells.
From these observations, we hypothesized (i) that DSB repair pathways are regulated in a hierarchical order and (ii) that mammalian cells can switch to alternative pathways if the preferred repair mode is impaired. In this study, we simultaneously examined the three major DSB pathways in mammalian cells using novel chromosomal reporter substrates. We found that Ku80 controls the accuracy of NHEJ and regulates the usage of the other two pathways. In addition, we present evidence that SSA can serve as a back-up mechanism for both NHEJ and GC. Furthermore, the current study reveals a novel mechanism by which Rad51 may regulate the genomic integrity in mammalian cells by controlling the ratio between NHEJ and SSA.
MATERIAL AND METHODS
Cells
The hamster cell lines CHO K1 (wild-type) and xrs5 (Ku80-deficient) were grown in Alpha-Medium (Gibco-Invitrogen, Karlsruhe, Germany) supplemented with 5% fetal calf serum, 100 U/ml penicillin and 100 µg/ml streptomycin at 37°C with 5% CO2. For complementation of Ku80-deficiency, 40 μg of pcDNA3.1-hKu80 were electroporated, thereby transiently expressing human KU80 (kindly provided by P. A. Jeggo).
Plasmids
Three novel green fluorescent protein (GFP)-based repair substrates were cloned using pEGFP-N1 (Invitrogen) and pBlueskriptII-KS (Stratagene, Amsterdam, the Netherlands) as backbones: pEJ to monitor NHEJ, pEJSSA for SSA together with end-joining and pGC for homology-directed GC.
The end-joining substrate, pEJ, is designed similar to the previously described substrate pPHW2 (32,33) containing two I-SceI sites inserted into the 5′ untranslated region of the GFP transcript. Between both I-SceI sites, an artificial start codon (ATGart) was placed out of frame with the original open reading frame (ORF), hence preventing translation of GFP. The Kozak sequence flanking the original GFP start codon of the pEGFP-N1 was modified by site-directed mutagenesis (QuikChange, Stratagene) replacing the A preceding the ATG with a G. The sequence now reads as -CCGCCATGG-. The modification gradually weakens the translational enhancer (34). In four subsequent steps, the following oligonucleotides were inserted into the multiple cloning site of pEGFP-N1: (i) XhoI-TCCACCGAGACATCTACTTGATCAATCGAACACT GCG-EcoRI, (ii) EcoRI-TAGGGATAACAGGGTAATTAAGCTT-PstI, (iii) PstI-ACCATGGAGATTACCCTGTTATCCCTACCCCGGGGATACTGAC-KpnI, (iv) KpnI-TCAATAATCCGATCGAAGTCTACTGATCGC-BamHI. The oligonucleotide (ii) contains the first I-SceI site and (iii) the artificial ATG and the second I-SceI site. Splice donor or acceptor sites were avoided.
To create pEJSSA, the combined substrate for end-joining and SSA, two 50 bp repetitive sequences named SSA1 (XhoI-GCAACCGCTCATACGACCGACAACCGACCGCGCATCACGCCGCAAGATCT-BclI) and SSA2 (PvuI-GCAACCGCTCATACGACCGACAACCGACCGCGCATCACGCCGCAGTCGAC-AgeI) were inserted as XhoI-BclII and PvuI-AgeI fragments, respectively. After DSB induction repair can proceed via NHEJ or SSA, as both will result in reconstituted translation of the GFP.
For the GC substrate pGC, the 18-bp I-SceI recognition site was inserted into the unique BcgI site of the pEGFP-N1 thereby inactivating the GFP-coding sequence. This intermediate, named pGC-intermedI and linked to a modified fragment of pBluescriptII-KS, was previously generated as follows: The sequences for the puromycin resistance gene and the BSH-polyA-signal were amplified by PCR from pPHW1 (32) and subsequently inserted into the pBluescriptII-KS as HindIII/EcoRV- and EcoRV-EcoRI fragments, respectively. A 526-bp fragment of the pEGFP-N1 was amplified by PCR and inserted as XbaI/NotI fragment into the modified pBluescriptII-KS. This fragment spans the terminal 30 bp of the Cytomegalovirus (CMV) promoter and 398 N-terminal bp of the GFP-cDNA. This intermediate was cleaved by ClaI/AflIII and ligated with the ClaI/AflIII fragments of pGC-intermed1. By this approach, the neomycin resistance gene was replaced with the puromycin resistance gene, which is now under control of the SV40 promoter. The truncated GFP donor copy is located 2.2 kb downstream of the mutated GFP acceptor sequence and placed in the same orientation. The 2.2 kb intervening sequence carries the puromycin resistance gene. The homologies between both inactive GFP-copies amount to 219 bp upstream and 301 bp downstream of the I-SceI recognition site.
After linearization of pEJ or pEJSSA with AflIII or pGC with XmnI, 0.5 μg of either vector was electroporated into the hamster cells as described (32). For stable integration of pEJ and pEJSSA, CHO cells were grown in 14 µg/ml and xrs5 cells in 6 µg/ml of G418 for two weeks. For pGC integration, puromycin was used at concentrations of 1.5 mg/ml for CHO and xrs5 cells. Thirty to sixty individual colonies were picked for each cell line and repair substrate and further analyzed as described (24,32).
RAD51 down-regulation by short interfering RNA (siRNA)
The siRNA duplexes were custom designed and synthesized by Qiagen, Hilden, Germany. The sequences were the following: 5′-GCUGGUUUCCAUACGGUGG-3′, to target the hamster Rad51 transcript, and 5′-UAGGCAUUGCGCGUGUGUC-3′ (scrambled control). Cells were transfected using Trans-IT-TKO (Mirus, Madison, WI, USA) according to the manufacturer's protocol, with a final oligonucleotide concentration of 200 nM. Clonogenic cell survival was not affected by Rad51 knockdown. Protein expression was monitored by Western blotting following standard procedures (32, anti-Rad51 mAb-51Rad01, Calbiochem, Darmstadt, Germany).
