Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome (original) (raw)
Abstract
Bile salt hydrolases (BSHs) catalyze the “gateway” reaction in a wider pathway of bile acid modification by the gut microbiota. Because bile acids function as signaling molecules regulating their own biosynthesis, lipid absorption, cholesterol homeostasis, and local mucosal defenses in the intestine, microbial BSH activity has the potential to greatly influence host physiology. However, the function, distribution, and abundance of BSH enzymes in the gut community are unknown. Here, we show that BSH activity is a conserved microbial adaptation to the human gut environment with a high level of redundancy in this ecosystem. Through metagenomic analyses we identified functional BSH in all major bacterial divisions and archaeal species in the gut and demonstrate that BSH is enriched in the human gut microbiome. Phylogenetic analysis illustrates that selective pressure in the form of conjugated bile acid has driven the evolution of members of the Ntn_CGH-like family of proteins toward BSH activity in gut-associated species. Furthermore, we demonstrate that BSH mediates bile tolerance in vitro and enhances survival in the murine gut in vivo. Overall, we demonstrate the use of function-driven metagenomics to identify functional anchors in complex microbial communities, and dissect the gut microbiome according to activities relevant to survival in the mammalian gastrointestinal tract.
Keywords: bile modification, microbiota, functional anchor, GI survival
The human gut harbors a complex microbial ecosystem with many emergent properties that directly benefit the host (1–3). This community encodes ≈100 times as many genes as the human genome and humans can be considered as “superorganisms” with the gut microbiota as a “virtual organ” that has coevolved with the host (1–4). Sequence-based metagenomic analyses have begun to reveal the core metabolic functions of the human gut microbiota and determine the extant properties of this community that impact on human health (2, 5–8). However, many questions remain concerning the selective pressures governing evolution of a stable microbiota, the forces influencing transmission of the microbiota to new hosts, and the factors that distinguish evolution of the human microbiota from that of other mammals (3).
Conjugated bile acids (CBAs) are cholesterol derivatives synthesized in the liver that consist of a steroid ring component that is conjugated with either glycine or taurine before secretion (glyco-CBA and tauro-CBA, respectively) (9, 10). CBAs facilitate lipid absorption and act as signaling molecules regulating systemic endocrine functions including triglyceride, cholesterol, and possibly glucose homeostasis (9, 11, 12). In addition, CBAs have been suggested to repress bacterial growth in the small intestine either through direct antimicrobial effects, up-regulation of host mucosal defenses, or synergistic action of both mechanisms (9, 13–15).
The initial “gateway” reaction in the bacterial metabolism of CBAs is mediated by bile salt hydrolase (BSH; also referred to as choloylglycine hydrolase), which catalyzes the deconjugation of CBAs to liberate free primary bile acids (BAs; cholic acid or chenodeoxycholic acid) and amino acids (9, 10, 16, 17) (supporting information (SI) Fig. S1). Free primary BAs are subsequently open to a wider pathway of BA modification encoded by the gut microbiota, which generates secondary and tertiary forms through dehydroxylation, dehydrogenation, and sulfation (9, 10, 16, 17). Deconjugated and modified BAs have been associated with acquisition of gallstones and colorectal cancer (18–20), and recent studies indicate that the mammalian gut microbiota can modulate host bile acid synthesis and metabolism, which may influence host lipid metabolism (21). BSH activity and subsequent BA modification in this community could significantly impact host physiology through perturbations of BA-controlled endocrine functions and influence risk of metabolic diseases such as obesity, diabetes, and atherosclerosis (11, 12, 21).
Although BAs clearly play a significant role in regulating physiological and antimicrobial responses in the host, the extent of CBA metabolism by the gut microbiota remains unclear. Therefore such evaluation of the distribution, abundance, and function of BSH in this community is required.
Results
Distribution of BSH Activity in the Human Gut Microbiota.
We first examined the distribution of BSH activity among the constituent members of the human gut community. Because this ecosystem is composed of hundreds of species with the majority being uncultured (1–3), we used a function-driven metagenomic approach. A metagenomic library from fecal microbiota was constructed and screened to identify fosmid clones expressing BSH activity (Fig. S1). In total 89,856 metagenomic clones representing ≈3.6 Gb of metagenomic DNA were screened and yielded 142 positive clones, of which 101 were stable and characterized further.
