Effects of Codon Distributions and tRNA Competition on Protein Translation (original) (raw)
Abstract
Translation is a central cellular process and the complexity of its mechanism necessitates mathematical frameworks to better understand system properties and make quantitative predictions. We have developed a gene sequence-specific mechanistic model for translation which accounts for all the elementary steps of translation elongation. Included in our model is the nonspecific binding of tRNAs to the ribosomal A site, and we find that the competitive, nonspecific binding of the tRNAs is the rate-limiting step in the elongation cycle for every codon. By introducing our model in terms of the Michaelis-Menten kinetic framework, we determine that these results are due to the tRNAs that do not recognize the ribosomal A site codon acting as competitive inhibitors to the tRNAs that do recognize the ribosomal A site codon. We present the results of a sensitivity analysis to determine the contribution of elongation cycle kinetic parameters of each codon on the overall translation rate, and observe that the translation rates of mRNAs are controlled by segments of rate-limiting codons that are sequence-specific. Along these lines, we find that the relative position of codons along the mRNA determines the optimal protein synthesis rate.
INTRODUCTION
Translation, or protein synthesis, is a process that is central to cellular function. It is essentially a template polymerization process (1) consisting of initiation, elongation, and termination phases. Messenger RNA (mRNA), composed of a sequence of codons coding for amino acids, carries genetic information. Initiation occurs with binding of the ribosome to the ribosomal binding site near the 5′ end of the mRNA. During the elongation phase the ribosome facilitates assembly of the polypeptide chain with one amino acid (aa) added per elongation cycle at each codon. Amino acids are delivered to the ribosome by transfer RNAs (tRNAs) in the form of ternary complexes that serve as adaptor molecules between the amino acid and the codon present in the ribosomal A site. Termination involves release of the completed peptide from the ribosome near the 3′ end of the mRNA. Multiple proteins can be synthesized simultaneously on a single mRNA molecule, forming a structure called the polysome (or polyribosome) consisting of several ribosomes simultaneously translating the same mRNA. Polysome size is the number of ribosomes bound to a single mRNA molecule. Hence, the higher the polysome size, the greater the coverage of the mRNA due to ribosomes translating it. Polysomes have been observed experimentally (2), and modern techniques have allowed the quantification of polysome size for almost every mRNA in yeast cells (3).
The sheer complexity of the translation mechanism necessitates mathematical, mechanistic frameworks to better understand the system properties of translation and make quantitative predictions. Several studies have been conducted involving investigating the kinetics of protein synthesis that take into account the ribosome movement on mRNAs (4–6), and other studies (7,8) have involved the effects of competition for ribosomes between mRNAs on cell-wide mapping between mRNA and protein levels. An assumption in these studies is that the elongation kinetics at each codon depends on a single rate constant that is the same for all codon species at all positions along the length of the mRNA. In reality, codons have varying elongation kinetics due to different tRNA availabilities (9) and codon-anticodon compatibilities (10–12), and the multiple elementary steps and translational components involved in the elongation cycle at every codon. Therefore, a better understanding of the properties of translation requires the consideration of the translation elongation phase, accounting for all elongation cycle intermediate steps. In previous work (13), we developed such a kinetic model of the translational machinery that is deterministic and sequence-specific, and accounts for all the elementary steps of the translation mechanism. We also performed a sensitivity analysis to determine the effects of the kinetic parameters and concentrations of the translational components on the protein synthesis rate.
A finding from our model was that tRNA concentrations have almost no impact on protein synthesis rate. However, experimental evidence suggests tRNA concentrations are significant to translation kinetics. The work by Ikemura (14) shows a correlation between tRNA abundances and codon frequencies. Other work demonstrates that synonymous codons (different codons coding for the same amino acid) are not translated at the same rate (11), with higher translation rates for more abundant or major codons (15). Given the difference between experimental results and those determined from our computational studies (13), it is important to note that a simplifying assumption made in our model is that only ternary complexes that recognize the A site codon can bind to the ribosome. In reality, ternary complexes initially bind nonspecifically to the ribosomal A site, which means that both ternary complexes recognizing and not recognizing the ribosomal A site codon can bind to the ribosome in the first intermediate step of the elongation cycle of each codon. The experimentally observed importance of tRNA concentration to protein synthesis kinetics, coupled with our observation that tRNA concentrations are not scarce enough to modulate translation rate, motivates questions about the role the competition between ternary complexes for ribosomal A site binding plays in protein synthesis kinetics.
Hence, in this work we expand our mechanistic framework to account for ternary complex competitive binding to the ribosomal A site. We also expand our sensitivity analysis to make it codon-specific, meaning that we account for the contribution of kinetic parameters and translational component concentrations of each codon on the overall protein synthesis rate. We find that our expanded mechanistic framework predicts lower protein synthesis rates than our previous framework (13). Our sensitivity analysis predicts that, at low polysome sizes, the codons near the 5′ end of the mRNA control protein synthesis rate, at intermediate polysome sizes different configurations of codons along the length of the mRNA control protein synthesis rate, and at high polysome sizes the codons near the 3′ end of the mRNA control protein synthesis rate. Moreover, our sensitivity analysis identifies the competitive, nonspecific binding of the tRNAs to the ribosomal A site as rate-limiting to the elongation cycle for every codon. By introducing our previous (13) and current mechanistic models in terms of the Michaelis-Menten kinetic framework, we determine that these results are due to the tRNAs that do not recognize the ribosomal A site codon acting as competitive inhibitors to the tRNAs that do recognize the ribosomal A site codon. We also observe that the relative position of codons along the mRNA determines the optimal protein synthesis rate, and that the translation rates of mRNAs are controlled by segments of rate-limiting codons that are sequence-specific.
METHODS
Elementary steps of the elongation cycle
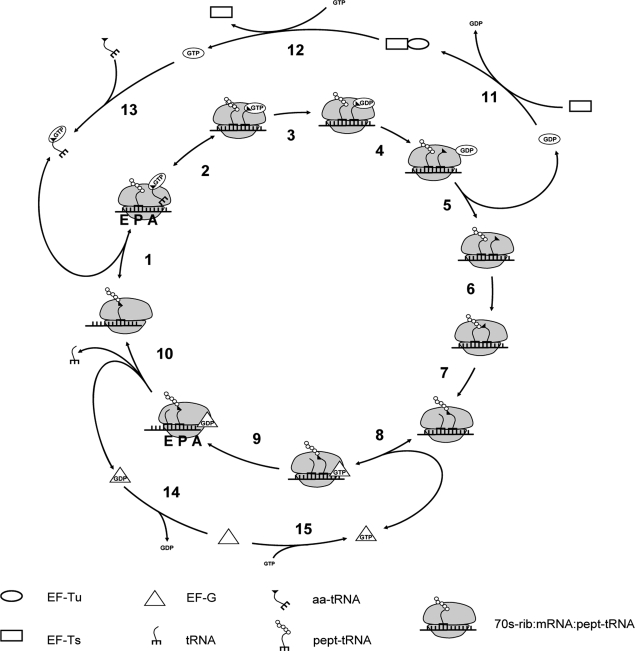
The translation elongation phase is a cyclic process that involves codons, ribosomes, amino acids, tRNAs, elongation factors Tu, Ts, and G, and leads to the assembly of polypeptide chains (Fig. 1). Each amino acyl-tRNA (aa-tRNA) binds to Ef-Tu:GTP, forming a ternary complex (step 13). The ternary complex then binds reversibly to the ribosomal A site in a codon-independent manner (step 1). After finding the correct codon match and reversible codon-dependent binding (step 2), GTP is hydrolyzed (step 3), Ef-Tu:GDP changes position on the ribosome (step 4) and is released (step 5). In a two-step process, Ef-Ts catalyzes regeneration of Ef-Tu:GTP (steps 11 and 12). During accommodation the aa-tRNA undergoes a conformation change and enters the A site (step 6). Transpeptidation then occurs (step 7), where the peptide chain is transferred from the peptidyl-tRNA to the aa-tRNA, resulting in the elongation of the polypeptide chain by one amino acid. Reversible binding of Ef-G:GTP (step 8) facilitates translocation (step 9). During translocation the P site tRNA and codon move to the E site of the ribosome and the A site tRNA and codon move to the P site, resulting in the complex moving toward the 3′ end of the mRNA by one codon. The tRNA in the E site is released along with Ef-G:GDP (step 10), and Ef-G:GTP is recycled in a two-step process (steps 14 and 15).
FIGURE 1.
The elementary mechanistic steps of the translation elongation process. Ribosomal A, P, and E sites indicated on the intermediates between steps 1 and 2 and steps 9 and 10. Step 1: Reversible, codon-independent binding of the ternary complex to the ribosomal A site. Step 2: Reversible, codon-dependent binding of the ternary complex to the ribosomal A site. Step 3: GTP hydrolysis. Step 4: Ef-Tu:GDP position change on the ribosome. Step 5: Ef-Tu:GDP release. Step 6: aa-tRNA accommodation. Step 7: Transpeptidation. Step 8: Reversible binding of Ef-G:GTP. Step 9: Translocation. Step 10: E site tRNA release. Steps 11 and 12: Ef-Ts catalyzed regeneration of Ef-Tu:GTP. Step 13: Ef-Tu:GTP binding to the aa-tRNA. Steps 14 and 15: Regeneration of Ef-G:GTP.
Mathematical model
In this section, we introduce a mechanistic framework that incorporates the kinetics of all the intermediate steps of the translation elongation cycle occurring at a given codon in a single expression. A summary of the assumptions made in this formulation, along with descriptions of the variables and parameters, can be found in Appendix A. A detailed description of this model can be found in our previous study (13).
The initiation rate is described as
 |
(1) |
|---|
where _k_I,r is the initiation rate constant of mRNA r, R(f) is the free ribosome concentration, and 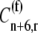 is the concentration of mRNA r having a free ribosomal binding site.
is the concentration of mRNA r having a free ribosomal binding site.