DSB-repair reporter assay
The cells containing single stably integrated copies of either reporter construct were electroporated with 50 μg of I-SceI expression vector (pCMV3xnls-I-SceI, a kind gift of M. Jasin) as described (32) to induce DSB or with pCMV-neo as a control. Forty eight hours posttransfection the cells were assessed for green fluorescence by flow cytometry (FACScan, BD Bioscience). For further analysis of the individual NHEJ or SSA events, positive cells were sorted (FACS Calibur, BD Bioscience, Heidelberg, Germany) and processed. First, cells were reseeded, raised to individual colonies, and further expanded for DNA isolation (DNeasy Tissue Kit, Qiagen) and PCR amplification and sequencing using the primers P1 and P2 flanking the I-SceI cleavage sites (forward 5′-GCAAATGGGCGGTAGGCGTGTA-3′, reverse 5′-TCGGGCATGGCGGACTTGAA-3′). PCR conditions were 96°C for 10 min for lysis of the bacterial cells, then 35 cycles at 96°C for 20 s, at 68°C for 20 s and at 72°C for 80 s, and a postamplification extension for 7 min at 72°C. Second, total genomic DNA of the entire sorted cell population was isolated and subjected to PCR. The mixtures of those PCR products were ligated into the TOPO-cloning vector (TOPO TA cloning kit, Invitrogen) according to the manufacturer's protocol and transformed into bacteria (one shot TOP10 competent Escherichia coli, Invitrogen). Single bacterial clones were scraped off the plate, directly subjected to PCR using the primers P1 and P2 and subsequently sequenced (ABI 3100, Applied Biosystems-Hitachi, Foster City, CA, USA). PCR products were analyzed on 2% agarose gels stained with ethidium bromide. For two clones (one of each CHO K1 and xrs5 carrying pEJSSA), TOPO cloning was directly compared to the method of raising positive cells individually, which gave identical values for the ratio between NHEJ and SSA events.
To assess transfection efficiencies, 30 μg of pEGFP-N1 (Clontech, Saint-Germain-en-Laye, France) was electroporated into both strains and analyzed by fluorescent-activated cell sorting (FACS) 24 h later. CHO K1 and xrs5 cells showed 70.8 ± 3.9% and 64.2 ± 8.4% of green fluorescent cells, respectively. All repair results were corrected for the 1.1-fold lower transfection efficiency of xrs5 cells.
RESULTS
Repair substrates
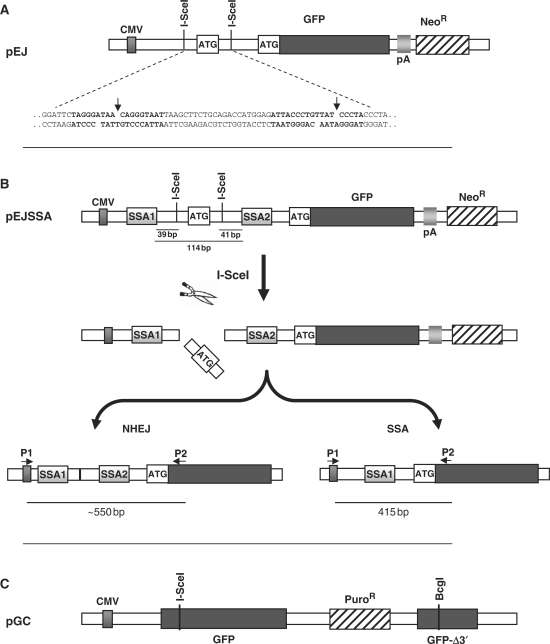
Three novel GFP-based repair substrates were designed to investigate the relationship between the main DSB repair pathways, NHEJ, GC and SSA in the chromosomal context. All constructs rely on reactivation of GFP expression upon repair of single I-SceI-endonuclease-induced DSBs. The substrate to monitor NHEJ, pEJ, makes use of the same principle as the previously introduced pPHW2 (32). It contains two recognition sites for the rare-cutting I-SceI restriction enzyme inserted in opposite directions into the 5′-untranslated region of the GFP gene (Figure 1A). Between the two I-SceI sites, an artificial start codon is integrated out of frame with the original start codon, thus preventing GFP translation. I-SceI cleavage removes the artificial ATG and subsequent rejoining of the free DNA ends reactivates translation leading to green fluorescence. The advantage of this system is that it is independent of how the ends are rejoined as long as any repair-associated deletions are no longer than 160 bp. The critical distance for base loss is reached at the transcription initiation site and the GFP open reading frame (86 bp upstream of the first and 76 bp downstream of the second I-SceI site, respectively).
Figure 1.
Reporter constructs. (A) Schematic structure of the NHEJ substrate pEJ. Translation of GFP is prevented by an insert between the CMV promoter and the ORF, which is flanked by two inverted repeat I-SceI recognition sequences. Repair of the I-SceI-induced DSB by NHEJ restores GFP translation (see text for details). The insert illustrates the sequences flanking the two I-SceI sites (bold). (B) The pEJSSA substrate for monitoring repair either by NHEJ or by SSA. PEJSSA is identical to pEJ, except for the presence of two additional 50-bp direct repeats (SSA1 and SSA2) flanking the I-SceI recognition sites (top). Simultaneous cleavage of both I-SceI sites leads to pop out of the artificial ATG (middle). Repair by either pathway leads to green fluorescence. PCR analysis of NHEJ events using the primers P1 and P2 (bottom) generates a fragment of about 550 bp. SSA events result in loss of one of the two SSA-cassettes and yields a PCR fragment size of exactly 415 bp. (C) The pGC reporter with two nonfunctional GFP copies that share 520 bp of homology. DSB repair proceeds by GC resulting in functional GFP. Constructs are not drawn to scale.
pEJSSA is identical to pEJ except for the insertion of two homologous direct repeats (SSA1 and SSA2) flanking the break site, thereby enabling the use of SSA in addition to NHEJ (Figure 1B). These SSA1 and SSA2 repeats are 50 bp in length and located 39 and 41 bp away from the DSB ends, respectively. This arrangement leaves sufficient space for NHEJ mechanisms to occur before sequences suitable for SSA become available. PCR amplification of the repair junctions using P1 and P2 primers (Figures 1B and 2B) were used to differentiate between the two repair pathways as NHEJ events produce bands of about 550 bp while SSA events lead to a 415-bp band.
