Kinesin-II Is Preferentially Targeted to Assembling Cilia and Is Required for Ciliogenesis and Normal Cytokinesis in Tetrahymena (original) (raw)
Abstract
We cloned two genes, KIN1 and KIN2, encoding kinesin-II homologues from the ciliate Tetrahymena thermophila and constructed strains lacking either KIN1 or KIN2 or both genes. Cells with a single disruption of either gene showed partly overlapping sets of defects in cell growth, motility, ciliary assembly, and thermoresistance. Deletion of both genes resulted in loss of cilia and arrests in cytokinesis. Mutant cells were unable to assemble new cilia or to maintain preexisting cilia. Double knockout cells were not viable on a standard medium but could be grown on a modified medium on which growth does not depend on phagocytosis. Double knockout cells could be rescued by transformation with a gene encoding an epitope-tagged Kin1p. In growing cells, epitope-tagged Kin1p preferentially accumulated in cilia undergoing active assembly. Kin1p was also detected in the cell body but did not show any association with the cleavage furrow. The cell division arrests observed in kinesin-II knockout cells appear to be induced by the loss of cilia and resulting cell paralysis.
INTRODUCTION
Kinesins-II are microtubule-dependent motors that exist as heterotrimeric complexes in diverse organisms (Cole et al., 1993, 1998; Yamazaki et al., 1995). Members of the kinesin-II family have been implicated in axonal transport, assembly of cilia and flagella, and transport of pigment granules (Walther et al., 1994; Yamazaki et al., 1995; Morris and Scholey, 1997; Rogers et al., 1997). Kinesin-II is essential for assembly of cilia and flagella. A mutation in the Chlamydomonas kinesin-II motor, FLA10, resulted in a loss of flagella (Walther et al., 1994). Injection of anti-kinesin-II antibodies into blastula-stage sea urchin embryos partially blocked assembly of cilia (Morris and Scholey, 1997). Furthermore, a knockout of essential murine kinesin-II genes leads to a complete block in the assembly of monocilia on embryonic nodal cells (Nonaka et al., 1998; Marszalek et al., 1999; Takeda et al., 1999). In Chlamydomonas, video-enhanced microscopy showed bidirectional movement of particles inside flagella. These membraneless particles, named rafts, were proposed to be involved in delivery of flagellar subunits from the cell body to the tips of growing flagella (Kozminski et al., 1993; Rosenbaum et al., 1999). Importantly, FLA10 activity is required to maintain intraflagellar transport (Kozminski et al., 1995). Kinesin-II motor subunits supported microtubule plus end-directed motility in vitro (Cole et al., 1993; Kondo et al., 1994), and kinesin-II was found highly enriched in cilia and flagella (Walther et al., 1994; Cole et al., 1998; Nonaka et al., 1998). Recently, a green fluorescent protein (GFP)-tagged subunit of a kinesin-II complex was observed to move inside the chemosensory cilia of living Caenorhabditis elegans, primarily in the anterograde direction toward the distal tips of axonemes (Orozco et al., 1999). Because incorporation of new axonemal subunits is known to take place at the tips of assembling flagella, which correspond to the plus ends of axonemal microtubules (Johnson and Rosenbaum, 1992), kinesin-II is a strong candidate for an anterograde motor transporting flagellar components, possibly in association with rafts, from the cell body to the tips of flagella. The retrograde flagellar movement appears to be essential for flagellar assembly (Piperno et al., 1998) and is most likely powered by a cytoplasmic dynein complex (Pazour et al., 1998; Porter et al., 1999).
In nerve cells kinesin-II is associated with membrane-bounded organelles (Yamazaki et al., 1995; Muresan et al., 1998; Yang and Goldstein, 1998). Thus, it is most likely that kinesin-II complexes transport different types of cellular cargo in axons and flagella. Most kinesin-II motors exist as heterotrimeric complexes containing two nonidentical motor subunits and a third accessory nonmotor subunit. Because kinesin-II is the only kinesin-related protein complex known to contain nonidentical motor subunits, heterodimerization may be used to create combinatorial motors that have different cargo specificities. In mouse and rat, kinesin-II was found in two complexes containing one common motor subunit (KIF3A) and one of two variable motor subunits (KIF3B or KIF3C) (Muresan et al., 1998; Yang and Goldstein, 1998). Furthermore, three isoforms of the kinesin-II-associated subunit KAP3 were identified in mouse (Yamazaki et al., 1995, 1996). In C. elegans three kinesin-II motor subunits were identified, which form at least one heterotrimeric complex and one dimeric complex (Signor et al., 1999). Combinatorial kinesin-II complexes could be formed by mixing and matching of different motor and nonmotor subunits that produce functionally distinct motors in different cells.
We and others have recently identified three kinesin-II homologous genes (KIN1, KIN2, and KIN5) in the unicellular organism the ciliate Tetrahymena thermophila (this study; Bernstein, personal communication). Thus, combinatorial interactions may be used to generate multiple, functionally distinct variants of kinesin-II within a single cell. Here we describe the cloning and functional analysis of two members of the kinesin-II family of T. thermophila, KIN1 and KIN2. Our analyses show that KIN1 and KIN2 genes have overlapping but nonidentical functions. Either KIN1 or KIN2 is required for assembly and maintenance of cilia, and kinesin-II encoded by the KIN1 gene preferentially accumulates in cilia that undergo active assembly. Surprisingly, the mutants lacking both KIN1 and KIN2 genes frequently fail to complete cytokinesis. Multiple lines of evidence indicate that the cytokinesis phenotype in kinesin-II mutants is induced by the loss of cilia and resulting cell paralysis.
MATERIALS AND METHODS
Strains, Culture Growth, and Conjugation
Strains used are described in Table 1. Tetrahymena cell cultures were grown in 50 ml of either SPP (1% proteose peptone, 0.2% glucose, 0.1% yeast extract, 0.003% EDTA·ferric sodium salt) (Gorovsky, 1973) or MEPP medium (2% proteose peptone, 2 mM Na3 citrate·2 H2O, 1 mM ferric chloride, 12.5 μM cupric sulfate, 1.7 μM folinic acid, Ca salt) (Orias and Rasmussen, 1976) supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin, and 0.25 μg/ml amphotericin B in 250-ml Erlenmayer flasks with moderate shaking at 30°C. To induce conjugation, two strains of different mating types were grown to midlogarithmic phase, and 50 ml of each strain were washed two times and left in the starvation medium (10 mM Tris-Cl, pH 7.5) in the original volume. After 16–20 h, equal numbers of cells (1.5 × 107 cells of each strain) were mixed in a total volume of 100 ml in a 2-l Erlenmeyer flask and left unshaken at 30°C.
