Differences in the length of the carboxyl terminus mediate functional properties of neurokinin-1 receptor (original) (raw)
Abstract
The neurokinin-1 receptor (NK1R) has two naturally occurring forms that differ in the length of the carboxyl terminus: a full-length receptor consisting of 407 aa and a truncated receptor consisting of 311 aa. We examined whether there are differential signaling properties attributable to the carboxyl terminus of this receptor by using stably transfected human embryonic kidney (HEK293) cell lines that express either full-length or truncated NK1R. Substance P (SP) specifically triggered intracellular calcium increase in HEK293 cells expressing full-length NK1R but had no effect in the cells expressing the truncated NK1R. In addition, in cells expressing full-length NK1R, SP activated NF-κB and IL-8 mRNA expression, but in cells expressing the truncated NK1R, SP did not activate NF-κB, and it decreased IL-8 mRNA expression. In cells expressing full-length NK1R, SP stimulated phosphorylation of PKCδ but inhibited phosphorylation of PKCδ in cells expressing truncated NK1R. There are also differences in the timing of SP-induced ERK activation in cells expressing the two different forms of the receptor. Full-length NK1R activation of ERK was rapid (peak within 1–2 min), whereas truncated NK1R-mediated activation was slower (peak at 20–30 min). Thus, the carboxyl terminus of NK1R is the structural basis for differences in the functional properties of the full-length and truncated NK1R. These differences may provide important information toward the design of new NK1R receptor antagonists.
Keywords: G protein-coupled receptor, cell signaling, Substance P, truncated neurokinin-1, calcium mobilization
The biological responses to the peptide Substance P (SP) (1), a major regulatory peptide, are mediated primarily by its preferred receptor neurokinin-1 receptor (NK1R). The human NK1R gene (2–5), localized to chromosome 2, has been cloned. NK1R is a member of the G protein-coupled receptor (GPCR) superfamily and has a structure of seven transmembrane domains, an extracellular amino terminus and an intracellular carboxyl terminus (2). The full-length NK1R is the predominant form expressed at selected sites in the human brain, and truncated NK1R is widespread throughout the central nervous system and in peripheral tissues (6). Kage et al. (7) reported that functional differences in two forms of NK1R present in rat submaxillary glands are caused by differences in the length of their carboxyl termini. Many studies have suggested differences in NK1R function by using an approximation of the truncated form; these studies, for the most part, however, have not used the naturally occurring variant. The naturally occurring splice variant of NK1R mRNA has been cloned from humans (2) and guinea pigs (8) and identified. This natural variant of truncated NK1R, expressed in COS cells, has a binding affinity that is at least 10-fold less than that of the full-length receptor and elicits only weak electrophysiological responses in Xenopus oocytes (2) and has reduced maximal current responses compared with full-length NK1R (9). We propose that the NK1R carboxyl terminus is the structural basis for SP-induced calcium mobilization, activation of NF-κB, and up-regulation of NF-κB-targeted gene expression (such as IL-8). In this work, we compare several different biologic responses of the full-length NK1R with the naturally occurring truncated form.
Results
NK1R Carboxyl Terminus Is Required for SP-Induced Calcium Mobilization.
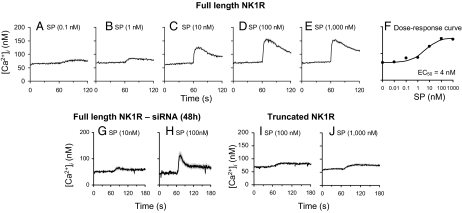
In cells that express full-length NK1R, SP increased intracellular calcium levels in a concentration-dependent relationship (0.1–1,000 nM) (Fig. 1 A–E) with EC50 of 4 nM (Fig. 1F). After reduction of the expression of the full-length NK1R by siRNA, the ability of SP to induce intracellular calcium increases was impaired (Fig. 1 G and H), which demonstrated that the expression of full-length NK1R is required for SP to induce intracellular calcium increases. In contrast, no intracellular Ca2+ increase was observed in cells that express truncated NK1R in response to SP, even at 10−6 M concentration (Fig. 1 I and J).
Fig. 1.
Full-length NK1R mediates intracellular calcium increase, whereas truncated NK1R signals through calcium-independent mechanisms. (A–E) Representative tracings recorded while HEK293 cells expressing full-length NK1R were exposed to increasing concentrations of SP. (F) Dose–response curve to SP in HEK293 cells stably expressing the full-length NK1R. Peak calcium responses were determined, concentration–response curves were constructed, and a logistic curve was fitted to the data. The EC50 value was derived from the fitted curve. (G and H) Effect of siRNA targeting full-length NK1R on SP-induced intracellular calcium responses. NK1R expression was reduced by using siRNA as described under Materials and Methods, and the effect of siRNA was confirmed by detection of NK1R mRNA with real-time RT-PCR. Individual tracings were recorded 48 h after siRNA transfection, during which the cells were exposed to 10 or 100 nM SP and are shown. (I and J) HEK293 cells stably expressing the truncated NK1R do not respond with an intracellular calcium increase in response to SP. Representative tracings recorded with SP concentrations of 100 nM and 1,000 nM are shown. Each data point on the tracings shown in A–E and G–J corresponds to the mean values measured in four to eight individual cells ± SEM. Dose-dependent response to SP in HEK293 cells expressing full-length NK1R or truncated NK1R on intracellular calcium levels is shown.
