Constitutive Activation of the PrfA Regulon Enhances the Potency of Vaccines Based on Live-Attenuated and Killed but Metabolically Active Listeria monocytogenes Strains (original) (raw)
Abstract
Recombinant vaccines derived from the facultative intracellular bacterium Listeria monocytogenes are presently undergoing early-stage clinical evaluation in oncology treatment settings. This effort has been stimulated in part due to preclinical results that illustrate potent activation of innate and adaptive immune effectors by L. monocytogenes vaccines, combined with efficacy in rigorous animal models of malignant and infectious disease. Here, we evaluated the immunologic potency of a panel of isogenic vaccine strains that varied only in prfA. PrfA is an intracellularly activated transcription factor that induces expression of virulence genes and encoded heterologous antigens (Ags) in appropriately engineered vaccine strains. Mutant strains with PrfA locked into a constitutively active state are known as PrfA* mutants. We assessed the impacts of three PrfA* mutants, G145S, G155S, and Y63C, on the immunologic potencies of live-attenuated and photochemically inactivated nucleotide excision repair mutant (killed but metabolically active [KBMA]) vaccines. While PrfA* substantially increased Ag expression in strains grown in broth culture, Ag expression levels were equivalent in infected macrophage and dendritic cell lines, conditions that more closely parallel those in the immunized host. However, only the prfA(G155S) allele conferred significantly enhanced vaccine potency to KBMA vaccines. In the KBMA vaccine background, we show that PrfA*(G155S) enhanced functional cellular immunity following an intravenous or intramuscular prime-boost immunization regimen. These results form the basis of a rationale for including the prfA(G155S) allele in future live-attenuated or KBMA L. monocytogenes vaccines advanced to the clinical setting.
Recognition of the advantages of recombinant _Listeria monocytogenes_-based vaccines compared to those of other recombinant-vaccine platforms has facilitated the ongoing development and current evaluation of the former in early-phase clinical trials. These advantages include practical considerations, such as straightforward fermentation methods for manufacturing, and other desirable features, such as the ability to repeat administer even in the presence of protective _L. monocytogenes_-specific immunity (6, 40, 41). One compelling rationale for this vaccine platform is based on the well-known correlates of protection in the mouse listeriosis model: long-lived functional CD4+ and CD8+ memory T cells induced in response to a single immunization with L. monocytogenes (19, 28). There are now numerous publications that demonstrate the striking efficacy of recombinant L. monocytogenes vaccines in several animal models due to robust innate and adaptive cellular immunity (9, 10, 29). Recombinant _L. monocytogenes_-based vaccines represent an emerging approach to addressing an acute global need for effective vaccines that elicit functional cellular immunity to prevent or treat infections such as human immunodeficiency virus infection, hepatitis C virus infection, tuberculosis, and malaria as well as cancer.
As L. monocytogenes is a food-borne pathogen having increased virulence among immunocompromised individuals, attenuated vaccine platforms are a prerequisite for advancement to evaluation with humans (23). We have previously described both live-attenuated and photochemically inactivated vaccine platforms derived from the wild-type (WT) strain 10403S (8, 9). The live-attenuated vaccine strain is deleted of both the actA and the inlB virulence genes (L. monocytogenes Δ_actA_ Δ_inlB_ vaccine strain), which in combination limit growth in the liver, a principal target organ of infection by the WT organism. Liver toxicity in mice, as measured by serum liver function tests for alanine transaminase and aspartate transaminase, is dramatically lower in mice injected intravenously (i.v.) with the L. monocytogenes Δ_actA_ Δ_inlB_ strain than in those injected i.v. with WT L. monocytogenes. Furthermore, liver toxicity was minimal and not dose limiting in two toxicology studies performed under good laboratory practice guidelines with cynomolgus monkeys given escalating doses of L. monocytogenes Δ_actA_ Δ_inlB_-based strains (unpublished data). The L. monocytogenes Δ_actA_ Δ_inlB_ vaccine strain forms the basis for two ongoing FDA-approved phase 1 clinical trials being conducted with adult subjects with advanced cancers. The second vaccine platform, termed “killed but metabolically active” (KBMA), is derived from the L. monocytogenes Δ_actA_ Δ_inlB_ vaccine strain and is deleted of both uvrA and uvrB, genes encoding the DNA repair enzymes of the nucleotide excision repair pathway. KBMA vaccines (L. monocytogenes Δ_actA_ Δ_inlB_ Δ_uvrAB_ vaccine strains) are exquisitely sensitive to photochemical inactivation by combined treatment with the synthetic psoralen S-59 and long-wave UV light. While killed, KBMA L. monocytogenes vaccines can transiently express their gene products, allowing them to escape the phagolysosome and induce functional cellular immunity and protection against WT L. monocytogenes and vaccinia virus challenge (8).
While studies of _L. monocytogenes_-based vaccines have to date been encouraging, all-too-frequently promising preclinical results do not necessarily forecast clinical success. There is a continued need for refinements that either enhance the potency or reduce the toxicity of _L. monocytogenes_-based vaccines in order to facilitate their ultimate clinical development. Here, we evaluated the impact of constitutively active PrfA on the vaccine potencies of both live-attenuated and KBMA vaccines. PrfA is a transcription factor activated intracellularly that acts as a central virulence regulator, serving to enable what has been described as the “Dr. Jekyll and Mr. Hyde” dichotomy of L. monocytogenes growth lifestyles in mammals or as a saprophyte (16). PrfA knockout strains are avirulent (43). In WT L. monocytogenes, PrfA is expressed upon infection of host cells and, in turn, induces expression of the prfA regulon including the hly and plcA genes, encoding listeriolysin O (LLO) and phospholipase C, respectively. In combination, these gene products mediate escape of the bacterium from the harsh microenvironment of the phagolysosome. PrfA-dependent promoters can be utilized to drive antigen (Ag) expression in recombinant L. monocytogenes vaccines by linking the heterologous gene to either the hly or the actA promoters (references 17 and 36 and this study). Amino acid substitutions in PrfA that result in the constitutive activation of PrfA-dependent genes are known collectively as PrfA* mutants (34, 35). A number of WT L. monocytogenes strains with a hyperhemolytic phenotype have a mutation in prfA, most commonly G145S, which can result in increased virulence compared to that of laboratory-adapted strains, such as 10403S (34). Similarly, other prfA mutants with increased expression of PrfA-dependent genes that were selected by a chemical mutagenesis approach also had increased virulence in mice (38). We hypothesized that induction of PrfA-dependent genes prior to immunization would enhance the efficiency of vaccines through diverse mechanisms, including increase of phagolysosomal escape and expression of PrfA-dependent encoded Ags in the cytosol of the host cell, leading to more-potent CD4+ and CD8+ T-cell responses.
In this study, we assessed the impacts of three prfA mutations on the potencies of isogenic live-attenuated and KBMA vaccine strains. While Ag expression levels were increased in PrfA* vaccine strains grown in broth culture, no differences in Ag expression level between PrfA* vaccine strains and vaccine strains with native prfA could be detected in the cytosol from infected macrophage or dendritic cell (DC) lines. Strikingly, however, the magnitude and functionality of vaccine-induced CD8+ T cells, as measured by protection against bacterial or viral challenge, were significantly improved among recombinant KBMA L. monocytogenes vaccine strains with PrfA*(G155S) compared to those of vaccines with all other prfA alleles tested. These findings indicate that activation of the prfA regulon and induction of Ag expression prior to immunization enhance the potency of _L. monocytogenes_-based vaccines.
MATERIALS AND METHODS
Mice.
Six- to 12-week-old female C57BL/6 and BALB/c mice were obtained from Charles River Laboratories (Wilmington, MA). Studies were performed under animal protocols approved by the Anza Institutional Animal Care and Use Committee.
Bacterial strains.
