Dissecting β-ring assembly pathway of the mammalian 20S proteasome (original) (raw)
Abstract
The 20S proteasome is the catalytic core of the 26S proteasome. It comprises four stacked rings of seven subunits each, α1–7β1–7β1–7α1–7. Recent studies indicated that proteasome-specific chaperones and β-subunit appendages assist in the formation of α-rings and dimerization of half-proteasomes, but the process involved in the assembly of β-rings is poorly understood. Here, we clarify the mechanism of β-ring formation on α-rings by characterizing assembly intermediates accumulated in cells depleted of each β-subunit. Starting from β2, incorporation of β-subunits occurs in an orderly manner dependent on the propeptides of β2 and β5, and the C-terminal tail of β2. Unexpectedly, hUmp1, a chaperone functioning at the final assembly step, is incorporated as early as β2 and is required for the structural integrity of early assembly intermediates. We propose a model in which β-ring formation is assisted by both intramolecular and extrinsic chaperones, whose roles are partially different between yeast and mammals.
Keywords: assembly, chaperone, propeptide, proteasome, ubiquitin
Introduction
The ubiquitin–proteasome system is the main pathway for ATP-dependent non-lysosomal degradation of intracellular proteins in eukaryotes (Coux et al, 1996; Baumeister et al, 1998). Protein degradation achieved by this system is involved in various cellular processes, including cell cycle regulation, stress response, intracellular signalling, transcription regulation, and acquired immunity (Glickman and Ciechanover, 2002). Proteins involved in such regulatory processes as well as damaged proteins are recognized by the ubiquitin system and are marked for degradation by covalent attachment of polyubiquitin chains. Polyubiquitinated proteins are then recognized and degraded by the 26S proteasome, a 2.5-MDa multisubunit protease.
The 26S proteasome is composed of a catalytic core particle, called the 20S proteasome, bound at one or both ends by a 19S regulatory particle. The 20S proteasome is a cylindrical structure comprised of 28 subunits arranged in four stacked seven-membered rings. Each ring contains seven different subunits, whereby the two outer rings are formed by non-catalytic α-type subunits, named α1–α7, and the two inner rings are formed by the β-type subunits, β1–β7, three of which are catalytic (β1, β2, and β5) (Baumeister et al, 1998). Each of the 14 different proteins occupies a defined position within the 20S proteasome (Groll et al, 1997; Unno et al, 2002). Vertebrates encode four additional catalytic β-subunits; three interferon-γ-inducible β1i, β2i, and β5i and one thymus-specific β5t, which are incorporated in place of their most closely related β-subunits, thus forming distinct subtypes of proteasomes with altered catalytic activities called immunoproteasome and thymoproteasome (Tanaka and Kasahara, 1998; Murata et al, 2007).
The integrity of the 20S proteasome is assured by correct assembly of the 14 α-subunits and 14 β-subunits. All of the active β-subunits as well as non-catalytic β6 and β7 are synthesized with N-terminal propeptides, which are removed autocatalytically at the final step of the assembly to expose the catalytic threonine residues of β1, β2, and β5. The N-terminal active sites of β-subunits are on the inner surface of the β-rings, whereas the C termini of β-subunits are on the outer surface of the 20S proteasome (Groll et al, 1997). It has been shown that efficient assembly of the 20S proteasome is orchestrated by proteasome-specific chaperones such as PAC1 (Pba1 or POC1 in yeast), PAC2 (Pba2 or POC2 in yeast), PAC3 (Pba3, Dmp2, or POC3 in yeast), PAC4 (Pba4, Dmp1, or POC4 in yeast), hUmp1 (also known as POMP, proteassemblin in mammals and as Ump1 in yeast), the N-terminal propeptides of β-subunits, and C-terminal tails of β-subunits, which provide specific subunit interactions with _cis_- and _trans_-β-rings (Heinemeyer et al, 2004; Ramos et al, 2004; Hirano et al, 2005, 2006; Murata, 2006; Le Tallec et al, 2007; Li et al, 2007; Kusmierczyk et al, 2008; Yashiroda et al, 2008).
Proteasome assembly proceeds through distinct assembly intermediates. The earliest intermediate observed in mammalian cells is an α-ring that is comprised of all seven α-subunits, a PAC1–PAC2 heterodimer, and a PAC3–PAC4 complex (Hirano et al, 2005, 2006; Le Tallec et al, 2007). Both PAC1–PAC2 and PAC3–PAC4 are involved in the formation of α-rings. Recently, Pba3–Pba4 or Dmp1–Dmp2, yeast orthologues of PAC3–PAC4, has shown to catalyse correct subunit orientation of an α-ring (Kusmierczyk et al, 2008; Yashiroda et al, 2008). PAC1–PAC2 prevents non-productive dimerization of α-rings. The α-ring serves as a scaffold for the assembly of β-subunits. Another intermediate is the 13S complex composed of one α-ring, unprocessed β2, β3, β4, and (h)Ump1, both in yeast and mammals (Frentzel et al, 1994; Nandi et al, 1997; Li et al, 2007). Recent studies in yeast showed that the addition of the other β-subunits except β7 form the subsequent intermediate referred to as a ‘half-mer' precursor complex (Li et al, 2007; Marques et al, 2007). The 16S complex containing all the subunits and hUmp1 has been described also in mammalian cells (Schmidtke et al, 1997; Witt et al, 2000). A ‘half-proteasome' is often used as a general term for assembly intermediates containing unprocessed β-subunits and (h)Ump1. Studies in yeast have shown that dimerization of the half-mer is driven by the propeptide of β5 and the C-terminal tail of β7, whose incorporation into the half-proteasome is coupled with the dimerization, where the role of Ump1 is proposed to be an assembly checkpoint factor that inhibits the dimerization until a full set of β-subunits are recruited on the α-ring (Ramos et al, 2004; Li et al, 2007). Removal of β-propeptides and degradation of Ump1 coincide with completion of proteasome maturation, followed by degradation of PAC1–PAC2 (Ramos et al, 1998; Hirano et al, 2005). PAC3 is released from the intermediates during the maturation process (Hirano et al, 2006).
