UNC-11, a Caenorhabditis elegans AP180 Homologue, Regulates the Size and Protein Composition of Synaptic Vesicles (original) (raw)
Abstract
The unc-11 gene of Caenorhabditis elegans encodes multiple isoforms of a protein homologous to the mammalian brain-specific clathrin-adaptor protein AP180. The UNC-11 protein is expressed at high levels in the nervous system and at lower levels in other tissues. In neurons, UNC-11 is enriched at presynaptic terminals but is also present in cell bodies. unc-11 mutants are defective in two aspects of synaptic vesicle biogenesis. First, the SNARE protein synaptobrevin is mislocalized, no longer being exclusively localized to synaptic vesicles. The reduction of synaptobrevin at synaptic vesicles is the probable cause of the reduced neurotransmitter release observed in these mutants. Second, unc-11 mutants accumulate large vesicles at synapses. We propose that the UNC-11 protein mediates two functions during synaptic vesicle biogenesis: it recruits synaptobrevin to synaptic vesicle membranes and it regulates the size of the budded vesicle during clathrin coat assembly.
INTRODUCTION
Exocytosis and endocytosis are interdependent processes in synaptic transmission. Synaptic vesicles filled with neurotransmitter fuse at the active zone in a calcium-regulated signaling process (for reviews see Matthew et al., 1981; Südhof, 1995). After the fusion event, synaptic vesicle membrane components rapidly and selectively undergo endocytosis. Recycling of synaptic vesicle membranes and proteins occurs at extrasynaptic sites via clathrin-mediated endocytosis (Cremona and De Camilli, 1997). Recycling is the local source of synaptic vesicle components and hence is essential for maintaining the homeostasis of neuronal membranes required for efficient neurotransmitter release. Furthermore, recycling provides an efficient mechanism to remove synaptic vesicle proteins such as neurotransmitter transporters from the plasmalemma, where their function may be deleterious to the cell.
A combination of genetic, cellular, and biochemical studies has provided a general outline of the mechanisms likely to mediate endocytosis of synaptic membranes (reviewed by Cremona and De Camilli, 1997). Endocytosis of membranes through clathrin-coated pits is initiated when clathrin is recruited and organized into lattices by the heterotetrameric adaptor protein AP-2 and the monomeric adaptor protein AP180 (for review see Schmid, 1997). The AP-2 complex associates with synaptic membranes through its interactions with synaptotagmin and phosphoinositides (Beck and Keen, 1991; Zhang et al., 1994; Gaidarov et al., 1996; Rapoport et al., 1997). The analysis of Drosophila α-adaptin mutants implicates AP-2 and clathrin coats in the recycling of synaptic vesicle components (González-Gaitan and Jackle, 1997). Defects in this AP-2 component disrupt clathrin-mediated endocytosis and result in animals with nerve terminals lacking synaptic vesicles.
Monomeric AP180 (a.k.a. NP185, F1–20, and AP-3) shares many biochemical properties with the tetrameric adaptor complexes. Like the tetrameric complexes, AP180 binds clathrin, assembles clathrin lattices (Ahle and Ungewickell, 1986; Ye and Lafer, 1995b), and binds phosphoinositides (Norris et al., 1995; Ye et al., 1995; Hao et al., 1997). AP180 also interacts with AP-2 (Wang et al., 1995), suggesting that AP-2 and AP180 might coordinately regulate clathrin-mediated endocytosis of synaptic vesicle components.
In mammals there are at least two AP180 homologues. The related protein CALM is ubiquitously expressed (Dreyling et al., 1996), whereas AP180 is expressed specifically in neurons and localizes to synapses (Puszkin et al., 1992; Morris et al., 1993; Zhou et al., 1993). In spite of the large number of studies that suggest that AP180-like proteins function in endocytosis, yeast mutants lacking two AP180 homologues exhibit no detectable abnormalities in endocytosis (Wendland and Emr, 1998). Thus, a role for AP180 in the recycling of synaptic vesicles or a role for AP180-like proteins in endocytosis has yet to be identified.
We demonstrate here that the Caenorhabditis elegans gene unc-11 encodes a homologue of AP180. Mutations in unc-11 were first identified by Brenner (1974) based on their uncoordinated locomotory phenotype: the mutant animals coil and jerk, particularly when going backward. We demonstrate that mutants homozygous for null alleles of unc-11 are viable, but are defective for the release of multiple neurotransmitters. Furthermore, the UNC-11 protein is required to localize synaptobrevin, but not other synaptic vesicle proteins, to synaptic sites. Finally, in unc-11 mutants, synaptic membrane still undergoes endocytosis but vesicle diameter is significantly larger. We conclude that AP180 provides dual functions for the biogenesis of synaptic vesicles: to recruit synaptobrevin to synaptic vesicle membranes and to regulate vesicle diameter during clathrin assembly.
MATERIALS AND METHODS
Genetic Manipulations and Nematode Strains
Growth and culture of nematodes were performed using standard techniques (Brenner, 1974; Sulston and Hodgkin, 1988). The Bristol strain N2 was used as the wild-type strain. Several independent genetic screens have led to the isolation of at least 15 unc-11 alleles. The unc-11 alleles e47, e511, and e1054 were isolated in screens for locomotion defects (Brenner, 1974). Seven alleles of unc-11, designated md179, md1009, md1182, md261, md244, md306, and md315, were isolated in screens for aldicarb-resistant mutants (Nguyen et al., 1995; Miller et al., 1996). unc-11(ad571) was isolated in a screen for mutants defective in pharyngeal pumping (Avery, 1993b). unc-11(n2954) was isolated in a screen for jerky forward motion (Jorgensen, unpublished data). q358 and q359 were isolated in a screen for deletions removing the fog-1 gene closely linked to unc-11 (Barton and Kimble, 1990). q359 has a deletion identical to that in e47 and likely represents a reisolate of the mutation. unc-11(ic9) was recovered during outcrossing of the strain CB384 (Nguyen et al., 1995). jsIs1 and jsIs219 were used in all experiments as the source of synaptobrevin tagged-green fluorescent protein (GFP) and synaptogyrin-tagged GFP, respectively (Nonet, 1999). Immunohistochemical and GFP localization experiments were performed on the unc-11 alleles e47, md1182, n2954, and q358. Other strains were provided by the Caenorhabditis Genetics Center.
Germline Transformation
Transgenic nematode strains were created as previously described (Mello et al., 1991) using the plasmid pRF4 (50 μg/ml) containing the rol-6(e1006) dominant mutation as a cotransformation marker (Kramer et al., 1990). Cosmid C32E8 was isolated using standard methods and injected at 20 μg/ml. pFB11 (injected at 50 μg/ml) was obtained by screening a λZAPII library prepared with _Xba_I-digested C32E8 DNA. The 8.8-kilobase (kb) _Xba_I-containing plasmid was recovered by conversion as indicated by the manufacturer (Stratagene, La Jolla, CA). pFM544 (injected at 10 μg/ml) consisting of the fusion of GFP to unc-11 in exon 3 at amino acid 77 was created by inserting the 1.8-kb GFP fragment from pPD95.77 (a gift of Andy Fire) into 5.5-kb _Msc_I-_Apa_I fragment of pFB11.
Molecular Analysis of unc-11
Southern analysis, screening of libraries, and isolation of genomic subclones were performed using standard procedures (Sambrook et al., 1989). Sequencing was performed either using an ABI Cycle Sequencing kit (Perkin Elmer-Cetus, Norwalk, CT) or a Sequenase II kit (United States Biochemical, Cleveland, OH). Genomic DNA and RNA were isolated from nematodes as previously described (Alfonso et al., 1994). Poly-A+–enriched RNA was isolated from mixed or L1-enriched cultures as described previously (Nonet and Meyer, 1991). First-strand cDNA was created from mixed staged poly A+-selected RNA using random hexanucleotide primers as described by Innis et al. (1990).
Insertion and deletion mutations were localized to genomic fragments by Southern analysis. The end points of the deletion and the sites of the insertions were determined from sequencing appropriate PCR products. The Tc1 insertions in md1009 and md1182 are within codon 254 and after codon 232, respectively. e47 deletes 210 base pairs (bp) including portions of exon 1 and 2 as well as the intron between them (GAAAGCGCTGCATCAA Δ CCAACTTGGCAAGA). q358 deletes 247 bp including the end of exon 2 and ∼90% of the intron between exons 2 and 3 (TACCGAAGAAGTTATt Δ aAATTAGGAAATTTTT). Two additional bases, TA, are flanking the deletion**.** n2954 represents an in-frame deletion of 222 bp within exon 4 and retains the potential to encode an UNC-11 protein with 74 amino acids deleted within the conserved N-terminal domain (GTGAAACGC Δ GATATGAACCA).
Isolation of cDNAs
A 2.7-kb _Eco_RI/_Bam_HI DNA fragment derived from cosmid C32E8 was used to screen a mixed-stage λZAP cDNA library (Barstead and Waterston, 1989). Three partial cDNAs corresponding to unc-11 were isolated and sequenced. JC2, JC13, and JC21 cDNAs were truncated but included exon 6. One of them, JC2, had the two additional amino acids at exon 8 present in the “B” isoform (see below). To identify the 5′-end of the unc-11 messages, primers corresponding to the SL1 and SL2 trans-spliced leaders found commonly at the 5′-end of C. elegans messages (Blumenthal, 1995), and coding sequences in exon 5, were used in a PCR to amplify sequences from first-strand cDNA. Products were cloned and sequenced to determine the sites of trans-spliced leader addition. The most abundant product contained the SL1 trans-spliced leader added 33 bp upstream of the ATG codon. A less abundant product contained the SL2 trans-spliced leader added after codon 39. To isolate cDNA clones containing the entire UNC-11-coding region, oligonucleotides corresponding to the region around the initiation codon (5′-ccg gga tcc atg cag act atc gag aaa gcg) and corresponding to a region 3′ of the termination codon (5′-cag ggt acc tta cga gag aga tag aga gaa ata g) were used in a PCR to amplify unc-11 cDNA sequences from first-strand cDNA. The PCR products were cloned into pRSETA (Invitrogen, San Diego, CA). Analysis of 37 clones by restriction digests identified five classes of cDNAs (3 of “A” represented by pAP112, 2 of “B” represented by pAP114; 29 of “C” represented by pAP111, 2 of “D” represented by pAP113, and 1 of “E” represented by pAP110). The “F” class is represented by JC13. Two (if available) clones of each class were sequenced.