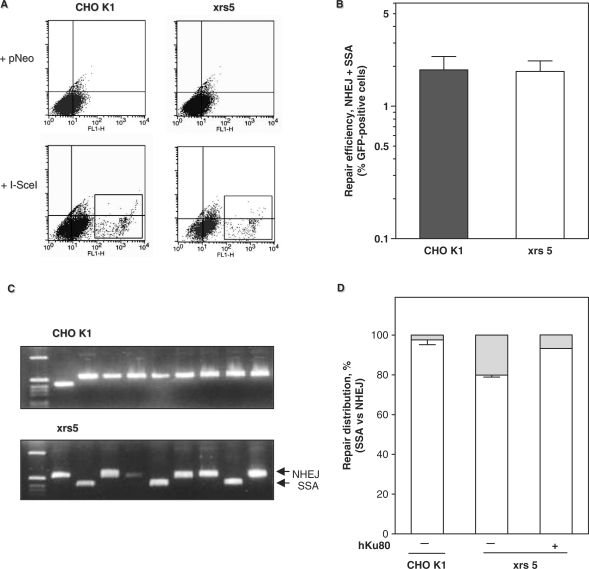
Figure 2.
Ku80 deficiency reduces NHEJ and promotes SSA. (A) Flow cytometric analysis of CHO K1 and xrs5 cells containing pEJSSA 48 h after transfection of a control vector (+pNeo) or the I-SceI expression vector (+I-SceI). Green fluorescent cells are highlighted by the R2 gate. (B) Total repair efficiency corresponds to the fraction of green fluorescent cells produced 48 h after transfection of the I-SceI expression plasmid. Shown are the means based on seven CHO K1- and four xrs5-independent clones. Differences between CHO K1 and xrs5 clones were not significant (Mann–Whitney, P = 0.97). (C) Examples of PCR amplification of repair junctions using flanking primers P1 and P2. CHO K1 cells (top) lane 1: size marker; lane 2: a 415-bp fragment corresponding to an SSA event: lanes 3–8: ∼550-bp fragments corresponding to NHEJ events. Xrs5 cells (bottom) lane 1: size marker; lanes 3, 6, 9: SSA events, lanes 2, 4, 5, 7, 8, 10: NHEJ events. (D) Relative fractions of SSA (gray) and NHEJ events (white). Mean values are given for two independent CHO K1 and two xrs5 clones. For complementation, pcDNA3.1-hKu80 was cotransfected with the I-SceI expression plasmid.
The GC substrate, pGC, carries a single I-SceI site within the GFP sequence, which is hence disrupted (Figure 1C). After DSB induction, only GC from the 3′-truncated donor copy (520-bp shared homology) but neither NHEJ, SSA, nor crossing-over can restitute a functional GFP.
Ku80-mediated NHEJ suppresses SSA, which can serve as a back-up mechanism for impaired NHEJ in Ku80-deficient cells
The Ku heterodimer mediates classical NHEJ (19,20). In the absence of Ku, however, repair might be shuttled into alternative pathways. To address this possibility, we stably integrated single copies of pEJSSA (Figure 1B) into CHO K1 cells with wild-type Ku80 and Ku80-deficient-xrs5 cells (1). Transient transfection of the I-SceI expression vector into either strain led to numerous green fluorescent cells (Figure 2A). Notably, the mean repair frequency across seven independent clones from each cell line revealed no difference between CHO K1 and xrs5 cells (1.88 ± 0.49% and 1.83 ± 0.37% of green fluorescent cells, respectively; Mann–Whitney test P = 0.927). This indicates that DSB repair was equally efficient with or without functional Ku80, in excellent agreement with our previous findings in MEFs (24).
Next, we wished to elucidate the contributions of NHEJ and SSA to DSB repair in the pEJSSA substrate. Green fluorescent cells were sorted, plated and expanded. PCR across the repair junctions revealed many SSA events in xrs5 but only few in CHO K1 cells (Figure 2C). CHO K1 cells showed a repair distribution of 97.6% NHEJ and 2.4% SSA events while the respective ratio for xrs5 cells was 79.9% and 20.1%, thus representing an ∼8-fold relative increase in SSA (Fisher's Exact test, P < 0.001). To verify that the shift towards SSA required the absence of Ku, we ‘added back’ hKu80 into the xrs5 cells and found that NHEJ was rescued (Figure 2D). Together, these data indicate that NHEJ is only mildly impaired in the absence of Ku80 (∼1.2-fold reduction) and that the SSA repair mode using two nearby repetitive sequences can efficiently replace NHEJ. This observation implies that functional Ku is required to restrict mutagenic SSA. Of note, NHEJ remains the principal DSB repair pathway in xrs5 cells, which may reflect the activity of the remaining NHEJ core proteins or the presence of an alternative end-joining pathway (25,26,35).