Of these, 90 clones were affiliated with phylogenetic divisions based on fosmid end sequence data (Table S1 and associated text). The majority of those that could be affiliated with a phylogenetic division were members of the Firmicutes (30%), followed by Bacteroidetes (14.4%) and Actinobacteria (8.9%) (Table S1). The unaffiliated clones (43.3%) potentially include representatives of novel and uncultured members of the human gut microbiota. When clones were tested against glyco-CBA, tauro-CBA, and human bile, a group-specific trend in substrate range was observed (Table S2). Clones assigned as Firmicutes and Actinobacteria were capable of degrading all tested CBAs and human bile, whereas clones assigned to the Bacteroidetes were generally only active against tauro-CBA. This result is in keeping with substrate ranges reported for BSH of various cultivated representatives of these phylogenetic groups (9, 22, 23) (Table S2).
The genetic basis for CBA deconjugation was elucidated in a subset of 19 clones affiliated to various phylogenetic groups by in vitro transposon mutagenesis or subcloning. All were found to encode BSH genes and comparison of amino acid sequences from complete ORFs revealed 9 distinct BSH types, designated BSH type A-I (Table S3). BlastP analysis revealed that all recovered ORFs were members of the Ntn_PVA family of enzymes (COG3049) and indicated that novel BSHs had been captured (Table S3). Greatest homology was exhibited to proteins from draft genomes of gut bacteria recently sequenced as part of the human gut microbiome initiative and represented all major bacterial divisions in the gut (3) (Table S3). In all cases, the homologies of the recovered bsh genes, along with partial ORFs in sequences flanking bsh genes, were found to match the phylogenetic groups to which clones were affiliated based on end sequence data.
BSH Activity of Methanobrevibacter smithii.
One BSH type was found to exhibit high identity to a protein encoded by the archaeon M. smithii (BSH type F, 56%). Further investigation revealed that both archaeal species identified in the human gut microbiota (M. smithii and Methanosphera stadmanae) encode proteins with high identity to bacterial BSH enzymes (62% to Listeria monocytogenes str. 4b F2365, 61% to L. monocytogenes EGDe, respectively). Cloning and expression of the M. smithii BSH in Escherichia coli demonstrated that this archaeal species encodes a functional BSH capable of degrading both tauro- and glyco-CBA (Fig. S2, Table S2). Furthermore, the high identity of archaeal BSH with bacterial BSH strongly suggests a role for their acquisition through horizontal gene transfer (HGT) although the direction of gene flow is uncertain. Additional searches of a further 46 complete or draft genomes from non-gastrointestinal (GI) archaeal species did not reveal any potential BSH enzymes. Thus, we demonstrate that functional BSHs are distributed among two domains of life and all major bacterial divisions of the human gut microbiota. Such widespread distribution indicates excellent functional redundancy of this activity in the gut community.
Comparative Metagenomic Analyses of BSHs in Human Gut and Other Environmental Metagenomes.
To examine the abundance and conservation of BSH activity among the microbiota of different hosts, a comparative metagenomic strategy was used to identify BSH and homologous enzymes in other human gut and environmental metagenomes. Amino acid sequences homologous to those of our functional BSH were identified in 14 of 15 other human gut metagenomes searched, demonstrating the general conservation of this enzyme in the gut microbiomes of widely dispersed and geographically isolated individuals (American, Japanese, European) (6, 7) (Fig. 1, Fig. S3). Of the 193 complete and partial BSH sequences retrieved from these metagenomes, identity to our functional BSH ranged from 25 to 100% with the majority exhibiting >60% identity (Fig. S3). All showed >50% identity to proteins encoded by gut-associated bacteria or archaea, with the majority (75%) exhibiting >70% identity (Fig. S3).
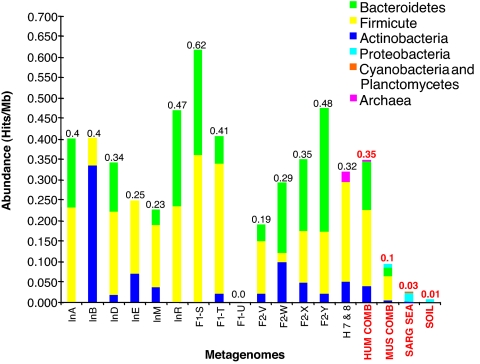
Fig. 1.
