The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors (original) (raw)
Abstract
Almost 2000 drought-responsive genes were identified in Arabidopsis thaliana under progressive soil drought stress using whole-genome oligonucleotide microarrays. Most of the drought-regulated genes recovered to normal expression levels by 3 h after rewatering. It has previously been shown that the abscisic acid (ABA) analogue (+)-8′-acetylene-ABA (PBI425) hyperinduces many ABA-like changes in gene expression to reveal a more complete list of ABA-regulated genes, and it is demonstrated here that PBI425 produced a correspondingly increased drought tolerance. About two-thirds of drought-responsive genes (1310 out of 1969) were regulated by ABA and/or the ABA analogue PBI425. Analysis of promoter motifs suggests that many of the remaining drought-responsive genes may be affected by ABA signalling. Concentrations of endogenous ABA and its catabolites significantly increased under drought stress and either completely (ABA) or partially (ABA catabolites) recovered to normal levels by 3 h after rehydration. Detailed analyses of drought transcript profiles and in silico comparisons with other studies revealed that the ABA-dependent pathways are predominant in the drought stress responses. These comparisons also showed that other plant hormones including jasmonic acid, auxin, cytokinin, ethylene, brassinosteroids, and gibberellins also affected drought-related gene expression, of which the most significant was jasmonic acid. There is also extensive cross-talk between responses to drought and other environmental factors including light and biotic stresses. These analyses demonstrate that ABA-related stress responses are modulated by other environmental and developmental factors.
Keywords: ABA analogue, ABA metabolite profiling, abscisic acid, drought, gene expression, microarray, rehydration
Introduction
Drought is a major factor limiting crop productivity in North America and worldwide (Boyer, 1982; Bartels and Nelson, 1994). To improve crop productivity, it is necessary to understand the mechanism of plant responses to drought conditions with the ultimate goal of improving crop performance in the vast areas of the world where rainfall is limiting or unreliable.
Recently many efforts have been focused on the molecular response of plants to water deficit stress using the model plant Arabidopsis thaliana (Ingram and Bartels, 1996; Shinozaki and Yamaguchi-Shinozaki, 1997). Many genes respond to drought at the transcriptional level, and their products are thought to function in drought tolerance and response (Bohnert et al., 1995; Ingram and Bartels, 1996; Bray, 1997; Shinozaki et al., 1999; Shinozaki and Yamaguchi-Shinozaki, 2000). Some stress-inducible genes have been used to improve the stress tolerance of plants by gene transfer (Umezawa et al., 2006). Although hundreds of genes have been found to be involved in abiotic stress responses and a number of them have been well characterized (Shinozaki et al., 1999; Shinozaki and Yamaguchi-Shinozaki, 2000, 2007; Zhu, 2002), the functions of the majority of the genes remain unknown and there are probably more genes yet to be discovered.
In recent years, microarray technology has been applied to the identification of stress-responsive genes in Arabidopsis (Seki et al., 2001, 2002; Chen et al., 2002; Kreps et al., 2002; Kawaguchi et al., 2004; Swindell, 2006; Ma and Bohnert, 2007). However, since the microarrays used in these studies only represented parts of the Arabidopsis genome, there are likely to be many drought-responsive genes not included. Comparison of the lists of drought-inducible genes from various studies revealed that only 27 genes were commonly induced in these studies (Bray, 2004). This striking lack of commonality is probably due to the fact that different sets of genes were probed in the various microarray platforms utilized and varying conditions of plant growth and stress treatments were employed.
The phytohormone (+)-abscisic acid (ABA) plays a key role in plant adaptation to adverse environmental conditions including drought stress. Numerous studies have shown that ABA accumulation is a key factor in controlling downstream responses essential for adaptation to stress. However, molecular and genomic analyses have suggested that both ABA-dependent and ABA-independent regulatory systems are involved in stress-responsive gene expression (Shinozaki and Yamaguchi-Shinozaki, 1997, 2000; Bray, 1997; Riera et al., 2005). In a previous study, it was established that at least 14% of Arabidopsis genes were ABA regulated (Huang et al., 2007). In that study, an ABA analogue, 8′-acetylene ABA (PBI425), was used to reveal and confirm weakly ABA-regulated genes based on its ability to hyperinduce ABA-related gene expression. Therefore, PBI425 is a useful tool for revealing the full extent of ABA-related gene expression in plants.
In order to identify a full spectrum of drought-responsive genes and to gain more understanding of drought stress responses in relation to other developmental and environmental signalling systems, a genome-wide investigation of drought-responsive genes was performed in Arabidopsis using oligonucleotide microarrays. Large numbers of drought-regulated genes including many novel genes were identified. The relationships between drought, rehydration, plant hormones, and other environmental factors were investigated by microarray analysis, in silico comparisons, and ABA metabolite profiling.
Materials and methods
Plant growth and treatments
Wild-type Arabidopsis plants, ecotype Columbia, were germinated and grown in a mixture of sand and soil (2:1) in a growth chamber at 22 °C with a 16 h light/8 h dark cycle with a light intensity of 150 mmol m−2 s−1. Plants were watered every 3 d with 0.5× Hoagland solution, ensuring that the soil remained moist. Watering was stopped from 20 d after germination until the soil was dry, with relative water content ∼5% (measured in a separate experiment), which typically took 5 d. After this dehydration treatment, some plants were rewatered. At 3 h after rewatering, the aerial tissues of control (no dehydration treatment), drought, and rewatered plants were collected and frozen in liquid nitrogen for RNA extraction or hormone metabolite profiling. Two biological replicates from plants grown under identical conditions at different times were prepared for drought versus control and for rehydration versus drought. Each biological replicate was hybridized twice with dyes reversed (technical replicates). Three biological replicates were prepared for ABA metabolite profiling. Each biological replicate contained material pooled from 24 plants.
Treatment of plants with (+)-ABA and PBI425 (chemical synthesis of this compound is described in Rose et al., 1997) is described in detail in Huang et al. (2007). Briefly, plants were treated with 20 μM of the appropriate compound by imbibition and all above-ground plant parts were harvested at 3, 6, 24, and 48 h after application.
Microarray analysis
Protocols for total RNA extraction, cDNA synthesis, dye labelling, microarray hybridization, and scanning, as well as data acquisition and analyses were described in Huang et al. (2007). Data were normalized using RobustSplines in Bioconductor, and GeneSpring software was used for data visualization, analysis of promoter motifs, and hierarchical clustering. Spotted glass microarray slides were obtained from the University of Arizona (http://ag.arizona.edu/microarray/) and are based on 70mer Arabidopsis probes produced by Qiagen. Similar arrays were also obtained from the University of Alberta Microarray and Proteomics Facility (http://www.biology.ualberta.ca/facilities/microarray/).
Quantitative real-time PCR analyses
To validate the expression profiles obtained from microarray hybridizations, the relative expression of 15 selected genes in response to drought and rehydration treatments was measured using quantitative real-time PCR. Quantitative real-time PCR and data normalization and quantification were performed as described in Huang et al. (2007). The 15 genes and their primers are listed in Supplementary Table 1 available at JXB online.
Quantification of ABA, ABA metabolites, and PBI425 by LC-MS-MS
Extraction and quantification of ABA, ABA metabolites, and PBI425 were described in Huang et al. (2007) and Feurtado et al. (2004). Synthesis and use of the deuterium-labelled internal standards were described by Abrams et al. (2003), Chiwocha et al. (2003), and Zaharia et al. (2005).
Results
Drought responsive genes
Genome-wide detection of drought-responsive genes was performed by comparing the transcriptome in Arabidopsis aerial tissues under progressive soil drought stress with that of control plants growing under normal conditions using oligonucleotide microarrays containing a total of 26 090 elements. Using the statistical method of significance analysis of microarray (SAM; Tusher et al., 2001) with a false discovery rate (FDR) <0.05, a total of 1969 (∼7.5% of the Arabidopsis genome) differentially expressed genes were identified, of which 923 were up-regulated and 728 down-regulated (Supplementary Table 2 at JXB online).
Responses of drought-regulated genes to rewatering
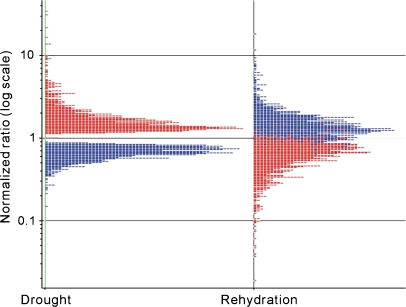
To investigate how the expression of drought-regulated genes is altered during recovery from drought, the transcripts of Arabidopsis aerial tissues during rehydration and under drought stress were compared. The results showed predominantly opposite regulatory effects of rehydration on drought-regulated genes (Fig. 1) which indicated that most drought-regulated genes have recovered to the normal expression level by 3 h after rewatering. This is consistent with the fact that rehydration produces a reversal of the drought stress responses. The opposite regulatory effects of rehydration on drought-regulated genes provided a large-scale validation of the identity of drought-regulated genes.
Fig. 1.
Responses of 1969 drought-regulated genes to drought (left panel) and rehydration (right panel). Each spot corresponds to an individual gene, and the number of genes with a given ratio is plotted on the _x_-axis. The colour of each spot is based on the relative expression in drought versus control (left panel); red, up-regulated; blue, down-regulated.
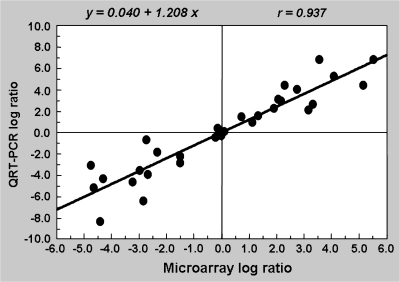
Validation of microarray data by quantitative real-time PCR (QRT-PCR)
To confirm further the reliability of microarray results, the expression of 15 genes from the drought and rehydration treatments were compared using both microarray data and QRT-PCR. The log ratios calculated from both approaches were highly correlated (r = 0.937) and were described by a simple linear regression equation as y = 0.040 + 1.208_x_ (Fig. 2), thereby indicating good consistency between the two methods. The regression coefficient of 1.208 suggested that the QRT-PCR ratio is slightly higher than that calculated from microarray results.
Fig. 2.
Scatter diagram of log ratios of 15 selected genes in drought and rehydration treatments calculated from quantative RT-PCR and microarray data. The regression equation and correlation coefficient (r) between log ratios are presented.