NHEJ is error-prone in Ku80-deficient cells
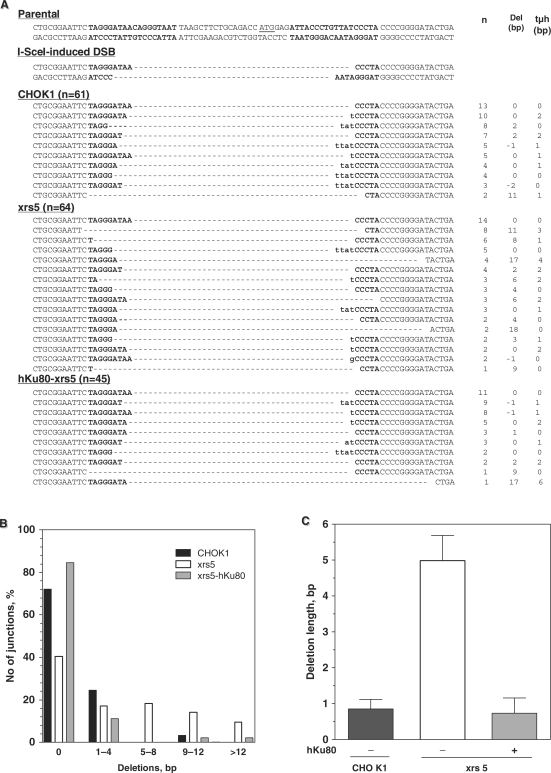
Loss of Ku80 has been reported to affect repair fidelity (22,24,36–38). To address this, 125 different NHEJ breakpoints (61 in CHO K1 and 64 in xrs5 cells) were sequenced (Figure 3A). Cleavage of both I-SceI sites creates noncompatible ends with four-base overhangs that require modification before rejoining. The most straightforward processing involves removal of the terminal A on either side, annealing along TA/AT microhomologies and fill-in synthesis of the missing Ts. In fact, 16.4% of junctions in CHO K1 cells were formed in this way (Figure 3A, see second row line of CHO K1 sequences). In contrast, only 3.1% of junctions were aligned along terminal TA/AT microhomologies in xrs5 cells (Figure 3A, third line from the bottom of xrs5 sequences), indicating that in the absence of Ku, cells repair DSB with more complex end modifications, including long deletions (Figure 3A and B). 41.8% of junctions in xrs5 cells showed a loss of >4 bp, i.e. error-prone repair with end resection extending beyond the 4-nt overhangs, indicating instability of the ends. In contrast, CHO K1 cells rejoined the vast majority of ends via a high-fidelity mechanism that was restricted to ≤4 bp deletions (59/61 junctions) (Figure 3B). Accordingly, in xrs5 cells deletions were significantly longer than in CHO K1 cells (Figure 3C), i.e. 5.0 ± 0.7 bp versus 0.9 ± 0.3 bp, respectively (Fisher's Exact test, P < 0.0001). Error-prone repair events were almost completely abolished in xrs5 cells upon complementation with hKu80 (Figure 3A–C).
Figure 3.
Loss of Ku80 leads to error-prone end-joining (A) Sequences of NHEJ repair junctions based on pEJSSA obtained from CHO K1, xrs5 and xrs5 cells complemented with hKu80. For the parental and the cleaved sequences, both strands are shown. For the repair products, only the sense strand is displayed. I-SceI recognition sites are depicted in bold. The artificial start codon is underlined in the parental sequence. Del indicates the net loss or gain of base pairs, whereby both single-stranded overhangs were regarded as 4-bp double-stranded DNA. tμh indicates the number of terminal microhomologies used for rejoining. (B) Distribution of the length of deletions generated upon NHEJ. (C) Mean deletion length. Differences between CHO K1 and xrs5 cells were statistically significant (Mann–Whitney, P = 0.03).
Interestingly, two common features of NHEJ-deficient cells were not observed with the pEJSSA substrate: (i) extended sequence loss at the break site and (ii) use of longer microhomologies for end-joining (2,23,24,36,37). In a preceding study, we found an average deletion length of 31 and 30 bp in Ku80 and XRCC4−/− MEF, respectively (24), compared to 5 bp observed here for xrs5 cells. This raises the possibility that the presence of the long flanking sequence repeats restricts uncontrolled nucleolytic activity (see Discussion section). The observed limited end-degradation would also explain why the small built-in microhomologies flanking the DSB were not reached.
Presence of Ku80 suppresses GC in addition to SSA
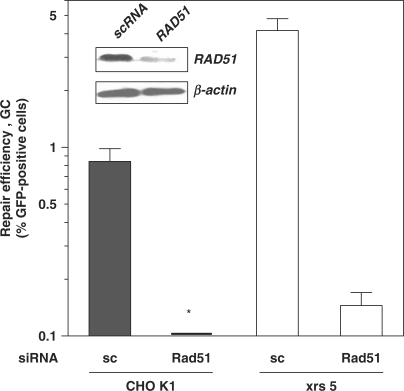
Next, we wished to ascertain whether the observed increase in SSA in Ku80-deficient cells was a reflection of a general increase in homology-mediated repair activities. To measure GC, we stably integrated the pGC substrate into the two CHO strains. Transfection of the I-SceI expression vector into CHO K1 and xrs5 cells resulted in 0.8 ± 0.1% and 4.6 ± 0.7% of green fluorescent cells, respectively (Mann–Whitney test, P = 0.02) (Figure 4). This almost 6-fold increase in the GC frequency in xrs5 cells was inhibited by preceding knockdown of Rad51 (Figure 4, inset). Together, these data suggest that the presence of Ku80 suppresses both SSA and GC mechanisms from acting on DSB.
Figure 4.
Ku80 suppresses Rad51-mediated GC. Repair efficiency by GC assessed by the substrate pGC. Cells were pretreated for 16 h with siRNA and then transfected with the I-SceI expression plasmid. Shown is the mean fraction (± SE) of green fluorescent cells derived from three independent clones after 48 h. Cells not subjected to siRNA yielded essentially the same results as the scambled control (sc) (data not shown). Asterisk: actual value is 0.02%. Inset: Western blot for Rad51 expression in CHO K1 cells 48 h after transfection of siRNA. Anti-Rad51 siRNA (Rad51) reduced protein expression by 80% compared to scrambled control (sc).