Abundance of putative BSH and related proteins in the human gut metagenome. Amino acid sequences of putative BSH and related amidases described in Fig. S3, from 15 human gut metagenomes (6, 7), Sargasso sea (24), soil (25), and the combined gut metagenomes of lean and obese mice (8) were used to calculate and compare the relative abundance of these proteins in each metagenome. Abundance was calculated as hits per megabase (Mb) for individual and combined metagenomes and revealed variation between individuals and a general enrichment of BSH and related proteins in the human gut metagenome. Japanese individuals: InA, male 45years; InB, male 6 months; InD, male 35 years; InE, male 3 months; InM, female 4 months; F1-S, T, U, Family I, male 30 years, female 28 years, female 7 months, respectively; F2-V, W, X, Y, Family II, male 37 years, female 36 years, male 3 years, female 1.5 years, respectively. American metagenomes: H 7 & 8, female 28 years combined with male 37 years; HUM COMB, all human metagenomes combined; MUS COMB, all murine metagenomes combined; SARG SEA, Sargasso sea metagenome; SOIL, soil metagenome. Colors indicate putative origin of BSH homologues retrieved from each metagenome or combined metagenomes. Values above bars represent overall hits per Mb for each category.
Considerable variation in overall abundance and origin of these proteins was observed between individual human gut metagenomes, but no correlation with age, sex, or weaning, or between family groups was observed (Fig. 1). In light of the differing substrate ranges among clones affiliated with different bacterial divisions in our functional metagenomic analysis, such interindividual variations may indicate differences in the capacity for bile acid modification between the microbiota of individuals. In particular, M. smithii can account for a significant fraction of the colonic microbial population, but may not be present or reach high levels in all individuals (2, 3, 6, 7).
Sequences with homology to our functional BSH types were also identified in metagenomes derived from lean and obese mice (8). Our analysis demonstrated distinct differences between the BSH complement of the murine and human microbiotas (Fig. S3), which most likely reflects the differing composition of BA pools in each host species, because murine bile is composed predominantly of tauro-CBA, as well as murocholic acid, which is not synthesized in humans (21). We also examined non-GI metagenomes from Sargasso sea (24) and soil (25) for the presence of BSH. However, only a single sequence was retrieved from the soil metagenome, whereas >90% of the sequences retrieved from the Sargasso sea metagenome exhibited <40% identity to our functional BSH sequences (Fig. S3).
Evolution of the Ntn_CGH-like Family of Proteins to BSH Activity in the Gut.
Sequences with low identity to our functional BSH may represent BSH enzymes, but it is likely that they are also derived from related members of the wider Ntn_CGH-like family of proteins. In particular, penicillin V amidase (PVA) is closely related to BSH and exhibits conservation of putative critical catalytic amino acids (23). It has previously been suggested that BSH has diverged from PVA during its evolution (23). To further investigate the evolution of BSH activity, and examine whether this activity is a specific adaptation to the mammalian gut, we conducted a phylogenetic analysis by using amino acid sequences exhibiting residues conserved among both BSH and PVA (23), including sequences from various metagenomes. This analysis illustrated a distinct evolutionary trend among these proteins relating to microbial habitat (Fig. 2). This trend was highly conspicuous among the three archaeal sequences included in this analysis, which did not display phylogenetic congruity but clustered according to environmental origin (Fig. 2).
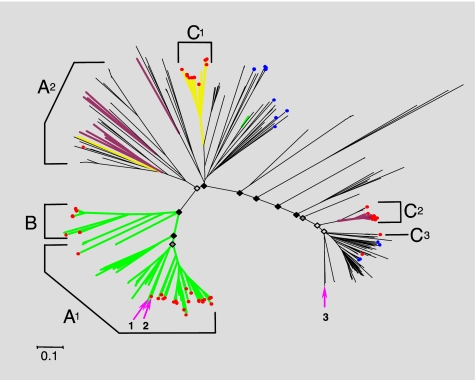
Fig. 2.
Phylogenetic analysis of putative BSH and PVA amino acid sequences. A set of 367-aa sequences belonging to the Ntn_CGH-like family (Ntn_PVA and Ntn_CGH families), and displaying all putative conserved critical catalytic amino acids common to both BSH and PVA (based on alignments of our functional BSH and previous analysis of BSH and PVA crystal structures) (23) were used to construct phylogenetic trees and examine the evolution of BSH activity. Sequences originated from a diverse range of bacterial divisions and full-length sequences retrieved from human (7) and Sargasso sea (24) metagenomes were also included. Supporting bootstrap values for deep branching nodes are indicated with black filled diamonds identifying nodes supported by bootstrap values of 50 or over, dark-gray filled diamonds identifying nodes with bootstraps values of 40 or over, and light-gray filled diamonds nodes with bootstrap values <40. Green branches represent clades containing sequences with proven activity against glyco-CBA and tauro-CBA, whereas yellow branches indicate clades containing sequences proven to exhibit activity against tauro-CBA only. Purple branches represent clades with sequences that have been proven to exhibit no BSH activity. Red circles indicate positions of sequences derived from human gut metagenomes. Blue circles indicate sequences retrieved from the Sargasso sea metagenome. Arrows show positions of Archaeal sequences included in this analysis: 1→, Methanosphera stadmanae (human gut); 2→, Methanobrevibacter smithii (human gut); 3→, Methanosarcina acetovorans (marine sediment). Brackets indicate regions dominated by major bacterial divisions comprising the human gut microbiota, and are generally composed of sequences from gut-associated genera. A1, Firmicutes: Eubacterium, Coprococcus, Clostridium, Ruminococcus, Dorea, Lactobacills, Enterococcus, Listeria, Lactococcus; A2, Firmicutes: Lactobacillus, Clostridium, Listeria, Staphylococcus, Oenococcus, Bacillus; B, Actinobacteria: Bifidobcterium, Collinsella; C1, C2, and C3, Bacteroidetes: Parabacteroides, Bacteroides. (Scale bar, 0.1 aa substitution per site.)
