SNP-specific extraction of haplotype-resolved targeted genomic regions (original) (raw)
Abstract
The availability of genotyping platforms for comprehensive genetic analysis of complex traits has resulted in a plethora of studies reporting the association of specific single-nucleotide polymorphisms (SNPs) with common diseases or drug responses. However, detailed genetic analysis of these associated regions that would correlate particular polymorphisms to phenotypes has lagged. This is primarily due to the lack of technologies that provide additional sequence information about genomic regions surrounding specific SNPs, preferably in haploid form. Enrichment methods for resequencing should have the specificity to provide DNA linked to SNPs of interest with sufficient quality to be used in a cost-effective and high-throughput manner. We describe a simple, automated method of targeting specific sequences of genomic DNA that can directly be used in downstream applications. The method isolates haploid chromosomal regions flanking targeted SNPs by hybridizing and enzymatically elongating oligonucleotides with biotinylated nucleotides based on their selective binding to unique sequence elements that differentiate one allele from any other differing sequence. The targeted genomic region is captured by streptavidin-coated magnetic particles and analyzed by standard genotyping, sequencing or microarray analysis. We applied this technology to determine contiguous molecular haplotypes across a ∼150 kb genomic region of the major histocompatibility complex.
INTRODUCTION
Genetic variation between individuals has become a focal point for genomic re-sequencing and mapping technologies, particularly since the widespread use of single nucleotide polymorphism (SNP) markers in association studies (1,2). Ultimately, the goal is to understand how genetic variation affects disease at an individual level, as well as phenotypes in general. In addition to its value for public health, the ability to efficiently study genetic variation in individuals and in larger populations has fundamental significance for our understanding of evolution in humans and model organisms (3,4). Recent technical advances for determining genetic variation have focused on the development of methods that streamline DNA analysis in parallel and automated ways as well as approaches to address limitations of high-throughput genotyping and statistical haplotype data interpretation (5,6).
Despite continuously improving tools for studying genetic variation and disease, association studies based on unrelated individuals have been less successful than expected in uncovering genetic risk factors for common diseases (7,8). An increasingly large number of genome-wide association studies (9–14) report specific SNP associations that indicate the importance of certain genomic regions for disease susceptibility. However, the detailed characterization of these genomic regions that would elucidate the connection between genetic variation and phenotype can not be adequately addressed by the current technologies. Next generation DNA sequencing methods, thus far, lack the capability of focusing on specific DNA fragments. Other methods that have this capability have been developed primarily for providing haplotyping information and are not easily applicable to the specific problem due to a number of different limitations that characterize these methods. Allele-specific PCR (15,16) is generally not robust enough for amplifying stretches of DNA at the range of about 100 kb, and therefore is not practical for long-range haplotype analysis. Clone-based strategies (17,18) or somatic hybrid cell lines (19) are laborious and not amenable to the degree of automation required for population studies. Methods involving single DNA molecule PCR amplification (20,21) are typically limited by the number of markers that can be studied with just one molecule and may require special instrumentation.
Here, we present a method for fast and efficient characterization of DNA segments. This method has the capability of focusing on a particular region based on previous knowledge of SNPs or other polymorphisms. It provides phase information for polymorphic markers across a target region in a simple, automated and scalable manner. Polymorphic phase information can be determined over arbitrary distances by tiling. To demonstrate the method, we selected to work with the major histocompatibility complex (MHC) region for which haplotypes were already known based on family pedigree analysis. Furthermore, the selection of the site was influenced by the availability of good performing TaqMan (Applied Biosystems, Foster City, CA, USA) assays and their relative density. The selection of the MHC was also due to the significance of this region for the immune response and the need for mapping haplotype variation of a region that is known for its association with many immune-related diseases.
Here, we describe a magnetic particle-based sample preparation method that selectively purifies specific, unmodified segments of genomic DNA for the purpose of long-range haplotype analysis. The method extends the capabilities of existing genetic analysis systems by converting the input material, diploid genomic DNA, into its haploid components, without requiring cloning or PCR. Any resulting typing information of this material is therefore also haploid. Large segments of haploid DNA can be typed and assembled into extended molecular haplotype blocks linked by overlapping heterozygous markers, such as SNPs.