Ku80 and Rad51 act together in suppressing SSA
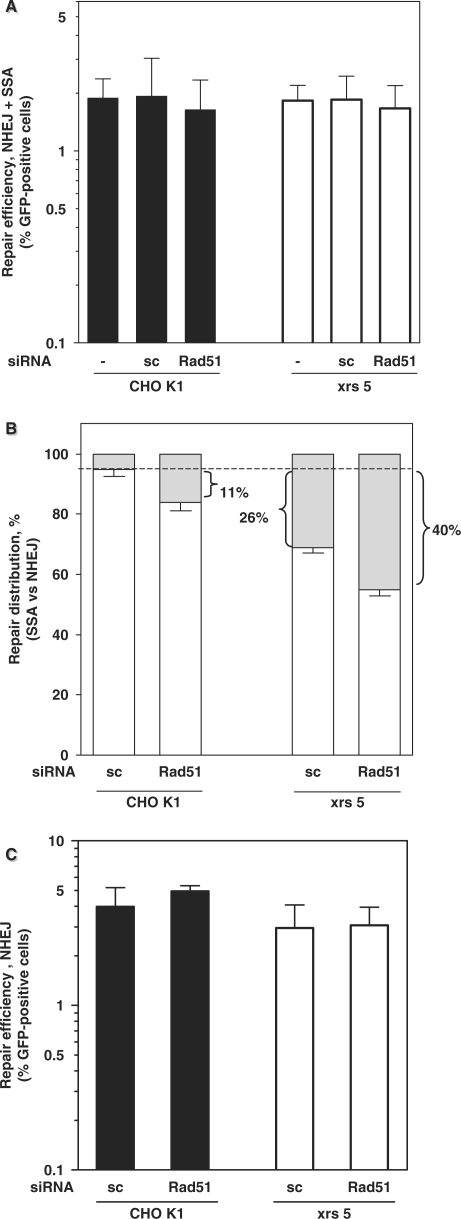
Having established the dominance of Ku80-mediated NHEJ over GC and SSA, we next asked whether Rad51-dependent GC is preferred over SSA. To address this, we knocked down Rad51 in CHO K1 and xrs5 cells carrying pEJSSA. No significant change in the total repair efficiency was observed compared to untreated cells or cells transfected with control siRNA (Figure 5A). To assess the contributions of NHEJ and SSA to I-SceI repair, we applied a TOPO-TA subcloning strategy, which proved to give the same results as raising individual clones (data not shown). Interestingly, Rad51 knockdown in CHO K1 cells resulted in a small yet robust increase in the frequency of SSA by 11% (Figure 5B; second bar), consistent with the idea that SSA is normally restricted in the presence of Rad51. Similarly, in Ku80-deficient xrs5 cells, which already exhibit an increased SSA frequency (Figures 2 and 5B, third bar), Rad51 knockdown raised the fraction of SSA-mediated repair events up to 40% (Figure 5B, fourth bar), suggesting that Ku80 and Rad51 suppress this mutagenic pathway in an additive manner, i.e. 26% due to Ku80 plus 11–14% due to Rad51. These results also illustrate that SSA can be an efficient alternative repair pathway in case NHEJ or GC is impaired.
Figure 5.
Impact of Rad51 function on the NHEJ–SSA balance. (A) Total repair efficiency (NHEJ plus SSA) using pEJSSA in CHO K1 and xrs5 cells without or with 16 h of pretreatment with Rad51 siRNA. Experiments were carried out as before. Neither the individual values nor the means of all CHO K1 versus all xrs five data were significantly different from each other (Mann–Whitney, P = 0.97). siRNA experiments were performed with two independent clones of each strain. Data represent the means of three independent repeat experiments. (B) Relative distribution between SSA (gray) and NHEJ (open bars). Indicated is the difference in the respective fractions of SSA as compared to CHO K1 cells (dashed line). (C) Total NHEJ efficiency using pEJ in CHO K1 and xrs5 cells with or without 16 h of pretreatment with Rad51 or sc siRNA. Data represent the mean (±SE) of three clones each and two independent repeat experiments. Notably, the repair efficiency of clones harboring the pEJ was slightly higher compared to those with the pEJSSA.
Unexpectedly, inactivation of GC in cells carrying pEJSSA by knockdown of Rad51 led to a relative decrease in the NHEJ frequency with an associated rise in SSA, which was not reported previously (28,29). We hypothesized that this NHEJ decrease required the availability of the SSA pathway. We thus knocked down Rad51 in cells carrying the pEJ substrate, which lacks the 50-bp repeat sequences that are substrates for SSA (Figure 1A). We found that the NHEJ frequency was unaffected (Figure 5C) indicating that Rad51-dependent GC does not by itself compete with NHEJ but rather suppresses SSA.
DISCUSSION
The NHEJ pathway is dominant over GC and SSA
In the current study, we investigated the relationship between the three main DSB repair pathways, NHEJ, GC and SSA, using novel chromosomal reporter substrates that stably integrated in hamster cell lines. We found that functional Ku80 acts as a suppressor of GC and SSA (Figures 4 and 2D), thus rendering NHEJ the dominant DSB repair pathway. It has been shown previously that loss of Ku70/80, XRCC4 or DNA–PKcs function (27,28,39–43) increases the frequency of GC. These results indicate that inactivation of any NHEJ core component permits DSBs to be repaired via GC. We now show for the first time that loss of Ku80 also promotes SSA (Figure 2D). GC and SSA commonly require long 3′ single-strands for homology search and strand-annealing, and the initial resection of DSB ends has been suggested to guide the choice between these pathways (29,44–46). However, the Ku protein ‘hides’ DNA ends, protects them from degradation (47–49) and hence prevents channelling repair towards recombination. In addition, Ku may compete with Rad52 for DNA binding. Rad52 has been shown biochemically to bind to and mediate ligation of blunt and cohesive ends similar to Ku (30). Under identical conditions, however, Ku preferably bound to ends with short protrusions while Rad52 strongly favoured long single-stranded overhangs (31). The absence of Ku might significantly facilitate access of Rad52 to double-stranded DNA ends, in particular, as these become prone to nucleolytic attack. In yeast, a single-stranded overhang of only 8–10 bp is sufficient to initiate a Rad52-mediated recombination process (31), which could be either SSA or GC. We observed for the pEJSSA substrate in Ku80-deficient xrs5 cells a mean deletion length of only 5 bp. The end degradation never reached into any homologous repeat sequences although several microhomologies were offered to mediate end-joining (i.e. several copies of CCGC and others, see SSA1/2 sequences depicted in Material and Methods section). In contrast, Ku deficiency in MEFs or xrs5 hamster cells leads to extensive base loss in pure end-joining substrates such as pPHW2 and pEJ (24, and Mansour et al., unpublished results). We conclude that not only the overhang length but also the presence of the nearby tandem copies are recognized and actively guide the repair towards SSA. A similar switch from the end-joining mode to SSA has been described in a cell-free system if the length of homologies extended beyond 25 nt (37).