L. monocytogenes vaccine strains were constructed in the L. monocytogenes Δ_actA_ Δ_inlB_ Δ_uvrAB_ strain (8). PrfA* variants were constructed by cloning the various prfA alleles with 800 bp to 1 kb of flanking homology into the temperature-sensitive allelic exchange vector pKSV7 and used to replace the WT allele by using standard procedures (11). Strains with activated PrfA phenotypes were screened for increased zones of both phospholipase activity on egg yolk overlay plates (47) and hemolysis on horse blood agar (Remel). Phenotypically correct clones were confirmed by sequencing the genomic prfA locus. An Ag expression cassette termed the Quadvac construct, expressing four vaccinia CD8+ T-cell epitopes of various levels of strength and the ovalbumin (OVA)-derived CD8+ T-cell epitope SIINFEKL from a single synthetic gene, was designed in silico where the epitopes were strung together and spaced with a linker sequence. The amino acid sequence was codon optimized for expression in L. monocytogenes by using Gene Designer software (45), synthesized (DNA2.0; Menlo Park, CA), and cloned downstream of the actA promoter and in-frame with the amino terminus of the actA gene. The construct was cloned into a derivative of the pPL2 integration vector and stably integrated at the tRNAArg locus of the bacterial chromosome in the various prfA* strain backgrounds, as described previously (21). All molecular constructs were confirmed by DNA sequencing.
Western blot detection of heterologous Ag expression.
Western blot analyses of broth culture were performed on equivalent amounts of trichloroacetic acid-precipitated supernatants of bacterial cultures grown in yeast extract medium to an optical density at 600 nm of 0.7 (late log). For Western blots of _L. monocytogenes_-infected host cells, J774 cells or DC2.4 cells were inoculated at a multiplicity of infection of 50 or 100 for 1 h, and the cells were washed three times with phosphate-buffered saline and Dulbecco's modified Eagle's medium supplemented with 50 μg/ml gentamicin. For early time points, DC2.4 cells were harvested at 1.5 or 2.5 h postinfection. For late time points, J774 cells were harvested at 7 h. Cells were lysed with sodium dodecyl sulfate sample buffer, collected, and run on 4 to 12% polyacrylamide gels and transferred to nitrocellulose membranes for Western blot analysis. All Western blots utilized a polyclonal antibody raised against the mature N terminus of the ActA protein.
Immunizations.
Live-attenuated bacteria were prepared for immunization from overnight cultures grown in yeast extract medium. KBMA L. monocytogenes strains were S59-psoralen and UVA treated as previously described (8). Photochemically inactivated bacteria (KBMA L. monocytogenes) were washed once with Dulbecco's phosphate-buffered saline, resuspended in 8% dimethyl sulfoxide, and then stored at −80°C. Bacteria were diluted in Hanks' balanced salt solution (HBSS) for injection. Injection stocks of live-attenuated bacteria were plated to confirm CFU counts. Totals of 5 × 106 CFU live-attenuated bacteria and 1 × 108 particles of KBMA bacteria were administered either i.v. into the tail vein at a 200-μl volume or intramuscularly (i.m.) into a single tibialis cranialis muscle at a 30 μl volume, with a boost vaccination given in an alternate tibialis cranialis muscle.
Detection of serum cytokines and chemokines.
Serum was collected from mice by retro-orbital bleed at 2, 4, and 8 h postvaccination with KBMA or 4, 8, and 24 h with live-attenuated L. monocytogenes. Cytokines/chemokines were measured using a mouse inflammation Cytometric Bead Array kit (BD Biosciences, San Jose, CA) according to the manufacturer's instructions. Samples were acquired using a FACSCanto flow cytometer (BD Biosciences). Data were analyzed using Cytometric Bead Array software (BD Biosciences).
Peptides.
The OVA257-264 (SIINFEKL), LLO190-201 (NEKYAQAYPNVS), HSV-gB2 (SSIEFARL), B8R20-27 (TSYKFESV), K3L6-15 (YSLPNAGDVI), C4L125-132 (LNFRFENV), and A42R88-96 (YAPVSPIVI) (24) peptides were synthesized by Synthetic Biomolecules (San Diego, CA).
Reagents for flow cytometry.
CD3 fluorescein isothiocyanate (FITC) or phycoerythrin (PE)-Cy7 (clone 145-2C11), CD4 FITC (clone GK1.5), CD8 PE-Cy7 or Ag-presenting cell (APC)-Cy7 (clone 53-6.7), CD19 FITC (clone MB19-1), tumor necrosis factor (TNF) PE (clone MP6-XT22), and gamma interferon (IFN-γ) APC (clone XMG1.2) were purchased from eBioscience (San Diego, CA). CD8α PerCP (clone 53-6.7) was purchased from BD Biosciences (San Jose, CA).
In vivo cytotoxicity assay.
Splenocytes from naïve recipients were pulsed with a 1 μM concentration of either control (HSV-gB2) or target (B8R or A42R) peptide. Cells were then labeled with concentrations of carboxyfluorescein diacetate succinimidyl ester (CFSE; Molecular Probes, Eugene, OR) of 0.2 μM (CFSElo), 1 μM (CFSEmed), or 5 μM (CFSEhi). A total of 3 × 106 labeled spleen cells of each population were mixed and injected i.v. Spleens were harvested 16 h later, and the proportion of target to control populations was determined and the percentage of killing calculated.
Intracellular staining of Ag-specific T cells.
Splenocytes were stimulated for 5 h with the relevant peptide in the presence of brefeldin A for intracellular cytokine staining, as previously described (9). Stimulated cells were surface stained for CD4 and CD8 and then fixed and permeabilized using a cytofix/cytoperm kit (BD Biosciences, San Jose, CA). Cells were then stained for IFN-γ, TNF-α, and/or interleukin 2 (IL-2). Samples were acquired using a FACSCanto flow cytometer (BD Biosciences). Data were gated to include exclusively CD4+ or CD8+ events, and then the percentage of these cells expressing IFN-γ was determined. Data were analyzed using FlowJo software (Treestar, Ashland, OR).
L. monocytogenes protection studies.
To assess protective immunity, BALB/c mice were vaccinated with the indicated strains and challenged at 14 days postvaccination with a 2× 50% lethal dose (LD50) of WT L. monocytogenes (1 × 105 CFU), and CFU counts in the spleen in organ homogenates were measured 3 days later, as described previously (4). Median lethality (LD50) values were determined as described previously (9).
Vaccinia virus protection studies.
C57BL/6 mice were given prime and boost vaccinations separated by 27 days with L. monocytogenes Quadvac strains, and 28 days later, mice were challenged intraperitoneally with 1 × 107 PFU of vaccinia virus. Protection was evaluated by measuring viral titer in the ovaries 5 days after virus challenge, as described previously (1).
Statistical analysis.
Differences in protection against vaccinia virus or L. monocytogenes challenges were determined by Student's t test. Unless otherwise indicated, all experiments were conducted at least twice.
RESULTS
Construction and characterization of isogenic PrfA* L. monocytogenes vaccine strains.
We hypothesized that induction of the PrfA regulon prior to immunization would increase the immunologic potencies of recombinant live-attenuated and KBMA L. monocytogenes vaccines. To test this hypothesis, we constructed a panel of isogenic strains on an L. monocytogenes background that contained complete coding region deletions of actA, inlB, uvrA, and uvrB (L. monocytogenes Δ_actA_ Δ_inlB_ Δ_uvrAB_ strains) that varied only in prfA. We selected three prfA mutants that were generated by chemical mutagenesis or were spontaneous mutants and encoded a constitutively active PrfA* protein (34, 38). Strains with PrfA*(G155S), PrfA*(G145S), or PrfA*(Y63C) have previously been shown to either increase the expression level of an actA promoter-dependent β-glucouronidase reporter protein relative to those of isogenic strains with the native prfA gene or, in some cases, increase the virulence of WT L. monocytogenes (25, 34, 38; M. D. Miner, G. C. Port, and N. E. Freitag, submitted for publication).