Extraction of haploid DNA based on associated SNPs
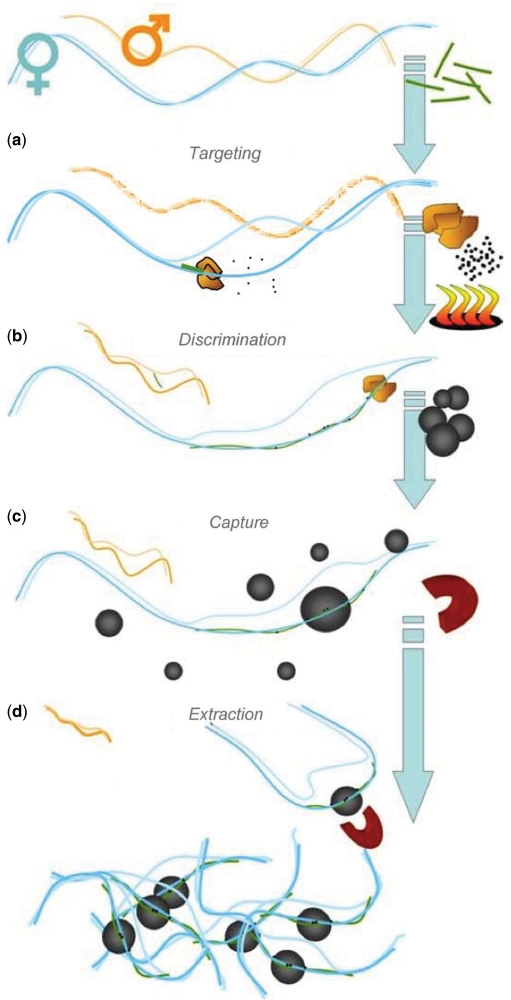
Haplotype-specific extraction exploits sequence variation to distinguish between different alleles or genomic regions and selectively isolates only those regions that contain the targeted sequence element (22). This variation can be as small as a single base-pair difference between allelic forms. For the purpose of molecular haplotyping, heterozygous SNPs can be used to separate diploid chromosomal segments into their maternal and paternal components (23,24). Standard genotyping methods, performed on haploid DNA, then provide definitive haplotype information for the targeted sequence. SNP-specific extraction consists of a four-stage process that involves identifying a heterozygous SNP, targeting an allele with a sequence-specific oligo, discrimination of the targeted region from other fragments by enzymatic incorporation of biotin tags and purification of the targeted DNA. SNPs, as well as other polymorphisms or unique sequences, can be exploited to isolate only such fragments that contain this particular sequence element.
Targeting
A SNP-specific oligonucleotide is designed with its 3′-end sequence overlapping the targeted SNP. A diploid DNA sample is heat-denatured and the extraction oligo is hybridized to the target sequence (Figure 1a). For heterozygous polymorphisms, the exact sequence of the extraction oligo will be matched only by one of the two alleles of the diploid sample. The bound extraction oligo is enzymatically elongated with biotinylated nucleotides (Figure 1b), which results in highly efficient tagging of only the targeted allele.
Figure 1.
Extraction oligos are incubated with denatured genomic DNA (a). Hybridization and conditional extension of the SNP-specific oligo incorporates biotinylated nucleotides (b), which are used to selectively capture the targeted allele by attachment to streptavidin-coated magnetic microparticles (c). The targeted haploid DNA is magnetically separated from the rest of the sample (d).
Extraction
The tagged allele is then captured, along with flanking genomic DNA, from the diploid sample by attachment to streptavidin-coated magnetic microparticles (Figure 1c). The haploid DNA/magnetic particle complex is washed twice to remove nontargeted, nonspecifically bound DNA from the surface (Figure 1d), leaving the targeted allele of interest isolated for further analysis.
Tiling
Neighboring haploid genomic fragments are characterized and overlapping sequences are used to determine in-phase sequences. This forms the basis for extended molecular haplotyping.
MATERIALS AND METHODS
DNA preparation
DNA was purified from blood by conventional isolation methods such as the Qiagen EZ1 magnetic separation kit from 350 μl of whole blood or ethanol precipitation (25) from 5–7 ml of whole blood.
Genotyping
A segment of DNA within the MHC that includes the HLA-C, HLA-B and MICA genes was selected to test whether a combination of SNP-based haploseparation and downstream characterization can determine extended, molecular haplotype information across this region. Low-resolution genotyping of diploid, genomic DNA samples at HLA-B and HLA-C was determined using Olerup SSPTM HLA-A-B-C combi tray (Qiagen, GmbH, Hilden, Germany) according to manufacturer's protocol. High-resolution typing at HLA-C and HLA-B was determined with Olerup SSP kits (Qiagen). MICA was typed with sequencing. Each 25 μl PCR reaction contained 1× Qiagen PCR buffer, 200 μM each dNTP, 0.3 μM each forward and reverse primers (26), and 2.5 U of Hot Star polymerase (Qiagen). The PCR thermal cycling conditions began with a 15 min denaturation at 95°C followed by 35 cycles of 96°C for 30 s, 64°C for 30 s and 72°C for 2 min with an additional 10 min final extension at 72°C. PCR products were cleaned prior to sequencing reactions by adding 4 μl ExoSAP-IT® (USB) to 10 μl PCR products, incubating at 37°C for 45 min and inactivating the enzyme with heat at 80°C for 15 min. BigDye chemistry was used for cycle sequencing; however, each 10 μl sequencing reaction included 0.25× BigDye Terminator v1.1 Cycle Sequencing RR-100 (ABI), 0.75× BigDye buffer, 0.16 μM primer (26) and 2 μl (∼100 ng) cleaned PCR products. Thermal cycling conditions consisted of a 1 min denaturation at 96°C followed by 25 cycles of 96°C for 10 s, 50°C for 30 s and 60°C for 2 min. The sequencing reactions were cleaned with CleanSEQ beads (Agencourt, Beverly, MA, USA) according to manufacturer's protocol and capillary electrophoresis was performed on an ABI 3130 Sequencing System.