Several studies in yeast reported that the propeptides and the C-terminal tails of certain β-subunits have important roles in proteasome biogenesis. The propeptide of β5 is crucial for the incorporation of β5 during proteasome formation and is thus essential for life (Chen and Hochstrasser, 1996). The propeptides of β1 and β2 are dispensable for cell viability but are known to protect the N-terminal catalytic threonine residue against Nα-acetylation. In addition, mutants lacking these two propeptides displayed modest defects in proteasome biogenesis (Arendt and Hochstrasser, 1999). The C-terminal tail of β2, which wraps around β3 within the same β-ring, is also essential for proteasome biogenesis in yeast (Groll et al, 1997; Ramos et al, 2004). The C-terminal tail of β7, which is inserted into a groove between β1 and β2 in the opposite ring, also has an important function in dimerization of half-proteasomes as well as stabilization of active conformation of β1 (Groll et al, 1997; Ramos et al, 2004). In mammals, analysis of propeptides has been mainly conducted in the context of immunoproteasome formation, but there is little or no information on the C-terminal tails of β2 and β7, whose location in the mammalian proteasome closely resembles those of yeast β2 and β7 in the yeast proteasome (Unno et al, 2002).
Here, we describe a series of biochemical experiments employing RNA interference of each β-subunit, which resulted in the accumulation of distinct assembly intermediates. By characterizing these intermediates, we clarified the order of β-subunit incorporation on the α-ring. We also assessed the roles of propeptides and C-terminal tails of β-subunits in mammalian proteasome biogenesis, which revealed that these appendages mostly function in a manner similar to yeast counterparts but also displayed some phenotypes not observed in yeast. Furthermore, we identified a novel function of hUmp1 in stabilizing assembly intermediate of proteasomes that has not been appreciated in yeast.
Results
Ordered assembly of _β_-subunits on _α_-ring
During the assembly pathway from the α-ring through the half-proteasome, each β-subunit assembles on the α-ring. To clarify the order of incorporation of β-subunits, we used the strategy of small interfering RNA (siRNA)-mediated knockdown of each β-subunit, which was expected to result in arrest of the assembly process before the incorporation of the targeted subunit and accumulation of a specific intermediate.
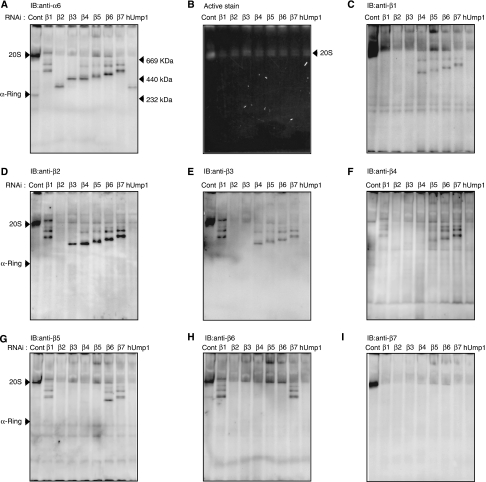
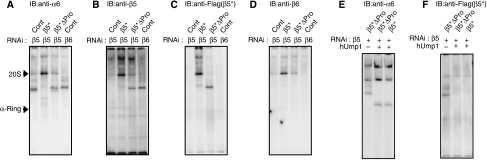
The total level of the different subunits as well as proteasome activity assessed by the peptide-hydrolysing activity of HEK293T cells transfected with siRNA targeting each β-subunit or hUmp1 was markedly reduced compared with those of control cells, suggesting that the biogenesis of proteasomes is severely impaired in each knockdown cell (Supplementary Figure S1). Each cell extract was resolved by native PAGE, followed by active staining or immunoblot analyses for α6- and all β-subunits (Figure 1). Immunoblot for α6 revealed accumulation of different complexes (molecular weight, 232–669 kDa) in each knockdown cell, as well as the normal α-ring in control cells, which have been shown to be a distinct assembly intermediate comprising all the seven α-subunits, PAC1–PAC2, and PAC3 (Hirano et al, 2006) (Figure 1A). Besides the major, fast-migrating band, a more slowly migrating minor band was observed for each knockdown cell except β2, β3, and hUmp1 knockdown (Figure 1A). Both the major and minor species did not show any peptide-hydrolysing activity, which was observed only in the complex of approximately 700-kDa, that is, 20S proteasomes (Figure 1B), indicating that they are assembly intermediates of 20S proteasomes.
Figure 1.
Accumulation of distinct assembly intermediates in each β-subunit knockdown cells. The cell extracts (40 μg) used in Supplementary Figure S1 were separated by native PAGE. Assembly intermediates were detected by immunoblotting using the indicated antibodies (A, C–I) The bands corresponding to α-ring and the 20S proteasome as well as the locations of molecular size markers are depicted by arrowheads. (B) The peptide-hydrolysing activity was assayed by active staining of the gel using Suc-LLVY-MCA in the presence of SDS. Note that the 26S proteasome did not move inside the native PAGE gel.
Among the seven β-subunits, β2 was the first assembled on the α-ring based on the finding that β2 was detected in all the intermediates except for that in its own knockdown (Figure 1D) and the intermediate that accumulated in β2-knockdown cells did not contain any β-subunit (Figure 1C–I, lanes for β2 RNAi). The assembly of β3 followed that of β2; β3 was detected in the intermediates observed in β1-, β4-, β5-, β6-, and β7-knockdown cells, and thus the incorporation of β3 should precede these subunits (Figure 1E). This view was further confirmed by the fact that the intermediate in β3-knockdown cells contained only β2 among the β-subunits (Figure 1C–I, lanes for β3 RNAi). β3 assembly was followed by β4 incorporation, which would result in the formation of the 13S complex, comprising the α-ring plus β2, β3, and β4, as suggested by the presence of β4 in the intermediate in β1, β5, β6, and β7 knockdown (Figure 1F), consistent with the previous reports that identified the 13S complex as a distinct entity of proteasome precursors (Frentzel et al, 1994; Nandi et al, 1997; Li et al, 2007).