To enable us to distinguish immunologic potency differences between isogenic L. monocytogenes vaccine strains, we constructed an Ag expression cassette that encoded five well-defined H-2b-restricted major histocompatibility complex (MHC) class I epitopes that have previously been shown to elicit a range of CD8+ T-cell responses in mice. A construct encoding four tandemly spaced vaccinia virus (A42R, C4L, K3L, and B8R) epitopes and the chicken OVA (SL8) epitope was synthesized and then cloned under the control of the PrfA-regulated actA promoter as a fusion protein with the 100 N-terminal amino acids of ActA. The construct known as Quadvac was cloned into a derivative of pPL2 and then integrated at the tRNAArg locus in four isogenic strains (L. monocytogenes Δ_actA_ Δ_inlB_ Δ_uvrAB_ strains), each harboring the WT, G145S, G155S, or Y63C prfA allele, as described previously (Fig. 1A) (21). The four isogenic vaccine strains all grew equivalently in yeast extract or brain heart infusion broth culture (data not shown). Vaccine strains grown in yeast extract broth culture were either used directly as a live-attenuated L. monocytogenes vaccine (9) or photochemically inactivated with the synthetic psoralen S-59 and long-wave UV light to yield a KBMA L. monocytogenes vaccine, as described previously (8).
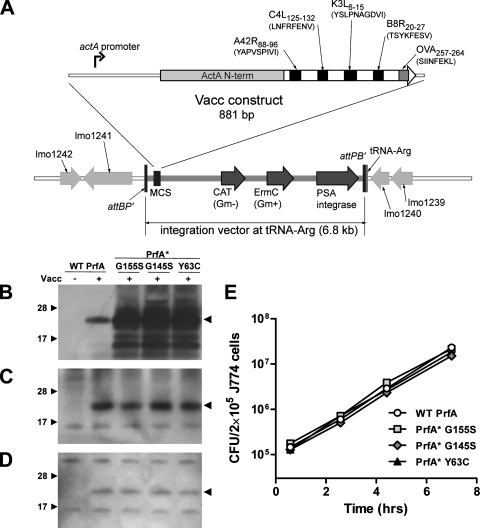
FIG. 1.
Characterization of L. monocytogenes PrfA* vaccine strains. (A) Construction of the L. monocytogenes Quadvac strain expressing four vaccinia virus T-cell epitopes (A24R, C4L, K3L, and B8R) and the OVA SL8 epitope spaced with linker sequences and fused to the first 100 amino acids of ActA (ActAN100) by use of a derivative of the pPL2 site-specific integration vector. CAT, chloramphenicol acetyltransferase; Gm−, gram negative; Gm+, gram positive. (B) Expression of the heterologous protein in yeast extract broth. (C) Expression of the heterologous Ag at 7 h postinfection in infected J774 macrophage cells. (D) Expression of the heterologous Ag at 2.5 h postinfection in infected DC2.4 DCs. (E) Intracellular growth of isogenic L. monocytogenes vaccine strains in J774 macrophages.
We compared the levels of Ag expression and secretion in broth culture from the four live-attenuated L. monocytogenes vaccines, and as expected, higher expression levels were observed in PrfA* strains than in the strain with a WT prfA allele (Fig. 1B). While overexpression of PrfA-dependent genes in PrfA* strains grown in broth culture, combined with enhanced invasion of epithelial cells, has been well described (25, 31, 38, 44), little is known regarding whether the PrfA* phenotype is recapitulated in infected, cultured mammalian cells. We evaluated Ag expression from the L. monocytogenes vaccine strains in the phagocytic mouse macrophage and DC lines J774 and DC2.4, respectively, rather than in nonphagocytic cell lines, such as HepG2 (liver) or PtK2 (epithelial). The L. monocytogenes Δ_actA_ Δ_inlB_ strain cannot mediate InlB-dependent infection of liver cells expressing the hepatocyte growth factor receptor, and epithelial tissues do not represent a major target that is relevant to the i.m. and i.v. immunization routes used in this investigation. Surprisingly, Ag expression levels in J774 macrophages infected with the isogenic L. monocytogenes vaccine strains were equivalent regardless of prfA allele (Fig. 1C). Notably, Ag expression levels were also equivalent at early time points in DC2.4 DCs (Fig. 1D). Infection (uptake) and intracellular growth of all L. monocytogenes vaccine strains in J774 (Fig. 1E) and DC2.4 (not shown) cell lines were equivalent and not dependent on prfA allele.
PrfA* minimally increases the virulence of L. monocytogenes Δ_actA_ Δ_inlB_ Δ_uvrAB_ strains.
It has previously been reported that PrfA* mutants have virulence levels increased up to 10-fold in BALB/c mice. For example, prfA(G155S) decreased the LD50 value of its isogenic WT strain from 2 × 104 CFU to 3 × 103 CFU (38). However, the impact of PrfA* mutants on the virulence of attenuated strains is unknown. As the immunization dose used for L. monocytogenes vaccine studies in mice is typically 1/10 the LD50 value, we measured the impacts that the three PrfA* mutants used in this investigation had on the virulence of the L. monocytogenes Δ_actA_ Δ_inlB_ Δ_uvrAB_ strain, a strain that is rapidly cleared from the liver following i.v. administration and is attenuated in mice by more than 3 logs compared to the level for WT L. monocytogenes (5, 8, 9). PrfA* marginally increased the virulence of the L. monocytogenes Δ_actA_ Δ_inlB_ Δ_uvrAB_ strain, with the LD50 value decreased only 2.1-fold (3.5 × 107 CFU versus 7.3 × 107 CFU) in isogenic strains harboring the prfA(G145S) or prfA(Y63C) allele and 1.4-fold (5.2 × 107 CFU versus 7.3 × 107 CFU) in the isogenic strain harboring the prfA(G155S) allele (Table 1).
TABLE 1.
Virulence of live-attenuated L. monocytogenes strains
| Strain | Genotype | prfA mutation | LD50 | |
|---|---|---|---|---|
| No. of CFU | Fold change relative to WT level | |||
| BH1299 | Δ_actA_ Δ_inlB_ Δ_uvrAB_ | None (WT) | 7.3 × 107 | |
| BH1371 | Δ_actA_ Δ_inB_ Δ_uvrAB prfA_(G155S) | G155S | 5.2 × 107 | 1.4 |
| BH1375 | Δ_actA_ Δ_inB_ Δ_uvrAB prfA_(G145S) | G145S | 3.5 × 107 | 2.1 |
| BH1379 | Δ_actA_ Δ_inB_ Δ_uvrAB prfA_(Y63C) | Y63C | 3.5 × 107 | 2.1 |
L. monocytogenes_-based vaccines, including attenuated L. monocytogenes Δ_actA Δ_inlB_-based strains, are potent activators of innate immunity, as reflected by the Th1-polarizing, proinflammatory serum cytokine profile induced in response to i.v. administration (4). As activation of innate immunity is related to the quality of the L. monocytogenes vaccine-induced immune response, we measured the serum cytokine levels at several time points during the first 24 h following i.v. administration of the isogenic vaccine strains. C57BL/6 mice were injected i.v. with 5 × 106 CFU, a dose that approximated the 0.1-LD50 value for the four isogenic vaccine strains. All three PrfA* vaccine strains induced statistically significantly higher levels of the proinflammatory cytokines/chemokines, such as IL-6 and MCP-1, within 8 hours of administration compared to the vaccine strain with native prfA (Fig. 2A). Additionally, PrfA*(G155S) and PrfA*(G145S) induced significantly more TNF-α and IFN-γ. No significant differences between the three PrfA* strains were observed; however, increased mouse-to-mouse variability was observed in mice given the PrfA*(Y63C) vaccine strain.
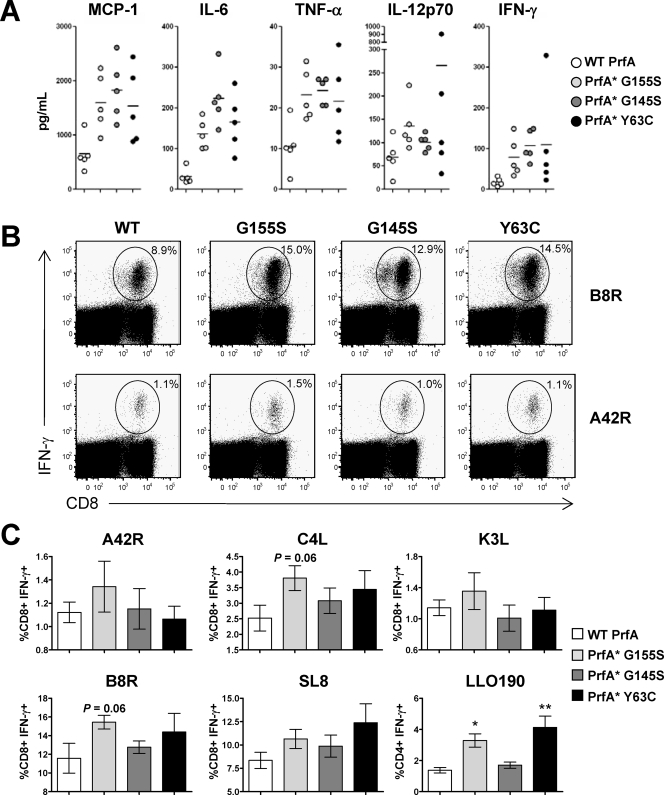
FIG. 2.