Table 1.
Strains used in this study
| Strain | Micronuclear genotype | Macronuclear genotype | Macronuclear phenotype (known drug resistances, mating types) |
|---|---|---|---|
| B2086.1 | WT | WT | II |
| CU427.3 | chx1-1/chx1-1 | WT | cy-s, VI |
| CU428.1 | mpr1-1/mpr1-1 | WT | mp-s, VII |
| B*VII | nonfunctional mic (star) | WT | VII |
| UG1 | kin1∷neo2/kin1∷neo2 | kin1∷neo2 | pm-s |
| UG2 | kin1∷neo2/kin1∷neo2 | kin1∷neo2 | pm-s |
| UG3 | WT | WT | |
| UG4 | WT | WT | |
| UG7 | kin2∷bsr1/kin2∷bsr1 | kin2∷bsr1 | bs-r |
| UG8 | kin2∷bsr1/kin2∷bsr1 | kin2∷bsr1 | bs-r |
| UG9 | WT | WT | |
| UG10 | WT | WT | |
| UG11 | WT | kin2∷neo2 | pm-r |
| UG12 | WT | kin2∷neo2 | pm-r |
| UG13 | kin1∷neo2/kin1∷neo2, kin2∷bsr1/kin2∷bsr1, | KIN2/kin2∷bsr1 | bs-r, pm-s, mp-s |
| UG14 | kin1∷neo2/kin1∷neo2, kin2∷bsr1/kin2∷bsr1, mpr1-1/mpr1-1 | WT | bs-s, pm-s, mp-s |
Gene Cloning and Sequence Analysis
PCR was used to amplify kinesin-related protein (KRP) sequences of T. thermophila. Total genomic DNA isolated using the fast urea extraction method (Gaertig et al., 1994b) was used as a template. Three types of degenerate primers were used to amplify sequences encoding the most conserved peptides of motor domains present in other KRPs. Primer A, 5′-AT(T/C)TT(T/C)GC(T/C)TA(T/C)GG(T/A)(T/C)A(A/G)AC-3′, encodes a sense strand of IFAYGQT. Primer B, 5′-G(A/T)(A/T)CC(A/G)GC(T/A)A(A/G)(A/G)TC(A/G)AC-3′, encodes an antisense sequence of LVDLAGSE. Primer C, 5′-CTTAGA(G/A)T(T/C)TCT(G/A)(T/A)A(G/A)GG(A/G)AT(G/A)TG-3′, encodes an antisense sequence of the peptide HIP(Y/F)RDSK. Total genomic DNA was amplified using primers A and C and amplified again with primers A and B. Final products of ∼400 bp were cloned. To clone genomic fragments of KIN1 and KIN2, total T. thermophila DNA was digested with restriction endonucleases and used to prepare a Southern blot. The PCR-generated KIN1 and KIN2 fragments were labeled with [α32P]dATP using random hexamer primers and used as probes. The KIN1 gene was cloned as a 3.5-kb _Hin_dIII fragment and a partially overlapping 2.5-kb _Eco_RI–_Xba_I fragment. The majority of KIN2 was cloned as partly overlapping 2.5-kb _Hin_dIII and 2-kb Csp45 I–_Bgl_II restriction fragments. The 3′ end of the KIN2 gene was cloned using rapid amplification of cDNA ends (Frohman, 1990). Protein secondary structure was predicted using NNPREDICT (Kneller et al., 1990). The probability of coiled coil formation was calculated using COILS (Lupas et al., 1991). Sequence homology searches were done using BLAST from the National Center for Biotechnology Information (Bethesda, MD; Altschul et al., 1990). Protein sequence comparison was done using BESTFIT, COMPARE, and DOTPLOT programs of the University of Wisconsin Genetics Computer Group (UWGCG; Madison, WI) Wisconsin Package. Alignments of multiple sequences were prepared using PILEUP; evolutionary distances between sequences were calculated using DISTANCES; and an evolutionary tree was made using GROWTREE of the UWGCG package (Devereux et al., 1984).
Germ Line and Somatic Gene Knockouts
To construct a targeting fragment for disruption of the KIN1 gene, plasmid pKIN17-7 containing the 3.5-kb _Hin_dIII fragment of KIN1 was linearized at its single _Bgl_II site within the region encoding a motor domain. The protruding _Bgl_II ends were filled in using T4 DNA polymerase. The neo2 disruption cassette (Gaertig et al., 1994a) was inserted into the _Bgl_II site of pKIN17-7 to give pKIN17-7neo. To construct a gene disruption fragment for KIN2, plasmid pCS4, containing a 5-kb fragment of KIN2, was linearized at the Csp45 I site present in the coding region. The bsr1 gene cassette was inserted into the Csp45 I site. This cassette is a derivative of the neo2 gene cassette in which a blasticidin S (bs) resistance gene (Sutoh, 1993) is inserted between the Tetrahymena histone HHF1 promoter and the BTU2 transcription terminator (Gaertig et al., 1994a). To disrupt either the KIN1 or KIN2 gene in the germline micronucleus (MIC) we introduced transforming DNA into early mating cells using the biolistic gun (Cassidy-Hanley et al., 1997). For disruption of KIN1, 4 μg of pKIN17-7neo DNA that had been digested with _Hin_dIII were used to coat 1 mg of tungsten M10 (Bio-Rad, Hercules, CA) particles (Sanford et al., 1991). For disruption of KIN2, 4 μg of pCS4bsr1 plasmid DNA that had been linearized with _Kpn_I and _Sac_I were used to coat gold particles (1 μm, Bio-Rad). Strains CU428.1 and B2086.1 were allowed to conjugate for 2.5–3.5 h before bombardment. For KIN1 disruption, bombarded cells were incubated in SPP for 12 h at room temperature, and transformants were selected in SPP with 120 μg/ml paromomycin. For KIN2 disruption, bombarded cells were incubated for 7 h at 30°C in SPP, left for 14–16 h at room temperature, and selected in SPP with 60 μg/ml bs. Transformants heterozygous for KIN1/kin1::neo2 or KIN2/kin2::bsr1 were identified and brought to homozygosity in the MIC as described (Cassidy-Hanley et al., 1997). To construct strains lacking all copies of both KIN1 and KIN2 in their MICs, we crossed a strain homozygous for the kin1::neo2 gene to a strain homozygous for the kin2::bsr1 gene and selected double heterozygotes resistant to both paromomycin and bs. These were crossed to the B*VII strain (Orias and Bruns, 1976) to generate micronuclear homozygotes, and the exconjugants from this cross were reisolated, grown, and crossed to a CU427.3 strain. Double knockout heterokaryons strains were identified (UG13 and UG14) that have different mating types.