Haplotype-specific extraction
SNP specific
Each 30 μl extraction contained 300–500 ng genomic DNA, 5 μM SNP-specific oligos, 1× H-Buffer, which contains a polymerase, dNTPs and biotinylated dNTPs (Qiagen, Cat. # 4340004, 2× initial concentration) and DNAse free water. All extractions were performed as separate reactions and placed on an external heat block with a heated lid (HybexTM SciGene, Sunnyvale, CA, USA) to denature the DNA at 95°C for 15 min. The samples were then transferred to a BioRobot® EZ1, which completes a 20 min incubation at 64°C during which the allele-specific oligos anneal and are extended, incorporating biotinylated dNTPs. The targeted genomic DNA is captured with streptavidin-coated magnetic microparticles by continuous relative motion of the beads through the fluid. The targeted, haploid DNA on the particles is then washed twice with wash buffer by gentle mixing within the tip. The particles carrying the targeted DNA are then collected with a magnet and resuspended in 50 μl EB buffer by the EZ1. Reagent cartridges, including streptavidin-coated magnetic particles and buffers, are commercially available (Qiagen, Cat. # 4340004).
In this experiment, genomic DNA was separated at a SNP intermediate to HLA-B and HLA-C, rs1634789 (A/G). Each allele was extracted using oligos which selectively target alleles based on a single base difference. Positions of oligos are shown in Table 1. DNA was also separated at a SNP intermediate to HLA-B and MICA, rs2507984 (A/G). Again, each allele was extracted using oligos which selectively target alleles based on a single base change (Table 1).
Table 1.
Positions of oligonucleotides and the polymorphisms they detect
| SNP | Targeted polymorphism/position | 5′ base | 3′ base |
|---|---|---|---|
| rs1634789a | rs1634789-A/31385066 | 31385081 | 31385065 |
| rs1634789-T/31385066 | 31385044 | 31385067 | |
| rs1634789-G/31385066 | 31385081 | 31385065 | |
| rs1634789-C/31385066 | 31385044 | 31385067 | |
| rs2507984 | rs2507984-A/31453575 | 31453592 | 31453574 |
| rs2507984-G/31453575 | 31453591 | 31453574 |
HLA-allele specific
For each extraction at HLA-C and HLA-B, genomic DNA was separated as described above with 1.5 μl of commercially available allele-specific oligos in 30 μl total reaction volume (HaploPrep HLA B539A, B355A, C102C and C419T; Qiagen).
Typing of haploid DNA
The isolated haploid DNA bound to the magnetic microparticles was directly substituted for diploid DNA in standard genotyping assays or sequencing reactions. It is practical and economical to leave the DNA on the particles; however, at high concentrations the particles may interfere with subsequent fluorescent detection steps. When typing with TaqMan assays, the captured DNA was removed from the particles by incubating the eluted sample for 10 min at 80°C (27), removing the magnetic particles and transferring the captured DNA in solution to a new tube.
Sequencing
For HLA and MICA sequence-based typing, haploid DNA was kept associated with the beads. Five microliters of the haploid DNA from these extractions were used in all sequencing reactions. HLA-B was sequenced with commercially available kits (Atria Genetics, South San Francisco, CA, USA and Genome Diagnostics, Utrecht, NL, USA) and the number of cycles in PCR was increased to 45. MICA was sequenced as described above, increasing the number of PCR cycles from 35 to 37.
HLA-C was amplified using PCR primers, 5′-AGATGGGGAAGGCTCCCCACT-3′ and 5′-CGAGGKGCCCKCCCGGCGC-3′ to delineate exons 2 and 3 of the gene. Each 25 μl PCR reaction contained 1× Qiagen PCR buffer, 2 mM MgCl2, 200 μM each dNTP, 0.25 μM each primer and 0.75× Q–solution (Qiagen). The PCR cycles included a 10 min denaturation at 95°C followed by 45 cycles of 96°C for 20 s, 65°C for 30 s and 72°C for 2 min with a 4 min extension step at 72°C. PCR product cleanup and cycle sequencing was performed as described above under MICA typing. Sequencing primers 2F (5′-GGAGCCGCGCAGGGAG-3′), 2R (5′-AGGGGTCGTGACCTGCG-3′), 3F (GGGCTGACCRCGGGGG-3′) and 3R (AAGGCTCCCCACTGCCC-3′) were used to sequence both strands of HLA-C exons 2 and 3. Primers were from Integrated DNA Technologies Inc., Coralville, IA, USA. ‘K’ in the nucleotide sequence denotes a ‘G,T’ mixed base position; ‘R’ an ‘A,G’ mixed base position.
Quantitative PCR
The samples were typed at three SNPs, rs1634789, rs2507984 and rs1131896 using TaqMan® real-time PCR system (ABI 7300, Applied Biosystems). All TaqMan® assays used standard 2× Master Mix (no AmpErase® UNG) and TaqMan® probes from ABI. Reactions were 20 µl (9 µl of DNA sample, 10 μl TaqMan master mix and 1 μl TaqMan probe), and were run for 40 cycles using absolute quantitation with a serially diluted diploid standard ranging from ∼15 000 to 50 copies. SNP typing was performed on extracted haploid DNA removed from the magnetic particles as described above. Allele typing was determined based on copy number with the ABI PRISM Sequence Detection System software package by interpolation to a plot of Ct values versus the logarithm of copy number of the serially diluted genomic DNA standard.
