SopB promotes phosphatidylinositol 3-phosphate formation on Salmonella vacuoles by recruiting Rab5 and Vps34 (original) (raw)
Abstract
Salmonella colonizes a vacuolar niche in host cells during infection. Maturation of the _Salmonella_-containing vacuole (SCV) involves the formation of phosphatidylinositol 3-phosphate (PI(3)P) on its outer leaflet. SopB, a bacterial virulence factor with phosphoinositide phosphatase activity, was proposed to generate PI(3)P by dephosphorylating PI(3,4)P2, PI(3,5)P2, and PI(3,4,5)P3. Here, we examine the mechanism of PI(3)P formation during Salmonella infection. SopB is required to form PI(3,4)P2/PI(3,4,5)P3 at invasion ruffles and PI(3)P on nascent SCVs. However, we uncouple these events experimentally and reveal that SopB does not dephosphorylate PI(3,4)P2/PI(3,4,5)P3 to produce PI(3)P. Instead, the phosphatase activity of SopB is required for Rab5 recruitment to the SCV. Vps34, a PI3-kinase that associates with active Rab5, is responsible for PI(3)P formation on SCVs. Therefore, SopB mediates PI(3)P production on the SCV indirectly through recruitment of Rab5 and its effector Vps34. These findings reveal a link between phosphoinositide phosphatase activity and the recruitment of Rab5 to phagosomes.
Introduction
Salmonella enterica serovar typhimurium (S. Typhimurium) is a facultative intracellular pathogen that can cause disease in a variety of hosts (Tsolis et al., 1999). These bacteria have the ability to invade host cells and grow intracellularly (Knodler and Steele-Mortimer, 2003). S. typhimurium establish a niche in host cells known as _Salmonella_-containing vacuoles (SCVs). The ability of S. typhimurium to modulate SCV trafficking is essential for its virulence, but the underlying molecular mechanisms are incompletely understood (Brumell and Grinstein, 2004).
S. typhimurium manipulates many aspects of host cell physiology through a special class of virulence factors known as effectors. These proteins are translocated directly into host cells via type III secretion systems (T3SSs), needle-like apparati on the bacterial surface (Galan and Wolf-Watz, 2006). S. typhimurium is endowed with two separate T3SSs encoded on Salmonella pathogenicity island (SPI)-1 and SPI-2 of the bacterial chromosome (Schlumberger and Hardt, 2006). The SPI-1–encoded T3SS delivers effectors across the host cell plasma membrane during invasion. These initiate actin rearrangements by activating Rho family GTPases or by directly interacting with actin (Schlumberger and Hardt, 2006). Modulation of phosphoinositide signaling also plays a key role in S. typhimurium invasion (Drecktrah et al., 2004; Hilbi, 2006). Expression of the SPI-2 T3SS is stimulated several hours after invasion and allows intracellular survival and growth of the bacteria (Schlumberger and Hardt, 2006).
In addition to its role in bacterial entry, recent studies suggest that the SPI-1 T3SS also dictates the early intracellular traffic of the SCV. Accordingly, the intracellular growth of an SPI-1 T3SS–defective mutant was found to be impaired (Steele-Mortimer et al., 2002). Among the SPI-1 effectors, SopB (also called SigD) was found to play a role in SCV maturation (Hernandez et al., 2004). SopB was shown to be required for the generation of phosphatidylinositol (PI) 3-phosphate (PI(3)P) on the SCV and to promote lysosomal-associated membrane protein-1 (LAMP-1) acquisition by this compartment (Hernandez et al., 2004). A sopB deletion mutant had attenuated intracellular growth, implying that its role in SCV maturation is critical for the establishment of a replicative niche in host cells (Hernandez et al., 2004).
In vitro SopB can hydrolyze a variety of inositol phosphates and phosphoinositides, including the PI3-kinase products PI(3,4)P2, PI(3,5)P2, and PI(3,4,5)P3 (Norris et al., 1998; Marcus et al., 2001). However, the in vivo substrates of SopB remain unclear. Hernandez et al. (2004) have proposed that SopB generates PI(3)P on the SCV in a direct manner via the dephosphorylation of PI(3,4)P2 and PI(3,4,5)P3. These authors also proposed that the PI(3,5)P2 phosphatase activity of SopB may affect the progression of SCVs down the endocytic pathway, preventing their fusion with lysosomes (Hernandez et al., 2004). This model for the SopB-mediated generation of PI(3)P is in good accord with the broad in vitro substrate specificity reported for the phosphatase. However, other observations apparently conflict with such a model. First, SopB is required for the activation of Akt in S. _typhimurium_–infected cells (Steele-Mortimer et al., 2000). Akt is recruited to the membrane and activated, at least in part, by PI(3,4)P2 and PI(3,4,5)P3, products of class I and II PI3-kinases (Dummler and Hemmings, 2007). Dephosphorylation of PI3-kinase products should therefore inhibit Akt activation rather than promote it. Second, otherwise unstimulated cells have very low levels of PI(3,4)P2 and PI(3,4,5)P3, and recent studies suggest that the main in vivo substrate of SopB is the much more abundant PI(4,5)P2 (Terebiznik et al., 2002; Mason et al., 2007). Thus, despite clear evidence that SopB contributes to S. typhimurium invasion and early SCV trafficking, the mechanisms by which it acts are not completely understood. Here, we examine in more detail the mechanism of PI(3)P formation by SopB.
Results
SopB is required for PI(3)P localization to SCVs and an elevation of cellular PI phosphate (PIP)
To investigate the localization of 3′-phosphorylated phosphoinositides generated during S. typhimurium infection, we used chimeras consisting of GFP or RFP fused to the phosphoinositide-binding domains of various proteins. These included the pleckstrin homology (PH) domain of Akt, which binds PI(3,4)P2 and PI(3,4,5)P3 (Rong et al., 2001), and a construct with two tandem FYVE domains from the early endosome autoantigen 1, which binds PI(3)P (Vieira et al., 2001). HeLa cells expressing the different phosphoinositide probes were infected with either wild type (WT) or an isogenic sopb deletion mutant of S. typhimurium. Changes in phosphoinositide metabolism during infection were examined by time-lapse confocal fluorescence microscopy of live cells.
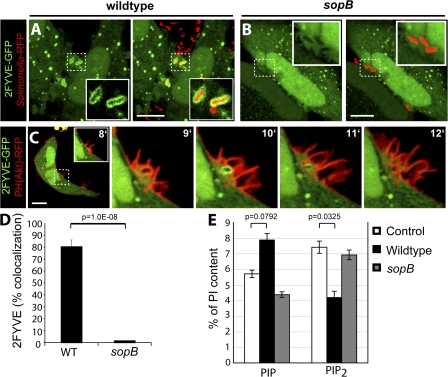
The PI(3)P-specific 2FYVE-GFP probe labeled SCVs containing WT S. typhimurium (Fig. 1 A and Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200804131/DC1) immediately after bacterial entry, which is consistent with previous findings (Pattni et al., 2001; Scott et al., 2002; Hernandez et al., 2004). PI(3)P persisted on the SCV for ∼1–5 min. However, the 2FYVE-GFP probe was not detected in the ruffling areas of the plasma membrane where invasion occurred. Cells infected with the sopB mutant did not display recruitment of 2FYVE-GFP to the SCV at any time (Fig. 1, B and D; and Video 2). Similar results were seen with phox homology (PX)–GFP, a different probe for PI(3)P (Scott et al., 2002; unpublished data). To simultaneously examine the production of PI(3)P and of its putative precursors, PI(3,4)P2 and/or PI(3,4,5)P3, during invasion, cells were cotransfected with 2FYVE-GFP and PH(Akt)-RFP. As shown in Fig. 1 C, PH(Akt)-RFP was recruited exclusively to invasion ruffles that project from the plasma membrane at the site of bacterial entry during infection but not to nascent SCVs. Conversely, 2FYVE-GFP was found only on newly formed vacuoles where PH(Akt)-RFP was absent. Analysis of the dynamics of both signaling events, which is presented in Video 3, revealed that PI(3,4)P2 and/or PI(3,4,5)P3 was present at invasion ruffles before PI(3)P became apparent at the SCV. Jointly, these findings are consistent with the notion introduced by Hernandez et al. (2004) that PI(3)P accumulation on SCVs derives from the dephosphorylation of PI(3,4)P2 and/or PI(3,4,5)P3 and that SopB is required for this conversion.
Figure 1.
SopB is required for PI(3)P localization to SCVs and an elevation of cellular PIP. (A and B) Cells were transfected with 2FYVE-GFP and infected with WT S. typhimurium (A) or a sopb deletion mutant (B). Left panels indicate 2FYVE-GFP relative to the RFP-expressing bacteria (red) in the merged images in the right panels. Insets are enlarged from the indicated areas (dashed boxes). Cells were analyzed using confocal microscopy. These images correspond to <15 min of infection. (C) Cells were cotransfected with PH(Akt)-RFP and 2FYVE-GFP before infection by WT S. typhimurium. Images were acquired at 1-min intervals. The time after infection of each frame is indicated. Insets are enlarged from the dashed boxes. Bars, 10 μm. (D) Cells were transfected with 2FYVE-GFP, infected with WT or sopB mutant, and analyzed as in A and B. Images were acquired at 1-min intervals for at least 1 h. Colocalization of the RFP-expressing bacteria with 2FYVE-GFP during the course of infection is shown. Data are means ± SEM of four separate experiments for WT bacteria (137 SCVs analyzed) and three separate experiments for sopB mutant (76 SCVs analyzed). (E) Phosphoinositide levels were quantified before (control) and after infection with either WT or sopB mutant. Lipids were extracted and analyzed by HPLC. The amount of the phosphoinositides is given as the percentage of PI in the same sample. PIP refers to the sum of PIPs. PIP2 refers to the sum of PIP2s. Data are means ± SEM of five separate experiments. The p-values are shown.