Phenotypic Analyses
Growth rates were measured in SPP medium without shaking by counting cells periodically using a Coulter Electronics (Hialeah, FL) model ZF counter. Dead cells were identified by trypan blue exclusion test by adding 30 μl of 0.4% trypan blue (Sigma, St. Louis, MO) to 70 μl of cells in culture medium.
To measure the rate of cell movement, 40 μl of cells from a growing culture were placed on a slide and analyzed in a hanging drop, using an Olympus Optical (Tokyo, Japan) inverted microscope and 4× phase-contrast objective. Images of moving cells were recorded using a video camera and image capture software. Distances traveled by moving cells were measured using NIH Image version 1.62. To analyze the rate of ciliary regeneration, cells were grown to a density of 3 × 105 cells/ml, starved for 24 h in 10 mM Tris-HCl, pH 7.5, and deciliated (Calzone and Gorovsky, 1982). Cilia regeneration was monitored by determining the fraction of motile cells.
To bring the double knockout phenotype to expression, heterokaryon strains UG13 and UG14 were crossed to each other. Nine to 10 h after mixing, individual pairs were isolated into drops of SPP or MEPP medium and left for 13–15 h at room temperature. As controls, wild-type exconjugants or parental cells were isolated. The cell number in each drop was determined by counting live cells under an inverted microscope. Observations of live dividing cells were done on the Zeiss (Thornwood, NY) Axioscope microscope using differential interference contrast optics with the Plan-NeoFluar 40× (numerical aperture, 0.75) lens. The images were recorded on the DAGE-MTI (Michigan City, IN) DC330 charge-coupled device camera. The s-video output of the camera was fed directly into the Macintosh G3/AV All-in-one computer using the Apple (Cupertino, CA) Video Player software.
Immunocytochemistry and Electron Microscopy
For staining double knockout cells, ∼500–1000 cells were isolated into 30 μl of 10 mM Tris, pH 7.5, on a coverslip. An equal volume of 2% paraformaldehyde, 0.5% Triton X-100, and 1 μM paclitaxel in PHEM (60 mM PIPES, pH 6.9, 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2) was added, and samples were air dried at 30°C. Coverslips were processed for immunofluorescent labeling as described (Gaertig et al., 1995), using rabbit polyclonal SG serum raised against Tetrahymena total tubulin (Guttman and Gorovsky, 1979) at 1:100 dilution. Cells expressing the epitope-tagged Kin1p were double labeled using the TAP952 mouse monoclonal antibodies directed against the monoglycylated isoforms of tubulins (Callen et al., 1994) and the anti-GFP rabbit polyclonal antibodies (Clontech, Palo Alto, CA) at 1:100 and 1:400 dilution, respectively. Secondary antibodies were goat anti-mouse FITC (Sigma), goat anti-rabbit-Cy3, and goat-anti-rabbit-FITC (Zymed, San Francisco, CA) conjugates, and all were used at 1:100 dilution. Cells were viewed using a Bio-Rad MRC 600 confocal microscope. The length of axonemes either on cells or isolated was determined on confocal sections using NIH Image software version 1.62. To increase the consistency of analysis of ciliary axoneme lengths on whole cells, for each cell analyzed a single confocal section was chosen, which included the widest cross-section of the macronucleus that could be found in that z-series.
For electron microscopy, Cells were washed with 10 mM Tris, pH 7.5, and fixed in 2% glutaraldehyde in 100 mM cacodylate buffer at 4°C for 1 h, incubated in 2.5% sucrose in 100 mM cacodylate for 20 min, and postfixed in 1% osmium tetroxide in 100 mM cacodylate for 1.5 h at 4°C. Cells were embedded in Epon after dehydration in graded steps from 30 to 100% ethanol. Sections were stained with uranyl acetate and lead citrate and were visualized on a JEOL (Tokyo, Japan) 100CXII transmission electron microscope.
Construction of Epitope-tagged Targeting Fragments and Rescue Transformation
The Muta-Gene phagemid in vitro mutagenesis kit (Bio-Rad) was used to create _Mlu_I (5′) and _Nco_I (3′) sites near the N terminus of the KIN1 coding sequence on the plasmid pKIN17-7 to construct pKIN17NM. The sequence of the mutagenic oligonucleotide, KINL-MC, is 5′-TTT ACT ATT TTT TTC CAT GGC TTC TAC GCG TTT GCT CAT TAT ACT T-3′. The 5xMyc insert was prepared by amplifying the plasmid pJR1265 (kindly provided by Dr. K. Kozminski, University of California, Berkeley, CA) with the primers MYC-ML, 5′-GGA CGC GTC TTT AAA GCT ATG GAG CAA AAG-3′, and SK, 5′-TCT AGA ACT AGT TGG ATC-3′. After digesting with _Mlu_I and _Nco_I it was inserted into the corresponding sites of pKIN17NM to construct pKIN17myc-6. To prepare a GFP-tagged Kin1 gene, the GFP sequence was amplified from pH4.GFP1 plasmid (kindly provided by Dr. A. Turkewitz, University of Chicago, Chicago, IL; Haddad and Turkewitz, 1997) using primers GFP5′Ml, 5′-GACGCGTAATGAGTAAAGGAGAAGAAC-3′, and GFP3′Bsp, 5′-GTCATGATTTTGTATAGTTCATCCATGC. The PCR product was digested with _Mlu_I and _Bsp_HI and subcloned between the _Mlu_I and _Nco_I sites of the pKIN17myc-6 in place of the Myc epitope tag to give pKIN17gfp plasmid. For preparation of rescuing DNA, pKIN17-7, pKIN17myc-6, and pKIN17gfp were digested with _Hin_dIII, and pCS4 was digested with _Sac_I and _Kpn_I. Double knockout heterokaryon strains UG13 and UG14 were starved, mixed, and transformed by biolistic bombardment at 10 h after mating. Rescued conjugation progeny were selected with 15 μg/ml 6-methylpurine.