The elongation rate at codon n along the length of the mRNA species r is described as
 |
(2) |
|---|
where the subscript i denotes the P site codon species, the subscript j denotes the A site codon species, the subscript n denotes the position of the ribosomal P site codon, 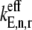 is the effective elongation rate constant, and _S_ij,n,r is the fraction of the mRNA species r concentration with codon position n occupied by the P site of a translating ribosome. Ribosome movement along the length of the sequence is dependent on the conditional probability that the codon adjacent to the codon occupied by the front of the ribosome is free, given that the previous codon is occupied by the front of the ribosome, _U_n,r, and _M_r is the concentration of mRNA r.
is the effective elongation rate constant, and _S_ij,n,r is the fraction of the mRNA species r concentration with codon position n occupied by the P site of a translating ribosome. Ribosome movement along the length of the sequence is dependent on the conditional probability that the codon adjacent to the codon occupied by the front of the ribosome is free, given that the previous codon is occupied by the front of the ribosome, _U_n,r, and _M_r is the concentration of mRNA r.
The effective elongation rate constant at codon position n, 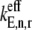 (Eq. 3), is comprised of terms representing the kinetics of each of the translation elongation cycle intermediate steps occurring at that codon, and these terms depend on the reaction rate constants corresponding to the elongation cycle intermediate steps (13):
(Eq. 3), is comprised of terms representing the kinetics of each of the translation elongation cycle intermediate steps occurring at that codon, and these terms depend on the reaction rate constants corresponding to the elongation cycle intermediate steps (13):
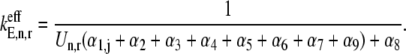 |
(3) |
|---|
A summary of the effective elongation rate constant terms is included in Table 1 and Zouridis and Hatzimanikatis (13). In this work, we investigate effects of ternary complex competition for ribosomal A site binding, so it is important to note that the first effective elongation rate constant term, _α_1,j, corresponds to the reversible, codon-independent binding of the ternary complex to the ribosomal A site codon species j. The expression for _α_1,j is
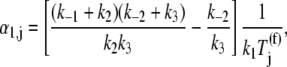 |
(4) |
|---|
where _k_1, _k_−1, _k_2, _k_−2, and _k_3 are reaction rate constants corresponding to ternary complex binding. Free ternary complex concentrations 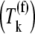 are of species k, with k ∈ K, where K is the set of ternary complex species. Hence,
are of species k, with k ∈ K, where K is the set of ternary complex species. Hence,  is the free ternary complex concentration of species j recognizing A site codon species j. Equation 4 was derived assuming that only ternary complexes recognizing the ribosomal A site codon bind to the ribosome during nonspecific binding. In reality, all ternary complexes species can bind to the ribosome during the codon-independent binding intermediate step, regardless of whether or not they recognize the A site codon. Hence, in this work we relax our original assumption by allowing all ternary complex species to be able to bind to the ribosomal A site at this step, yielding the following expression for the nonspecific ternary complex binding term of the effective elongation rate constant,
is the free ternary complex concentration of species j recognizing A site codon species j. Equation 4 was derived assuming that only ternary complexes recognizing the ribosomal A site codon bind to the ribosome during nonspecific binding. In reality, all ternary complexes species can bind to the ribosome during the codon-independent binding intermediate step, regardless of whether or not they recognize the A site codon. Hence, in this work we relax our original assumption by allowing all ternary complex species to be able to bind to the ribosomal A site at this step, yielding the following expression for the nonspecific ternary complex binding term of the effective elongation rate constant,
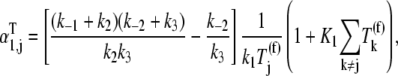 |
(5) |
|---|
where the term  accounts for ternary complex competitive binding and _K_1 = _k_1/_k_−1. By replacing _α_1,j with
accounts for ternary complex competitive binding and _K_1 = _k_1/_k_−1. By replacing _α_1,j with 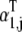 in the expression for the effective elongation rate constant (Eq. 3), we define
in the expression for the effective elongation rate constant (Eq. 3), we define 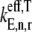 to be the effective elongation rate constant accounting for ternary complex competitive binding:
to be the effective elongation rate constant accounting for ternary complex competitive binding:
 |
(6) |
|---|
The termination rate is described as
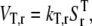 |
(7) |
|---|
where _k_T,r is the termination rate constant of mRNA r and  is the total concentration of ribosomes on mRNA r that have completed the translation elongation phase.
is the total concentration of ribosomes on mRNA r that have completed the translation elongation phase.
TABLE 1.
Effective elongation rate constant terms
The dynamics describing the transition between the states of the elongation phase are as follows:
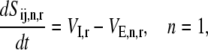 |
(8) |
|---|
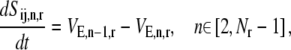 |
(9) |
|---|
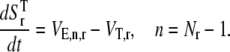 |
(10) |
|---|
The total ribosome and codon concentrations are expressed by Eqs. 11 and 12, respectively,
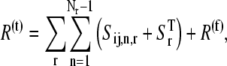 |
(11) |
|---|
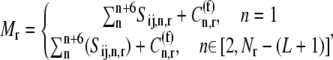 |
(12) |
|---|
where  is the concentration of free codons at position n of mRNA r.
is the concentration of free codons at position n of mRNA r.
Sensitivity analysis
We investigate the effects of elongation cycle kinetics at each codon along the length of the mRNA on the steady-state protein synthesis rate by examining the flux control coefficients, 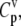 which are defined as fractional flux changes with respect to fractional input parameter changes (16). Similar to the Summation Theorem (16), we can show that the sum of the control coefficients with respect to the reaction rate constants for an mRNA species that is not competing for translational resources with other mRNA species is equal to one,
which are defined as fractional flux changes with respect to fractional input parameter changes (16). Similar to the Summation Theorem (16), we can show that the sum of the control coefficients with respect to the reaction rate constants for an mRNA species that is not competing for translational resources with other mRNA species is equal to one,
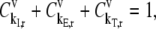 |
(13) |
|---|
where 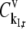 and
and 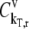 are the fractional changes in flux with respect to fractional changes in the initiation and termination rate constants, respectively. A detailed derivation of the Summation Theorem is included in section 5.3.1 of Heinrich and Schuster (17). The control coefficient
are the fractional changes in flux with respect to fractional changes in the initiation and termination rate constants, respectively. A detailed derivation of the Summation Theorem is included in section 5.3.1 of Heinrich and Schuster (17). The control coefficient 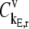 is the fractional change in flux with respect to the simultaneous fractional change in the elongation rate constant,
is the fractional change in flux with respect to the simultaneous fractional change in the elongation rate constant, 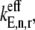 of every codon expressed as
of every codon expressed as
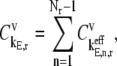 |
(14) |
|---|
where  is the control coefficient corresponding to the elongation step occurring at the codon at position n on the mRNA and is the fractional change in flux with respect to the fractional change in the effective elongation rate constant at position n. The control coefficient with respect to the effective elongation rate constant at codon position n,
is the control coefficient corresponding to the elongation step occurring at the codon at position n on the mRNA and is the fractional change in flux with respect to the fractional change in the effective elongation rate constant at position n. The control coefficient with respect to the effective elongation rate constant at codon position n, 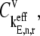 is equal to the sum of the control coefficients with respect to the reaction rate constants of the elongation cycle intermediate steps at codon position n, where
is equal to the sum of the control coefficients with respect to the reaction rate constants of the elongation cycle intermediate steps at codon position n, where
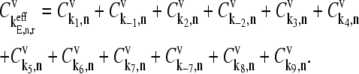 |
(15) |
|---|
Equations 14 and 15 are also applied to determine control coefficients of the elongation steps along the length of the mRNA with under ternary complex competitive binding conditions, 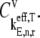 Details of the flux control coefficient derivation are included in previous work (13).
Details of the flux control coefficient derivation are included in previous work (13).
COMPUTATIONAL STUDIES
We utilize our mathematical model of protein synthesis and the sensitivity analysis to investigate the steady-state translation properties of Escherichia coli mRNAs as functions of polysome size with and without accounting for ternary complex competitive binding to the ribosomal A site. Polysome size is the number of ribosomes bound to a single mRNA molecule, so the higher the polysome size, the greater the coverage of the mRNA by ribosomes. Hence, we define ρ to be the fraction of the mRNA molecule covered by translating ribosomes, as
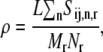 |
(16) |
|---|
where L = 12 is the number of codons covered by the ribosome (18–20) and _N_r is the number of codons of mRNA r. The ribosomal fractional coverage, ρ, varies between zero (no ribosomes translating the mRNA) and one (full coverage of the mRNA by translating ribosomes). The values for the concentration of each mRNA species r, _M_r, the free ribosome concentration, R(f), the free ternary complex concentrations, 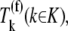 and the free Ef-G concentration, G(f), applied in these studies, along with the reaction rate constants, _k_1, _k_−1, _k_2, _k_−2, _k_3, _k_4, _k_5, _k_6, _k_7, _k_−7, _k_8, and _k_9, are the same as those used in previous work (13). It is important to note that the concentrations of the translational machinery and the reaction rate constants are derived from experimental data. The mRNA concentration can be found in Bremer and Dennis (21), the ribosome concentration can be found in the literature (21,22), the ternary complex concentrations can be found in Dong et al. (23), and the Ef-G concentration can be found in Hershey (1). The reaction rate constants listed above can be found in the literature (24–27), and the rate constants for translation initiation, _k_I,r, and translation termination, _k_T,r, are allowed to vary in our mechanistic framework. Also, the method used to calculate steady-state translation rate as a function of polysome size is the same as that from previous work (13). The obtained steady-state translation rates are applied to the sensitivity analysis to determine the flux control coefficients.
and the free Ef-G concentration, G(f), applied in these studies, along with the reaction rate constants, _k_1, _k_−1, _k_2, _k_−2, _k_3, _k_4, _k_5, _k_6, _k_7, _k_−7, _k_8, and _k_9, are the same as those used in previous work (13). It is important to note that the concentrations of the translational machinery and the reaction rate constants are derived from experimental data. The mRNA concentration can be found in Bremer and Dennis (21), the ribosome concentration can be found in the literature (21,22), the ternary complex concentrations can be found in Dong et al. (23), and the Ef-G concentration can be found in Hershey (1). The reaction rate constants listed above can be found in the literature (24–27), and the rate constants for translation initiation, _k_I,r, and translation termination, _k_T,r, are allowed to vary in our mechanistic framework. Also, the method used to calculate steady-state translation rate as a function of polysome size is the same as that from previous work (13). The obtained steady-state translation rates are applied to the sensitivity analysis to determine the flux control coefficients.