Improved innate and adaptive immunity induced by PrfA* vaccine strains. (A) Serum cytokine/chemokine levels determined 8 h following a single i.v. administration of 5 × 106 CFU of L. monocytogenes Δ_actA_ Δ_inlB_ WT prfA, PrfA*(G155S), PrfA*(G145S), and PrfA*(Y63C) strains. Cytokine/chemokine levels were determined by a cytometric bead array. Each symbol represents a single animal. Data are from a single experiment, representative of at least two experiments. The levels of monocyte chemoattractant protein 1 (MCP-1), IL-6, TNF-α, and IFN-γ induced by the PrfA*(G155S) and the PrfA*(G145S) strains were significantly increased over the WT levels (P < 0.05); PrfA*(Y63C) was significantly increased for MCP-1 (P < 0.05) and IL-6 (P < 0.005). (B, C) Live-attenuated PrfA* vaccine strains induced Ag-specific immunity of a higher magnitude. C57BL/6 mice were immunized i.v. with 5 × 106 CFU of L. monocytogenes Δ_actA_ Δ_inlB_ WT prfA, PrfA*(G155S), PrfA*(G145S), and PrfA*(Y63C) strains. Ag-specific T-cell responses were determined by intracellular cytokine staining at the peak of the response 7 days following vaccination. (B) Dot blots from a representative animal from each group are shown. (C) The means ± standard deviations are shown for each group of five animals. *, P < 0.05; **, P < 0.005 for comparison to WT prfA.
PrfA* increases the immunogenicity of live-attenuated L. monocytogenes vaccines.
We evaluated the immunogenicity of the isogenic vaccine strains to assess the impact of PrfA* on vaccine potency. To facilitate comparison, isogenic vaccine strains expressed a common heterologous Ag (termed Quadvac) composed of multiple defined, H-2b-restricted MHC class I vaccinia virus epitopes (A42R, C4L, K3L, and B8R) that have been shown to elicit high-, intermediate-, and low-frequency T-cell responses following vaccinia virus infection and, in addition, the strong OVA epitope SL8. This strategy allowed us both to rank the magnitudes of vaccine-induced CD8+ T-cell responses over a dose range of immunization and to evaluate the quality of the response by challenge with vaccinia virus.
Groups of female C57BL/6 mice were immunized i.v. with 5 × 106 CFU of the four isogenic L. monocytogenes Quadvac strains, and the CD8+ and (_L. monocytogenes_-specific) CD4+ T-cell frequencies were determined by intracellular cytokine staining at the peak of the response (9), 7 days following a single immunization. The PrfA*(G155S) L. monocytogenes vaccine induced Ag-specific T-cell responses of greater magnitude than the G145S and Y63C PrfA* vaccine strains and the strain with WT prfA (Fig. 2B and C). The increased magnitude of the Ag-specific IFN-γ-positive T cells in PrfA*(G155S) L. monocytogenes vaccine-immunized mice was greater not only with the immunodominant SL8 and B8R epitopes but, importantly, also with the intermediate- and low-frequency epitopes, A42R, C4L, and K3L. The LLO-specific CD4+ T-cell response was increased twofold in mice immunized with the PrfA*(G155S) vaccine strain compared to the level for the other vaccine strains.
PrfA*(G155S) increases the immunogenicity of KBMA L. monocytogenes vaccines.
We hypothesized that constitutive activation of PrfA and induction of the PrfA regulon might improve the immunogenicity of KBMA L. monocytogenes through a variety of mechanisms, including increased escape from the vacuole as well as an increased expression level of the heterologous Ag in the cytosol of APCs. We demonstrated previously that CD8+ T-cell potency and protective immunity require that the immunizing L. monocytogenes strain access the cytosol (4). As KBMA vaccine strains are unable to expand in the cytosol, increased efficiency of escape from the phagosome through increased expression of LLO and phospholipase C might enhance vaccine potency.
To assess the innate as well as adaptive immunity to KBMA L. monocytogenes strains, C57BL/6 mice were immunized i.v. with 1 × 108 particles, a well-tolerated dose. As described previously, we used photochemical inactivation conditions that resulted in 10-log killing of L. monocytogenes vaccine preparations (8). Thus, individual mice had a 10−2 chance of receiving a single live L. monocytogenes Δ_actA_ Δ_inlB_ Δ_uvrAB_ bacterium, a nonimmunizing dose. Serum cytokine/chemokine levels were measured during the first 8 h of infection, which we had observed in previous experiments to include the peak of the response, which then returned to background levels within 24 h (5). KBMA PrfA* vaccine strains induced higher levels of MCP-1, IL-12p70, and IFN-γ than the KBMA vaccine with WT prfA; the differences were significant for some of the cytokines (Fig. 3A). No significant differences between the three PrfA* strains were observed, but the levels of cytokines induced by the KBMA PrfA*(G155S) vaccine tended to be higher than the levels induced by the KBMA PrfA*(G145S) or PrfA*(Y63C) vaccine.
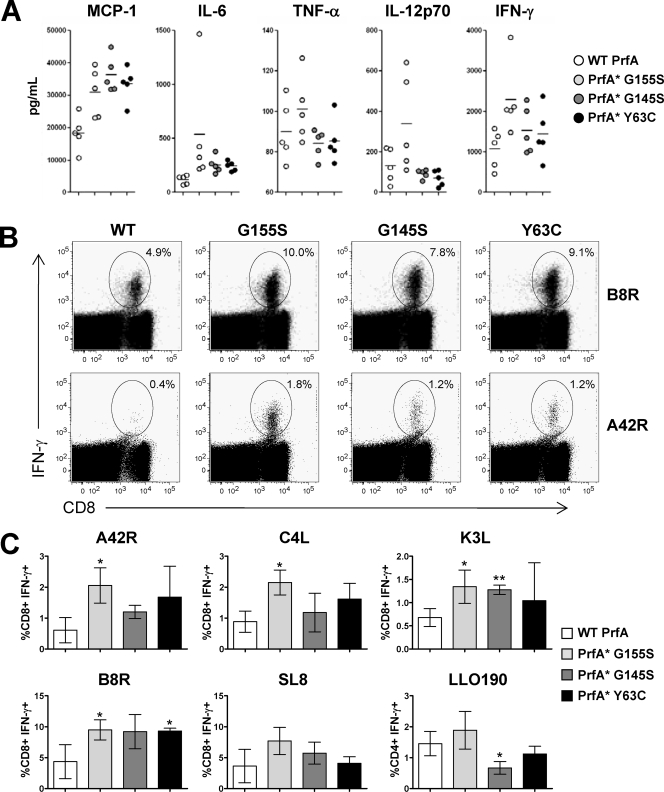
FIG. 3.
