The Caenorhabditis elegans unc-64 Locus Encodes a Syntaxin That Interacts Genetically with Synaptobrevin (original) (raw)
Abstract
We describe the molecular cloning and characterization of the unc-64 locus of Caenorhabditis elegans. unc-64 expresses three transcripts, each encoding a molecule with 63–64% identity to human syntaxin 1A, a membrane- anchored protein involved in synaptic vesicle fusion. Interestingly, the alternative forms of syntaxin differ only in their C-terminal hydrophobic membrane anchors. The forms are differentially expressed in neuronal and secretory tissues; genetic evidence suggests that these forms are not functionally equivalent. A complete loss-of-function mutation in unc-64 results in a worm that completes embryogenesis, but arrests development shortly thereafter as a paralyzed L1 larva, presumably as a consequence of neuronal dysfunction. The severity of the neuronal phenotypes of C. elegans syntaxin mutants appears comparable to those of Drosophila syntaxin mutants. However, nematode syntaxin appears not to be required for embryonic development, for secretion of cuticle from the hypodermis, or for the function of muscle, in contrast to Drosophila syntaxin, which appears to be required in all cells. Less severe viable unc-64 mutants exhibit a variety of behavioral defects and show strong resistance to the acetylcholinesterase inhibitor aldicarb. Extracellular physiological recordings from pharyngeal muscle of hypomorphic mutants show alterations in the kinetics of transmitter release. The lesions in the hypomorphic alleles map to the hydrophobic face of the H3 coiled-coil domain of syntaxin, a domain that in vitro mediates physical interactions with similar coiled-coil domains in SNAP-25 and synaptobrevin. Furthermore, the unc-64 syntaxin mutants exhibit allele-specific genetic interactions with mutants carrying lesions in the coiled-coil domain of synaptobrevin, providing in vivo evidence for the significance of these domains in regulating synaptic vesicle fusion.
INTRODUCTION
Communication between neurons at synaptic contacts occurs through the regulated release of chemical neurotransmitters. This release process is mediated by fusion of transmitter-filled vesicles with the plasma membrane. Through both biochemical and genetic studies, several dozen proteins have been implicated in this fusion process in neurons (Bennett and Scheller, 1994; Sollner and Rothman, 1994; Sudhof, 1995). A central player is the integral membrane protein syntaxin (Bennett et al., 1992). In neurons, syntaxin is found in abundance on the plasma membrane and also in low levels in synaptic vesicles (Bennett et al., 1992; Galli et al., 1995; Walch-Solimena et al., 1995). A vast array of experiments all indicate that syntaxin plays a critical role in regulating neurotransmitter release in neurons. For example, serotype C neurotoxin from Clostridium botulinum, which is a potent inhibitor of synaptic transmission, is a metalloprotease with high specificity for cleaving syntaxin (Blasi et al., 1993; Schiavo et al., 1995) Moreover, physiological recordings from Drosophila syntaxin mutants reveal the absence of both induced and spontaneous fusion events, suggesting an absolute requirement for syntaxin function (Broadie et al., 1995). Despite these and many other studies, the precise role of syntaxin in vesicle fusion remains mysterious. Specifically, is syntaxin an integral component of the fusion machinery (i.e., capable of fusing two lipid bilayers) or does it regulate that machinery?
A reason to suspect that syntaxin is a critical component of the release apparatus per se is that in vitro syntaxin associates with numerous synaptic proteins. First, syntaxin forms an extremely stable ternary complex with the synaptic vesicle-associated membrane protein synaptobrevin (also called VAMP) and SNAP-25, a protein associated with the plasma membrane (Sollner et al., 1993; Hayashi et al., 1994). This and other related complexes between vesicle-associated and plasma membrane-associated proteins have been hypothesized to represent intermediates in the fusion process. However, this hypothesis remains effectively untested as biochemical studies of the ternary complex have been performed in the absence of membrane. Furthermore, a multitude of other proteins show strong interactions with syntaxin. These include Munc-13, Munc-18, calcium channels, synaptotagmin, and several other proteins (Bennett et al., 1992; Hata et al., 1993; Sollner et al., 1993; Garcia et al., 1994; Pevsner et al., 1994; Sheng et al., 1994; Betz et al., 1997; Edwardson et al., 1997). Although genetic and perturbation experiments suggest many of these proteins also play pivotal roles in regulating the release process, it is unclear whether the interactions of these proteins with syntaxin are even functionally significant in vivo. Elucidating the roles of each of these proteins in regulating vesicle fusion remains a complex task.
Caenorhabditis elegans is a powerful model system with which to study mechanisms of neurotransmission (Rand and Nonet, 1997). Genetic selection for mutants with transmission defects is feasible in this organism using the acetylcholinesterase inhibitor aldicarb (Rand and Russell, 1985; Miller et al., 1996). This drug kills wild-type animals, presumably through hyperstimulation of muscle, but spares mutants with reduced cholinergic transmission. Distinct approaches aimed at identifying synaptic regulators, including biochemistry in vertebrates and genetics in C. elegans, have identified a largely overlapping set of genes (Sudhof, 1995; Rand and Nonet, 1997). At least five genes identified by mutation in C. elegans, namely snb-1 (synaptobrevin), snt-1 (synaptotagmin), unc-2 (calcium channel α-subunit), unc-13, and unc-18, encode homologs of proteins that interact in vitro with vertebrate syntaxin (Maruyama and Brenner, 1991; Gengyo-Ando et al., 1993; Nonet et al., 1993; Schafer and Kenyon, 1995; Nonet et al., 1998). Loss-of-function mutations in each of these C. elegans genes result in severe behavioral and synaptic function defects (Brenner, 1974; Nonet et al., 1993, 1998). Although, the C. elegans proteins have not yet been reported as interacting with syntaxin as occurs in vertebrates, the extensive conservation of these molecules makes it likely that such interactions exist. Disrupting, through mutation, specific interactions between syntaxin and its many interacting partners will disclose in vivo functions of the proteins that require syntaxin interaction.
In this study, we describe the isolation and characterization of mutants defective in syntaxin function. We demonstrate that C. elegans syntaxin is encoded by the unc-64 gene. Three distinct gene products derive from the locus; all share an identical cytoplasmic domain but contain different C-terminal membrane anchors and are expressed in distinct but overlapping secretory tissues. unc-64 syntaxin appears to be essential for neuronal function, as mutants lacking the gene product arrest as paralyzed larvae. However, the gene is not required for embryogenesis or secretion of the cuticle, suggesting that C. elegans syntaxin is not required for secretion in all cells. Viable hypomorphic mutants carrying lesions in the coiled-coil domain show milder dysfunction of synaptic transmission. These mutants exhibit a variety of behavioral deficits and alterations in the kinetics of transmission. Furthermore, these mutants show specific genetic interactions with mutants carrying lesions in the coiled-coil domain of synaptobrevin (Nonet et al., 1998). Our studies provide in vivo evidence for a critical role for the coiled-coil domains of both syntaxin and synaptobrevin in regulated secretion.
MATERIALS AND METHODS
Growth and Culture of C. elegans
C. elegans was grown at 22.5°C on solid medium as described by Sulston and Hodgkin (1988). All mapping, complementation, and deficiency testing were performed using standard genetic methods (Herman and Horvitz, 1980). Aldicarb, 2-methyl-2-[methylthio]proprionaldehyde _O_-[methylcarbamoyl]oxime, was obtained from Chem Services (West Chester, PA) and was prepared as a 100 mM stock solution in 70% ethanol. Aldicarb was added to the agar growth medium after autoclaving or added directly to plates.
DNA and RNA Manipulations
C. elegans genomic DNA was isolated as described by Sulston and Hodgkin, (1988). cDNA was synthesized by reverse transcribing RNA using random hexanucleotide primers as described by Sambrook et al., (1989). Poly (A)+ selected RNA was isolated from a mixed-stage culture of the wild-type strain N2 as previously described (Nonet and Meyer, 1991). Manipulations of DNA and RNA including electrophoresis, blotting, and probing of blots was performed using standard procedures except where noted (Sambrook et al., 1989). Analyses of DNA sequences and amino acid sequences were performed on a SPARC station using the GCG program package (Devereux et al., 1984).
Isolation of the C. elegans Syntaxin Gene
Oligonucleotide primers TX1 (5′-TTY TTY GAR CAR GTN GAR GAR AT) and TX5 (5′-CAT NGC CAT RTC NTN RAA CAT) corresponding to regions of syntaxin conserved between rat syntaxin 1A and 1B (Bennett et al., 1992) were used to amplify a specific sequence from cDNA made as described by Nonet et al., (1993). The fragment was cloned, sequenced, and used as a probe to isolate three cDNA clones from an embryonic cDNA library (Lichtsteiner and Tjian, 1993). The largest cDNA (_Not_I fragment) was subcloned into pBluescript KS(−) to create pTX4. Sequencing of the cDNA revealed it began with the trans-spliced leader sequence found on many C. elegans messages and ended with a poly A sequence (accession number AF047885). The full-length clone was used as a probe on a Northern blot of mixed-staged poly (A)+ selected RNA and identified three transcripts. Sequencing of a genomic clone of the locus revealed the presence of potential alternative terminal exons. Two additional transcripts were amplified from cDNA using PCR and confirmed that the exons are utilized in vivo (accession numbers AF047886 and AF047887). We have not confirmed the structure of the noncoding portion of the larger messages identified on Northern blots because the region between the exons encoding transmembrane regions contains numerous high-copy repetitive elements (rep A, C, H, and W). The unc-64 probe hybridized to YAC clones Y39E4, Y43F4, Y106H1, and Y109C2 on chromosome III of the physical map of C. elegans (Coulson et al., 1988). As cosmid clones were not available for this genomic region, clone λTXg.1 was obtained by probing a genomic lambda library (Barstead and Waterston, 1989), and clone H20G23 was obtained by probing a fosmid library with assistance from the Genome Sequencing Center, Washington University School of Medicine, St. Louis, MO. pTX20 contains a 16.2-kilobase (kb) _Pst_I fragment from H20G23 inserted into pBluescript KS(−). pTX6 was created by inserting an 8.2-kb _Nco_I–_Nhe_I fragment of λTXg.1 into pRSETC. An unc-64::lacZ fusion plasmid, pTX9, was created by inserting the 3.1-kb _Xba_I–SpeI lacZ fragment from pPD95.03 (kindly provided by A. Fire, J. Ahnn, G. Seydoux, and S. Xu) into the _Nhe_I site of pTX6. pTX12, containing a functional syntaxin minigene, was created by inserting the 5.2-kb _Xho_I-_Nhe_I fragment of pTX6 into pTX4. Three GFPS65T-tagged transmembrane domain constructs were created by amplifying sequences from pTX20 with an oligonucleotide in exon 5 (TX-17 5′-GCGCAAGACGAACCCAGAG) and oligonucleotides (TX-52 5′-ATCGGCCGACCGGTTTGAAAAAATGTATAATACTTTC, TX-53 5′-ATCGGCCGACCGGTATGCCAGGAATATACTGAATGAG) that replaced the stop codon in the exon with a linker sequence containing both an _Age_I and _Eag_I restriction site. The amplified fragments were introduced into pTX12 after cleavage at the _Nhe_I and _Eag_I restriction sites, replacing the cDNA sequences of pTX12. Green fluorescent protein (GFP) from pPD93.65 (kindly provided by A. Fire, J. Ahnn, G. Seydoux, and S. Xu) was added subsequently using the _Age_I and _Eag_I sites to create pTX34 and pTX37 (Figure 1). Using a similar amplification scheme as above (TX-55 5′-TTACCGGTCGAACACGAAAACGTTGAGAAGAC), pTX38 was created in a single step by replacing the _Nhe_I–_Age_I fragment of pTX34 with the smaller fragment tagging syntaxin C.