In this work, we consider three cases with respect to ternary complex binding to the ribosomal A site codon. These cases differ by how the free ternary complex concentrations are applied for the quantification of the effective elongation rate constants 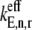 and
and 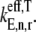 In Table 2 we list the free ternary complex concentrations and corresponding magnitudes for
In Table 2 we list the free ternary complex concentrations and corresponding magnitudes for 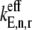 and
and 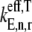 for all the ternary complex species. The effective elongation rate constant magnitudes shown correspond to _U_n,r = 1. The following assumptions were employed for each case:
for all the ternary complex species. The effective elongation rate constant magnitudes shown correspond to _U_n,r = 1. The following assumptions were employed for each case:
TABLE 2.
Effective elongation rate constant magnitudes for each E. coli ternary complex species
Case I: Noncompetitive binding
Only the ternary complex species recognizing the ribosomal A site codon are allowed to participate in the nonspecific binding step of the elongation cycle. In studies considering Case I, the ternary complex concentrations recognizing the A site codons, 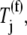 are set equal to the median concentration,
are set equal to the median concentration, 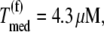 in the effective elongation rate constant expressions
in the effective elongation rate constant expressions 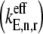 of every codon. Although variations in ternary complex concentrations cause variations in effective elongation rate constant magnitudes, we have observed that these differences are negligibly small under noncompetitive binding conditions (13).
of every codon. Although variations in ternary complex concentrations cause variations in effective elongation rate constant magnitudes, we have observed that these differences are negligibly small under noncompetitive binding conditions (13).
Case II: Uniform competitive binding
All ternary complex species are allowed to participate in the nonspecific binding step of the elongation cycle. Similar to Case I, in studies considering Case II, the ternary complex concentrations recognizing the A site codons, 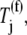 are set equal to the median concentration,
are set equal to the median concentration, 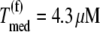 in the effective elongation rate constant expressions
in the effective elongation rate constant expressions 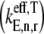 of every codon. Although we observe in this work that variations in ternary complex concentrations cause significant variations in elongation rate-constant magnitudes, Case II allows us to study the effects of ternary complex competitive binding in a codon-independent manner. Moreover, because all codons are treated uniformly in Case II, we study the effects of ternary complex competitive binding in a sequence-independent manner.
of every codon. Although we observe in this work that variations in ternary complex concentrations cause significant variations in elongation rate-constant magnitudes, Case II allows us to study the effects of ternary complex competitive binding in a codon-independent manner. Moreover, because all codons are treated uniformly in Case II, we study the effects of ternary complex competitive binding in a sequence-independent manner.
Case III: Nonuniform competitive binding
Similar to Case II, all ternary complex species are allowed to participate in the nonspecific binding step of the elongation cycle. However, in studies considering Case III, the ternary complex concentrations recognizing the A site codons,  are set equal to their respective physiological levels in the effective elongation rate constant expression
are set equal to their respective physiological levels in the effective elongation rate constant expression 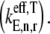 Because codons are not treated uniformly in Case III, we study the effects of ternary complex competitive binding in both a codon- and sequence-specific manner.
Because codons are not treated uniformly in Case III, we study the effects of ternary complex competitive binding in both a codon- and sequence-specific manner.
Effects of ternary complex competitive binding on the relationships between protein synthesis properties and polysome size
In these studies we apply Cases I and II to investigate the translation properties of the trpR gene of E. coli in both a codon- and sequence-independent manner.
Effects of ternary complex competitive binding on the relationship between translation rate and polysome size
We observe that as ribosomal fractional coverage increases, the protein synthesis rate increases, reaches a maximum, and then decreases under both competitive (Fig. 2, curves ii and iii) and noncompetitive (Fig. 2, curve i) binding conditions. Included in Fig. 2 are results for Cases I and II (curves i and iii, respectively), and Case II with all the codons in the sequence recognized by the ternary complex species having the maximum free concentration of 23.1 _μ_M (curve ii). The translation rates determined under Case I are higher at each polysome size than those determined under Case II. This result is due to the large difference in the effective elongation rate constant magnitudes under the two cases. For _U_n,r = 1, under Case I, 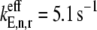 (curve i), while under Case II,
(curve i), while under Case II, 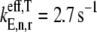 (curve ii) and
(curve ii) and 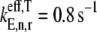 (curve iii). Because the effective elongation rate constant magnitudes under Case I are higher than those under Case II, the translation rates observed under Case I are higher than those observed under Case II.
(curve iii). Because the effective elongation rate constant magnitudes under Case I are higher than those under Case II, the translation rates observed under Case I are higher than those observed under Case II.
FIGURE 2.
Relationship between translation rate and polysome size for (i) Case I, and Case II with all codons recognized by the ternary complex species of (ii) maximum and (iii) median concentrations.
Rate-limiting steps and polysome size
We applied the control analysis framework to the model to determine if translation is initiation-, elongation-, or termination-limited under different polysome sizes. We observe that under both Cases I and II, translation is initiation-limited for ρ < 0.5; elongation-limited for 0.5 < _ρ_ < 0.99, with elongation control maximal at the same ribosomal fractional coverage that specific protein production rate is maximal; and termination-limited for _ρ_ > 0.99.
Relationship between codon-specific control of protein translation rate and polysome size
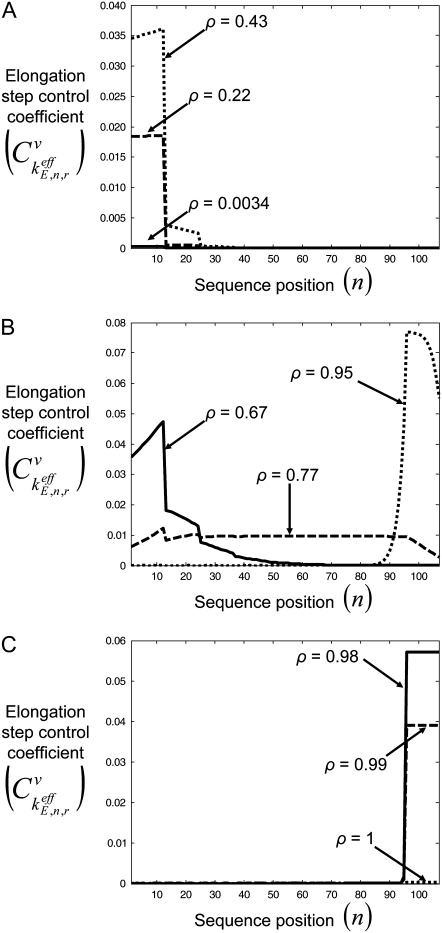
We investigated how control of the elongation phase over translation rate 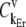 is distributed with respect to the codons along the length of the mRNA at different polysome sizes by examining the control coefficients corresponding to the effective elongation rate constants,
is distributed with respect to the codons along the length of the mRNA at different polysome sizes by examining the control coefficients corresponding to the effective elongation rate constants,  (Fig. 3). We observe that at low polysome sizes the elongation phase control over translation rate lies in the codons near the 5′ end of the mRNA. This result is in agreement with early experimental results demonstrating that point mutations near the start codon of the mRNA cause dramatic changes in protein expression levels (28,29). Also, at intermediate polysome sizes the control is distributed along the length of the mRNA in different configurations, and at high polysome sizes the control lies in the codons near the 3′ end of the mRNA. We observe the same results under both Cases I and II. These results are expected because at low polysome sizes kinetics are initiation-limited (see previous paragraph for discussion), which means that the initiation process limits the progress of protein translation. Hence, the more efficiently the codons near the 5′ end of the mRNA can be translated, the more ribosomes can be transferred to downstream codons along the length of the sequence. Faster transfer of ribosomes due to more efficient translation of these codons elevates protein synthesis rate by increasing the probability of an initiation event occurring without changes to the initiation process being made. The converse is true for termination-limited conditions.
(Fig. 3). We observe that at low polysome sizes the elongation phase control over translation rate lies in the codons near the 5′ end of the mRNA. This result is in agreement with early experimental results demonstrating that point mutations near the start codon of the mRNA cause dramatic changes in protein expression levels (28,29). Also, at intermediate polysome sizes the control is distributed along the length of the mRNA in different configurations, and at high polysome sizes the control lies in the codons near the 3′ end of the mRNA. We observe the same results under both Cases I and II. These results are expected because at low polysome sizes kinetics are initiation-limited (see previous paragraph for discussion), which means that the initiation process limits the progress of protein translation. Hence, the more efficiently the codons near the 5′ end of the mRNA can be translated, the more ribosomes can be transferred to downstream codons along the length of the sequence. Faster transfer of ribosomes due to more efficient translation of these codons elevates protein synthesis rate by increasing the probability of an initiation event occurring without changes to the initiation process being made. The converse is true for termination-limited conditions.
FIGURE 3.
Elongation step control coefficients, 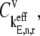 with respect to sequence position under initiation (A), elongation (B), and termination (C) limited conditions for Cases I and II.
with respect to sequence position under initiation (A), elongation (B), and termination (C) limited conditions for Cases I and II.