In further support of their hypothesis, Hernandez et al. (2004) analyzed the lipid composition of infected cells by electrospray ionization mass spectrometry (ESI-MS). As anticipated, they demonstrated an increase in PI phosphate (PIP) at the expense of PI bisphosphate (PIP2). Although powerful, ESI-MS cannot distinguish among the different isomers of PIP (i.e., PI(3)P, PI(4)P, and PI(5)P) or those of PIP2 (PI(3,4)P2, PI(3,5)P2, and PI(4,5)P2; Wenk et al., 2003). To more directly identify the phosphoinositides altered by S. typhimurium, we labeled HeLa cells metabolically with [3H]inositol and analyzed the lipids extracted before and after infection by HPLC (see Materials and methods). The cells were infected with either WT or sopB mutant bacteria to assess the effects of the phosphatase. When considered jointly, the PIP isomers increased upon infection, although the change did not attain statistical significance in five experiments (Fig. 1 E). In parallel, the PIP2 species decreased significantly. Importantly, SopB was required to produce both changes. These data are in good agreement with the findings of Hernandez et al. (2004) and are, in principle, consistent with their hypothesis that PI(3)P is generated by dephosphorylation of polyphosphoinositides by SopB. It must be borne in mind, however, that their reported analysis by ESI-MS failed to single out PI(3)P as the species elevated in infected cells.
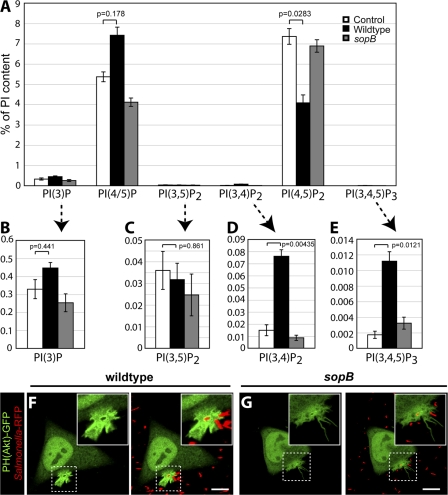
To more precisely define the lipid species affected by SopB, we separately analyzed the contribution of the individual PIP and PIP2 isomers to the global changes reported in Fig. 1 E. The results of the more detailed analysis are summarized in Fig. 2. The HPLC system we used was able to resolve PIP into two distinct and separately quantifiable peaks: one containing PI(3)P and a separate peak containing PI(4)P and/or PI(5)P, hereafter referred to as PI(4/5)P. As shown in Fig. 2 A, PI(4/5)P constitutes the vast majority (>90%) of the PIP present in control (uninfected) cells, with comparatively very low levels of PI(3)P. Contrary to our expectation, PI(3)P increased only modestly after infection by WT bacteria (Fig. 2, A and B), whereas PI(4/5)P increased markedly (Fig. 2 A). The increase in PI(4/5)P was observed in cells infected with WT bacteria but not the sopB mutant and was accompanied by a commensurate decrease in PI(4,5)P2, supporting our earlier contention that SopB dephosphorylates the latter at the 4′ and/or 5′ position (Terebiznik et al., 2002; Mason et al., 2007).
Figure 2.
SopB mediates PI3-kinase activation at the plasma membrane during Salmonella invasion. (A) Lipids were extracted and analyzed by HPLC. The amount of phosphoinositides is given as the percentage of PI in the same sample. PI(4/5)P refers to the sum of PI(4)P and PI(5)P. (B–E) Insets are provided (arrows) for each 3′-phosphorylated phosphoinositide. Data are means ± SEM of five separate experiments. Levels of significance are indicated by p-values. (F and G) Cells were transfected with PH(Akt)-GFP and infected with WT (F) or sopB mutant (G). Cells were analyzed using confocal microscopy. Left panels indicate the PH(Akt)-GFP localization relative to the RFP- expressing bacteria (red) in the merged images in the right panels. Insets are enlarged from dashed boxes. These images correspond to <15 min of infection. Bars, 10 μm.
The absolute change in PI(4/5)P measured by HPLC, calculated as a percentage of PI that is generally considered to be invariant, was >10-fold greater than that of PI(3)P. These findings imply that the ESI-MS data obtained by Hernandez et al. (2004) reflect primarily the changes in PI(4/5)P and not those in PI(3)P. In addition, the bulk of the decrease in PIP2 was attributable to degradation of PI(4,5)P2 and not PI(3,4)P2 or PI(3,5)P2 (Fig. 2, A, C, and D), as implied by Hernandez et al. (2004). In fact, the content of PI(3,4)P2 increased severalfold upon infection (Fig. 2 D), whereas that of PI(3,5)P2 was invariant (Fig. 2 C). Therefore, the loss of PIP2 reported by ESI-MS cannot be construed as evidence in favor of the formation of PI(3)P by SopB-mediated dephosphorylation of PI(3,4)P2 and/or PI(3,5)P2.
SopB mediates PI(3,4)P2 and PI(3,4,5)P3 production at the plasma membrane during Salmonella invasion
In view of these apparent inconsistencies, we analyzed the origin of the changes in 3′-phosphoinositides in more detail. Cells infected with WT bacteria displayed a significant increase in the cellular levels of PI(3,4,5)P3 (Fig. 2 E). Although the source of this increase has not been identified, the model of Hernandez et al. (2004) posits that PI(3)P is generated by SopB-mediated dephosphorylation of PI(3,4)P2 and/or PI(3,5)P2, which are in turn generated by dephosphorylation of PI(3,4,5)P3. This model predicts that the level of PI(3,4,5)P3 would be higher in cells infected with the sopB mutant. This prediction was not supported by the detailed HPLC analysis. As shown in Fig. 2 E, the accumulation of PI(3,4,5)P3 in infected cells was in fact obliterated by omission of SopB. Consistent with this, we observed recruitment of PH(Akt)-GFP to invasion ruffles in cells infected by WT bacteria (Fig. 2 F and Video 4, available at http://www.jcb.org/cgi/content/full/jcb.200804131/DC1) but not in sopB mutant–infected cells (Fig. 2 G and Video 5). Analysis of infected cells by live imaging revealed that 100% of WT-infected cells (50 ruffles analyzed in three separate experiments) had recruitment of PH(Akt)-GFP to invasion ruffles, whereas 0% (19 in four separate experiments) of the sopB mutant–infected cells did so. These findings indicate a strict requirement of SopB for the accumulation of PI(3,4)P2 and/or PI(3,4,5)P3 at invasion ruffles. Together, our analysis reveals that SopB initiates two discernible signaling events during S. typhimurium infection: (1) the generation of PI3-kinase products PI(3,4)P2 and PI(3,4,5)P3 at invasion ruffles and (2) the formation of PI(3)P on SCVs.
PI(3)P localization to the SCV is sensitive to pharmacologic inhibitors of PI3-kinase, but PI(3,4)P2 and PI(3,4,5)P3 production at invasion ruffles is not
We were intrigued by the observation that SopB is required for the formation of PI(3,4)P2 and PI(3,4,5)P3. The finding that a bacterial phosphatase can lead to the accumulation of PI3-kinase products is remarkable but not without precedent: Niebuhr et al. (2002) reported that IpgD, a type III secreted effector expressed by Shigella flexneri with homology to SopB, is required for the accumulation of PI3-kinase products during S. flexneri invasion. These authors proposed that the phosphatase activity of IpgD leads to the activation of class I PI3-kinase at the plasma membrane (Pendaries et al., 2006). To determine whether SopB is similarly mediating the activation of class I PI3-kinases, we treated cells with two pharmacologic inhibitors of these enzymes, LY294002 and wortmannin (Vlahos et al., 1994; Woscholski et al., 1994). Remarkably, treatment of cells with LY294002 had no effect on recruitment of PH(Akt)-GFP to invasion ruffles (Fig. 3 A). Analysis of infected cells by live imaging revealed that 100% of WT infected cells (49 ruffles analyzed in three separate experiments) had recruitment of PH(Akt)-GFP to invasion ruffles in the presence of LY294002. Similar results were obtained in wortmannin-treated cells (unpublished data). In agreement with the fluorescence data, analysis of lipid content by HPLC revealed that LY294002 had little effect on the generation of PI(3,4)P2 (Fig. 3 B) and PI(3,4,5)P3 (Fig. 3 C). Class I PI3-kinases are sensitive to wortmannin/LY294002, and independent experiments verified that the concentrations used were sufficient to impair these enzymes. We therefore concluded that class I PI3-kinases are not involved in the SopB-dependent production of PI(3,4)P2 and PI(3,4,5)P3. The mechanism of PI3-kinase activation by SopB or its products is the subject of ongoing investigation.
Figure 3.
PI(3)P formation on the SCV is sensitive to pharmacologic inhibitors of PI3-kinase, but PI(3,4)P2 and PI(3,4,5)P3 production at invasion ruffles is not. (A) Cells were transfected with PH(Akt)-GFP and infected with WT S. typhimurium in the presence of LY294002. (B, C, and E) Phosphoinositide levels were quantified by HPLC in control cells and after infection with WT in the absence or presence of LY294002, as indicated. The amount of PI(3,4)P2 (B), PI(3,4,5)P3 (C), and PI(3)P (E) is given as the percentage of PI in the same sample. (D) Cells were transfected with 2FYVE-GFP and infected with WT in the presence of LY294002. The inset is enlarged from the indicated area (dashed box). Data in B, C, and E are means ± SEM of five separate experiments. Levels of significance are indicated by p- values. Cells in A and D were analyzed by confocal microscopy and correspond to <15 min of infection. Bars, 10 μm.