RESULTS
Diploid DNA from a four-member family (parent 1, parent 2 and two siblings) was genotyped at HLA-C, SNP rs1634789, HLA-B, SNP rs2507984, MICA and SNP rs1131896 by sequencing and/or TaqMan assays (Table 2, panel a). All family genotypings were successful except that of the MICA gene. As shown later (Figure 3), the difficulty was due to a single base difference within one of two tandem repeat alleles in the diploid sample, which resulted in out-of-phase electropherograms. We used a single sample (parent 1) with heterozygous positions at these five loci for long-range molecular haplotyping. Based on the family's genotyping, the haplotype for this sample was derived (Table 2, panel b). The objective was to determine whether the haplotypes derived molecularly by haploseparation were equivalent to the known haplotypes, derived from family genotyping.
Table 2.
Family genotype information (panel a), haplotype for parent 1 predicted from familial data (panel b) and molecular haplotype for parent 1 of 146.6 kb region (HLA-C to MICA) obtained by haploseparation (panel c)
| HLA-C | SNP rs1634789 | HLA-B | SNP rs2507984 | MICA | SNP rs1131896 | |
|---|---|---|---|---|---|---|
| Panel a | ||||||
| Parent 1 | 0702/0401 | A/G | 0801/3501 | A/G | – | A/G |
| Parent 2 | 0102/0202 | A/A | 2705/1501 | G/G | – | A/A |
| Child 1 | 0102/0401 | A/A | 2705/3501 | G/G | – | A/A |
| Child 2 | 0102/0401 | A/A | 2705/3501 | G/G | – | A/A |
| Panel b | ||||||
| Haplotype 1 (Parent 1) | 0401 | A | 3501 | G | – | A |
| Haplotype 2 (Parent 1) | 0702 | G | 0801 | A | – | G |
| Panel c | ||||||
| Haplotype 1 (Parent 1) | 0401 | A | 3501 | G | 016/A5 | A |
| Haplotype 2 (Parent 1) | 0702 | G | 0801 | A | 008/A5.1 | G |
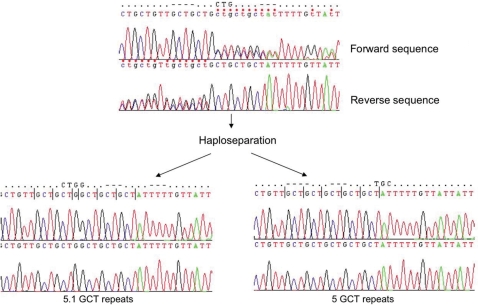
Figure 3.
A region located in exon 5 of the MICA gene contains STRs, which often resist sequence-based typing of heterozygous samples due to out-of-phase extension products. When a separation of the alleles is carried out before sequencing, the typing of the sample becomes possible. Sequencing electropherograms are shown for forward (upper panels) and reverse orientation (lower panels).
SNP-specific separation of alleles and haplotype tiling
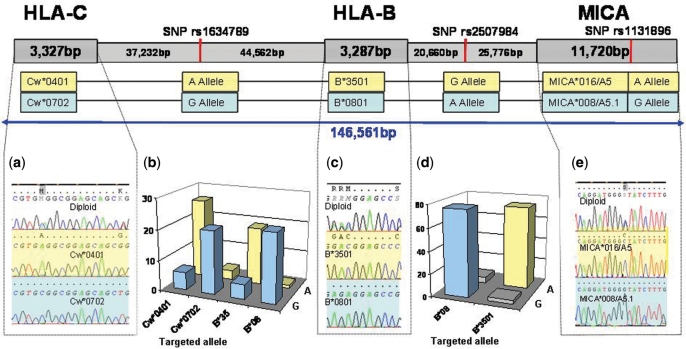
We designed oligos to target the A (rs1634789-A/T) and G (rs1634789-G/C) alleles of rs1634789 (Table 1), located between HLA-Cw and HLA-B, to separate diploid genomic DNA from parent 1 into its haploid components based on this heterozygous SNP (Figure 2). The extracted samples were then sequenced at HLA-Cw and HLA-B. The haploid DNA extracted with the oligo targeting the G allele typed as Cw*0702 and B*0801, while the DNA extracted with the oligo that targeted the A allele typed as Cw*0401 and B*3501. Both sets of experiments confirmed the known familial haplotype data (Figure 2a and c: A-allele in yellow, G-allele in blue).
Figure 2.
Molecular haplotype mapping of HLA-C, HLA-B and MICA. Each contiguous haplotype is denoted by color: haplotype 1, yellow; haplotype 2, blue. Oligos were designed to separate the A and G alleles at SNP rs1634789 for subsequent sequencing at HLA-C (a) and HLA-B (c). Sequencing electropherograms are shown for both alleles. The alleles Cw*0401 and Cw*0702 were separated at HLA-C and typed at SNP rs1634789 by quantitative PCR (b; _y_-axis, copy number per µl after extraction). The alleles B*3501 and B*0801 were separated at HLA-B and typed at SNPs rs1634789 (b) and rs2507984 (d). The A and G alleles at SNP rs2507984 were separated with oligos targeting this SNP and sequenced at HLA-B (c) and MICA (e).