Relationship between codon-specific elongation cycle intermediate step control of protein translation rate and polysome size
We investigated how the elongation phase control is distributed with respect to the elongation cycle intermediate steps at each codon along the length of the mRNA by examining the control coefficients: 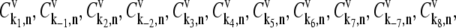 and
and 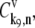 along with the control coefficients corresponding to free ternary complex concentration,
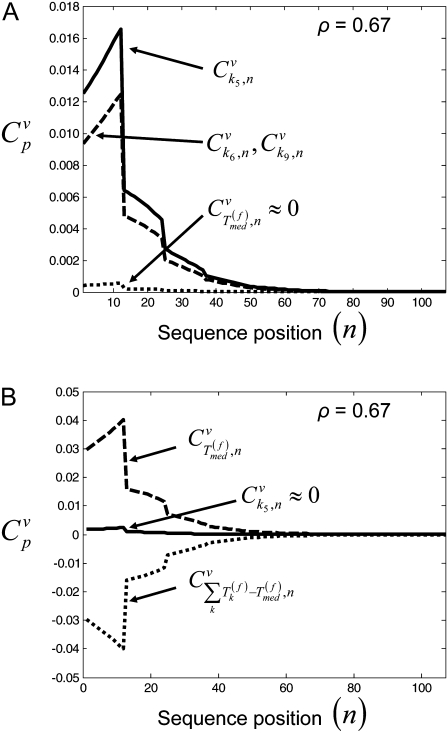
along with the control coefficients corresponding to free ternary complex concentration, 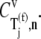 We observe that the rate-limiting step at each codon along the length of the mRNA is different between Cases I and II. Under Case I, we observe that the control coefficient with respect to the Ef-Tu:GDP release rate constant,
We observe that the rate-limiting step at each codon along the length of the mRNA is different between Cases I and II. Under Case I, we observe that the control coefficient with respect to the Ef-Tu:GDP release rate constant,  is the highest of the control coefficients corresponding to elongation cycle intermediate steps at every sequence position and polysome size (Fig. 4 A, results shown only for ρ = 0.67), indicating that this intermediate step is rate-limiting to the elongation cycle. This result is consistent with experimental reports which identify Ef-Tu:GDP release as one of the rate-limiting steps of the elongation cycle at a given codon (26). Control coefficients for A site tRNA accommodation
is the highest of the control coefficients corresponding to elongation cycle intermediate steps at every sequence position and polysome size (Fig. 4 A, results shown only for ρ = 0.67), indicating that this intermediate step is rate-limiting to the elongation cycle. This result is consistent with experimental reports which identify Ef-Tu:GDP release as one of the rate-limiting steps of the elongation cycle at a given codon (26). Control coefficients for A site tRNA accommodation  and E site tRNA release
and E site tRNA release 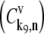 are equal to each other and also high (Fig. 4 A, results shown only for ρ = 0.67) at every sequence position and polysome size. The remaining elongation cycle intermediate steps have low control coefficients, including that for the free ternary complex concentration control coefficient
are equal to each other and also high (Fig. 4 A, results shown only for ρ = 0.67) at every sequence position and polysome size. The remaining elongation cycle intermediate steps have low control coefficients, including that for the free ternary complex concentration control coefficient 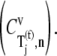
FIGURE 4.
Elongation cycle intermediate control coefficients with respect to sequence position under Case I (A) and Case II (B) binding conditions. Results shown are for elongation-limited conditions (ρ = 0.67).
However, under Case II we observe that the control coefficient with respect to the free ternary complex concentration, 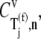 is highest at every sequence position and polysome size (Fig. 4 B, results shown only for ρ = 0.67), indicating that ternary complex nonspecific binding is rate-limiting to the elongation cycle. The remaining elongation cycle control coefficients are close to zero, indicating that the intermediate steps after ternary complex nonspecific binding have very little influence on elongation cycle kinetics. Moreover, the rate-limiting effects of ternary complex nonspecific binding are much higher under Case II than the rate-limiting effects of Ef-Tu:GDP release under Case I, with free ternary complex concentration control coefficients
is highest at every sequence position and polysome size (Fig. 4 B, results shown only for ρ = 0.67), indicating that ternary complex nonspecific binding is rate-limiting to the elongation cycle. The remaining elongation cycle control coefficients are close to zero, indicating that the intermediate steps after ternary complex nonspecific binding have very little influence on elongation cycle kinetics. Moreover, the rate-limiting effects of ternary complex nonspecific binding are much higher under Case II than the rate-limiting effects of Ef-Tu:GDP release under Case I, with free ternary complex concentration control coefficients  under Case II, and more than twice as high as Ef-Tu:GDP release control coefficients
under Case II, and more than twice as high as Ef-Tu:GDP release control coefficients  under Case I. We also observe that the concentrations of ternary complexes that do not recognize the A site codon,
under Case I. We also observe that the concentrations of ternary complexes that do not recognize the A site codon, 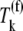 (k ≠ j), have an inhibitory effect on translation kinetics because the corresponding control coefficients for the combined concentration of the incorrect ternary complexes,
(k ≠ j), have an inhibitory effect on translation kinetics because the corresponding control coefficients for the combined concentration of the incorrect ternary complexes,  are negative (Fig. 4 B, results shown only for ρ = 0.67), meaning that an increase in this concentration would cause a decrease in translation rate.
are negative (Fig. 4 B, results shown only for ρ = 0.67), meaning that an increase in this concentration would cause a decrease in translation rate.
It is important to note that it is the relative magnitudes of the terms in the effective elongation rate constant,  that play a significant role in the distribution of control with respect to the elongation cycle intermediate steps at each codon. The influence each elongation cycle intermediate step has over the overall kinetics of the elongation cycle at a given codon is proportionate to the magnitude of its corresponding term in the effective elongation rate constant. Under noncompetitive binding conditions, Ef-Tu:GDP release is rate-limiting, with _α_5 = 0.067 (Table 1) being the largest term in
that play a significant role in the distribution of control with respect to the elongation cycle intermediate steps at each codon. The influence each elongation cycle intermediate step has over the overall kinetics of the elongation cycle at a given codon is proportionate to the magnitude of its corresponding term in the effective elongation rate constant. Under noncompetitive binding conditions, Ef-Tu:GDP release is rate-limiting, with _α_5 = 0.067 (Table 1) being the largest term in 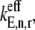 and nonspecific ternary complex binding has almost no influence over elongation cycle kinetics, with _α_1,j = 6 × 10−4 − 0.04 (Table 1). However, under competitive binding conditions, the magnitude of the nonspecific ternary complex binding term in the effective elongation rate constant is much higher than _α_5, with
and nonspecific ternary complex binding has almost no influence over elongation cycle kinetics, with _α_1,j = 6 × 10−4 − 0.04 (Table 1). However, under competitive binding conditions, the magnitude of the nonspecific ternary complex binding term in the effective elongation rate constant is much higher than _α_5, with 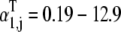 (Table 1), making nonspecific binding rate-limiting.
(Table 1), making nonspecific binding rate-limiting.
To further understand the relationship between the magnitudes of the effective elongation rate constant terms and the control the corresponding elongation cycle intermediate steps have over translation rate, we introduce the elasticities of the elongation rate at codon n with respect to the free ternary complex concentration, 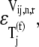 and the reaction rate constant for Ef-Tu:GDP release,
and the reaction rate constant for Ef-Tu:GDP release, 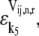 under competitive binding conditions:
under competitive binding conditions:
 |
(17) |
|---|
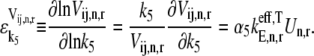 |
(18) |
|---|
Elasticity is defined as the differential change in the rate of a single reaction step, i.e., in this case, _V_ij,n,r. Unlike the control coefficients, which pertain to the overall translation rate of the mRNA, elasticity is therefore a property local to that reaction step and not a systemic property. However, due to the compactness of the elasticity expressions, they are useful for obtaining general quantitative insight into the impact of the individual reaction rate constants and translational components on their respective control coefficient magnitudes. It is evident from the above expressions that the elasticity of _V_ij,n,r with respect to a given parameter is dependent on the effective elongation rate constant term to which the parameter pertains, and not only on that parameter. Along these lines, the relative magnitudes of the elasticities are proportionate to the relative magnitudes of the corresponding effective elongation rate constant terms, and similar relationships are obtained between the remaining effective elongation rate constant terms and their corresponding elasticities. Consequently, the control the elongation cycle intermediate steps have over the translation rate of each codon is strongly influenced by the magnitudes of their respective effective elongation rate constant terms.
Overall, in these studies we observe that ternary complex competitive binding to the ribosomal A site introduces changes to translation rate (Fig. 2). However, competitive binding does not cause changes to the distribution of overall initiation, elongation, and termination control with respect to polysome size. Moreover, competitive binding does not affect the codon-specific distribution of control with respect to polysome size (Fig. 3), but instead introduces changes to the distribution of control with respect to elongation cycle intermediate step at each codon (Fig. 4).
Ternary complexes not recognizing the ribosomal A site codon act as competitive inhibitors to elongation cycle kinetics
To further investigate the inhibitory effects of ternary complexes not recognizing the ribosomal A site codon, we derive our mechanistic framework in the context of Michaelis-Menten enzyme kinetics. Treating all the translating ribosomes in a single E. coli cell having codon species j occupying the A site as the enzyme, and the ternary complex species j recognizing the A site codon as the substrate, it can be shown that, in the absence of ternary complex competitive binding,
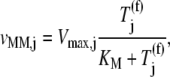 |
(19) |
|---|
where the Michaelis-Menten constant is
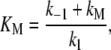 |
(20) |
|---|
and the maximum reaction rate is
 |
(21) |
|---|
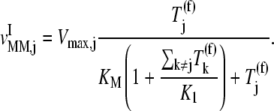
In the above expression, _R_j is the cellular concentration of ribosomes in the cell participating in translation with codon species j occupying the A site. By accounting for ternary complex competitive binding, it can be shown that
 |
(22) |
|---|
Details of the derivation of the above equations, along with the estimation of _R_j, are included in Appendix B.