Cell Fractionation and Western Blotting
Cells were grown to 4 × 105/ml at 30°C in 300 ml of SPP, washed once with 10 mM Tris, pH 7.5, and suspended in 15 ml of 10 mM Tris, pH 7.5, containing protease inhibitors (1 mM PMSF, 1 μg/ml pepstatin A, 1 μg/ml leupeptin, 10 μg/ml chymostatin, 10 μg/ml antipain, 5 μg/ml E-64; all inhibitors from Sigma). Dibucaine (Sigma) was added at a final concentration of 3 mM. When most cells had stopped moving (∼2 min), 5 vol of ice-cold 10 mM Tris, pH 7.5, were added. All subsequent procedures were performed at 4°C. Cell bodies were sedimented by centrifugation at 1100 × g for 5 min. The supernatant was collected, and the centrifugation was repeated. Cilia were collected by centrifugation at 14,000 × g for 20 min, washed once in wash buffer (10 mM HEPES, pH 7.4, 100 mM NaCl, 4 mM MgCl2, 0.1 mM EGTA) containing protease inhibitors, and recentrifuged at 14,000 × g. Cell bodies were washed once in 10 mM Tris, pH 7.5, and recentrifuged at 1100 × g. Pellets containing cell bodies or cilia were weighed and resuspended at 100 mg of wet pellet/ml in either 10 mM Tris, pH 7.5, plus protease inhibitors (cell bodies) or wash buffer plus protease inhibitors (cilia). To obtain axonemes, 10 mg of cilia were extracted with wash buffer containing 0.5% NP-40 on ice for 10 min and centrifuged at 14,000 × g. The supernatant, containing membranes, was retained. The axonemal fraction was resuspended in the original volume of wash buffer plus protease inhibitors. Either 500 μg of cilia or axonemes or NP-40-soluble fraction (each obtained from 500 μg of cilia) were subjected to 8% SDS-PAGE and transferred onto a 0.45-μm Trans-Blot nitrocellulose membrane (Bio-Rad) by semidry electroblotting using the Trans-Blot SD cell (Bio-Rad) at 20 V for 2 h. Filters were blocked in 5% dry milk, 0.1% Tween 20, and 1× PBS for 1.5 h and incubated overnight at 4°C in reaction buffer (1% dry milk, 0.1% Tween 20 in 1× PBS) with anti-Myc hybridoma supernatant (clone 9E10; American Type Culture Collection, Manassas, VA) at 1:2.5 dilution, rabbit anti-histone hv1 serum at 1:10,000 dilution, or mouse AXO49 antibodies at 1:3000 dilution. Membranes were washed in PBST (1× PBS, 0.1% Tween 20) at room temperature, incubated in reaction buffer with 1:1500 dilution of either anti-mouse immunoglobulin G or anti rabbit immunoglobulin HRP-linked goat antibodies (Amersham, Arlington Heights, IL) for 1.5 h, and washed once for 15 min, two times for 5 min each in PBST, and then three times for 5 min each in 1× PBS. Blots were developed using the ECL Western blotting analysis system (Amersham).
RESULTS
Cloning of KIN1 and KIN2, Members of the Kinesin-II Family, in T. thermophila
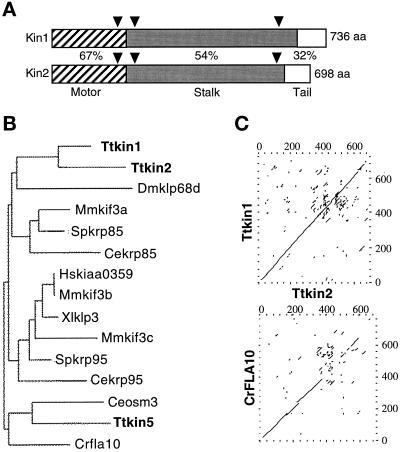
We identified fragments of four putative KRP genes (KIN1–KIN4) using PCR of total genomic DNA of T. thermophila. We cloned genomic restriction fragments of KIN1 and KIN2, and sequence analysis revealed open reading frames encoding proteins of 735 and 697 amino acids, respectively, that were most similar to kinesin-II. The predicted KIN1-encoded protein (Kin1p) has a calculated molecular mass of 85.04 kDa and a pI of 7.61, whereas Kin2p has a calculated molecular mass of 82 kDa and a pI of 6.88. By comparing genomic sequences with the corresponding cDNAs, we identified three small introns in each gene. Two of the three introns of KIN1 and KIN2 are located at the same positions within the coding sequences (Figure 1A). The two genes are genetically linked in the micronuclear genome (our unpublished results). Both predicted proteins have an expected structure of a kinesin-II heavy chain, with a central region of α-helical coiled coil flanked by globular N- and C-terminal motor and tail regions, respectively (Figure 1A). By comparing the Kin1p and Kin2p sequences with kinesin heavy chain of Drosophila melanogaster (Yang et al., 1989), we identified a kinesin motor domain within the N-terminal globular domains of both proteins. Phylogenetic analysis showed that the motor domains of Kin1p and Kin2p are more related to each other (67% identical) than to any other known kinesin-II (60–63%) (Figure 1B). KIN1 and KIN2 also showed significant homology (54%) between their rod domains and lower levels of homology with the rod domains of other kinesins-II (Figure 1C). The tails of KIN1 and KIN2 are very basic but showed only weak homology to each other (32% sequence identity).
Figure 1.