Enriched gene ontology (GO) terms
To understand the functional significance of drought-induced gene expression, the web-based tool Athena (O'Connor et al., 2005; http://www.bioinformatics2.wsu.edu/cgi-bin/Athena/cgi/analysis_select.pl) was used to find statistically over-represented functional terms (Table 1). A number of GO terms involved in stress and ABA responsiveness were highly enriched in the drought-up-regulated gene list. GO terms such as responses to stress, ABA, desiccation, and water deprivation were ranked highest in drought-up-regulated genes. Other stress-related GO terms such as salinity response, response to cold, response to oxidative stress, and reactive oxygen species (ROS) were also over-represented in the drought-up-regulated genes. The presence of genes functionally associated with other abiotic stresses among the drought-up-regulated genes suggested that different stresses share some common signalling pathways. GO attributes such as proline biosynthesis, proline metabolism, sucrose biosynthesis, and alkaloid metabolism were highly enriched in the drought-up-regulated genes, suggesting that those metabolic pathways are important in responses to drought stress. Indeed the importance of many of these pathways to drought tolerance has been empirically supported by transgenic experiments (Umezawa et al., 2006). Terms such as aldehyde dehydrogenase, antioxidant activity, oxidoreductase activity, ubiquitin-conjugating enzyme activity, and ion homeostasis (copper homeostasis, transition metal ion homeostasis) were also over-represented in the drought-up-regulated gene list. Previous studies reported that enzymes with these functions play important roles under certain stresses (Zhu, 2002; Kirch et al., 2005).
Table 1.
Enriched GO terms in drought-regulated genes
| Up-regulated | Down-regulated | ||
|---|---|---|---|
| GO terms | _P_-value | GO terms | _P_-value |
| Response to stress | <10−9 | Structural constituent of ribosome | <10−10 |
| Response to abscisic acid stimulus | <10−9 | Intracellular | <10−10 |
| Response to desiccation | <10−7 | Ribosome | <10−10 |
| Response to water deprivation | <10−7 | Cytosolic small ribosomal subunit (sensu eukarya) | <10−10 |
| Cytoplasm | <10−5 | Protein biosynthesis | <10−10 |
| Protein amino acid dephosphorylation | <10−5 | Chloroplast | <10−10 |
| Proline biosynthesis | <10−5 | Large ribosomal subunit | <10−10 |
| Response to water | <10−5 | Ribosome biogenesis | <10−9 |
| Catalytic activity | <10−4 | Thylakoid lumen (sensu viridiplantae) | <10−9 |
| Aldehyde dehydrogenase activity | <10−4 | Structural constituent of cytoskeleton | <10−8 |
| Lipid transport | <10−4 | Nucleosome | <10−6 |
| Salinity response | <10−4 | Cytosolic large ribosomal subunit (sensu bacteria) | <10−6 |
| Alkaloid metabolism | <10−4 | Nucleosome assembly | <10−5 |
| 1-Pyrroline-5-carboxylate dehydrogenase activity | <10−3 | Protein folding | <10−5 |
| 4-Aminobutyrate transaminase activity | <10−3 | Chromosome organization and biogenesis (sensu eukarya) | <10−5 |
| NADPH-haemoprotein reductase activity | <10−3 | Chloroplast stroma | <10−5 |
| Adenosylmethionine-8-amino-7-oxononanoate transaminase activity | <10−3 | Chaperone activity | <10−4 |
| Aminopeptidase activity | <10−3 | Peptidyl-prolyl cis-trans isomerase activity | <10−4 |
| Lactoylglutathione lyase activity | <10−3 | FK506 binding | <10−4 |
| Ubiquitin-conjugating enzyme activity | <10−3 | Translational elongation | <10−4 |
| Cysteine protease inhibitor activity | <10−3 | Steroid biosynthesis | <10−4 |
| Lipid transporter activity | <10−3 | Porphyrin biosynthesis | <10−4 |
| Mitochondrial matrix | <10−3 | Thylakoid membrane (sensu viridiplantae) | <10−4 |
| Vacuolar membrane | <10−3 | Water channel activity | <10−4 |
| Cytosol | <10−3 | Small ribosomal subunit | < 10−4 |
| Sucrose biosynthesis | <10−3 | Phosphoric ester hydrolase activity | <10−4 |
| Protein modification | <10−3 | Tubulin | <10−4 |
| High affinity iron ion transport | <10−3 | Plastid large ribosomal subunit | <10−3 |
| Amino acid transport | <10−3 | Sterol 24-c-methyltransferase activity | <10−3 |
| Copper ion homeostasis | <10−3 | 3-β-Hydroxy-Δ5-steroid dehydrogenase activity | <10−3 |
| Protein serine/threonine phosphatase complex | <10−3 | Uroporphyrinogen decarboxylase activity | <10−3 |
| Lipid binding | <10−3 | Extracellular | <10−3 |
| Response to cold | <10−3 | Glycolysis | <10−3 |
| Oxidoreductase activity | <10−3 | Cytoskeleton organization and biogenesis | <10−3 |
| Oxidoreductase activity, acting on NADH or NADPH, disulphide as acceptor | <10−3 | Dihydrodipicolinate reductase activity | <10−3 |
| Oxidoreductase activity, acting on sulphur group of donors, disulphide as acceptor | <10−3 | Lysine biosynthesis via diaminopimelate | <10−3 |
| ATPase activity | <10−3 | Nucleotide metabolism | <10−3 |
| Δ1-Pyrroline-5-carboxylate synthetase activity | <10−3 | Response to auxin stimulus | <10−3 |
| Removal of superoxide radicals | <10−3 | Carboxylic ester hydrolase activity | <10−3 |
| Response to reactive oxygen species | <10−3 | Fructose-bisphosphatase activity | <10−3 |
A number of depleted GO terms were also associated with drought-up-regulated genes. For example, ribosome, non-membrane-bound organelle, protein biosynthesis, kinase activity, and nucleotide binding were significantly under-represented. Similarly, among drought-down-regulated genes there was enrichment of terms related to structural molecule activity, ribosome, biosynthesis, metabolism, water channel activity, photosynthesis, and porphyrin metabolism. These results are consistent with a previous report that water stress inhibits photosynthesis and plant growth (Tezara et al., 1999).
_Cis_-elements associated with drought-responsive genes
_Cis_-acting regulatory elements are key determinants of transcriptional regulation, and the identification of a comprehensive set of drought-regulated genes in this study provided an opportunity to search for sequences common to their promoter regions. In previous studies, the dehydration-responsive element (DRE) with the core sequence A/GCCGAC was identified as a _cis_-acting promoter element in regulating gene expression in response to drought, high salinity, and cold stresses in Arabidopsis (Yamaguchi-Shinozaki and Shinozaki, 1994). Similar motifs were identified as CRT (C-repeat) and LTRE (low-temperature-responsive element) in cold-inducible genes (Baker et al., 1994; Jiang et al., 1996). Promoters of many drought-, cold stress-, and salinity-inducible genes contain the ABA-responsive element or ABRE (PyACGTGGC). These ABRE motifs are known target-binding sites for the basic leucine zipper (bZIP) transcription factors (Shinozaki and Yamaguchi-Shinozaki, 2000). The ACTCAT _cis_-acting element was identified as being involved in rehydration-, proline-, and low osmolarity-inducible gene expression (Satoh et al., 2002).
These known promoter motifs were searched for in 1000 bp regions upstream of the 5′ termini of drought- and rehydration-regulated genes (using Genespring). The ABA-responsive _cis_-acting regulatory elements ABRE and ABF-binding site motifs were highly over-represented among drought-regulated genes (Table 2), and DRE and LTRE promoter motifs were also enriched.
Table 2.
Enriched promoter motifs in drought-regulated genes
| Up-regulated genes | Down-regulated genes | ||
|---|---|---|---|
| Enriched promoter motif | _P_-value | Enriched promoter motif | _P_-value |
| ABFs-binding site | <10−10 | HY5AT | <10−3 |
| ABRE-binding site | <10−10 | Ibox promoter motif | <10−5 |
| ABRE-like binding | <10−10 | ||
| ABREATRD22 | <10−10 | ||
| ACGTABREMOTIFA2OSE | <10−10 | ||
| CACGTGMOTIF | <10−10 | ||
| DRE core motif | <10−5 | ||
| DREB1A/CBF3 | <10−6 | ||
| Evening element promoter motif | <10−5 | ||
| GADOWNAT | <10−10 | ||
| GBF1/2/3 BS in ADH | <10−3 | ||
| GBOXLERBCS | <10−10 | ||
| LTRE promoter motif | <10−3 | ||
| TGA1-binding site | <10−4 | ||
| UPRMOTIFIAT | <10−4 | ||
| UPRMOTIFIIAT | <10−4 | ||
| Z-box promoter motif | <10−10 |
A search was carried out for the occurrence of all known _cis_-acting promoter elements (based on AGRIS and PLACE promoter database) in selected gene sets by analysis tools in Athena (http://wyrick.sbs.wsu.edu/cgi-bin/Athena/cgi/home.pl), and the results are summarized in Table 2. There were more enriched motifs found in drought-up-regulated gene sets than in promoters of drought-down-regulated genes. However, the light-responsive motifs Ibox and HY5AT were enriched in the promoters of drought-down-regulated genes. The consensus sequence of each motif is available at the AtcisDB (http://arabidopsis.med.ohio-state.edu/AtcisDB/bindingSiteContent.jsp) and PLACE (http://www.dna.affrc.go.jp/PLACE/) databases.