Under competitive binding conditions, the ternary complexes that do not recognize the A site codon 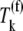 (k ≠ j) bind to the ribosome as the ternary complexes do
(k ≠ j) bind to the ribosome as the ternary complexes do 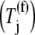 during the nonspecific binding step of the elongation cycle, but do not proceed to the subsequent intermediate steps. The ternary complexes
during the nonspecific binding step of the elongation cycle, but do not proceed to the subsequent intermediate steps. The ternary complexes 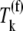 (k ≠ j) occupying the ribosomal A site prevent the ternary complexes
(k ≠ j) occupying the ribosomal A site prevent the ternary complexes 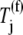 from binding to the ribosome, so the apparent affinity the ternary complexes have in recognizing the A site codon
from binding to the ribosome, so the apparent affinity the ternary complexes have in recognizing the A site codon 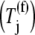 for the ribosome decreases. This decrease is due to the term multiplied by the Michaelis-Menten constant (_K_M) in the expression for
for the ribosome decreases. This decrease is due to the term multiplied by the Michaelis-Menten constant (_K_M) in the expression for 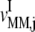 (Eq. 22), which represents the inhibitory effects of the ternary complexes
(Eq. 22), which represents the inhibitory effects of the ternary complexes 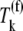 (k ≠ j) on translation rate. However, the maximum reaction rate
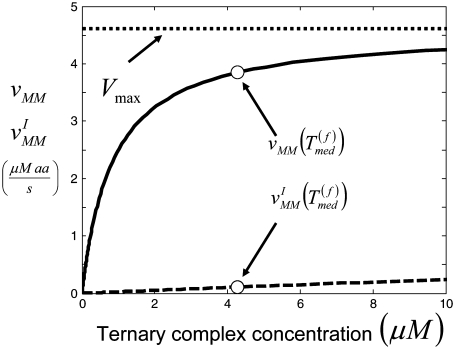
(k ≠ j) on translation rate. However, the maximum reaction rate 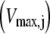 is the same under both noncompetitive and competitive binding conditions. Fig. 5 shows the relationship between translation rates
is the same under both noncompetitive and competitive binding conditions. Fig. 5 shows the relationship between translation rates 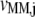 and
and 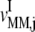 as functions of the ternary complex concentration recognizing the A site codon,
as functions of the ternary complex concentration recognizing the A site codon, 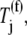 for the median ternary complex concentration,
for the median ternary complex concentration, 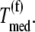 The maximum reaction rate
The maximum reaction rate  is proportional to the concentration of translating ribosomes having the codon recognized by the ternary complex species of median concentration present in the A site, and the ternary complex concentration is allowed to vary. When evaluating the translation rate expressions with and without competitive binding at the median ternary complex concentration
is proportional to the concentration of translating ribosomes having the codon recognized by the ternary complex species of median concentration present in the A site, and the ternary complex concentration is allowed to vary. When evaluating the translation rate expressions with and without competitive binding at the median ternary complex concentration 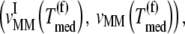 we observe a much lower translation rate under competitive binding conditions than under noncompetitive binding conditions (Fig. 5). Similar results are observed for the remaining ternary complex species.
we observe a much lower translation rate under competitive binding conditions than under noncompetitive binding conditions (Fig. 5). Similar results are observed for the remaining ternary complex species.
FIGURE 5.
Amino-acid rate of incorporation as a function of ternary complex concentration for Case I (solid line) and Case II (dashed line). Results shown are at the E. coli cellular level for the ternary complex species of median concentration. Similar results are observed for all ternary complex species.
We examined the expressions for the elasticities, 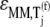 and
and 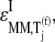 of the reaction rates
of the reaction rates 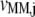 and
and 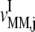 with respect to the ternary complex concentration recognizing the ribosomal A site, i.e., the ratios of the proportional changes in
with respect to the ternary complex concentration recognizing the ribosomal A site, i.e., the ratios of the proportional changes in 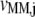 and
and 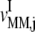 with respect to the proportional change in
with respect to the proportional change in 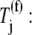
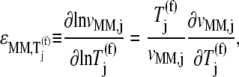 |
(23) |
|---|
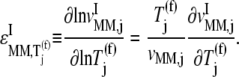 |
(24) |
|---|
Evaluating Eqs. 23 and 24 yields
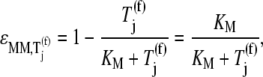 |
(25) |
|---|
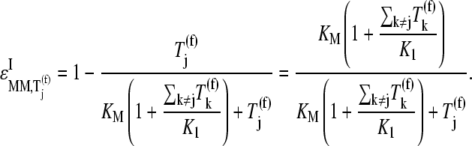 |
(26) |
|---|
We observe that the elasticities determined under competitive binding conditions 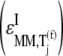 are much greater than those determined under noncompetitive binding conditions
are much greater than those determined under noncompetitive binding conditions 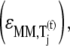 with
with 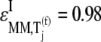 and
and  for the median ternary complex concentration,
for the median ternary complex concentration, 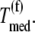 Equations 25 and 26 suggest that the lower the ternary complex concentration recognizing the ribosomal A site codon,
Equations 25 and 26 suggest that the lower the ternary complex concentration recognizing the ribosomal A site codon, 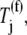 the stronger the sensitivity to change under competitive binding conditions
the stronger the sensitivity to change under competitive binding conditions 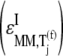 than noncompetitive binding conditions
than noncompetitive binding conditions 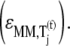 Similar results are observed for the remaining ternary complex species. As we observed in the results above relating to reaction rates
Similar results are observed for the remaining ternary complex species. As we observed in the results above relating to reaction rates 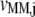 and
and 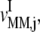 the increased elasticities under competitive binding conditions are observed because of the term multiplied by the Michaelis-Menten constant (_K_M) in the expression for
the increased elasticities under competitive binding conditions are observed because of the term multiplied by the Michaelis-Menten constant (_K_M) in the expression for 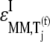 (Eq. 26) that represents the inhibitory effects of the ternary complexes
(Eq. 26) that represents the inhibitory effects of the ternary complexes 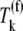 (k ≠ j) on translation rate. The results in this section support our results discussed in previous sections pertaining to ternary complex competitive binding lowering translation rate and causing the nonspecific binding intermediate step to be rate-limiting to the elongation cycle at each codon. However, the results presented in this section suggest that the effects of competitive binding are due to the ternary complexes not recognizing the A site codon
(k ≠ j) on translation rate. The results in this section support our results discussed in previous sections pertaining to ternary complex competitive binding lowering translation rate and causing the nonspecific binding intermediate step to be rate-limiting to the elongation cycle at each codon. However, the results presented in this section suggest that the effects of competitive binding are due to the ternary complexes not recognizing the A site codon 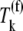 (k ≠ j) acting as competitive inhibitors to elongation cycle kinetics.
(k ≠ j) acting as competitive inhibitors to elongation cycle kinetics.
The relative position of codons along the mRNA determines the optimal protein synthesis rate and the rate-limiting effect of the individual codons
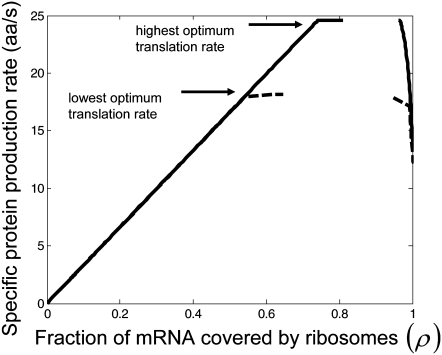
In these studies, we apply Case III to investigate the translation properties of mRNAs in both a codon and sequence-dependent manner. We applied our mechanistic framework to 100 randomly permuted sequences having identical codon frequencies representative of those of the E. coli genome. Each sequence is 361-codons-long, approximately the average length of an E. coli mRNA (22). Similar to our results in previous sections, we observe that the translation rate increases, reaches a maximum, and then decreases as polysome size increases (Fig. 6). However, optimum protein synthesis rates vary with sequence (Fig. 6, results shown only for sequences producing highest and lowest optimum translation rates). We also observe that the optimum rate occurs at multiple polysome sizes for each sequence and that there are regimes of polysome sizes for which translation properties are highly sensitive to the input parameters of our model (Fig. 6). Because all the sequences in this study have the same codon frequencies, the results presented in this section emphasize that the relative positions of codons along the length of the mRNA can influence protein synthesis properties.
FIGURE 6.
Relationship between translation rate and polysome size under Case III conditions.
Relationship between effective elongation rate constant magnitudes and polysome size
To investigate the overall relationship between translation rate and polysome size with Case III conditions, we examined changes in the effective elongation rate constant magnitudes with polysome size. We scaled the effective elongation rate constants by dividing them by the effective elongation rate constant, 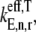 evaluated at _U_n,r = 1 and
evaluated at _U_n,r = 1 and 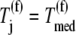 that has a magnitude of 0.8 s−1. In the absence of ribosomal crowding on the mRNA, i.e., when _U_n,r = 1, the scaled effective elongation rate constant magnitudes vary between 0.10 and 3.44 due to differences in the nonspecific binding term,
that has a magnitude of 0.8 s−1. In the absence of ribosomal crowding on the mRNA, i.e., when _U_n,r = 1, the scaled effective elongation rate constant magnitudes vary between 0.10 and 3.44 due to differences in the nonspecific binding term,  (Eq. 5). Using effective elongation rate constants determined with Case II conditions as a reference, we scaled them the same way by dividing them by the effective elongation rate constant,
(Eq. 5). Using effective elongation rate constants determined with Case II conditions as a reference, we scaled them the same way by dividing them by the effective elongation rate constant, 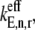 evaluated at _U_n,r = 1 and
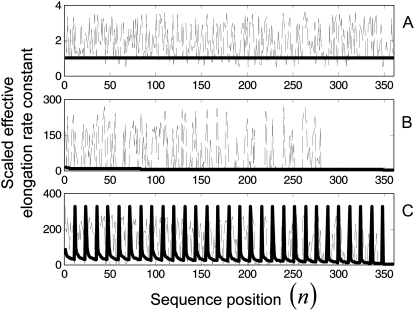
evaluated at _U_n,r = 1 and 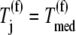 that has a magnitude of 5.1 s−1. Included in Fig. 7 are the distributions of scaled effective elongation rate constant magnitudes as functions of sequence position under initiation (A), elongation (B), and termination (C) limited conditions for one of the sequences used in this section (similar results are observed for the other sequences). The dashed lines represent magnitudes under Case III conditions, and the solid lines represent magnitudes under Case II conditions.
that has a magnitude of 5.1 s−1. Included in Fig. 7 are the distributions of scaled effective elongation rate constant magnitudes as functions of sequence position under initiation (A), elongation (B), and termination (C) limited conditions for one of the sequences used in this section (similar results are observed for the other sequences). The dashed lines represent magnitudes under Case III conditions, and the solid lines represent magnitudes under Case II conditions.
FIGURE 7.
Scaled effective elongation rate constant magnitudes under initiation (A), elongation (B), and termination (C) limited conditions for one of the sequences used in this section (similar results are observed for the other sequences). The dashed lines represent magnitudes determined with Case III conditions, while the solid lines represent magnitudes determined with Case II conditions.