KIN1 and KIN2 are kinesin-II homologous genes in Tetrahymena. (A) Diagram of the alignment of predicted KIN1 and KIN2 encoded protein sequences. Domains are indicated as follows: stripes, motor domain; gray, coiled coil stalk; white, globular tail. Percent identity between Kin1p and Kin2p in each domain is indicated. Triangles represent positions of introns in the corresponding genomic DNA. (B) Phylogram of kinesin-II proteins. Alignments of multiple sequences were prepared using PILEUP; evolutionary distances between sequences were calculated using DISTANCES; and an evolutionary tree was made using GROWTREE of the UWGCG system (Devereux et al., 1984). (C) Sequence comparison between KIN1 and KIN2 and between KIN2 and Chlamydomonas reinhardtii FLA10 (Walther et al., 1994). Dot matrix analysis was done using COMPARE and DOTPLOT programs of UWGCG. Sequence data used for comparisons are available from European Molecular Biology Laboratory, GenBank, and DNA Data Bank of Japan under accession numbers AJ244020 (KIN1), AJ244021 (KIN2), L33697 (Crfla10), D14968 (Ceosm3), AB002357 (Hskiaa0359), A57107 (Mmkif3b), C48835 (Xlklp3), AF013116 (Mmkif3c), U00996 (Spkrp95), U15974 (Dmklp68d), D12645 (Mmkif3a), and L16993 (Spkrp85). The sequence of Ttkin5 was provided by M. Bernstein (personal communication). The Cekrp85 and Cekrp95 sequences were identified by the C. elegans sequencing project (Signor et al., 1999).
Cells Lacking Either KIN1 or KIN2 Show Deficiencies in Cell Growth, Motility, Ciliary Assembly, and Thermoresistance
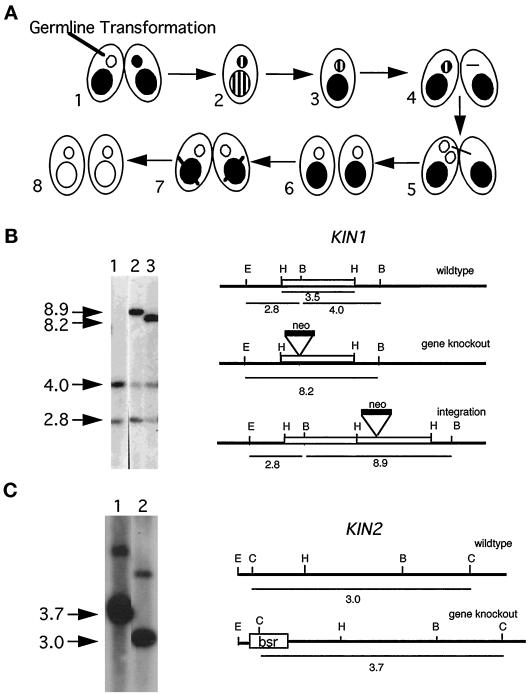
Tetrahymena cells, like most other ciliates have two nuclei, the germ line, transcriptionally silent MIC and the somatic, transcriptionally active macronucleus (MAC). To disrupt KIN1 in both the MIC and MAC, we used a fragment in which the neo2 gene was inserted into the motor domain. We biolistically transformed the MICs of early conjugating cells (Figure 2A; see MATERIALS AND METHODS). Southern blotting showed that one transformant contained a disrupted KIN1 gene (Figure 2B), whereas in the second transformant, the kin1::neo2 fragment integrated into the 3′ flanking region of KIN1. Homozygotes for the disrupted allele in the MIC and MAC were constructed using two rounds of “genomic exclusion” (Figure 2A; see MATERIALS AND METHODS). We used essentially the same strategy to disrupt KIN2, except that we used the bsr1 gene as a disruption cassette instead of neo2 (Figure 2C).
Figure 2.
Germ line disruption of KIN1 and KIN2. (A). Genetic scheme for creation of homozygous single knockout strains. (1 and 2) During mating between two wild-type strains the targeted gene is disrupted in the MIC by biolistic transformation. The resulting progeny are heterozygous for the null allele in both the MAC and the MIC. (3) During vegetative growth heterozygous clones become homozygous for the wild-type allele in the MAC because of random segregation of alleles in the amitotic MAC (phenotypic assortment). (4–6) A strain heterozygous for the disrupted allele in the MIC is crossed to a star strain lacking a functional MIC, resulting in the transfer of a haploid MIC containing only a disrupted allele of the target gene. Endoreplication produces a diploid MIC yielding knockout heterokaryon cells. (7 and 8) Conjugation of two knockout heterokaryons leads to formation of a new MAC containing only disrupted copies of the targeted gene. (B) Analysis of germ line KIN1::neo2 transformants. Left panel, Southern blot of total genomic DNAs digested with _Eco_RI and _Bgl_II and probed with a radiolabeled KIN1 fragment. Right panel, diagram of the KIN1 locus. Lane 1, control strain DNA; lanes 2 and 3, independent germline transformants. The endogenous locus gives two fragments of 2.8 and 4 kb. A gene knockout is expected to give a single 8.2-kb fragment, and a 3′ integration should give 2.8- and 8.9-kb fragments. Note that the restriction patterns of transformant DNA analyzed in lane 3 is consistent with gene replacement, whereas in the transformant DNA analyzed in lane 2 the KIN1::neo2 fragment integrated into the 3′ flanking region of KIN1 gene. The endogenous 4.0-kb fragment is found in the original germ line transformants, indicating that these clones are heterozygous for KIN1/kin1::neo2. (C) Left panel, Southern blot of total genomic DNAs digested with Csp45 I and probed with a radiolabeled KIN2 genomic fragment. Right panel, diagram of the KIN2 locus. Lane 1, a transformant strain homozygous for KIN2::bsr1 gene; lane 2, wild-type control. The endogenous KIN2 gives a fragment of 3.0 kb. Disrupted KIN2 gives a fragment of 3.7 kb. The minor upper band present in both lanes most likely represents an incompletely digested KIN2 fragment.
Strains lacking either KIN1 or KIN2 were viable, morphologically normal, and motile but multiplied more slowly than wild-type (WT) strains, with the doubling times that were 23 and 86% longer than WT, respectively (Table 2). Furthermore, single knockout strains showed decreased survival rate when exposed to higher temperatures. After 45–47 h of incubation at 39–40°C, ΔKIN1 and ΔKIN2 cells had viabilities that were 50 and 73% of WT viabilities, respectively (Table 2).
Table 2.