Rad51 controls the balance between NHEJ and SSA
Both homologous recombination pathways (GC and SSA) may be used if sequences of sufficient homology are available (50–52). In line with the model of a competition between GC and SSA, lack of functional Rad51, Rad54, BARD or BRCA2 decreases the rate of GC and increased SSA (28,52,53). The repair substrate used here, pEJSSA, was not designed to monitor both homology-directed processes but instead addressed the relationship between SSA and NHEJ. We not only found a competition between those two pathways, but also discovered a hitherto unknown role for Rad51 controlling this balance. Knockdown of Rad51 increased the usage of SSA at the expense of NHEJ even in Ku80-proficient cells (Figure 5B). Rad51 may normally hinder Rad52 from interfering with end-joining as it physically associates with Rad52 after DNA damage (54,55). This Rad51–Rad52 interaction simultaneously promotes strand-exchange during GC (56,57) and inhibits Rad52-mediated strand annealing for SSA (55). In the absence of Rad51, Rad52 is freely available to compete with Ku for end binding and may promote SSA as it retains its Rad51-independent strand-annealing function (57). These results suggest that Rad51 indirectly supports NHEJ via interference with SSA. In contrast, using the pure end-joining substrate pEJ, we verified that Rad51 does not interfere with NHEJ in the absence of a SSA mechanism (Figure 5C).
Features of the chromosomal repair substrates
The use of I-SceI-based repair assays has greatly advanced our understanding of the molecular mechanisms and genetic determinants of homologous recombination (8–11,28,43,51), and more recently, several investigators have begun to successfully employ NHEJ substrates (21,22,32,58–60). However, several caveats need to be recognized. For example, studying the impact of a genetic manipulation on the functional repair readout may only produce evidence of an indirect relationship. It also remains to be defined to which extent I-SceI-type ends are models for DSB generated during normal DNA metabolism or after exposure to DNA-damaging agents. By their nature, I-SceI assays select DSB induction and processing events that trigger the reporter/selection signal. As a result, repair efficiency and pathway utilization may be to a certain degree assay specific. With regard to the NHEJ assays employed here, recombination events require a pop-out of the sequence between the two tandem I-SceI sites. Thus, factors that influence simultaneous I-SceI cleavage and synapsis of the cleaved ends can principally affect the repair readout.
Of note, however, we and Lopez and colleagues recently reported a strikingly different phenotype of XRCC4- and Ku80-deficient cells with regard to the repair of I-SceI-induced DSBs, which mirrored the embryonic lethality of XRCC4 knock-out mice as opposed to the viability of the Ku80 knock-out (23,24). These findings suggest that I-SceI ends may be representative of DSB generated during normal cell development.
CONCLUSION
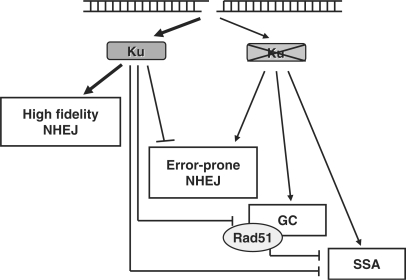
We propose a hierarchical model for DSB repair (Figure 6), which is dominated by the Ku protein. Due to its abundance and high affinity (2), Ku occupies all DNA ends and initiates high fidelity end-joining. In the absence of Ku, NHEJ shifts to an error-prone mode, which either relies on the remaining NHEJ core proteins or involves an alternative end-joining pathway (25,26). In addition, homology-mediated recombinational repair pathways (SSA and GC) can partly substitute for NHEJ and rescue repair proficiency. Yet, the reverse does not occur, as loss of Rad51 function is not compensated by an increased repair via NHEJ. This is consistent with the notion that the 3′-ssDNA ends generated as substrates for homologous recombination can no longer be channelled towards NHEJ. Together, the data imply that the most mutagenic pathway, the SSA pathway, is the least desirable option for the cell, as it is suppressed by both Ku80 and Rad51.
Figure 6.
Hierarchical DSB repair model. Illustration of the main findings based on the I-SceI assays and genetic analysis employed here. Proposed promoting and inhibitory effects of Ku80 are indicated. High-fidelity and error-prone NHEJ are operationally defined. See text for details.
ACKNOWLEDGEMENTS
This work was supported by the German Cancer Aid (Deutsche Krebshilfe, Dr Mildred Scheel Stiftung, Grant No. Da-10-1510 to JDD) and by the German Federal Ministry of Education and Research (Grant No. 02S8427 to JDD). H.W. was in part supported by a grant from the Susan G. Komen for the Cure Breast Cancer Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wyman C, Kanaar R. DNA double-strand break repair: all's well that ends well. Annu. Rev. Genet. 2006;40:363–383. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- 2.Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 3.Haber JE, Leung WY. Lack of chomosome territoriality in yeast: promiscous rejoining of broken chromosome ends. Proc. Natl Acad. Sci. 1996;93:13949–13954. doi: 10.1073/pnas.93.24.13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris T, Thacker J. Formation of large deletions by illegitimate recombination in the HPRT gene of primary human fibroblasts. Proc. Natl Acad. Sci. 1993;90:1392–1396. doi: 10.1073/pnas.90.4.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Difilippantonio MJ, Zhu J, Chen HT, Meffre E, Nussenzweig MC, Max EE, Roed T, Nussenzweig A. DNA repair protein Ku80 suppress chromosomal aberrations and malignant transformation. Nature. 2000;404:510–514. doi: 10.1038/35006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rothkamm K, Kühne M, Jeggo PA, Löbrich M. Radiation-induced genomic rearrangements formed by nonhomologous end-joining of DNA double-strand breaks. Cancer Res. 2001;61:3886–3893. [PubMed] [Google Scholar]