Genes commonly regulated by drought and ABA
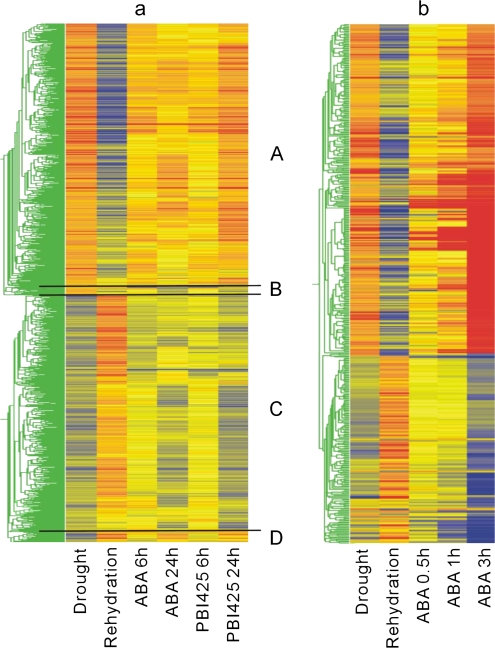
The above analyses suggested that many drought-regulated genes are ABA-responsive genes. In a previous study, ABA analogues—in particular PBI425—were used to reveal transient and weakly ABA-responsive genes (Huang et al., 2007). Comparing the drought- (Supplementary Table 2 at JXB online) and ABA-responsive gene lists (from Huang et al., 2007; Supplementary Tables 1–8), as expected, substantial overlap was found. Specifically, 1310 out of 1969 drought-regulated genes (67% of drought-regulated genes) (Supplementary Table 3 at JXB online) were significantly regulated by ABA and/or ABA analogues. Of these genes, 1306 were regulated by (+)-ABA and/or PBI425. Hierarchical clustering analysis on these overlapping genes revealed that most ABA- or drought- responsive genes are regulated by drought and ABA in the same direction—they are commonly up-regulated or down-regulated by ABA and drought (Fig. 3a). This analysis suggests that ABA plays the pivotal role in drought stress responses.
Fig. 3.
(a) Clustering of 1310 drought and ABA overlapping genes according to their expression after drought, rehydration, ABA, and PBI425 treatments. The colour scale indicates the degree of gene expression change. ABA and PBI425 data are from Huang et al. (2007). (b) Clustering of 496 drought and ABA overlapping genes. ABA data are from Nemhauser et al. (2006).
Many of the drought- and ABA-regulated genes have previously been shown to be induced or repressed by ABA treatment or by various stresses such as drought, cold, and high salinity. For example, well-known ABA- and stress-inducible genes KIN1, KIN2, RD29B, LTP3, LTP4, ERD10, RD20, COR413, RAB18, P5CS1, LEA14, VSP2, ABF3, ABI1, and ABI2 were commonly up-regulated by ABA and drought. A number of genes encoding proteins responsive to gibberellic acid (GA), ethylene or auxin were identified as drought-down-regulated genes, suggesting that these hormones may modulate drought stress signalling by acting antagonistically to ABA.
As noted above, most genes common to drought and ABA treatments were regulated in the same direction (Fig. 3a, groups A and C). However, some genes displayed anomalous expression patterns (Fig. 3a, groups B and D). Group B genes were up-regulated by drought and down-regulated by ABA, including a WRKY family transcription factor (At2g38470) and several zinc finger transcription factors including ZAT10 (At2g28200) as well as At3g55980 and ADOF1 (At1g51700). The functional significance of genes in this cluster requires further study.
Group D genes are up-regulated by ABA but repressed by drought. Several group D genes are related to other hormones such as GASA1 [At1g75750, responsive to ABA, GA, and brassinosteroid (BR)], an auxin-responsive gene (At3g15450), 1-aminocyclopropane-1-carboxylate oxidase (involved in ethylene biosynthesis, At1g05010), and several genes encoding cell wall-related proteins such as xyloglucan endotranglycosylases (At5g65730, At2g06850, At4g03210) and glycosyl hydrolase family 1 (At3g60130 and At1g49750). These genes appear to correspond to a subset of ABA-induced genes that are associated with cell expansion. Decreased expression of these genes under drought stress is probably associated with decreased cell expansion—possibly related to the reduced turgor pressure associated with stress.
‘ABA-independent’ drought-responsive genes
About a third (659 out of 1969) of drought-regulated genes were not significantly regulated by ABA or ABA analogues. It has been suggested that there are ABA-independent stress signalling pathways (Shinozaki and Yamaguchi-Shinozaki, 2000; Yamaguchi-Shinozaki and Shinozaki, 2005). To determine if the functional role of these genes was similar to that of ABA-regulated genes, GO annotations were analysed. As expected, many functional terms were found to be related to stresses (response to heat, cold, salt, oxidative, osmotic stress) and ABA, but terms related to other plant hormones were also found, including auxin, cytokinin, jasmonic acid (JA), salicylic acid, ethylene, and gibberellins (Supplementary Table 4 at JXB online). In addition, promoter motif analyses were performed on these ‘drought-specific’ genes, and it was found that ABA-responsive promoter motifs (e.g. ABRE-like, ACGTABREMOTIFA2OSEM, and GADOWNAT) were significantly over-represented in the promoters of these genes. This suggests that expression of many of these genes is at least partially linked to the ABA-dependent stress signal pathway. In addition, other plant hormones might also affect or modulate drought responses.
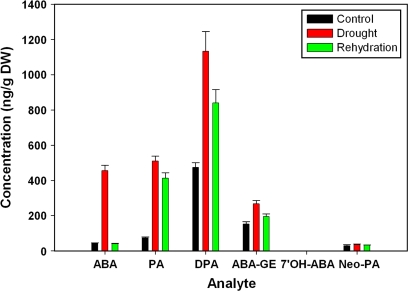
ABA metabolite profiling in response to drought and rehydration
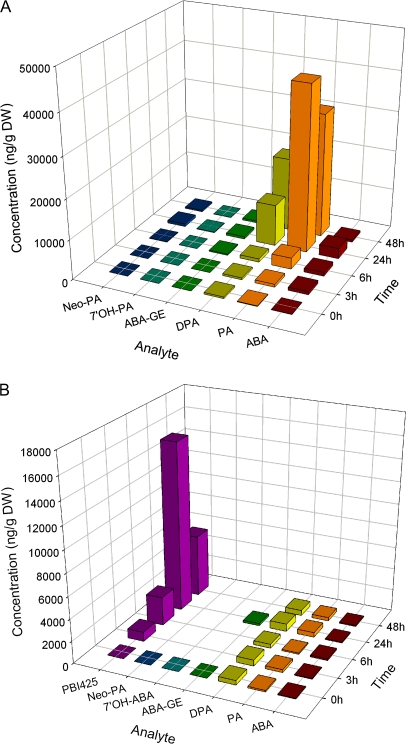
To understand better the relationship of ABA metabolism to stress responses, the contents of ABA and ABA metabolites were measured in plants under drought and rehydration conditions. The results (Fig. 4) showed first that the endogenous ABA concentration increased dramatically under drought stress and recovered to normal levels after rehydration, which is consistent with previous reports in excised leaf blades of Xanthium strumarium (Zeevaart, et al., 1980). Secondly, the ABA metabolites phaseic acid (PA), dihydrophaseic acid (DPA), and abscisic acid glucose ester (ABA-GE) significantly increased under the drought condition and partially recovered at 3 h after rewatering; neophaseic acid increased slightly after drought and rehydration treatment, and 7′-hydroxyABA (7′OH-ABA) was not detectable. These results indicated that drought enhances both ABA biosynthesis and catabolism, resulting in an increase in both ABA and ABA metabolites, while rehydration reversed the effect of drought on ABA biosynthesis. These results confirm the close coupling between drought and ABA metabolism.
Fig. 4.
Amounts of endogenous ABA and ABA metabolites in plants under control conditions, drought treatment, and after rewatering.
ABA metabolite profiling after treatments with ABA and PBI425
The ABA analogue PBI425 produces stronger effects on induction of ABA-related genes than ABA itself (Huang et al., 2007) and also exhibits ABA hyperactivity in germination assays (Cutler et al., 2000). The concentrations of ABA, PBI425, and ABA metabolites in treated plants were measured at intervals after ABA and PBI425 treatments. After ABA treatment, tissue ABA was maximal at 24 h and then declined (Fig. 5A). The ABA metabolite PA was also maximal at 24 h, whereas DPA increased substantially until 48 h, suggesting that applied ABA was rapidly metabolized to PA, which was then more slowly reduced to DPA. Comparing Figs 4 and 5A, it is apparent that the ratio of PA to ABA is much higher after exogenous ABA treatment than during drought or rehydration. This indicates that ABA catabolism via 8′-hydroxylation is greater in unstressed plants. Since the DPA to PA ratio is lower in Fig. 5A than in Fig. 4, it may also be deduced that the PA to DPA conversion is rate limiting in unstressed plants.
Fig. 5.
(a) Changes in ABA and ABA metabolite concentrations in ABA-treated plants. (b) Changes in ABA, ABA metabolites, and PBI425 concentrations in PBI425-treated plants.
After application of PBI425, the quantity detected in the plants increased substantially until 24 h and then declined. At all time points, the amount of PBI425 in plants was much higher than that of ABA (Fig. 5B), indicating slower turnover of PBI425, which is consistent with a previous report showing that PBI425 is resistant to 8′-hydroxylation (Cutler et al., 2000). The amount of PBI425 was also higher than ABA levels in drought-stressed plants (Figs 4 and 5B). The levels of ABA, and the ABA metabolites PA and DPA were slightly increased in PBI425-treated plants and a low level of ABA-GE was also detected at 24 h. This suggests that PBI425 slightly enhanced endogenous ABA metabolism.
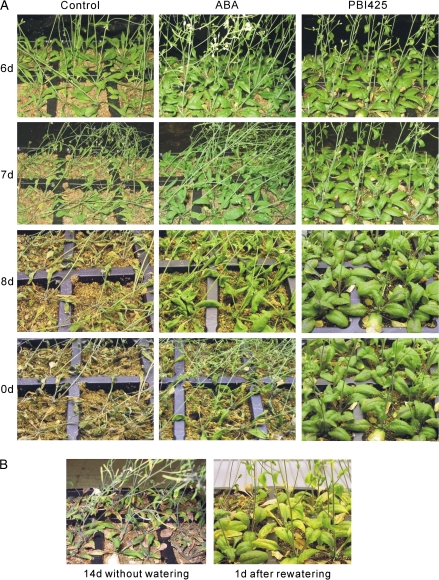
PBI425 further increased drought tolerance in Arabidopsis relative to ABA
As indicated in the Fig. 6A, drought tolerance was significantly increased in both ABA- and PBI425-treated Arabidopsis plants, although the latter was more effective. Control plants started to wilt after 6 d without watering, whereas ABA-treated plants started to wilt at day 8. At day 10, both control and ABA-treated plants were dead, while PBI425-treated plants remained healthy. By day 14, the appearance of purple leaf colour and some wilting indicated that PBI425-treated plants were stressed; however, more than half of these plants recovered after re-watering (Fig. 6B). Therefore, the ability of PBI425 to accumulate to relatively high levels in plants and to hyperinduce ABA-related genes (Huang et al., 2007) also results in substantially increased drought tolerance.