Analysis of single knockout phenotypes
| Phenotype | Δ_KIN1_ | Control for KIN1 | Δ_KIN2_ | Control for KIN2 |
|---|---|---|---|---|
| Doubling time 30°C (min) | 447 ± 36 | 362 ± 31 | 608 ± 144 | 327 ± 50 |
| Viability 38–40°C (%) | 27 ± 8 | 53 ± 22 | 69 ± 8 | 94 ± 4 |
| Swimming rate 30°C (μm/sec) | 93 ± 29 | 139 ± 39 | 158 ± 38 | 166 ± 38 |
| Cilia regeneration t1/2 30°C (min) | 86 ± 17 | 74 ± 22 | 50 ± 20 | 52 ± 30 |
| Cilia regeneration t1/2 38°C (min) | 128 ± 23 | 70 ± 13 | 71 ± 37 | 60 ± 32 |
ΔKIN1 cells swim more slowly than control WT cells (Table 2). When grown for 1 d before measurement at 30°C, the average speed of ΔKIN1 was 67% of WT. ΔKIN2 cells did not show any defect in motility rate (Table 2). At 30°C, ΔKIN1 cells showed a nearly normal rate of cilia formation after deciliation (Table 2). However, at 38–40°C, ΔKIN1 cells regenerated cilia with a half-time of recovery almost twice that of WT cells. ΔKIN2 showed no difference from WT at 30 or 39°C.
Either KIN1 or KIN2 Is Required for Assembly of Cilia and Normal Cytokinesis
To test for synthetic interactions between KIN1 and KIN2, we created double knockout heterokaryons lacking KIN1 and KIN2 in their MICs and having WT alleles in their MACs. To bring the double knockout (ΔKIN1ΔKIN2) phenotype to expression in the MACs, we crossed heterokaryon strains to each other. As a control, we also mated cells having only WT copies. ΔKIN1ΔKIN2 pairs separated at the proper time, indicating that zygotic expression of neither Kin1p nor Kin2p is required for the completion of conjugation. When refed ΔKIN1ΔKIN2 cells multiplied more slowly than WT controls. By 36 h after pair isolation, the average number of cells per drop (cells derived from a single mating pair) was 27.9 ± 32.0 and 91.6 ± 25.0 for ΔKIN1ΔKIN2 and WT cells, respectively (n = 41 and 47, respectively). On a standard culture medium (SPP), WT cells continued to grow to maximal density, whereas after 84 h, all ΔKIN1ΔKIN2 cells failed to multiple further and died out within a few days.
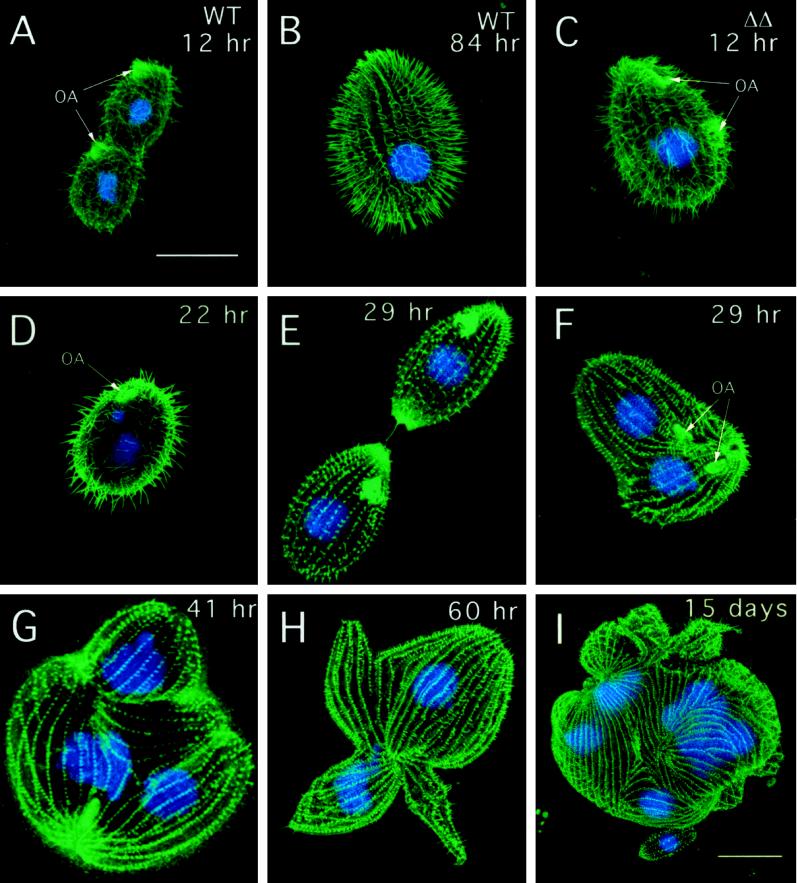
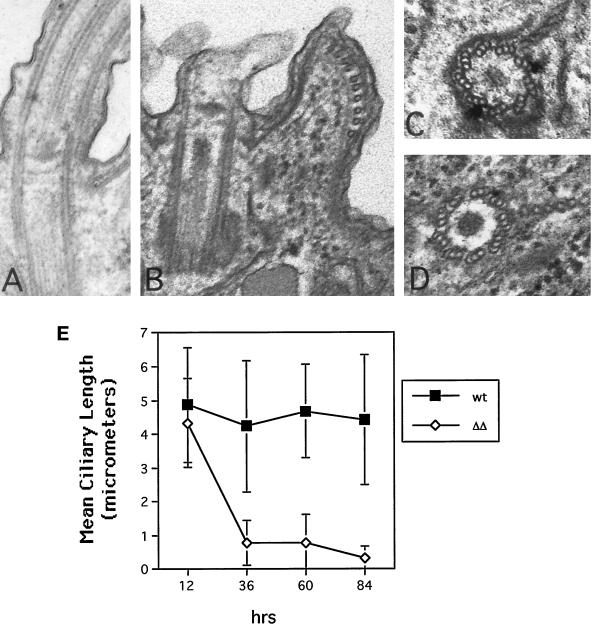
The most striking difference in the ΔKIN1ΔKIN2 cells compared with single knockouts was their progressive loss of motility. By 36 and 84 h after pair isolation, 71.4 and 100% of ΔKIN1ΔKIN2 drops (n = 41) contained no motile cells, respectively, compared with only 2% of control isolates at 84 h (n = 47). Confocal analysis revealed that ΔKIN1ΔKIN2 cells contained fewer normal-length ciliary axonemes (Figure 3), with most of the decrease occurring between 12 and 36 h (Figure 4E). During the Tetrahymena cell cycle, locomotory cilia are not resorbed before cell division. Instead, full-length cilia are retained, whereas new cilia are assembled from new basal bodies that appear near the existing basal bodies (Dippell, 1968). At 22 h after pair isolation we observed ΔKIN1ΔKIN2 cells with axonemes that were highly heterogeneous in length (Figure 3D). Further axoneme shortening occurred between 22 and 84 h, leading to axonemes with a mean length of 7.7% of the length in WT (Figures 3, E–H, and 4E). By 29 h most cells have undergone only zero to four cell divisions and therefore would normally contain 100–6.25% of the preexisting cilia transmitted from the exconjugants. However, at 29 h, many cells already completely lacked cilia of normal length (Figure 3, E and F). Thus, kinesin-II is required for both assembly of new cilia and maintenance of preexisting cilia. Transmission electron micrographs revealed that at 84 h most cilia contained only extremely short remnants of outer doublets and were covered by a small bulge of the plasma membrane (Figure 4, A and B). Cross-sections, in contrast, showed normal structure of basal bodies in mutants (Figure 4, C and D).