We observe that under initiation-limited conditions the scaled effective elongation rate constants for both Case II and Case III are approximately equal to the values they take on when _U_n,r = 1, and this result is expected because the polysome size is low and hence the mRNA is not crowded. Under elongation-limited conditions, the level of crowding on the mRNA is higher as reflected in the conditional probability term, _U_n,r, of the effective elongation rate constant decreasing, which results in the scaled effective elongation rate constant magnitudes and translation rates increasing. The level of ribosomal crowding on the mRNA determines the magnitudes under Case II conditions (see (13) for more discussion), while the complex interplay between the level of ribosomal crowding on the mRNA and the level of ternary complex competition for the ribosomal A site at each codon determines the scaled effective elongation rate constant magnitudes under Case III conditions. Codons that experience a lot of ternary complex competitive inhibition have lower effective elongation rate constants and are hence translated more slowly than codons that do not, causing high ribosome density upstream on the mRNA and large variation in the conditional probability term, _U_n,r, that is not observed under Case II conditions. Consequently the scaled effective elongation rate constants determined with Case III are much higher than those determined with Case II. Under termination limited conditions the polysome size is high, so crowding on the mRNA is maximal and _U_n,r ≈ 0, regardless of whether the binding conditions are uniform (Case II) or nonuniform (Case III). Due to the ribosomal queuing that occurs along the length of the mRNA at high polysome size (see (13) for more discussion), the effective elongation rate constants at positions spaced one-ribosome-length apart are approximately equal to the translocation rate constant, _k_8 (see (13) for more discussion).
Effects of rate-limiting codon segments on the relationship between optimum translation rate and polysome size
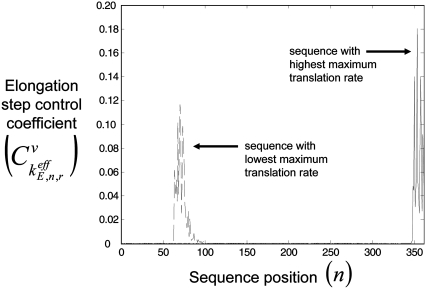
To investigate translation properties occurring in the regimes of polysome sizes associated with optimum rates, we obtained the elongation step control coefficients  of each sequence at its respective optimum translation rate (Fig. 6). Similar to previous results (13), at the optimum rate the kinetics are completely elongation-limited, with
of each sequence at its respective optimum translation rate (Fig. 6). Similar to previous results (13), at the optimum rate the kinetics are completely elongation-limited, with 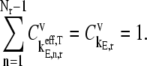 The control over rate is dominated by segments of codons that have high elongation step control coefficients (Fig. 8, results shown only for sequences producing highest and lowest optimum translation rates). For all of the polysome sizes that the translation kinetics are completely elongation-limited, we observe that the configuration of elongation step control coefficients
The control over rate is dominated by segments of codons that have high elongation step control coefficients (Fig. 8, results shown only for sequences producing highest and lowest optimum translation rates). For all of the polysome sizes that the translation kinetics are completely elongation-limited, we observe that the configuration of elongation step control coefficients  does not change, and therefore the segments of rate-limiting codons do not change.
does not change, and therefore the segments of rate-limiting codons do not change.
FIGURE 8.
Elongation step control coefficients, 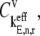 with respect to sequence position under Case III conditions.
with respect to sequence position under Case III conditions.
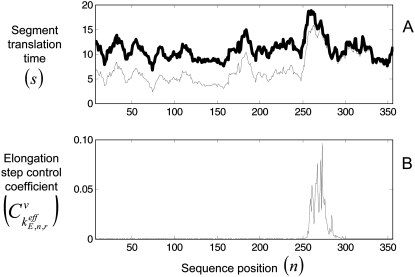
The positions of rate-limiting codon segments are expected because they correspond to segments of high translation time (Fig. 9). We define the translation time of the codon segments to be
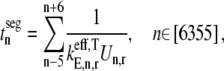 |
(27) |
|---|
where the codon segments are equal to one ribosome length, and the translation time of the segment corresponding to codon n is equal to the combined translation time of that codon along with the five upstream and six downstream codons. We consider the codon segments of one ribosome length in this way because n denotes the position of the ribosomal P site codon, and in this work and in our previous work (13) we assume the front and back ends of the ribosome are on the sides closest to the 3′ and 5′ ends of the mRNA, respectively, with the P site covering the seventh codon relative to the front of the ribosome. We estimated the segment translation times with _U_n,r = 1 for all of the codons in the sequence and with _U_n,r values corresponding to the ribosome distribution at the optimum translation rate (Fig. 9 A). The part of the sequence having the highest translation times (Fig. 9 A) corresponds to the rate-limiting codon segment (Fig. 9 B). Additionally, the segment translation times with _U_n,r = 1 can be interpreted as the translation times in the absence of ribosomal crowding. At the optimum translation rate the ribosome density is high, resulting in low values for _U_n,r for all of the codons in the sequence. Consequently, the translation times at high ribosome densities are higher than those at low ribosome densities. However, the part of the sequence having the highest segment translation times remains the same at both high and low ribosome densities—consistent with the rate-limiting codon segment remaining unchanged for the polysome sizes in which the translation kinetics are completely elongation-limited.
FIGURE 9.
(A) Segment translation times, 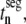 with respect to sequence position for one of the sequences used in this study (similar results are observed for the other sequences). The thin line represents the segment translation times with _U_n,r = 1 for all codons in the sequence, while the thick line represents the segment translation times with _U_n,r values corresponding to the ribosome distribution at the optimum translation rate. (B) Elongation step control coefficients,
with respect to sequence position for one of the sequences used in this study (similar results are observed for the other sequences). The thin line represents the segment translation times with _U_n,r = 1 for all codons in the sequence, while the thick line represents the segment translation times with _U_n,r values corresponding to the ribosome distribution at the optimum translation rate. (B) Elongation step control coefficients, 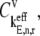 with respect to sequence position under Case III conditions for the same sequence used in Fig. 9 A.
with respect to sequence position under Case III conditions for the same sequence used in Fig. 9 A.
Because the rate-limiting codon segments correspond to regions of high translation time, they also lead to nonuniform ribosome distributions along the length of the mRNA. Defining p to be the sequence position of the codon at the 3′ end of the rate-limiting codon segment, the ribosome density upstream of p is as
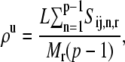 |
(28) |
|---|
while the ribosome density downstream of p is as
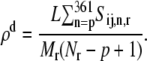 |
(29) |
|---|
The ribosome density upstream of p, _ρ_u, corresponds to the part of the sequence between the 5′ end of the mRNA and the 3′ end of the rate-limiting codon segment, while the ribosome density downstream of p, _ρ_d, corresponds to the remainder of the sequence. For all of the sequences studied at the optimum translation rate _ρ_u > _ρ_d, with 0.61 ≤ _ρ_u ≤ 0.89 and 0.0091 ≤ _ρ_d ≤ 0.76. Ribosomes translate the codons in the rate-limiting segments more slowly than those in the remainder of the sequence, leading to higher ribosome densities upstream of the rate-limiting codon segments than downstream. It is important to note that, at the regime of polysome sizes corresponding to optimum translation rate, the translation kinetics shift from elongation- to termination-limited. Under elongation-limited conditions we observe nonuniform ribosome densities, while under termination-limited conditions we observe uniform queuing of the ribosomes along the length of the mRNA (see (13) for more discussion). In this transitional regime, the ribosome density becomes very sensitive to the input parameters of our model, making it difficult to obtain data. Hence, in Fig. 6 there are regimes of polysome sizes for which we do not show translation rates.
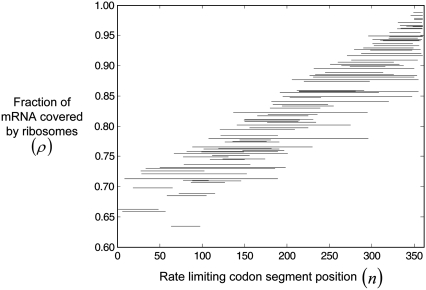
Furthermore, the positions of the rate-limiting codon segments determine the minimum polysome size at which the optimum translation rate occurs (Fig. 10). The closer to the 3′ end of the sequence the rate-limiting segment is, the more ribosomes are accommodated on the mRNA, the higher the polysome size, and the higher the protein synthesis rate. This result indicates that the positioning of the rate-limiting codon segment influences the optimum translation rate. Translation of the sequence with the highest optimum rate is limited by a codon segment near the 3′ end of the mRNA (Fig. 8). Consequently this sequence can accommodate the most ribosomes, maximizing the probability of a translation termination event occurring and hence maximizing the optimum protein synthesis rate. The converse is true for the sequence with the lowest optimum rate, because its translation is limited by a codon segment near the 5′ end of the mRNA (Fig. 8).
FIGURE 10.
Relationship between the positions of the rate-limiting codon segments and ribosomal fractional coverage.
DISCUSSION
We presented a theoretical analysis of protein synthesis that includes all the elementary steps of the translation mechanism and accounts for ternary complex competitive binding to the ribosomal A site. Considering protein synthesis kinetics in the context of ternary complex competitive binding provides insights into quantifying the systemic contributions of ternary complex concentrations to the translational output of genes. Moreover, our codon-specific sensitivity analysis allows us to separately quantify the influence the concentration of the ternary complex recognizing each codon along the length of the mRNA has on the overall protein synthesis rate. We find that the expanded mechanistic framework predicts lower protein synthesis rates than the previously developed framework (13) (Fig. 2), the configuration of codons that have the most control over protein synthesis rate changes with polysome size (Fig. 3), and competitive, nonspecific binding of the ternary complexes to the ribosomal A site is rate-limiting to the elongation cycle for every codon (Fig. 4). These results suggest that the ternary complexes that do not recognize the ribosomal A site codon act as competitive inhibitors to the ternary complexes recognizing the A site codon (Fig. 5). Considering this model in the context of a Michaelis-Menten mechanistic framework demonstrates that translation rates are lower and more sensitive to ternary complex concentrations under competitive binding conditions than under noncompetitive binding conditions, which is consistent with what Michaelis-Menten kinetics predicts under competitive inhibition conditions where a substrate and inhibitor are competing for access to the active site of an enzyme.
In these studies the same set of reaction rate constants were used for the elongation cycle intermediate steps at every codon along the length of the sequence. Hence, the results suggest that it is the interplay between the level of ternary complex competition for the ribosomal A site at each codon and the level of ribosomal crowding on the mRNA that determines the effective elongation rate constant magnitudes at each codon and polysome size (Fig. 6 B). This configuration at a given polysome size determines the corresponding protein synthesis properties (Fig. 6). However, given that codon-anticodon compatibilities affect translation rates (10–12,30), in future studies it will be important to incorporate anticodon specific kinetic and thermodynamic (31) parameters into our model to investigate how translational behavior is affected.