Generation of Polyclonal Antibodies
A UNC-11C/glutathione-_S_-transferase (GST) fusion was created by subcloning an _Alw_NI/_Bam_HI fragment from pAP111 into pGEX2T. UNC-11C-GST was purified using glutathione agarose in a batch purification procedure as specified by the manufacturer (Pharmacia, Piscataway, NJ) from BL21 cells expressing the construct and induced with IPTG for 2 h. New Zealand white rabbits were bled to collect a preimmune serum sample and then injected with 600 μg each of the UNC-11C/GST fusion protein in glutathione elution buffer (10 mM glutathione in 50 mM Tris-HCl, pH 8) mixed with Freund’s complete adjuvant. After 4 wk, rabbits were boosted with an additional 500 μg each of the protein (in Freund’s incomplete adjuvant). After 4 additional weeks, serum was collected from each rabbit, affinity purified against the fusion protein, immunodepleted against GST, and tested for immunohistochemistry. Sera from both rabbits show specific staining of the nervous system that is not detectable in the preimmune sera (our unpublished data). Polyclonal antibodies from rabbit 3838 are functional both in Western blots and immunohistochemistry and were used in these studies. Affinity purification of UNC-11 antibodies included an enrichment step for antibodies against the fusion protein and a depletion step for antibodies against GST according to published procedures (Smith and Fisher, 1984; Duerr et al., 1999).
Immunohistochemistry
Immunohistochemistry using α-synaptotagmin, α-synaptobrevin, and α-RAB-3 antisera was performed as previously described (Nonet et al., 1997, 1998). Immunohistochemistry using α-UNC-11 antisera was performed fixing freeze-cracked worms in 100% methanol according to published procedures (Albertson, 1984; Duerr et al., 1999). Affinity-purified UNC-11 antibody was used at a final 1:500 dilution in PBS with 1% BSA, whereas α-synaptotagmin antibody (Nonet et al., 1993) was used at a 1:100 dilution. Incubation with the primary antibodies was overnight at 4°C. Sheep α-rabbit immunoglobulin G (IgG) rhodamine and sheep α-mouse IgG FITC (Boehringer Mannheim, Indianapolis, IN), were used at a 1:100 dilution. Images were captured with a 63× objective in a Sensys charge-coupled device camera or a 40× objective in the Confocal LSM510 (Carl Zeiss, Thornwood, NY).
Western Blots
Mixed-stage nematodes were sonicated in PBS and the sample diluted in an equal volume of 2× Laemmli buffer with protease inhibitors (1× = 0.1 M Tris-HCl, pH 6.8, 2% glycerol, 4% SDS, 1% β-mercaptoethanol, 1 μg/ml pepstatin A, 2 μg/ml aprotinin, 2 μg/ml leupeptin). Samples were then centrifuged, and protein concentration was determined for the supernatant using the BCA Protein Assay kit (Pierce Chemical, Rockford, IL). Total protein (150 μg) was resolved in an 8% SDS-PAGE gel and transferred to Sequi Blot polyvinylidene fluoride membranes (Bio-Rad, Richmond, CA) at 230 mA for 4.5 h according to manufacturer’s instructions. Blocking and antibody incubations were done according to standard methods. Primary α-UNC-11 antibody was used at a 1:750 final dilution. Secondary sheep α-rabbit IgG HRP (Promega) was used at a 1:5000 dilution. Immunoreactivity was detected with an ECL kit (Amersham).
Electron Microscopy
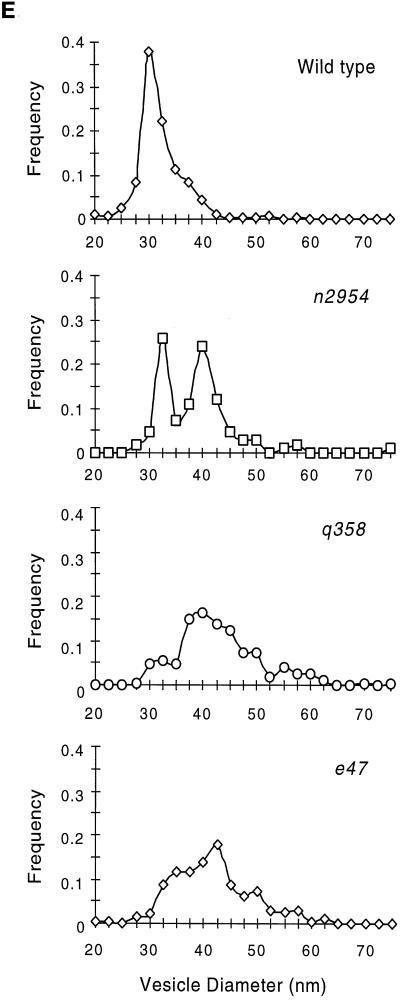
Nematodes were cut in 0.7% glutaraldehyde and 0.7% osmium tetroxide in 0.1 M cacodylate, pH 7.4, on ice. After 2 h on ice, they were moved to 2% osmium in 0.1 M cacodylate, pH 7.4, and left at 4°C overnight. Processing and sectioning were as previously described (McIntire et al., 1992), except that sections were cut ultrathin (30–40 nm). Serial sections were photographed, and vesicle diameters were measured in sections containing active zones. Twenty-eight representative sections from two worms of the wild type, nine representative sections from three worms of q358, seven representative sections from three worms for e47, and four representative sections from two worms for n2954 were analyzed. All vesicles present in each cross-section were measured. The precise number of vesicles measured is listed in Figure 9.
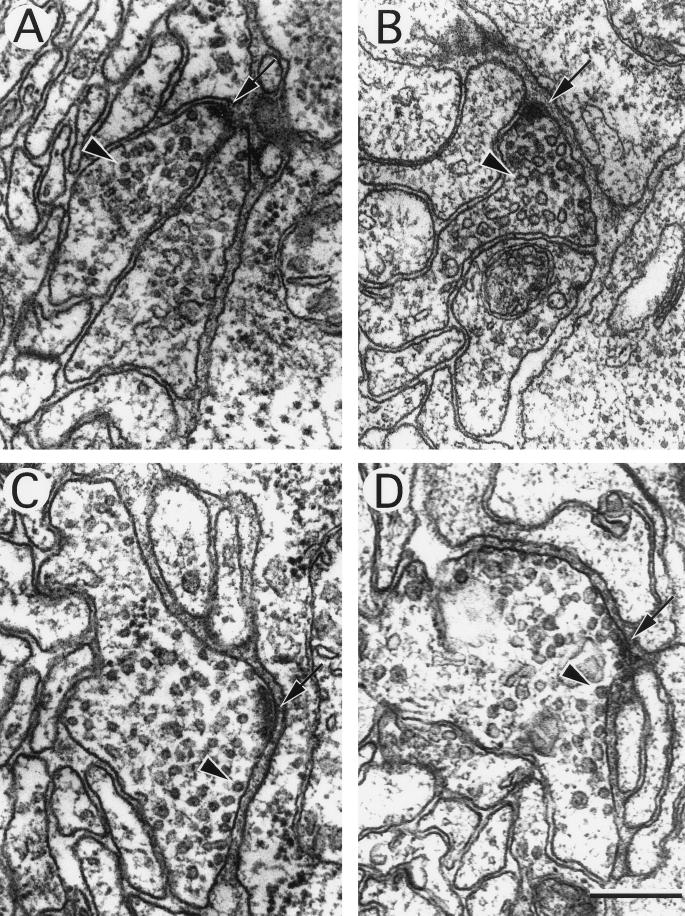
Figure 9.
Ultrastructure of neuromuscular junctions from wild-type and unc-11 mutants. Electron micrographs of the ventral nerve cord of young adult animals. The neuronal cell type was deduced from examination of serial sections. A thick electron-dense presynaptic specialization is visible at the apposition of nerve and muscle in each image (arrow), and a single synaptic vesicle is indicated (arrowhead) in each micrograph. (A) Cholinergic neuromuscular junction in a wild-type animal. (B) Cholinergic neuromuscular junction in an unc-11(q358) mutant. (C) GABAergic neuromuscular junctions in a wild-type animal. (D) GABAergic neuromuscular junction in an unc-11(e47) mutant. Note the increase in vesicles in contact with the membrane adjacent to the postsynaptic muscle. (E) Quantitation of vesicle diameter from wild-type and unc-11 neuromuscular junctions. Vesicle diameters were measured in sections containing active zones and binned into 2.5-nm intervals. The fraction of vesicles in each size class is plotted at the midpoint of each binning interval (wild-type, n = 305; unc-11(e47), n = 214; unc-11(q358), n = 197; unc-11(n2954), n = 109). Bar, 200 nm.
Electrophysiology
Young adult hermaphrodites were washed extensively in M9 (Brenner, 1974) to remove bacteria and incubated in Ascaris saline without serotonin. Recordings were essentially as described by Raizen and Avery (1994). Recording electrodes were pulled in a Brown/Flaming microelectrode puller (Sutter Instruments, San Francisco, CA), and the tip sizes varied according to the size of the worm. Signals were amplified with an Axoclamp 2A (Axon Instruments, Burlingame, CA) in current clamp mode and recorded on both videotape and a Gould (Valley View, OH) strip chart recorder. Eight to ten individuals of each genotype were analyzed on the same day.