Correlation of the substrate ranges of sequences with proven function from this and previous studies revealed a striking distribution of substrate specificity, with activity among enzymes from human gut microbes shifted toward BSH activity (Fig. 2). This analysis also confirmed that the majority of sequences retrieved from human metagenomes during our comparative metagenomic analysis were BSH, but PVA and associated amidases had also been recovered, primarily from representatives of gut-associated Bacteroidetes (Fig. 2). Many gut-associated species (particularly among the Bacteroidetes and Firmicutes) encode multiple but not identical copies of PVA/BSH proteins, and from the location of these sequences in our phylogenetic tree we predicted that some species encode both activities, whereas others were confined to BSH alone (Fig. S4). Cloning and in vitro assays were used to confirm these predictions for L. monocytogenes and B. vulgatus (Fig. S4). The varying copy number of bsh genes and related amidases present in different gut-associated species also has implications for the capacity of BA modification between the gut microbiotas of individuals, and indicates that this capacity may be related to the species composition of the microbiota. Although the widespread distribution of this activity will ensure functional stability overall, population shifts may lead to fluctuations in capacity for BA modification during the lifetime of the host.
Analysis of the Role of Microbial BSH Activity.
Having identified BSH as a conserved microbial adaptation to the human GI tract, we next sought to elucidate the function of BSH in the human gut microbiota. CBAs exhibit direct antimicrobial activity and during colonization of the human GI tract microbes are exposed to inhibitory levels of CBA (9, 10, 13, 15). Furthermore, traits that enhance survival in the human gut would also be expected to be appropriated by pathogens (26) and, in the case of BSH activity, previous studies have shown that this enzyme facilitates colonization of the gut by L. monocytogenes and Brucella abortus (15, 27, 28). Therefore, we hypothesized that BSH may facilitate colonization of the GI tract by mediating resistance to CBA.
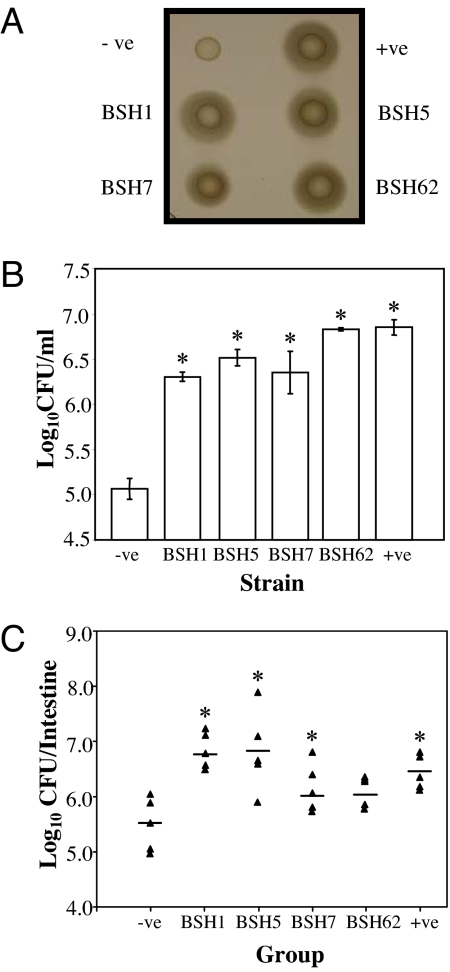
To test this, bsh genes isolated from our metagenomic library were subcloned and expressed in Listeria innocua, which does not encode BSH. Kill curves demonstrated a dramatic increase in survival of populations harboring BSH compared with the wild type when exposed to bile in vitro (Fig. 3). When ability to survive in the murine intestinal tract was assessed, a reproducible increase in survival was observed in clones expressing BSH compared with the wild type (Fig. 3). Our data indicate that the presence of cloned BSH in L. innocua provided a distinct survival advantage in the murine intestinal tract by mitigating the toxic effects of CBA. Although the observed increase was low, even small increases in survival are likely to have a significant impact during initial colonization of pristine gut habitats generated when new hosts are born.