The set of elongation cycle reaction rate constants used in these studies were the same as those used in our previous studies (13), which did not account for ternary complex competitive binding, and they predict higher translation rates, suggesting that Ef-Tu:GDP release is the rate-limiting step of the elongation cycle for every codon. The expanded mechanistic framework in this study predicts lower translation rates and indicates that ternary complex nonspecific binding to the ribosome is the rate-limiting step of the elongation cycle for every codon. It has been shown experimentally that the ternary complexes not recognizing the ribosomal A site codon do not inhibit translation rate (30), and that Ef-Tu:GDP release is one of the rate-limiting steps of the elongation cycle (26). Although these experimental results are consistent with the results of our previous study (13), these experiments were performed in vitro and consequently do not reflect in vivo conditions. The results from Bilgin et al. (30) were obtained by examining the competitive binding effects 1.3 _μ_M Phe ternary complex experiences from Leu2 and Leu4 varying from 0 _μ_M to 16 _μ_M during poly(Phe) synthesis. By increasing the Leu2 and Leu4 concentrations from 0 _μ_M to 16 _μ_M, the authors observe that the translation rates per ribosome decrease from 4.0 s−1 to 3.0 s−1. Hence, they conclude that ternary complex species not recognizing the ribosomal A site codon have almost no inhibitory effects in vitro. However, the total concentration of tRNA in E. coli is roughly 332 _μ_M (22), so in vivo a ternary complex species having a concentration of 1.3 _μ_M would experience much higher competitive effects than predicted in Bilgin et al. (30). By rearranging Eq. 2 we can express the translation rate per ribosome, _v_ij,n,r, evaluated at _U_n,r = 1 as
 |
(30) |
|---|
Applying 1.3 _μ_M to  and 0–16 _μ_M to
and 0–16 _μ_M to 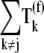 in the above expression, we observe that the translation rate per ribosome decreases from 5.0 s−1 to 2.4 s−1, which is close to the range of translation rates per ribosome observed in Bilgin et al. (30). Similar to Bilgin et al. (30), our model predicts low inhibitory effects of ternary complexes not recognizing the ribosomal A site codon on translation rate. On the contrary, when we apply 1.3 _μ_M to
in the above expression, we observe that the translation rate per ribosome decreases from 5.0 s−1 to 2.4 s−1, which is close to the range of translation rates per ribosome observed in Bilgin et al. (30). Similar to Bilgin et al. (30), our model predicts low inhibitory effects of ternary complexes not recognizing the ribosomal A site codon on translation rate. On the contrary, when we apply 1.3 _μ_M to 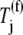 and 332 _μ_M to
and 332 _μ_M to 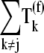 in Eq. 30, we obtain a translation rate per ribosome of 0.2 s−1, which is much lower than what is observed in Bilgin et al. (30) and what is predicted using our model above. This result indicates that ternary complexes have significant competitive effects in vivo. Moreover, our model predicts a two-to-ninefold reduction in optimal translation rate due to ternary complex competitive binding (Fig. 2), which is consistent with estimates in previous experimental work (25). Hence, the difference in the results from our previous work (13) and current mechanistic frameworks further suggests that ternary complexes have a significant effect on translation kinetics by acting as competitive inhibitors.
in Eq. 30, we obtain a translation rate per ribosome of 0.2 s−1, which is much lower than what is observed in Bilgin et al. (30) and what is predicted using our model above. This result indicates that ternary complexes have significant competitive effects in vivo. Moreover, our model predicts a two-to-ninefold reduction in optimal translation rate due to ternary complex competitive binding (Fig. 2), which is consistent with estimates in previous experimental work (25). Hence, the difference in the results from our previous work (13) and current mechanistic frameworks further suggests that ternary complexes have a significant effect on translation kinetics by acting as competitive inhibitors.
We applied our mechanistic framework to randomly permuted sequences having codon frequencies representative of that of the E. coli genome, and this study provides insight into the protein synthesis properties of genes with codons recognized by ternary complex species of varying concentrations. In ongoing work we apply this mechanistic framework to the protein-coding regions of the E. coli genome. For example, we investigate from a mechanistic perspective how codon usage patterns have been correlated with patterns of gene expression levels (32), with position within a gene (33–35), and correlated with gene length (36) to better understand the complex, nonlinear interplay between codon usage and protein synthesis properties, to characterize how these properties relate to patterns such as gene expression levels and function.
While some of the conclusions drawn from our studies might be as expected to those experienced with protein synthesis, the proposed computational framework provides a quantitative verification and allows the formulation of hypotheses for the origins of the observed phenomena that mental simulations alone cannot offer. In this investigation we expanded our mechanistic framework from our previous work (13) to incorporate information about ternary complex competitive binding to the ribosome and make quantitative predictions about the translation mechanism. These mathematical models allow us to consider each part of the complex biological process and to develop a more complete understanding of translation at the systems level.
CONCLUSIONS
In this work we expanded our mechanistic framework from Zouridis and Hatzimanikatis (13) to account for ternary complex competitive binding to the ribosomal A site. We also performed a sensitivity analysis to determine the effects of the kinetic parameters and concentrations of the translational components on the protein synthesis rate. We determined the following:
- Translation rates are lower under ternary complex competitive binding conditions than under noncompetitive binding conditions. This result is due to the tRNAs that do not recognize the ribosomal A site codon acting as competitive inhibitors to the tRNAs that do recognize the ribosomal A site codon. Along these lines, the competitive, nonspecific binding of the tRNAs to the ribosomal A site is rate-limiting to the elongation cycle for every codon.
- At low polysome sizes the codons near the 5′ end of the mRNA control protein synthesis rate, at intermediate polysome sizes different configurations of codons along the length of the mRNA control protein synthesis rate, and at high polysome sizes the codons near the 3′ end of the mRNA control protein synthesis rate.
- The relative position of codons along the mRNA determines the optimal protein synthesis rate. Optimal translation rates of mRNAs are controlled by segments of rate-limiting codons that are sequence-specific. The segments of rate-limiting codons correspond to regions of high translation time that cause nonuniform ribosome distributions on mRNAs.
Acknowledgments
The authors thank Dr. Olke Uhlenbeck, Dr. Amit Mehra, and Kevin Keegan for the helpful, in-depth discussions. The authors also thank the two anonymous reviewers for their constructive comments.
This research has been supported by the National Science Foundation through the Quantitative Systems Biotechnology Initiative (grant No. CMMI 0425833), and DuPont through a DuPont Young Professor Award to V.H.
APPENDIX A: MECHANISTIC FRAMEWORK ASSUMPTIONS
We have applied the following assumption in our current mechanistic formulation: All ternary complex species can bind to the ribosomal A site during the codon-independent binding intermediate step of the elongation cycle, regardless of the codon species present in the ribosomal A site.
Introducing the ternary complex subscript k to the fluxes and state corresponding to nonspecific binding yields 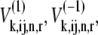 and
and 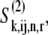 and denotes the nonspecific binding between each ternary complex species k and A site codon species j. Detailed descriptions of ribosomal states and fluxes can be found in our previous work (13). The equations describing the dynamics of the transitions of state 1, the state existing before ternary complex binding, are
and denotes the nonspecific binding between each ternary complex species k and A site codon species j. Detailed descriptions of ribosomal states and fluxes can be found in our previous work (13). The equations describing the dynamics of the transitions of state 1, the state existing before ternary complex binding, are
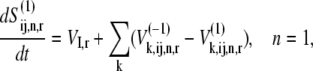 |
(31) |
|---|
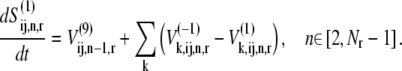 |
(32) |
|---|
We assume that the ternary complexes that do not recognize the A site codon cannot proceed past the nonspecific binding intermediate step of the elongation cycle, while ternary complexes recognizing the A site codon can continue on to the remaining steps of the elongation cycle. These assumptions yield the following expressions for the dynamics of the transitions of state 2:
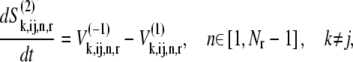 |
(33) |
|---|
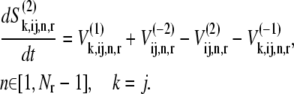 |
(34) |
|---|
The expressions describing the dynamics of the transitions between the remaining elongation cycle intermediate states are the same as those described in previous work (13). Equations 31–34, together with the expressions for the remaining intermediate states, are used to derive the expression for the effective elongation rate constant accounting for ternary complex competitive binding (Eq. 6) in the same manner that the original effective elongation rate constant (Eq. 3) was derived in previous work (13).
APPENDIX B: MICHAELIS-MENTEN REACTION RATE EXPRESSION DERIVATION
In the absence of ternary complex competitive binding, we consider the following reaction scheme for the elongation cycle occurring at a given codon:
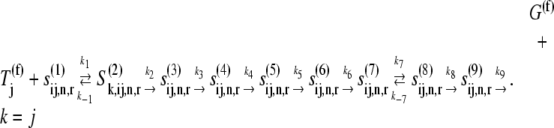 |
(35) |
|---|
The states 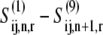 represent the intermediate elongation cycle ribosomal states that are described in detail in previous work (13). The first state,
represent the intermediate elongation cycle ribosomal states that are described in detail in previous work (13). The first state,  represents the ribosomal state that exists before ternary complex binding with the A site empty, and the remaining states have the A site occupied by the ternary complex. We allow
represents the ribosomal state that exists before ternary complex binding with the A site empty, and the remaining states have the A site occupied by the ternary complex. We allow 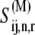 to be the grouped ribosomal state including all the intermediate elongation cycle states having the ternary complex bound to the ribosomal A site, where
to be the grouped ribosomal state including all the intermediate elongation cycle states having the ternary complex bound to the ribosomal A site, where
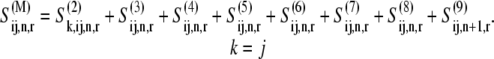 |
(36) |
|---|
By introducing the grouped state, 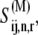 the reaction scheme in Eq. 35 simplifies to
the reaction scheme in Eq. 35 simplifies to
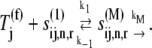 |
(37) |
|---|
Since our studies are performed at steady state, we obtain the expression for _k_M as we obtained the expression for the effective elongation rate constant previously (13), yielding
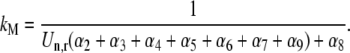 |
(38) |
|---|
In this work, _k_M is evaluated with _U_n,r = 1.