In contrast to the formation of PI(3,4)P2 and PI(3,4,5)P3, LY294002 treatment completely blocked the recruitment of 2FYVE-GFP to SCVs (Fig. 3 D and Video 6, available at http://www.jcb.org/cgi/content/full/jcb.200804131/DC1), which is consistent with previous observations (Scott et al., 2002). LY294002 treatment also blocked the increase in cellular PI(3)P levels measured by HPLC (Fig. 3 E). Similar results were obtained in wortmannin-treated cells (unpublished data). These findings suggest that PI(3)P generation on SCVs may be independent of PI(3,4)P2 and PI(3,4,5)P3 production at invasion ruffles during S. typhimurium infection.
PI(3)P localization to the SCV is not dependent on PI(3,4)P2 and PI(3,4,5)P3 production at invasion ruffles
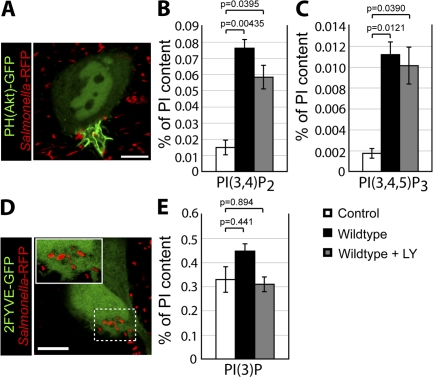
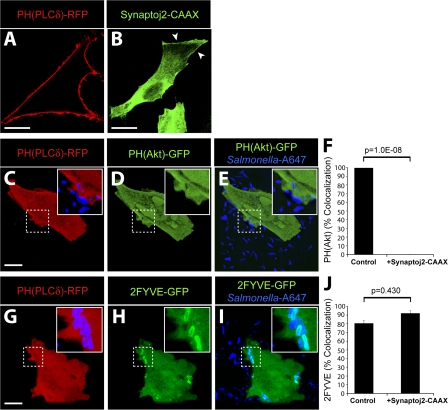
Because pharmacologic inhibitors did not affect the SopB-dependent generation of PI(3,4)P2 and PI(3,4,5)P3 at the plasma membrane, we used an alternative strategy to test the role of these lipids in PI(3)P formation. We hypothesized that depletion of PI(4,5)P2, the substrate of PI3-kinases at the plasma membrane (Anderson and Jackson, 2003; Lindmo and Stenmark, 2006), would block the formation of PI(3,4,5)P3 and PI(3,4)P2. For these experiments, we transfected cells with the catalytic domain of synaptojanin-2, a mammalian 5-phosphatase that selectively dephosphorylates PI(4,5)P2 to generate PI(4)P (Malecz et al., 2000; Rusk et al., 2003). This construct contained a carboxyl-terminal CAAX motif to target synaptojanin-2 to the plasma membrane and promote its activity at this subcellular location (Fig. 4 B, arrowheads). To detect PI(4,5)P2 at the plasma membrane, we transfected cells with a construct encoding the PH domain of phospholipase Cδ fused to RFP (PH(PLCδ)-RFP). In the absence of synaptojanin-2, the PH(PLCδ)-RFP probe was localized to the plasma membrane (Fig. 4 A), as previously described (Varnai and Balla, 1998). However, upon cotransfection with synaptojanin-2–CAAX, PH(PLCδ)-RFP was displaced to the cytosol, indicating that PI(4,5)P2 was successfully depleted from the plasma membrane (Fig. 4, C and G).
Figure 4.
PI(3)P formation on the SCV is not dependent on PI(3,4)P2 and PI(3,4,5)P3 production at invasion ruffles. (A and B) Localization of PH(PLCδ)-RFP (A) and synaptojanin-2–CAAX construct (B) in control cells. Arrowheads indicate plasma membrane localization of synaptojanin-2–CAAX construct. (C–F) Cells were cotransfected with synaptojanin-2–CAAX, PH(PLCδ)-RFP (C), and PH(Akt)-GFP (D). (E) Merged image showing localization of bacteria (labeled with Alexa Fluor 647) relative to signal for PH(Akt)-GFP. (F) Cells were cotransfected as in C–E, infected with WT S. typhimurium, and analyzed by confocal microscopy. Images were acquired at 1-min intervals for ∼1 h. Colocalization of the bacteria with PH(Akt)-GFP during infection is shown. As a control, cells were cotransfected with PH(PLCδ)-RFP and PH(Akt)-GFP but not synaptojanin-2–CAAX. Data are means ± SEM of three separate experiments for synaptojanin-2–CAAX-expressing cells (20 ruffles analyzed) and two separate experiments for control cells (13 ruffles analyzed). The p-value is shown. (G–J) Cells were cotransfected with synaptojanin-2–CAAX, PH(PLCδ)-RFP (G), and 2FYVE-GFP (H). (I) Merged image showing localization of bacteria (labeled with Alexa Fluor 647) relative to signal for 2FYVE-GFP. Insets are enlarged from dashed boxes. (J) Cells were cotransfected as in G–I, infected with WT bacteria, and analyzed by confocal microscopy. Images were acquired at 1-min intervals for at least 1 h. Colocalization of the bacteria with 2FYVE-GFP during infection is shown. As a control, cells were cotransfected with PH(PLCδ)-RFP and 2FYVE-GFP but not synaptojanin-2–CAAX. Data are means ± SEM of three separate experiments for synaptojanin-2–CAAX- expressing cells (111 SCVs analyzed) and two separate experiments for control cells (44 SCVs analyzed). The p-value is shown. Bars, 10 μm.
After depletion of PI(4,5)P2, cells were infected with WT S. typhimurium, and the synthesis of PI3-kinase products was examined. As expected, cells expressing synaptojanin-2–CAAX did not recruit PH(Akt)-GFP to invasion ruffles (Fig. 4, D–F). This demonstrates that depletion of PI(4,5)P2 effectively blocked the generation of PI(3,4)P2 and/or PI(3,4,5)P3 at the plasma membrane. Remarkably, 2FYVE-GFP recruitment to the SCV persisted in PI(4,5)P2-depleted cells (Fig. 4, H–J). These results demonstrate that the production of PI(3,4)P2 and/or PI(3,4,5)P3 at invasion ruffles is not essential for the production of PI(3)P on the SCV and suggest that SopB-mediated dephosphorylation of PI3-kinase products is not the main pathway for the generation of PI(3)P on the SCV.
Vps34 expression is required for PI(3)P localization to the SCV
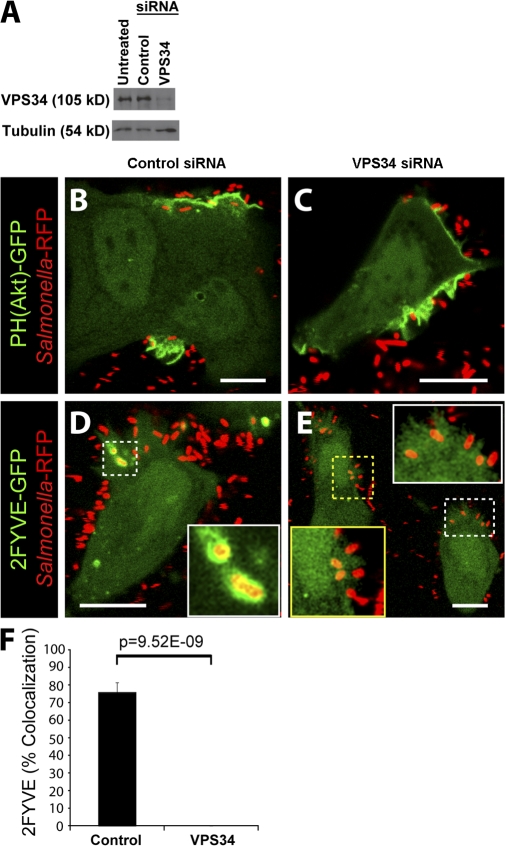
The main pathway for the formation of PI(3)P on endosomes and early phagosomes is via phosphorylation of PI by Vps34, a class III PI3-kinase (Vieira et al., 2001; Shin et al., 2005). Because our results were not compatible with the earlier notion that PI(3)P is generated in the SCV by dephosphorylation of PI(3,4)P2 and/or PI(3,4,5)P3, we considered the possible role of Vps34 in vacuolar PI(3)P generation. To examine this possibility, we used siRNA to silence the expression of Vps34. As shown in Fig. 5 A, an almost complete inhibition of Vps34 expression was achieved with targeted siRNA, whereas control siRNA had no effect (Fig. 5 A). Next, we infected cells with WT bacteria and analyzed PI3-kinase products using fluorescent probes. Cells treated with control siRNA displayed normal recruitment of PH(Akt)-GFP to invasion ruffles (Fig. 5 B) and 2FYVE-GFP to the SCV (Fig. 5 D). Treatment of cells with Vps34 siRNA had no effect on PH(Akt)-GFP recruitment to invasion ruffles (Fig. 5 C). However, the recruitment of 2FYVE-GFP to the SCV was abolished in cells depleted of Vps34 (Fig. 5, E and F). These findings demonstrate that PI(3)P formation on the SCV is dependent on the expression of Vps34.
Figure 5.