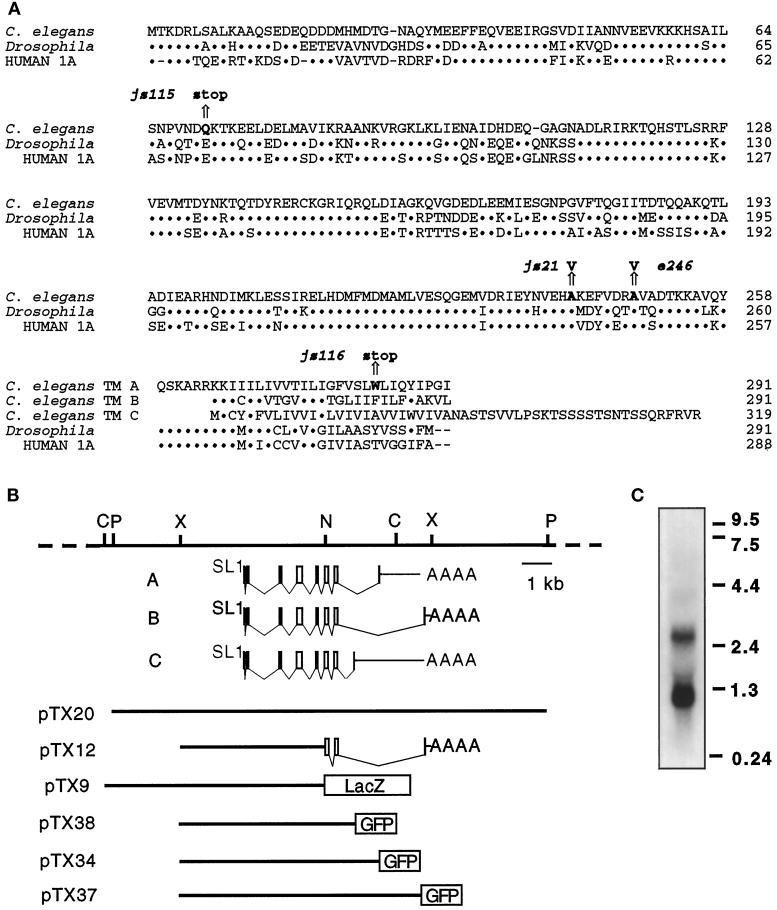
Figure 1.
The unc-64 syntaxin locus. (A) Similarity among syntaxin molecules. An alignment of syntaxin family proteins from C. elegans, Drosophila (Schulze et al., 1995), and human (Osborne et al., 1997). The position of the molecular lesions identified in selected mutations in the unc-64 syntaxin gene (bold) and the resulting change in amino acid sequence are indicated above an arrow. The standard single-letter amino acid code is used. Dots represent identity with the C. elegans sequence. Amino acid number appears at the right. (B) Restriction map of the genomic unc-64 gene. Below the restriction map is a schematic diagram showing the intron–exon structure of three unc-64 products and the sequences contained in plasmid constructs used in this study. Gray represents predicted message structures. C, _Nco_I; X, _Xho_I; N, _Nhe_I; P, _Pst_I restriction endonuclease sites. (C) Autoradiograph of a Northern blot probed with a syntaxin cDNA fragment; 10 μg of mixed-staged poly A(+) selected RNA isolated from wild-type hermaphrodites was loaded on the gel. RNA size standards (kb) are on the right.
Production of Syntaxin Antibodies
Sequences from pTX4 coding for amino acids 1–266 were amplified by PCR using oligonucleotides TX-7 (5′-GCTCTAGAATGACTAAGGACAGATTG) and TX-8 (5′-CAAGATCTACTTCTTCCTTCGCGCCTTC) and cloned into pRSETB (Invitrogen, San Diego, CA) to create the plasmid pTX10. The plasmid expresses a 32-kDa fusion protein containing a six-histidine tag on the amino terminus of a 30.5- kDa cytoplasmic domain of the UNC-64 protein. The protein was purified and used to immunize rabbits as described by Nonet et al. (1993). UNC-64 antiserum was affinity purified using the a method described previously (Smith and Fisher, 1984). Affinity-purified antibodies were used at 1:50 dilution, and sera were used at 1:1000 dilution. Immunocytochemistry was performed as previously described using Bouin’s fixative (Nonet et al., 1993).
Genetic Analysis of unc-64
Wild-type males were mutagenized with 50 mM ethyl methanesulfonate for 4 h and crossed to dpy-18(e364) unc-64(e246); xol-1(y9) flu-2(e1003), and the cross-progeny was screened for lethargic non-Dpy animals. Two additional alleles of unc-64 were isolated in a screen of 5200 genomes. Each mutant was outcrossed to wild-type and subsequently balanced with bli-5(e518). The lethal unc-64 alleles are balanced effectively by the bli-5 mutant as no recombinants between bli-5(e518) and unc-64(e246) were isolated in more than 1500 animals examined. The entire coding region and all intron–exon junctions of e246, js21, js115, js116, md130, and md1259 were sequenced to characterize the molecular lesions. The identified lesions are C to T in the second base of codon 248 in e246, C to T in the second base of codon in 241 in js21, C to T in the first base of codon 71 in js115, G to A in the second base of codon 283 (transmembrane exon A) in js116, a G to A in the base after exon 6 in md130, and a G to A in the first base after exon 3 in md1259.
To examine the phenotype of animals harboring js21 and e246 mutations in trans to a deficiency, unc-64/+ males were crossed to eDf2; eDp6 hermaphrodites and eDf2/unc-64 animals were examined. In the case of js115, which confers a lethal phenotype, single js115/+ males were mated to single tra-2(q122)/+;eDf2/+ female animals isolated from a cross of tra-2(q122) males with e246/eDf2. js115/eDf2 animals segregating from js115/eDf2; eDp6 were also analyzed. The phenotypes of e246, js21 and js115 in trans to the deficiency eDf2 in each case were very similar to that of animals homozygous for the respective mutation. In addition, js115/e246 animals were phenotypically indistinguishable to eDf2/e246 animals. In concert, these results indicate that js115 behaves like a null mutant. unc-64; snb-1 mutants were created by crossing dpy-11(e224)/snb-1 males to unc-64; dpy-11(e224) yielding unc-64/+; dpy-11/snb-1 animals. After selecting animals homozygous for unc-64, unc-64; snb-1 animals were identified as animals that failed to segregate Dpy animals. The unc-64 genotype in the double mutants was confirmed by PCR analysis.
Transformation and RNA Injections of C. elegans
Germline transformation was accomplished by coinjecting the plasmid of interest with either rol-6 (pRF4 at 150 μg/ml) as a dominant marker or lin-15 (pJM23 at 50 μg/ml) injected into lin-15(n765) as a recessive transformation marker (Mello et al., 1991; Huang et al., 1994). The dominant marker rol-6 was used to rescue e246 and js115 mutants by the introduction of pTX20 (20 μg/ml). js115 and j_s116_ were also rescued by the minigene construct pTX12 (20 μg/ml). LacZ (pTX9) and GFP (pTX34, 37, 38) expression constructs were introduced at 10 μg/ml into lin-15(n765) using lin-15 as marker. For RNA interference experiments, full-length antisense RNA was synthesized in vitro using T7 RNA polymerase from a PCR-amplified pTX4 insert DNA template and injected into the germline of wild-type animals at 0.8 mg/ml as described by Rocheleau et al. (1997).
Mosaic Analysis
To examine animals with no germline contribution, js115; jsEx136 animals were created. The jsEx136 extrachromosomal array was created by coinjection of the unc-64 (+) plasmid pTX20 (20 μg/ml), the dominant rol-6 marker pRF4 (150 μg/ml), and pMR1 (10 μg/ml) into js115/bli-5(e518). pMR1, an unc-54 promoter::GFPS65T construct that expresses high levels of GFPS65T in body wall muscle and sex muscles, was constructed by inserting a _Kpn_I-_-Apa_I GFPS65T fragment from pPD93.51 into pPD30.38 (both plasmids kindly provided by A. Fire, J. Ahnn, G. Seydoux, and S. Xu). More than 140 rollers were examined and none that segregated no progeny were identified. Among 354 nonroller mosaic animals, 10 that segregated no viable progeny were isolated. One produced no progeny, and the other 9 produced from 12 to greater than 100 progeny (a total of >740). Ten to 30 of the L1-arrested animals from each mother were cloned and examined for the presence of GFP in muscle and the general organization of structures including the cuticle, nerve ring ganglia, intestine, and pharynx. The animals appeared similar to js115 homozygotes from a _unc-64(js11_5)/bli-5(e518) heterozygote. The plates also were examined for arrested unhatched eggs. A few progeny of 4 of the 9 animals failed to hatch. Of the 10 eggs found, only 3 were arrested before threefold stage. Finally, of approximately 2500 progeny screened under a fluorescent dissecting microscope for progeny lacking fluorescence in muscle, we identified none lacking fluorescence in all muscle and analyzed the locomotion of 14 lacking GFP in large sections of muscle.
Behavioral Assays
Locomotion was assayed by imaging L4-staged animals shortly after they were deposited onto fresh agar plates containing an Escherichia coli lawn. Charge-coupled device camera images were collected for 1 min with an LG3 frame grabber (Scion) at 1-s intervals at a magnification between 1.6× and 2.5×. Distance traveled was calculated by summing displacements in the position of the tail. Defecation was observed under a dissecting microscope and cycles were recorded using a simple computer program (Liu and Thomas, 1994). Pharyngeal pumping of mutant animals with slow pumping rates was assayed by counting the number of pumps in a 1-min interval. Pharyngeal pumping of animals with faster rates (>120 pumps/min) was assayed by counting pumps from digital images captured at 15 frames/s for 20-s intervals.
Resistance to Aldicarb and Levamisole
Mutants were assayed for acute exposure to aldicarb and levamisole. Aldicarb resistance was examined by transferring individual animals to plates containing aldicarb and assaying for paralysis 4 h after exposure. Animals were considered paralyzed if they failed to move even if prodded with a platinum wire. Levamisole resistance was examined by placing 25 worms in S-basal medium (Sulston and Hodgkin, 1988) with 100 μM levamisole and scoring for movement by examining video images of the animals at different time points.