RESULTS
unc-11 Encodes a Homologue of Vertebrate AP180
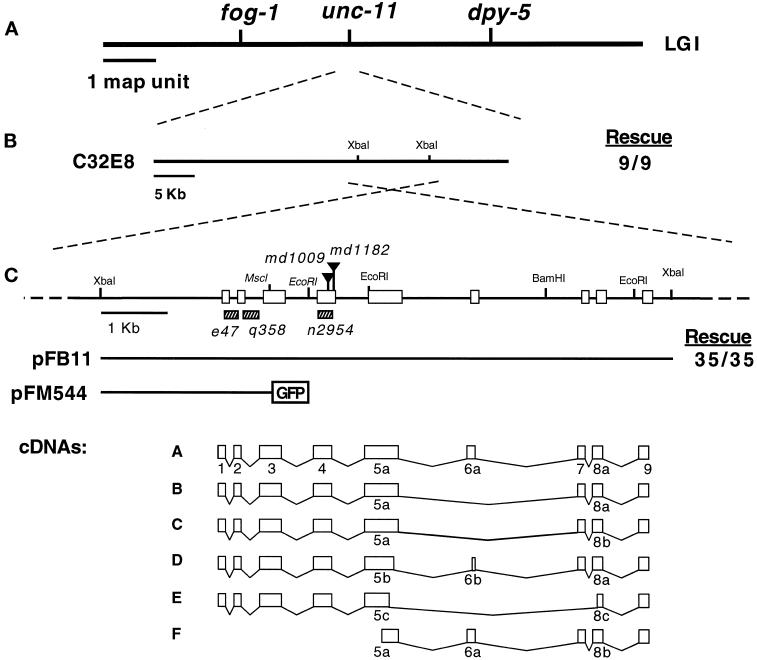
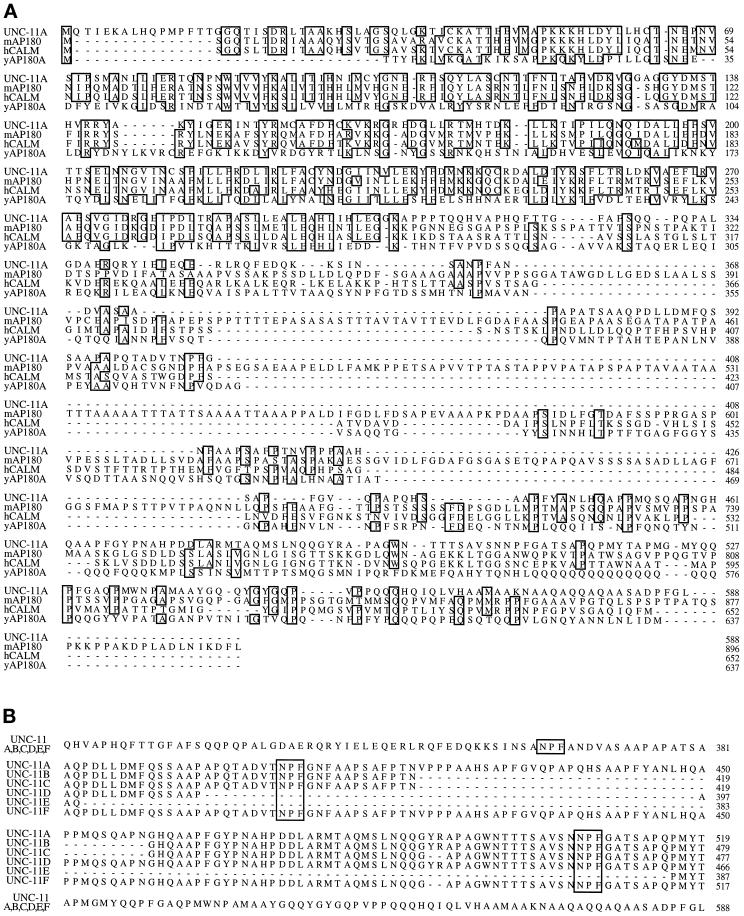
The C. elegans unc-11 was cloned by transformation rescue of its mutant phenotype with cosmid clones. unc-11 maps genetically between the fog-1 and dpy-5 loci on chromosome I, and a cosmid contained within this interval, C32E8, rescued the unc-11 locomotion phenotype (Figure 1). Subsequently, we narrowed the rescuing activity to an 8.8-kb _Xba_I fragment from this cosmid that contains a single predicted open reading frame (Figure 1, B and C). Furthermore, we determined the molecular defect for five unc-11 alleles, and each contained a mutation within the single open reading frame encoded in the rescuing fragment (see below), thus demonstrating that we cloned the correct gene. cDNAs representing six transcripts derived from the locus were isolated by screening cDNA libraries and by RT-PCR amplification (see MATERIALS AND METHODS for details). Analysis of the DNA sequences revealed that the cDNA clones were splicing isoforms (called A–F), the largest of which was predicted to encode a protein of 588 amino acids (Figure 2). The deduced UNC-11 products are most similar to the mammalian neuronal-specific assembly protein AP180 (Figure 2A; Ahle and Ungewickell, 1986; Kohtz and Puszkin, 1988). UNC-11 shares 44.7% identity with mouse AP180 (Morris et al., 1993; Zhou et al., 1993) and 41.4% identity with the related ubiquitously expressed human CALM protein (Dreyling et al., 1996). Two related yeast genes, yAP180A and yAP180B (Wendland and Emr, 1998), share 22.1% and 21.4% identity, respectively, with UNC-11A (Figure 2A). Alignments of UNC-11 with AP180 family members suggest the protein consists of at least two domains: an N-terminal 290-amino acid domain that is well-conserved among metazoans (UNC-11 shares 62.8% identity with mouse AP180 in this region), and a C-terminal domain that contains only minimally conserved motifs (27.1% identity with mouse AP180 in this region).
Figure 1.
Molecular cloning of the unc-11 locus. (A) Genetic map of chromosome I of C. elegans. (B) Transgenic rescue of unc-11 mutants with DNA clones. Rescue data are provided as a fraction of stable lines that gave complete rescue of the unc-11 phenotype. (C) Organization of the unc-11 locus. A restriction map of the genomic unc-11 locus. Selected restriction sites are indicated on the linear map. Triangles delineate the positions of Tc1 transposons inserted in the unc-11 alleles listed in italics. Sequences deleted in unc-11 alleles are shown as hatched rectangles. Exons are represented as boxes. The structure of constructs used in this study are illustrated below the restriction map. A schematic representation of the six classes of unc-11 cDNAs is also shown. Exons are numbered 1–9. The a, b, or c suffixes represent alternative versions of these exons. GenBank accession numbers for the six cDNA forms are AF144257–AF144262.
Figure 2.
Sequence comparison of UNC-11 and its vertebrate homologues. (A) An alignment of the UNC-11A isoform with mouse AP180 (Zhou et al., 1993), human CALM (Dreyling et al., 1996), and yeast AP180A protein sequences (Wendland and Emr, 1998). The alignment was obtained using the program ClustalW 1.5. Sequences identical in three or four of the proteins are boxed. (B) An alignment of the six UNC-11 isoforms illustrating the diversity in the C-terminal domains of products from this locus. NPF tripeptide motifs are boxed.
In addition, UNC-11 contains several domains that have been shown to have important biochemical properties in AP180. The conserved N-terminal domain of mammalian AP180 is known from biochemical studies to bind to inositol polyphosphates in vitro (Norris et al., 1995; Ye et al., 1995; Hao et al., 1997), and both AP180 and UNC-11 share three adjacent lysine residues in this region. This KKK motif is shared with the phosphoinositide-binding domain of the α-adaptin subunit (Gaidarov et al., 1996), and stretches of lysines have been shown to be required for the binding of inositol phosphates in arrestin (Gaidarov et al., 1999). The C-terminal domain of the mouse AP180 molecule interacts with clathrin and by itself can promote the assembly of clathrin cages (Ye and Lafer, 1995b). Likewise, the C-terminal region of UNC-11 promotes clathrin assembly in vitro despite minimal sequence similarity with AP180 (Golan, Prasad, Lafer, and Alfonso, unpublished data). The C terminus of yeast AP180A, although not well conserved, also binds clathrin (Wendland and Emr, 1998). These data indicate that the C-terminal domains are functionally conserved despite minimal sequence similarity.
unc-11 Expresses Distinct Isoforms
The genomic unc-11 locus is composed of nine coding exons. We identified six isoforms of unc-11 (referred to as UNC-11A-F) by analyzing cDNAs and RT-PCR products, these isoforms are identical in the conserved N-terminal domain that is encoded by exons 1–4 (Figure 1C). Differential usage of exons 5–8 accounts for the differences in the cDNAs derived from the unc-11 locus (Figures 1C and 2B). All transcripts share the ninth exon, which encodes the last 59 identical amino acids of the C terminus. cDNAs representing the UNC-11C form were the most abundant of the species isolated by RT-PCR (see MATERIALS AND METHODS for details), but this may not be representative of the relative abundance of the messages in vivo.
One sequence motif found in the C terminus of both the yeast AP180s and human CALM is the tripeptide NPF, which has been implicated in binding to EH domains such as those found in eps15 (Salcini et al., 1997; Wendland and Emr, 1998). Interestingly, the splice isoforms of UNC-11 differ in the number of NPF motifs they contain (Figure 2B).
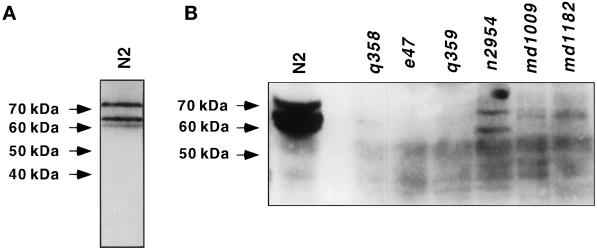
A polyclonal antibody generated against a bacterially expressed UNC-11C fusion protein detected three abundant proteins of ∼60, 65, and 75 kDa on Western blots of mixed-staged wild-type animals (Figure 3A). These proteins were undetectable in extracts derived from strains harboring the alleles e47, q358, and q359 (Figure 3B). Products of 58 and 66 kDa were detected in extracts from n2954, a mutant that results from a small in-frame deletion in exon 4. Similarly, only low abundance protein bands were detected in the Tc1 insertion alleles md1009 and md1182 and the deletion allele q358 (Figure 3B). The predicted molecular masses for the UNC-11 isoforms (50.7–64.4 kDa) are slightly smaller than the observed proteins in wild-type extracts; posttranslational modifications such as phosphorylation might account for the altered mobility of the isoforms. Several potential protein kinase C phosphorylation sites are present in the UNC-11 sequences. In summary, unc-11 expresses several distinct isoforms, which all have identical N-terminal domains but different C-terminal domains.
Figure 3.
C. elegans expresses at least three UNC-11 isoforms. Western blots of protein extracts isolated from the wild type and unc-11 mutants. UNC-11 immunoreactivity was detected as specified in MATERIALS AND METHODS. The position of molecular weight markers is indicated on the left of the panels. (A) Total protein extracts (63 μg) from a mixed-staged culture of the wild type showing three UNC-11 isoforms. (B) Total protein extracts (150 μg) from mixed-stage cultures of the indicated strains. All UNC-11 iso-forms are severely reduced or absent in unc-11 mutants e47, q358, q359, md1009, and md1182. Two products of altered mobility are visible in n2954.