- 7.Soutoglou E, Dorn JF, Sengupta K, Jasin J, Nussenzweig A, Ried T, Danuser G, Misteli T. Positional stability of single double-strand breaks in mammalian cells. Nat. Cell Biol. 2007;9:675–682. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Golding SE, Rosenberg E, Khalil A, McEwen A, Holmes M, Neill S, Povirk LF, Valerie K. Double-strand break repair by homologous recombination is regulated by cell-cycle-independent signaling via ATM in human glioma cells. J. Biol. Chem. 2004;279:15402–15410. doi: 10.1074/jbc.M314191200. [DOI] [PubMed] [Google Scholar]
- 9.Saleh-Gohari N, Helleday TH. Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells. Nucleic Acids Res. 2004;32:3683–3688. doi: 10.1093/nar/gkh703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moynahan ME, Jasin M. Loss of heterozygosity induced by a chromosomal double-strand break. Proc. Natl Acad. Sci. 1997;94:8988–8993. doi: 10.1073/pnas.94.17.8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richardson C, Mohynahan ME, Jasin M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lasko D, Cavenee W, Nordenskjöld M. Loss of constitutional heterozygocity in human cancer. Annu. Rev. Genet. 1991;25:281–314. doi: 10.1146/annurev.ge.25.120191.001433. [DOI] [PubMed] [Google Scholar]
- 13.Sugawara N, Ira G, Haber JE. DNA length dependence of the single-strand annealing pathway and the role of saccharomyces cerevisiae RAD59 in double-strand break repair. Mol. Cell. Biol. 2000;20:5300–5320. doi: 10.1128/mcb.20.14.5300-5309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivanow EL, Sugawara N, Fishman-Loebell J, Haber JE. Genetic requirements for single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.International human genome sequencing consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 16.Strout MP, Marcucci G, Bloomfield CD, Caligiuri MA. The partial tandem duplication of ALL1 (MLL) is consistently generated by Alu-mediated homologous recombination in acute myeloid leukemia. Proc. Natl Acad. Sci. USA. 1998;95:2390–2395. doi: 10.1073/pnas.95.5.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elliott B, Richardson C, Jasin M. Chromosomal translocation mechanisms at intronic Alu elements in mammalian cells. Mol. Cell. 2005;17:885–894. doi: 10.1016/j.molcel.2005.02.028. [DOI] [PubMed] [Google Scholar]
- 18.Weinstock DM, Elliott B, Jasin M. A model of oncogenic rearrangements: differences between chromosomal translation mechanisms and simple double-strand break repair. Blood. 2006;107:777–780. doi: 10.1182/blood-2005-06-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mari PO, Florea BI, Persengiev SP, Verkaik NS, Brüggenwirth HT, Modesti M, Giglia-Mari G, Bezstarosti K, Demmers JAA, Luider TM, et al. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc. Natl Acad. Sci. USA. 2006;103:18597–18602. doi: 10.1073/pnas.0609061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Downs J, Jackson SA. A means to DNA end: the many roles of Ku. Nat. Rev., Mol. Cell Biol. 2004;5:367–378. doi: 10.1038/nrm1367. [DOI] [PubMed] [Google Scholar]
- 21.Ma JL, Kim EM, Haber JE, Lee SE. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping sequences. Mol. Cell. Biol. 2003;23:8820–8828. doi: 10.1128/MCB.23.23.8820-8828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guirouilh-Barbat J, Huck S, Bertrand P, Pirzio L, Desmaze C, Sabatier L, Lopez BS. Impact of the Ku80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol. Cell. 2004;14:611–623. doi: 10.1016/j.molcel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 23.Guirouilh-Barbat J, Rass E, Plo I, Bertrand P, Lopez BS. Defects in XRCC4 and Ku80 differentially affect the joining of distal non-homologous ends. Proc. Natl Acad. Sci. USA. 2007;104:20902–20907. doi: 10.1073/pnas.0708541104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schulte-Uentrop L, El Awady RA, Schliecker L, Willers H, Dahm-Daphi J. Distinct roles of XRCC4 and Ku80 in non-homologous endjoining of enzyme- and radiation-induced DNA double-strand breaks. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn094. [Epub ahead of print, 10 March] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang M, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G. PARP-1 and Ku compete for repair of DNa double-strand breaks by distict NHEJ pathways. Nucleic Acids Res. 2007;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III is an alternative route for DNA double-strand breaks rejoining. J. Biol. Chem. 2004;279:55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 27.Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stark JM, Pierce AJ, Oh J, Pastink A, Jasin M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol. Cell. Biol. 2004;24:9305–9316. doi: 10.1128/MCB.24.21.9305-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frank-Vaillant M, Marcand S. Transient stability of DNA ends allows nonhomologous end joining to precede homologous recombination. Mol. Cell. 2002;10:1189–1199. doi: 10.1016/s1097-2765(02)00705-0. [DOI] [PubMed] [Google Scholar]
- 30.Van Dyck E, Stasiak AZ, Stasiak A, West SC. Binding of double-strand breaks in DNA by human RAD52 protein. Nature. 1999;398:728–731. doi: 10.1038/19560. [DOI] [PubMed] [Google Scholar]
- 31.Ristic D, Modesti M, Kanaar R, Wyman C. Rad52 and Ku bind to different DNA structures produced early in double-strand break repair. Nucleic Acids Res. 1997;31:5229–5237. doi: 10.1093/nar/gkg729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dahm-Daphi J, Hubbe P, Horvath F, El-Awady RA, Bouffard KE, Powell SN, Willers H. Non-homologous end-joining of site-specific but not of radiation-induced DNA double-strand breaks is reduced in the presence of wild-type p53. Oncogene. 2005;24:1663–1672. doi: 10.1038/sj.onc.1208396. [DOI] [PubMed] [Google Scholar]
- 33.Willers H, Husson J, Lee LW, Hubbe P, Horvath F, Powell SN, Dahm-Daphi J. Distinct mechanisms of non-homologous end-joining in the repair of site-directed chromosomal breaks with non-complementary and complementary ends. Radiat. Res. 2006;166:567–574. doi: 10.1667/RR0524.1. [DOI] [PubMed] [Google Scholar]