Electrophysiology
Electropharyngeograms (EPGs) were recorded using a AC preamplifier (designed by David Brumley, University of Oregon, Eugene, OR) and LabView Acquisition software (National Instruments, Austin, TX) as previously described (Avery et al., 1995). Bath solution consisted of M9 or Dent’s saline (Avery et al., 1995) with 2.5 mM serotonin to stimulate pumping. Only recordings from young adult hermaphrodites with at least 10 pharyngeal pumps were analyzed. Analysis of the records was performed as described by Iwasaki et al. (1997) except that amplitudes were calculated as peak-to-peak, and the threshold was set at 5% (rather than 10%) of the mean amplitude of the R-phase transients to identify M3 transients in a record.
RESULTS
unc-64 Encodes a Syntaxin Homolog
unc-64 was first described by Brenner (1974) as a mutant with locomotory defects. unc-64 was suspected to encode a component regulating synaptic transmission because it shares a common phenotype with many other synaptic mutants in C. elegans: resistance to the acetylcholinesterase inhibitor aldicarb (Alfonso et al., 1993, 1994; Nonet et al., 1993, 1997, 1998; Nguyen et al., 1995; Miller et al., 1996; Iwasaki et al., 1997). We isolated a C. elegans gene with strong similarity to rat syntaxin 1 that mapped to a region of chromosome III on the physical map, roughly corresponding to the genetic map position of the unc-64 gene (see Figure 1 and MATERIALS AND METHODS for details). We tested whether the syntaxin-like gene was encoded by unc-64 using germline transformation (Mello et al., 1991). unc-64(e246) mutants harboring a genomic clone of the C. elegans syntaxin gene were phenotypically rescued to wild-type, as determined by both behavioral and aldicarb resistance assays. Additionally, sequencing of the coding region of the candidate syntaxin gene from unc-64 mutant alleles in each case revealed a molecular lesion that could account for the genetic defect (Figure 1A). Thus, we conclude that unc-64 encodes a C. elegans syntaxin homolog.
Three Distinct Forms of Syntaxin Are Encoded by unc-64
We isolated cDNA clones derived from the unc-64 syntaxin locus. Several complete cDNA clones representing one transcript from the C. elegans syntaxin locus were isolated from an embryonic cDNA library, and partial cDNAs representing two additional transcripts were isolated from first-strand cDNA using the PCR. Each transcript is predicted to encode a product with 63% or 64% identity to human syntaxin 1A. The three transcripts differ only in the last of eight coding exons and thus encode products with identical cytoplasmic domains but different transmembrane anchor domains (Figure 1, A and B). We refer to these three products as the syntaxin A, B, and C products of the unc-64 locus. At least three distinct transcripts of approximately 1.1 kb, 2.6 kb, and 3.8 kb are present on Northern blots of mixed-staged RNA (Figure 1C). The size of the full-length syntaxin B cDNA isolated from the cDNA library is of the appropriate size to correspond with the smaller transcript. The larger transcripts are of an appropriate length to represent the syntaxin A and C cDNAs if they utilize the same polyadenylation site. The splicing of the transmembrane domain of syntaxin in C. elegans is reminiscent of the splicing pattern observed for the rat syntaxin 2 and the mouse syntaxin 3 genes (Bennett et al., 1993; Ibaraki et al., 1995).
unc-64 Syntaxin Is Expressed in the Nervous System and Secretory Tissues
The C. elegans syntaxin sequences are most similar to the vertebrate neuronal-specific syntaxin 1 molecules and hence are likely to be expressed in the nematode nervous system. To examine the expression pattern, antisera were raised against bacterially expressed C. elegans syntaxin fusion protein. An affinity-purified anti-syntaxin antiserum was incubated with fixed whole adult animals, and the antibodies were detected using FITC-conjugated secondary antisera. As expected, syntaxin was detected in the nervous system (Figure 2). Syntaxin immunoreactivity was very strong and distributed uniformly along the major process bundles: the nerve ring (Figure 2A), ventral cord (Figure 2B), and dorsal cord (Figure 2A). Syntaxin immunoreactivity also was detected in the vast majority of neuronal cell bodies as well as in commissural and dendritic processes (Figure 2B). By contrast, the vesicle-associated markers synaptobrevin, synaptotagmin, and RAB-3 are restricted to the synaptic-rich regions of the nervous system where they show a punctate, rather than uniform, appearance (Nonet et al., 1993, 1997, 1998). Syntaxin was also detected in the pharyngeal nervous system (Figure 2A). Furthermore, syntaxin was detected outside the nervous system. Immunoreactivity was strong in the uv1 cells of the vulva, in the intestine (Figure 2C), and in the spermatheca (Figure 2D). Detectable levels of syntaxin protein were not observed in other tissues. Our protein expression data suggest that unc-64 encodes the homolog of the vertebrate syntaxin 1 gene (Bennett et al., 1992, 1993).
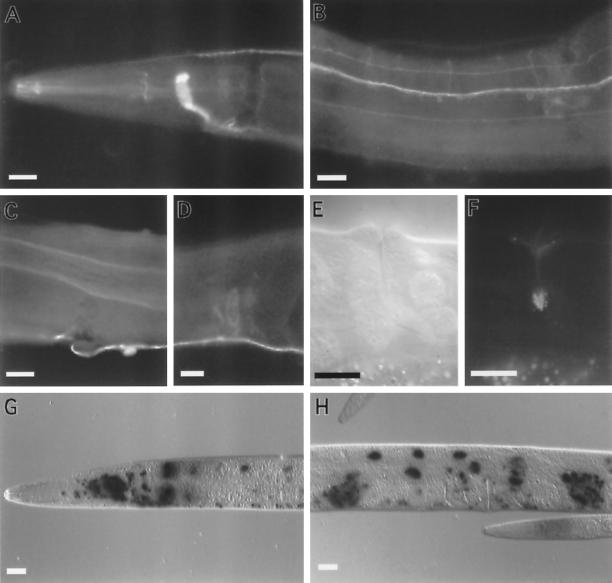
Figure 2.
Expression of unc-64 syntaxin. (A–D) Whole adult wild-type hermaphrodite worms fixed and stained with α-UNC-64 primary antibodies and visualized with FITC-conjugated antibodies. (A) Lateral view of the head region showing immunoreactivity in the nerve ring, dorsal cord, and pharyngeal nervous system. (B) Ventral view of the midbody region showing expression in ventral cord axons, neuronal cell bodies, and in commissural and sublateral processes. (C) A lateral view of the midbody showing UNC-64 immunoreactivity on the basolateral surface of the intestine. (D) A lateral view of the midbody showing immunoreactivity in the hermaphrodite spermatheca. The male vas deferens also stains. (E) A lateral view of the vulva viewed using differential interference contrast microscopy. (F) Same plane of focus as panel E showing fluorescence from the UNC-64 A::GFP product in the uv1 cells of the vulva. (G–H) Whole adult wild-type animals fixed and stained for LacZ gene activity expressed under the control of the unc-64 promoter. LacZ activity assayed using X-gal as a substrate. The lacZ gene contains a nuclear localization signal. (G) Lateral view of the head region showing expression of unc-64 ::lacZ in neuronal and intestinal nuclei. (H) Lateral view of the midbody of an adult hermaphrodite showing unc-64::lacZ expression in intestinal nuclei, neuronal nuclei, and the spermatheca. Scale bar, in all panels, 20 μm.
As it was not feasible using immunohistochemistry to determine which neurons expressed syntaxin, the expression pattern of the SNARE gene was examined using a translational lacZ fusion. A genomic syntaxin fragment (Figure 1C) was inserted into a plasmid containing a nuclear-localized lacZ reporter gene (Fire et al., 1990). As expected, syntaxin was expressed in the vast majority of all neurons (Figure 2, G and H). In addition, syntaxin expression was high in intestinal nuclei, in the spermatheca, and in the uv1 cells of the vulva (Figure 2H). In summary, C. elegans syntaxin is expressed in both secretory and neuronal tissues. In neurons, the protein is not specifically associated or concentrated at synaptic release sites; rather, it is ubiquitously distributed.
The three identified products deriving from the C. elegans syntaxin locus differ only in those sequences that are predicted to lie in the lipid bilayer and act as transmembrane anchors. Because production of antibodies capable of distinguishing these products did not seem feasible, we engineered constructs that tagged each product by appending the GFP-coding region to the last codon of each predicted transmembrane domain. The constructs were introduced into wild-type animals by germline transformation, and the expression pattern of the GFP-tagged syntaxins was examined. Syntaxin A and C were expressed in neurons and in five nonneuronal tissues. By contrast, expression of syntaxin B was limited to the five nonneuronal cell types. In neurons, syntaxin A and C were found ubiquitously on neuronal cell bodies, axons, and dendrites. Also stained were the uv1 secretory cells of the vulva, the excretory gland cells, the distal tip cells, the spermatheca, and the intestinal muscle (Figure 2, E and F). In the uv1 cells, GFP fluorescence was concentrated subcellularly (Figure 2, E and F). The expression pattern from the GFP, lacZ, and antibody studies were consistent, except that GFP reporter expression was observed in the intestine only with syntaxin C fusions. This expression was limited to the posterior half of the intestine. It is possible that sequences that direct gut expression are absent from the GFP clones since sequences further upstream of the initiation codon are present in the lacZ constructs. In summary, the alternative UNC-64 syntaxin products are expressed in distinct but overlapping neuronal and nonneuronal secretory cells.
Isolation of Additional unc-64 Syntaxin Mutants
Additional alleles of the unc-64 gene were isolated in three independent screens. md130 and md1259 were isolated in a screen for aldicarb- resistant mutants occurring spontaneously in a strain with high levels of Tc1 transposition (Miller et al., 1996). js21 was isolated in a general screen for aldicarb-resistant mutants using EMS as a mutagen (M.L.N., unpublished data). Finally, two lethal alleles, js115 and js116, were isolated in a noncomplementation screen (see MATERIALS AND METHODS). Molecular characterization revealed that the e246 and js21 lesions are missense mutations that result in alanine-to-valine substitutions at codons 241 and 248, respectively. These codons are in the H3 domain of syntaxin, a region thought to assume an amphipathic helical structure capable of forming coiled-coil interactions with other proteins (Hardwick and Pelham, 1992; Calakos et al., 1994; Fasshauer et al., 1997). The point mutations in md130 and md1259 disrupt splicing sites (see MATERIALS AND METHODS for details). The js116 mutation results in premature termination in the transmembrane domain of the syntaxin A product, while the js115 lesion results in premature termination at codon 71. Immunohistochemical examination of whole fixed animals revealed that syntaxin product remained in the nervous system of all viable mutants, although the level of syntaxin was reproducibly lower in the md1259 mutant. UNC-64 staining was not detected in js115. All available evidence suggests that js115 represents the null phenotype (see MATERIALS AND METHODS).