β5 was the next β-subunit incorporated into the 13S complex because β5 was detected only in the intermediates in β1-, β6-, and β7-knockdown cells (Figure 1G). The assembly of β6 followed that of β5, as evidenced by the presence of β6 in the intermediates of β1 and β7 knockdown (Figure 1H). β7 was likely the last β-subunit incorporated in the precursor proteasomes because β7 was not found in any of the intermediate complexes (Figure 1I) and because the intermediate observed in β7-knockdown cells contained all the β-subunits with the exception of β7 (Figure 1C–I, lanes for β7 RNAi). The behaviour of β1 was rather elusive. The intermediate in β1-knockdown cells contained β2, β3, β4, β5, and β6 (Figure 1C–I, lanes for β1 RNAi), whereas β1 was already included in the intermediates of β4, β5, β6, and β7 knockdown (Figure 1C). The former observation suggests that β1 was incorporated following β2, β3, β4, β5, and β6, and that β1 is required for β7 incorporation. The latter observation suggests that the presence of β2 and β3 is sufficient for the incorporation of β1 and that β1 can be incorporated anytime during the maturation pathway from the complex containing both β2 and β3 through the half-mer.
Association of PA28, Hsp90, and Hsc70 with 20S proteasome precursors
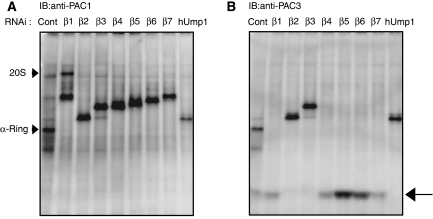
When the same panel was probed for PAC1, the major assembly intermediate bands were associated with PAC1 (Figure 2A), which has been shown to be retained in the proteasome precursor until the completion of the assembly (Hirano et al, 2005). However, the slowly migrating minor bands above the major bands did not contain PAC1, whereas the composition of each major and minor bands in terms of α- and β-subunits is identical (Figures 1 and 2A). It is also curious that the intermediate in β2-knockdown cells was apparently larger than the α-ring (Figure 1A), although the subunit composition is supposed to be identical to that of the α-ring. To address the identity of these bands, we tested whether PA28, PA200, Hsp90α, and Hsc70, which have been reported to be involved in proteasome biogenesis (Schmidtke et al, 1997; Preckel et al, 1999; Fehlker et al, 2003; Imai et al, 2003; Marques et al, 2007), associate with the intermediates.
Figure 2.
Release of PAC3 is coupled with β3 incorporation. The same panels in Figure 1 were probed with anti-PAC1 (A) and -PAC3 (B) antibodies. The arrow indicates PAC3 species dissociated from proteasome precursors (B).
PA28 was associated with the slow-migrating minor bands but not with the primary bands, different from PAC1 and Hsp90α, which were detected only in the major bands (Supplementary Figure S2A and B). Hsc70 was observed in both the major and the minor bands (Supplementary Figure 2C). Neither Hsp90α nor Hsc70 was detected in the α-ring. By contrast, PA200, whose yeast orthologue Blm10 was shown to associate with nascent proteasomes (Fehlker et al, 2003; Marques et al, 2007), was not observed in the intermediates, whereas its association with 20S proteasomes was detected (Supplementary Figure S2D, arrowhead). However, we cannot conclude that PA200 is not bound to assembly intermediates as free forms of PA200, which probably dissociated from 20S or nascent proteasomes during native PAGE analysis, were found (Supplementary Figure S2D, arrow).
The association of Hsp90 and Hsc70 with the assembly intermediates accounts for the increased size of the intermediate in β2-knockdown cells and suggests that recruitment of these chaperones precedes β2 and hUmp1 incorporation. The minor bands are characterized by the association of PA28, a 200-kDa heterohexameric complex. At present, we do not know whether these molecules really have some functions in the proteasome biogenesis or are associated with the intermediates as an experimental artefact. Further studies are needed to answer this question.
Release of PAC3 upon incorporation of _β_3
We previously showed that precursor proteasomes purified with tagged hUmp1 did not contain PAC3 and demonstrated that PAC3 is released during the maturation pathway of the mammalian proteasome (Hirano et al, 2006). To elucidate the step where PAC3 was released, we took advantage of the knockdown experiments in which distinct assembly intermediates accumulated depending on which β-subunit was targeted (Figure 1).
The same panel in Figure 1 was probed with anti-PAC1 and -PAC3 antibody. All the assembly intermediates as well as the α-ring were associated with PAC1 (Figure 2A) (Hirano et al, 2005). PAC3 is also associated with the α-ring in control cells as reported previously (Hirano et al, 2006). However, PAC3 was associated with intermediates of β2-, β3-, and hUmp1-knockdown cells but not with those of others, where PAC3 was found as fast migrating species, presumably as a free complex (Figure 2B, arrow). Considering the order of incorporation of β-subunits shown in Figure 1, the release of PAC3 is apparently coupled with the incorporation of β3.
A new role of hUmp1 in the assembly pathway
One intriguing difference in the phenotypes of loss of Ump1 orthologues between yeast and mammals is that knockdown of hUmp1 in mammalian cells did not result in the accumulation of intermediates containing unprocessed β-subunits (Figure 1, lanes for hUmp1 RNAi), whereas deletion of Ump1 in yeast caused apparent accumulation of such intermediates (Ramos et al, 1998). This observation in hUmp1-knockdown cells has also been shown in previous studies (Hirano et al, 2005, 2006). This finding raises the possibility that the role of Ump1 orthologues is different between yeast and mammals.
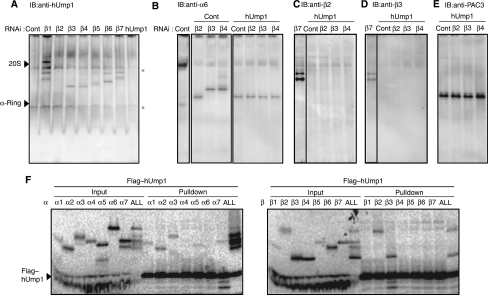
To determine the step at which hUmp1 is incorporated, the same panel in Figure 1 was probed with anti-hUmp1 antibody. hUmp1 was included in a complex other than that in β2-knockdown cells (Figure 3A), indicating that the incorporation of hUmp1 precedes that of β3. On the other hand, the intermediate in the hUmp1 knockdown complex did not contain any of the β-subunits, including β2, a finding closely resembling that in β2-knockdown cells with regard to size and composition (Figures 1 and 2; compare lanes for β2 RNAi to lanes for hUmp1 RNAi). These results suggest that incorporations of β2 and hUmp1 are coupled with each other and that loss of either result in dissociation of the other.