PrfA* enhances the immunogenicity of KBMA L. monocytogenes vaccines. (A) Serum cytokine/chemokine levels determined 8 h following a single i.v. administration of 1 × 108 particles of KBMA L. monocytogenes Δ_actA_ Δ_inlB_ WT prfA, PrfA*(G155S), PrfA*(G145S), and PrfA*(Y63C) strains. Cytokine/chemokine levels were determined by cytometric bead analysis. Each symbol represents a single animal. The levels of monocyte chemoattractant protein 1 (MCP-1) were significantly increased compared to the WT levels for PrfA*(G155S) (P < 0.05), PrfA*(G145S), and PrfA*(Y63C) (P < 0.005). PrfA*(G155S) increased the level of IFN-γ (P < 0.05). The levels of IL-6 were increased in PrfA*(G145S) (P < 0.05) and PrfA*(Y63C) (P < 0.005). (B, C) KBMA L. monocytogenes PrfA* strains induced Ag-specific immunity of a higher magnitude. C57BL/6 mice were immunized i.v. with 1 × 108 particles of KBMA L. monocytogenes Δ_actA_ Δ_inlB_ WT prfA, PrfA*(G155S), PrfA*(G145S), and PrfA*(Y63C) strains. Ag-specific T-cell responses were determined by intracellular cytokine staining at the peak of the response 7 days following vaccination. (B) Dot blots from a representative animal from each group are shown. (C) The means ± standard deviations are shown for each group of five animals. *, P < 0.05; **, P < 0.005 for comparison to WT prfA.
We compared the immunogenicities of the isogenic KBMA vaccine strains in C57BL/6 mice, each given two vaccinations separated by 2 weeks. The highest magnitude of the secondary Ag-specific CD8+ T-cell response specific for the five Quadvac epitopes was observed in mice immunized with KBMA PrfA*(G155S), and the four vaccinia epitopes were significantly higher than those for WT prfA (Fig. 3B and C). While the magnitude of the CD8+ response was generally higher in mice immunized with the other two KBMA PrfA* vaccine strains than in those immunized with KBMA WT prfA, this was not the case with all CD8 T-cell epitopes evaluated, with only one epitope from each strain reaching statistical significance (Fig. 3B and C). Interestingly, in contrast to what was found for the live-attenuated vaccine strains, LLO-specific CD4+ T-cell responses were not higher in magnitude among mice immunized with KBMA L. monocytogenes PrfA* strains. Thus, the prfA(G155S) allele conferred the highest immunogenicities to both live-attenuated and KBMA vaccine strains.
KBMA PrfA* vaccines have increased immune potency.
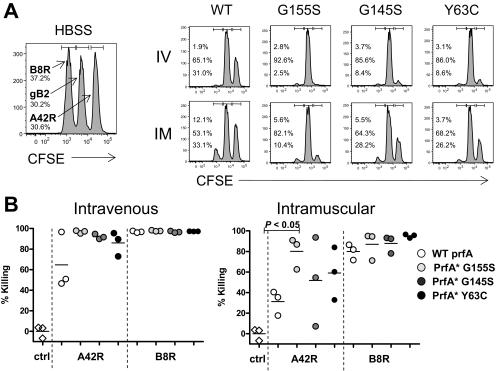
It is well established that the magnitude of an induced T-cell response is not necessarily representative of the potency of the response. To assess the potency of the vaccine-induced CD8+ T-cell response, we assessed the in vivo cytolytic activities specific for two vaccinia epitopes, A42R and B8R. C57BL/6 mice were immunized twice with the four isogenic KBMA L. monocytogenes strains. We evaluated immunogenicity, following two alternative immunization routes: i.v. and i.m. We evaluated i.m. immunization to assess the potency of KBMA PrfA* vaccine strains administered by a conventional vaccination route. Mice were immunized i.v. or i.m. twice, 2 weeks apart, with KBMA L. monocytogenes Δ_actA_ Δ_inlB_ WT prfA, PrfA*(G155S), PrfA*(G145S), and PrfA*(Y63C) strains or vehicle (HBSS) as a control. In vivo cytolytic activity was determined 7 days later by challenging mice with gB2 (negative control; middle peak), A42R-loaded splenocytes (right peak), or B8R-loaded splenocytes (left peak). Robust responses specific for the strong B8R epitope were observed in all groups immunized with KBMA PrfA* or WT prfA vaccines. The extent of target killing was slightly higher in mice that were immunized i.v. (Fig. 4A and B). Potent killing activity was also elicited against the A42R vaccinia virus epitope in mice immunized i.v. or i.m. with KBMA PrfA*(G155S). However, the killing activity against A42R induced by KBMA vaccines based on WT PfrA* and given i.m. was reduced twofold compared to that for PrfA*(G155S). KBMA vaccines based on G145S or Y63C were of intermediate potency when given by the i.m. route (Fig. 4B).
FIG. 4.
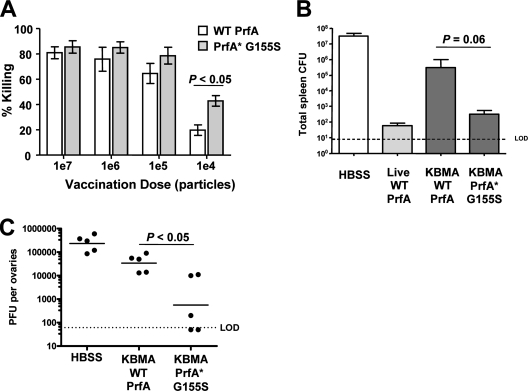
Improved potency of T-cell responses elicited by KBMA L. monocytogenes PrfA* strains. (A, B) C57BL/6 mice were immunized i.v. or i.m. (as indicated in the figure) twice, 2 weeks apart, with HBSS (left) or with 1 × 108 particles of KBMA L. monocytogenes Δ_actA_ Δ_inlB_ WT prfA, PrfA*(G155S), PrfA*(G145S), and PrfA*(Y63C) strains. In vivo cytolytic activity was determined 7 days later by challenging mice with gB2 (control; middle peak), A24R-loaded targets (right peak), or B8R-loaded targets (left peak). (A) A histogram for a representative animal is shown for each group. (B) In vivo cytolytic activity specific to A42R and B8R is shown for mice vaccinated i.v. or i.m. Each symbol represents an individual animal. The A42R response in i.m. vaccinated mice was significantly higher for the PrfA*(G155S) strain than for the WT (P < 0.05).
KBMA PrfA*(G155S) vaccines elicit protective immunity.
While all three of the mutant prfA alleles tested conferred increased immune potency to KBMA vaccines compared to native prfA, the highest magnitude and degree of in vivo killing specific for A42R, which was the weakest epitope evaluated, was observed with KBMA PrfA*(G155S) vaccines (Fig. 4B). To better characterize the functionality of the CD8+ T-cell immunity elicited by KBMA PrfA*(G155S) vaccines, we determined its ability to establish protection against WT L. monocytogenes or virus challenge compared to that of KBMA vaccines with native prfA. In a dose-response experiment, the superior potency of KBMA PrfA*(G155S) for inducing B8R responses could be seen compared to levels for mice immunized with KBMA harboring WT prfA (Fig. 5A).
FIG. 5.
The PrfA*(G155S) vaccine strain induces protective immunity. (A) In vivo cytolytic activity specific to B8R is shown following two vaccinations 2 weeks apart at various doses of the KBMA L. monocytogenes Δ_actA_ Δ_inlB_ WT prfA or PrfA*(G155S) strain. The means ± standard deviations are shown for groups with five animals each. The difference between WT prfA and PrfA*(G155S) vaccine strains is statistically significant at the 1 × 104-CFU vaccination dose. (B) Protective immunity to a 2× LD50 challenge with WT L. monocytogenes is shown. BALB/c mice were immunized i.v. once with 1 × 108 particles of the KBMA L. monocytogenes Δ_actA_ Δ_inlB_ WT prfA or PrfA*(G155S) strain. HBSS served as a control. Spleens were harvested 3 days after challenge and plated for CFU. (C) Viral titers in ovaries following an intraperitoneal challenge with 1 × 107 PFU of vaccinia virus. C57BL/6 mice were vaccinated twice i.v. with 1 × 108 particles of the KBMA L. monocytogenes Δ_actA_ Δ_inlB_ WT prfA or PrfA*(G155S) strain. Viral titers were determined 5 days after vaccinia virus challenge. Each symbol represents an individual animal. The log difference in protection between WT prfA and PrfA*(G155S) is statistically significant.