Behavioral Defects of unc-64 Syntaxin Mutants
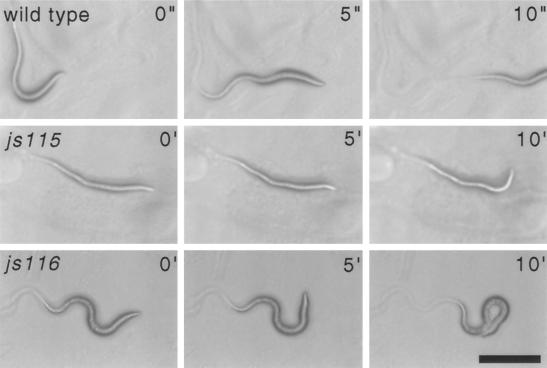
All six unc-64 mutants exhibit locomotory abnormalities. The lethal mutants exhibit the most severe motor defects. js115 animals are virtually completely paralyzed and rarely maintain a sinusoidal posture, although they occasionally will make slow head movements (Figure 3). js116 animals exhibit very slow motor movements (Figure 3) and tend to adopt a coiled position. Both js115 and js116 animals arrest development just after hatching and eventually die as L1 larvae. Behaviorally, the viable js21 and e246 mutants are very lethargic, although they retain the ability to move briefly when prodded. Locomotory abnormalities were quantified by examining the rate of movements of animals imaged using a charge-coupled device camera (Table 1). Despite exhibiting severe locomotory defects, other behaviors of the hypomorphic mutants were only weakly affected (Table 1). The rate of pharyngeal pumping was relatively normal in the hypomorphic mutants e246 and js21, although pumping was completely abolished in the js115 null mutant. The motor defecation program was only slightly abnormal in hypomorphs, but completely abolished in js115. It is interesting to note that, compared with other synaptic mutants with similarly severe locomotory defects (e.g., unc-18 and unc-13), the unc-64(e246) mutant has much milder deficiencies in other behaviors. unc-64(e246) mutants also exhibit a tendency to form enduring dauer larvae (Iwasaki et al., 1997), a developmental decision regulated by sensory input (Bargmann and Horvitz, 1991). Two other mutants, md130 and md1259, also exhibit milder behavioral defects, but minimal analysis was performed on these alleles due to the difficulty in determining, with certainty, the resulting gene products. Finally, at the morphological level, muscle, the intestine, the pharynx, the cuticle, and the organization of the nervous system appear normal in both the hypomorphs and the null mutant. In summary, a variety of genetic lesions in the syntaxin gene result in behavioral defects of varying severity. Presumably, all these phenotypes represent manifestations of an underlying impairment of synaptic transmission.
Figure 3.
Phenotype of lethal unc-64 syntaxin mutants. Bright field images of first larval stage animals of wild-type, unc-64(js115), and unc-64(js116) animals on an E. coli bacterial lawn at the indicated time intervals (wild-type in seconds and mutants in minutes). Scale bar, 100 μm.
Table 1.
Behavioral defects unc-64 mutants
| Strain | Locomotion | Pharyngeal pumps/min | Defecation | ||
|---|---|---|---|---|---|
| Speed (μm/sec) | Efficiencya | Cycle time (s) | EMCb | ||
| N2c | 161 ± 36 | 0.63 | 250 ± 30 | 42.0 ± 3.3 | 98% |
| e246 | 13 ± 8 | 0.18 | 209 ± 20 | 39.3 ± 3.3 | 87% |
| js21 | 51 ± 22 | 0.43 | 234 ± 10 | 48.8 ± 9.2 | 80% |
| js115d | 0 | NA | 0.2 ± 0.6 | None | NA |
| js116d | 0 | NA | 3 ± 2 | None | NA |
Late Arrest of unc-64 Syntaxin Mutants Is Not the Result of Maternal Contribution
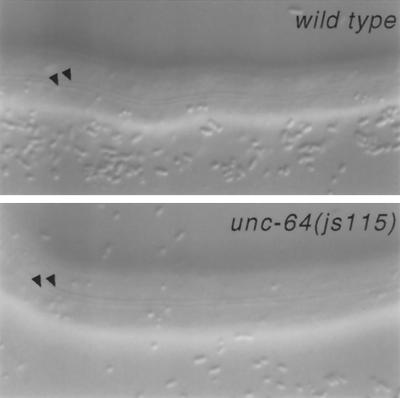
In D. melanogaster, mosaic clones that lack syntaxin cannot be isolated, suggesting the molecule is required in all cells (Schulze and Bellen, 1996). Furthermore, syntaxin is also required early in development during cellularization of the Drosophila embryo (Burgess et al., 1997). As our most severe mutant arrests development after completing embryogenesis, we examined whether unc-64 syntaxin mutants completed embryogenesis using maternally derived products. We constructed a strain homozygous for js115, which also harbored an extrachromosomal array bearing a visible marker (rol-6), a muscle-specific unc-54::GFP construct, and the wild-type unc-64 syntaxin gene. These animals segregate both wild-type and mutant progeny as the array is not transmitted to all progeny. We examined the progeny of nine mosaic animals lacking syntaxin product in the germline. These animals were identified on the basis of their failure to produce viable progeny. Nearly 99% (n = 740) of the progeny of these mosaic animals completed embryogenesis and arrested as first larval stage animals with a phenotype indistinguishable from the js115 progeny of heterozygous mothers. The secreted cuticle of these animals appeared normal, containing well organized alae (Figure 4). Most of the remaining 1% of progeny arrested late in embryonic development as threefold embryos. None of the progeny of the mosaic animals we examined expressed GFP, confirming that the unc-64(+) array was absent from the germline. We also examined viable mosaic animals that lacked unc-64 expression in muscle. Three mosaics that lacked GFP in muscle anterior of the vulva and 11 mosaics that lacked GFP in both dorsal body wall muscle quadrants all moved normally, suggesting that syntaxin is not required in muscle. Additionally, most of the severely uncoordinated mosaic animals we examined retained GFP expression in muscle, consistent with a neuronal site of action for syntaxin.
Figure 4.
Cuticle of L1 js115 syntaxin mutant animals. Lateral view of the cuticle structures (alae) of the L1 larvae of wild-type and homozygous unc-64(js115) animals segregating from a mosaic animal lacking unc-64(+) in the germline. Arrowheads point to the alae (cuticular specializations) observed in L1 larvae. Scale bar, 20 μm.
In separate experiments, we injected antisense RNA into the germline of wild-type animals in an attempt to inactivate the unc-64 gene by RNA-mediated interference (Guo and Kemphues, 1995; Rocheleau et al., 1997). This technique phenocopies the null phenotype of most genes that provide maternal contributions to the embryo, but is much less efficacious in phenocopying late-acting zygotic genes. Our injections failed to confer the js115 phenotype or an aldicarb-resistant phenotype. In summary, we find no evidence for either a maternal contribution of unc-64 or for a requirement for zygotic syntaxin in the early embryo.
Synaptic Transmission Defects in unc-64 Mutants
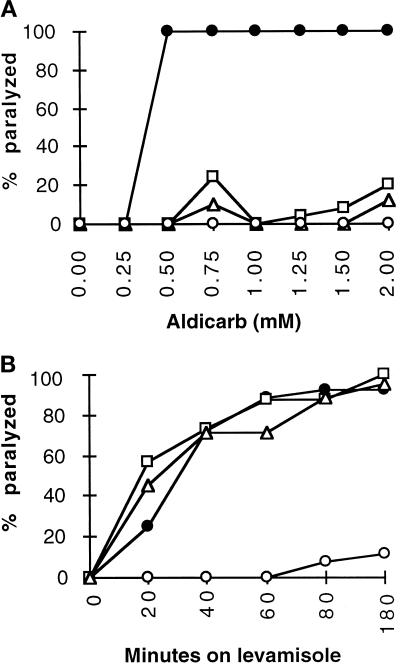
To assess neurotransmission in unc-64 mutants, we began by examining the effect of cholinergic pharmacological agents. We quantified the sensitivity of unc-64 mutants to aldicarb, a potentiator of released acetylcholine, and to levamisole, an agonist of nicotinic acetylcholine receptors in the nematode (Lewis et al., 1980a,b). All of the viable unc-64 mutants examined were resistant to high levels of aldicarb (Figure 5A), as was the acetylcholine receptor mutant unc-29 (Fleming et al., 1997). This strongly suggests that cholinergic transmission is reduced in these mutants. However, unlike unc-29, the unc-64 syntaxin mutants were sensitive to levamisole (Figure 5B), suggesting an intact postsynaptic apparatus and thus a presynaptic defect. Finally, examination of the lethal mutations revealed that aldicarb enhanced js116 movement but failed to stimulate movement of js115 animals, providing evidence that cholinergic synaptic transmission may be completely abolished in the null mutant.
Figure 5.
Pharmacological properties of unc-64 mutants. (A) Aldicarb sensitivity of unc-64 mutants and the wild type. Shown is the percentage of adult animals paralyzed after a 4-h exposure to various concentrations of aldicarb on plates seeded with E. coli wild-type (•), js21(□), e246 (Δ), and unc-29 (○). (B) Levamisole sensitivity of unc-64 mutants and the wild type. Shown is the percentage of adult animals paralyzed after exposure to 100 μM levamisole at various times. Wild-type (•), js21(□), e246 (Δ), and unc-29 (○).
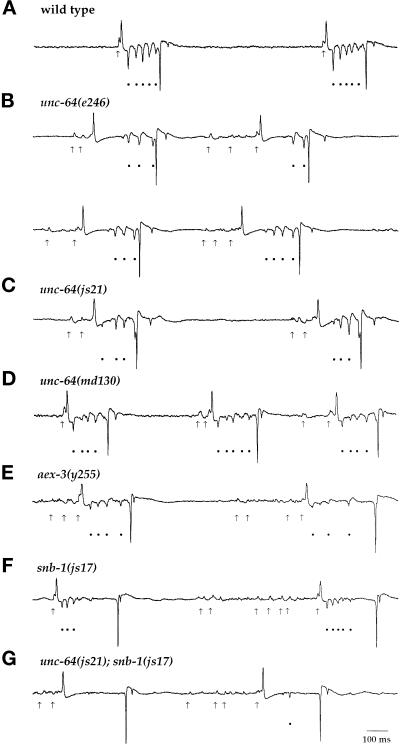
To provide direct evidence of synaptic transmission defects in C. elegans syntaxin mutants, we used an extracellular recording technique developed by Raizen and Avery (1994). This method permits the detection of postsynaptic potentials in pharyngeal muscle resulting from the action of both excitatory and inhibitory motor neurons. MC is an excitatory motor neuron thought to stimulate pharyngeal muscle contraction (Raizen et al., 1995). Its activation is visualized as a single depolarizing transient preceding many pumps. In unc-64 syntaxin mutants, MC neuronal function appeared defective. Specifically, we observed a series of multiple excitatory postsynaptic potentials before initiation of pharyngeal pumps (arrows, Figure 6), which may reflect nonsynchronous transmitter release from MC. A similar phenotype has previously been observed in aex-3, rab-3, and snb-1 mutants (Iwasaki et al., 1997; Nonet et al., 1997, 1998).
Figure 6.
Pharyngeal recordings from wild-type and unc-64 mutant animals. Shown are characteristic electrophysiological recordings from the wild-type strain N2 (A), unc-64(e246) (B), unc-64(js21) (C), unc-64(md130) (D), aex-3(y255) (E), snb-1(js17) (F), and unc-64(js21); snb-1(js17) (G). Arrows indicate MC-induced transients, and filled circles indicate M3-induced transients. All traces are millivolts versus time.