Vps34 expression is required for PI(3)P formation on the SCV. (A) Western blot analysis of HeLa cell lysates. Tubulin expression was assessed as a loading control. (B–E) Cells were treated with control (B and D) or Vps34 (C and E) siRNA before transfection with either PH(Akt)-GFP (B and C) or 2FYVE-GFP (D and E). Cells were then infected with WT bacteria (_Salmonella-_RFP) and analyzed by confocal microscopy. Insets in D and E are enlarged from dashed boxes. These images correspond to <15 min of infection. Bars, 10 μm. (F) Cells were treated with control or Vps34 siRNA and infected with WT bacteria expressing RFP. Infected cells were analyzed using confocal microscopy. Images were acquired at 1-min intervals for ∼1 h. Colocalization of the bacteria with 2FYVE-GFP during infection is shown. Data are means ± SEM of four separate experiments for control siRNA–treated cells (102 SCVs analyzed) and three separate experiments for Vps34 siRNA–treated cells (170 SCVs analyzed). The p-value is shown.
Rab5 recruitment to the SCV is SopB dependent
In early endosomes and phagosomes, Vps34 binds to and is seemingly activated by GTP-bound (active) Rab5 (Christoforidis et al., 1999). Accordingly, Rab5 is known to promote formation of PI(3)P on endosomes (Shin et al., 2005) and phagosomes (Vieira et al., 2001). Previous observations have demonstrated the presence of Rab5 on SCVs and its requirement for normal SCV maturation (Steele-Mortimer et al., 1999; Baldeon et al., 2001; Scott et al., 2002; Smith et al., 2007). We therefore wondered whether a Rab5-dependent pathway also contributes to the production of PI(3)P on the SCV and whether SopB is involved in this process. To test this possibility, we visualized the dynamics of Rab5 recruitment to the SCV during infection. Cells were transfected with GFP-Rab5A, subjected to infection, fixed, and differentially stained for the identification of intracellular bacteria (see Materials and methods). The localization of GFP-Rab5A with respect to the intracellular bacteria was then determined microscopically. As shown in Fig. 6 A, Rab5 association with SCVs was maximal ∼10 min after infection and declined thereafter. To further examine the dynamics of Rab5 recruitment to SCVs, we monitored live cells by spinning disk confocal microscopy (Fig. 6 B, top; and Video 7, available at http://www.jcb.org/cgi/content/full/jcb.200804131/DC1). We observed rapid delivery of Rab5 to the SCV, occurring within as little as 1 min of vacuole formation. Rab5 delivery appeared to occur largely via fusion of the SCV with Rab5-containing vesicles (Fig. 6 B, arrowheads). Although it is possible that soluble Rab5 is recruited to the SCV from the cytosol, our image analysis suggests that this potential contribution is minimal.
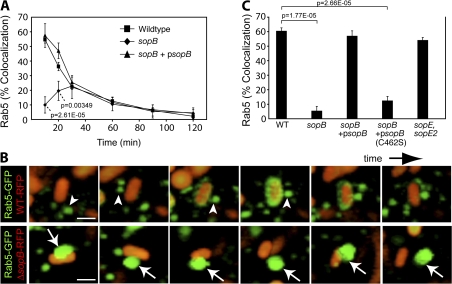
Figure 6.
Rab5 recruitment to the SCV requires the phosphatase activity of SopB. (A) Cells expressing Rab5A-GFP were infected for designated times and were fixed and stained for bacteria. Colocalization of Rab5A-GFP with intracellular bacteria was quantified. Data are means ± SEM of three separate experiments. Levels of significance are indicated by p-values compared with WT-infected cells. (B) Cells were transfected with Rab5A-GFP and infected with WT bacteria (WT-RFP, top) or an sopB mutant expressing RFP (sopB-RFP, bottom). Live imaging was performed on a spinning disk confocal microscope. Total sequence time for both series is ∼3 min. Arrowheads indicate Rab5A+ endosomes that dock and fuse with WT SCVs. Arrows indicate Rab5A+ endosomes that dock but do not fuse with sopB SCVs. Bar, 2.5 μm. (C) Cells were transfected with Rab5A-GFP and infected with either WT or sopB_-deficient bacteria. In parallel, cells were infected with sopB mutant bacteria expressing either WT sopB (sopB + p_sopB) or a catalytically inactive mutant (sopB + p_sopB_[C462S]) on a plasmid. A mutant lacking sopE and sopE2 was also analyzed. At 10-min after infection, infected cells were fixed and stained as in A. Colocalization of Rab5A-GFP with intracellular bacteria was quantified. Data are means ± SEM of three separate experiments. The p-values are shown.
To test the role of SopB in Rab5 recruitment to the SCV, we infected cells with the _sopB-deficient mutant. Remarkably, deletion of sopB impaired recruitment of Rab5 (Fig. 6, A and C). Transformation of the sopb deletion mutant with WT sopB on a plasmid (p_sopB) was sufficient to complement Rab5 recruitment to SCVs (Fig. 6, A and C). In contrast, expression of the catalytically inactive mutant of SopB (C462S) did not complement the sopb deletion mutant (Fig. 6 C), showing that its phosphatase activity is important for Rab5 recruitment to the SCV. Rab5 recruitment was largely unaffected in cells infected with sopE and _sopE2_-deficient bacteria (Fig. 6 C). This mutant is partially defective for invasion (Zhou et al., 2001), suggesting that invasion efficiency does not influence Rab5 recruitment.
We examined the fate of Rab5 in live cells during the course of infection with the _sopB_-deficient bacteria using spinning disk confocal microscopy (Fig. 6 B, bottom; and Video 8, available at http://www.jcb.org/cgi/content/full/jcb.200804131/DC1). As expected, Rab5-positive vesicles did not fuse with SCVs containing this mutant. However, we routinely observed a close association of these vesicles with SCVs containing the sopB mutant. Rab5-bearing vesicles appeared to dock with these SCVs for extended periods of time but failed to fuse with them (Fig. 6 B, arrows). Our data suggest that docking of Rab5-positive vesicles to the SCV precedes a fusion step catalyzed by the phosphatase activity of SopB.
Rab5 expression is required for PI(3)P localization to the SCV
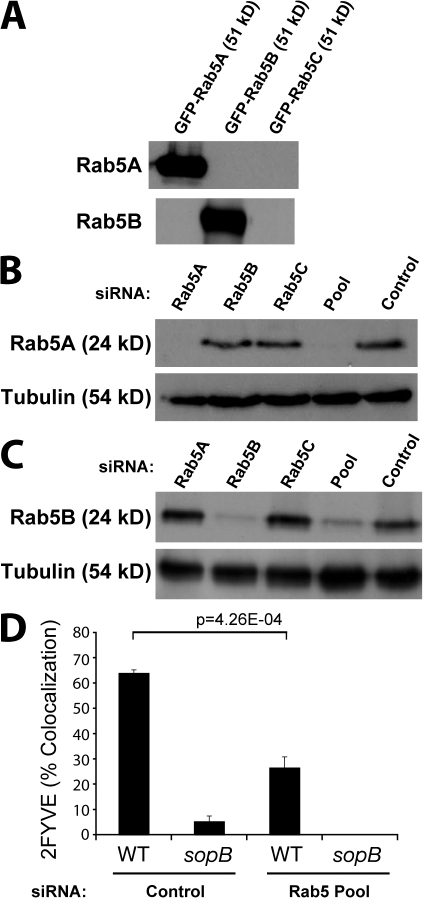
There are three Rab5 proteins expressed in mammalian cells: Rab5A, Rab5B, and Rab5C. A previous study demonstrated that the three Rab5 isoforms share similar subcellular localization and regulatory functions in the early endocytic pathway (Bucci et al., 1995). These Rab5 isoforms are thought to be at least partially redundant because down-regulation of expression of any single Rab5 isoform using siRNA has no effect on EGF or transferrin internalization, but down-regulation of all three isoforms has a significant effect (Huang et al., 2004). Similarly, silencing of all three Rab5 isoforms is required to impair Akt activation after insulin treatment (Su et al., 2006). With this in mind, we tested the role of Rab5 in PI(3)P synthesis on the SCV by silencing expression of all three Rab5 isoforms using siRNA, as previously described (Huang et al., 2004). Isoform-specific antibodies to Rab5A and Rab5B detected their respective GFP-tagged proteins in lysates from transfected cells (Fig. 7 A) and the endogenous protein in cell lysates (Fig. 7, B and C). Treatment with Rab5A siRNA eliminated expression of this isoform (Fig. 7 B) but did not affect expression of Rab5B (Fig. 7 C). Similarly, Rab5B siRNA blocked expression of this isoform (Fig. 7 C) but did not affect expression of Rab5A (Fig. 7 B). Rab5C siRNA did not affect expression of either Rab5A or Rab5B. This demonstrates the isoform specificity of the individual siRNAs. When cells were treated with the three siRNAs pooled together, Rab5A (Fig. 7 B) and Rab5B (Fig. 7 C) expression was depressed by ≥90%. We were unable to verify the silencing of Rab5C because the commercially available Rab5C antibody failed to detect endogenous Rab5C and even heterologously overexpressed GFP-tagged Rab5C (unpublished data).
Figure 7.
Rab5 expression is required for PI(3)P localization to the SCV. (A) Cells were transfected with GFP-tagged Rab5A, Rab5B, or Rab5C as indicated. Cell lysates were then immunoblotted for either Rab5A or Rab5B. (B) Cells were treated with siRNA to the indicated Rab5 isoform, all three Rab5 isoforms (Pool), or control siRNA that does not affect Rab5 expression. Cell lysates were immunoblotted for Rab5A or tubulin (loading control). (C) Samples from B were immunoblotted for Rab5B or tubulin. (D) Cells were treated with either control siRNA or Rab5 Pool siRNA and transfected with 2FYVE-GFP. Cells were then infected with either WT or sopb mutant bacteria, fixed at 10-min after infection, and stained for bacteria. Intracellular bacteria colocalizing with 2FYVE-GFP was then quantified. Data are means ± SEM of three separate experiments (>100 bacteria analyzed per experiment). The p-value is shown.