Activation of NK1R and Threonine Phosphorylation of PKCδ.
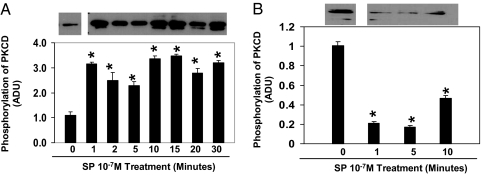
In cells that express full-length NK1R, SP potently induced threonine-505 phosphorylation of PKCδ (Thr-505) by 1 min, and this activation remained through 30 min of incubation (Fig. 2A). In contrast, SP reduced threonine phosphorylation of PKCδ (Thr-505) in human embryonic kidney (HEK293) cells that express truncated NK1R (Fig. 2B).
Fig. 2.
Time course of SP-induced differential effects on threonine phosphorylation of PKCδ in HEK293 cells expressing full-length NK1R or truncated NK1R. The HEK293 cells that express full-length NK1R (A) or truncated NK1R (B) were treated with SP at 10−7 M for different time periods as indicated in the figure or untreated as controls (0 min). Phospho-PKCδ (Thr-505) was detected by anti-PKCδ (Thr-505) antibody (representative Western blot analysis from three separate experiments). Densitometry analysis of SP-induced threonine phosphorylation is shown. Values are expressed as mean ± SD (n = 3) and are expressed in arbitrary densitometry units (ADU). *, P < 0.05 untreated control cells vs. SP-treated HEK293 cells expressing full-length NK1R or truncated NK1R.
Activation of NK1R and Phosphorylation of ERK1/2.
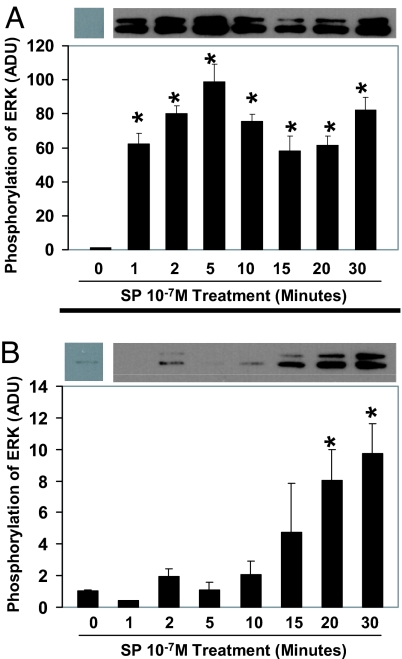
In cells that express full-length NK1R, SP potently and rapidly activated ERK1/2 as early as 1 min, and this activation remained through 30 min of incubation (Fig. 3A). In the cells that express truncated NK1R, however, the kinetics of the SP-induced phosphorylation of ERK1/2 differed from that observed in the cells expressing full-length NK1R. The activation was slower for truncated NK1R and peaked at 20–30 min in response to SP (Fig. 3B).
Fig. 3.
Time course of SP-induced phosphorylation of ERK1/2 in HEK293 cells expressing full-length NK1R or truncated NK1R. The HEK293 cells that express full-length NK1R (A) or truncated NK1R (B) were treated with SP at 10−7 M for different time intervals as indicated in the figure or untreated as controls (0 min). Phospho-ERK1/2 was detected by anti-phospho-ERK1/2 antibody. These results are representative of three independent Western blot analyses. Densitometry analysis of phosphorylation of ERK1/2 in the presence or absence of 10−7 M SP is shown. Values are expressed as mean ± SD (n = 3) and are expressed in arbitrary densitometry units (ADU). *, P < 0.05 untreated control cells vs. SP-treated HEK293 cells expressing full-length NK1R or truncated NK1R.
Carboxyl Terminus of NK1R Is the Structural Basis for SP to Activate NF-κB.