Fig. 3.
Elucidation of BSH function in the human gut microbiota. bsh genes from functional metagenomic clones FM1, FM5, FM7, and FM62 (Table S2), corresponding to BSH types A, D, E, F, respectively (Table S3) were cloned in L. innocua FH2333 by using plasmid pNZ44. The resulting clones were designated BSH1, BSH5, BSH7, and BSH62, respectively. Empty plasmid and plasmid-containing bsh from Lactobacillus plantarum were used as negative (−ve) and positive (+ve) controls, respectively. (A) Confirmation of expression of cloned BSH. An agar plate assay (22, 33) was used to examine the bile hydrolase activity of strains. Cultures that were grown in brain heart infusion (BHI) broth were spotted (10 μl) onto LB bile agar supplemented with 0.5% bile acids. Strains with bile salt hydrolase activity were surrounded with white precipitate halos of deconjugated bile acids. (B) Bile survival assays. Overnight cultures were inoculated (3%) into broth supplemented with 25% (wt/vol) bovine bile (Oxgall, B3883 Sigma). Viable cell counts were performed after 3 h by diluting cultures in one-quarter-strength Ringer's solution and enumeration on BHI. Statistical differences from the negative control were determined by Student's t test. *, P < 0.01. (C) Murine experiments. Spontaneous rifampicin-resistant mutants of strains were isolated as described (15). For each strain, five BALB/c mice were administered with ≈109 cells by oral gavage on two consecutive days. On the third day the small and large intestines were removed and homogenized in PBS and bacterial numbers were determined by spread plating homogenized organs BHI on agar supplemented with 50 μg/ml rifampicin. Horizontal bars indicate the mean. Statistical differences were determined by Student's t test. *, P < 0.05
Discussion
This study demonstrates the use of metagenomics to identify functional anchors (the functional equivalent of phylogenetic anchors such as 16S rRNA) in complex microbial communities, and the subsequent use of these genes in a phylogenetic approach to dissect the gut microbiome according to activities relevant to survival in the GI tract (29). This is a powerful approach for the assessment of distribution and redundancy of functions within a community, and identification of functional anchors will underpin and enhance interpretation of current sequence-based metagenomic projects.
Bacteria face numerous challenges when colonizing the mammalian GI tract, and for the development of a beneficial microbiota (undertaking commensal, mutualistic, or symbiotic relationships with the human host) (1, 3), these must be overcome without reducing host fitness. Up-regulation of mucosal defenses through BA signaling and the antimicrobial properties of BA themselves are considered to be major mechanisms by which bacterial overgrowth in the small intestine is prevented (13, 14).
Our data clearly demonstrate that BSH activity benefits bacteria by enhancing resistance to CBA and increasing survival in the GI tract, and we propose that this facilitates colonization and development of the gut microbiota. However, the host will also benefit indirectly through acquisition of a gut microbial community, which ultimately provides advantages such as protection against gut pathogens and an increase in calories acquired from the diet. The general conservation and enrichment of BSH activity in the human gut metagenomes of healthy individuals suggests that BSH can mediate resistance to CBA in the small intestine without leading to bacterial overgrowth and compromising the host. Local mucosal defenses most likely act in synergy with the inhibitory properties of BA to control bacterial colonization and prevent overgrowth of BSH-producing organisms in the small intestine (9, 13–15).
The wide phylogenetic distribution and high level of redundancy we observed for BSH activity in the human gut microbiota is indicative of selection for this activity at the host level (3), and recent studies hint at direct benefits of BSH activity on host health. Studies of the intestinal pathogen Clostridium difficile, showed that products of microbial BA metabolism inhibit the growth of vegetative cells in vitro, and a direct role for BA modification in the protection of the host from C. difficile and other gut pathogens was proposed (30). In addition, through modulation of host lipid metabolism and reduction in cholesterol levels BSH activity and additional BA modification may reduce the risk of diseases such as obesity, arthrosclerosis, and cardiac disease (5, 10, 11, 21). These potential beneficial effects of BSH activity and BA modification support the possibility that BSH activity has been selected for at the host level because of a positive effect on host fitness.