To further confirm the molecular haplotypes derived from separating the sample at SNP rs1634789, genomic DNA from parent 1 was haploseparated at HLA-Cw with oligos C102C and C419T (Qiagen) targeting the Cw*0702 and Cw*0401 alleles respectively, and at HLA-B with oligos B539A and B355A (Qiagen) that target the B*0801 and B*3501, respectively. All four haploseparations were performed as separate reactions. The haploid DNA from each reaction was then assayed by quantitative PCR at SNP rs1634789 (Figure 2b). The haploseparated DNA which used oligos targeting Cw*0401 and B*3501 resulted in SNP rs1634789 typing as A, while the haploseparated DNA which used oligos targeting Cw*0702 and B*0801 typed this SNP as G confirming the molecular haplotypes derived from the haplo-separations with oligos rs1634789-A/T and rs1634789-G/C and assembling a molecular haplotype across an 85 kb region. The haploseparated DNA obtained by targeting the HLA-B alleles in the two separate reactions. These extractions were typed with quantitative PCR at SNP rs2507984 located between HLA-B and MICA (20.6 kb from HLA-B and 25.7 kb from MICA) (Figure 2d). Both haploseparated samples amplified in this assay, linking B*3501 to the G allele and B*0801 to the A allele of SNP rs2507984. This extended the assembled haplotype to ∼106 kb.
Oligos rs2507984-G and rs2507984-A (Table 1) were then used to separate the G and A alleles at SNP rs2507984. The haploid samples were tested for separation at the extraction site with a quantitative PCR assay that targets SNP rs2507984. Having confirmed the haploid nature of the DNA, the samples were tested at SNP rs1131896 in exon 3 of MICA and also sequenced at MICA (Figure 2e). The haploid DNA derived from targeting the G allele of rs2507984 typed as A at SNP rs1131896 and sequenced as MICA allele 016/A5, while the DNA derived from targeting the A allele of rs2507984 typed as G at SNP rs1131896 and sequenced as MICA allele 008/A5.1. Data from the SNP typing of rs1131896 by quantitative PCR are not shown in Figure 2. Again, both sets of experiments confirmed the familial haplotype data.
With this approach it was demonstrated that haploid DNA can be obtained whether we target HLA or SNP polymorphisms and that the haplotypes determined by haploseparation for parent 1 (Table 2, panel c) are identical to those derived from family genotyping (Table 2b); therefore confirming the utility of the method. The size of the region that was linked by this molecular haplotyping approach is 146.6 kb.
In a related set of experiments (data not shown), a sample with unknown HLA-B/MICA typing was haploseparated at SNP rs2507984 with oligos rs2507984-G and rs2507984-A (Table 1). The extracted haploid material was sequenced at HLA-B and MICA. The sequence-based typing of the haploid DNA as HLA-B*5001/MICA*01201/A4 and HLA-B*4002/MICA*027/A5 matched subsequent parental HLA-B/MICA typing, confirming that extended molecular haplotypes can be determined at two or more separate gene locations with a single SNP-specific separation.
Resolution of short-tandem repeat (STR) by haploseparation and sequencing
During the typing of the MICA region, a separate problem was encountered and resolved: the MICA gene contains an STR element in exon 5, which is particularly difficult to sequence due to a variable number of GCT repeats in heterozygous samples. This site is also a key-differentiating factor for allelic determination at MICA, with differences in the number of repetitive elements as small as single base changes. Extension products of samples with different numbers of GCT repeats often become unreadable when the samples are no longer in phase with each other and must be subsequently resolved by size differentiation with fragment analysis. In our experiments, the initially unreadable sequence of the diploid sample for parent 1 was fully resolved to 5 and 5.1 repeats after sequencing SNP extracted haploid DNA (Figure 3).
Assay performance over distance
In the first part of the results, we demonstrated our strategy and the performance of the method for targeting particular haplotypes, characterization by TaqMan assays or DNA sequencing and tiling to generate extended haplotypes. Here, we present documentation of the performance, reproducibility and limits of this technology in its current stage.
A total of 371 TaqMan assays were performed on 244 haplospecific extractions using eight different genomic DNA samples as input. The genomic DNA for these experiments was routinely extracted by standard protocols described in the Materials and methods section. Eighty-five of the haplospecific extractions targeted 15 SNP polymorphisms, while the other 159 extractions targeted 14 polymorphic sites within the HLA-B, -C genes and seven polymorphic sites within MICA.
Of the 85 haplospecific extractions that targeted SNP polymorphisms, 76 (89%) were considered successful as they provided DNA that was at least 100% enriched for the targeted allele, thereby allowing successful determination of haplotypes by TaqMan assays. Although there is some fluctuation in the percentage of successful haploseparations (59–100%), the ratio of targeted to nontargeted allelic forms is high (110 and 270) within 5 kb of the extraction point and decreases to about 30 between 10 and 30 kb (Table 3). When the distance from the capture point is over 30 kb and up to 43 kb, a gradual drop of both the percentage of successful haploseparations and of the ratio of targeted versus nontargeted regions is observed.
Table 3.