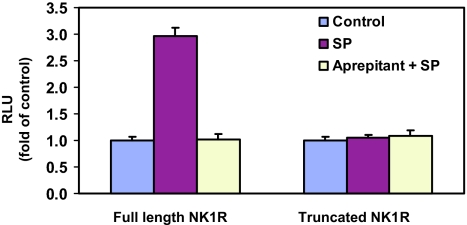
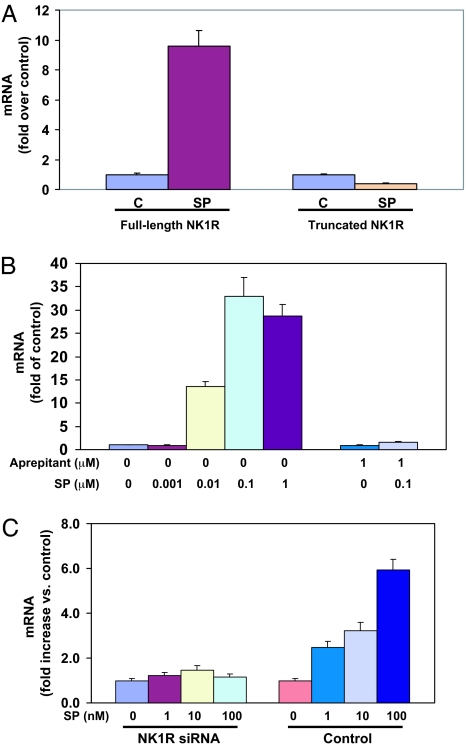
SP enhanced NF-κB-driven luciferase gene expression in the cells coexpressing NF-κB-luc and full-length NK1R (Fig. 4). This enhancement was antagonized by pretreatment of the cells with the NK1R antagonist aprepitant (10−6 M) (Fig. 4). In contrast, SP had no effect on NF-κB-driven luciferase gene expression in the cells that coexpress NF-κB-luc and truncated NK1R (Fig. 4). Therefore, the NK1R carboxyl terminus is required for SP to activate NF-κB.
Fig. 4.
Differential effect of SP on stimulation of NF-κB in HEK293 cells coexpressing NF-κB-luc and full-length NK1R or truncated NK1R. HEK293 cells that coexpress NF-κB-luc and full-length NK1R or truncated NK1R were preincubated with or without 10−6 M aprepitant for 30 min followed by stimulation with 10−7 M SP for 6 h. Untreated cells were used as controls. The results are presented as relative fold of RLU of the untreated controls, which are defined as 1.0.
NK1R Carboxyl Terminus Is Required for SP-Induced IL-8 mRNA Expression.
SP potently induced the expression of IL-8 mRNA in the cells that express full-length NK1R (Fig. 5A), but the expression level of IL-8 mRNA was slightly reduced by SP treatment in the cells that express truncated NK1R (Fig. 5A). We validated that the expression of IL-8 mRNA is an indicator of NF-κB activation and investigated the dose response and specificity of SP to stimulate the expression of IL-8 mRNA in the cells that express full-length NK1R. SP increased the expression of IL-8 mRNA in a dose-dependent manner (from 10−9 to 10−6 M SP) (Fig. 5B), with the peak at 10 −7 M SP. Increased IL-8 expression was mediated by NK1R, and pretreatment of the cells with the NK1R antagonist aprepitant abrogated the SP-induced effect (Fig. 5B). Consistent with the reduced expression of full-length NK1R by NK1R siRNA, the ability of SP to increase the expression of IL-8 mRNA was significantly decreased (Fig. 5C).
Fig. 5.
Full-length NK1R is required for SP-induced expression of IL-8 in HEK293 cells. (A) Differential effect of SP on the expression of IL-8 mRNA in cells expressing full-length NK1R or truncated NK1R. HEK293 cells expressing full-length NK1R or truncated NK1R were incubated in the presence or absence (as control) of 10−7 M SP for 3 h. The levels of IL-8 mRNA are presented as fold of untreated control, which is defined as 1.0. The data are averaged from three independent experiments. C, untreated controls. (B) SP-induced dose-dependent increases of IL-8 mRNA in HEK293 cells expressing full-length NK1R. HEK293 cells that express full-length NK1R were preincubated in the presence or absence of 1 μM aprepitant for 30 min followed by treatment with different concentrations of SP as indicated for 3 h. The untreated sample was used as the control. Aprepitant and SP-treated sample was used as specificity control. The expression levels of IL-8 mRNA are presented as fold of untreated control, which is defined as 1.0. (C) Reduction of NK1R expression by siRNA inhibits SP-stimulated expression of IL-8 mRNA in HEK293 cells expressing full-length NK1R. Forty-eight hours after siRNA or control siRNA transfection, the HEK293 cells were incubated in the presence or absence (as controls) of 1–100 nM SP for 3 h. The expression levels of IL-8 mNA are presented as fold of the untreated control, which is defined as 1.0.
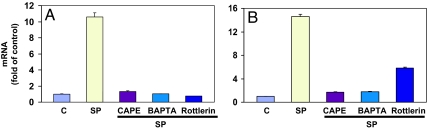
The HEK293 cells that express full-length NK1R were pretreated with CAPE (20 μM), a specific NF-κB inhibitor, or BAPTA/AM (20 μM), an intracellular calcium chelator, or rottlerin (10 μM), a PKCδ-specific inhibitor, for 40 min at 37°C and then treated with 10−7 M SP for an additional 3 h. Inhibition of NF-κB or the calcium signal almost completely inhibited SP-induced expression of IL-8 mRNA expression, and rottlerin strongly prevented SP induction of IL-8 mRNA expression (Fig. 6A). To establish this finding further, the human astrocytoma cell line U373MG that expresses endogenous full-length NK1R (10) was studied. Patterns similar to HEK293 were observed: inhibition of NF-κB or chelation of calcium inhibited SP-induced expression of IL-8 mRNA; rottlerin, a PKCδ inhibitor, dramatically attenuated the expression of SP-induced IL-8 mRNA (Fig. 6B). NF-κB, calcium, and PKCδ are required for SP-mediated IL-8 synthesis.