Figure 3.
Role of hUmp1 in the structural integrity of early assembly intermediates. (A) The same panel in Figure 1 was probed with anti-hUmp1 antibody. (B–E) Extracts of HEK293T cells transfected with the indicated combinations of siRNAs were separated by native PAGE. Intermediate complexes were detected by immunoblotting using the indicated antibodies. The left lane representing β7 RNAi serves as a positive control for immunoblotting (C, D). Asterisks indicate nonspecific bands (A). (F) Flag–hUmp1 and each 20S subunit were co-translated and radio-labelled in reticulocyte lysates, immunoprecipitated with M2 agarose, and analysed by SDS–PAGE and autoradiography. ‘ALL' represents co-translation of all β-subunits together with hUmp1.
To confirm this concept, hUmp1 was knocked down concurrently with β2, β3, or β4, and the resultant assembly intermediates were compared with those in β2, β3, or β4 single-knockdown cells. Intriguingly, the size of intermediates observed in the simultaneous knockdown of hUmp1 with β3 or β4 was similar to that of the complex in β2 single-knockdown cells (Figure 3B). Notably, the intermediate found in β3–hUmp1 double-knockdown cells lacked β2 (Figure 3C), which was found in the complex of β3 single-knockdown cells (Figure 1D). The β4 and hUmp1 double knockdown was associated with loss of both β2 and β3 in the intermediate (Figure 3C and D), which were clearly detected in the β4 single knockdown complex (Figure 1D and E). Furthermore, the complexes manifested in the double knockdown cells were associated with PAC3, consistent with the absence of β3, whose incorporation would detach PAC3 from the precursor proteasome (Figures 2B and 3E).
To gain mechanistic insight into the early function of hUmp1, we tested the interactions between hUmp1 and each 20S proteasome subunit. hUmp1 could directly bind to β2 and β3 as well as some of the α-subunits (α2, α3, α5, and α7) (Figure 3F). This observation raises the possibility that hUmp1, either alone or as a complex with β2 and β3, is recruited on the α-ring through direct interaction between hUmp1 and certain α-subunits.
Taken together, these results demonstrate that β2 is unable to associate with the α-ring without hUmp1 and suggest the important role of hUmp1 in promoting the maturation process beyond the α-ring, either by stabilizing the complex or by recruiting β2.
Propeptides of _β_1, _β_6, and _β_7 are dispensable for proteasome maturation
It has been reported that the propeptides and C-terminal tails of β-subunits have important roles in proteasome biogenesis in yeast (Chen and Hochstrasser, 1996; Ramos et al, 2004; Li et al, 2007; Marques et al, 2007), but little is known about their roles in the maturation of proteasomes in mammals.
To elucidate the role of propeptides of β1, β2, β5, β6, and β7, and the C-terminal tails of β2 and β7 in mammals, we first established cell lines stably transfected with constructs encoding wild-type subunits (β1*, β2*, β5*, β6*, and β7*), mature subunits whose propeptides were replaced with ubiquitin (β1*Δpro, β2*Δpro, β5*Δpro, β6*Δpro, and β7*Δpro), and β2 and β7 lacking their C-terminal tails (β2*Δtail and β7*Δtail) (Supplementary Figure S3A). Synonymous mutations were introduced into these constructs so that they were not sensitive to siRNAs against each β-subunit used in Figure 1. These constructs were attached with C-terminal Flag tag to distinguish the expressed proteins from endogenous proteins. We first confirmed the expression of both precursor and mature forms of β-subunits in HEK293T cells transfected with constructs encoding wild-type and Δtail β-subunits (Supplementary Figure S3B). On the other hand, as expected, only mature forms of β-subunits were detected in cells transfected with ubiquitin-fused Δpro constructs (Supplementary Figure S3B). These ubiquitin-fused proteins are known to be cleaved rapidly by cellular deubiquitinating enzymes to generate free ubiquitin and the mature moiety of the proteasome subunit, so that the exposure and integrity of the N-terminal residue are ensured (Chen and Hochstrasser, 1996; Arendt and Hochstrasser, 1999; Jager et al, 1999). siRNA-mediated knockdown of endogenous subunits in these cells allowed us to determine the precise roles of propeptides and C-terminal tails.
Expression of constructs encoding each wild-type subunit (β1*, β2*, β5*, β6*, and β7*) restored production of 20S proteasomes and rescued cells from death caused by siRNA treatment (data not shown), verifying that exogenously expressed constructs worked appropriately (Figures 4A, 5A and 6A; Supplementary Figures S4 and S5). Among the Δpro constructs, cells expressing β1*Δpro, β6*Δpro, and β7*Δpro grew apparently normal and produced 20S proteasomes at an amount comparable to wild-type expressing cells (Supplementary Figures S4 and S5; Figure 6A). These findings indicate that the propeptides of β1, β6, and β7 are not prerequisite for proteasome maturation in mammals, similar to the results in yeast. On the other hand, cells expressing β2*Δpro, β5*Δpro, β2*Δtail, and β7*Δtail were non-viable in the absence of their endogenous counterparts (data not shown), suggesting the indispensable roles of these propeptides and C-terminal tails in proteasome biogenesis in mammals.
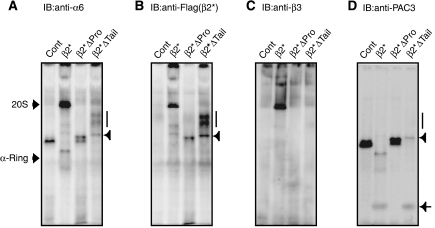
Figure 4.