A relevant measure of vaccine-induced T-cell potency is protective immunity against challenge with a live pathogen. We evaluated T-cell potency in mice immunized with KBMA PrfA*(G155S) or WT prfA by challenge with WT L. monocytogenes or vaccinia virus. Strains of L. monocytogenes that fail to escape from the phagolysosome fail to induce protective immunity, although Ag-specific T-cell responses that can be expanded upon secondary challenge are elicited (4, 18). We previously described that KBMA L. monocytogenes_-based vaccination results in transient protection against a lethal WT L. monocytogenes challenge. The induced T-cell response wanes over time, reminiscent of a T-cell response induced in the absence of CD4+ T-cell help (42; K. S. Bahjat, unpublished data). Immunization of mice with the KBMA L. monocytogenes Δ_actA Δ_inlB_ Δ_uvrAB_ strain resulted in a 2-log protection at 14 days. To evaluate the potency of the KBMA PrfA*(G155S) vaccine-induced adaptive response, mice were immunized once with 1 × 108 particles and challenged with a 2× LD50 with WT L. monocytogenes 14 days later. CFU counts in spleen and liver were determined (data not shown) 3 days later. As shown in Fig. 5B, protection from WT L. monocytogenes was improved by 3 logs in mice immunized with KBMA PrfA*(G155S) compared to levels for mice immunized with the KBMA strain with WT prfA.
We then determined whether the enhanced immunologic potency of KBMA PrfA*(G155S) also extended to vaccinia virus challenge. C57BL/6 mice were immunized twice by an i.m. route 2 weeks apart with KBMA PrfA*(G155S) or WT prfA vaccines encoding the Quadvac Ag. To evaluate protective memory immunity, mice were challenged with 1 × 107 PFU of vaccinia virus 30 days following the last immunization and viral titers in the ovaries of the mice were determined 5 days later. Consistent with the improved magnitude of the induced T-cell response to the various vaccinia virus epitopes, we observed a statistically significantly improved protection by 2 logs against viral challenge in mice that received the PrfA*(G155S) mutant on the background of the KBMA L. monocytogenes Δ_actA_ Δ_inlB_ strain (Fig. 5C).
These results demonstrate that PrfA*(G155S) confers increased immunologic potency to live-attenuated and KBMA L. monocytogenes vaccines, notably providing the ability of KBMA vaccines to elicit protective immunity in rigorous infectious-disease challenge models following a conventional immunization route.
DISCUSSION
Recombinant vectors derived from viruses or bacteria represent an attractive but as-yet-unproven approach to stimulating functional immune responses in human prophylactic or therapeutic disease settings. Despite the challenges that face the multiple technologies under development, efforts in the field of recombinant vector platforms continue to be substantial due to their potential to address significant unmet medical needs. Recent high-profile failures of vaccine clinical trials, such as reports detailing the failure of an adenovirus serotype 5-based preventative vaccine in advanced testing to protect against human immunodeficiency virus infection (this vaccine may have even augmented the infection), underline the acute need for effective new vaccine platforms (12, 13, 33). Vaccine platforms based on live-attenuated L. monocytogenes are being developed and evaluated clinically due to an inherent property of stimulating potent innate immunity and acquired cellular immunity (identifiers NCT00327652 and NCT00585845 [http://www.clinicaltrials.gov/]). In this investigation, we show that activating the PrfA regulon prior to vaccinating mice enhanced the levels of innate and cellular immunity induced by live-attenuated and KBMA L. monocytogenes vaccines, which correlated with improved protection against challenge with the cognate WT bacterial pathogen or vaccinia virus. These results form the basis of a rationale for including the prfA(G155S) allele in future _L. monocytogenes_-based vaccines advanced to the clinical setting.
The immune potencies for both live-attenuated and KBMA vaccines were enhanced by activation of the PrfA regulon prior to immunization. PrfA* significantly enhanced the immune potency of KBMA vaccines. We have shown previously that, although killed, KBMA vaccines still escape the phagolysosome, a necessary step toward inducing IFN-β and other activating signals in APCs required to elicit protective cellular immunity against challenge with WT L. monocytogenes (4, 8, 27). However, KBMA L. monocytogenes vaccines can elicit protective immunity after a single immunization only when administered in combination with surrogate help provided by anti-CD40 antibody or when a homologous prime and boost immunization regimen is used (4). These results demonstrate a reduced immune potency for KBMA vaccines compared to that for live-attenuated L. monocytogenes Δ_actA_ Δ_inlB_ vaccine strains, which, like WT L. monocytogenes, can elicit protective immunity after a single immunization. Live L. monocytogenes strains expanded 100-fold over 7 h in the cytoplasm of infected macrophages in vitro (Fig. 1E), resulting in full activation of PrfA and induced expression of _prfA_-dependent genes, including encoded Ags which were driven from the actA promoter. In contrast, KBMA vaccines are unable to propagate in cells of the immunized host, and under conditions of a nonreplicating vector, induction of the PrfA regulon and expression of an encoded Ag prior to vaccination significantly enhanced immune potency.
In a recently published study, the immunogenicity of WT L. monocytogenes strain 10403 was compared with that of a different WT L. monocytogenes strain, 43251, which contains an unknown activating prfA mutation (39). While the latter L. monocytogenes strain elicited enhanced LLO- and p60-specific immunity and increased protection against WT bacterial challenge, because this study utilized different WT L. monocytogenes strains, it is difficult to draw conclusions regarding underlying mechanisms and possible application to recombinant attenuated vaccine platforms that are appropriate for testing with humans. Furthermore, the impact of constitutive PrfA activation on the immunogenicity of expressed heterologous Ags in recombinant L. monocytogenes vaccine strains was not evaluated in this study.
The enhanced immune potency of KBMA PrfA*-based vaccines could be due to several independent mechanisms. Contributing factors may include increased efficiency of escape from the phagolysosome and increased Ag expression and secretion in the cytosol, ultimately resulting in a higher density of epitopes displayed on MHC class I molecules and more efficient priming of CD8+ T cells. While overexpression of PrfA-dependent virulence genes can increase cytotoxicity, resulting in decreased virulence of WT strains and decreased vaccine potency (9, 15, 43), this mechanism does not appear to have affected the relative immunogenicities of the vaccine strains used in this study, as shown by equivalent intracellular growth in J774 cells (Fig. 1E). Other possibilities may include enhanced migration of DCs to the lymph nodes of animals immunized with PrfA* vaccines, due to an improved proinflammatory cytokine milieu at the site of infection or increased InlA-mediated disruption of E-cadherin DC-DC adhesions (20). Although binding of InlA to mouse E-cadherin is diminished compared to binding to its human homolog (22, 46), increased levels of InlA from PrfA* strains may still enhance this process. On the other hand, it seems unlikely that the enhanced immune potency of KBMA PrfA* vaccines was due to either an increased host range or an enhanced infection of target cells. Notably, while important for oral infection, for the i.m. or i.v. routes used in this study, InlA does not play the same role in mediating infection of nonphagocytic cells. Furthermore, InlB-mediated infection of hepatocytes via the hepatocyte growth factor receptor is not relevant, since this virulence determinant was deleted from the vaccine strains used in this study. Supporting this notion are the combined observations that infections of cultured macrophages or DCs were indistinguishable between all of the vaccine strains tested (Fig. 1C and D) and that the virulence of L. monocytogenes Δ_actA_ Δ_inlB_ PrfA* strains was increased only approximately twofold over that of the L. monocytogenes Δ_actA_ Δ_inlB_ parent strain, which is attenuated by more than 3 logs compared to WT L. monocytogenes (9) (Table 1). We are currently investigating whether any of these mechanisms contribute to the increased potency of KBMA PrfA* vaccines.