Next, cells were treated with control siRNA or the Rab5 siRNA pool, and PI(3)P was visualized using the 2FYVE-GFP probe. In control cells, 2FYVE-GFP localized to the cytosol and endosomes. As expected, depletion of all three Rab5 isoforms caused a loss of endosomal 2FYVE-GFP, as did depletion of Vps34 (unpublished data). Depletion of Rab5 isoforms also caused a marked decrease in the association of 2FYVE-GFP with the SCV (Fig. 7 D). Together, these findings demonstrate that, as in endosomes, Rab5 is involved in PI(3)P acquisition by the SCV.
Depletion of PI(4,5)P2 partially complements Rab5 and PI(3)P localization to sopB mutant SCVs
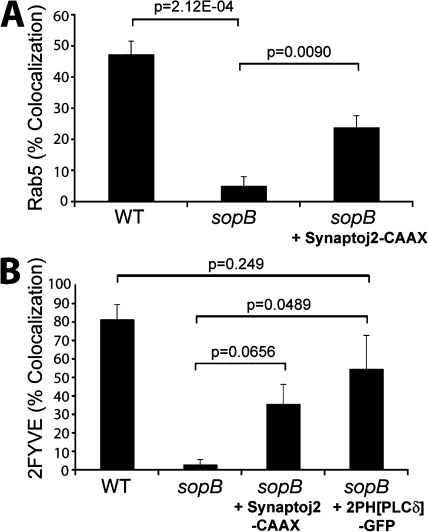
We proceeded to explore the mechanism by which SopB mediates Rab5 recruitment to the SCV. The data in Fig. 6 C suggested that the phosphatase activity of SopB is essential for Rab5 recruitment. Because the major substrate of SopB in host cells is PI(4,5)P2 (Fig. 2 A), we hypothesized that dephosphorylation of this lipid may mediate the effects of the phosphatase on Rab5 recruitment and PI(3)P formation. If this were indeed the case, depletion of PI(4,5)P2 by other means should complement Rab5 recruitment to SCVs formed by _sopB_-deficient bacteria. To test this possibility, we transfected cells with synaptojanin-2–CAAX to dephosphorylate PI(4,5)P2 as in Fig. 4. Cotransfection with the PH(PLCδ)-RFP probe confirmed that the construct effectively depleted PI(4,5)P2 from the plasma membrane. After depletion of PI(4,5)P2, cells were infected with S. typhimurium, and the colocalization of SCVs with GFP-Rab5A was determined. As shown in Fig. 8 A, depletion of PI(4,5)P2 caused a significant increase in the association of Rab5 with SCVs containing _sopB_-deficient bacteria.
Figure 8.
Depletion of PI(4,5)P2 partially complements Rab5 and PI(3)P localization to sopB mutant SCVs. (A) Cells were cotransfected with Rab5A-GFP and PH(PLCδ)-RFP or synaptojanin-2–CAAX, PH(PLCδ)-RFP, and Rab5A-GFP as indicated. Cells were then infected with either WT or sopB mutant expressing RFP, fixed at 10-min after infection, and stained for extracellular bacteria. Intracellular bacteria colocalizing with Rab5A-GFP was then determined by microscopic analysis. Data are means ± SEM of three separate experiments (>100 bacteria analyzed per experiment). (B) Cells were transfected with the indicated construct and then infected with either WT or sopB mutant labeled covalently with Alexa Fluor 647. Infected cells were analyzed using a spinning disk confocal microscope. Images were acquired at 1-min intervals for ∼1 h. Localization of PI(3)P to SCVs (colocalization with 2FYVE-GFP or PX-mCherry) during the course of infection is shown. Data are means ± SEM of three separate experiments for WT- (94 SCVs analyzed) or sopB mutant– (68 SCVs analyzed) infected cells expressing 2FYVE-GFP. Five separate experiments were analyzed for sopb mutant–infected cells expressing synaptojanin 2-CAAX, PH(PLCδ)-RFP, and 2FYVE-GFP (121 SCVs analyzed), and three separate experiments were analyzed for sopb mutant–infected cells expressing 2PH(PLCδ)-GFP and PX-mCherry-GFP (120 SCVs analyzed). The p-values are shown.
We also examined the formation of PI(3)P on SCVs under conditions in which PI(4,5)P2 was depleted. To this end, cells were cotransfected with synaptojanin-2–CAAX, PH(PLCδ)-RFP, and 2FYVE-GFP. Recruitment of the PI(3)P probe to SCVs containing _sopB_-deficient bacteria was partially complemented by depletion of plasmalemmal PI(4,5)P2 (Fig. 8 B), which is in agreement with results obtained for Rab5 (Fig. 8 A). These findings suggest that the recruitment to SCVs of Rab5 and its effector Vps34 (as indicated by the formation of its product, PI(3)P) is mediated, at least in part, by the SopB-mediated dephosphorylation of PI(4,5)P2.
In principle, SopB could be acting through its ability to reduce PI(4,5)P2 levels or by generation of its hydrolysis products, PI(4/5)P. To differentiate these possibilities, we sought to reduce the concentration of available PI(4,5)P2 without generating PI(4)P or PI(5)P. This was accomplished by transfecting cells with a construct bearing two tandem PH domains of phospholipase Cδ fused to GFP (2PH(PLCδ)-GFP). This construct has an enhanced avidity for PI(4,5)P2 compared with the single PH domain probe and, when overexpressed, can mask a significant fraction of the phosphoinositide at the plasma membrane (Mason et al., 2007). As an indicator of Vps34 recruitment to SCVs, PI(3)P was visualized by cotransfecting the cells with a construct bearing the PX domain of p40phox fused to mCherry (PX-mCherry). As shown in Fig. 8 B, expression of 2PH(PLCδ)-GFP caused a significant increase in PI(3)P localization to SCVs formed by _sopB_-deficient bacteria. In fact, PI(3)P accumulation was not significantly different in SCVs containing WT or _sopB_-deficient bacteria. Thus, diminution of available PI(4,5)P2 without concomitant generation of PI(4)P or PI(5)P mimics the effects of SopB and synaptojanin-2. These data suggest that a localized decrease in PI(4,5)P2, rather than the accumulation of the products of its hydrolysis, is critical for the SopB-mediated recruitment of Rab5 and subsequent generation of PI(3)P on SCVs.
Discussion
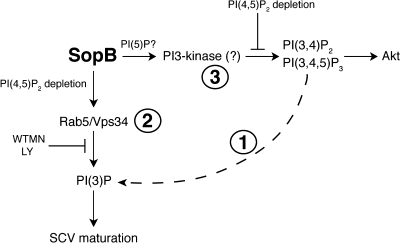
In the present study, we confirmed the findings of Hernandez et al. (2004), demonstrating that SopB is required for the generation of PI(3)P on SCVs. Based on their observations, these authors concluded that SopB generates PI(3)P via the dephosphorylation of polyphosphorylated products of PI3-kinase (Fig. 9, pathway 1; Hernandez et al., 2004; Dukes et al., 2006). However, multiple lines of evidence presented here suggest that this model does not fully account for the mechanism by which SopB activity leads to PI(3)P formation on the SCV: (1) the main PIP isomer formed during infection is PI(4/5)P, not PI3P as surmised from ESI-MS determinations (Hernandez et al., 2004); (2) the production of PI(3)P is sensitive to wortmannin/LY294002, whereas production of PI(3,4)P2 and PI(3,4,5)P3 is not; (3) blocking production of PI(3,4)P2 and PI(3,4,5)P3 at invasion ruffles using the synaptojanin-2–CAAX construct does not prevent PI(3)P formation at SCVs; and (4) expression of Vps34 is essential for the formation of PI(3)P on SCVs but not for the accumulation of PI(3,4)P2 and PI(3,4,5)P3 at invasion ruffles. It therefore appears unlikely that SopB mediates PI(3)P formation by dephosphorylation of PI(3,4)P2 and/or PI(3,4,5)P3.
Figure 9.
Model of SopB action in host cells during Salmonella infection. During invasion, S. typhimurium injects SopB into host cells. At the plasma membrane, SopB mediates dephosphorylation of PI(4,5)P2 to generate PI(4/5)P. Through this event, SopB causes the accumulation of PI(3,4)P2 and PI(3,4,5)P3 at invasion ruffles by a wortmannin/LY294002-insensitive mechanism. SopB-mediated dephosphorylation of PI(4,5)P2 also allows SCV fission from the plasma membrane and the recruitment of Rab5 to nascent SCVs via their fusion with early endosomes. The Rab5 effector Vps34 catalyzes phosphorylation of PI to generate PI(3)P on SCVs, leading to downstream SCV maturation events such as the recruitment of LAMP-1.