Fig. 6.
NF-κB, calcium, and PKCδ are required for SP-induced expression of IL-8 mRNA. HEK293 cells (A) that express full-length NK1R and U373MG cells (B) that express endogenous full-length NK1R were pretreated with an NF-κB inhibitor, CAPE (20 μM) or a calcium chelator, BAPTA/AM (20 μM), or a PKCδ inhibitor, rottlerin (10 μM), or untreated as control for 30 min followed by 10−7 M SP stimulation for 3 h. C, untreated control. Expression levels of IL-8 mRNA are represented as fold of untreated control, which is defined as 1.0. These results are representative of three independent experiments.
Discussion
The data reported in this work demonstrated that the NK1R carboxyl terminus is the structural basis for the SP-induced intracellular calcium increase and the activation of NF-κB and hence, the increase of IL-8 mRNA expression. The carboxyl terminus of NK1R is also required for SP treatment to activate PKC-δ. SP activation of truncated NK1R (Δ311-NK1R) has different effects on the phosphorylation of PKC-δ and ERK1/2 and on the expression levels of IL-8 mRNA, but it did not trigger an intracellular calcium increase or activate NF-κB (see Table 1).
Table 1.
Differences in the length of the carboxyl terminus mediate functional properties of NK1R
| Effect | Full-length NK1R | Truncated NK1R |
|---|---|---|
| Length of NK1R | 407 aa | 311 aa, no carboxyl terminus |
| Calcium mobilization | Yes | No |
| PKCδ phosphorylation | Increased | Decreased |
| Activation of ERK | Fast (<1 min) and sustained | Slow (peaked at 20–30 min) |
| Activation of NF-κB | Yes | No |
| IL-8 mRNA expression | Increased | Decreased |
Increasing [Ca2+]i is a mechanism involved in complex intracellular signaling events, for example, the activation of NF-κB, which is mediated by many GPCRs, including NK1R (11). Our data demonstrated that the NK1R carboxyl terminus is required for SP-induced intracellular calcium increases in HEK293 cells (Fig. 1). This difference in SP-induced intracellular calcium increase resulting from the full-length NK1R and truncated NK1R is directly related to the NK1R carboxyl terminus. The truncation of the NK1R carboxyl terminus could theoretically impair the ability of the G protein (Gαq) to interact properly with the receptor, thus resulting in impaired capacity to induce calcium mobilization.
NF-κB is a ubiquitous transcription factor that governs the expression of genes involved in cell-to-cell communication, cell-to-cell interaction, cell cycle, and cell growth regulation (12). SP-NK1R interaction stimulates PKCδ and activates NF-κB, resulting in up-regulation of IL-8 expression (10, 11, 13–17). SP induces IL-8 expression via NK1R in different cell systems, including mast cells (18), NCM 460 cells, a nontransformed human colonic epithelial cell line (13, 14, 19), human mesenteric preadipocytes (15), human corneal epithelial cells (20), human astrocytoma cell lines (10, 21, 22), but not in human blood mononuclear cells (22), which express only the truncated NK1R (6). The SP activation of NF-κB requires mobilization of calcium (Gαq signal) (10, 11).
SP stimulated expression of IL-8 mRNA in cells expressing full-length NK1R but decreased the expression of the IL-8 gene in cells expressing the truncated NK1R as determined by real-time PCR assay. SP decreased the expression of IL-8 through truncated NK1R, which suggests that SP activation of truncated NK1R results in an inhibitory effect on NF-κB activity. A possible interpretation of this observation is that the third intracellular loop of the truncated NK1R interacts with β-arrestin2 (23), which results in stabilization of IKBα protein, thus inhibiting the expression of NF-κB-targeted genes, including IL-8 (24).
In nontransformed human colonic epithelial NCM 460 cells overexpressing NK-1R (full-length NK1R), SP rapidly induces threonine phosphorylation of PKCδ, and this activation is critical for SP-induced NF-κB activation and IL-8 gene expression (14). Our work demonstrates that the activation of truncated NK1R affects threonine phosphorylation of PKCδ differently than activation of the full-length NK1R. There are timing differences in the activation of ERK1/2 between full-length NK1R and truncated NK1R. SP activation of full-length NK1R induces phosphorylation of ERK1/2 through stimulating the formation of a scaffolding complex comprising the internalized receptor, β-arrestin, src, and ERK1/2 (25). Truncated NK1R does not have the carboxyl terminus but does have the third intracellular loop that can interact with β-arrestin2 (23). β-arrestin2 plays an important role in angiotensin II type 1A-receptor-mediated and G protein-independent activation of ERK1/2. This β-arrestin2 (G protein-independent)-mediated activation of ERK1/2 is slower compared with that mediated through G protein (26). The slower or delayed activation of ERK1/2 has been reported in cells expressing an experimentally truncated (Δ325) NK1R (25). The activation of truncated NK1R may lead to activation of ERK1/2 via the interaction of the third intracellular loop and β-arrestin2, resulting in the different timing pattern of ERK1/2 activation.