Fig. 6.
(a) Drought tolerance of control, ABA-, and PBI425-treated plants. Three-week-old plants grown on soil were subjected to drought stress. Water was withheld for the number of indicated days after treatment, with no additions (control), ABA, and PBI425. Soil dried at an almost identical rate in all treatments. (b) PBI425-treated plants under drought and rewatering conditions. Drought stress was imposed by withholding water for 14 d after PBI425 treatment (left). One day after rewatering following 14 d drought stress (right).
Genes regulated by drought and multiple plant hormones
From GO analyses it was observed that many drought-regulated genes are annotated as responsive to other plant hormones including auxin, cytokinin, BR, ethylene, gibberellins, and JA (Supplementary Table 5 at JXB online). These included both drought-up-regulated and drought-down-regulated genes. Several genes were responsive to more than one hormone, suggesting they might be the nodes in a regulatory network. For example, GASA1 was reported to be induced by GA and repressed by BR (Bouquin et al., 2001). In this study, GASA1 was found to be up-regulated by ABA and rehydration, and down-regulated by drought, as discussed earlier (group D, Fig. 3a).
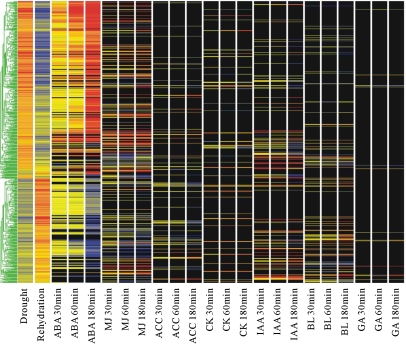
To investigate further the involvement of other plant hormones in drought responses, a set of processed microarray data that listed genes responsive to seven plant hormones was downloaded from Nemhauser et al. (2006). The present drought-regulated gene list was compared with the lists of genes regulated by seven hormones [ABA, methyl jasmonate (MJ), indole-3-acetic acid (IAA), brassinolide (BL), 1-amino-cyclopropane-1-carboxylic acid (ACC; ethylene precursor), zeatin (CK), and GA]. It was immediately apparent that, in addition to a great number of ABA-responsive genes, many drought-regulated genes were also regulated by other plant hormones. In total, 641 hormone-responsive genes were involved in the drought response. The numbers of overlapping genes are 491, 197, 95, 85, 60, 46, and 13 between drought and ABA, MJ, IAA, BL, ACC, CK, and GA, respectively. The regulation of these genes by drought and hormones is summarized in Supplementary Table 6 at JXB online. Clustering analysis on these 641 overlapping genes is shown in Fig. 7. As shown in Supplementary Table 7 at JXB online, a total of 228 genes were regulated by drought and at least two plant hormones; 84 were regulated by at least three plant hormones; 28 were regulated by at least four plant hormones; and five were regulated by at least five plant hormones, indicating strong cross-talk between drought and multiple plant hormone response signalling networks. Genes that are regulated by drought and at least three plant hormones are listed in Table 3. Hierarchical clustering analysis demonstrated that many of these genes have unique expression patterns, suggesting specific roles in the stress and hormonal signalling pathways, while some grouped in clusters of similar expression patterns (Supplementary Fig. 1 at JXB online). For example, a group of auxin-responsive genes (IAA1, IAA4, At4g34760, At4g38860, and At1g29430) were down-regulated by drought, ABA, and MJ, and up-regulated by IAA, BL, CK, and ACC. On the other hand, AR781 (At4g04840) and ZAT10 (At2g26530; discussed above as a member of group B, Fig, 3a) were up-regulated by both MJ and IAA.
Fig. 7.
Clustering of 641 drought and plant hormone overlapping genes to show commonalities between treatments. Hormone data are from Nemhauser et al. (2006).
Table 3.
Drought-responsive genes that are regulated by at least three plant hormones
| Gene ID | Annotations | Drought (fold change) |
|---|---|---|
| At1g01470 | Late embryogenesis abundant protein, putative/LEA protein, putative [(LEA14)] | 5.621 |
| At1g03870 | Fasciclin-like arabinogalactan-protein (FLA9) [(FLA9)] | 0.367 |
| At1g04240 | Auxin-responsive protein/indoleacetic acid-induced protein 3 (IAA3) [(SHY2)] | 1.339 |
| At1g05010 | 1-Aminocyclopropane-1-carboxylate oxidase/ACC oxidase/ethylene-forming enzyme (ACO) (EAT1) [(ATEAT1)] | 0.465 |
| At1g06830 | Glutaredoxin family protein | 0.83 |
| At1g07720 | β-Ketoacyl-CoA synthase family protein | 2.45 |
| At1g08930 | Early-responsive to dehydration stress protein (ERD6)/sugar transporter family protein [(ERD6)] | 0.536 |
| At1g09530 | Phytochrome-interacting factor 3 (PIF3) [(PAP3, PIF3)] | 1.536 |
| At1g10140 | Expressed protein | 1.377 |
| At1g13110 | Cytochrome P450 71B7 (CYP71B7) [(CYP71B7)] | 0.665 |
| At1g21000 | Zinc-binding family protein | 2.753 |
| At1g23040 | Hydroxyproline-rich glycoprotein family protein | 1.606 |
| At1g27730 | Zinc finger (C2H2 type) family protein (ZAT10)/salt-tolerance zinc finger protein (STZ) [(ZAT10)] | 1.512 |
| At1g29430 | Auxin-responsive family protein | 0.661 |
| At1g29660 | GDSL-motif lipase/hydrolase family protein | 0.445 |
| At1g30360 | Early-responsive to dehydration stress protein (ERD4) | 1.449 |
| At1g48320 | Thioesterase family protein | 1.418 |
| At1g48750 | Protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 4.466 |
| At1g51500 | ABC transporter family protein | 1.718 |
| At1g55330 | Arabinogalactan-protein (AGP21) [(AGP21)] | 0.344 |
| At1g58270 | Meprin and TRAF homology domain-containing protein/MATH domain-containing protein [(ZW9)] | 4.059 |
| At1g62570 | Flavin-containing monooxygenase family protein/FMO family protein | 1.342 |
| At1g63840 | Zinc finger (C3HC4-type RING finger) family protein | 1.484 |
| At1g64950 | Cytochrome P450, putative [(CYP89A5)] | 1.628 |
| At1g69530 | Expansin, putative (EXP1) [(ATEXPA1)] | 0.273 |
| At1g72450 | Expressed protein | 0.769 |
| At1g72510 | Expressed protein | 1.515 |
| At1g76580 | SPL1-related3 protein (SPL1R3) | 1.175 |
| At1g77000 | F-box family protein | 1.409 |
| At2g26530 | Expressed protein [(AR781)] | 0.685 |
| At2g28950 | Expansin, putative (EXP6) [(ATEXPA6)] | 0.463 |
| At2g33330 | 33 kDa secretory protein-related | 0.655 |
| At2g34510 | Expressed protein | 0.621 |
| At2g35940 | Homeodomain-containing protein [(BLH1)] | 2.244 |
| At2g44210 | Expressed protein | 0.566 |
| At2g46650 | Cytochrome _b_5, putative [(B5 #1)] | 1.195 |
| At2g47600 | Magnesium/proton exchanger (MHX1) [(ATMHX, ATMHX1)] | 1.349 |
| At3g02170 | Expressed protein | 0.748 |
| At3g06770 | Glycoside hydrolase family 28 protein/polygalacturonase (pectinase) family protein | 0.727 |
| At3g15530 | Expressed protein | 1.614 |
| At3g21670 | Nitrate transporter (NTP3) | 1.724 |
| At3g26510 | Octicosapeptide/Phox/Bem1p (PB1) domain-containing protein | 2.433 |
| At3g28180 | Glycosyl transferase family 2 protein [(ATCSLC04)] | 0.576 |
| At3g29575 | Expressed protein | 3.503 |
| At3g45160 | Expressed protein | 1.322 |
| At3g50340 | Expressed protein | 1.693 |
| At3g52155 | Expressed protein | 0.766 |
| At3g57520 | Alkaline α galactosidase, putative | 6.314 |
| At3g58120 | bZIP transcription factor family protein | 0.756 |
| At3g61460 | Zinc finger (C3HC4-type RING finger) family protein (BRH1) [(BRH1)] | 1.945 |
| At3g62700 | Glutathione-conjugate transporter, putative [(ATMRP10)] | 1.522 |
| At3g63210 | Expressed protein [(MARD1)] | 1.637 |
| At4g00880 | Auxin-responsive family protein | 1.353 |
| At4g04840 | Methionine sulphoxide reductase domain-containing protein/SeIR domain-containing protein | 0.453 |
| At4g09890 | Expressed protein | 1.856 |
| At4g14560 | Auxin-responsive protein/indoleacetic acid-induced protein 1 (IAA1) [(IAA1)] | 1.455 |
| At4g18130 | Phytochrome E (PHYE) [(PHYE)] | 1.254 |
| At4g24780 | Pectate lyase family protein | 0.549 |
| At4g26080 | Protein phosphatase 2C ABI1/PP2C ABI1/abscisic acid-insensitive 1 (ABI1) [(ABI1)] | 3.253 |
| At4g28240 | Wound-responsive protein-related | 2.485 |
| At4g28780 | GDSL-motif lipase/hydrolase family protein | 0.622 |
| At4g30490 | AFG1-like ATPase family protein | 1.662 |
| At4g34760 | Auxin-responsive family protein | 0.755 |
| At4g36540 | Basic helix–loop–helix (bHLH) family protein | 0.667 |
| At4g37080 | Expressed protein | 0.759 |
| At4g37560 | Formamidase, putative/formamide amidohydrolase, putative | 1.354 |
| At4g37790 | Homeobox-leucine zipper protein 22 (HAT22)/HD-ZIP protein 22 [(HAT22)] | 1.521 |
| At4g38060 | Expressed protein | 1.551 |
| At4g38860 | Auxin-responsive protein, putative | 0.514 |
| At4g39940 | Adenylylsulphate kinase 2 (AKN2) [(AKN2)] | 1.348 |
| At5g01520 | Zinc finger (C3HC4-type RING finger) family protein | 3.431 |
| At5g02760 | Protein phosphatase 2C family protein/PP2C family protein | 0.556 |
| At5g11060 | Homeobox protein knotted-1 like 4 (KNAT4) [(KNAT4)] | 1.307 |
| At5g11160 | Adenine phosphoribosyltransferase, putative | 0.817 |
| At5g11420 | Expressed protein | 0.489 |
| At5g16110 | Expressed protein | 1.433 |
| At5g25190 | Ethylene-responsive element-binding protein, putative | 0.642 |
| At5g43700 | Auxin-responsive protein/indoleacetic acid-induced protein 4 (IAA4)/auxin-induced protein (AUX2-11) [(ATAUX2-11] | 0.807 |
| At5g46790 | Expressed protein | 0.722 |
| At5g47370 | Homeobox-leucine zipper protein 2 (HAT2)/HD-ZIP protein 2 [(HAT2)] | 1.622 |
| At5g49480 | Sodium-inducible calcium-binding protein (ACP1)/sodium-responsive calcium-binding protein (ACP1) [(ATCP1)] | 0.717 |
| At5g57240 | Oxysterol-binding family protein | 1.219 |
| At5g59820 | Zinc finger (C2H2 type) family protein (ZAT12) [(RHL41)] | 0.671 |
| At5g62280 | Expressed protein | 0.766 |
Drought and ABA:
A total of 491 out of 641 drought and hormone overlapping genes (Nemhauser et al., 2006) were found to be ABA responsive in this comparison. However, 92 out of the remaining 150 ‘non-ABA-regulated genes’ were regulated by ABA and the analogue PBI425 in Huang et al. (2007) (Supplementary Table 2 at JXB online). Therefore, >90% of the drought and hormone overlapping genes were significantly regulated by ABA, showing the predominant role of ABA signalling in drought responses. Clustering analysis (Fig. 3b) on genes responding to both drought and ABA (Nemhauser et al., 2006) showed very similar results to the above analysis on genes commonly regulated by drought and ABA and/or PBI425 (Fig. 3a)—most of these overlapping genes were regulated similarly by drought and ABA. They were either commonly up-regulated or down-regulated, with a few exceptions, and the effect of ABA on expression of most genes increased with time. Indeed, 489 out of the 641 overlapping genes were included in the 1310 drought and ABA (and/or ABA analogues) overlapping gene list (Supplementary Table 2 at JXB online).