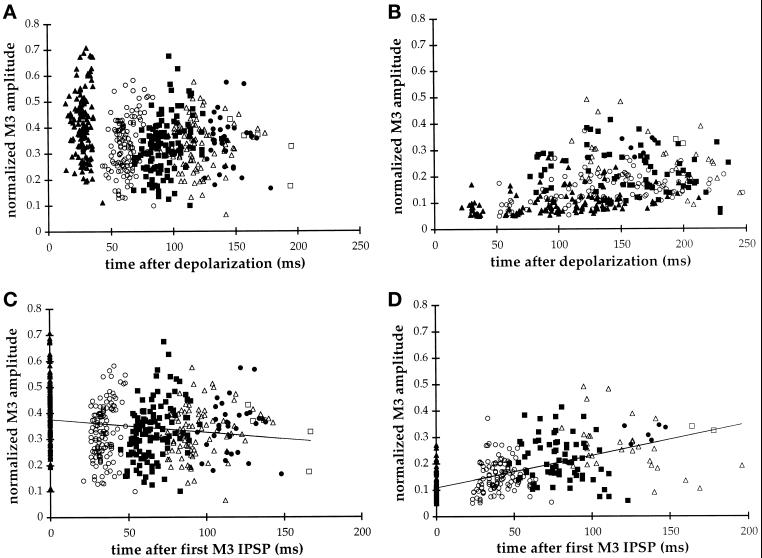
Our analysis concentrated on the examination of inhibitory postsynaptic potentials (IPSPs) produced by the motor neuron M3, a glutaminergic neuron that regulates the duration of pharyngeal muscle contractions (Avery, 1993; Dent et al., 1997). In wild-type animals, M3 transients were regularly spaced and initiated 28 ± 5 ms after depolarization of the pharynx. In unc-64 (e246) animals, the first detectable IPSP often was greatly delayed (102 ± 43 ms; Figure 6). This is most easily illustrated by examining the distribution of IPSPs as a function of time after the initiation of the pharyngeal pump (Figure 7, A and B). While IPSPs occurred relatively synchronously in wild-type, they were broadly distributed in e246 mutants. However, normalizing the time of the IPSP relative to the first IPSP within the pump reveals that, although the initial IPSP was variably delayed in the mutant, subsequent IPSPs were temporally synchronized (Figure 7, C and D). A second difference between wild-type and mutant was that the amplitude of M3 IPSPs was relatively constant in wild-type but increased substantially in e246. While in wild-type, the initial IPSP was slightly larger than the final IPSP, in e246 there was a threefold increase in IPSP amplitude by the end of the pump (Figure 7, C and D). js21 exhibited a similar but less severe defect (our unpublished data). These particular defects in M3 transmission are specific to the syntaxin helical mutants. The splicing mutants md130 and md1259 do not exhibit these alterations but instead resemble aex-3(y255), rab-3(y250), and snb-1(js17) in that their IPSPs were consistently decreased in amplitude yet initiated without delay after contraction of the pharynx (Figure 6). In summary, lesions in the H3 domain of syntaxin affect both the kinetics of transmitter release and the amplitude of postsynaptic potentials.
Figure 7.
Analysis of M3 transmission in wild-type and unc-64(e246) animals. The amplitude of the first ([triof]), second (○), third (▪), fourth (Δ), fifth (•), and six (□) M3-induced IPSPs within each pump (see Figure 6) is plotted as a function of time of occurrence. M3 amplitudes are normalized relative to the mean R-phase amplitude for each record. (A and B) Time of occurrence is plotted relative to initiation of pharyngeal muscle excitation. (C and D) Time of occurrence is normalized to the first M3 IPSP of the same pump. (A) Each group of M3 IPSPs cluster showing the high temporal synchrony of M3 transmitter release in wild-type (pump duration 148 ± 23 ms [mean ± S.D.], N = 130 pumps). (B) In unc-64(e246) temporal synchrony is lost as the timings of each group of M3 IPSPs are distributed over a larger time span. The first M3 IPSP occurs from 22 to 212 ms after muscle excitation (pump duration 209 ± 44 ms [mean ± S.D.], N = 130 pumps). (C) Aligning the IPSPs by timing of the first M3 IPSP again shows temporal clustering in wild-type. Mean IPSP spacings are [1st to 2nd] 34 ms, [2nd to 3rd] 31 ms, [3rd to 4th] 32 ms, [4th to 5th] 32 ms, and [5th to 6th] 36 ms. The downward sloping linear regression reveals that subsequent wild-type IPSP amplitudes tend to decrease slightly. (D) Similar alignment to the first M3 IPSP in e246 reveals that subsequent IPSPs are, in fact, regularly spaced at [1st to 2nd] 41 ms, [2nd to 3rd] 42 ms, [3rd to 4th] 48 ms, [4th to 5th] 42 ms, and [5th to 6th] 43-ms mean intervals. The upward sloping regression line demonstrates that mutant IPSP amplitudes increase up to threefold with successive M3 firings.
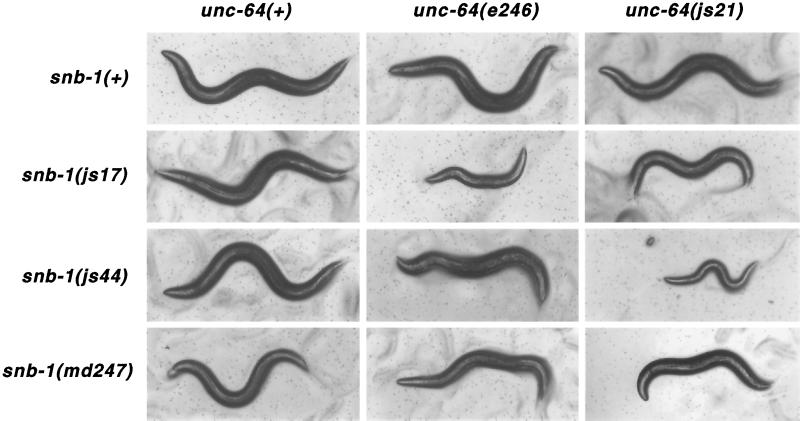
Genetic Interactions between Syntaxin and Synaptobrevin Mutants
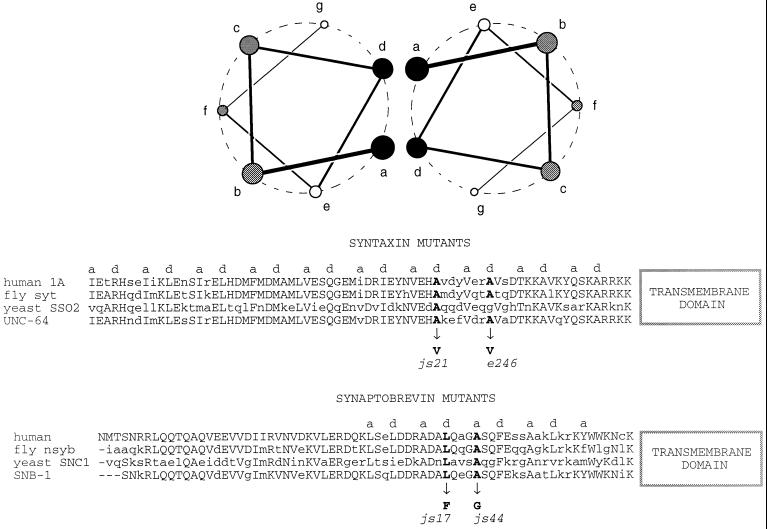
Syntaxin interacts tightly with synaptobrevin and SNAP-25 to form a stable ternary complex. This complex is proposed to represent an intermediate in the synaptic vesicle fusion cycle (Sollner et al., 1993). In each of these proteins, the domains participating in the interaction are predicted to contain α-helices capable of assembling into coiled-coil structures (Hardwick and Pelham, 1992; Fasshauer et al., 1997). Lesions that lie on the hydrophobic face of the presumed coiled helix region exist in both C. elegans syntaxin and synaptobrevin (Nonet et al., 1998). We constructed double mutants between the synaptobrevin mutants snb-1(js17), snb-1(js44), and snb-1(md247) and the two missense helical domain syntaxin mutants js21 and e246 (Figure 8). When analyzing the phenotypes of all six double mutants, we observed strong synergistic effects with certain allelic combinations. The size of young adult animals (Figure 9) and the rates of pharyngeal pumping (Table 2) were drastically reduced in certain double-mutant combinations. For example, when the weakest synaptobrevin mutant, js44, was combined with the weakest syntaxin mutant, js21, the result was a more severe phenotype than when js44 was paired with the stronger e246 syntaxin lesion. Conversely, snb-1(js17) interacted strongly with e246, but not with js21. Neither syntaxin allele showed interactions with snb-1(md247), whose lesion is outside the helical domain and alters the transmembrane domain of synaptobrevin (Nonet et al., 1998). EPG analysis provided no additional insight, as even the healthier double mutants displayed more severe EPG phenotypes than the singles and failed to exhibit any significant M3 activity (Figure 6). This was not unexpected, as this assay examining M3 activity is very sensitive and is able to detect defects in mutants that exhibit only very mild behavioral changes (Nonet et al., 1997).
Figure 8.
Mutations in snb-1 and unc-64 on the hydrophobic faces of α-helices proposed to mediate interactions between synaptobrevin and syntaxin. Two model amphipathic α-helices composed of a repeating seven-amino acid pattern. The residues are labeled a through g. Residues in a and d positions are usually hydrophobic and are proposed to mediate interactions between binding partners. Below, a portion of the sequences of human, fly, yeast, and worm syntaxin and synaptobrevin (Archer et al., 1990; DiAntonio et al., 1993; Gerst et al., 1992) are aligned. Hydrophobic residues in the a and d positions of the predicted α-helices are labeled. The site of lesions in C. elegans syntaxin and synaptobrevin (Nonet et al., 1998) are also labeled. The two sequences are oriented assuming they will interact in a parallel manner (Hanson et al., 1997; Lin and Scheller, 1997).
Figure 9.
Phenotypes of unc-64; snb-1 double- mutant animals. Bright field photographs of young adult animals singly and double mutant for unc-64 syntaxin and snb-1 synaptobrevin mutations. Animals were isolated as L4 larvae and then incubated 1 d before examination. Specific combinations of alleles cause severe behavioral, growth, and pharyngeal pumping defects.
Table 2.