In contrast, modified BAs have been implicated as carcinogens and cocarcinogens and considerable evidence exists for a role of modified BA in colorectal cancer (CRC) (19, 20). However, because CRC is predominantly diagnosed in individuals >50 years of age (31), the effect of CRC on reproductive fitness in the ancestors of modern humans would have been negligible, and as such would not pose a significant counter selection to the shorter-term beneficial effects of BSH activity and subsequent BA modification (18). It has also been suggested that BSH activity may protect the colonic epithelium from the genotoxic effects of CBA that reach this compartment (32). Deconjugation of CBA has been proposed to prevent their accumulation in colonic epithelial cells and subsequent DNA damage (32).
However, although BSH activity appears generally enriched in the human gut microbiota compared with metagenomes from the murine gut and other environments, the proportion of species that encode this activity remains to be established, and it is likely that many well adapted members of this community do not encode BSH. Additional investigation is required to identify these species and elucidate the mechanisms by which they mitigate the toxic effects of CBA or contribute to BA modification.
The activation of mucosal defenses and other signaling functions of BA are mediated through a variety of receptors for which BAs are the natural ligands, such as TGR5 and the farnesoid X receptor α (FXRα) (11, 12, 14). Activation of FXRα (which is expressed in the liver and intestine) plays a key role in mucosal defense against bacterial invasion in the small intestine BA, cholesterol, and triglyceride homeostasis, whereas TGR5 activation is involved in energy and possibly glucose homeostasis (11, 12, 14). The effect of modified BA on activation of these receptors is unclear, but there is potential for the gut microbiota to exert a major influence on host physiology and the etiology of metabolic diseases via modulation of BA signaling. Further investigation of the potential impact of microbial BA modification on host metabolic processes is required.
Overall, we have shown that BSH activity is distributed among all major bacterial divisions and two domains of life (Bacteria and Archaea) and conserved among the microbiota of individual human hosts. In conjunction with our comparative metagenomic and phylogenetic analyses, results from animal experiments show that BSH activity is a common microbial adaptation to the toxic effects of CBA, distributed among diverse microbial lineages in the human gut microbiota. The conservation, high level of redundancy, and general enrichment of BSH and associated Ntn_CGH-like family proteins in the gut microbiomes of geographically isolated individuals indicate that BSH activity is an ancient adaptation to the human gut environment, and the result of a deep coevolutionary relationship between host and microbiota.
Further investigation of BSH activity and BA modification by the gut microbial community, and its impact on host health and physiology, has the potential to provide insight into the coevolution of humans and their gut microbiota, and may lead to strategies to manipulate this community for the treatment of disease and the enhancement of human health.
Materials and Methods
Metagenomic Library Construction.
Metagenomic DNA was prepared from faecal samples as described in ref. 33. A fosmid library was generated by using the Epicentre copy control fosmid library kit according to manufacturer's instructions. Fosmid clones were picked into 384-well microtiter plates by using a Genetix QPIX2-XT and stored at −80°C.
End Sequence Analysis, Transposon Mutagenesis, Subcloning, and Sequencing.
End sequences of fosmid clones were obtained by using standard T7 and M13R sequencing primers (GATC Biotech). The results of tBlastX searches with an average minimum of 1,466 bp of sequence derived from both ends of the fosmid insert were used to assign clones to broad phylogenetic groups. Assignments were made by using criteria previously described (6), and sequences generating hits with a minimum 35% identity over 50 aa or more and an e-value of 1−15 or lower were used to assign clones to a phylogenetic group based on species hit. In vitro transposon mutagenesis was performed by using the Epicentre EZ-Tn_5_ Kan2 OriV element according to the manufacturer's instructions, and sequence data were assembled by using SeqMan (DNASTAR). For subcloning, fosmids were partially digested with Sau3A (Roche), and cloned into BamHI-digested pCI372 that was treated with shrimp alkaline phosphatase (Roche). The resulting ligation mix was transformed into chemically competent E. coli TOP10 (Invitrogen) and transformants were selected on LB containing 10 μg/ml chloramphenicol and subsequently patched onto LB bile agar (10 g of tryptone, 10 g of NaCl, 5 g of yeast extract, 10 g of glucose, 0.35 g of CaCl2·2H2O, 15 g of agar) containing 0.5% taurodeoxycholic acid (TDCA; Sigma T0875). All bile hydrolase positive transformants were checked by PCR to examine the insert size and sequenced. All sequencing was performed by GATC Biotech.
Comparative Metagenomic and Phylogenetic Analyses.