Drought and jasmonic acid:
A total of 197 genes were commonly regulated by drought and MJ. Among them, 122 were also regulated by ABA (Nemhauser et al., 2006) and 156 were regulated by ABA and/or an ABA analogue (Huang et al., 2007). In total, 176 (∼90%) were ABA responsive. As expected, GO terms and promoter motif analysis also showed that ABA-responsive promoter motifs and GO terms were highly enriched in these 197 genes. Many of these genes respond similarly to ABA and MJ, although some of them show opposite responses (Supplementary Fig. 2a at JXB online), indicating both synergistic and antagonistic interactions between the two hormones under drought stress.
Drought and IAA:
A total of 95 genes were commonly regulated by drought and IAA. Seventy-seven of them (>80%) were also regulated by ABA either in Nemhauser et al. (2006) or in Huang et al. (2007).
Drought and BL:
A total of 85 genes were commonly regulated by drought and BL. Thirty-four of them were regulated by ABA in Nemhauser et al. (2006) and 58 of them were regulated by ABA in Huang et al. (2007). In total, 64 are ABA responsive. Most of these genes were regulated in an opposite manner by ABA (and drought) and BL.
Drought and ACC:
Sixty drought-responsive genes were regulated by ACC. Forty-seven of them were regulated by ABA in Huang et al. (2007) and 30 were regulated by ABA in Nemhauser et al. (2006). Many of these genes were commonly down-regulated by drought, ABA, and ACC.
Drought and GA:
Only 13 genes were found to be commonly regulated by drought and GA. Of these, 12 out of 13 were also regulated by ABA. This small number of genes is somewhat surprising since ABA and GA have a well-known antagonistic relationship, especially with regard to seed development and germination (Ho et al., 2003), and there is evidence that ABA regulates GA metabolism (Seo et al., 2006).
Drought-regulated genes in relation to other environmental factors
GO term analysis showed that drought stress not only induced many genes involved in responses to desiccation, water, and water deprivation, but also activated a number of genes related to other stress responses such as cold, high salt, oxidative stress, high light intensity, heat, wounding, and others (Supplementary Table 4 at JXB online), indicating cross-talk between responses to diverse stresses. It is notable that many drought-regulated genes were also responsive to light (KNAT3, KNAT4, SEN1, DIN9, DIN10, and ACP4) and/or circadian rhythm (e.g. CCA1, WNK1, and FSD1), suggesting that drought might affect a plant's light and circadian cycles and/or vice versa.
Some drought-regulated genes were responsive to multiple stresses and stimuli. For example, RD29B, COR47, and ERD14 were responsive to both water deprivation and cold stress; RD20 and RD22 were responsive to both desiccation and salinity; VSP2 was responsive to heat, oxidative stress, desiccation, insect, salinity, and wounding; and RHL41 (At5g59820) was responsive to salinity, wounding, cold, heat, oxidative stress, chitin, and light stimulus. Recently, Ma and Bohnert (2007) applied the fuzzy K-means clustering method to publicly available microarray data from the AtGenExpression project to compare the response of Arabidopsis genes with a variety of abiotic and biotic stresses, and found a cluster (cluster N12 in Ma and Bohnert's K-mean clustering tree) including 197 genes that responded to all stress treatment conditions. Swindell (2006) identified 67 genes that exhibited responses to all nine stress treatments (cold, osmotic stress, salt, drought, genotoxic stress, ultraviolet light, oxidative stress, wounding, and high temperature). However, comparing the gene lists from these two papers, only two genes [_ZAT10_ (group D in Fig. 3a) and _VSP28_] were common to both of them. Comparing the drought-regulated genes in this study, it was found that six were in common with those of Swindell and 23 genes overlapped with Ma and Bohnert's cluster N12 genes (Ma and Bohnert, 2007). The overlapping genes are listed in Table 4 and include several zinc finger and other transcription factors.
Table 4.
Drought-regulated genes that are also responsive to multiple stresses
| Gene ID | Annotations |
|---|---|
| Twenty-three genes overlapping with N12 cluster (common to all stresses) from Ma and Bohnert (2007) | |
| At2g16720 | Myb family transcription factor [(ATMYB7, MYB7)] |
| At4g18880 | Heat shock transcription factor 21 (HSF21) [(AT-HSFA4A)] |
| At3g52800 | Zinc finger (AN1-like) family protein |
| At3g60420 | Expressed protein |
| At1g19380 | Expressed protein |
| At5g20230 | Plastocyanin-like domain-containing protein [(ATBCB)] |
| At3g17800 | Expressed protein |
| At3g47420 | Glycerol-3-phosphate transporter, putative/glycerol 3-phosphate permease, putative |
| At5g54940 | Eukaryotic translation initiation factor SUI1, putative |
| At5g42050 | Expressed protein |
| At5g16830 | Syntaxin 21 (SYP21)/PEP12 homologue [(SYP21)] |
| At5g22000 | Zinc finger (C3HC4-type RING finger) family protein [(CIC7E11, RHF2A)] |
| At5g65140 | Trehalose-6-phosphate phosphatase, putative |
| At5g59820 | Zinc finger (C2H2 type) family protein (ZAT12) [(RHL41)] |
| At4g31550 | WRKY family transcription factor [(WRKY11)] |
| At2g38470 | WRKY family transcription factor [(WRKY33)] |
| At4g21560 | Vacuolar protein sorting-associated protein 28 family protein/VPS28 family protein |
| At2g41410 | Calmodulin, putative |
| At4g30210 | NADPH-cytochrome P450 reductase, putative/NADPH-ferrihaemoprotein reductase, putative [(AR2, ATR2)] |
| At3g55980 | Zinc finger (CCCH-type) family protein |
| At1g27730 | Zinc finger (C2H2 type) family protein (ZAT10)/salt-tolerance zinc finger protein (STZ) [(ZAT10)] |
| At4g37790 | Homeobox-leucine zipper protein 22 (HAT22)/HD-ZIP protein 22 [(HAT22)] |
| At5g24590 | Turnip crinkle virus-interacting protein/TCV-interacting protein (TIP) [(TIP)] |
| Six genes also responsive to nine abiotic stresses in Swindell (2006) | |
| At1g27730 | Zinc finger (C2H2 type) family protein (ZAT10)/salt-tolerance zinc finger protein (STZ) [(ZAT10)] |
| At5g23970 | Transferase family protein |
| At1g72900 | Disease resistance protein (TIR-NBS class), putative |
| At4g21560 | Vacuolar protein sorting-associated protein 28 family protein/VPS28 family protein |
| At3g17810 | Dihydroorotate dehydrogenase family protein/dihydroorotate oxidase family protein |
| At1g06830 | Glutaredoxin family protein |
Discussion
Genome-wide identification of drought-regulated genes under progressive soil drought stress
Bray (2004) compared several studies that reported drought-inducible genes and found only 27 that were commonly induced. More recently, Swindell (2006) identified 67 genes that are responsive to nine abiotic stress treatments, and Ma and Bohnert (2007) reported that 197 Arabidopsis genes are found to be induced by a broad range of diverse stress conditions. Again, only two genes were found to be common between these last two studies, suggesting that the full extent of overlap between stress responses has not been established yet.
The number of genes identified as drought responsive in a microarray experiment largely depends on the criteria used in selecting genes. In earlier studies, differentially expressed genes were selected based on expression ratios instead of statistical thresholds. Since expression ratio thresholds of up to 5-fold were used, many weakly responsive genes were ignored, and thus the proportion of differentially expressed genes was underestimated.
An additional factor producing discrepancies between different studies is the methods used for plant growth and stress treatments. To understand responses to drought, experimental conditions should approximate stress development in the field. However, some published experiments are based on a rapid, severe water deficit treatment (Bray, 2004; Kilian et al., 2007), which plants rarely experience in the field. Rapid drought protocols may produce changes in ‘shock’-induced genes that are unrelated to adaptive responses and not expressed under progressive drought. In this study, conditions were used that produced progressive soil drought stress, which approximates the development of drought conditions in the field. A total of 1969 drought-regulated genes (7.6% of the Arabidopsis genome) were identified using microarrays, which represented the whole Arabidopsis genome. Relative to previous studies, many more drought-regulated genes were identified since statistical analysis was relied on rather than arbitrary thresholds, and a full genome array was used. These genes included many known drought-, ABA-, and many other stress-responsive genes, together with numerous novel genes. Among the 277 drought-inducible genes identified by Seki et al. (2002), more than half of them were significantly regulated in the present study, although the drought treatments were very different.