Severe Alleles of unc-11 Disrupt the Shared N-Terminal Domain and Eliminate All Identified UNC-11 Isoforms
We characterized 15 independently isolated unc-11 alleles by Southern analysis, and five showed restriction fragment length polymorphisms within the coding region shared by all potential isoforms. We determined the sequences of these five polymorphic alleles. unc-11(md1009) and unc-11(md1182) consist of Tc1 transposable element insertions (Rosenzweig et al., 1983) into distinct sites of the fourth exon (Figure 1C). The alleles e47, q358, and n2954 contain small deletions of sequences coding for portions of the conserved N-terminal domain (Figure 1C). The deletions in e47 and q358 each result in frame shifts and premature termination. The deduced unc-11 translation products from e47 and q358 would encode only 10 and 49 amino acids, respectively, of the native UNC-11 sequence. The n2954 lesion results in an in-frame deletion of 74 amino acids from exon 4. Since the first four exons are shared by all alternatively spliced isoforms, we expect all of the isoforms to be missing or altered in structure in these five alleles. The five mutant strains exhibit very similar behavioral phenotypes and, with the possible exception of n2954, all these mutants are likely to display the unc-11 null phenotype.
unc-11 Expression Is Localized to the Nervous System and Is Enriched at Synaptic Sites
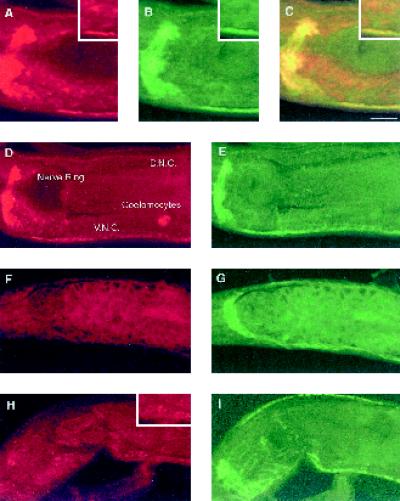
Like mammalian AP180, UNC-11 protein is enriched in synaptic regions. We stained whole worms using polyclonal antibodies raised against UNC-11C. Affinity-purified antibodies detected immunoreactivity in the nervous system of wild-type animals (Figure 4A and D) that was absent in unc-11 worms harboring the null allele e47 (Figure 4F). No neuronal immunoreactivity is detected in the mutant strains q358, md1009, and md1182 (our unpublished results). By contrast, the mutant allele n2954 had low but detectable UNC-11 immunoreactivity that was largely restricted to the cell bodies, as shown for those of the motor neurons in the ventral nerve cord (Figure 4H and inset). Immunostaining in wild-type animals was punctate and enriched at synapses. Specifically, we observed punctate immunoreactivity in the ventral and dorsal nerve cords and the nerve ring (Figure 4, A and D, and our unpublished results). The staining pattern is quite similar to synaptotagmin, a synaptic vesicle-associated marker, suggesting that UNC-11 is closely associated with release sites. However, the unc-11 pattern is more diffuse than synaptotagmin (Figure 4, B, E, G, and I) and staining was observed in cell somas throughout the nervous system (Figure 4D). UNC-11 is much more abundant in the soma than the vesicle-associated markers synaptotagmin and the vesicular acetylcholine transporter (our unpublished results). Finally, the antibodies failed to detect UNC-11 in motor neuron commissures or in the dendrites of sensory neurons in the head (our unpublished results).
Figure 4.
UNC-11 is enriched in synaptic regions of C. elegans. Confocal images of adult nematodes prepared for immunohistochemistry using both affinity-purified rabbit α-UNC-11 antisera and affinity-purified mouse α-synaptotagmin antisera. Animals were double stained for UNC-11 with rhodamine-conjugated secondary antibody and for synaptotagmin with FITC-conjugated secondary antibody: wild-type (A–E), unc-11(e47) (F and G), and unc-11 (n2954) (H and I). Anterior is left in all images. Panel C is the merge of panels A and B. A representative nerve ring, ventral nerve cord (V. N. C.), dorsal nerve cord (D. N. C.), and coelomocyte are labeled in panel D. Insets in panels A–C and H depict localization of UNC-11 staining in neuronal cell bodies. Bar, 10 μm.
We also detected UNC-11 expression in the coelomocytes (Figure 4D) and diffuse staining in the intestine (Figure 4, A, C, and D). Thus, immunohistochemistry indicates that UNC-11 is preferentially expressed in neurons. However, promoter fusion data (see below) and the nonneuronal staining are consistent with low-level expression in other tissues that may be too low to detect with our polyclonal UNC-11 antisera. The presence of AP180 homologues in yeast (Wendland and Emr, 1998) and a ubiquitously expressed AP180 homologue in humans (Dreyling et al., 1996) suggest that all cell types may express an AP180-like molecule. It is unlikely that a second homologue of AP180 is expressed in C. elegans because analyses of the complete genome (consortium, 1998) and EST data bases have not revealed another AP180 homologue in C. elegans.
The expression pattern of an unc-11 promoter-GFP fusion supports the observation that unc-11 is transcribed primarily in the nervous system and is also expressed in other nonneuronal tissues. We fused GFP to the unc-11 promoter (Figure 1C) and examined the pattern of fluorescence in transgenic animals. The promoter expressed GFP at high levels in the vast majority of the nervous system, including the nerve ring and ventral nerve cord (our unpublished results). Strong expression of the promoter fusion was also seen in two nonneuronal cell types: the coelomocytes and the uv1 secretory cells of the vulva (our unpublished results). Finally, weak expression was observed in the intestine (our unpublished results).
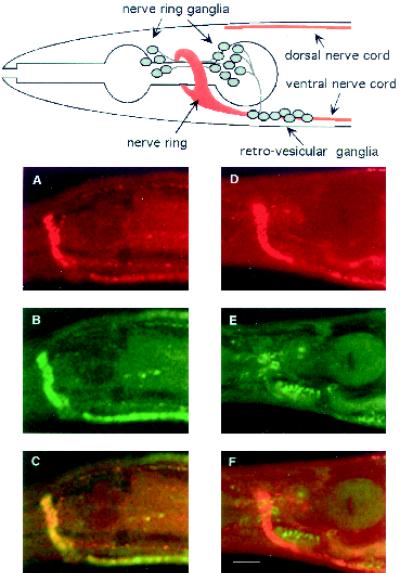
Further experiments analyzing the distribution of UNC-11 in kinesin mutants indicated that synaptic localization was probably not mediated via interactions with synaptic vesicles. Previous ultrastructural analysis of unc-104 animals revealed that synaptic vesicles are largely confined to the soma of this mutant (Hall and Hedgecock, 1991). Null alleles of unc-104 confer a larval lethal phenotype. Thus, we examined the distribution of UNC-11 in two viable mutants that differ in the degree of severity of the mutant phenotypes. In unc-104(rh43) animals, synaptotagmin is localized to cell bodies as expected of a synaptic vesicle protein (Figure 5E). By contrast, UNC-11 protein remains abundant in the neuropil (Figure 5D). Similar results were obtained in the less severe allele unc-104(e1265) (our unpublished results). In summary, UNC-11 is detected both in the cell bodies and presynaptic sites of neurons. Furthermore, UNC-11 is transported down the axon independently of the kinesin-like motor protein UNC-104.
Figure 5.
Subcellular localization of UNC-11 in wild-type and unc-104 mutant nematodes. Confocal images of freeze-cracked nematodes double stained with affinity-purified rabbit α-UNC-11 antisera (first row); affinity purified mouse α-synaptotagmin antisera (second row), and merge (third row). (A–C) Wild-type. (D–F) unc-104(rh43). A diagram depicting the organization of nervous tissue in the C. elegans head including the position of neuronal cell bodies, the nerve ring neuropil, and the ventral and dorsal nerve cords is positioned above the confocal images. Synaptic-rich regions are depicted in red. Commissures, dendrites, minor process bundles, and many neuronal cell bodies of the ganglia were omitted for clarity. Bar, 10 μm.
unc-11 Function Is Required by Multiple Neurotransmitters
unc-11 mutants originally were isolated in a screen for behaviorally uncoordinated animals (Brenner, 1974). Numerous additional alleles have been isolated as mutants resistant to the acetylcholinesterase inhibitor aldicarb (Miller et al., 1996). In C. elegans, resistance to aldicarb is well correlated with synaptic defects (reviewed by Rand and Nonet, 1997). The unc-11 gene is thought to function presynaptically because acetylcholine levels are elevated in the mutant (Hosono and Kamiya, 1991; Nguyen et al., 1995) and because the animals respond normally to acetylcholine receptor agonists (Miller et al., 1996). In addition to a defect in cholinergic transmission, unc-11 mutants have a defect in γ-aminobutyric acid (GABA)-ergic transmission. Contraction of the enteric muscles requires GABA (McIntire et al., 1993), and unc-11 mutants display decreased enteric muscle contractions (Miller et al., 1996). These data suggest that the unc-11 defect is caused by a general disruption of synaptic function rather than a specific defect in cholinergic function.
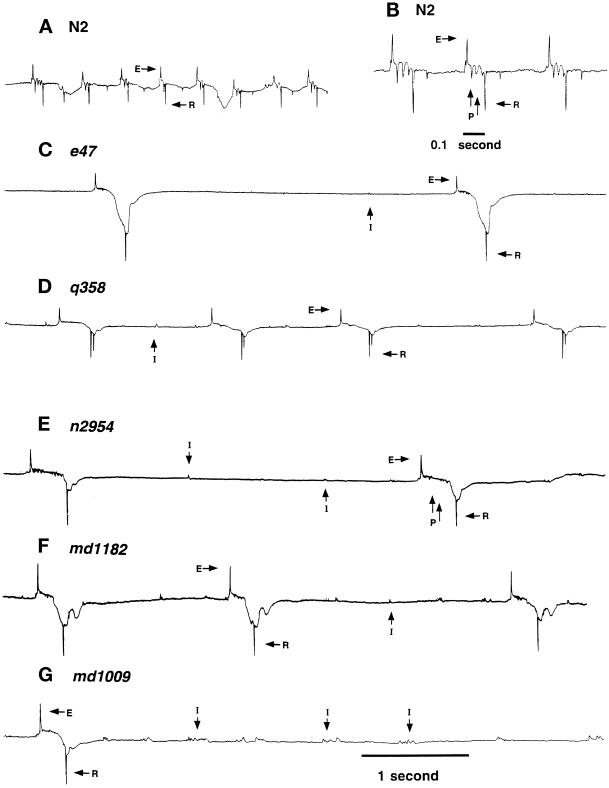
To determine whether glutamatergic transmission is also defective and to directly demonstrate a defect in synaptic transmission, we analyzed synaptic currents in the pharyngeal muscle using an extracellular recording technique developed by Raizen and Avery (1994). These recordings, or electropharyngeograms (EPGs), reveal both muscle-derived and synaptic currents (Raizen and Avery, 1994); Figure 6, A and B). We analyzed 15 unc-11 alleles, and all but the mildly uncoordinated allele ic9 behaved similarly (Nguyen et al., 1995). Representative traces for the wild-type and five mutants are shown (Figure 6, C–G). In the strong mutants, we observed two abnormalities that indicate that synaptic transmission is disrupted. First, function of the cholinergic MC motor neuron is diminished. MC is known to stimulate depolarization of pharyngeal muscle (Raizen et al., 1995). We also observed an increased frequency of interpump transients (labeled I; Figure 6, C–G). These interpump transients represent subthreshold synaptic activity of the MC motor neuron (Raizen et al., 1995). Pharyngeal pumping also was less frequent and interpump intervals were longer than in wild-type, both of which are consistent with a reduction in MC activity. Second, function of the glutamatergic neuron M3 also was reduced. M3 generates inhibitory transients that accelerate the repolarization of pharyngeal muscle (Avery, 1993a; Li et al., 1997). We observed a decreased number of inhibitory transients between the depolarization and repolarization of the muscle (labeled P in Figure 6, B and E). The absence is particularly noticeable in e47 and q358 (Figure 6, C and D). Since M3 induces repolarization of pharyngeal muscle, the reduction in M3 activity also is consistent with the observed increase in mean pump duration (Figure 6). In summary, the pharyngeal physiology of unc-11 mutants is consistent with a decrease in both cholinergic and glutamatergic synaptic function.