Instead, we propose that SopB mediates PI(3)P formation on SCVs primarily via phosphorylation of PI by Vps34 (Fig. 9, pathway 2). This notion is consistent with the wortmannin and LY294002 sensitivity of the process and with the inhibitory effects of Vps34 siRNA. Furthermore, we suggest that Vps34 is recruited to the SCV by fusion with Rab5-positive vesicles. Rab5 is thought to recruit and/or activate Vps34 on early endosomes, and siRNA directed to all three Rab5 isoforms blocked PI(3)P localization to SCVs (Fig. 7 D). Little is known about the mechanisms that regulate Rab5 recruitment to phagosomes and invasion vacuoles. Our analysis revealed that Rab5-containing vesicles are recruited to the SCVs in a SopB-dependent manner. The phosphatase activity of SopB was required for Rab5 recruitment (Fig. 6 C), suggesting that dephosphorylation of a host cell substrate allows endosomes to acquire Rab5. Indeed, we were able to partially restore Rab5 recruitment to _sopB_-deficient SCVs by depleting PI(4,5)P2 with synaptojanin-2, another inositide phosphatase. The mechanism whereby PI(4,5)P2 dephosphorylation promotes Rab5 recruitment to SCVs is unclear but may involve electrostatic interactions that are known to regulate the localization of Rab GTPases (Heo et al., 2006; Yeung and Grinstein, 2007; Yeung et al., 2008). The negative surface charge contributed by PI(4,5)P2, a tetravalent anion at physiological pH, may preclude Rab5 recruitment to endosomes/phagosomes. In support of this notion, we observed that masking the headgroups of PI(4,5)P2 could partially complement PI(3)P localization to _sopB_-deficient SCVs (Fig. 8 B). Together, our findings reveal a link between phosphoinositide phosphatase activity and the recruitment of Rab5 to phagosomes.
An unexpected finding was the observation that SopB mediates the accumulation of PI(3,4)P2 and PI(3,4,5)P3 at invasion ruffles (Fig. 2, F and G; and Fig. 9, pathway 3). How might SopB lead to the accumulation of these lipids? This effector can efficiently dephosphorylate a wide variety of phosphoinositides and inositol phosphates in vitro (Norris et al., 1998; Marcus et al., 2001). Recent evidence suggests that the main phosphoinositide substrate of SopB in vivo is PI(4,5)P2 (Fig. 2 A; Terebiznik et al., 2002; Mason et al., 2007) and that SopB generates PI(5)P (Mason et al., 2007). It is possible that this rare phosphoinositide leads to the activation of a PI3-kinase at invasion ruffles. Indeed, an important role for PI(5)P in intracellular signaling was recently suggested by Pendaries et al. (2006). These authors demonstrated that PI(5)P generation by IpgD is sufficient to activate class I PI3-kinase at the plasma membrane (Pendaries et al., 2006). Here we show that SopB is similarly involved in the generation of PI3-kinase products. However, the fact that wortmannin and LY294002 did not affect the formation of PI(3,4)P2 and PI(3,4,5)P3 during S. typhimurium infection implies that this is not the result of activation of class I PI3-kinases, which are sensitive to these agents. It is possible that members of the class II family of PI3-kinases, which are least sensitive to pharmacologic inhibition (Falasca et al., 2007), mediate PI(3,4)P2 and PI(3,4,5)P3 generation during infection. These findings suggest a potential difference in the mechanisms used by S. typhimurium and S. flexneri to invade host cells.
An alternative mechanism of action of PI(5)P is suggested by the work of Carricaburu et al. (2003), who found that accumulation of PI(3,4,5)P3 is antagonized by type II PI(5)P 4-kinase. These data were interpreted to mean that PI(5)P promotes the accumulation PI(3,4,5)P3 by exerting an inhibition on polyphosphoinositide phosphatases. In support of this hypothesis, PI(5)P was reported to exert an inhibitory effect on the PI(3,4,5)P3 5-phosphatase activity of SHIP2 (Pendaries et al., 2006). Thus, the marked accumulation of PI(5)P induced by SopB may result in inhibition of the phosphatases that normally eliminate the products of constitutively active PI3-kinases, perhaps those members of the class II family that are least sensitive to pharmacologic inhibition.
In summary, SopB appears to regulate different classes of PI3-kinases in distinct cellular compartments during S. typhimurium infection. At the invasion ruffle, SopB induces the accumulation of PI(3,4)P2 and PI(3,4,5)P3 by an incompletely understood process involving its phosphatase activity. At the SCV, SopB promotes the formation of PI(3)P by recruitment of Vps34 on fusion with Rab5-positive vesicles. Dephosphorylation of PI(4,5)P2 appears to be the critical event that allows Rab5 recruitment to the SCV. The relationship between the events occurring at the ruffle and SCV and the precise mechanism underlying polyphosphoinositide induction remain incompletely understood and require additional study.
Materials and methods
DNA constructs and bacteria
The vectors pEGFP∷AktPH and pEGFP∷PLCδPH encode the PH domain of Akt (GFP-PH-Akt; Weernink et al., 2000) and the PH domain of PLCδ (PH-PLCδ-GFP; Stauffer et al., 1998), respectively. The 2PH(PLCδ)-GFP construct was made by combining two tandem PH domains of PLCδ to GFP and was provided by M. Rebecchi (State University of New York, Stony Brook, NY). The construct expressing the catalytic domain of synaptojanin-2 with a membrane-targeting domain (synaptojanin-2–CAAX) was previously described (Malecz et al., 2000) and provided by M. Symons (North Shore Research Institute, Manhasset, NY). The construct mRFP-PH-Akt was made by digestion of GFP-PH-Akt at the XhoI and EcoRI sites followed by subcloning of the fragment into the mRFP-C1 vector using the same sites. PH-PLCδ-mRFP was subcloned in mRFP-N1 by digestion of PH-PLCδ-GFP at the XhoI and BamHI sites. The plasmids encoding the chimeras of GFP with the tandem FYVE domains (2FYVE-GFP) were described previously (Vieira et al., 2001). The plasmids encoding the PX domain of p40_phox_ and Rab5A-GFP (Scott et al., 2002), Rab5B-GFP, and Rab5C-GFP (Heo et al., 2006) were described previously. The mRFP vectors were provided by R. Tsien (University of California at San Diego, La Jolla, CA; Campbell et al., 2002). The source and properties of WT and _sopB_-deficient S. typhimurium SL1344 were described previously (Hoiseth and Stocker, 1981; Steele-Mortimer et al., 2000). The Cys462Ser mutant of SopB (C462S) was described previously (Marcus et al., 2001). WT bacteria expressing mRFP (Birmingham et al., 2006) were also used for these studies.
Cell culture and bacterial infection
HeLa cells were maintained in DME with 10% FBS, transfected with FuGene 6 (Roche) according to the manufacturer's instructions, and used between 12 and 16 h after transfection. Where indicated, cells were pretreated for 45 min with either 100 μM LY294002 (EMD) or 100 nM wortmannin (Sigma-Aldrich), and the drug was maintained in the medium for the duration of the experiment. Late-log S. typhimurium cultures used to infect cells were prepared as described previously (Steele-Mortimer et al., 1999). To label bacteria for live imaging experiments, bacterial cultures were washed three times with PBS with calcium and magnesium (PBS2+; by centrifugation at 10,000 rpm for 45 s) and resuspended in PBS2+ containing 25 μg/ml Alexa Fluor 647 carboxylic acid and succinimidyl ester dye (Invitrogen). The bacterial suspension was incubated in a shaking incubator for 5 min at 37°C. Bacteria were then washed once with PBS2+, resuspended in RPMI 1640, and used immediately for invasion of live HeLa cells.
Lipid extraction and phosphoinositide analysis
Lipid labeling and extraction were performed essentially as described previously (Carricaburu et al., 2003). In brief, HeLa cells were labeled with 20 μCi/ml [3H]myoinositol for 24–48 h in inositol-free medium. After labeling, the cells were infected with the indicated strain of S. typhimurium for 15 min and immediately lysed in 1 M HCl. Lipids were extracted in chlorofom-methanol (1:1 vol/vol) and deacylated as described previously (Serunian et al., 1991). Deacylated lipids were separated by anion exchange HPLC, detected by an online Radiomatic detector (PerkinElmer), and quantified relative to PI using the Pro FSA analysis program. Individual peaks in the chromatogram were identified using in vitro synthesized internal standard lipids. Data from these experiments are presented in two formats. In Fig. 1 F, PIP (the sum of PI(3)P, PI(4)P, and PI(5)P) and PIP2 (the sum of PI(3,4)P2, PI(3,5)P2, and PI(4,5)P2) are presented. In Fig. 2 A, all phosphoinositides are presented separately, and the data for PI(3,4,5)P3 has been included.
Immunofluorescence
For immunofluorescence, cells were fixed with 2.5% paraformaldehyde in PBS for 10 min at 37°C. To distinguish between intracellular and extracellular S. typhimurium, extracellular and total bacteria were differentially stained as previously described (Smith et al., 2007). Rabbit anti-Salmonella (group B) was purchased from Difco Laboratories. Mouse anti-GFP was purchased from Invitrogen. Quantifications of colocalization were performed using an epifluorescence microscope (DMIRE2; Leica) equipped with a 100× oil objective, as previously described (Smith et al., 2007). Antibodies to the HA epitope used for the detection of the synaptojanin-2–CAAX construct were purchased from Covance.