Number of TaqMan assays performed at different distances from the point of haplospecific extraction, percent of successful extractions (A successful extraction is defined as providing at least 100% increase in TaqMan copy number of the targeted allele over the copy number of the non-targeted allele) and average of enrichment ratios (defined as copy number of the targeted allele per one copy of the non-targeted allele)
| Distance to target (kb) | n | Percent success (%) | Average (target/non-target ratio) |
|---|---|---|---|
| 0 | 76 | 88 | 110 |
| 5 | 18 | 100 | 270 |
| 11.6 | 4 | 100 | 29 |
| 12.5 | 61 | 98 | 50 |
| 20.5 | 31 | 74 | 29 |
| 21.3 | 18 | 100 | 10 |
| 27.8 | 14 | 86 | 6.7 |
| 28.3 | 16 | 81 | 4.7 |
| 31.6 | 15 | 100 | 20 |
| 32.4 | 2 | 100 | 30 |
| 37.8 | 16 | 75 | 9.4 |
| 42.2 | 22 | 64 | 4.8 |
| 42.4 | 18 | 83 | 2.7 |
| 42.7 | 34 | 59 | 2.8 |
| 47.2 | 26 | 88 | 4.0 |
| Total | 371 | 86 |
This effect is to be expected based on the average size distribution of the available DNA fragments containing the targeted locus: a larger number of copies from various captured DNA fragments are available near the targeted point since here both short and large fragments contribute as template. As the distance from this point increases, a reduction of targeted copies is observed. The total size of the region characterized around the capture point is often larger than the individual DNA fragments captured and depends on whether a sufficient number of captured fragments contain flanking sequences which extend in either direction. This is confirmed by the ability to characterize DNA after haplopreparation up to at least 42 kb to either side of the capture point. However, the TaqMan assays performed at these distances tend to be less robust and show reduced enrichment due to the decreased number of template available.
DISCUSSION
The selective targeting of unique genomic variations and the isolation of large fragments of haploid DNA provide a valuable tool for addressing a number of current problems, including, but not limited to the characterization of particular genomic segments pointed to by genome-wide association SNP analysis studies, the evaluation of regions with variable copy numbers and the generation of molecular haplotypes when familial or computational approaches are limiting or not possible. The method presented here is a simple, robust tool for extracting large genomic regions including their surrounding sequence context (22–24). By selectively targeting unique genomic variation, including SNPs, individual molecular haplotypes can be isolated and characterized. This method has been used extensively to resolve short-range HLA allele ambiguities and determine new HLA alleles (25,29,30).
Here, a segment of about 150 kb within the MHC that includes HLA-C, HLA-B and MICA was characterized, demonstrating the ability of this method for SNP-specific targeting of genomic regions from individual samples by an automated method, which can generate phased sequences by DNA sequencing or TaqMan assays. The information is assembled based on the overlap of multiple haploid DNA fragments at shared heterozygous positions. Complex and potentially unknown genotyping problems may be resolved, such as the one encountered while typing the MICA gene (described in Results section). Sequencing failures caused by frame shifting are avoided by the separation of repetitive elements into their respective alleles (Table 2, panel c and Figure 3).
The performance of SNP-specific extractions is influenced by the selection of the targeting oligos and it is useful to evaluate multiple oligos to determine, which best increases the enrichment level of the targeted allele. Any single polymorphic site is typically placed at the 3′-end _n_−1 position of the oligo. If multiple polymorphic sites are available, these positions are all placed towards the 3′-end of the oligo. Certain types of base mismatches and combinations of mismatch and preceding sequence are exploited where possible due to their greater ability to disrupt enzymatic extension (31). Any given SNP can be separated by four different types of oligos, with each one targeting one of the two possible alleles on either the forward or reverse strand.
Example: for an A/G SNP, forward-oriented oligos can be designed that perfectly match either the ‘T’ or ‘C’ present on the respective reverse strand of the genomic template, and reverse-oriented oligos can be designed to match the ‘A’ or ‘G’ present on the forward strand of the genomic template. It is also possible to combine both forward- and reverse-oriented oligos that target the same allele, but we have not seen any consistent difference in overall performance compared to the use of single oligos. Generally, our experience is that of the 32 oligos we have designed for targeting SNPs, 27 (84%) performed well, providing allelic discrimination. Our current practice is to synthesize four different oligos for every SNP, to target each of the alleles in the forward and reverse direction. These are evaluated by quantitative PCR at the extraction site and the best performers are selected for future extractions.
The length of haploseparated DNA fragments depends primarily on the size of the diploid input DNA, i.e. on the genomic DNA extraction method used. Automated and commercially available DNA preparation methods consistently provide diploid DNA fragments with an average size limit of about 40 kb. The ability to characterize in forward and reverse directions from the extraction point is based on the capture of the original genomic template itself, along with the enzymatically extended portion of the targeting oligo. The enzymatically extended portion is simply an anchor point for capturing the haploid DNA. The conditional, SNP-specific extension of a perfectly matched oligo with multiple biotins topologically locks large genomic fragments to the bead surface for extraction.
Given the fact that heterozygous SNPs in a typical sample occur with an average of about 18 kb (32,33), and since DNA can routinely be haploseparated at these distances from the capture point, this means that extended haplotypes can reliably be derived for individual samples based on multiple overlapping haploseparations and the overall linkage distance that can be achieved with this approach is therefore independent of the DNA fragment length that is available for any given region. Haplotype blocks in the human genome rarely exceed 100 kb (34) and therefore this method is an appropriate step to characterize such regions after they have been identified in an association study.