While the results presented here demonstrate the importance of activation of the PrfA regulon for increasing the potency of L. monocytogenes vaccines, increased Ag expression in cultured APCs infected with PrfA* vaccine strains was not observed. These results were somewhat surprising since previous studies showed increased actin polymerization in PtK2 cells infected with PrfA* mutants on a WT L. monocytogenes background, potentially due to increased or earlier expression of ActA, although ActA protein levels were not directly measured (38). Nevertheless, prfA(G155S), prfA(G145S), and prfA(Y63C) mutations all conferred high (and equivalent) levels of overexpression of PrfA-dependent Ags in vaccine strains grown in broth culture, but only PrfA*(G155S) vaccine strains had significantly increased immunologic potency. Interestingly, the live-attenuated PrfA*(G155S) and PrfA(G145S) [but not the PrfA*(Y63C)] vaccine strains elicited higher-magnitude T-cell responses to p60, a non-PrfA-dependent L. monocytogenes Ag, indicating that improved immunogenicity may not only be a consequence of improved Ag expression levels (data not shown). Thus, it is not surprising that enhanced PrfA-dependent expression did not necessarily correlate with optimal immunogenicity, since host-pathogen interactions are by definition a complex multifactorial process. Temporal regulation of PrfA-dependent genes provides expression of particular bacterial proteins in appropriate cellular compartments to facilitate pathogenesis. For example, ActA expression is induced 200-fold in the cytoplasm to promote host cell actin polymerization and cell-to-cell spread (32, 37). The Ags in this study were expressed as an N-terminal fusion with the first 100 amino acids of ActA and driven from a native actA promoter. In the case of KBMA vaccines, prfA(G155S) provided the appropriate level of PrfA-dependent induction to augment potency but afforded a sufficient balance of metabolic economy for the photochemically inactivated bacterium. In a recent study utilizing prfA(L104F) to characterize the PrfA-dependent L. monocytogenes secretome (31), several proteins whose expression was not known previously to be related to activated PrfA were identified. These data provide evidence for the multiple bacterial proteins involved in the pathogenesis of WT L. monocytogenes and illustrate that PrfA* mutants may have a complex impact on the potency of L. monocytogenes vaccines.
The overwhelming majority of preclinical studies with L. monocytogenes vaccines have utilized either i.v. or intraperitoneal administration. There have been few reports examining i.m. and subcutaneous immunization routes (9, 14). However, oral immunization has been explored in studies with mice and nonhuman primates and a single study with humans (2, 7, 26, 30). The three clinical trials with L. monocytogenes conducted to date or ongoing have utilized i.v. administration. While KBMA L. monocytogenes vaccines may have an improved risk-to-benefit profile compared to live-attenuated vaccines, a traditional immunization route of administration may be necessary for broad adoption and/or approval, particularly in prophylactic settings. Here, we show that with a prime-boost immunization regimen following i.m. immunization, KBMA PrfA*(G155S) vaccines elicited functional cellular immunity that was comparable to that elicited by live-attenuated L. monocytogenes vaccines.
By evaluating the immune potency of a panel of isogenic live-attenuated and KBMA L. monocytogenes vaccine strains varying only in prfA, we have shown that constitutive induction of the PrfA regulon prior to immunization enhances the ability of live-attenuated KBMA vaccines to elicit functional cellular immunity, using a conventional immunization route. We are presently evaluating this platform with various infectious-disease applications, advancing toward eventual testing of KBMA PrfA*(G155S) vaccines in the clinical setting.
Acknowledgments
We thank David Cook and Dan Portnoy for their critical review of the manuscript. We also thank Ellyn Shocron for technical assistance, along with Gary Bolton and Steve Killian for assistance with animal procedures.
All authors except for N. E. Freitag are employees of Anza Therapeutics, which owns intellectual property covering the compositions and methods described in the manuscript. Anza Therapeutics employees hold stock and/or stock options in the company. N. E. Freitag has no known financial interest in Anza Therapeutics.
Footnotes
▿
Published ahead of print on 9 June 2008.
REFERENCES
- 1.Alexander-Miller, M. A., G. R. Leggatt, and J. A. Berzofsky. 1996. Selective expansion of high- or low-avidity cytotoxic T lymphocytes and efficacy for adoptive immunotherapy. Proc. Natl. Acad. Sci. USA 934102-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angelakopoulos, H., K. Loock, D. Sisul, E. Jensen, J. F. Miller, and E. L. Hohmann. 2002. Safety and shedding of an attenuated strain of Listeria monocytogenes with a deletion of actA/plcB in adult volunteers: a dose escalation study of oral inoculation. Infect. Immun. 703592-3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reference deleted.
- 4.Bahjat, K. S., W. Liu, E. E. Lemmens, S. P. Schoenberger, D. A. Portnoy, T. W. Dubensky, Jr., and D. G. Brockstedt. 2006. Cytosolic entry controls CD8+-T-cell potency during bacterial infection. Infect. Immun. 746387-6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bahjat, K. S., R. A. Prell, H. E. Allen, W. Liu, E. E. Lemmens, M. L. Leong, D. A. Portnoy, T. W. Dubensky, Jr., D. G. Brockstedt, and M. A. Giedlin. 2007. Activation of immature hepatic NK cells as immunotherapy for liver metastatic disease. J. Immunol. 1797376-7384. [DOI] [PubMed] [Google Scholar]
- 6.Bouwer, H. G., H. Shen, X. Fan, J. G. Miller, R. A. Barry, and D. J. Hinrichs. 1999. Existing antilisterial immunity does not inhibit the development of a _Listeria monocytogenes_-specific primary cytotoxic T-lymphocyte response. Infect. Immun. 67253-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyer, J. D., P. C. Maciag, R. Parkinson, L. Wu, M. G. Lewis, D. B. Weiner, and Y. Paterson. 2006. Rhesus macaques with high levels of vaccine induced IFN-gamma producing cells better control viral set-point following challenge with SIV239. Vaccine 244498-4502. [DOI] [PubMed] [Google Scholar]
- 8.Brockstedt, D. G., K. S. Bahjat, M. A. Giedlin, W. Liu, M. Leong, W. Luckett, Y. Gao, P. Schnupf, D. Kapadia, G. Castro, J. Y. Lim, A. Sampson-Johannes, A. A. Herskovits, A. Stassinopoulos, H. G. Bouwer, J. E. Hearst, D. A. Portnoy, D. N. Cook, and T. W. Dubensky, Jr. 2005. Killed but metabolically active microbes: a new vaccine paradigm for eliciting effector T-cell responses and protective immunity. Nat. Med. 11853-860. [DOI] [PubMed] [Google Scholar]
- 9.Brockstedt, D. G., M. A. Giedlin, M. L. Leong, K. S. Bahjat, Y. Gao, W. Luckett, W. Liu, D. N. Cook, D. A. Portnoy, and T. W. Dubensky, Jr. 2004. Listeria-based cancer vaccines that segregate immunogenicity from toxicity. Proc. Natl. Acad. Sci. USA 10113832-13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruhn, K. W., N. Craft, and J. F. Miller. 2007. Listeria as a vaccine vector. Microbes Infect. 91226-1235. [DOI] [PubMed] [Google Scholar]
- 11.Camilli, A., L. G. Tilney, and D. A. Portnoy. 1993. Dual roles of plcA in Listeria monocytogenes pathogenesis. Mol. Microbiol. 8143-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen, J. 2007. AIDS research. Did Merck's failed HIV vaccine cause harm? Science 3181048-1049. [DOI] [PubMed] [Google Scholar]
- 13.Cohen, J. 2007. AIDS research. Promising AIDS vaccine's failure leaves field reeling. Science 31828-29. [DOI] [PubMed] [Google Scholar]
- 14.Datta, S. K., S. Okamoto, T. Hayashi, S. S. Shin, I. Mihajlov, A. Fermin, D. G. Guiney, J. Fierer, and E. Raz. 2006. Vaccination with irradiated listeria induces protective T cell immunity. Immunity 25143-152. [DOI] [PubMed] [Google Scholar]