Confocal microscopy
Cells plated on 25-mm coverslips were grown in DME media supplemented with serum without any antibiotics. Live samples were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope (LSM 510; Carl Zeiss, Inc.). Frames were taken every minute for almost 60 min; however, all of the figures here include frames from ≤15 min after infection. GFP, RFP, and far red fluorescence were examined using the conventional laser excitation and filter sets. As indicated, live imaging with spinning disk was performed on a Quorum confocal microscope (DMIRE2; Leica) with Volocity software (Improvision). Z-stack images were acquired at ∼15-s intervals with z steps of 0.5 μm with a back-thinned EM-CCD camera (Hamamatsu Photonics). Fast iterative deconvolution was performed with Volocity software. Confocal images were imported into Photoshop (Adobe) and assembled in Illustrator (Adobe).
siRNA oligonucleotides and transfections
The siRNA used to inhibit expression of vps34 (5′-ACUCAACACUGGCUAAUUAUU-3′) was provided by J. Backer (Albert Einstein College of Medicine, New York, NY). Cells were maintained in DME with 10% FBS and transfected with Oligofectamine (Invitrogen). siSTABLE nontargeting siRNA #1 (Thermo Fisher Scientific) was used as a control. This nontargeting siRNA was designed to have comparable guanine-cytosine content to that of the Vps34 siRNA but lacks identity with known gene targets. Both siRNAs were used at a final concentration of 200 nM. The cells were infected and used for confocal microscopy after 4 d. Where necessary, cells were treated with siRNA for 72 h before transfection with PH(Akt)-GFP or 2FYVE-GFP using FuGene 6. Cells were used the next day for confocal live studies.
The siRNAs used to inhibit expression of rab5A (5′-AGGAATCAGTGTTGTAGTA-3′), rab5B (5′-GCTATGAACGTGAATGATC-3′), or rab5C (5′-CAATGAACGTGAACGAAAT-3′) were purchased from Thermo Fisher Scientific. These siRNAs were previously shown to successfully inhibit expression of their respective Rab5 isoform by Huang et al. (2004). Cells were transfected with 160 nM of an individual siRNA or ∼50 nM of all three isoform-specific siRNAs in the pooled experiments using Oligofectamine. Where necessary, cells were treated with siRNA for 48 h before transfection with 2FYVE-GFP using GeneJuice reagent (VWR Scientific).
Western blot analysis
Samples were separated on 10% SDS-PAGE gels, transferred to polyvinylidene fluoride membranes, and blocked in 5% milk solution overnight. Primary and secondary antibodies were incubated in 5% milk solution overnight. Rabbit anti-Vps34 antibodies (provided by J. Backer; Siddhanta et al., 1998) were used at a dilution of 1:300. Mouse monoclonal antibodies to Rab5A (1:1,000 dilution) were purchased from BD Transduction Laboratories. Rabbit polyclonal antibodies to Rab5B (1:250 dilution) and goat polyclonal antibodies to Rab5C were purchased from Santa Cruz Biotechnology, Inc. Equal protein loading was confirmed by blotting with an anti–α-tubulin monoclonal antibody (1:1,000 dilution; Sigma-Aldrich). Horseradish peroxidase–conjugated anti–rabbit secondary antibody (Jackson ImmunoResearch Laboratories) was used at a dilution of 1:1,000. The ECL detection system (GE Healthcare) was used according to the manufacturer's instructions.
Statistical analysis
Statistical analyses were performed using a two-tailed unpaired t test. P-values <0.05 were considered statistically significant.
Online supplemental material
Video 1 shows localization of 2FYVE-GFP during WT S. typhimurium invasion. Video 2 shows localization of 2FYVE-GFP during infection by an S. typhimurium sopB deletion mutant. Video 3 shows localization of PH(Akt)-RFP and 2FYVE-GFP during WT S. typhimurium invasion. Video 4 shows localization of PH(Akt)-GFP during WT S. typhimurium invasion. Video 5 shows localization of PH(Akt)-GFP during infection by an S. typhimurium sopB deletion mutant. Video 6 shows localization of 2FYVE-GFP during invasion by WT S. typhimurium in the presence of LY294002. Video 7 shows localization of Rab5-GFP during WT S. typhimurium invasion. Video 8 shows localization of Rab5-GFP during infection by an S. typhimurium sopB deletion mutant. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200804131/DC1.
Acknowledgments
We are grateful to colleagues who provided reagents used in this study. We thank M. Woodside and P. Paroutis for assistance with confocal microscopy and T. Yeung, D. Mason, and members of the Brumell laboratory for critical review of the manuscript.
This work was supported by grants from the Canadian Institutes of Health Research (CIHR) to J.H. Brumell (151733), S. Grinstein, and B.B. Finlay. B.B. Finlay is a Distinguished Investigator of the CIHR, and S. Grinstein is the current holder of the Pitblado Chair in Cell Biology. G.V. Mallo is the recipient of a postdoctoral fellowship provided by the Reverend T.K. Todd. J.H. Brumell holds an Investigators in Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund.
Abbreviations used in this paper: ESI-MS, electrospray ionization mass spectrometry; PH, pleckstrin homology; PI, phosphatidylinositol; PI(3)P, PI 3-phosphate; PIP, PI phosphate; PIP2, PI bisphosphate; SCV, _Salmonella_-containing vacuole; SPI, Salmonella pathogenicity island; T3SS, type III secretion system; WT, wild type.
References
- Anderson, K.E., and S.P. Jackson. 2003. Class I phosphoinositide 3-kinases. Int. J. Biochem. Cell Biol. 35:1028–1033. [DOI] [PubMed] [Google Scholar]
- Baldeon, M.E., B.P. Ceresa, and J.E. Casanova. 2001. Expression of constitutively active Rab5 uncouples maturation of the _Salmonella_-containing vacuole from intracellular replication. Cell. Microbiol. 3:473–486. [DOI] [PubMed] [Google Scholar]
- Birmingham, C.L., A.C. Smith, M.A. Bakowski, T. Yoshimori, and J.H. Brumell. 2006. Autophagy controls Salmonella infection in response to damage to the _Salmonella_-containing vacuole. J. Biol. Chem. 281:11374–11383. [DOI] [PubMed] [Google Scholar]
- Brumell, J.H., and S. Grinstein. 2004. Salmonella redirects phagosomal maturation. Curr. Opin. Microbiol. 7:78–84. [DOI] [PubMed] [Google Scholar]
- Bucci, C., A. Lutcke, O. Steele-Mortimer, V.M. Olkkonen, P. Dupree, M. Chiariello, C.B. Bruni, K. Simons, and M. Zerial. 1995. Co-operative regulation of endocytosis by three Rab5 isoforms. FEBS Lett. 366:65–71. [DOI] [PubMed] [Google Scholar]
- Campbell, R.E., O. Tour, A.E. Palmer, P.A. Steinbach, G.S. Baird, D.A. Zacharias, and R.Y. Tsien. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA. 99:7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carricaburu, V., K.A. Lamia, E. Lo, L. Favereaux, B. Payrastre, L.C. Cantley, and L.E. Rameh. 2003. The phosphatidylinositol (PI)-5-phosphate 4-kinase type II enzyme controls insulin signaling by regulating PI-3,4,5-trisphosphate degradation. Proc. Natl. Acad. Sci. USA. 100:9867–9872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoforidis, S., M. Miaczynska, K. Ashman, M. Wilm, L. Zhao, S.C. Yip, M.D. Waterfield, J.M. Backer, and M. Zerial. 1999. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat. Cell Biol. 1:249–252. [DOI] [PubMed] [Google Scholar]
- Drecktrah, D., L.A. Knodler, and O. Steele-Mortimer. 2004. Modulation and utilization of host cell phosphoinositides by Salmonella spp. Infect. Immun. 72:4331–4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukes, J.D., H. Lee, R. Hagen, B.J. Reaves, A.N. Layton, E.E. Galyov, and P. Whitley. 2006. The secreted Salmonella dublin phosphoinositide phosphatase, SopB, localizes to PtdIns(3)P-containing endosomes and perturbs normal endosome to lysosome trafficking. Biochem. J. 395:239–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dummler, B., and B.A. Hemmings. 2007. Physiological roles of PKB/Akt isoforms in development and disease. Biochem. Soc. Trans. 35:231–235. [DOI] [PubMed] [Google Scholar]
- Falasca, M., W.E. Hughes, V. Dominguez, G. Sala, F. Fostira, M.Q. Fang, R. Cazzolli, P.R. Shepherd, D.E. James, and T. Maffucci. 2007. The role of phosphoinositide 3-kinase C2alpha in insulin signaling. J. Biol. Chem. 282:28226–28236. [DOI] [PubMed] [Google Scholar]
- Galan, J.E., and H. Wolf-Watz. 2006. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 444:567–573. [DOI] [PubMed] [Google Scholar]
- Heo, W.D., T. Inoue, W.S. Park, M.L. Kim, B.O. Park, T.J. Wandless, and T. Meyer. 2006. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science. 314:1458–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez, L.D., K. Hueffer, M.R. Wenk, and J.E. Galan. 2004. Salmonella modulates vesicular traffic by altering phosphoinositide metabolism. Science. 304:1805–1807. [DOI] [PubMed] [Google Scholar]
- Hilbi, H. 2006. Modulation of phosphoinositide metabolism by pathogenic bacteria. Cell. Microbiol. 8:1697–1706. [DOI] [PubMed] [Google Scholar]