- 34.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 35.Nussenzweig A, Nussenzweig MC, McGlynn AP, Wasson G, O’Connor J, McKelvey-Martin VJ, Downes CS. A backup DNA repair pathway moves to the front. Cell. 2007;131:223–225. doi: 10.1016/j.cell.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 36.Kabotyanski EB, Gomelsky L, Han J-O, Stamato TD, Roth DB. Double-strand break repair in Ku86- and XRCC4-deficient cells. Nucleic Acids Res. 1998;26:5333–5342. doi: 10.1093/nar/26.23.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuhfittig-Kulle S, Feldmann E, Odersky A, Kuliczkowska A, Goedecke W, Eggert A, Pfeiffer P. The mutagenic potential of non-homologous endjoining in the absence of the NHEJ core factors Ku70/80, DNA-PKcs and XRCC4-LigIV. Mutagenesis. 2007;22:217–233. doi: 10.1093/mutage/gem007. [DOI] [PubMed] [Google Scholar]
- 38.Feldmann E, Schmiemann V, Goedecke W, Reichenberger S, Pfeiffer P. DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic Acids Res. 2000;28:2585–2596. doi: 10.1093/nar/28.13.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delacote F, Han M, Stamato TD, Jasin M, Lopez BS. An XRCC4 defect or Wortmannin stimulates homologous recombination specifically induced by double-strand breaks in mammalian cells. Nucleic Acids Res. 2002;30:3454–3463. doi: 10.1093/nar/gkf452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clikeman JA, Khalsa J, Barton SL, Nickoloff JA. Homologous recombinational repair of double-strand breaks in yeast is enhanced by MAT through yKu-dependent and -independent mechanisms. Genetics. 2001;157:579–588. doi: 10.1093/genetics/157.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fukushima T, Takata M, Morrison C, Araki R, Fujimori A, Abe M, Tatsumi K, Jasin M, Dhar PK, Sonoda E, et al. Genetic analysis of the DNA-dependent protein kinase reveals an inhibitory role of Ku in late S-G2 phase DNA double-strand break repair. J. Biol. Chem. 2001;276:44413–44418. doi: 10.1074/jbc.M106295200. [DOI] [PubMed] [Google Scholar]
- 42.Cui X, Yu Y, Gupta S, Cho YM, Lees-Miller SP, Meek K. Autophosphorylation of DNA-dependent protein kinase regulates DNA end-processing and may also alter double-strand breaks repair pathway choice. Mol. Cell. Biol. 2005;25:10842–10852. doi: 10.1128/MCB.25.24.10842-10852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allen C, Halbrook J, Nickoloff JA. Interactive competition between homologous recombination and non-homologous endjoining. Mol. Cancer Res. 2003;1:913–920. [PubMed] [Google Scholar]
- 44.Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, et al. DNA end resection, homologous recombination and DNA damage require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aylon Y, Liefshitz B, Kupiec M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO. 2004;23:4868–4675. doi: 10.1038/sj.emboj.7600469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scully R, Xie A. In my end is my beginning: control of end resection and DSBR pathway ‘choice'by cyclin-dependent kinases. Oncogene. 2005;24:2871–2876. doi: 10.1038/sj.onc.1208609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walker JR, Corpina RA, Goldberg J. Structure of the ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 48.Mimori T, Hardin JA. Mechanism of interaction between Ku protein and DNA. Genetic analysis of ionising radiation sensitive mutants of cultured mammalian cell lines. J. Biol. Chem. 1986;261:10375–10379. [PubMed] [Google Scholar]
- 49.Liang F, Jasin M. Ku80-deficient cells exhibit excess degradation of extrachromosomal DNA. J. Biol. Chem. 1996;271:14405–14111. doi: 10.1074/jbc.271.24.14405. [DOI] [PubMed] [Google Scholar]
- 50.Fishman-Lobell J, Rudin N, Haber JE. Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Mol. Cell. Biol. 1992;12:1292–1303. doi: 10.1128/mcb.12.3.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schildkraut E, Miller CA, Nickoloff JA. Gene conversion and deletion frequencies during double-strand break repair in human cells are controlled by the distance between direct repeats. Nucleic Acids Res. 2005;33:1574–1580. doi: 10.1093/nar/gki295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dronkert MLG, Beverloo HB, Johnson RD, Hoeijmakers JHJ, Jasin M, Kanaar R. Mouse RAD54 affects DNA double-strand break repair and sister chromatid exchange. Mol. Cell. Biol. 2000;20:3147–3156. doi: 10.1128/mcb.20.9.3147-3156.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tutt A, Bertwistle D, Valentine J, Gabriel A, Swift S, Ross G, Griffin C, Thacker J, Ashworth A. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J. 2001;20:4704–4716. doi: 10.1093/emboj/20.17.4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen G, Yuan SSF, Liu W, Xu Y, Trujillo K, Song B, Cong F, Goff S, Wu Y, Arlinghaus R, et al. Radiation-induced assembly of Rad51 and Rad52 recombination complex requires ATM and cABL. J. Biol. Chem. 1999;274:12748–12752. doi: 10.1074/jbc.274.18.12748. [DOI] [PubMed] [Google Scholar]
- 55.Wu Y, Kantake N, Sugiyama T, Kowalczykowski SC. Rad51 protein controls rad52-mediated DNA Annealing. J. Biol. Chem. 2008 doi: 10.1074/jbc.M801097200. [Epub ahead of print, 12 March 2008] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Benson FE, Baumann P, West SC. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature. 1998;391:401–404. doi: 10.1038/34937. [DOI] [PubMed] [Google Scholar]
- 57.Krejci L, Song B, Bussen W, Rothstein R, Mortensen UH, Sung P. Interaction with Rad51 is indispensible for recombination mediator function of Rad52. J. Biol. Chem. 2002;42:40132–40141. doi: 10.1074/jbc.M206511200. [DOI] [PubMed] [Google Scholar]
- 58.Lin Y, Waldman BC, Waldman AS. Suppression of high-fidelity double-strand break repair in mammalian chromosomes by pifithrin-.symbol.a, a chemical inhibitor of p53. DNA Repair. 2003;2:1–11. doi: 10.1016/s1568-7864(02)00183-0. [DOI] [PubMed] [Google Scholar]
- 59.van Heemst D, Brugmans L, Verkaik NS, van Gent DC. End-joining of blunt DNA double-strand breaks in mammalian fibroblasts is precise and requires DNA-PK and XRCC4. DNA Repair. 2004;3:43–50. doi: 10.1016/j.dnarep.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 60.Xie A, Hartlerode A, Stucki M, Odate S, Puget N, Kwok A, Nagaraju G, Yan C, Alt FW, Chen J, et al. Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol. Cell. 2007;28:1045–1057. doi: 10.1016/j.molcel.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]