Both the propeptide and C-terminal tail of β2 are indispensable for β3 incorporation. Stable cell lines expressing the indicated mutant β2-subunits were treated with the siRNA targeting endogenous β2. Intermediate complexes were detected by immunoblotting using the indicated antibodies following native PAGE (A–D). Intermediates observed in β2*ΔTail cells can be divided into two species; faster migrating ones (arrowheads) and slower migrating ones (vertical bars). The free complex of PAC3 is depicted by an arrow (D).
Figure 5.
β5 propeptide is required for β6 incorporation. Stable cell lines expressing the indicated mutant β5-subunits were treated with the siRNA(s) for endogenous β5 or β6 (A–D), or for the indicated combinations (E, F). Cell extracts were resolved by native PAGE, followed by immunoblot analysis for the indicated antibodies.
Figure 6.
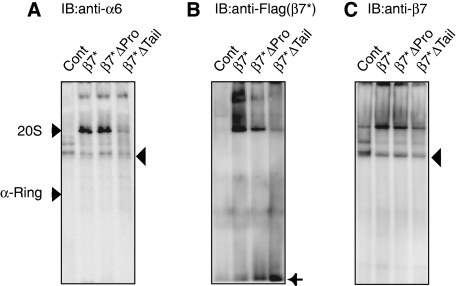
C-terminal tail of β7 is essential for the incorporation of β7 and dimerization of half-mers. Stable cell lines expressing the indicated mutant β7-subunits were treated with siRNA targeting endogenous β7. Cell extracts were resolved by native PAGE, followed by immunoblot analysis using the indicated antibodies. The free form of β7*Δtail is depicted by an arrow (B). The ‘half-mer' assembly intermediates are depicted by arrowheads (A, C).
Both the propeptide and C-terminal tail of _β_2 are required for incorporation of _β_3
To clarify the role of the propeptide and C-terminal tail of β2, extracts resolved by native PAGE were probed with several antibodies (Figure 4). β2*Δpro- and β2*Δtail-expressing cells showed accumulation of intermediates that included α6 and β2, indicating that β2 propeptide and β2 tail are not required for the incorporation of β2 itself (Figure 4A and B). However, these intermediates did not contain β3 (Figure 4C), suggesting that the assembly pathway is arrested before β3 incorporation and that both β2 propeptide and β2 tail are required for the incorporation of β3. PAC3 was also found in the intermediates in β2*Δpro cells and the faster migrating intermediates in β2*Δtail cells (indicated by an arrowhead), consistent with lack of β3 (Figure 4D). However, the slower migrating intermediates in β2*Δtail cells (indicated by the bar) did not include PAC3 (Figure 4D). These species did not contain any β-subunits other than β2Δtail (Supplementary Figure S6), precluding the possibility that loss of β2 tail and PAC3 causes disordered incorporation of β-subunits. Rather, it is likely that they represent either aggregation of intermediates or association of other molecules, which would be prevented in the presence of β2 tail or PAC3.
_β_5 propeptide is required for incorporation of _β_6 but not hUmp1-dependent proteasome maturation
Next, we examined the role of β5 propeptide. As expected from its lethality, β5*Δpro-expressing cells could not produce mature 20S proteasomes but showed accumulation of an intermediate that included α6 and β5, indicating that β5 propeptide is not essential for the incorporation of β5 itself (Figure 5A–C). This intermediate did not contain the next subunit β6 (Figure 5D) and represented the same size as the intermediate observed in β6-knockdown cells (Figure 5A and B), suggesting that β5 propeptide is required for the incorporation of β6 in mammalian cells.
In yeast, deletion of β5 propeptide, which is fatal, was rescued by concomitant loss of Ump1, suggesting the role of β5 propeptide in Ump1-dependent maturation of the yeast proteasome (Ramos et al, 1998). In the next series of experiment, siRNAs for both hUmp1 and β5 were transfected into β5*Δpro-expressing mammalian cells. Unlike the observation in yeast, however, simultaneous loss of β5 propeptide and hUmp1 was still lethal in mammalian cells, with the accumulation of an intermediate closely resembling to that observed in hUmp1 single-knockdown cells that did not contain β5* (Figure 5E and F). Although these results do not exclude the checkpoint function of hUmp1 as proposed in yeast Ump1 (Li et al, 2007), they confirm the important role of hUmp1 in the integrity of early assembly intermediates, which is a function independent of β5 propeptide.
Importance of C-terminal tail of _β_7 for stable incorporation of _β_7
β7*Δpro-expressing cells grew normal, incorporated β7*, and produced 20S proteasomes similar to wild-type β7*-expressing cells (Figure 6A–C). However, deletion of the C-terminal tail could not rescue loss of endogenous β7, and the cells could hardly produce 20S proteasomes (Figure 6A–C). In these cells, β7*Δtail failed to be incorporated in the assembly intermediates, presumably half-mers (Li et al, 2007), as suggested by the similar size as those in β7-knockdown cells and by the presence of β6 (Figure 6A and C, arrowheads) and accumulation of β7*Δtail as a free subunit (Figure 6B, arrow). These observations indicate that the C-terminal tail, which directly associates with the trans-β-ring, is essential for its stable incorporation into proteasomes and supports the model where β7 incorporation is tightly coupled with dimerization of half-mers, thus forming 20S proteasomes, as proposed in previous reports in yeast (Li et al, 2007; Marques et al, 2007).
Discussion
In the present study, we investigated in detail the assembly pathway of mammalian 20S proteasome and found a strictly ordered β-subunit incorporation, which was supported by propeptides of certain β-subunits. Starting from recruitment of β2 and hUmp1 on the α-ring, the adjacent β-subunit within the same β-ring, except β1, appears to assemble one after another, β7 being the last subunit whose incorporation is tightly coupled with dimerization of half-mers (see the model illustrated in Figure 7). This view is also supported by the observation that the size of the assembly intermediate in each knockdown cells increased as the maturation process proceeded (Figure 1A). The order of the β-subunit recruitment clarified in this study is entirely consistent with previous findings where β-subunits were separated into two categories; β2, β3, and β4 as ‘early' subunits and others as ‘late' subunits (Heinemeyer et al, 2004). In both yeast and mammals, the 13S complex containing β2, β3, β4, and (h)Ump1 was observed as a distinct assembly intermediate, suggesting that the following step, that is, incorporation of β5, is one of the rate-limiting steps in proteasome biogenesis. This may account for the intriguing observation that sole overexpression of β5 in mammalian cells increased the amount of assembled proteasomes (Chondrogianni et al, 2005). Although our data imply a stepwise addition of β-subunits on the α-ring, we cannot preclude the possibility that some β-subunits are incorporated as a group, for example as a β2–β3–β4–hUmp1 complex, under normal conditions as we observed intermediate species that can be seen only when a certain β-subunit is depleted.