Pharyngeal pumping in unc-64 snb-1 double mutants
| unc-64(+) | e246 | js21 | |
|---|---|---|---|
| snb-1(+) | 250 ± 30 | 209 ± 20 | 234 ± 10 |
| js17 | 206 ± 24 | 65 ± 24 | 127 ± 25 |
| js44 | 215 ± 23 | 102 ± 21 | 27 ± 10 |
| md247 | 64 ± 20 | 49 ± 10 | 73 ± 19 |
DISCUSSION
We have cloned and characterized the unc-64 locus of C. elegans and demonstrated that it encodes a gene with high similarity to the vertebrate neuronal synaptic protein syntaxin 1. The C. elegans syntaxin null mutant is paralyzed and arrests development after completing embryogenesis. Although this phenotype is consistent with a complete block in neurotransmitter release, physiological techniques to document this directly are still lacking in C. elegans. The unc-64 mutant phenotype is the most severe of all known synaptic mutants in C. elegans. Mutants in three genes encoding syntaxin-interacting proteins, namely unc-13, unc-18, and snt-1 (synaptotagmin), all are viable although they exhibit severe behavioral defects (Maruyama and Brenner, 1991; Gengyo-Ando et al., 1993; Nonet et al., 1993). Although nematode synaptobrevin mutants also arrest developmentally as first larval stage animals, they remain capable of some movement (Nonet et al., 1998). This is consistent with the phenotypes of syntaxin mutants in Drosophila, which also show the most severe synaptic transmission defect; both evoked and spontaneous transmitter release are blocked (Broadie et al., 1995). Drosophila synaptotagmin and synaptobrevin mutants also show reduced evoked transmitter release, but do not eliminate spontaneous release (DiAntonio and Schwarz, 1994; Littleton et al., 1994; Broadie et al., 1995). Thus, among the molecular components of the release apparatus, absence of syntaxin is most disruptive to secretion.
unc-64 syntaxin expression is restricted to neurons and to several other tissues with secretory functions. Our expression data indicate that syntaxin is widely expressed in the nervous system, probably in all neurons, and is present in both axons as well as dendrites. In vertebrates, syntaxin is found predominantly on the plasma membrane, although its presence on synaptic vesicles has been reported recently (Bennett et al., 1992; Galli et al., 1995; Walch-Solimena et al., 1995). C. elegans syntaxin does not appear to be concentrated at release sites, as detected by either antibodies or a GFP tag. This ubiquitous distribution of syntaxin on axons is inconsistent with the presence of large steady-state concentrations in C. elegans synaptic vesicles. Expression of syntaxin is also observed in several other tissues that have either a secretory or neuronal character, including the intestine, spermatheca, excretory gland cells, distal tip cells, and uv1 cells. The excretory gland cells are known to contain extensive populations of dense core vesicles and have been proposed to perform several functions, including osmoregulation and hormone secretion, in C. elegans (Nelson et al., 1983; Nelson and Riddle, 1984). The uv1 cells exhibit characteristics of neurosecretory cells, as they contain neuropeptides of the FMRF-amide family (Schinkman and Li, 1992) and express other synaptic protein markers (Nonet et al., 1993). The spermatheca is thought to be involved in secretion of the egg shell and expresses at least one other synaptic vesicle marker (Nonet et al., 1993). Evidence also supports a secretory role for the distal tip cells. These cells extend processes around the end of the germline (Fitzgerald and Greenwald, 1995), express multiple other neuronal markers including synaptobrevin (Gao, Crittenden, and Kimble, personal communication), and are the source of signals that regulate germline function (reviewed by Schedl, 1997). Finally, yolk in C. elegans is synthesized in the intestine and secreted from the basolateral surface into the pseudocoelom (Kimble and Sharrock, 1983). Thus, with the lone exception of the intestinal muscle, a secretory role can be ascribed to all cells expressing syntaxin.
Analysis of the defects in syntaxin mutants also suggests that unc-64 syntaxin function is required only in neurons. For example, some cells have extensive secretory functions, yet do not require syntaxin. Most notably, secretion of first larval cuticle from the hypodermis is normal in the syntaxin null mutant. By contrast, Drosphila syntaxin mutants have severe cuticle defects (Schulze et al., 1995). Furthermore, in Drosophila, syntaxin is also required during cellularization of the embryo (Burgess et al., 1997). Indeed, Drosophila mosaic patches that lack syntaxin are not observed (Schulze and Bellen, 1996; Burgess et al., 1997), suggesting that the protein is required for cell viability. In contrast, C. elegans animals lacking both maternal and zygotic contributions of syntaxin complete embryogenesis normally and arrest only when neuronal function is required for locomotion and feeding. Furthermore, mosaic animals lacking syntaxin in large patches of muscle show entirely normal locomotion. All of our results indicate that C. elegans syntaxin is required only in neuronal tissues. Why is C. elegans syntaxin not required in other tissues? In addition to the unc-64 gene, at least three other divergent syntaxin homologs are found in C. elegans. Therefore, we propose that secretory function in different cell types has been segregated to different syntaxins. This is analogous to the situation in vertebrates in which at least 16 different syntaxins have been described with expression patterns often limited to selected tissues (Bennett et al., 1993; Bock and Scheller, 1997). By contrast, Drosophila appears to utilize one syntaxin gene to regulate secretion in more varied cell types.
The C. elegans syntaxin gene expresses three distinct isoforms that differ only in sequences comprising the protein’s transmembrane domain. Similar alternative isoforms of the vertebrate syntaxin 2 and syntaxin 3 genes have also been described (Bennett et al., 1993; Ibaraki et al., 1995). In fact, the position of four of eight splicing sites in the unc-64 gene are precisely conserved within both the vertebrate syntaxin 1 and 3 genes (Ibaraki et al., 1995; Osborne et al., 1997). Paradoxically, the coding region of the more widely expressed Drosophila gene appears to be encoded within a single exon (although 6 different messages are transcribed from the locus [Schulze and Bellen, 1996]). Due to its assumed sole function as membrane anchor, more exotic roles of the transmembrane domain have been largely neglected. Furthermore, the vast majority of syntaxin biochemistry has been done in the absence of any transmembrane domain, because of the difficulty of working in vitro with protein containing a hydrophobic domain. However, evolutionary conservation of this diversity of membrane anchors from worm to man suggests that these different anchors are important for syntaxin function. Our data suggest that the transmembrane domains play distinct roles in the organism. First, the js116 lesion in transmembrane domain A results in lethality despite the fact that the UNC-64C is expressed in the same cell types. Although we have not demonstrated that syntaxin A and C are coexpressed in all neurons, our data suggest that the functions of these proteins are not identical. Second, the js116 lesion leaves 16 hydrophobic amino acids to act as a hydrophobic domain. This size domain should be sufficient to anchor a protein in the membrane (Whitley et al., 1996), suggesting that syntaxin’s hydrophobic domain may need to span both lipid layers of the membrane to function properly. Third, expression of syntaxin B (normally not expressed in neurons) under the control of the syntaxin promoter using a minigene construct is capable of rescuing the lethality of both js115 (early stop codon) and js116 mutants (our unpublished data). However, these rescued animals are not phenotypically wild-type, in contrast to animals rescued using a genomic construct (our unpublished data). This suggests that the functions of syntaxin A and B overlap significantly, but not completely. Our data indicate that the alternative forms of syntaxin do not play equivalent roles in the secretion process. Furthermore, our work suggests that the transmembrane domain is more than simply a membrane anchor for syntaxin. Isolation of lesions in the other transmembrane domains will probably be required to further dissect the role of the individual forms of syntaxin.
Examination of the hypomorphic lesions in C. elegans syntaxin has provided the most direct evidence that syntaxin acts to regulate secretion of neurotransmitter in this organism. Our simple extracellular physiological assay demonstrates that M3 synaptic transmission to pharyngeal muscle is defective in syntaxin mutants. Specifically, the syntaxin mutants seem to alter the kinetics of release rather than the ability to release neurotransmitter per se. Although M3-induced transients in wild-type appear within 25–30 ms after depolarization of the pharyngeal muscle, the first transient in e246 takes four times as long to appear. This defect cannot simply represent a slowing of the fusion process, because subsequent transients appear with a much more regular, although slightly increased, interspike interval. In wild-type animals, M3 transients are detected at a remarkably regular 30-ms cycle, whereas in e246 this inter-M3 interval remains regular, but expands to 40 ms. The amplitudes of the M3 transients increase within a given pump. Since increasing amplitude on an EPG represents an increase in the rate of change of pharyngeal muscle membrane potential (Raizen and Avery, 1994; Avery et al., 1995), this can be regarded as more efficacious signaling between M3 and the muscle. Therefore, after overcoming the initial delay, efficacy of M3 transmission improves with each firing, but with a longer latency. This physiological defect is observed only in syntaxin mutants with lesions in the H3 domain, not in the md130 or md1259 splicing mutants (Figure 6 and our unpublished data). Furthermore, this defect is distinct from defects observed in rab-3 and aex-3 (RAB-3 nucleotide exchange protein) and certain synaptobrevin mutants. In these mutants the M3 transients appear uniformly smaller and less synchronous.
One model to explain the unc-64(e246) EPG defect would be that interactions between syntaxin (or more likely a complex containing syntaxin) and calcium channels is altered in the mutant. Syntaxin and N-type calcium channels are known to interact via the syntaxin H3 domain; moreover, this interaction is calcium dependent (Sheng et al., 1994, 1996). Disruption of the syntaxin/calcium channel interaction alters the calcium dependence of transmitter release (Rettig et al., 1997). Phosphorylation of the syntaxin- binding domain on calcium channels also inhibits the interaction with syntaxin, which could permit quick temporal regulation of release (Yokoyama et al., 1997). In our mutants, transmitter release from M3 may be minimal during the first several attempted firings due to alterations in calcium sensitivity of the release machinery. Short-term modulation of the release apparatus may then account for the subsequent increase in release. While this highly speculative model is only one of numerous explanations for our recordings, it is consistent with our behavioral observations. Specifically, unc-64(e246) mutants are unusual in that they are differentially defective in distinct behaviors. Differences in the typical firing frequency of different synapses could account for this varied behavioral severity. Rapid firings, such as those required for normal pumping, would be less affected, while lower frequency excitations governing locomotion may be more severely impaired.
The most interesting lesions in unc-64 are the point mutations that alter the hydrophobic face of the H3 coiled-coil domain of syntaxin. This domain is required in vitro for assembly of a ternary complex (Hayashi et al., 1994; Kee et al., 1995). Kee et al. (1995) also demonstrated that double-point mutations in hydrophobic residues (a and d; see Figure 8) of the coiled-coil domain of syntaxin led to reduced binding to synaptobrevin. One double mutant analyzed (A240V;V244A) includes the identical lesion found in the js21 mutant. This double mutant retained the ability to bind to SNAP-25, α-SNAP, and n-sec1 (unc-18 equivalent). However, all the point mutations in residues on the hydrophobic face that Kee et al. (1995) analyzed failed to block ternary complex formation. These observations are consistent with the fact that the js21 mutant retains significant fusion activity. Furthermore, they suggest that complex formation may not serve as a sensitive assay for assessing functionality of syntaxin. In vivo, we have examined the interactions of the js21 lesion with lesions in the proposed coiled-coil domain of synaptobrevin. The fact that combining js21 with the weak snb-1(js44) mutant yields a much more severe behavioral phenotype than combining the mutant with the stronger snb-1(js17) mutant provides a genetic argument for functional interaction of these proteins. unc-64(e246) exhibits a reciprocal set of interactions with snb-1 alleles and neither unc-64 lesion significantly interacts with the snb-1(md247) lesion that maps outside the H3 domain. In a simplified coiled-coil model placing the two proteins in a parallel orientation (Hanson et al., 1997; Lin and Scheller, 1997), the allelic combinations resulting in the most severe phenotypes are those in which lesions in each of the molecules are at positions in the coiled coil more distant from one another (Figure 8). These combinations of lesions may disrupt formation of interactions in a more extended region of the coiled-coil leading to synergy. It is hoped that an atomic level structure of the ternary complex will provide a more definitive molecular hypothesis regarding the perturbation of synaptic transmission in the coiled-coil lesions in both snb-1 and unc-64. Regardless of the molecular defects, our mutants in both synaptobrevin and syntaxin provide strong in vivo evidence for the importance of the helical domains in synaptic transmission. Furthermore, the isolation of extragenic suppressors of these mutants may identify proteins that normally inhibit syntaxin function. Greater than 10 suppressors of unc-64 lesions defining at least 4 different complementation groups have already been identified (Saifee and Nonet, unpublished results). These genetic approaches provide an alternative means of isolating novel molecules that regulate syntaxin function at the synapse.