Compared with full-length NK1R, activation of the truncated NK1R had different effects on phosphorylation of PKCδ and ERK1/2, regulation of IL-8 mRNA expression, which may reflect a preference of truncated NK1R for a different G protein, an interaction with other GPCRs, or G protein-independent signaling pathways. NK1R couples to Gα(q), (s), and (o) (27), whereas the truncation of the NK1R carboxyl terminus may change or impair its coupling capacity to one or more of the G proteins (28) by inducing conformational constraints. This change of coupling to G protein could also change the interaction of G protein with effector systems, resulting in an effect that is different from that of the full-length NK1R. Truncated NK1R may also interact with other GPCR in other cell functions. We have demonstrated, for example, that SP activation of the natural endogenous truncated NK1R primes CCL5-mediated calcium increases in undifferentiated THP-1 cells that express only the truncated NK1R (29), indicating an independent downstream signaling pathway for truncated NK1R. Activation of the truncated NK1R may also transduce its signals through G protein-independent mechanisms as demonstrated by Ahn et al. (26) in cells that express angiotensin II type 1A receptor.
Truncated receptor forms participate in regulatory signals and signal transduction. A full-length form of follicle-stimulating hormone (FSH) receptor couples to the Gαs and is a GPCR; in contrast, a truncated form of FSH receptor (generated by exclusion of exons 9 and 10 and use of additional exon 11) belongs to the superfamily of growth factor receptors (30). The unique absence of the carboxyl terminus occurs in the gonadotropin-releasing hormone receptors (GnRH) during evolution of the vertebrate lineage from sea lamprey to more complex vertebrates. The truncated tailless GnRH receptor is functional and capable of activating the inositol phosphate signaling system (31), the ERK system (32), and of affecting ERK compartmentalization probably mediated by scaffolds other than arrestins (33). Truncated forms of receptors also possess downstream pathways of signal transduction that may or may not be involved in full-length receptor signal transduction (34). In undifferentiated THP-1 cells that express only the truncated NK1R, SP does not trigger calcium increases in these cells but primes CCL5-mediated calcium increases, indicating that the truncated NK1R has the capacity to interact with other GPCR and has its own unique downstream signal pathway(s) (29).
Materials and Methods
Cells.
HEK293 cells and the human astrocytoma cell line U373MG were purchased from American Type Culture Collection and grown in DMEM supplemented with 10% FCS, glutamine, and antibiotics at 37°C in 5% CO2. Cells were plated for SP treatment in the presence or absence of SP and/or inhibitors as described in the figure legends. Medium was changed 1 day before SP treatment for RNA extraction.
Reagents.
Full-length NK1R plasmid was a gift from Norma Gerard, Harvard University, Boston, MA; the truncated NK1R (Δ311) cDNA3.1Hygro(+) plasmids were constructed from Lenti6/V5-D-Topo NK1R plasmids (19), which contain full-length wild-type NK1R. Using the lentiviral plasmids as templates, we introduced a premature stop codon at position 312 of the full-length NK1R by using site-directed mutagenesis [forward primer, 5-GCCTCAATGACAGGTGACGTCTGGGCTTCAAGC-3′; reverse primer, 5′-GCTTGAAGCCCAGACGTCACCTGTCATTGAGGC-3′ (QuikChange site-directed mutagenesis kit, Stratagene)]. The truncated NK1R sequence was then extracted from the modified lentiviral plasmids by using endonuclease digestion and ligated into pcDNA3.1Hygro(+) plasmid (Invitrogen). Truncation and proper orientation were confirmed by sequencing in both the lentiviral and pcDNA vectors (Harvard Core Facility, Boston MA). The pNF-κB-luc multimer-driven luciferase construct (35) was generously provided by Randy Q. Cron (Children's Hospital of Philadelphia) (35). The luciferase assay system and reporter lysis buffer were obtained from Promega. Nucleofector II and the Cell Line Nucleofector kit V were purchased from Amaxa.
Antibodies.
Phospho-PKCδ (Thr-505) and phospho-ERK1/2 antibodies were obtained from Cell Signaling. Goat anti-rabbit IgG-horseradish peroxidase (HRP) and SuperSignal West Pico chemiluminescent substrate were purchased from Pierce. SP was purchased from Sigma. The NK1R antagonist, aprepitant (Emend), manufactured by Merck, was purchased through the Children's Hospital of Philadelphia pharmacy and was extracted and purified by chromatography.
Establishment of Stable NK1R Expression in HEK293 Cells.
NK1R plasmids (full-length or truncated) were introduced into HEK293 cells by using Nucleofector II and the Cell Line Nucleofector kit V according to the manufacturer's instructions. Transfected cells were selected with antibiotic (200 μg/ml hygromycin was used for selection of the cells that express truncated NK1R; 1 mg/ml G418 for selection of the cells that express full-length NK1R) for 4–6 weeks. The antibiotic-resistant cells were further selected for by limited dilution in 96-well plates for another 3–5 weeks. The antibiotic-resistant clones were selected for further characterization by real-time RT-PCR quantitation of NK1R mRNA and by flow cytometry staining of the surface expression of NK1R.