Figure 3.
Cytological analysis of ΔKIN1ΔKIN2. The phenotype was brought to expression by mating of double knockout heterokaryon strains (UG13 and UG14). Cells shown in A–H were grown in SPP medium, whereas the cell shown in I was grown in MEPP medium. Individual conjugation progeny cells were isolated and prepared for immunofluorescent confocal microscopy by staining with anti-tubulin antibodies (SG) and DAPI. (A) Dividing WT control 12 h after pair isolation. (B) WT control (84 h). (C) ΔKIN1ΔKIN2 cell (12 h) undergoing an early stage of cell division. The oral apparatus is already duplicated. (D) ΔKIN1ΔKIN2 cell (22 h) showing cilia heterogeneous in length. (E) ΔKIN1ΔKIN2 cell (29 h) at a final stage of cytokinesis. (F) ΔKIN1ΔKIN2 cell (29 h) with unseparated daughter cells and two sets of nuclei. (G and H) ΔKIN1ΔKIN2 cells (41 and 60 h) with uniformly short cilia, multiple nuclei, and multiple cortical subcells. (I) ΔKIN1ΔKIN2 cell 15 d after isolation of pairs grown in MEPP medium. Bars, 25 μm (bar in A shows scale for A–H). OA, oral apparatus.
Figure 4.
(A–D) Electron microscopic analysis of WT and ΔKIN1ΔKIN2 cells. (A) Longitudinal section through WT axoneme. (B) Longitudinal section through axonemal remnant of a ΔKIN1ΔKIN2 cell. (C and D) Cross-sections through WT and double knockout basal bodies, respectively. Cells were processed for thin-section electron microscopy 84 h after pair isolation. (E) Analysis of the length of ciliary axonemes in ΔKIN1ΔKIN2 and WT cells, measured on confocal sections of cells labeled for tubulin by the SG serum using the NIH Image software package. Values are mean ± SD (n = 110 and 120 for wild-type and ΔKIN1ΔKIN2 cells, respectively).
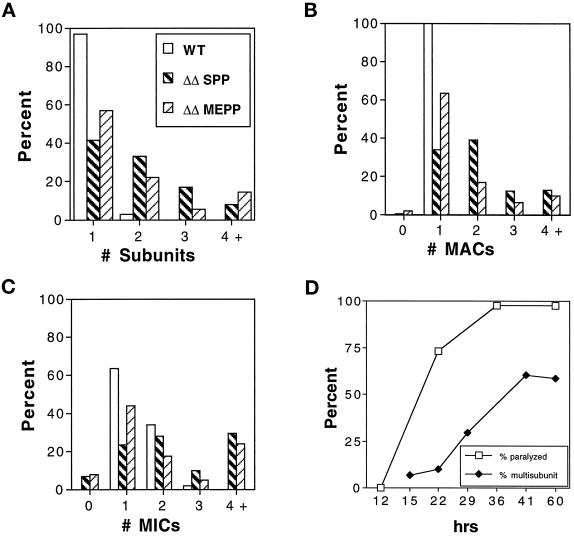
Surprisingly, many ΔKIN1ΔKIN2 cells grew large in size and had increased numbers of nuclei. A WT cell undergoing cytokinesis is shown in Figure 3A. Many ΔKIN1ΔKIN2 cells contained multiple “subcells” and multiple nuclei, indicating that they failed to complete cytokinesis after nuclear division (Figure 3, F–H). At 60 h in the standard SPP medium 58.5% of ΔKIN1ΔKIN2 cells had more than one subcell, compared with 3% of WT cells (Figure 5A). Along with the increase in the number of subcells, we observed an increase in the number of nuclei in ΔKIN1ΔKIN2 cells (Figure 5, B and C). Importantly, the two major phenotypic traits of ΔKIN1ΔKIN2 mutants, cell paralysis and arrest in cytokinesis, have different times of onset. Specifically, the cell paralysis appears to occur before the cytokinesis arrests. For example, at 22 h, the majority of double knockout isolates already contained paralyzed cells, whereas only a few percent of cells contained multiple subunits or nuclei (Figure 5D). Furthermore, virtually all live multinucleated “monsters” that we observed were completely paralyzed. These observations raise the intriguing possibility that the observed cytokinesis defects are caused by the loss of cilia or simply by the loss of cell motility.
Figure 5.
Quantitative analysis of cell morphology and subunit composition. ΔKIN1ΔKIN2 and WT cells were prepared by mating of appropriate parental strains and isolation of individual mating pairs into either SPP or MEPP medium. Cells prepared for immunofluorescence microscopy were scored for the number of cortical subcells (A), number of MACs (B), and number of MICs (C). Wild-type and ΔKIN1ΔKIN2 cells grown in SPP were scored 60 h after isolation of pairs. ΔKIN1ΔKIN2 cells grown in MEPP were scored 15 d after pair isolation. Histograms show percentages of cells with the indicated numbers of cortical subunits or nuclei (n = 135). (D) Compiled data from observations on living cells and parallel immunofluorescence studies. Paralysis is measured as the percentage of drops containing paralyzed cells (n = 41), and cytokinesis failures are estimated as the percentage of cells prepared as in A–C containing multiple subunits (n = 10–135).