- 15.Glomski, I. J., A. L. Decatur, and D. A. Portnoy. 2003. Listeria monocytogenes mutants that fail to compartmentalize listerolysin O activity are cytotoxic, avirulent, and unable to evade host extracellular defenses. Infect. Immun. 716754-6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray, M. J., N. E. Freitag, and K. J. Boor. 2006. How the bacterial pathogen Listeria monocytogenes mediates the switch from environmental Dr. Jekyll to pathogenic Mr. Hyde. Infect. Immun. 742505-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gunn, G. R., A. Zubair, C. Peters, Z.-K. Pan, T.-C. Wu, and Y. Paterson. 2001. Two Listeria monocytogenes vaccine vectors that express different molecular forms of human papilloma virus-16 (HPV-16) E7 induce qualitatively different T cell immunity that correlates with their ability to induce regression of established tumors immortalized by HPV-16. J. Immunol. 1676471-6479. [DOI] [PubMed] [Google Scholar]
- 18.Hamilton, S. E., V. P. Badovinac, A. Khanolkar, and J. T. Harty. 2006. Listeriolysin O-deficient Listeria monocytogenes as a vaccine delivery vehicle: antigen-specific CD8 T cell priming and protective immunity. J. Immunol. 1774012-4020. [DOI] [PubMed] [Google Scholar]
- 19.Harty, J. T., A. R. Tvinnereim, and D. W. White. 2000. CD8+ T cell effector mechanisms in resistance to infection. Annu. Rev. Immunol. 18275-308. [DOI] [PubMed] [Google Scholar]
- 20.Jiang, A., O. Bloom, S. Ono, W. Cui, J. Unternaehrer, S. Jiang, J. A. Whitney, J. Connolly, J. Banchereau, and I. Mellman. 2007. Disruption of E-cadherin-mediated adhesion induces a functionally distinct pathway of dendritic cell maturation. Immunity 27610-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lauer, P., M. Y. Chow, M. J. Loessner, D. A. Portnoy, and R. Calendar. 2002. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 1844177-4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lecuit, M., S. Dramsi, C. Gottardi, M. Fedor-Chaiken, B. Gumbiner, and P. Cossart. 1999. A single amino acid in E-cadherin responsible for host specificity towards the human pathogen Listeria monocytogenes. EMBO J. 183956-3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lorber, B. 1997. Listeriosis. Clin. Infect. Dis. 241-9. [DOI] [PubMed] [Google Scholar]
- 24.Moutaftsi, M., B. Peters, V. Pasquetto, D. C. Tscharke, J. Sidney, H. H. Bui, H. Grey, and A. Sette. 2006. A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nat. Biotechnol. 24817-819. [DOI] [PubMed] [Google Scholar]
- 25.Mueller, K. J., and N. E. Freitag. 2005. Pleiotropic enhancement of bacterial pathogenesis resulting from the constitutive activation of the Listeria monocytogenes regulatory factor PrfA. Infect. Immun. 731917-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neeson, P., J. Boyer, S. Kumar, M. G. Lewis, L. Mattias, R. Veazey, D. Weiner, and Y. Paterson. 2006. A DNA prime-oral Listeria boost vaccine in rhesus macaques induces a SIV-specific CD8 T cell mucosal response characterized by high levels of alpha4beta7 integrin and an effector memory phenotype. Virology 354299-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Riordan, M., C. H. Yi, R. Gonzales, K. D. Lee, and D. A. Portnoy. 2002. Innate recognition of bacteria by a macrophage cytosolic surveillance pathway. Proc. Natl. Acad. Sci. USA 9913861-13866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pamer, E. G. 2004. Immune responses to Listeria monocytogenes. Nat. Rev. Immunol. 4812-823. [DOI] [PubMed] [Google Scholar]
- 29.Paterson, Y., and P. C. Maciag. 2005. Listeria-based vaccines for cancer treatment. Curr. Opin. Mol. Ther. 7454-460. [PubMed] [Google Scholar]
- 30.Peters, C., X. Peng, D. Douven, Z. K. Pan, and Y. Paterson. 2003. The induction of HIV Gag-specific CD8+ T cells in the spleen and gut-associated lymphoid tissue by parenteral or mucosal immunization with recombinant Listeria monocytogenes HIV Gag. J. Immunol. 1705176-5187. [DOI] [PubMed] [Google Scholar]
- 31.Port, G. C., and N. E. Freitag. 2007. Identification of novel Listeria monocytogenes secreted virulence factors following mutational activation of the central virulence regulator, PrfA. Infect. Immun. 755886-5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Portnoy, D. A., V. Auerbuch, and I. J. Glomski. 2002. The cell biology of Listeria monocytogenes infection: the intersection of bacterial pathogenesis and cell-mediated immunity. J. Cell Biol. 158409-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rappuoli, R. 2007. Bridging the knowledge gaps in vaccine design. Nat. Biotechnol. 251361-1366. [DOI] [PubMed] [Google Scholar]
- 34.Ripio, M. T., G. Dominguez-Bernal, M. Lara, M. Suarez, and J. A. Vazquez-Boland. 1997. A Gly145Ser substitution in the transcriptional activator PrfA causes constitutive overexpression of virulence factors in Listeria monocytogenes. J. Bacteriol. 1791533-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scortti, M., H. J. Monzo, L. Lacharme-Lora, D. A. Lewis, and J. A. Vazquez-Boland. 2007. The PrfA virulence regulon. Microbes Infect. 91196-1207. [DOI] [PubMed] [Google Scholar]
- 36.Shen, H., M. K. Slifka, M. Matloubian, E. R. Jensen, R. Ahmed, and J. F. Miller. 1995. Recombinant Listeria monocytogenes as a live vaccine vehicle for the induction of protective anti-viral cell-mediated immunity. Proc. Natl. Acad. Sci. USA 923987-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shetron-Rama, L. M., H. Marquis, H. G. Bouwer, and N. E. Freitag. 2002. Intracellular induction of Listeria monocytogenes actA expression. Infect. Immun. 701087-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shetron-Rama, L. M., K. Mueller, J. M. Bravo, H. G. Bouwer, S. S. Way, and N. E. Freitag. 2003. Isolation of Listeria monocytogenes mutants with high-level in vitro expression of host cytosol-induced gene products. Mol. Microbiol. 481537-1551. [DOI] [PubMed] [Google Scholar]
- 39.Smithey, M. J., S. Brandt, N. E. Freitag, D. E. Higgins, and H. G. Bouwer. 2008. Stimulation of enhanced CD8 T cell responses following immunization with a hyper-antigen secreting intracytosolic bacterial pathogen. J. Immunol. 1803406-3416. [DOI] [PubMed] [Google Scholar]
- 40.Starks, H., K. W. Bruhn, H. Shen, R. A. Barry, T. W. Dubensky, D. Brockstedt, D. J. Hinrichs, D. E. Higgins, J. F. Miller, M. Giedlin, and H. G. Bouwer. 2004. Listeria monocytogenes as a vaccine vector: virulence attenuation or existing antivector immunity does not diminish therapeutic efficacy. J. Immunol. 173420-427. [DOI] [PubMed] [Google Scholar]
- 41.Stevens, R., A. Lavoy, S. Nordone, M. Burkhard, and G. A. Dean. 2005. Pre-existing immunity to pathogenic Listeria monocytogenes does not prevent induction of immune responses to feline immunodeficiency virus by a novel recombinant Listeria monocytogenes vaccine. Vaccine 231479-1490. [DOI] [PubMed] [Google Scholar]
- 42.Tvinnereim, A. R., S. E. Hamilton, and J. T. Harty. 2002. CD8+-T-cell response to secreted and nonsecreted antigens delivered by recombinant Listeria monocytogenes during secondary infection. Infect. Immun. 70153-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vázquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Domínguez-Bernal, W. Goebel, B. González-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vega, Y., M. Rauch, M. J. Banfield, S. Ermolaeva, M. Scortti, W. Goebel, and J. A. Vazquez-Boland. 2004. New Listeria monocytogenes prfA* mutants, transcriptional properties of PrfA* proteins and structure-function of the virulence regulator PrfA. Mol. Microbiol. 521553-1565. [DOI] [PubMed] [Google Scholar]
- 45.Villalobos, A., J. E. Ness, C. Gustafsson, J. Minshull, and S. Govindarajan. 2006. Gene Designer: a synthetic biology tool for constructing artificial DNA segments. BMC Bioinformatics 7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wollert, T., B. Pasche, M. Rochon, S. Deppenmeier, J. van den Heuvel, A. D. Gruber, D. W. Heinz, A. Lengeling, and W. D. Schubert. 2007. Extending the host range of Listeria monocytogenes by rational protein design. Cell 129891-902. [DOI] [PubMed] [Google Scholar]
- 47.Wong, K. K., and N. E. Freitag. 2004. A novel mutation within the central Listeria monocytogenes regulator PrfA that results in constitutive expression of virulence gene products. J. Bacteriol. 1866265-6276. [DOI] [PMC free article] [PubMed] [Google Scholar]