Figure 6.
Extracellular recording from wild-type and unc-11 animals. Representative traces of electropharygeogram recordings of the wild-type strain N2 (A and B), unc-11(e47) (C), unc-11(q358) (D), unc-11(n2954) (E), unc-11(md1182) (F), and unc-11(md1009) (G). Representative interpump transients (I), excitatory phase transients (E), relaxation phase transients (R), and plateau phase transients (P) are labeled. The wild-type and unc-11 mutants had the following mean pump lengths: wild-type (N2), 124 ± 10 ms; unc-11(e47), 310 ± 23 ms; unc-11(q358), 240 ± 11 ms; unc-11(n2954), 284 ± 11 ms; unc-11(md1182), 202 ± 5 ms; unc-11(md1009), 233 ± 7 ms; unc-11(e47), 310 ± 23 ms. All traces are millivolts versus time. All traces except for panel B share the same time scale indicated at the bottom of the trace in panel G.
Synaptobrevin, but Not Other Synaptic Vesicle Proteins, Is Mislocalized in unc-11 Mutants
Mutants lacking UNC-11 fail to efficiently localize the synaptic vesicle protein synaptobrevin to synaptic sites, although they efficiently target all other synaptic proteins we examined. We characterized the localization of a GFP fusion to the synaptic vesicle protein synaptobrevin (Nonet et al., 1998; Nonet, 1999). In wild-type worms, the GFP-tagged synaptobrevin was localized in a punctate pattern in the neuropil of the nervous system similar to the distribution of synaptic varicosities (Nonet, 1999) (Figure 7A). However, in the four unc-11 mutants characterized, the GFP fusion was diffuse and more broadly distributed in the nervous system, resembling the distribution of the plasma membrane protein syntaxin (Saifee et al., 1998) (Figure 7B). Moreover, commissural processes of motor neurons, which were rarely detectable in the wild type (Figure 7A), were clearly visible in unc-11 (Figure 7B). Finally, synaptobrevin-GFP fluorescence is absent from the dendritic processes of the amphid sensory neurons in the wild type, but fluorescence was present in these dendrites in the mutant (our unpublished results). Western blots containing equivalent amounts of total protein derived from wild-type and an unc-11 mutant line expressing a SNB-1::GFP fusion protein confirm that the levels of SNB-1 protein are equivalent in both backgrounds (our unpublished results). Thus, changes in the localization pattern in UNC-11 mutants are not the result of alterations in synaptobrevin expression.
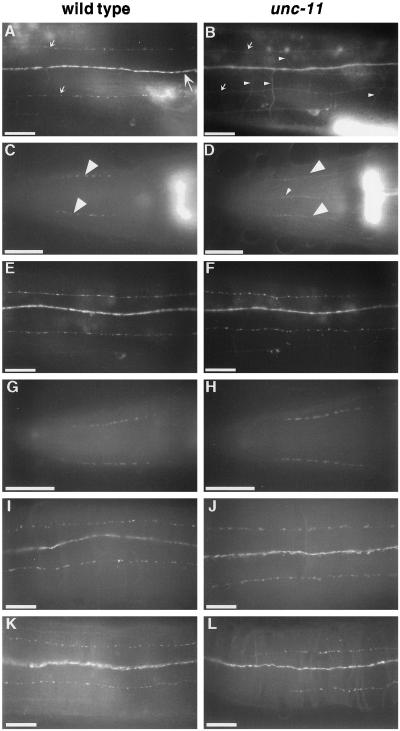
Figure 7.
Synaptobrevin, but not other synaptic vesicle proteins, is mislocalized in unc-11 mutants. The localization of vesicle proteins was examined in the wild type (first column) and in unc-11 animals (second column) using both GFP-tagged proteins and antibodies. (A and B) Images of the dorsal nerve cord of live immobilized adult animals expressing GFP-tagged synaptobrevin. The dorsal cord (large arrow), dorsal sublateral processes (small arrows), and commissural processes of motor neurons (small arrowheads) are labeled. The out-of-focus fluorescence in the lower right of each panel derives from the spermatheca that expresses synaptobrevin. (A) A wild-type animal. (B) unc-11(e47) animal. (C and D) Images of heads of adult worms fixed and stained with α-synaptobrevin antibodies and visualized with FITC-conjugated secondary antibodies. Synaptic varicosities of the SAB motor neuron processes are labeled with large arrowheads in a wild-type animal (C) and an unc-11(e47) animal (D). Note the appearance of synaptobrevin immunoreactivity in ventral processes (probably AVM and VA1) in panel D. (E and F) Images of the dorsal nerve cord of live immobilized adult animals expressing GFP-tagged synaptogyrin in a wild-type animal (E) and a unc-11(e47) animal (F). (G–J) Images of adult worms fixed andstained with α-RAB-3 antibodies and visualized with FITC-conjugated secondary antibodies in the head of a wild-type animal (G), and an unc-11(e47) animal (H); and a portion of the dorsal cord of a wild-type animal (I) and an unc-11(e47) animal (J). (K and L) Images of a portion of the dorsal cord of adult hermaphrodite worms fixed and stained with α-synaptotagmin antibodies and visualized with FITC-conjugated secondary antibodies in a wild-type animal (K) and an unc-11(n2954) animal (L). Anterior is left in all images. Bar, 10 μm.
To confirm synaptobrevin was missorted, we examined the localization of the native protein using immunohistochemical techniques. Antisera directed against synaptobrevin demonstrated that native synaptobrevin also was more diffusely localized in unc-11 mutants than in the wild type (Figure 7, C and D). For example, in the wild type, synaptobrevin immunoreactivity was tightly clustered in varicosities in SAB motor neurons (Figure 7C), but in unc-11 mutants immunoreactivity was distributed along the axon (Figure 7D). Axonal and dendritic processes lacking synaptobrevin immunoreactivity in the wild type were visible in unc-11 animals. For example, the AVM and VA1 processes anterior of the nerve ring and the amphid dendritic processes were undetectable using synaptobrevin antiserum in the wild type, but were visible in unc-11 mutants (Figure 7, C and D, and our unpublished results). In summary, in unc-11 mutants, the distribution of the synaptic vesicle protein synaptobrevin resembled that of a ubiquitously distributed plasma membrane marker rather than the distribution of a synaptic protein.
Our data also indicate that the localization defect in unc-11 mutants is specific for synaptobrevin. We determined the localization of other synaptic vesicle proteins in unc-11 mutants. First, we examined the distribution of a GFP fusion to the neuronally expressed sng-1 gene (Nonet, 1999), a C. elegans locus encoding a homologue of the integral membrane synaptic vesicle protein synaptogyrin (Stenius et al., 1995). In the wild type, GFP-tagged synaptogyrin colocalizes with other C. elegans synaptic markers and parallels the neuronal distribution of synaptobrevin (Figure 7E). In unc-11 mutants, the localization of GFP-tagged synaptogyrin remained indistinguishable from that in the wild type (Figure 7F). We also examined the distribution of the synaptic vesicle-associated proteins synaptotagmin (Nonet et al., 1993) and RAB-3 (Nonet et al., 1997) using antisera directed against these proteins. RAB-3 and synaptotagmin immunoreactivity in the wild type are similar to that of synaptobrevin, punctate and restricted to synaptic-rich regions of the nervous system (Figure 7, G, I, and K). In unc-11 animals, both the levels and distribution of RAB-3 and synaptotagmin immunoreactivity were indistinguishable from those in the wild type (Figure 7, H, J, and L). In all four severe unc-11 alleles we examined, RAB-3 and synaptotagmin immunoreactivity appeared normal, but synaptobrevin immunoreactivity was diffuse and aberrantly localized. Thus, synaptobrevin is the only synaptic vesicle component we have examined that is mislocalized in unc-11 animals.
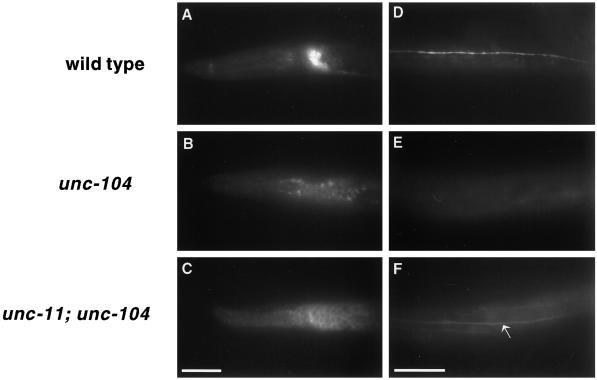
UNC-11 probably functions in the sorting of synaptobrevin in the cell body as well as at the synapse. This conclusion arises from experiments in which synaptic vesicles were retained in the soma. As discussed earlier, unc-104 encodes a kinesin-like molecule required for the transport of synaptic vesicles from the soma to synapses. In unc-104 animals, GFP-tagged synaptobrevin (Nonet, 1999; and Figure 8B) and other synaptic vesicle components were restricted to cell bodies (Nonet et al., 1993, 1997, 1998), and fluorescence was distributed within the intracellular space. Additionally, as predicted, GFP-tagged synaptobrevin was completely absent from the dorsal cord synaptic region (Figure 8E). However, in the unc-11; unc-104 double mutant, GFP-tagged synaptobrevin accumulated in the cell periphery rather than intracellularly in the soma of neurons (Figure 8C). Additionally, the GFP-tagged synaptobrevin was present and diffusely distributed in the dorsal cord and neighboring axonal processes (Figure 8F). Thus, in unc-11; unc-104 animals, synaptobrevin appears to be localized to the plasma membrane. The simplest interpretation of these experiments is that synaptobrevin-containing vesicles are fusing with the plasma membrane in the soma, but not being internalized by endocytosis.
Figure 8.