Comparative metagenomic analysis was performed by using tBlastn or BlastP searches of metagenomic datasets to identify sequences homologous to functional BSH obtained from our metagenomic analysis. For construction of phylogenetic trees, amino acid sequences homologous to functional BSH were obtained by BlastP searches and general database searches. All sequences were aligned and screened for presence of critical catalytic amino acids conserved among BSH and PVA family proteins (which were identified based on alignments of our functional metagenomic clones and previously analyzed crystal structures) (23). Putative critical catalytic amino acids identified and conserved between all functional BSH, PVA, and crystal structures were: C1, R17, D/E20, N172, R225, R/K228 with positions indicated relative to the Bifidobacterium longum BSH crystal structure (23). Potentially truncated sequences, and those which did not display the conserved amino acids, were removed from the dataset. Alignment accuracy was checked by calculating the overall mean distances by using the _p_-distances model and Minimum Evolution trees with 500 bootstrap replicates were constructed. Alignments, alignment accuracy, and tree analysis were conducted by using MEGA3 (34).
BSH Activity Assays.
BSH activity was examined by using a plate assay as previously described (22, 35). In brief, cultures that were grown overnight in LB were spotted (10 μl) onto LB bile agar supplemented with either 0.5% (wt/vol) TDCA, 0.5% (wt/vol) glycodeoxycholic acid (GDCA; Sigma G9910), or 3% (vol/vol) human bile that was obtained after laparascopic cholecystectomy. BSH activity was indicated by halos of precipitated deconjugated bile acids (Fig. S1).
Bile Survival Experiments.
Bile broth assays were carried out as previously described (36). In brief, overnight cultures of L. innocua were inoculated (3%) into broth supplemented with oxgall (Sigma B3883). Cell growth was measured by performing viable cell counts by diluting cultures in one-quarter-strength Ringer's solution and enumeration on BHI.
Murine Experiments.
Spontaneous rifampicin-resistant (rifR) mutants of strains were isolated as described (15). In brief, 10 ml of overnight cultures were centrifuged, cell pellets were resuspended in 200 μl of PBS, and plated onto BHI supplemented with 50 μg/ml rifampicin (BHI/rif). RifR strains demonstrated identical growth characteristics to non-rif-marked strains under in vitro conditions. Murine experiments were carried out according to institutional guidelines. For each strain, five BALB/c mice were administered with ≈109 cells by oral gavage on two consecutive days. On the third day the small and large intestines were removed and homogenized in PBS and bacterial numbers were determined by spread plating homogenized organs onto BHI/rif.
Heterologous Expression of Bile Salt Hydrolase Genes.
bsh genes of interest were amplified by PCR from E. coli fosmid-containing strains after lysis of cells in IGEPAL (Sigma CA-630). The proofreading enzyme KOD Hotstart DNA polymerase (Invitrogen) was used. PCR products were purified by using the Qiagen PCR purification kit, cut with the appropriate restriction enzymes and ligated to similarly digested plasmid pNZ44 (37). The resulting ligation mix was added to chemically competent E. coli TOP10 cells (Invitrogen) and transformants were selected on LB plates containing 10 μg/ml chloramphenicol. Plasmids containing inserts of correct size were selected by PCR. Plasmids were extracted and sequenced. L. innocua FH2333 were transformed by using standard procedures (38).
Supplementary Material
Supporting Information
Acknowledgments.
We thank Caroline Jones, Funing Sun, and Karen Daly for technical assistance and discussions and Pat Casey for technical assistance with murine experiments. This work was supported by Science Foundation Ireland through a Centre for Science, Engineering, and Technology award to the Alimentary Pharmabiotic Centre.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 2.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 4.O'Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rawls JF, Mahowald MA, Ley RE, Gordon JI. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. 2006;127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gill SR, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kurokawa K, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 9.Begley M, Hill C, Gahan CG. Bile salt hydrolase activity in probiotics. Appl Environ Microbiol. 2006;72:1729–1738. doi: 10.1128/AEM.72.3.1729-1738.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 11.Houten SM, Watanabe M, Auwerx J. Endocrine functions of bile acids. EMBO J. 2006;25:1419–1425. doi: 10.1038/sj.emboj.7601049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe M, et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439:484–489. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- 13.Hofmann AF, Eckmann L. How bile acids confer gut mucosal protection against bacteria. Proc Natl Acad Sci USA. 2006;103:4333–4334. doi: 10.1073/pnas.0600780103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inagaki T, et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci USA. 2006;103:3920–3925. doi: 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Begley M, Sleator RD, Gahan CG, Hill C. Contribution of three bile-associated loci, bsh, pva, and. btlB, to gastrointestinal persistence and bile tolerance of Listeria monocytogenes. Infect Immun. 2005;73:894–904. doi: 10.1128/IAI.73.2.894-904.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Batta AK, et al. Side chain conjugation prevents bacterial 7-dehydroxylation of bile acids. J Biol Chem. 1990;265:10925–10928. [PubMed] [Google Scholar]