Even though this method thus far has primarily been used for targeting genomic regions within the MHC, it can easily be extended to other regions. Also, its potential to generate short-range haplotype information has been confirmed and utilized successfully for characterizing new HLA alleles and resolve HLA typing ambiguities (22,24,25,29,30). We demonstrate that by haploseparating and tiling neighboring regions we can generate phased sequence information of almost 150 kb. Overlapping haploid segments can in principle be joined together to derive extended haplotypes over arbitrary distances.
Obtaining long-range, molecular haplotype information for numerous individuals will improve disease association studies and aid in better characterizing the genetics of complex disease. Statistical approaches do not always present a full alternative because they are not able to derive with certainty molecular linkage information for individual samples. SNP-specific extracted DNA overcomes limitations in current technologies by its near universal compatibility with common genotyping systems and provides an efficient method of examining potentially rare haplotype signatures for preexisting, individual samples.
The ability to reduce genomic complexity by selectively isolating specific regions from a given sample and determine molecular linkage across broad genomic regions in a high-throughput format could also be a useful tool to help deconvolute copy number and structural genomic variation in next-generation sequencing applications.
ACKNOWLEDGEMENTS
We gratefully acknowledge discussions and help in the preparation of this manuscript and certain experiments by A. Akkapeddi, K. Lennartz, K. Mackiewicz, V. McCarro, N. Murphy, M. Nagy, G. Scavello, R. Stacy and C. Turino. This work was supported by the National Institute of Allergy and Infectious Disease (R43 AI51036, R44 AI51036), the National Human Genome Research Institute (R43 HG004096) to J.D. and by the Children's Hospital of Philadelphia (Institutional funds to D.M.). Funding to pay the Open Access publication charges for this article was provided by the Children's; Hospital of Philadelphia department of pathology.
Conflict of interest statement. J.D. and D.F. are employees of Generation Biotech. D.M. is an employee of the University of Pennsylvania and the Children's; Hospital of Philadelphia and a collaborator of Generation Biotech under this grant. At the time of their contribution to this work, MK was an employee of Generation Biotech and E.M. was affiliated with the Children's; Hospital of Philadelphia. The Children's; Hospital of Philadelphia is a subcontractor of Generation Biotech under this grant. Qiagen, Inc. has licensed the HSE technology for tissue typing from Generation Biotech.
REFERENCES
- 1.West M, Ginsburg GS, Huang AT, Nevins JR. Embracing the complexity of genomic data for personalized medicine. Genome Res. 2006;16:559–566. doi: 10.1101/gr.3851306. [DOI] [PubMed] [Google Scholar]
- 2.Brookes AJ. The essence of SNPs. Gene. 1999;234:177–186. doi: 10.1016/s0378-1119(99)00219-x. [DOI] [PubMed] [Google Scholar]
- 3.Paabo S. The mosaic that is our genome. Nature. 2003;421:409–412. doi: 10.1038/nature01400. [DOI] [PubMed] [Google Scholar]
- 4.Reich DE, Schaffner SF, Daly MJ, McVean G, Mullikin JC, Higgins JM, Richter DJ, Lander ES, Altshuler D. Human genome sequence variation and the influence of gene history, mutation and recombination. Nat. Genet. 2002;32:135–142. doi: 10.1038/ng947. [DOI] [PubMed] [Google Scholar]
- 5.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen Y, Chen Z. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin PI, Vance JM, Pericak-Vance MA, Martin ER. No gene is an island: the flip-flop phenomenon. Am. J. Hum. Genet. 2007;80:531–538. doi: 10.1086/512133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Terwilliger JD, Weiss KM. Confounding, ascertainment bias, and the blind quest for a genetic ‘fountain of youth’. Ann. Med. 2003;35:532–544. doi: 10.1080/07853890310015181. [DOI] [PubMed] [Google Scholar]
- 8.Kaplan N, Morris R. Issues concerning association studies for fine mapping a susceptibility gene for a complex disease. Genet. Epidemiol. 2001;20:432–457. doi: 10.1002/gepi.1012. [DOI] [PubMed] [Google Scholar]
- 9.Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, Gallins P, Spencer KL, Kwan SY, Noureddine M, Gilbert JR, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 10.Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat. Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 11.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hakonarson H, Grant SFA, Bradfield JP, Marchand L, Kim CE, Glessner JT, Grabs R, Casalunovo T, Taback SP, Frackelton EC, et al. A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature. 2007;448:591–594. doi: 10.1038/nature06010. [DOI] [PubMed] [Google Scholar]