UNC-11 functions in the soma of neurons. Images of L1 larvae expressing synaptobrevin-GFP. A lateral view of the head of a wild-type animal (A), an unc-104(e1265) animal (B), and an unc-11(e47); unc-104(e1265) animal (C). A dorsal view of the body of (D) a wild-type animal (D), an unc-104(e1265) animal (E), and and unc-11(e47); unc-104(e1265) animal (F). Note that GFP fluorescence is visible in the dorsal cord (arrow) of the double mutant. Anterior is to the left in all photographs. Bar, 10 μm.
unc-11; snb-1 Double Mutants Resemble Synaptobrevin Null Mutants
Synaptobrevin is essential for the regulated fusion of synaptic vesicles at the synapse (Schiavo et al., 1992; Broadie et al., 1995; Nonet et al., 1998). Both the behavioral and physiological defects in unc-11 mutants are consistent with a decrease in exocytosis caused by depletion of synaptobrevin from synaptic vesicles. The behavioral defects of unc-11 null mutants are intermediate in phenotype between those of the viable hypomorphic snb-1(md247) mutant and the lethal snb-1(js124) null mutant (Nonet et al., 1998). Although synaptobrevin is mislocalized in unc-11 animals, synaptic vesicles in the mutants likely retain low levels of synaptobrevin. Indeed, complete absence of synaptobrevin on vesicles would be expected to result in a lethal phenotype similar to that of synaptobrevin null mutants; rather unc-11 null mutants are viable. We tested if further reduction of synaptobrevin function would enhance the defects associated with lack of AP180 activity. In fact, the double mutants unc-11(e47); snb-1(md247) and unc-11(n2954); snb-1(md247) arrest development in the first larval stage similar to snb-1(js124) null mutants. This observation is consistent with the hypothesis that residual synaptobrevin in vesicles contributes to the viability of unc-11 null mutants.
Synaptic Vesicle Diameter Is Larger in unc-11 Null Mutants
An analysis of unc-11 synaptic ultrastructure revealed that synaptic vesicle diameter is altered. The general morphology of neuromuscular synapses in three independent unc-11 mutants was similar to that of the wild-type (Figure 9). A thick presynaptic density was present, and synaptic vesicles were clustered around this site. We quantified vesicle diameter in the wild-type, unc-11(e47), unc-11(q358), and unc-11(n2954) animals (Figure 9). In the wild type, vesicles were relatively uniform in size and had a mean diameter of 29.5 ± 5.4 nm. In the two null mutants, mean vesicle diameter was 33% larger (unc-11(e47) 39.1 ± 7.3 nm; unc-11(q358) 40.4 ± 8.0 nm). This 33% increase in diameter translates into a 77% increase in mean vesicle membrane area for vesicles in the mutant. Vesicular diameter does not seem to be random in the mutant. The diameter of vesicles in the unc-11 mutants peaks at around 40 nm in each allele. Furthermore, unc-11(n2954), which contains a small in-frame deletion in the N terminus of the protein, has a bimodal distribution of normal (31 nm) and large vesicles (40 nm) (Figure 9E).
We noted that vesicles tend to accumulate at the plasma membrane in the unc-11 mutants. Because the analysis was done using serial sections, it was possible to unambiguously identify synapses of both excitatory cholinergic synapses (Figure 9, A and B) and inhibitory GABAergic synapses (Figure 9, C and D). GABAergic neuromuscular junctions typically are larger and less packed with vesicles than cholinergic synapses, and hence it is easier to evaluate membrane association of vesicles in these synapses. At GABAergic junctions from unc-11 animals, we noticed an accumulation of uncoated vesicles in close apposition to the membrane (Figure 9D). This accumulation of docked vesicles might be caused by the depletion of synaptobrevin from vesicles. Specifically, docking of vesicle proceeds, but exocytosis is reduced, which leads to an accumulation of vesicles associated with the membrane. This phenotype is consistent with experiments in which synaptobrevin was depleted by expressing toxins that specifically cleave this synaptic vesicle protein (Hunt et al., 1994; Broadie et al., 1995).
DISCUSSION
We demonstrated that the unc-11 locus of C. elegans encodes a neuronally expressed homologue of AP180, a protein previously implicated in the regulation of synaptic vesicle endocytosis. Members of this family are found throughout eukaryotes from yeast to humans. The UNC-11 isoforms we identified contain a 290-amino acid domain that is well conserved in members of this family. This domain has been implicated in phospholipid binding, an activity that is likely to mediate interactions between AP180 and membranes. The C-terminal half of the protein shows more limited similarity to AP180 and is alternatively spliced in the different UNC-11 isoforms. Despite the limited homology, this domain of the vertebrate, yeast, and C. elegans molecules binds clathrin (Ye and Lafer, 1995b; Wendland and Emr, 1998; Golan, Prasad, Lafer and Alfonso, unpublished data). Indeed, the vertebrate AP180 protein not only binds clathrin but also stimulates its assembly into organized lattices. These biochemical activities form the foundation for its proposed role in the assembly of clathrin into coated pits during the process of endocytosis at the nerve terminal. By contrast, we conclude from our studies of unc-11 mutants that AP180 is not an essential assembly protein for clathrin-mediated endocytosis. However, UNC-11 protein regulates the size of endocytosed vesicles, presumably through its interaction with clathrin. UNC-11 also is required for the efficient targeting of the synaptic vesicle protein synaptobrevin.
AP180 Is Not Essential for Endocytosis of Synaptic Membrane
Our analysis of the morphology of neuromuscular junctions in unc-11 mutants demonstrates that, in C. elegans, endocytosis of synaptic vesicle membrane does not require AP180. unc-11 mutants are viable and capable of some locomotion and hence must release transmitter through exocytosis. In the presence of continued exocytosis, an absolute block in endocytosis would be expected to lead to a chronic depletion of vesicle membrane, as is observed in Drosophila dynamin and α-adaptin mutants (Poodry and Edgar, 1979; Gonzalez-Gaitan and Jackle, 1997). However, we observe that synaptic vesicles, albeit of altered size, are present at neuromuscular junctions in unc-11 null mutants. It is unlikely that another AP180 homologue is directing endocytosis at neuromuscular junctions, because no other homologue has been identified in BLAST searches of the completed genomic sequence of C. elegans. Hence, our data show that endocytosis of synaptic vesicle membrane can occur in the absence of the UNC-11 AP180 homologue in C. elegans. Analysis of Drosophila AP180 mutants has revealed similar alterations in vesicle morphology without a complete block of endocytosis (Zhang et al., 1998). Drosophila AP180 null mutants have fewer synaptic vesicles and they are larger and heterogenous in size (Zhang et al., 1998). Interestingly, S. cerevisiae double mutants lacking two yeast AP180 homologues have no detectable defects in endocytosis (Wendland and Emr, 1998). However, vesicle diameters were not analyzed in the yeast mutants.
Our data suggest that UNC-11 is not required for clathrin-mediated endocytosis. However, this interpretation is complicated by the fact that synaptic vesicle endocytosis has not been demonstrated to occur via a clathrin-mediated pathway in C. elegans. Specifically, no C. elegans mutations have been isolated that disrupt the genes encoding either clathrin or AP-2 complex subunits. We cannot rule out that unc-11 is required for clathrin-mediated endocytosis, but that non-clathrin-mediated recycling mechanisms are also utilized and thus unmasked in the mutants.
Evidence in other systems suggests clathrin-mediated synaptic vesicle endocytosis predominates at the synapse. For example, analysis of mutants of Drosophila suggests that the α-adaptin subunit of AP-2 is essential for synaptic vesicle endocytosis; ultrastructural analysis of neuromuscular junctions from these lethal mutants were depleted of vesicles (Gonzalez-Gaitan and Jackle, 1997). As mentioned above, a potential complication in C. elegans is the possibility that other vesicle-trafficking pathways compensate in the absence of AP180. Faundez et al. (1998) have recently demonstrated that the tetrameric AP-3 complex, in conjunction with ARF1, is capable of budding synaptic vesicles from endosomal membrane in vitro in the absence of AP-2 or AP180 (Faundez et al., 1998). Thus, bulk endocytosis of synaptic membrane from the synapse followed by the action of the AP-3 complex could account for the replenishing of synaptic vesicles independently of an AP-2/AP180-dependent pathway. Alternatively, most exocytosis under modest stimulation might occur via a clathrin-independent kiss-and-run mechanism (Artalejo et al., 1995) that does not involve the complete fusion of the two membranes (Fesce et al., 1994; Palfrey and Artalejo, 1998). Analysis of the phenotypes of mutants in other components of the endocytic pathway will be required to resolve these questions.
AP180 Regulates Vesicle Size
In our working hypothesis one of the roles of AP180 is to regulate the size of clathrin lattice assemblies. Using an in vitro assembly assay, Ye and Lafer (1995a) demonstrated that clathrin assembles into random sized spheres; the addition of AP180 to such assays causes clathrin to assemble into spheres with uniform diameters. Vesicles at neuromuscular junctions are larger in unc-11 mutants than in the wild type. The size distribution of vesicles in these mutants is not random; rather, they are distributed around a 40-nm mean diameter. One explanation for the altered size distribution is that in the absence of AP180, vesicle diameter is unregulated by clathrin-assembly complexes; for example, the diameter of the vesicle might be determined by its lipid composition. Alternatively, proteins of the endocytic pathway, either clathrin-dependent or clathrin-independent, might confer a 40-nm diameter on the vesicles. Lastly, our data do not preclude the possibility that the large vesicles represent an accumulation of intermediates in synaptic vesicle genesis, rather than mature vesicles of an altered size. However, the fact that we observed docked vesicles of altered size (see Figure 9) suggests that these vesicles exhibit some features of mature synaptic vesicles. In summary, our ultrastructural analysis is consistent with an in vivo role for AP180 analogous to its previously defined biochemical function in regulating the size of clathrin cage assembly. Vesicles of differing diameter are found in different tissues. In vitro, the C-terminal domain of AP180 is capable of regulating the size of clathrin assemblies (Ye and Lafer, 1995b). C. elegans UNC-11 protein isoforms differ in the structure of this domain. These isoforms could provide a molecular explanation for how AP180 might regulate the differential size of secretory vesicles found in different neurons and secretory cells. Characterization of the clathrin assembly activities of these different isoforms combined with subcellular localization of various isoforms may address this issue.