Figure 7.
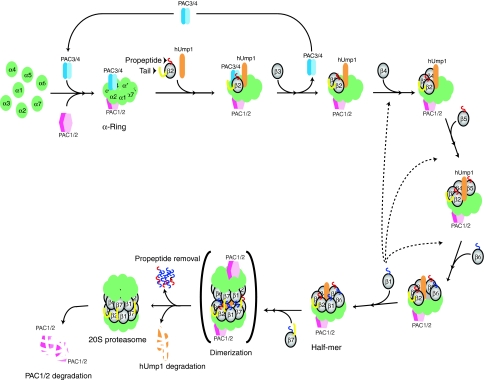
A model for β-ring formation in mammalian 20S proteasome assembly. The roles of PAC1–PAC2 and PAC3 in the formation of α-rings were described previously (Hirano et al, 2005, 2006). PAC4 was recently identified as a heterodimeric partner of PAC3 (Le Tallec et al, 2007). Sequential incorporation of β-subunits starts from the association of β2 and hUmp1 on the α-ring. hUmp1 is required for the association of β2 in the early assembly intermediates. PAC3–PAC4, whose release is coupled with association of β3, holds the structural integrity of the intermediates until β3 is incorporated on the α-ring. Subsequent orderly incorporation of other β-subunits is also assisted by intramolecular chaperones such as the propeptides of β2 and β5 and the C-terminal tail of β2. Although β1 can be incorporated at various steps (dotted lines), such incorporation most likely follows that of β6. Dimerization of half-mers is assisted by the C-terminal tail of β7. This is followed by removal of β-subunit propeptides (β1, β2, β5, β6, and β7) and hUmp1 degradation. Essential propeptides, non-essential propeptides, and essential C-terminal tails of β-subunits for mammalian 20S proteasome biogenesis are depicted in red, blue, and yellow, respectively. See text for more details.
β1 appears to be the last but one incorporated during proteasome assembly, which is a prerequisite for β7 incorporation. However, β1 can also be incorporated earlier than β4, β5, and β6 in the absence of these subunits, whereas the other subunits are assembled in a manner strictly dependent on the preceding incorporation of the neighbouring subunit. Although such β1-containing intermediates might be artefacts under non-physiological situations, this recalls the early assembly intermediate observed during immunoproteasome biogenesis, which contains β1i, β2i, β3, and β4 (Nandi et al, 1997). β1i is incorporated into the precursor earlier than β1, which has an important function in the immunoproteasome assembly (Griffin et al, 1998).
In the present study, we identified several new roles for the propeptides and the C-terminal tails of mammalian β-subunits that have not been appreciated in those of yeast β-subunits. The propeptide of yeast β2 is dispensable for efficient proteasome biogenesis (Arendt and Hochstrasser, 1999), but that of mammalian β2 contributes to the recruitment of its neighbouring subunit β3 (Figure 4). The propeptide of yeast β5 is reported to be required for its own incorporation (Chen and Hochstrasser, 1996), whereas that of mammalian β5 was not; instead, its loss caused failure of β6 recruitment (Figure 5). The role of the C-terminal tail of mammalian β7 appears to be quite similar to that of yeast β7, but it proved to be an essential component in proteasome biogenesis, unlike yeast β7 (Figure 6).
Our study also clarified the sequences leading to hUmp1 incorporation and release of PAC3 from proteasome precursors. Incorporation of hUmp1 was as early as that of β2, the first β-subunit assembled on the α-ring. In the early assembly intermediates, hUmp1 unexpectedly had an important function in the association of β2 with the precursor proteasomes and proved to be a prerequisite for the assembly of β-subunits on the α-ring (Figures 3 and 7). This function is not appreciated for yeast Ump1, which regulates dimerization of half-proteasomes and maturation of 20S proteasomes, presumably by acting as a checkpoint protein (Ramos et al, 1998; Li et al, 2007). As the phenotype of loss of hUmp1 emerged at a stage upstream of the dimerization, we are uncertain whether hUmp1 in mammalian cells also has a function similar to that in yeast.
In a previous study, we showed that PAC3 was scarcely included in the complex purified by a tag attached to hUmp1 (Hirano et al, 2006). In the present study, we clarified that PAC3 release was coupled with association of β3 with the assembly intermediate. This observation is consistent with the recent study showing that Dmp1–Dmp2, yeast orthologue of PAC3–PAC4, was copurified with β2, but not with other β-subunits (Yashiroda et al, 2008). Therefore, a complex containing both hUmp1 and PAC3, together with β2, is assumed to be present during the maturation pathway, and such a complex was indeed observed in β3-knockdown cells (Figure 1). As PAC3 is known to bind directly to β3 in vitro (Hirano et al, 2006), it may be argued that PAC3 facilitates the incorporation of β3, which might induce a conformational alteration of the assembly intermediate to release PAC3. Because PAC3 is also required for efficient α-ring formation (Hirano et al, 2006), we could not detect accumulation of an intermediate before the incorporation of β3 in PAC3-knockdown cells, such as a complex comprising α-ring and β2, which would be expected to occur on the assumption that PAC3 is required for β3 incorporation (Supplementary Figure S7). Although biochemical data both in yeast and mammals indicated that PAC3 is replaced when β3 enter, a model generated by superimposing Dmp1–Dmp2–α5 complex on the yeast 20S proteasome suggested steric hindrance of β4-subunit to Dmp1 (Yashiroda et al, 2008). The precise reason for this discrepancy is not clear at present. This may be simply because the model, which was based on the structure of Dmp1–Dmp2 determined in the absence of α-ring, does not represent bona fide structure of assembly intermediates. Further studies should be required for understanding the role of PAC3 in β-subunit incorporation.