The expression levels of NK1R mRNA in HEK293 cells that express full-length NK1R and truncated NK1R were similar (3.45 ± 0.11 × 105 copies per μg of RNA vs. 3.87 ± 0.07 × 105 copies per μg of RNA). By using flow cytometry, cell surface expression of NK1R in these cell lines was confirmed (data not shown).
SP-Induced Calcium Mobilization in HEK293 Cells.
Intracellular calcium measurements were performed in fura-2-loaded cells by using Tsien's ratiometric method (36) as described in ref. 29. Peak intracellular calcium increases were determined and used to construct concentration–response curves. Logistic curves were fitted to data and used to derive EC50 values for SP.
Western Blot Assays.
The cells that express full-length NK1R or truncated NK1R were treated with 10 −7 M SP for various time (see figure legends) or untreated as control. The cells were lysed in RIPA buffer (Sigma) containing proteinase inhibitors and phosphatase inhibitor mixture 1 and 2 (Sigma). Equal amounts (20 μg) of protein isolated from each sample were loaded and separated on 4–12% precast gels (Invitrogen) and transferred onto a nitrocellulose membrane. The membranes were probed with the primary antibody (1 μg/ml) directed against phosphorylated PKCδ (Thr-505) or phospho-ERK1/2 followed by a HRP-conjugated secondary antibody. The signals were detected by using an enhanced chemiluminescence detection system.
RNA Extraction.
Total RNA was extracted from cells that express either full-length NK1R or truncated NK1R by using a RNeasy kit (Qiagen), and the potential DNA contamination was eliminated by on-column DNase digestion as instructed by the manufacturer (Qiagen). RNA concentration was determined by Qubit fluorometer with a Quant-iT RNA assay kit (Invitrogen).
Reverse Transcription.
Total RNA (1 μg) was subjected to reverse transcription by using an AffinityScript qPCR cDNA synthesis kit (Stratagene) with random primer as instructed by the manufacturer. Reverse transcriptase negative controls were used to control for genomic DNA contamination. One-tenth (2 μl) of the resulting cDNA was used as a template for real-time PCR amplification.
Real-Time PCR Assay.
The primers and probe sequences of NK1R and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were reported (37, 38). The sequences of IL-8 primer pair were: sense, 5′-CTCTTGGCAGCCTTCCTGATTTCT-3′; antisense, 5′-TGTGGTCCACTCTCAATCACTCTC-3′. The levels of NK1R and IL-8 mRNA expression were determined by using real-time RT-PCR as described (37, 38). The specificity of the primer pair for IL-8 was confirmed by SYBR Green dye dissociation curve and agarose gel electrophoresis in our preliminary experiments. The magnitude of change of IL-8 mRNA expression in SP-treated compared with untreated control cells was calculated by using the standard 2−(ΔΔCt) method (39).
Luciferase Assay for NF-κB Activity.
pNF-κB-luc plasmid (5 μg) was transfected into HEK293 cells that express full-length NK1R or truncated NK1R (2 × 106 cells per transfection) by using Nucleofector II and Cell Line Nucleofector kit V following the manufacturer's instructions. Forty-eight hours after transfection, the cells were treated with 10−7 M SP in the presence or absence of 10−6 M aprepitant for 6 h. Untreated cells were used as controls. Cell lysates were collected in reporter lysis buffer and centrifuged at 10,000 × g for 1 min. The cell-free lysates were used to determine the NF-κB-driven luciferase activity by the luciferase assay system and a luminometer. The luciferase activity is expressed as relative light units (RLU). The results are presented as relative fold of RLU of the untreated controls, which are defined as 1.0.
Acknowledgments.