AP180 and Sorting of Vesicle Proteins
A more surprising phenotype of the unc-11 mutants is the specific mislocalization of the synaptic vesicle protein synaptobrevin. In these mutants, synaptobrevin was found at uniform low levels in axonal and dendritic processes. By contrast, all of the other synaptic vesicle-associated proteins that we examined were normally distributed in synaptic regions in unc-11 mutants. These include synaptogyrin, RAB-3, synaptotagmin (Figures 7 and 8), and the acetylcholine vesicular transporter (Holgado and Alfonso, unpublished results). Where is the mislocalized synaptobrevin? Most likely the mislocalized synaptobrevin is found in the plasma membrane. In the mutant, synaptobrevin localization is distinct from that of other vesicle proteins and resembles that of the neuronal plasma membrane protein syntaxin. Although this distribution is most consistent with synaptobrevin retention on the plasma membrane, it is possible that some synaptobrevin is segregated into internal membranous structures. Regardless of the exact cellular location of mislocalized synaptobrevin, these data indicate that UNC-11 is acting as a sorting protein as well as a clathrin assembly protein.
The specificity of the localization defect to synaptobrevin suggests this protein is sorted by a different mechanism than at least some other synaptic vesicle proteins. Why might this be the case? Interactions between synaptic vesicle proteins (Bennett et al., 1992) are likely to keep vesicle proteins clustered as “islands” in the plasma membrane and permit a battery of vesicle proteins to undergo selective endocytosis together. However, synaptobrevin is unusual among vesicle proteins because of its strong interaction with the plasma membrane proteins, syntaxin and SNAP-25 (Sollner et al., 1993a). Current models for vesicle fusion suggest that after fusion, synaptobrevin is assembled in an extremely stable trimeric 7S complex (Hanson et al., 1997; Weber et al., 1998). Since SNAP-25 and syntaxin are much less abundant than synaptobrevin in synaptic vesicles (Walch-Solimena et al., 1995), these complexes are probably disassembled before endocytosis. Hence, a more complex sorting machinery may be required to ensure the efficient recycling of synaptobrevin to vesicles after disassembly of the 7S fusion complex by _N_-ethylmaleimide-sensitive factor (Sollner et al., 1993b). UNC-11 may play a role in sorting synaptobrevin at the plasma membrane by either directly binding to synaptobrevin or indirectly sequestering synaptobrevin through interactions with other proteins. While localization of UNC-11 at synaptic sites would suggest that this sorting function occurs at the synapse, it could also occur in the soma during vesicle biogenesis.
Of the two cellular defects identified to date in unc-11 mutants, it is likely that the sorting defect, rather than the alteration in vesicle size, accounts for the severity of the behavioral defect observed in the mutants. The modest twofold increase in mean quantal size at unc-11 neuromuscular junctions that would be expected from the increase in vesicle size seems unlikely to account for the dramatic decrease in synaptic transmission in these mutants. Rather, we favor the notion that the depletion of synaptobrevin from mature synaptic vesicles reduces the efficiency of exocytosis. Synaptobrevin is an abundant synaptic vesicle protein found in ∼20 copies per synaptic vesicle (Walch-Solimena et al., 1995). Current molecular models of the fusion process suggest that a ring of several 7S complexes forming together are necessary for vesicle fusion (Weber et al., 1998). Thus, reduction of the number of synaptobrevin molecules in vesicles is probably sufficient to account for the behavioral and physiological defects we observe in the absence of AP180 in neurons.
ACKNOWLEDGMENTS
We thank H. Robert Horvitz in whose laboratory the electron microscopy was undertaken; the C. elegans genome sequencing consortium, Ron Ellis, and Thomas Barnes for narrowing down the physical location of unc-11; Yishi Jin for observations concerning the localization of SNB-1::GFP, John M. Patterson for the initial characterization of antibodies to UNC-11C; Kathy Barton and Judith Kimble for the alleles unc-11(q358) and unc-11(q359); Leon Avery for unc-11(ad571) and advice on EPG set up and interpretation; Bob Barstead for the cDNA library; Clayton Hollenback, Sheng-Hao Chao, and John Allen for technical support; Stephen Kelso for setting up the electrophysiology rig and helping in the interpretation of the EPG; Janet Duerr and James B. Rand for helpful hints on immunofluorescence and antibodies and Andy Fire for cloning vectors. Some of the strains used in this study were provided by the Caenorhabditis Genetics Center. These studies were initiated while A.A. was affiliated with the Department of Biological Sciences at the University of Iowa, Iowa City, IA 52242. This research was supported by grants to A.A. (NS-32449), E.J. (NS-34307), H. Robert Horvitz (GM-24663) and M.L.N. (NS-33535) from the United States Public Health Service.
REFERENCES
- Ahle S, Ungewickell E. Purification and properties of a new clathrin assembly protein. EMBO J. 1986;5:3143–3149. doi: 10.1002/j.1460-2075.1986.tb04621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson DG. Formation of the first cleavage spindle in nematode embryos. Dev Biol. 1984;101:61–72. doi: 10.1016/0012-1606(84)90117-9. [DOI] [PubMed] [Google Scholar]
- Alfonso A, Grundahl K, McManus JR, Rand JB. Cloning and characterization of the choline acetyltransferase structural gene (cha-1) from C. elegans. J Neurosci. 1994;14:2290–2300. doi: 10.1523/JNEUROSCI.14-04-02290.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artalejo CR, Henley JR, McNiven MA, Palfrey HC. Rapid endocytosis coupled to exocytosis in adrenal chromaffin cells involves Ca2+, GTP and dynamin, but not clathrin. Proc Natl Acad Sci USA. 1995;92:8328–8332. doi: 10.1073/pnas.92.18.8328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L. Motor neuron M3 controls pharyngeal muscle relaxation timing in Caenorhabditis elegans. J Exp Biol. 1993a;175:283–297. doi: 10.1242/jeb.175.1.283. [DOI] [PubMed] [Google Scholar]
- Avery L. The genetics of feeding in Caenorhabditis elegans. Genetics. 1993b;133:897–917. doi: 10.1093/genetics/133.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barstead RJ, Waterston RH. The basal component of the nematode dense-body is vinculin. J Biol Chem. 1989;264:10177–10185. [PubMed] [Google Scholar]
- Barton MK, Kimble J. fog-1, a regulatory gene required for specification of spermatogenesis in the germ line of Caenorhabditis elegans. Genetics. 1990;125:29–39. doi: 10.1093/genetics/125.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck KA, Keen JH. Interaction of phosphoinositide cycle intermediates with the plasma membrane-associated clathrin assembly protein AP-2. J Biol Chem. 1991;266:4442–4447. [PubMed] [Google Scholar]
- Bennett MK, Calakos N, Kreiner T, Scheller RH. Synaptic vesicle membrane proteins interact to form a multimeric complex. J Cell Biol. 1992;116:761–775. doi: 10.1083/jcb.116.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal T. Trans-splicing and polycistronic transcription in Caenorhabditis elegans. Trends Genet. 1995;11:132–136. doi: 10.1016/s0168-9525(00)89026-5. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadie K, Prokop A, Bellen HJ, O’Kane CJ, Schulze KL, Sweeney ST. Syntaxin and synaptobrevin function downstream of vesicle docking in Drosophila. Neuron. 1995;15:663–673. doi: 10.1016/0896-6273(95)90154-x. [DOI] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: a plaform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Cremona O, De Camilli P. Synaptic vesicle endocytosis. Curr Opin Neurobiol. 1997;7:323–330. doi: 10.1016/s0959-4388(97)80059-1. [DOI] [PubMed] [Google Scholar]
- Dreyling MH, Martinez-Climent JA, Zheng M, Mao J, Rowley JD, Bohlander SK. The t(10;11)(p13;q14) in the U937 cell line results in the fusion of the AF10 gene and CALM, encoding a new member of the AP-3 clathrin assembly protein family. Proc Natl Acad Sci USA. 1996;93:4804–4809. doi: 10.1073/pnas.93.10.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerr JS, Frisby DL, Gaskin JG, Duke A, Asermely K, Huddleston D, Eiden LE, Rand JB. The cat-1 gene of C. elegans encodes a vesicular monoamine transporter required for specific monoamine-dependent behaviors. J Neurosci. 1999;19:72–84. doi: 10.1523/JNEUROSCI.19-01-00072.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faundez V, Horng JT, Kelly RB. A function for the AP3 coat complex in synaptic vesicle formation from endosomes. Cell. 1998;93:423–432. doi: 10.1016/s0092-8674(00)81170-8. [DOI] [PubMed] [Google Scholar]
- Fesce R, Grohovaz F, Valtorta F, Meldolesi J. Neurotransmitter release: fusion or ’kiss-and-run’? Trends Cell Biol. 1994;4:1–4. doi: 10.1016/0962-8924(94)90025-6. [DOI] [PubMed] [Google Scholar]
- Gaidarov I, Chen Q, Falck JR, Reddy KK, Keen JH. A functional phosphatidylinositol 3,4,5-trisphosphate/phosphoinositide binding domain in the clathrin adaptor AP-2 alpha subunit. Implications for the endocytic pathway. J Biol Chem. 1996;271:20922–20929. doi: 10.1074/jbc.271.34.20922. [DOI] [PubMed] [Google Scholar]
- Gaidarov I, Krupnick JG, Falck JR, Benovic JL, Keen JH. Arrestin function in G protein-coupled receptor endocytosis requires phosphoinositide binding. EMBO J. 1999;18:871–881. doi: 10.1093/emboj/18.4.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Gaitan M, Jackle H. Role of Drosophila alpha-adaptin in presynaptic vesicle recycling. Cell. 1997;88:767–776. doi: 10.1016/s0092-8674(00)81923-6. [DOI] [PubMed] [Google Scholar]
- Hall DH, Hedgecock EM. Kinesin-related gene unc-104 is required for axonal transport of synaptic vesicles in C. elegans. Cell. 1991;65:837–847. doi: 10.1016/0092-8674(91)90391-b. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Morisaki H, Jahn R, Heuser JE. Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell. 1997;90:523–535. doi: 10.1016/s0092-8674(00)80512-7. [DOI] [PubMed] [Google Scholar]
- Hao W, Tan Z, Prasad K, Reddy KK, Chen J, Prestwich GD, Falck JR, Shears SB, Lafer EM. Regulation of AP-3 function by inositides. Identification of phosphatidylinositol 3,4,5-trisphosphate as a potent ligand. J Biol Chem. 1997;272:6393–6398. doi: 10.1074/jbc.272.10.6393. [DOI] [PubMed] [Google Scholar]
- Hosono R, Kamiya Y. Additional genes result in an elevation of acetylcholine levels by mutations in Caenorhabditis elegans. Neurosci Lett. 1991;128:243–244. doi: 10.1016/0304-3940(91)90270-4. [DOI] [PubMed] [Google Scholar]