Recently, the proteasome-specific inhibitor bortezomib was used clinically in refractory multiple myeloma, and a clinical trial of this agent in other malignant neoplasms is currently underway (Adams, 2004). Accordingly, the proteasome is now recognized as a potent target for cancer therapy (Adams, 2004). Assuming that inhibition of proteasome biogenesis is also beneficial in cancer therapy, it is important to understand the detailed mechanism of mammalian proteasome biogenesis. Our results could provide the foundation for the design and development of novel anticancer drugs that target proteasome biogenesis.
Materials and methods
DNA constructs
The cDNAs encoding β-subunits and those derivatives were cloned into pIRESpuro3 (Clontech) in frame with a C-terminal Flag tag. To make constructs for β-subunits resistant to siRNAs, synonymous mutagenesis in RNAi targeting sequences was performed using the QuikChange site-directed mutagenesis kit (Stratagene). Plasmids encoding β1*Δpro, β2*Δpro, β5*Δpro, β6*Δpro, and β7*Δpro were constructed by fusing human ubiquitin cDNA to the 5′ end of the cDNAs encoding mature forms of the corresponding subunits. In β2*Δtail and β7*Δtail, sequences encoding amino acids 244–277 and 248–264 were deleted, respectively. PCR was performed using Phusion DNA polymerase (Finnzymes), and all constructs were confirmed by sequencing.
Cell culture
HEK293T cell lines were cultured in Dulbecco's modified Eagle's medium (Sigma), supplemented with 10% fetal calf serum, 100 IU/ml penicillin G, 100 μg/ml streptomycin sulphate (all from Invitrogen). Stable transfection of HEK293T cells was performed using Fugene 6 (Roche), and the cells were selected with 5 μg/ml of puromycin (Sigma).
Protein extracts, immunological analysis, and antibodies
Cells were lysed in an ice-cold lysis buffer (50 mM Tris–HCl (pH 7.5), 0.5% (v/v) NP-40, 1 mM dithiothreitol, 2 mM ATP, and 5 mM MgCl2) and the extracts were clarified by centrifugation at 20 000 g for 15 min at 4°C. The supernatants were subjected to glycerol gradient analysis or native PAGE. Glycerol gradient analysis and assay of proteasome activity were described previously (Murata et al, 2001; Hirano et al, 2006). In vitro binding assay was described previously (Hirano et al, 2006). SDS–PAGE (12% Bis-Tris gel (Invitrogen)) and native PAGE (7% Tris–acetate gel (Invitrogen)) were performed according to the instructions provided by the manufacturer. The separated proteins were transferred onto polyvinylidene difluoride membrane and reacted with the indicated antibody. Anti-PAC1, PAC2, PAC3, hUmp1, β4 (55F8), β5 (P93250), β6 (P93199), and PA28α polyclonal antibodies were described previously (Tanahashi et al, 2000; Hirano et al, 2006). Antibodies against proteasome α6 (MCP20), β1 (MCP421), β2 (MCP168), β3 (MCP102), β7 (MCP205), and PA200 were purchased from BioMol. Anti-Hsp90α and anti-Hsc70 were obtained from MBL. Anti-FLAG M2 antibody was from Sigma.
RNA interference
The siRNAs targeting human β-subunits and hUmp1 (Supplementary Table S1) were transfected into HEK293T cells using Lipofectamine RNAi MAX (Invitrogen) at a final concentration of 50 nM in six-well dishes twice at a 12-h interval. The cells were analysed 24 h after the second transfection.
Supplementary Material
Supplementary Information
Acknowledgments
This study was supported by grants to SM and KT from the Ministry of Education, Science and Culture of Japan (MEXT), Target Protein Project of MEXT (to KT and KK), and the Takeda Science Foundation (KT).
References
- Adams J (2004) The proteasome: a suitable antineoplastic target. Nat Rev Cancer 4: 349–360 [DOI] [PubMed] [Google Scholar]
- Arendt CS, Hochstrasser M (1999) Eukaryotic 20S proteasome catalytic subunit propeptides prevent active site inactivation by N-terminal acetylation and promote particle assembly. EMBO J 18: 3575–3585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister W, Walz J, Zuhl F, Seemuller E (1998) The proteasome: paradigm of a self-compartmentalizing protease. Cell 92: 367–380 [DOI] [PubMed] [Google Scholar]
- Chen P, Hochstrasser M (1996) Autocatalytic subunit processing couples active site formation in the 20S proteasome to completion of assembly. Cell 86: 961–972 [DOI] [PubMed] [Google Scholar]
- Chondrogianni N, Tzavelas C, Pemberton AJ, Nezis IP, Rivett AJ, Gonos ES (2005) Overexpression of proteasome beta5 assembled subunit increases the amount of proteasome and confers ameliorated response to oxidative stress and higher survival rates. J Biol Chem 280: 11840–11850 [DOI] [PubMed] [Google Scholar]
- Coux O, Tanaka K, Goldberg AL (1996) Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem 65: 801–847 [DOI] [PubMed] [Google Scholar]
- Fehlker M, Wendler P, Lehmann A, Enenkel C (2003) Blm3 is part of nascent proteasomes and is involved in a late stage of nuclear proteasome assembly. EMBO Rep 4: 959–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frentzel S, Pesold-Hurt B, Seelig A, Kloetzel PM (1994) 20S proteasomes are assembled via distinct precursor complexes. Processing of LMP2 and LMP7 proproteins takes place in 13–16S preproteasome complexes. J Mol Biol 236: 975–981 [DOI] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A (2002) The ubiquitin–proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 82: 373–428 [DOI] [PubMed] [Google Scholar]