ACKNOWLEDGMENTS
We thank Barbara J. Meyer in whose laboratory this work was initiated, Larry Salkoff and Arthur Loewy for recording equipment, Stephanie Chisoe for isolating fosmid clones, Mariam Eshani for constructing pTX20, Anneliese Schaefer and Phyllis Hanson for critical comments regarding the manuscript, Anna Borodovski for mapping js21, Jim Rand for providing mutants, and Felisha Starkey for technical assistance. Several C. elegans strains were obtained from the Caenorhabditis Genetics Center (St. Paul, MN). This work was supported by grants to M.L.N. (NS33535) and Barbara J. Meyer from the Muscular Dystrophy Association. M.L.N. was supported during initial phases of this work by a public service award from the United States Public Health Service.
REFERENCES
- Alfonso A, Grundahl K, Duerr JS, Han HP, Rand JB. The Caenorhabditis elegans unc-17 gene: a putative vesicular acetylcholine transporter. Science. 1993;261:617–619. doi: 10.1126/science.8342028. [DOI] [PubMed] [Google Scholar]
- Alfonso A, Grundahl K, McManus JR, Rand JB. Cloning and characterization of the choline acetyltransferase structural gene (cha-1) from C. elegans. J Neurosci. 1994;14:2290–2300. doi: 10.1523/JNEUROSCI.14-04-02290.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer BI, Ozcelik T, Jahn R, Francke U, Sudhof TC. Structures and chromosomal localizations of two human genes encoding synaptobrevins 1 and 2. J Biol Chem. 1990;265:17267–17273. [PubMed] [Google Scholar]
- Avery L. Motor neuron M3 controls pharyngeal muscle relaxation timing in Caenorhabditis elegans. J Exp Biol. 1993;175:283–297. doi: 10.1242/jeb.175.1.283. [DOI] [PubMed] [Google Scholar]
- Avery L, Raizen D, Lockery S. Electrophysiological Methods. In: Epstein HF, Shakes DC, editors. Caenorhabditis elegans: Modern Biological Analysis of an Organism. San Diego, CA: Academic Press; 1995. pp. 251–269. [Google Scholar]
- Bargmann CI, Horvitz HR. Control of larval development by chemosensory neurons in Caenorhabditis elegans. Science. 1991;251:1243–1236. doi: 10.1126/science.2006412. [DOI] [PubMed] [Google Scholar]
- Barstead RJ, Waterston RH. The basal component of the nematode dense-body is vinculin. J Biol Chem. 1989;264:10177–10185. [PubMed] [Google Scholar]
- Bennett MK, Calakos N, Scheller RH. Syntaxin: a synaptic protein implicated in docking of synaptic vesicles at presynaptic active zones. Science. 1992;257:255–259. doi: 10.1126/science.1321498. [DOI] [PubMed] [Google Scholar]
- Bennett MK, Garcia-Arraras JE, Elferink LA, Peterson K, Fleming AM, Hazuka CD, Scheller RH. The syntaxin family of vesicular transport receptors. Cell. 1993;74:863–873. doi: 10.1016/0092-8674(93)90466-4. [DOI] [PubMed] [Google Scholar]
- Bennett MK, Scheller RH. A molecular description of synaptic vesicle membrane trafficking. Annu Rev Biochem. 1994;63:63–100. doi: 10.1146/annurev.bi.63.070194.000431. [DOI] [PubMed] [Google Scholar]
- Betz A, Okamoto M, Benseler F, Brose N. Direct interaction of the rat unc-13 homologue Munc-13–1 with the N-terminus of syntaxin. J Biol Chem. 1997;272:2520–2526. doi: 10.1074/jbc.272.4.2520. [DOI] [PubMed] [Google Scholar]
- Blasi J, Chapman ER, Yamasaki S, Binz T, Niemann H, Jahn R. Botulinum neurotoxin C1 blocks neurotransmitter release by means of cleaving HPC-1/syntaxin. EMBO J. 1993;12:4821–4828. doi: 10.1002/j.1460-2075.1993.tb06171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock JB, Scheller RH. Protein transport. A fusion of new ideas. Nature. 1997;387:133–135. doi: 10.1038/387133a0. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadie K, Prokop A, Bellen HJ, O’Kane CJ, Schulze KL, Sweeney ST. Syntaxin and synaptobrevin function downstream of vesicle docking in Drosophila. Neuron. 1995;15:663–673. doi: 10.1016/0896-6273(95)90154-x. [DOI] [PubMed] [Google Scholar]
- Burgess RW, Deitcher DL, Schwarz TL. The synaptic protein syntaxin1 is required for cellularization of Drosophila embryos. J Cell Biol. 1997;138:861–875. doi: 10.1083/jcb.138.4.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calakos N, Bennett MK, Peterson KE, Scheller RH. Protein-protein interactions contributing to the specificity of intracellular vesicular trafficking. Science. 1994;263:1146–1149. doi: 10.1126/science.8108733. [DOI] [PubMed] [Google Scholar]
- Coulson A, Waterston R, Kiff J, Sulston J, Kohara Y. Genome linking with yeast artificial chromosomes. Nature. 1988;335:184–186. doi: 10.1038/335184a0. [DOI] [PubMed] [Google Scholar]
- Dent JA, Davis MW, Avery L. avr-15 encodes a chloride channel subunit that mediates inhibitory glutamatergic neurotransmission and ivermectin sensitivity in Caenorhabditis elegans. EMBO J. 1997;16:5867–5879. doi: 10.1093/emboj/16.19.5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiAntonio A, Burgess RW, Chin AC, Deitcher DL, Scheller RH, Schwarz TL. Identification and characterization of Drosophila genes for synaptic vesicle proteins. J Neurosci. 1993;13:4924–4935. doi: 10.1523/JNEUROSCI.13-11-04924.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiAntonio A, Schwarz TL. The effect on synaptic physiology of synaptotagmin mutations in Drosophila. Neuron. 1994;12:909–920. doi: 10.1016/0896-6273(94)90342-5. [DOI] [PubMed] [Google Scholar]
- Edwardson JM, An S, Jahn R. The secretory granule protein syncollin binds to syntaxin in a Ca2+-sensitive manner. Cell. 1997;90:325–333. doi: 10.1016/s0092-8674(00)80340-2. [DOI] [PubMed] [Google Scholar]
- Fasshauer D, Otto H, Eliason WK, Jahn R, Brunger A. Structural changes are associated with soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor complex formation. J Biol Chem. 1997;272:28036–28041. doi: 10.1074/jbc.272.44.28036. [DOI] [PubMed] [Google Scholar]
- Fire A, Harrison SW, Dixon D. A modular set of lacZ fusion vectors for studying gene expression in Caenorhabditis elegans. Gene. 1990;93:189–198. doi: 10.1016/0378-1119(90)90224-f. [DOI] [PubMed] [Google Scholar]
- Fitzgerald K, Greenwald I. Interchangeability of Caenorhabditis elegans DSL proteins and intrinsic signalling activity of their extracellular domains in vivo. Development. 1995;119:4275–4282. doi: 10.1242/dev.121.12.4275. [DOI] [PubMed] [Google Scholar]
- Fleming JT, et al. Caenorhabditis elegans levamisole resistance genes lev-1, unc-29, and unc-38 encode functional nicotinic acetylcholine receptor subunits. J Neurosci. 1997;17:5843–5857. doi: 10.1523/JNEUROSCI.17-15-05843.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli T, Garcia EP, Mundigl O, Chilcote TJ, De Camilli P. v- and t-SNAREs in neuronal exocytosis: a need for additional components to define sites of release. Neuropharmacology. 1995;34:1351–1360. doi: 10.1016/0028-3908(95)00113-k. [DOI] [PubMed] [Google Scholar]
- Garcia EP, Gatti E, Butler M, Burton J, De Camilli P. A rat brain Sec1 homologue related to Rop and UNC18 interacts with syntaxin. Proc Natl Acad Sci USA. 1994;91:2003–2007. doi: 10.1073/pnas.91.6.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gengyo-Ando K, Kamiya Y, Yamakawa A, Kodaira K, Nishiwaki K, Miwa J, Hori I, Hosono R. The C. elegans unc-18 gene encodes a protein expressed in motor neurons. Neuron. 1993;11:703–711. doi: 10.1016/0896-6273(93)90080-b. [DOI] [PubMed] [Google Scholar]
- Gerst JE, Rodgers L, Riggs M, Wigler M. SNC1, a yeast homolog of the synaptic vesicle-associated membrane protein/synaptobrevin gene family: genetic interactions with the RAS and CAP genes. Proc Natl Acad Sci USA. 1992;89:4338–4342. doi: 10.1073/pnas.89.10.4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Kemphues KJ. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Morisaki H, Jahn R, Heuser JE. Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell. 1997;90:523–535. doi: 10.1016/s0092-8674(00)80512-7. [DOI] [PubMed] [Google Scholar]
- Hardwick KG, Pelham HR B. SED5 encodes a 39-KD integral membrane protein required for vesicular transport between the ER and Golgi complex. J Cell Biol. 1992;119:513–521. doi: 10.1083/jcb.119.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata Y, Slaughter CA, Sudhof TC. Synaptic vesicle fusion complex contains unc-18 homologue bound to syntaxin. Nature. 1993;366:347–351. doi: 10.1038/366347a0. [DOI] [PubMed] [Google Scholar]
- Hayashi T, McMahon H, Yamasaki S, Binz T, Hata Y, Sudhof TC, Niemann H. Synaptic vesicle membrane fusion complex: action of clostridial neurotoxins on assembly. EMBO J. 1994;13:5051–5061. doi: 10.1002/j.1460-2075.1994.tb06834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman RK, Horvitz HR. Nematodes as Biological Models, vol. 1: Behavioral and Developmental Models. B.M. Zuckerman, New York: Academic Press; 1980. Genetic analysis of Caenorhabditis elegans; pp. 227–262. [Google Scholar]
- Huang LS, Tzou P, Sternberg PW. The lin-15 locus encodes two negative regulators of Caenorhabditis elegans vulval development. Mol Biol Cell. 1994;5:395–411. doi: 10.1091/mbc.5.4.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibaraki K, Horikawa HP, Morita T, Mori H, Sakimura K, Mishina M, Saisu H, Abe T. Identification of four different forms of syntaxin 3. Biochem Biophys Res Commun. 1995;211:997–1005. doi: 10.1006/bbrc.1995.1910. [DOI] [PubMed] [Google Scholar]
- Iwasaki K, Staunton J, Saifee O, Nonet ML, Thomas J. aex-3 encodes a novel regulator of presynaptic activity in C. elegans. Neuron. 1997;18:613–622. doi: 10.1016/s0896-6273(00)80302-5. [DOI] [PubMed] [Google Scholar]