The equation describing the dynamics of the transitions between states is
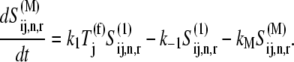 |
(39) |
|---|
Following from the pseudo steady-state approximation, the concentrations of the intermediates are assumed to reach steady state much faster than those of the product and substrate. Hence, we set the time derivative in the above equation equal to zero and rearrange it to obtain an expression for 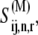 yielding
yielding
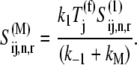 |
(40) |
|---|
By allowing 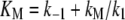 (Eq. 20), the above equation becomes
(Eq. 20), the above equation becomes
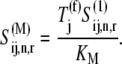 |
(41) |
|---|
The total concentration of translating ribosomes is equal to the sum of the concentration of ribosomes with the A site empty, 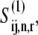 and the concentration of ribosomes with a ternary complex bound to the A site,
and the concentration of ribosomes with a ternary complex bound to the A site, 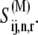 We assume that the concentration of translating ribosomes with codon species j in the A site, _R_j, is equal to the concentration of ribosomes participating in translation in an E. coli cell (estimated to be 24 _μ_M in previous work (13)) multiplied by the frequency of codon species j in the E. coli genome. Also, we assume that _R_j is constant and can be expressed as
We assume that the concentration of translating ribosomes with codon species j in the A site, _R_j, is equal to the concentration of ribosomes participating in translation in an E. coli cell (estimated to be 24 _μ_M in previous work (13)) multiplied by the frequency of codon species j in the E. coli genome. Also, we assume that _R_j is constant and can be expressed as
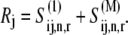 |
(42) |
|---|
Rearranging the above equation and applying it to Eq. 41 yields
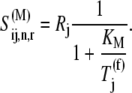 |
(43) |
|---|
Because the amino-acid incorporation rate is equal to 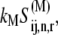 it can be shown that
it can be shown that
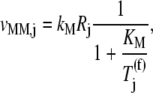 |
(44) |
|---|
which is equivalent to Eq. 19.
Under ternary complex competitive binding conditions we consider the following reaction scheme:
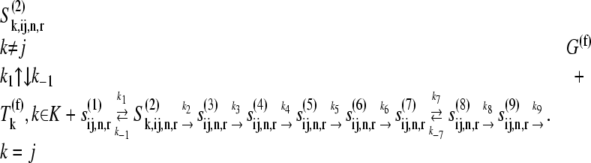 |
(45) |
|---|
A similar derivation to the one presented above for noncompetitive binding conditions yields Eq. 22 for competitive binding conditions.
Editor: Costas D. Maranas.
References
- 1.Hershey, J. 1987. Protein synthesis. In Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger, editor. American Society for Microbiology, Washington, DC.
- 2.Miller, O. L., Jr., B. A. Hamkalo, and C. A. Thomas, Jr. 1970. Visualization of bacterial genes in action. Science. 169:392–395. [DOI] [PubMed] [Google Scholar]
- 3.Arava, Y., Y. Wang, J. D. Storey, C. L. Liu, P. O. Brown, and D. Herschlag. 2003. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 100:3889–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.MacDonald, C. T., and J. H. Gibbs. 1969. Concerning kinetics of polypeptide synthesis on polyribosomes. Biopolymers. 7:707–725. [Google Scholar]
- 5.MacDonald, C. T., J. H. Gibbs, and A. C. Pipkin. 1968. Kinetics of biopolymerization on nucleic acid templates. Biopolymers. 6:1–5. [DOI] [PubMed] [Google Scholar]
- 6.Heinrich, R., and T. A. Rapoport. 1980. Mathematical modeling of translation of mRNA in eukaryotes; steady state, time-dependent processes and application to reticulocytes. J. Theor. Biol. 86:279–313. [DOI] [PubMed] [Google Scholar]
- 7.Mehra, A., K. H. Lee, and V. Hatzimanikatis. 2003. Insights into the relation between mRNA and protein expression patterns. I. Theoretical considerations. Biotechnol. Bioeng. 84:822–833. [DOI] [PubMed] [Google Scholar]
- 8.Mehra, A., and V. Hatzimanikatis. 2006. An algorithmic framework for genome-wide modeling and analysis of translation networks. Biophys. J. 90:1136–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varenne, S., J. Buc, R. Lloubes, and C. Lazdunski. 1984. Translation is a non-uniform process. Effect of tRNA availability on the rate of elongation of nascent polypeptide chains. J. Mol. Biol. 180:549–576. [DOI] [PubMed] [Google Scholar]
- 10.Curran, J. F., and M. Yarus. 1989. Rates of amino acyl-tRNA selection at 29 sense codons in vivo. J. Mol. Biol. 209:65–77. [DOI] [PubMed] [Google Scholar]
- 11.Soerensen, M. A., and S. Pedersen. 1991. Absolute in vivo translation rates of individual codons in Escherichia coli: the two glutamic acid codons GAA and GAG are translated with a threefold difference in rate. J. Mol. Biol. 222:265–280. [DOI] [PubMed] [Google Scholar]
- 12.Kruger, M. K., S. Pedersen, T. G. Hagervall, and M. A. Sorensen. 1998. The modification of the wobble base of tRNAGlu modulates the translation rate of glutamic acid codons in vivo. J. Mol. Biol. 284:621–631. [DOI] [PubMed] [Google Scholar]
- 13.Zouridis, H., and V. Hatzimanikatis. 2007. A model for protein translation: polysome self-organization leads to maximum protein synthesis rates. Biophys. J. 92:717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikemura, T. 1981. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J. Mol. Biol. 146:1–21. [DOI] [PubMed] [Google Scholar]
- 15.Ikemura, T. 1981. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J. Mol. Biol. 151:389–409. [DOI] [PubMed] [Google Scholar]
- 16.Kacser, H., and J. A. Burns. 1973. The control of flux. Symp. Soc. Exp. Biol. 27:65–104. [PubMed] [Google Scholar]
- 17.Heinrich, R., and S. Schuster. 1996. Metabolic control analysis. In The Regulation of Cellular Systems. Springer, New York.
- 18.Kazazian, H. H., Jr., and M. L. Freedman. 1968. The characterization of separated _α_- and _β_-chain polyribosomes in rabbit reticulocytes. J. Biol. Chem. 243:6446–6450. [PubMed] [Google Scholar]
- 19.Rose, J. K. 1977. Nucleotide sequences of ribosome recognition sites in messenger RNAs of vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA. 74:3672–3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Revel, M., and Y. Groner. 1978. Post-transcriptional and translational controls of gene expression in eukaryotes. Annu. Rev. Biochem. 47:1079–1126. [DOI] [PubMed] [Google Scholar]
- 21.Bremer, H., and P. P. Dennis. 1996. Modulation of chemical composition and other parameters of the cell by growth rate. In Escherichia coli and Salmonella. F. C. Neidhardt, editor. American Society for Microbiology, Washington, DC.
- 22.Sundararaj, S., A. Guo, B. Habibi-Nazhad, M. Rouani, P. Stothard, M. Ellison, and D. S. Wishart. 2004. The CyberCell Database (CCDB): a comprehensive, self-updating, relational database to coordinate and facilitate in silico modeling of Escherichia coli. Nucleic Acids Res. 32:D293–D295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong, H., L. Nilsson, and C. G. Kurland. 1996. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 260:649–663. [DOI] [PubMed] [Google Scholar]
- 24.Bilgin, N., F. Claesens, H. Pahverk, and M. Ehrenberg. 1992. Kinetic properties of Escherichia coli ribosomes with altered forms of S12. J. Mol. Biol. 224:1011–1027. [DOI] [PubMed] [Google Scholar]
- 25.Rodnina, M. V., T. Pape, R. Fricke, L. Kuhn, and W. Wintermeyer. 1996. Initial binding of the elongation factor Tu·GTP·aminoacyl-tRNA complex preceding codon recognition on the ribosome. J. Biol. Chem. 271:646–652. [DOI] [PubMed] [Google Scholar]
- 26.Pape, T., W. Wintermeyer, and M. V. Rodnina. 1998. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. EMBO J. 17:7490–7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Savelsbergh, A., V. I. Katunin, D. Mohr, F. Peske, M. V. Rodnina, and W. Wintermeyer. 2003. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol. Cell. 11:1517–1523. [DOI] [PubMed] [Google Scholar]
- 28.Chen, G. T., and M. Inouye. 1990. Suppression of the negative effect of minor arginine codons on gene expression; preferential usage of minor codons within the first 25 codons of the Escherichia coli genes. Nucleic Acids Res. 18:1465–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kozak, M. 1984. Point mutations close to the AUG initiator codon affect the efficiency of translation of rat preproinsulin in vivo. Nature. 308:241–246. [DOI] [PubMed] [Google Scholar]
- 30.Bilgin, N., M. Ehrenberg, and C. Kurland. 1988. Is translation inhibited by noncognate ternary complexes? FEBS Lett. 233:95–99. [DOI] [PubMed] [Google Scholar]
- 31.LaRiviere, F. D., A. D. Wolfson, and O. C. Uhlenbeck. 2001. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science. 294:165–168. [DOI] [PubMed] [Google Scholar]
- 32.Grantham, R., C. Gautier, M. Gouy, M. Jacobzone, and R. Mercier. 1981. Codon catalog usage is a genome strategy modulated for gene expressivity. Nucleic Acids Res. 9:r43–r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bulmer, M. 1988. Codon usage and intragenic position. J. Theor. Biol. 133:67–71. [DOI] [PubMed] [Google Scholar]
- 34.Bulmer, M. 1991. The selection-mutation-drift theory of synonymous codon usage. Genetics. 129:897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liljenstrom, H., and G. von Heijne. 1987. Translation rate modification by preferential codon usage: intragenic position effects. J. Theor. Biol. 124:43–55. [DOI] [PubMed] [Google Scholar]
- 36.Cameron, J. M., M. Kreitman, and M. Aguade. 1999. Natural selection on synonymous sites is correlated with gene length and recombination in Drosophila. Genetics. 151:239–249. [DOI] [PMC free article] [PubMed] [Google Scholar]