- 17.Van Eldere J, Celis P, De Pauw G, Lesaffre E, Eyssen H. Tauroconjugation of cholic acid stimulates 7 alpha-dehydroxylation by fecal bacteria. Appl Environ Microbiol. 1996;62:656–661. doi: 10.1128/aem.62.2.656-661.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernstein H, Bernstein C, Payne CM, Dvorakova K, Garewal H. Bile acids as carcinogens in human gastrointestinal cancers. Mutat Res. 2005;589:47–65. doi: 10.1016/j.mrrev.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Berr F, Kullak-Ublick GA, Paumgartner G, Münzing W, Hylemon PB. 7 alpha-dehydroxylating bacteria enhance deoxycholic acid input and cholesterol saturation of bile in patients with gallstones. Gastroenterology. 1996;111:1611–1620. doi: 10.1016/s0016-5085(96)70024-0. [DOI] [PubMed] [Google Scholar]
- 20.Hill MJ. Bile flow and colon cancer. Mutat Res. 1990;238:313–320. doi: 10.1016/0165-1110(90)90023-5. [DOI] [PubMed] [Google Scholar]
- 21.Martin FP, et al. A top-down systems biology view of microbiome-mammalian metabolic interactions in a mouse model. Mol Syst Biol. 2007;3:112. doi: 10.1038/msb4100153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dashkevicz MP, Feighner SD. Development of a differential medium for bile salt hydrolase-active Lactobacillus spp. Appl Environ Microbiol. 1989;55:11–16. doi: 10.1128/aem.55.1.11-16.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar RS, et al. Structural and functional analysis of a conjugated bile salt hydrolase from Bifidobacterium longum reveals an evolutionary relationship with penicillin V acylase. J Biol Chem. 2006;281:32516–32525. doi: 10.1074/jbc.M604172200. [DOI] [PubMed] [Google Scholar]
- 24.Venter JC, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 25.Tringe SG, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 26.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dussurget O, et al. Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. Mol Microbiol. 2002;45:1095–1106. doi: 10.1046/j.1365-2958.2002.03080.x. [DOI] [PubMed] [Google Scholar]
- 28.Delpino MV, et al. A bile salt hydrolase of Brucella abortus contributes to the establishment of a successful infection through the oral route in mice. Infect Immun. 2007;75:299–305. doi: 10.1128/IAI.00952-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Handelsman J. Metagenomics: Application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev. 2004;68:669–685. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sorg JA, Sonenshein AL. Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol. 2008;190:2505–2512. doi: 10.1128/JB.01765-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ries LAG, et al. SEER Cancer Statistics Review, 1975-2001. Bethesda, MD: National Cancer Institute; 2004. [Accessed July 1, 2008]. Available at http://seer.cancer.gov/csr/1975_2001. [Google Scholar]
- 32.Stamp DH. Three hypotheses linking bile to carcinogenesis in the gastrointestinal tract: Certain bile salts have properties that may be used to complement chemotherapy. Med Hypotheses. 2002;59:398–405. doi: 10.1016/s0306-9877(02)00125-1. [DOI] [PubMed] [Google Scholar]
- 33.Jones BV, Marchesi JR. Transposon-aided capture (TRACA) of plasmids resident in the human gut mobile metagenome. Nat Methods. 2007;4:55–61. doi: 10.1038/nmeth964. [DOI] [PubMed] [Google Scholar]
- 34.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 35.Christiaens H, Leer RJ, Pouwels PH, Verstraete W. Cloning and expression of a conjugated bile acid hydrolase gene from Lactobacillus plantarum by using a direct plate assay. Appl Environ Microbiol. 1992;58:3792–3798. doi: 10.1128/aem.58.12.3792-3798.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Begley M, Gahan CG, Hill C. Bile stress response in Listeria monocytogenes LO28: Adaptation, cross-protection, and identification of genetic loci involved in bile resistance. Appl Environ Microbiol. 2002;68:6005–6012. doi: 10.1128/AEM.68.12.6005-6012.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGrath S, Fitzgerald GF, van Sinderen D. Improvement and optimization of two engineered phage resistance mechanisms in Lactococcus lactis. Appl Environ Microbiol. 2001;67:608–616. doi: 10.1128/AEM.67.2.608-616.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park SF, Stewart GS. High-efficiency transformation of Listeria monocytogenes by electroporation of penicillin-treated cells. Gene. 1990;94:129–132. doi: 10.1016/0378-1119(90)90479-b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information