- Griffin TA, Nandi D, Cruz M, Fehling HJ, Kaer LV, Monaco JJ, Colbert RA (1998) Immunoproteasome assembly: cooperative incorporation of interferon gamma (IFN-gamma)-inducible subunits. J Exp Med 187: 97–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R (1997) Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature 386: 463–471 [DOI] [PubMed] [Google Scholar]
- Heinemeyer W, Ramos PC, Dohmen RJ (2004) The ultimate nanoscale mincer: assembly, structure and active sites of the 20S proteasome core. Cell Mol Life Sci 61: 1562–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano Y, Hayashi H, Iemura S, Hendil KB, Niwa S, Kishimoto T, Kasahara M, Natsume T, Tanaka K, Murata S (2006) Cooperation of multiple chaperones required for the assembly of mammalian 20S proteasomes. Mol Cell 24: 977–984 [DOI] [PubMed] [Google Scholar]
- Hirano Y, Hendil KB, Yashiroda H, Iemura S, Nagane R, Hioki Y, Natsume T, Tanaka K, Murata S (2005) A heterodimeric complex that promotes the assembly of mammalian 20S proteasomes. Nature 437: 1381–1385 [DOI] [PubMed] [Google Scholar]
- Imai J, Maruya M, Yashiroda H, Yahara I, Tanaka K (2003) The molecular chaperone Hsp90 plays a role in the assembly and maintenance of the 26S proteasome. EMBO J 22: 3557–3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager S, Groll M, Huber R, Wolf DH, Heinemeyer W (1999) Proteasome beta-type subunits: unequal roles of propeptides in core particle maturation and a hierarchy of active site function. J Mol Biol 291: 997–1013 [DOI] [PubMed] [Google Scholar]
- Kusmierczyk AR, Kunjappu MJ, Funakoshi M, Hochstrasser M (2008) A multimeric assembly factor controls the formation of alternative 20S proteasomes. Nat Struct Mol Biol 15: 237–244 [DOI] [PubMed] [Google Scholar]
- Le Tallec B, Barrault MB, Courbeyrette R, Guerois R, Marsolier-Kergoat MC, Peyroche A (2007) 20S proteasome assembly is orchestrated by two distinct pairs of chaperones in yeast and in mammals. Mol Cell 27: 660–674 [DOI] [PubMed] [Google Scholar]
- Li X, Kusmierczyk AR, Wong P, Emili A, Hochstrasser M (2007) Beta-subunit appendages promote 20S proteasome assembly by overcoming an Ump1-dependent checkpoint. EMBO J 26: 2339–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques AJ, Glanemann C, Ramos PC, Dohmen RJ (2007) The C-terminal extension of the beta 7 subunit and activator complexes stabilize nascent 20s proteasomes and promote their maturation. J Biol Chem 282: 34869–34876 [DOI] [PubMed] [Google Scholar]
- Murata S (2006) Multiple chaperone-assisted formation of mammalian 20S proteasomes. IUBMB Life 58: 344–348 [DOI] [PubMed] [Google Scholar]
- Murata S, Sasaki K, Kishimoto T, Niwa S, Hayashi H, Takahama Y, Tanaka K (2007) Regulation of CD8+ T cell development by thymus-specific proteasomes. Science 316: 1349–1353 [DOI] [PubMed] [Google Scholar]
- Murata S, Udono H, Tanahashi N, Hamada N, Watanabe K, Adachi K, Yamano T, Yui K, Kobayashi N, Kasahara M, Tanaka K, Chiba T (2001) Immunoproteasome assembly and antigen presentation in mice lacking both PA28alpha and PA28beta. EMBO J 20: 5898–5907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi D, Woodward E, Ginsburg DB, Monaco JJ (1997) Intermediates in the formation of mouse 20S proteasomes: implications for the assembly of precursor beta subunits. EMBO J 16: 5363–5375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preckel T, Fung-Leung WP, Cai Z, Vitiello A, Salter-Cid L, Winqvist O, Wolfe TG, Von Herrath M, Angulo A, Ghazal P, Lee JD, Fourie AM, Wu Y, Pang J, Ngo K, Peterson PA, Fruh K, Yang Y (1999) Impaired immunoproteasome assembly and immune responses in PA28−/− mice. Science 286: 2162–2165 [DOI] [PubMed] [Google Scholar]
- Ramos PC, Hockendorff J, Johnson ES, Varshavsky A, Dohmen RJ (1998) Ump1p is required for proper maturation of the 20S proteasome and becomes its substrate upon completion of the assembly. Cell 92: 489–499 [DOI] [PubMed] [Google Scholar]
- Ramos PC, Marques AJ, London MK, Dohmen RJ (2004) Role of C-terminal extensions of subunits beta2 and beta7 in assembly and activity of eukaryotic proteasomes. J Biol Chem 279: 14323–14330 [DOI] [PubMed] [Google Scholar]
- Schmidtke G, Schmidt M, Kloetzel PM (1997) Maturation of mammalian 20S proteasome: purification and characterization of 13S and 16S proteasome precursor complexes. J Mol Biol 268: 95–106 [DOI] [PubMed] [Google Scholar]
- Tanahashi N, Murakami Y, Minami Y, Shimbara N, Hendil KB, Tanaka K (2000) Hybrid proteasomes. Induction by interferon-gamma and contribution to ATP-dependent proteolysis. J Biol Chem 275: 14336–14345 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Kasahara M (1998) The MHC class I ligand-generating system: roles of immunoproteasomes and the interferon-gamma-inducible proteasome activator PA28. Immunol Rev 163: 161–176 [DOI] [PubMed] [Google Scholar]
- Unno M, Mizushima T, Morimoto Y, Tomisugi Y, Tanaka K, Yasuoka N, Tsukihara T (2002) The structure of the mammalian 20S proteasome at 2.75 Å resolution. Structure (Camb) 10: 609–618 [DOI] [PubMed] [Google Scholar]
- Witt E, Zantopf D, Schmidt M, Kraft R, Kloetzel PM, Kruger E (2000) Characterisation of the newly identified human Ump1 homologue POMP and analysis of LMP7(beta 5i) incorporation into 20S proteasomes. J Mol Biol 301: 1–9 [DOI] [PubMed] [Google Scholar]
- Yashiroda H, Mizushima T, Okamoto K, Kameyama T, Hayashi H, Kishimoto T, Niwa S, Kasahara M, Kurimoto E, Sakata E, Takagi K, Suzuki A, Hirano Y, Murata S, Kato K, Yamane T, Tanaka K (2008) Crystal structure of a chaperone complex that contributes to the assembly of yeast 20S proteasomes. Nat Struct Mol Biol 15: 228–236 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information