We acknowledge Donald E. Campbell, Ph.D., and the Stokes Flow Cytometry Core, and we thank Dr. Norma Gerard, Harvard University, for her gift of an NK1R full-length plasmid. We also thank Stephen Jasionowski, Joshua Taton, and Mary Calderone for editorial assistance. This work was supported by National Institutes of Health Grants P01-MH076388 and R01-MH049981 (to S.D.D.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Chang MM, Leeman SE. Isolation of a sialogogic peptide from bovine hypothalamic tissue and its characterization as Substance P. J Biol Chem. 1970;245:4784–4790. [PubMed] [Google Scholar]
- 2.Fong TM, Anderson SA, Yu H, Huang RR, Strader CD. Differential activation of intracellular effector by two isoforms of human neurokinin-1 receptor. Mol Pharmacol. 1992;41:24–30. [PubMed] [Google Scholar]
- 3.Gerard NP, et al. Human Substance P receptor (NK-1): Organization of the gene, chromosome localization, and functional expression of cDNA clones. Biochemistry. 1991;30:10640–10646. doi: 10.1021/bi00108a006. [DOI] [PubMed] [Google Scholar]
- 4.Hopkins B, Powell SJ, Danks P, Briggs I, Graham A. Isolation and characterisation of the human lung NK-1 receptor cDNA. Biochem Biophys Res Commun. 1991;180:1110–1117. doi: 10.1016/s0006-291x(05)81181-7. [DOI] [PubMed] [Google Scholar]
- 5.Takeda Y, Chou KB, Takeda J, Sachais BS, Krause JE. Molecular cloning, structural characterization and functional expression of the human Substance P receptor. Bio-chem Biophys Res Commun. 1991;179:1232–1240. doi: 10.1016/0006-291x(91)91704-g. [DOI] [PubMed] [Google Scholar]
- 6.Caberlotto L, et al. Neurokinin 1 receptor and relative abundance of the short and long isoforms in the human brain. Eur J Neurosci. 2003;17:1736–1746. doi: 10.1046/j.1460-9568.2003.02600.x. [DOI] [PubMed] [Google Scholar]
- 7.Kage R, Leeman SE, Boyd ND. Biochemical characterization of two different forms of the Substance P receptor in rat submaxillary gland. J Neurochem. 1993;60:347–351. doi: 10.1111/j.1471-4159.1993.tb05857.x. [DOI] [PubMed] [Google Scholar]
- 8.Baker SJ, Morris JL, Gibbins IL. Cloning of a C-terminally truncated NK-1 receptor from guinea pig nervous system. Brain Res Mol Brain Res. 2003;111:136–147. doi: 10.1016/s0169-328x(03)00002-0. [DOI] [PubMed] [Google Scholar]
- 9.Sasakawa N, Sharif M, Hanley MR. Attenuation of agonist-induced desensitization of the rat Substance P receptor by progressive truncation of the C terminus. FEBS Lett. 1994;347:181–184. doi: 10.1016/0014-5793(94)00532-x. [DOI] [PubMed] [Google Scholar]
- 10.Lieb K, Fiebich BL, Berger M, Bauer J, Schulze-Osthoff K. The neuropeptide Substance P activates transcription factor NF-κB and κB-dependent gene expression in human astrocytoma cells. J Immunol. 1997;159:4952–4958. [PubMed] [Google Scholar]
- 11.Williams R, Zou X, Hoyle GW. Tachykinin-1 receptor stimulates proinflammatory gene expression in lung epithelial cells through activation of NF-κB via a G(q)-dependent pathway. Am J Physiol. 2007;292:L430–L437. doi: 10.1152/ajplung.00475.2005. [DOI] [PubMed] [Google Scholar]
- 12.Chen F, Castranova V, Shi X, Demers LM. New insights into the role of nuclear factor-κB, a ubiquitous transcription factor in the initiation of diseases. Clin Chem. 1999;45:7–17. [PubMed] [Google Scholar]
- 13.Zhao D, et al. Substance P-stimulated interleukin-8 expression in human colonic epithelial cells involves Rho family small GTPases. Biochem J. 2002;368:665–672. doi: 10.1042/BJ20020950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koon HW, et al. Substance P-stimulated interleukin-8 expression in human colonic epithelial cells involves protein kinase Cδ activation. J Pharmacol Exp Ther. 2005;314:1393–1400. doi: 10.1124/jpet.105.088013. [DOI] [PubMed] [Google Scholar]
- 15.Karagiannides I, et al. Induction of colitis causes inflammatory responses in fat depots: Evidence for Substance P pathways in human mesenteric preadipocytes. Proc Natl Acad Sci USA. 2006;103:5207–5212. doi: 10.1073/pnas.0600821103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Page K, et al. Regulation of airway epithelial cell NF-κB-dependent gene expression by protein kinase Cδ. J Immunol. 2003;170:5681–5689. doi: 10.4049/jimmunol.170.11.5681. [DOI] [PubMed] [Google Scholar]
- 17.Yamaguchi K, Richardson MD, Bigner DD, Kwatra MM. Signal transduction through Substance P receptor in human glioblastoma cells: Roles for Src and PKCδ. Cancer Chemother Pharmacol. 2005;56:585–593. doi: 10.1007/s00280-005-1030-3. [DOI] [PubMed] [Google Scholar]