It should be noted that there is very little overlap (only 83 genes) between genes induced in the present study and in the drought treatment of Kilian et al. (2007). In that case, the authors employed a drastic 15 min water stress that reduced fresh weight by 10%, after which the plants were returned to their previous liquid growth medium and changes in gene expression were recorded at intervals of up to 24 h. It is interesting that the major functional terms (especially those related to abiotic stress) and promoter motifs (notably those associated with ABA) that are enriched in the present study (Tables 1 and 2) are also enriched in the very rapidly responsive genes (15 min to 3 h) in the Kilian et al. study (Supplementary Table 8 at JXB online) but not in genes responding at later time points (6–24 h). The changes in gene expression documented by Kilian et al. presumably reflect a transient induction of the drought response gene network that disappears as the plants recover.
Rehydration is a useful approach for large-scale validation of drought-regulated genes
The changes in gene expression associated with recovery from stress are likely to be just as important for plant survival as changes associated with the onset of stress. There are a few reports on recovery from drought stress (Kiyosue et al., 1996; Oono et al., 2003). Using cDNA arrays, Oono et al. (2003) identified 152 rehydration-inducible, 280 rehydration-repressed, 270 dehydration-inducible, and 168 dehydration-repressed genes. They found that 107 genes were regulated in opposite directions by drought and rehydration, whereas 16 genes were commonly up-regulated and 69 genes were commonly down-regulated. In the present study, many more drought- and rehydration-regulated genes were found, and the opposing effects of rehydration and drought on gene expression were more obvious. For example, most drought-regulated genes were reciprocally regulated by rehydration treatments. Examining the 16 genes commonly up-regulated and the 69 commonly down-regulated genes in Oono's study, it was found that most of them were either dehydration-repressed and rehydration-inducible genes such as GLUTAMATE DEHYDROGENASE 2 (At5g07440) and SUGAR TRANSPORTER 1 (At1g11260), or rehydration-repressed genes such as LIPID TRANSFER PROTEIN 3 (At5g59320) and SEED IMBIBITION 2 (At3g57520) in the present study. It is clear that rehydration results in a reversal of the changes taking place under drought stress; the expression of most drought-regulated genes returned to normal levels 3 h after rewatering. Therefore, rehydration provides a degree of cross-validation of the identity of drought-regulated genes.
ABA and drought stress responses
ABA and drought usually have similar effects on gene expression, but there are a few exceptions, one of which is cluster D in Fig. 3a; a subset of genes whose functions suggest ABA-promoted cell wall expansion. At least some of these genes are regulated by other plant hormones. Although ABA is normally considered to be a growth inhibitor, there are specific situations in which ABA is associated with growth. For example, ABA has been linked with secondary embryogenesis in carrot cultures (Ogata et al., 2005), and analysis of ABA-deficient mutants suggests that ABA acts as a growth promoter during vegetative development (Barrero et al., 2005). Furthermore, ABA has been associated with certain actively growing or differentiating cells (Peng et al., 2006). In root studies, exogenous application of ABA leads to swelling, root hair formation, and initiation of lateral root primordia in the tips of young, seminal rice roots (Chen et al., 2006), and there is evidence that ABA promotes lateral root growth as an aspect of stress responses (De Smet et al., 2006). Therefore, it appears that drought stress represses the expression of a subset of growth-promoting, ABA-induced genes.
ABA analogue PBI425 enhances drought tolerance via ABA responses
The level of endogenous ABA in plants increased ∼10 times under drought stress compared with that in control plants. In addition, the present studies showed that the levels of ABA metabolites also increased, indicating that catabolic metabolism remains active and that flux through the ABA pathway probably increases as biosynthesis increases.
A previous study (Huang et al., 2007) showed that exogenously applied ABA analogues induced the expression of large numbers of ABA-regulated genes including many that had not been associated with ABA before. Results from analogue treatments complemented results from ABA treatments in providing a more complete list of ABA-regulated genes. The most effective analogue was PBI425. In this study, 1310 genes were found to be commonly regulated by drought stress and ABA (and/or ABA analogues) treatment, and 1172 genes were overlapping between drought and PBI425. GO term and promoter motif analyses revealed that genes with functions in ABA signalling and ABA-responsive promoter motifs were highly enriched in genes regulated by drought and PBI425. It was also demonstrated that PBI425 produced greater drought tolerance than ABA, corresponding to its gene expression effects. Therefore, PBI425 enhances drought tolerance through ABA responses and the ABA signalling pathway.
Consistent with previous reports (Cutler et al., 2000; Huang et al., 2007), ABA metabolite profiling showed the accumulation of PBI425 to high concentration in plants whereas ABA was rapidly metabolized into PA and DPA. Because the endogenous ABA and ABA metabolites were only slightly increased in PBI425-treated plants, it is deduced that the higher concentration and longer persistence of the applied PBI425 is the major contribution to expression of more ABA-responsive genes and the significantly increased drought tolerance.
Are there any drought-stress responsive genes that are not affected by ABA?
It was noted earlier that >90% of the drought and hormone overlapping genes were significantly regulated by ABA. Previous studies based on promoter analyses of drought- and/or cold-inducible genes suggested the existence of both ABA-dependent and ABA-independent pathways for drought or stress signalling transduction (Yamaguchi-Shinozaki and Shinozaki, 2000, 2005). At least four independent regulatory systems for gene expression have been proposed for drought responsiveness. Two of them are ABA-dependent pathways, in which ABRE, and MYB and MYC recognition sites (MYBR and MYCR, respectively) were involved. The other two are ABA-independent pathways, in which the expression of genes in response to drought occurred through DREs/CRT _cis_-acting elements or a NAC recognition site (Yamaguchi-Shinozaki and Shinozaki, 2005).
Numerous drought-inducible genes with the ABRE, DRE, MYBR, and MYCR _cis_-elements in their promoter regions have been identified. However, as noted earlier, many genes were regulated by both drought stress and ABA treatment. Promoter motif analyses demonstrated that ABA-responsive elements (ABRE, ABF, and ABRE-like binding sites) were highly enriched not only in genes commonly regulated by drought and ABA but also in ‘drought only’ genes, indicating that expression of many of these apparently ‘drought only’ genes may be at least partially linked to ABA signalling.
Similarly, the DRE core motif is highly over-represented upstream of ABA-responsive genes (Huang et al., 2007). Many drought-inducible genes contained both DRE/CRT and ABRE motifs in their promoters. Genome-wide promoter analyses (data not shown) showed that 1314 Arabidopsis genes contained both ABRE or ABRE-like motifs and DRE core motifs in their promoter regions, suggesting that these genes could be activated through both signalling systems. The DRE/CRT and ABRE motifs were thought to function independently (Shinozaki and Yamaguchi-Shinozaki, 2000). However, precise analysis of these _cis_-acting elements in RD29A gene expression revealed that DRE/CRT functions cooperatively with ABRE as a coupling element in ABA-responsive gene expression in response to drought stress (Narusaka et al., 2003). A recent study demonstrated that ABA induced CBF gene transcription and subsequent induction of cold-regulated genes (COR genes) via the CRT/DRE promoter element, thereby providing a specific example of ABA-dependent signalling through the CRT/DRE element in Arabidopsis (Knight et al., 2004).
In maize, two DRE-like elements (DRE1 and DRE2), which are responsive to ABA, have been identified in the RAB17 gene promoter (Busk et al., 1997). A maize DRE-binding protein, DBF1 (a _trans_-acting factor that bind to DRE2), has been shown to function as a transcriptional activator of the RAB17 promoter by ABA (Kizis and Pages, 2002).
These observations suggest the existence of an ABA-dependent pathway for the regulation of stress-inducible genes that involves DRE/CRT. Furthermore, RD26, a dehydration-induced NAC protein, has been shown to be involved in a novel ABA-dependent stress signalling pathway (Fujita et al., 2004). Therefore, it is difficult to identify any genes whose expression is completely ABA independent. Although there must be ABA-independent stress signalling (perception response) pathways (Verslues and Zhu, 2005) since ABA biosynthesis is itself triggered by stress (Barrero et al., 2006), the promoters of individual genes integrate signals from both ABA- and stress-specific signalling pathways, so that their expression is not entirely ABA independent.
Complexity of drought-responsive gene networks
The large number of drought- or rehydration-regulated transcriptome changes underscores the difficulty in understanding the global context of the drought stress response. Various bioinformatic tools were used to identify the interactions of drought signalling pathways and to identify _cis_-acting elements in the promoters of drought- or rehydration-regulated genes. Strong cross-talk with other stress responses (cold, salt, heat, light, oxidative stress, reactive oxygen species, wounding, and pathogens) was found, which is consistent with previous reports (Chen et al., 2002; Kreps et al., 2002; Seki et al., 2002; Swindell, 2006; Ma and Bohnert, 2007).
Extensive overlaps also existed between drought response and several plant hormone responses including auxin, cytokinin, ethylene, the GAs, JA, the BRs, and especially ABA. Apart from ABA, little is known of the roles of the other hormones in relation to drought responses. Jasmonate (MJ) appears to be the most important of the other hormones affecting stress responses and has a well documented role in biotic stress and wounding responses. A recent paper describing responses of Arabidopsis to the pathogen Pythium irregulare suggested that ABA worked cooperatively with JA in promoting defence responses (Adie et al., 2007). Some reports also suggest an involvement of JA with stress responses; for example, JA was reported to be involved in the adaptation of barley to salt stress (Walia et al., 2007) and was associated with cold stress responses in the crucifer Thellungiella (Wong et al., 2006). Furthermore, there is evidence of an interaction between ABA and MJ that affects guard cell aperture (Munemasa et al., 2007). However, the idea that JA synergistically promotes stress responses with ABA is contradicted by a recent microarray study of JA-responsive genes in Arabidopsis which suggested that ‘…an antagonistic interaction occurs between the jasmonate and ABA signaling pathways in abiotic stress responses’ since MJ reduced expression of ABA-responsive stress-related genes (Jung et al., 2007). However, the same study also showed that jasmonate reduced expression of genes related to chlorophyll and photosynthesis, a response that is also produced by ABA. This latter observation can be rationalized as the common complement of responses to biotic or abiotic stress, which is to reduce photosynthesis and growth. It appears that there are both synergistic and antagonistic elements in the ABA–JA interaction and they have not been fully defined yet. Insofar as the primary role of JA appears to be related to biotic stress responses, it is suggested that the interaction between stress responses and JA may reflect the fact that biotic stress and wounding responses modulate responses to drought.