- Hunt JM, Bommert K, Charlton MP, Kistner A, Habermann E, Augustine GJ, Betz H. A postdocking role for synaptobrevin in synaptic vesicle fusion. Neuron. 1994;12:1269–1279. doi: 10.1016/0896-6273(94)90443-x. [DOI] [PubMed] [Google Scholar]
- Innis MA, Gelfand DH, Sninsky JJ, White TJ. PCR Protocols: A Guide to Methods and Applications. San Diego: Academic Press; 1990. 482. [Google Scholar]
- Kohtz DS, Puszkin S. A neuronal protein (NP185) associated with clathrin-coated vesicles. Characterization of NP185 with monoclonal antibodies. J Biol Chem. 1988;263:7418–7425. [PubMed] [Google Scholar]
- Kramer JM, French RP, Park EC, Johnson JJ. The Caenorhabditis elegans rol-6 gene, which interacts with the sqt-1 collagen gene to determine organismal morphology, encodes a collagen. Mol Cell Biol. 1990;10:2081–2089. doi: 10.1128/mcb.10.5.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Avery L, Denk W, Hess G. Identification of chemical synapses in the pharynx of Caenorhabditis elegans. Proc Natl Acad Sci USA. 1997;94:5912–5916. doi: 10.1073/pnas.94.11.5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews G. Neurotransmitter release. Annu Rev Neurosci. 1996;19:219–233. doi: 10.1146/annurev.ne.19.030196.001251. [DOI] [PubMed] [Google Scholar]
- McIntire SL, Garriga G, White J, Jacobson D, Horvitz HR. Genes necessary for directed axonal elongation or fasciculation in C. elegans. Neuron. 1992;8:307–322. doi: 10.1016/0896-6273(92)90297-q. [DOI] [PubMed] [Google Scholar]
- McIntire SL, Jorgensen E, Kaplan J, Horvitz HR. The GABAergic nervous system of Caenorhabditis elegans. Nature. 1993;364:337–341. doi: 10.1038/364337a0. [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KG, Alfonso A, Nguyen M, Crowell JA, Johnson CD, Rand JB. A genetic selection for Caenorhabditis elegans synaptic transmission mutants. Proc Natl Acad Sci USA. 1996;93:12593–12598. doi: 10.1073/pnas.93.22.12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SA, Schroder S, Plessmann U, Weber K, Ungewickell E. Clathrin assembly protein AP180: primary structure, domain organization and identification of a clathrin binding site. EMBO J. 1993;12:667–675. doi: 10.1002/j.1460-2075.1993.tb05700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M, Alfonso A, Johnson CD, Rand JB. Caenorhabditis elegans mutants resistant to inhibitors of acetylcholinesterase. Genetics. 1995;140:527–535. doi: 10.1093/genetics/140.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet, M.L. (1999). Visualization of synaptic specializations in live C. elegans using synaptic vesicle-GFP protein fusions. J. Neurosci. Methods (in press). [DOI] [PubMed]
- Nonet ML, Grundahl K, Meyer BJ, Rand JB. Synaptic function is impaired but not eliminated in C. elegans mutants lacking synaptotagmin. Cell. 1993;73:1291–1305. doi: 10.1016/0092-8674(93)90357-v. [DOI] [PubMed] [Google Scholar]
- Nonet ML, Meyer BJ. Early aspects of Caenorhabditis elegans sex determination and dosage compensation are regulated by a zinc-finger protein. Nature. 1991;351:65–68. doi: 10.1038/351065a0. [DOI] [PubMed] [Google Scholar]
- Nonet ML, Saifee O, Zhao H, Rand JB, Wei L. Synaptic transmission deficits in C. elegans synaptobrevin mutants. J Neurosci. 1998;18:70–80. doi: 10.1523/JNEUROSCI.18-01-00070.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet ML, Staunton J, Kilgard MP, Fergestad T, Hartweig E, Horvitz HR, Jorgensen E, Meyer BJ. C. elegans rab-3 mutant synapses exhibit impaired function and are partially depleted of vesicles. J Neurosci. 1997;17:8021–8073. doi: 10.1523/JNEUROSCI.17-21-08061.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris FA, Ungewickell E, Majerus PW. Inositol hexakisphosphate binds to clathrin assembly protein 3 (AP-3/AP180) and inhibits clathrin cage assembly in vitro. J Biol Chem. 1995;270:214–217. doi: 10.1074/jbc.270.1.214. [DOI] [PubMed] [Google Scholar]
- Palfrey HC, Artalejo CR. Vesicle recycling revisited: rapid endocytosis may be the first step. Neuroscience. 1998;83:969–989. doi: 10.1016/s0306-4522(97)00453-3. [DOI] [PubMed] [Google Scholar]
- Poodry C, Edgar L. Reversible alteration in the neuromuscular junctions of Drosophila melanogaster bearing a temperature-sensitive mutation. shibire. J Cell Biol. 1979;81:520–527. doi: 10.1083/jcb.81.3.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puszkin S, Perry D, Li S, Hanson V. Neuronal protein NP185 is developmentally regulated, initially expressed during synaptogenesis, and localized in synaptic terminals. Mol Neurobiol. 1992;6:253–283. doi: 10.1007/BF02780557. [DOI] [PubMed] [Google Scholar]
- Raizen DM, Avery L. Electrical activity and behavior in the pharynx of Caenorhabditis elegans. Neuron. 1994;12:483–495. doi: 10.1016/0896-6273(94)90207-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizen DM, Lee RYN, Avery L. Interacting genes required for pharyngeal excitation by motor neuron MC in Caenorhabditis elegans. Genetics. 1995;141:1365–1382. doi: 10.1093/genetics/141.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand JB, Nonet ML. Synaptic transmission. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 611–644. [PubMed] [Google Scholar]
- Rapoport I, Miyazaki M, Boll W, Duckworth B, Cantley LC, Shoelson S, Kirchhausen T. Regulatory interactions in the recognition of endocytic sorting signals by AP-2 complexes. EMBO J. 1997;16:2240–2250. doi: 10.1093/emboj/16.9.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig B, Liao LW, Hirsh D. Sequence of the C. elegans transposable element, Tc1. Nucleic Acids Res. 1983;11:4201–4209. doi: 10.1093/nar/11.12.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saifee O, Wei LP, Nonet ML. The C. elegans unc-64 gene encodes a syntaxin which interacts with genetically with synaptobrevin. Mol Biol Cell. 1998;9:1235–1252. doi: 10.1091/mbc.9.6.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcini A, Confalonieri S, Doria M, Santolini E, Tassi E, Minenkova O, Cesareni G, Pelicci P, Di Fiore P. Binding specificity and in vivo targets of the EH domain, a novel protein-protein interaction module. Genes Dev. 1997;11:2239–2249. doi: 10.1101/gad.11.17.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Book Molecular Cloning: A Laboratory Manual. 2nd edition. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Schiavo G, Benfenati F, Poulain B, Rossetto O, Polverino de Laureto P, DasGupta BR, Montecucco C. Tetanus and botulinum-B neurotoxins block neurotransmitter release by proteolytic cleavage of synaptobrevin. Nature. 1992;359:832–835. doi: 10.1038/359832a0. [DOI] [PubMed] [Google Scholar]
- Schmid SL. Clathrin-coated vesicle formation and protein sorting: an integrated process. Annu Rev Biochem. 1997;66:511–548. doi: 10.1146/annurev.biochem.66.1.511. [DOI] [PubMed] [Google Scholar]
- Smith D, Fisher PA. Identification, developmental regulation, and response to heat shock of two antigenically related forms of a major nuclear envelope protein in Drosophila embryos: applications of an improved method for affinity purification of antibodies using polypeptides immobilized on nitrocellulose blots. J Cell Biol. 1984;99:20–28. doi: 10.1083/jcb.99.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner T, Bennett MK, Whiteheart SW, Scheller RH, Rothman JE. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell. 1993b;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- Sollner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman JE. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993a;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- Stenius K, Janz R, Südhof TC, Jahn R. Structure of synaptogyrin (p29) defines novel synaptic vesicle protein. J Cell Biol. 1995;131:1801–1809. doi: 10.1083/jcb.131.6.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Südhof TC. The synaptic vesicle cycle: a cascade of protein-protein interactions. Nature. 1995;375:645–653. doi: 10.1038/375645a0. [DOI] [PubMed] [Google Scholar]
- Sulston J, Hodgkin J. Methods. In: Wood WB, editor. The Nematode Caenorhabditis elegans. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. pp. 587–606. [Google Scholar]
- Walch-Solimena C, Blasi J, Edelmann L, Chapman ER, von Mollard GF, Jahn R. The t-SNAREs syntaxin 1 and SNAP-25 are present on organelles that participate in synaptic vesicle recycling. J Cell Biol. 1995;128:637–645. doi: 10.1083/jcb.128.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L-H, Südhof TC, Anderson RGW. The appendage domain of α-adaptin is a high affinity binding site for dynamin. J Biol Chem. 1995;260:10079–10083. doi: 10.1074/jbc.270.17.10079. [DOI] [PubMed] [Google Scholar]
- Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Sollner TH, Rothman JE. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- Wendland B, Emr SD. Pan1p, yeast eps15, functions as a multivalent adaptor that coordinates protein-protein interactions essential for endocytosis. J Cell Biol. 1998;141:71–84. doi: 10.1083/jcb.141.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye W, Ali N, Bembenek ME, Shears SB, Lafer EM. Inhibition of clathrin assembly by high affinity binding of specific inositol polyphosphates to the synapse-specific clathrin assembly protein AP-3. J Biol Chem. 1995;270:1564–1568. [PubMed] [Google Scholar]
- Ye W, Lafer EM. Bacterially expressed F1–20/AP-3 assembles clathrin into cages with a narrow size distribution: implications for the regulation of quantal size during neurotransmission. J Neurosci Res. 1995a;41:15–26. doi: 10.1002/jnr.490410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye W, Lafer EM. Clathrin binding and assembly activities of expressed domains of the synapse-specific clathrin assembly protein AP-3. J Biol Chem. 1995b;270:10933–10939. doi: 10.1074/jbc.270.18.10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Koh YH, Beckstead RB, Budnik V, Ganetzky B, Bellen HJ. Synaptic vesicle size and number are regulated by a clathrin adaptor protein required for endocytosis. Neuron. 1998;21:1465–1475. doi: 10.1016/s0896-6273(00)80664-9. [DOI] [PubMed] [Google Scholar]
- Zhang JZ, Davletov BA, Südhof TC, Anderson RGW. Synaptotagmin I is a high affinity receptor for clathrin AP-2: implications for membrane recycling. Cell. 1994;78:751–760. doi: 10.1016/s0092-8674(94)90442-1. [DOI] [PubMed] [Google Scholar]
- Zhou S, Tannery NH, Yang J, Puszkin S, Lafer EM. The synapse-specific phosphoprotein F1–20 is identical to the clathrin assembly protein AP-3. J Biol Chem. 1993;268:12655–12662. [PubMed] [Google Scholar]