- Hoiseth, S.K., and B.A. Stocker. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 291:238–239. [DOI] [PubMed] [Google Scholar]
- Huang, F., A. Khvorova, W. Marshall, and A. Sorkin. 2004. Analysis of clathrin-mediated endocytosis of epidermal growth factor receptor by RNA interference. J. Biol. Chem. 279:16657–16661. [DOI] [PubMed] [Google Scholar]
- Knodler, L.A., and O. Steele-Mortimer. 2003. Taking possession: biogenesis of the _Salmonella_-containing vacuole. Traffic. 4:587–599. [DOI] [PubMed] [Google Scholar]
- Lindmo, K., and H. Stenmark. 2006. Regulation of membrane traffic by phosphoinositide 3-kinases. J. Cell Sci. 119:605–614. [DOI] [PubMed] [Google Scholar]
- Malecz, N., P.C. McCabe, C. Spaargaren, R. Qiu, Y. Chuang, and M. Symons. 2000. Synaptojanin 2, a novel Rac1 effector that regulates clathrin-mediated endocytosis. Curr. Biol. 10:1383–1386. [DOI] [PubMed] [Google Scholar]
- Marcus, S.L., M.R. Wenk, O. Steele-Mortimer, and B.B. Finlay. 2001. A synaptojanin-homologous region of Salmonella typhimurium SigD is essential for inositol phosphatase activity and Akt activation. FEBS Lett. 494:201–207. [DOI] [PubMed] [Google Scholar]
- Mason, D., G.V. Mallo, M.R. Terebiznik, B. Payrastre, B.B. Finlay, J.H. Brumell, L. Rameh, and S. Grinstein. 2007. Alteration of epithelial structure and function associated with PtdIns(4,5)P2 degradation by a bacterial phosphatase. J. Gen. Physiol. 129:267–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niebuhr, K., S. Giuriato, T. Pedron, D.J. Philpott, F. Gaits, J. Sable, M.P. Sheetz, C. Parsot, P.J. Sansonetti, and B. Payrastre. 2002. Conversion of PtdIns(4,5)P(2) into PtdIns(5)P by the S. flexneri effector IpgD reorganizes host cell morphology. EMBO J. 21:5069–5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris, F.A., M.P. Wilson, T.S. Wallis, E.E. Galyov, and P.W. Majerus. 1998. SopB, a protein required for virulence of Salmonella dublin, is an inositol phosphate phosphatase. Proc. Natl. Acad. Sci. USA. 95:14057–14059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattni, K., M. Jepson, H. Stenmark, and G. Banting. 2001. A PtdIns(3)P-specific probe cycles on and off host cell membranes during Salmonella invasion of mammalian cells. Curr. Biol. 11:1636–1642. [DOI] [PubMed] [Google Scholar]
- Pendaries, C., H. Tronchere, L. Arbibe, J. Mounier, O. Gozani, L. Cantley, M.J. Fry, F. Gaits-Iacovoni, P.J. Sansonetti, and B. Payrastre. 2006. PtdIns5P activates the host cell PI3-kinase/Akt pathway during Shigella flexneri infection. EMBO J. 25:1024–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong, S.B., Y. Hu, I. Enyedy, G. Powis, E.J. Meuillet, X. Wu, R. Wang, S. Wang, and A.P. Kozikowski. 2001. Molecular modeling studies of the Akt PH domain and its interaction with phosphoinositides. J. Med. Chem. 44:898–908. [DOI] [PubMed] [Google Scholar]
- Rusk, N., P.U. Le, S. Mariggio, G. Guay, C. Lurisci, I.R. Nabi, D. Corda, and M. Symons. 2003. Synaptojanin 2 functions at an early step of clathrin-mediated endocytosis. Curr. Biol. 13:659–663. [DOI] [PubMed] [Google Scholar]
- Schlumberger, M.C., and W.D. Hardt. 2006. Salmonella type III secretion effectors: pulling the host cell's strings. Curr. Opin. Microbiol. 9:46–54. [DOI] [PubMed] [Google Scholar]
- Scott, C.C., P. Cuellar-Mata, T. Matsuo, H.W. Davidson, and S. Grinstein. 2002. Role of 3-phosphoinositides in the maturation of _Salmonella_-containing vacuoles within host cells. J. Biol. Chem. 277:12770–12776. [DOI] [PubMed] [Google Scholar]
- Serunian, L.A., K.R. Auger, and L.C. Cantley. 1991. Identification and quantification of polyphosphoinositides produced in response to platelet-derived growth factor stimulation. Methods Enzymol. 198:78–87. [DOI] [PubMed] [Google Scholar]
- Shin, H.W., M. Hayashi, S. Christoforidis, S. Lacas-Gervais, S. Hoepfner, M.R. Wenk, J. Modregger, S. Uttenweiler-Joseph, M. Wilm, A. Nystuen, et al. 2005. An enzymatic cascade of Rab5 effectors regulates phosphoinositide turnover in the endocytic pathway. J. Cell Biol. 170:607–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddhanta, U., J. McIlroy, A. Shah, Y. Zhang, and J.M. Backer. 1998. Distinct roles for the p110α and hVPS34 phosphatidylinositol 3′-kinases in vesicular trafficking, regulation of the actin cytoskeleton, and mitogenesis. J. Cell Biol. 143:1647–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, A.C., W.D. Heo, V. Braun, X. Jiang, C. Macrae, J.E. Casanova, M.A. Scidmore, S. Grinstein, T. Meyer, and J.H. Brumell. 2007. A network of Rab GTPases controls phagosome maturation and is modulated by Salmonella enterica serovar Typhimurium. J. Cell Biol. 176:263–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauffer, T.P., S. Ahn, and T. Meyer. 1998. Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr. Biol. 8:343–346. [DOI] [PubMed] [Google Scholar]
- Steele-Mortimer, O., S. Meresse, J.P. Gorvel, B.H. Toh, and B.B. Finlay. 1999. Biogenesis of _Salmonella typhimurium_-containing vacuoles in epithelial cells involves interactions with the early endocytic pathway. Cell. Microbiol. 1:33–49. [DOI] [PubMed] [Google Scholar]
- Steele-Mortimer, O., L.A. Knodler, S.L. Marcus, M.P. Scheid, B. Goh, C.G. Pfeifer, V. Duronio, and B.B. Finlay. 2000. Activation of Akt/protein kinase B in epithelial cells by the Salmonella typhimurium effector sigD. J. Biol. Chem. 275:37718–37724. [DOI] [PubMed] [Google Scholar]
- Steele-Mortimer, O., J.H. Brumell, L.A. Knodler, S. Meresse, A. Lopez, and B.B. Finlay. 2002. The invasion-associated type III secretion system of Salmonella enterica serovar Typhimurium is necessary for intracellular proliferation and vacuole biogenesis in epithelial cells. Cell. Microbiol. 4:43–54. [DOI] [PubMed] [Google Scholar]
- Su, X., I.J. Lodhi, A.R. Saltiel, and P.D. Stahl. 2006. Insulin-stimulated interaction between insulin receptor substrate 1 and p85alpha and activation of protein kinase B/Akt require Rab5. J. Biol. Chem. 281:27982–27990. [DOI] [PubMed] [Google Scholar]
- Terebiznik, M.R., O.V. Vieira, S.L. Marcus, A. Slade, C.M. Yip, W.S. Trimble, T. Meyer, B.B. Finlay, and S. Grinstein. 2002. Elimination of host cell PtdIns(4,5)P(2) by bacterial SigD promotes membrane fission during invasion by Salmonella. Nat. Cell Biol. 4:766–773. [DOI] [PubMed] [Google Scholar]
- Tsolis, R.M., R.A. Kingsley, S.M. Townsend, T.A. Ficht, L.G. Adams, and A.J. Baumler. 1999. Of mice, calves, and men. Comparison of the mouse typhoid model with other Salmonella infections. Adv. Exp. Med. Biol. 473:261–274. [PubMed] [Google Scholar]
- Varnai, P., and T. Balla. 1998. Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J. Cell Biol. 143:501–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira, O.V., R.J. Botelho, L. Rameh, S.M. Brachmann, T. Matsuo, H.W. Davidson, A. Schreiber, J.M. Backer, L.C. Cantley, and S. Grinstein. 2001. Distinct roles of class I and class III phosphatidylinositol 3-kinases in phagosome formation and maturation. J. Cell Biol. 155:19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlahos, C.J., W.F. Matter, K.Y. Hui, and R.F. Brown. 1994. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002). J. Biol. Chem. 269:5241–5248. [PubMed] [Google Scholar]
- Weernink, P.A., Y. Guo, C. Zhang, M. Schmidt, C. Von Eichel-Streiber, and K.H. Jakobs. 2000. Control of cellular phosphatidylinositol 4,5-bisphosphate levels by adhesion signals and rho GTPases in NIH 3T3 fibroblasts involvement of both phosphatidylinositol-4-phosphate 5-kinase and phospholipase C. Eur. J. Biochem. 267:5237–5246. [DOI] [PubMed] [Google Scholar]
- Wenk, M.R., L. Lucast, G. Di Paolo, A.J. Romanelli, S.F. Suchy, R.L. Nussbaum, G.W. Cline, G.I. Shulman, W. McMurray, and P. De Camilli. 2003. Phosphoinositide profiling in complex lipid mixtures using electrospray ionization mass spectrometry. Nat. Biotechnol. 21:813–817. [DOI] [PubMed] [Google Scholar]
- Woscholski, R., T. Kodaki, M. McKinnon, M.D. Waterfield, and P.J. Parker. 1994. A comparison of demethoxyviridin and wortmannin as inhibitors of phosphatidylinositol 3-kinase. FEBS Lett. 342:109–114. [DOI] [PubMed] [Google Scholar]
- Yeung, T., and S. Grinstein. 2007. Lipid signaling and the modulation of surface charge during phagocytosis. Immunol. Rev. 219:17–36. [DOI] [PubMed] [Google Scholar]
- Yeung, T., G.E. Gilbert, J. Shi, J. Silvius, A. Kapus, and S. Grinstein. 2008. Membrane phosphatidylserine regulates surface charge and protein localization. Science. 319:210–213. [DOI] [PubMed] [Google Scholar]
- Zhou, D., L.M. Chen, L. Hernandez, S.B. Shears, and J.E. Galan. 2001. A Salmonella inositol polyphosphatase acts in conjunction with other bacterial effectors to promote host cell actin cytoskeleton rearrangements and bacterial internalization. Mol. Microbiol. 39:248–259. [DOI] [PubMed] [Google Scholar]