One effect of BRs is to increase stress tolerance (Krishna, 2003). However, a relatively small number of genes were co-regulated by BL and both ABA and drought, and most of these exhibited an antagonistic relationship. Therefore, the mechanism by which BL affects stress tolerance seems to be substantially distinct from the ABA-based mechanism. Much further investigation is required to answer the intriguing question of how BL produces stress tolerance. It is conceivable that a significant mechanism of stress tolerance remains that has not been revealed by gene expression studies carried out to date.
Promoter motif analyses also confirmed the complexity of drought-induced gene expression. In addition to the ABA- and drought-responsive _cis_-acting elements, many other known _cis_-acting elements were enriched in the promoters of drought-up-regulated genes or rehydration-down-regulated genes. Some of these elements suggested interaction between light and drought responses; for example, the ‘evening element’ promoter motif was originally identified from the promoter of genes involved in circadian rhythm (Harmer et al., 2000). Several G-box-containing _cis_-acting elements in light-regulated genes including GBOXLERBCS, CACGTGMOTIF, and GBF1/2/3 BS in ADH1, and other light-responsive elements such as the TGA1-binding site motif and Z-box promoter motif were significantly over-represented in drought-inducible genes, indicating strong cross-talk between drought response and light signalling pathways. A role for day length and spectral composition has been established in mediating cold acclimation (Gusta et al., 2005), and Kreps et al. (2002) observed that 68% of the known circadian-controlled genes are implicated in stress responses, suggesting that an important function of the circadian clock genes is to ‘anticipate’ predictable stresses such as cold nights.
Genome-wide promoter motif analysis showed high correlation between certain promoter motifs (data not shown here). For example, the above ‘G-box’-containing _cis_-acting elements usually appeared with an ABRE motif, suggesting that the two motifs—which are very similar—might work cooperatively. Most genes have several _cis_-acting elements in their promoter region. It seems likely that overall gene expression is determined by interactions between these elements rather than that each motif functions entirely independently.
Conclusions
The present results confirm a predominant role for ABA in mediating gene expression responses to drought stress. Although ABA-independent signalling mechanisms play a role in mediating responses, ABA is capable of inducing the expression of most genes involved in drought responses since the multiple sequence motifs present in upstream regions of stress-responsive genes allow integration of signals from multiple perception–response systems. These interacting signalling systems include not only ABA-dependent and ABA-independent drought responses, but light effects and other plant hormones, the most quantitatively significant of which (number of genes affected) appears to be JA. The interactions between signalling systems allow the physiological and developmental context to modulate stress responses. For example, it would be predicted that a wounded or pathogen-infected plant with elevated levels of JA will respond to drought stress somewhat differently from healthy plants. In other words, the universality of stress responses will be modulated by other factors in the environment as well as the history and stage of the plant in question. In contrast to Nemhauser et al. (2006), the present results and analysis suggest that overlapping transcriptional regulation is important for fine-tuning responses to environmental factors.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Table 1. Fifteen selected genes and their primers for QRT-PCR.
Supplementary Table 2. Genes significantly regulated by drought.
Supplementary Table 3. Drought and ABA (or ABA analogues) co-regulated genes.
Supplementary Table 4. ABA-‘independent’ drought-responsive genes in relation to stress and hormone responses.
Supplementary Table 5. Drought-regulated genes in relation to stress and hormonal responses.
Supplementary Table 6. Overlapping responses to drought and seven plant hormones.
Supplementary Table 7. Genes regulated by drought and at least two plant hormones.
Supplementary Table 8. Enriched GO terms and promoter motifs in genes responsive to ‘shock’ drought treatment (from Kilian et al., 2007).
Supplementary Fig. 1. Clustering analysis of genes regulated by drought and at least three plant hormones.
Supplementary Fig. 2a. Clustering analysis of genes commonly regulated by drought, ABA and JA.
Supplementary Fig. 2b. Clustering analysis of genes commonly regulated by drought, ABA, and IAA.
Supplementary Material
[Supplementary Material]
Acknowledgments
Funding for this study was provided by Genome Canada and Genome Prairie as part of the ‘Functional genomics of abiotic stress’ programme. Additional funding to DH was provided by the Saskatchewan Agriculture Development Fund and to both DH and WW by the China Scholar Council. We thank Steve Ambrose for analysis of metabolites, Mary Loewen for technical support, and Tony Cornish (Microarray and Proteomics Facility, University of Alberta) for some of the microarray slides employed. During the preparation of this paper helpful comments were provided by Mark Smith and Gopalan Selvaraj. This paper is NRCC number 48453.
References
- Abrams SR, Nelson K, Ambrose SJ. Deuterated abscisic acid analogs for mass spectrometry and metabolism studies. Journal of Labeled Compounds and Radiopharmaceuticals. 2003;46:273–283. [Google Scholar]
- Adie BA, Perez-Perez J, Perez-Perez MM, Godoy M, Sanchez-Serrano JJ, Schmelz EA, Solano R. ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. The Plant Cell. 2007;19:1665–1681. doi: 10.1105/tpc.106.048041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SS, Wilhelm KS, Thomashow MF. The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Molecular Biology. 1994;24:701–713. doi: 10.1007/BF00029852. [DOI] [PubMed] [Google Scholar]
- Barrero JM, Piqueras P, González-Guzmán M, Serrano R, Rodríguez PL, Ponce MR, Micol JL. A mutational analysis of the ABA1 gene of Arabidopsis thaliana highlights the involvement of ABA in vegetative development. Journal of Experimental Botany. 2005;56:2071–2083. doi: 10.1093/jxb/eri206. [DOI] [PubMed] [Google Scholar]
- Barrero JM, Rodríguez PL, Quesada V, Piqueras P, Ponce MR, Micol J. Both abscisic acid (ABA)-dependent and ABA-independent pathways govern the induction of NCED3, AAO3 and ABA1 in response to salt stress. Plant, Cell and Environment. 2006;29:2000–2008. doi: 10.1111/j.1365-3040.2006.01576.x. [DOI] [PubMed] [Google Scholar]
- Bartels D, Nelson D. Approaches to improve stress tolerance using molecular genetics. Plant, Cell and Environment. 1994;17:659–667. [Google Scholar]
- Bohnert HJ, Nelson DE, Jensen RG. Adaptations to environmental stresses. The Plant Cell. 1995;7:1099–1111. doi: 10.1105/tpc.7.7.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouquin T, Meier C, Foster R, Nielsen ME, Mundy J. Control of specific gene expression by gibberellin and brassinosteroid. Plant Physiology. 2001;127:450–458. [PMC free article] [PubMed] [Google Scholar]
- Boyer JS. Plant productivity and environment. Science. 1982;218:443–448. doi: 10.1126/science.218.4571.443. [DOI] [PubMed] [Google Scholar]
- Bray EA. Plant responses to water deficit. Trends in Plant Science. 1997;2:48–54. [Google Scholar]
- Bray EA. Genes commonly regulated by water-deficit stress in Arabidopsis thaliana. Journal of Experimental Botany. 2004;55:2331–2341. doi: 10.1093/jxb/erh270. [DOI] [PubMed] [Google Scholar]
- Busk PK, Jensen AB, Pages M. Regulatory elements in vivo in the promoter of the abscisic acid responsive gene rab17 from maize. The Plant Journal. 1997;11:1285–1295. doi: 10.1046/j.1365-313x.1997.11061285.x. [DOI] [PubMed] [Google Scholar]
- Chen CW, Yang YW, Lur HS, Tsai YG, Chang MC. A novel function of abscisic acid in the regulation of rice (Oryza sativa L.) root growth and development. Plant and Cell Physiology. 2006;47:1–13. doi: 10.1093/pcp/pci216. [DOI] [PubMed] [Google Scholar]
- Chen W, Provart NJ, Glazebrook J, et al. Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. The Plant Cell. 2002;14:559–574. doi: 10.1105/tpc.010410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiwocha SDS, Abrams SR, Ambrose SJ, Cutler AJ, Loewen M, Ross ARS, Kermode AR. Metabolic profiling of four classes of plant hormones and their major metabolites by liquid chromatography electrospray tandem mass spectrometry: an analysis of hormone turnover associated with thermodormancy and germination of lettuce (Lactuca sativa L.) seeds. The Plant Journal. 2003;35:405–417. doi: 10.1046/j.1365-313x.2003.01800.x. [DOI] [PubMed] [Google Scholar]
- Cutler AJ, Rose PA, Squires TM, Loewen MK, Shaw AC, Quail JW, Krochko JE, Abrams SR. Inhibitors of abscisic acid 8′-hydroxylase. Biochemistry. 2000;39:13614–13624. doi: 10.1021/bi0014453. [DOI] [PubMed] [Google Scholar]
- De Smet I, Zhang H, Inzé D, Beeckman T. A novel role for abscisic acid emerges from underground. Trends in Plant Science. 2006;11:434–459. doi: 10.1016/j.tplants.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Feurtado JA, Ambrose SJ, Cutler AJ, Ross AR, Abrams SR, Kermode AR. Dormancy termination of western white pine (Pinus monticola Dougl. Ex D. Don) seeds is associated with changes in abscisic acid metabolism. Planta. 2004;218:630–639. doi: 10.1007/s00425-003-1139-8. [DOI] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Tran LS, Yamaguchi-Shinozaki K, Shinozaki K. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. The Plant Journal. 2004;39:863–876. doi: 10.1111/j.1365-313X.2004.02171.x. [DOI] [PubMed] [Google Scholar]
- Gusta LV, Trischuk R, Weiser CJ. Plant cold acclimation: the role of abscisic acid. Journal of Plant Growth Regulation. 2005;24:308–318. [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- Ho THD, Gomez-Cadenas A, Zentella R, Casaretto J. Crosstalk between gibberellin and abscisic acid in cereal aleurone. Journal of Plant Growth Regulation. 2003;22:185–194. [Google Scholar]