- Kee Y, Lin RC, Hsu SC, Scheller RH. Distinct domains of syntaxin are required for synaptic vesicle fusion complex formation and dissociation. Neuron. 1995;14:991–998. doi: 10.1016/0896-6273(95)90337-2. [DOI] [PubMed] [Google Scholar]
- Kimble J, Sharrock WJ. Tissue-specific synthesis of yolk proteins in Caenorhabditis elegans. Dev Biol. 1983;96:189–196. doi: 10.1016/0012-1606(83)90322-6. [DOI] [PubMed] [Google Scholar]
- Lewis JA, Wu CH, Berg H, Levine JH. The genetics of levamisole resistance in the nematode Caenorhabditis elegans. Genetics. 1980;95:905–928. doi: 10.1093/genetics/95.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JA, Wu CH, Levine JH, Berg H. Levamisole-resistant mutants of the nematode Caenorhabditis elegans appear to lack pharmacological acetylcholine receptors. Neuroscience. 1980;5:967–989. doi: 10.1016/0306-4522(80)90180-3. [DOI] [PubMed] [Google Scholar]
- Lichtsteiner S, Tjian R. Cloning and properties of the Caenorhabditis elegans TATA-box-binding protein. Proc Natl Acad Sci USA. 1993;90:9673–9677. doi: 10.1073/pnas.90.20.9673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin RC, Scheller RH. Structural organization of the synaptic exocytosis core complex. Neuron. 1997;19:1087–1094. doi: 10.1016/s0896-6273(00)80399-2. [DOI] [PubMed] [Google Scholar]
- Littleton JT, Stern M, Perin M, Bellen HJ. Calcium dependence of neurotransmitter release and rate of spontaneous vesicle fusions are altered in Drosophila synaptotagmin mutants. Proc Natl Acad Sci USA. 1994;91:10888–10892. doi: 10.1073/pnas.91.23.10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DW, Thomas JH. Regulation of a periodic motor program in C. elegans. J Neurosci. 1994;14:1953–1962. doi: 10.1523/JNEUROSCI.14-04-01953.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama IN, Brenner S. A phorbol ester/diacylglycerol-binding protein encoded by the unc-13 gene of Caenorhabditis elegans. Proc Natl Acad Sci USA. 1991;88:5729–5733. doi: 10.1073/pnas.88.13.5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KG, Alfonso A, Nguyen M, Crowell JA, Johnson CD, Rand JB. A genetic selection for Caenorhabditis elegans synaptic transmission mutants. Proc Natl Acad Sci USA. 1996;93:12593–12598. doi: 10.1073/pnas.93.22.12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson FK, Albert PS, Riddle DL. Fine structure of the Caenorhabditis elegans secretory-excretory system. J Ultrastruct Res. 1983;82:156–171. doi: 10.1016/s0022-5320(83)90050-3. [DOI] [PubMed] [Google Scholar]
- Nelson FK, Riddle DL. Functional study of the Caenorhabditis elegans secretory-excretory system using laser microsurgery. J Exp Zool. 1984;231:45–56. doi: 10.1002/jez.1402310107. [DOI] [PubMed] [Google Scholar]
- Nguyen M, Alfonso A, Johnson CD, Rand JB. Caenorhabditis elegans mutants resistant to inhibitors of acetylcholinesterase. Genetics. 1995;140:527–535. doi: 10.1093/genetics/140.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet ML, Grundahl K, Meyer BJ, Rand JB. Synaptic function is impaired but not eliminated in C. elegans mutants lacking synaptotagmin. Cell. 1993;73:1291–1305. doi: 10.1016/0092-8674(93)90357-v. [DOI] [PubMed] [Google Scholar]
- Nonet ML, Meyer BJ. Early aspects of Caenorhabditis elegans sex determination and dosage compensation are regulated by a zinc-finger protein. Nature. 1991;351:65–68. doi: 10.1038/351065a0. [DOI] [PubMed] [Google Scholar]
- Nonet ML, Saifee O, Zhao H, Rand JB, Wei L. Synaptic transmission deficits in C. elegans synaptobrevin mutants. J Neurosci. 1998;18:70–80. doi: 10.1523/JNEUROSCI.18-01-00070.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet ML, Staunton J, Kilgard MP, Fergestad T, Hartweig E, Horvitz HR, Jorgensen E, Meyer BJ. C. elegans rab-3 mutant synapses exhibit impaired function and are partially depleted of vesicles. J Neurosci. 1997;17:8021–8073. doi: 10.1523/JNEUROSCI.17-21-08061.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne LR, Soder S, Shi XM, Pober B, Costa T, Scherer SW, Tsui LC. Hemizygous deletion of the syntaxin 1A gene in individuals with Williams syndrome. Am J Hum Genet. 1997;61:449–452. doi: 10.1086/514850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevsner J, Hsu SC, Scheller RH. n-Sec1: a neural-specific syntaxin-binding protein. Proc Natl Acad Sci USA. 1994;91:1445–1449. doi: 10.1073/pnas.91.4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizen DM, Avery L. Electrical activity and behavior in the pharynx of Caenorhabditis elegans. Neuron. 1994;12:483–495. doi: 10.1016/0896-6273(94)90207-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizen DM, Lee RYN, Avery L. Interacting genes required for pharyngeal excitation by motor neuron MC in Caenorhabditis elegans. Genetics. 1995;141:1365–1382. doi: 10.1093/genetics/141.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand JB, Nonet ML. Synaptic transmission. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 611–644. [PubMed] [Google Scholar]
- Rand JB, Russell RL. Molecular basis of drug-resistance mutations in the nematode Caenorhabditis elegans. Psychopharm Bull. 1985;21:623–630. [PubMed] [Google Scholar]
- Rettig J, Heinemann C, Ashery U, Sheng ZH, Yokoyama CT, Catterall WA, Neher E. Alteration of Ca2+ dependence of neurotransmitter release by disruption of Ca2+ channel/syntaxin interaction. J Neurosci. 1997;17:6647–6656. doi: 10.1523/JNEUROSCI.17-17-06647.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocheleau CE, Downs WD, Lin RL, Wittmann C, Bei YX, Cha YH, Ali M, Priess JR, Mello CC. Wnt signaling and an APC-related gene specify endoderm in early C. elegans embryos. Cell. 1997;90:707–716. doi: 10.1016/s0092-8674(00)80531-0. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Schafer WR, Kenyon CJ. A calcium-channel homologue required for adaptation to dopamine and serotonin in Caenorhabditis elegans. Nature. 1995;375:73–78. doi: 10.1038/375073a0. [DOI] [PubMed] [Google Scholar]
- Schedl T. Developmental genetics of the germ line. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 611–644. [PubMed] [Google Scholar]
- Schiavo G, Shone CC, Bennett MK, Scheller RH, Montecucco C. Botulinum neurotoxin type C cleaves a single Lys-Ala bond within the carboxyl-terminal region of syntaxins. J Biol Chem. 1995;270:10566–10570. doi: 10.1074/jbc.270.18.10566. [DOI] [PubMed] [Google Scholar]
- Schinkman K, Li C. Localization of FMRFamide-like peptides in Caenorhabditis elegans. J Comp Neurol. 1992;316:251–260. doi: 10.1002/cne.903160209. [DOI] [PubMed] [Google Scholar]
- Schulze KL, Bellen HJ. Drosophila syntaxin is required for cell viability and may function in membrane formation and stabilization. Genetics. 1996;144:1713–1724. doi: 10.1093/genetics/144.4.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze KL, Broadie K, Perin MS, Bellen HJ. Genetic and electrophysioloical studies of Drosophila syntaxin-1A demonstrate its role in nonneuronalsecretion and neurotransmission. Cell. 1995;80:311–320. doi: 10.1016/0092-8674(95)90414-x. [DOI] [PubMed] [Google Scholar]
- Sheng Z-H, Rettig J, Takahashi M, Catterall WA. Identification of a syntaxin-binding site on N-type calcium channels. Neuron. 1994;13:1303–1313. doi: 10.1016/0896-6273(94)90417-0. [DOI] [PubMed] [Google Scholar]
- Sheng ZH, Rettig J, Cook T, Catterall WA. Calcium-dependent interaction of N-type calcium channels with the synaptic core complex. Nature. 1996;379:451–454. doi: 10.1038/379451a0. [DOI] [PubMed] [Google Scholar]
- Smith D, Fisher PA. Identification, developmental regulation, and response to heat shock of two antigenically related forms of a major nuclear envelope protein in Drosophila embryos: applications of an improved method for affinity purification of antibodies using polypeptides immobilized on nitrocellulose blots. J Cell Biol. 1984;99:20–28. doi: 10.1083/jcb.99.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner T, Bennett MK, Whiteheart SW, Scheller RH, Rothman JE. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell. 1993;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- Sollner T, Rothman JE. Neurotransmission: harnessing fusion machinery at the synapse. Trends Neurosci. 1994;17:344–348. doi: 10.1016/0166-2236(94)90178-3. [DOI] [PubMed] [Google Scholar]
- Sudhof T. The synaptic vesicle cycle: a cascade of protein-protein interactions. Nature. 1995;375:645–653. doi: 10.1038/375645a0. [DOI] [PubMed] [Google Scholar]
- Sulston J, Hodgkin J. Methods. In: Wood W B, editor. The Nematode Caenorhabditis elegans. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. pp. 587–606. [Google Scholar]
- Walch-Solimena C, Blasi J, Edelmann L, Chapman ER, von Mollard GF, Jahn R. The t-SNAREs syntaxin 1 and SNAP-25 are present on organelles that participate in synaptic vesicle recycling. J Cell Biol. 1995;128:637–645. doi: 10.1083/jcb.128.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitley P, Grahn E, Kutay U, Rapoport TA, von Heijne G. A 12-residue-long polyleucine tail is sufficient to anchor synaptobrevin to the endoplasmic reticulum membrane. J Biol Chem. 1996;271:7583–7586. doi: 10.1074/jbc.271.13.7583. [DOI] [PubMed] [Google Scholar]
- Yokoyama CT, Sheng ZH, Catterall WA. Phosphorylation of the synaptic protein interaction site on N-type calcium channels inhibits interactions with SNARE proteins. J Neurosci. 1997;17:6929–6938. doi: 10.1523/JNEUROSCI.17-18-06929.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]