- 18.Kulka M, Sheen CH, Tancowny BP, Grammer LC, Schleimer RP. Neuropeptides activate human mast cell degranulation and chemokine production. Immunology. 2008;123:398–410. doi: 10.1111/j.1365-2567.2007.02705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tansky MF, Pothoulakis C, Leeman SE. Functional consequences of alteration of N-linked glycosylation sites on the neurokinin 1 receptor. Proc Natl Acad Sci USA. 2007;104:10691–10696. doi: 10.1073/pnas.0703394104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tran MT, Lausch RN, Oakes JE. Substance P differentially stimulates IL-8 synthesis in human corneal epithelial cells. Invest Ophthalmol Visual Sci. 2000;41:3871–3877. [PubMed] [Google Scholar]
- 21.Palma C, Manzini S. Substance P induces secretion of immunomodulatory cytokines by human astrocytoma cells. J Neuroimmunol. 1998;81:127–137. doi: 10.1016/s0165-5728(97)00167-7. [DOI] [PubMed] [Google Scholar]
- 22.Derocq JM, et al. Effect of Substance P on cytokine production by human astrocytic cells and blood mononuclear cells: Characterization of novel tachykinin receptor antagonists. FEBS Lett. 1996;399:321–325. doi: 10.1016/s0014-5793(96)01346-4. [DOI] [PubMed] [Google Scholar]
- 23.Schmidlin F, Roosterman D, Bunnett NW. The third intracellular loop and carboxyl tail of neurokinin 1 and 3 receptors determine interactions with β-arrestins. Am J Physiol. 2003;285:C945–C958. doi: 10.1152/ajpcell.00541.2002. [DOI] [PubMed] [Google Scholar]
- 24.Chen F. Arresting NF-κB by β-arrestin2. Cell Death Differ. 2004;11:1155–1156. doi: 10.1038/sj.cdd.4401496. [DOI] [PubMed] [Google Scholar]
- 25.DeFea KA, et al. The proliferative and antiapoptotic effects of Substance P are facilitated by formation of a β-arrestin-dependent scaffolding complex. Proc Natl Acad Sci USA. 2000;97:11086–11091. doi: 10.1073/pnas.190276697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahn S, Shenoy SK, Wei H, Lefkowitz RJ. Differential kinetic and spatial patterns of β-arrestin and G protein-mediated ERK activation by the angiotensin II receptor. J Biol Chem. 2004;279:35518–35525. doi: 10.1074/jbc.M405878200. [DOI] [PubMed] [Google Scholar]
- 27.Roush ED, Kwatra MM. Human Substance P receptor expressed in Chinese hamster ovary cells directly activates Gαq/11, Gαs, Gαo. FEBS Lett. 1998;428:291–294. doi: 10.1016/s0014-5793(98)00553-5. [DOI] [PubMed] [Google Scholar]
- 28.Holst B, Hastrup H, Raffetseder U, Martini L, Schwartz TW. Two active molecular phenotypes of the tachykinin NK1 receptor revealed by G protein fusions and mutagenesis. J Biol Chem. 2001;276:19793–19799. doi: 10.1074/jbc.M100621200. [DOI] [PubMed] [Google Scholar]
- 29.Lai JP, et al. Full-length and truncated neurokinin-1 receptor expression and function during monocyte/macrophage differentiation. Proc Natl Acad Sci USA. 2006;103:7771–7776. doi: 10.1073/pnas.0602563103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Babu PS, Krishnamurthy H, Chedrese PJ, Sairam MR. Activation of extracellular-regulated kinase pathways in ovarian granulosa cells by the novel growth factor type 1 follicle-stimulating hormone receptor: Role in hormone signaling and cell proliferation. J Biol Chem. 2000;275:27615–27626. doi: 10.1074/jbc.M003206200. [DOI] [PubMed] [Google Scholar]
- 31.Silver MR, Nucci NV, Root AR, Reed KL, Sower SA. Cloning and characterization of a functional type II gonadotropin-releasing hormone receptor with a lengthy carboxyl-terminal tail from an ancestral vertebrate, the sea lamprey. Endocrinology. 2005;146:3351–3361. doi: 10.1210/en.2005-0305. [DOI] [PubMed] [Google Scholar]
- 32.Lin X, Conn PM. Transcriptional activation of gonadotropin-releasing hormone (GnRH) receptor gene by GnRH: Involvement of multiple signal transduction pathways. Endocrinology. 1999;140:358–364. doi: 10.1210/endo.140.1.6452. [DOI] [PubMed] [Google Scholar]
- 33.Caunt CJ, et al. Arrestin-mediated ERK activation by gonadotropin-releasing hormone receptors: Receptor-specific activation mechanisms and compartmentalization. J Biol Chem. 2006;281:2701–2710. doi: 10.1074/jbc.M507242200. [DOI] [PubMed] [Google Scholar]
- 34.Smirnova OV, Bogorad RL. Short forms of membrane receptors: Generation and role in hormonal signal transduction. Biochemistry (Mosc) 2004;69:351–363. doi: 10.1023/b:biry.0000026189.92397.19. [DOI] [PubMed] [Google Scholar]
- 35.Petrak D, Memon SA, Birrer MJ, Ashwell JD, Zacharchuk CM. Dominant negative mutant of c-Jun inhibits NF-AT transcriptional activity and prevents IL-2 gene transcription. J Immunol. 1994;153:2046–2051. [PubMed] [Google Scholar]
- 36.Grynkiewicz G, Poenie M, Tsien RY. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J Biol Chem. 1985;260:3440–3450. [PubMed] [Google Scholar]
- 37.Lai JP, Douglas SD, Wang YJ, Ho WZ. Real-time reverse transcription-PCR quantitation of Substance P receptor (NK-1R) mRNA. Clin Diagn Lab Immunol. 2005;12:537–541. doi: 10.1128/CDLI.12.4.537-541.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lai JP, et al. Quantification of CCR5 mRNA in human lymphocytes and macrophages by real-time reverse transcriptase PCR assay. Clin Diag Lab Immunol. 2003;10:1123–1128. doi: 10.1128/CDLI.10.6.1123-1128.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]