- 13.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pettersson M, Bylund M, Alderborn A. Molecular haplotype determination using allele-specific PCR and pyrosequencing technology. Genomics. 2003;82:390–396. doi: 10.1016/s0888-7543(03)00177-0. [DOI] [PubMed] [Google Scholar]
- 16.Tost J, Brandt O, Boussicault F, Derbala D, Caloustian C, Lechner D, Gut IG. Molecular haplotyping at high throughput. Nucleic Acid Res. 2002;30 doi: 10.1093/nar/gnf095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raymond CK, Subramanian S, Paddock M, Qiu R, Deodato C, Palmieri A, Chang J, Radke T, Haugen E, Kas A, et al. Targeted, haplotype-resolved resequencing of long segments of the human genome. Genomics. 2005;86:759–766. doi: 10.1016/j.ygeno.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 18.Burgtorf C, Kepper P, Hoehe M, Schmitt C, Reinhardt R, Lehrach H, Sauer S. Clone-based systematic haplotyping (CSH): a procedure for physical haplotyping of whole genomes. Genome Res. 2003;13:2717–2724. doi: 10.1101/gr.1442303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan H, Papadopoulos N, Marra G, Perrera C, Jiricny J, Boland CR, Lynch HT, Chadwick RB, de la Chapelle A, Berg K, et al. Conversion of diploidy to haploidy. Nature. 2000;403:723–724. doi: 10.1038/35001659. [DOI] [PubMed] [Google Scholar]
- 20.Zhang K, Zhu J, Shendure J, Porreca GJ, Aach JD, Mitra RD, Church GM. Long-range polony haplotyping of individual human chromosome molecules. Nat. Genet. 2006;38:382–387. doi: 10.1038/ng1741. [DOI] [PubMed] [Google Scholar]
- 21.Xiao M, Gordon MP, Phong A, Ha C, Chan TF, Cai D, Selvin PR, Kwok PY. Determination of haplotypes from single DNA molecules: a method for single-molecule barcoding. Hum. Mutat. 2007;28:913–921. doi: 10.1002/humu.20528. [DOI] [PubMed] [Google Scholar]
- 22.Dapprich J, Cleary MA. 2001. Method for selectively isolating a nucleic acid. U.S. Patent application #20010031467: Australian Patent #785211. [Google Scholar]
- 23.Gabriel A, Dapprich J, Kunkel M, Gresham D, Pratt SC, Dunham MJ. Global mapping of transposon location. PLoS Genet. 2006;2:e212. doi: 10.1371/journal.pgen.0020212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dapprich J, Cleary MA, Gabel HW, Akkapeddi A, Iglehart B, Turino C, Beaudet L, Lian J, Murphy NB. A rapid, automatable method for molecular haplotyping. In: Hansen JA, editor. Proceedings of the 13th International Histocompatibility Workshop and Congress. Vol. II. Seattle, WA: IHWG Press; 2006. pp. 271–274. [Google Scholar]
- 25.Dapprich J, Magira E, Samonte MA, Rosenman K, Monos D. Identification of a novel HLA-DPB1 allele (DPB1*1902) by haplotype specific extraction and nucleotide sequencing. Tissue Antigens. 2007;69:282–224. doi: 10.1111/j.1399-0039.2006.00752.x. [DOI] [PubMed] [Google Scholar]
- 26.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zou Y, Han M, Wang Z, Stastny P. MICA allele-level typing by sequence-based typing with computerized assignment of polymorphic sites and short tandem repeats within the transmembrane region. Hum. Immunol. 2006;67:145–151. doi: 10.1016/j.humimm.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 28.Holmberg A, Blomstergren A, Nord O, Lukacs M, Lundeberg J, Uhlen M. The biotin-streptavidin interaction can be reversibly broken using water at elevated temperatures. Electrophoresis. 2005;26:501–510. doi: 10.1002/elps.200410070. [DOI] [PubMed] [Google Scholar]
- 29.Mrazek F, Fae I, Ambruzova Z, Raida L, Indrak K, Petrek M, Fischer GF. A novel HLA-B*420502 allele identified by PCR-SSO/SSP routine typing and confirmed by Sequencing-based typing. Tissue Antigens. 2005;65:275–277. doi: 10.1111/j.1399-0039.2005.00376.x. [DOI] [PubMed] [Google Scholar]
- 30.Nagy M, Entz P, Otremba P, Schoenemann C, Murphy N, Dapprich J. Haplotype-specific extraction: a universal method to resolve ambiguous genotypes and detect new alleles - demonstrated on HLA-B. Tissue Antigens. 2007;69:176–180. doi: 10.1111/j.1399-0039.2006.00741.x. [DOI] [PubMed] [Google Scholar]
- 31.Carver TE, Hochstrasser RA, Millar DP. Proofreading DNA: recognition of aberrant DNA termini by the Klenow fragment of DNA polymerase I. Proc. Natl Acad. Sci. USA. 1994;91:10670–10674. doi: 10.1073/pnas.91.22.10670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuzaki H, Dong S, Loi H, Di X, Liu G, Hubbell E, Law J, Berntsen T, Chadha M, Hui H, et al. Genotyping over 100,000 SNPs on a pair of oligonucleotide arrays. Nat. Methods. 2004;1:109–111. doi: 10.1038/nmeth718. [DOI] [PubMed] [Google Scholar]
- 33.Affymetrix. Part No. 702087, Rev.4, 1–4. 2005–2006. GeneChip®Human Mapping 500K Array Set Data Sheet. [Google Scholar]
- 34.Anderson EC, Novembre J. Finding haplotype block boundaries by using the minimum-description-length principle. Am. J. Hum. Genet. 2003;73:336–354. doi: 10.1086/377106. [DOI] [PMC free article] [PubMed] [Google Scholar]