- Huang D, Jaradat MR, Wu W, Ambrose SJ, Ross AR, Abrams SR, Cutler AJ. Structural analogs of ABA reveal novel features of ABA perception and signaling in Arabidopsis. The Plant Journal. 2007;50:414–428. doi: 10.1111/j.1365-313X.2007.03056.x. [DOI] [PubMed] [Google Scholar]
- Ingram J, Bartels D. The molecular basis of dehydration tolerance in plants. Annual Review of Plant Physiology and Plant Molecular Biology. 1996;47:377–403. doi: 10.1146/annurev.arplant.47.1.377. [DOI] [PubMed] [Google Scholar]
- Jiang C, Lu B, Singh J. Requirement of a CCGAC cis-acting element for cold induction of the BN115 gene from winter Brassica napus. Plant Molecular Biology. 1996;30:679–684. doi: 10.1007/BF00049344. [DOI] [PubMed] [Google Scholar]
- Jung C, Lyou SH, Yeu S, Kim MA, Rhee S, Kim M, Lee JS, Choi YD, Cheong JJ. Microarray-based screening of jasmonate-responsive genes in Arabidopsis thaliana. Plant Cell Reports. 2007;26:1053–1063. doi: 10.1007/s00299-007-0311-1. [DOI] [PubMed] [Google Scholar]
- Kawaguchi R, Girke T, Bray EA, Bailey-Serres JN. Differential mRNA translation contributes to gene regulation under non-stress and dehydration stress conditions in Arabidopsis thaliana. The Plant Journal. 2004;38:823–839. doi: 10.1111/j.1365-313X.2004.02090.x. [DOI] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D'Angelo C, Bornberg-Bauer E, Kudla J, Harter K. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. The Plant Journal. 2007;50:347–363. doi: 10.1111/j.1365-313X.2007.03052.x. [DOI] [PubMed] [Google Scholar]
- Kiyosue T, Yoshiba Y, Yamaguchi-Shinozaki K, Shinozaki K. A nuclear gene encoding mitochondrial proline dehydrogenase, an enzyme involved in proline metabolism, is upregulated by proline but down regulated by dehydration in Arabidopsis. The Plant Cell. 1996;8:1323–1335. doi: 10.1105/tpc.8.8.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirch HH, Schlingensiepen S, Kotchoni S, Sunkar R, Bartels D. Detailed expression analysis of selected genes of the aldehyde dehydrogenase (ALDH) gene superfamily in Arabidopsis thaliana. Plant Molecular Biology. 2005;57:315–332. doi: 10.1007/s11103-004-7796-6. [DOI] [PubMed] [Google Scholar]
- Kizis D, Pages M. Maize DRE-binding proteins DBF1 and DBF2 are involved in rab17 regulation through the drought-responsive element in an ABA-dependent pathway. The Plant Journal. 2002;30:679–689. doi: 10.1046/j.1365-313x.2002.01325.x. [DOI] [PubMed] [Google Scholar]
- Knight H, Zarka DG, Okamoto H, Thomashow MF, Knight MR. Abscisic acid induces CBF gene transcription and subsequent induction of cold-regulated genes via the CRT promoter element. Plant Physiology. 2004;135:1710–1717. doi: 10.1104/pp.104.043562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreps J, Wu Y, Chang HS, Zhu T, Wang X, Harper J. Transcriptome changes for Arabidopsis in response to salt, osmotic and cold stress. Plant Physiology. 2002;130:2129–2141. doi: 10.1104/pp.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna P. Brassinosteroid-mediated stress responses. Journal of Plant Growth Regulation. 2003;22:289–297. doi: 10.1007/s00344-003-0058-z. [DOI] [PubMed] [Google Scholar]
- Ma S, Bohnert HJ. Integration of Arabidopsis thaliana stress-related transcript profiles, promoter structures, and cell-specific expression. Genome Biology. 2007;8:R49. doi: 10.1186/gb-2007-8-4-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munemasa S, Oda K, Watanabe-Sugimoto M, Nakamura Y, Shimoishi Y, Murata Y. The coronatine-insensitive 1 mutation reveals the hormonal signaling interaction between abscisic acid and methyl jasmonate in arabidopsis guard cells. Specific impairment of ion channel activation and second messenger production. Plant Physiology. 2007;143:1398–1407. doi: 10.1104/pp.106.091298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narusaka Y, Nakashima K, Shinwari ZK, Sakuma Y, Furihata T, Abe H, Narusaka M, Shinozaki K, Yamaguchi-Shinozaki K. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. The Plant Journal. 2003;34:137–148. doi: 10.1046/j.1365-313x.2003.01708.x. [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126:467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- O'Connor TR, Dyreson C, Wyrick JJ. Athena: a resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences. Bioinformatics. 2005;21:4411–4413. doi: 10.1093/bioinformatics/bti714. [DOI] [PubMed] [Google Scholar]
- Ogata Y, Iizuka M, Nakayama D, Ikeda M, Kamada H, Koshiba T. Possible involvement of abscisic acid in the induction of secondary somatic embryogenesis on seed-coat-derived carrot somatic embryos. Planta. 2005;221:417–423. doi: 10.1007/s00425-004-1449-5. [DOI] [PubMed] [Google Scholar]
- Oono Y, Seki M, Nanjo T, et al. Monitoring expression profiles of Arabidopsis gene expression during rehydration process after dehydration using ca. 7000 full-length cDNA microarray. The Plant Journal. 2003;34:868–887. doi: 10.1046/j.1365-313x.2003.01774.x. [DOI] [PubMed] [Google Scholar]
- Peng YB, Zou C, Wang DH, Gong HQ, Xu ZH, Bai SN. Preferential localization of abscisic acid in primordial and nursing cells of reproductive organs of Arabidopsis and cucumber. New Phytologist. 2006;170:459–466. doi: 10.1111/j.1469-8137.2006.01683.x. [DOI] [PubMed] [Google Scholar]
- Riera M, Valon C, Fenzi F, Giraudat J, Leung J. The genetics of adaptive responses to drought stress: abscisic acid-dependent and abscisic acid-independent signalling components. Physiologia Plantarum. 2005;123:111–119. [Google Scholar]
- Rose PA, Cutler AJ, Irvine NM, Shaw AC, Squires TM, Loewen MK, Abrams SR. 8′-Acetylene ABA: an irreversible inhibitor of ABA 8′-hydroxylase. Bioorganic and Medicinal Chemistry Letters. 1997;7:2543–2546. [Google Scholar]
- Satoh R, Nakashima K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. ACTCAT, a novel cis-acting element for proline- and hypoosmolarity-responsive expression of the ProDH gene encoding proline dehydrogenase in Arabidopsis. Plant Physiology. 2002;130:709–719. doi: 10.1104/pp.009993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninci P, Hayashizaki Y, Shinozaki K. Monitoring the expression pattern of 1,300 Arabidopsis genes under drought and cold stresses using full-length cDNA microarray. The Plant Cell. 2001;13:61–72. doi: 10.1105/tpc.13.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, et al. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. The Plant Journal. 2002;31:279–292. doi: 10.1046/j.1365-313x.2002.01359.x. [DOI] [PubMed] [Google Scholar]
- Seo M, Hanada A, Kuwahara A, et al. Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. The Plant Journal. 2006;48:354–366. doi: 10.1111/j.1365-313X.2006.02881.x. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Molecular responses to drought and cold stress. Current Opinion in Biotechnology. 1997;7:161–167. doi: 10.1016/s0958-1669(96)80007-3. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Current Opinion in Plant Biology. 2000;3:217–223. [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. Journal of Experimental Botany. 2007;58:221–227. doi: 10.1093/jxb/erl164. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K, Liu Q, et al. Molecular responses to drought stress in plants: regulation of gene expression and signal transduction. In: Smallwood MF, Calvert CM, Bowles DJ, editors. Plant responses to environmental stress. Oxford: BIOS Scientific Publishers; 1999. pp. 133–143. [Google Scholar]
- Swindell WR. The association among gene expression responses to nine abiotic stress treatments in Arabidopsis thaliana. Genetics. 2006;174:1811–24. doi: 10.1534/genetics.106.061374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezara W, Mitchell VJ, Driscoll SD, Lawlor DW. Water stress inhibits plant photosynthesis by decreasing coupling factor and ATP. Nature. 1999;401:914–917. [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences, USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Fujita M, Fujita Y, Yamaguchi-Shinozaki K, Shinozaki K. Engineering drought tolerance in plants: discovering and tailoring genes to unlock the future. Current Opinion in Biotechnology. 2006;17:113–122. doi: 10.1016/j.copbio.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Verslues PE, Zhu JK. Before and beyond ABA: upstream sensing and internal signals that determine ABA accumulation and response under abiotic stress. Biochemical Society Transactions. 2005;33:375–379. doi: 10.1042/BST0330375. [DOI] [PubMed] [Google Scholar]
- Walia H, Wilson C, Condamine P, Liu X, Ismail AM, Close TJ. Large-scale expression profiling and physiological characterization of jasmonic acid-mediated adaptation of barley to salinity stress. Plant, Cell and Environment. 2007;30:410–421. doi: 10.1111/j.1365-3040.2006.01628.x. [DOI] [PubMed] [Google Scholar]
- Wong CE, Li Y, Labbe A, et al. Transcriptional profiling implicates novel interactions between abiotic stress and hormonal responses in Thellungiella, a close relative of Arabidopsis. Plant Physiology. 2006;140:1437–1450. doi: 10.1104/pp.105.070508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature or high-salt stress. The Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends in Plant Science. 2005;10:88–94. doi: 10.1016/j.tplants.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Zaharia I, Galka M, Ambrose SJ, Abrams SR. Preparation of deuterated abscisic acid metabolites for use in mass spectrometry and feeding studies. Journal of Labelled Compounds and Radiopharmaceuticals. 2005;48:435–445. [Google Scholar]
- Zeevaart JAD. Changes in the levels of abscisic acid and its metabolites in excised leaf blades of Xanthium strumarium during and after water stress. Plant Physiology. 1980;66:672–678. doi: 10.1104/pp.66.4.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Salt and drought stress signal transduction in plants. Annual Review of Plant Biology. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplementary Material]