Double Knockout Cells Can Be Rescued by Reintroduction of a Wild-Type KIN1 or KIN2 Gene
If the absence of both KIN1 and KIN2 is the cause of the phenotypes of ΔKIN1ΔKIN2 cells, reintroduction of either WT KIN1 or KIN2 should rescue the mutant cells. We mated cells that were not only double knockout heterokaryons but that also have the mpr1-1 gene in their MICs and not their MACs. The mpr1-1 gene confers resistance to 6-methylpurine (mp). Because the drug resistance allele is in the MIC and not in the MAC, only conjugation progeny cells can survive in the presence of mp, whereas all nonmating cells as well as cells that abort mating without developing a new MAC are killed (Orias and Bruns, 1976). When these were crossed and plated on SPP medium containing mp, no surviving cells were recovered in multiple experiments despite using large numbers of mating cells (1 × 107 cells per experiment; n = 4). In contrast, viable and motile clones were recovered (at a frequency of 1–50 per experiment) when ΔKIN1ΔKIN2 heterokaryons were bombarded with particles coated by either KIN1 or KIN2 fragments.
Epitope-tagged Kin1p Preferentially Accumulates in Cilia That Undergo Active Assembly
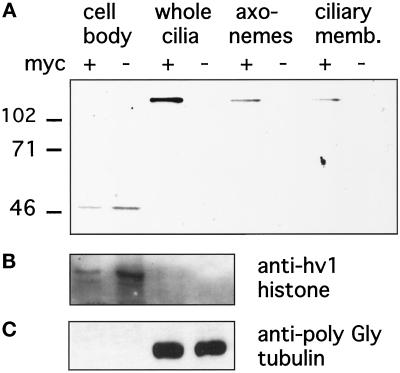
We subsequently rescued mating ΔKIN1ΔKIN2 heterokaryons using a genomic fragment of KIN1 modified by addition of a 5xMyc epitope tag to the N terminus of predicted Kin1p. The mechanism of rescue is based on replacement of the disrupted macronuclear copies of the target gene by the rescuing fragment, mediated by homologous recombination (Hai and Gorovsky, 1997). Thus, using the heterokaryon rescue approach, the epitope-tagged Kin1p is expressed at its normal locus using its own promoter. Using anti-Myc antibodies we detected a protein of the expected molecular mass of ∼100 kDa in cells rescued by a gene encoding 5xMyc-Kin1p but not in cells rescued by a WT KIN1 gene (Figure 6A). Western blotting analysis showed the presence of 5xMyc-Kin1p in the ciliary fraction, but no signal was detected in the equivalent amount of the cell bodies (Figure 6A). Purity of cell fractions was verified using anti-macronuclear histone, hv1 antibodies (Stargell et al., 1993), and AXO49 antibodies directed against hyperglycylated tubulin isoforms known to be specific to cilia (Bre et al., 1996). On Western blots, the anti-hv1 histone antibodies detected a single band of expected size in the cell body fraction but not in the ciliary fraction (Figure 6B), whereas the AXO49 cross-reacted only with the ciliary fraction (Figure 6C). Extraction of cilia with NP-40 showed that approximately half of 5xMyc-Kin1p was present in the membrane plus soluble matrix fraction, and half was in the insoluble axonemal fraction (Figure 6A). When fixed and permeabilized cells expressing 5xMyc-Kin1p were processed for immunofluorescence with anti-Myc antibodies, we repeatedly failed to detect any signal above the background observed in control cells.
Figure 6.
(A) Double knockout heterokaryon strains (UG13 and UG14) were crossed to express the ΔKIN1ΔKIN2 phenotype and rescued with gene fragments of either KIN1(−) or KIN1 containing an N-terminal 5xMyc epitope tag (+). Cells were selected with mp on SPP medium, and surviving, motile clones were analyzed by Western blotting of fractions prepared from cells rescued with either construct. The blot was probed with a monoclonal anti-myc antibody. A band of appropriate size (∼100 kDa) for 5xMyc-Kin1p is present only in ciliary fractions of cells transformed with epitope-tagged KIN1. (B and C) To verify the effectiveness of cell fractionation procedures, blots were probed with antibodies that recognize the macronuclear histone hv1 (B) or the AXO49 antibodies (C) directed against hyperglycylated tubulin isoforms, which are restricted to cilia (Bre et al., 1996).
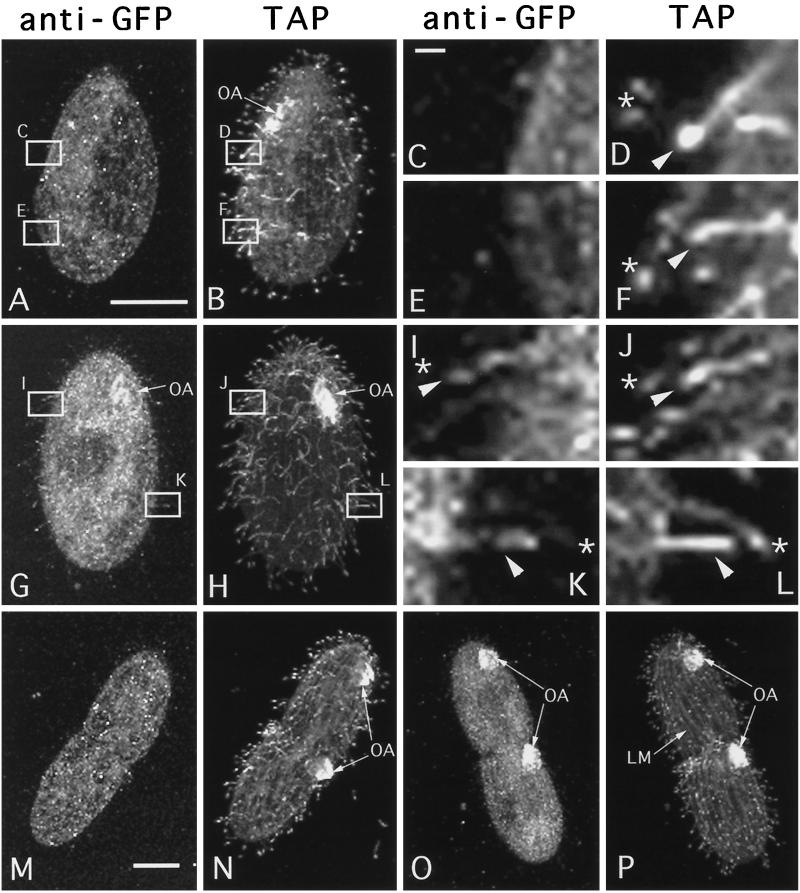
In an attempt to increase the sensitivity of detection of Kin1p, we rescued ΔKIN1ΔKIN2 cells with a gene encoding Kin1p fused to GFP. The GFP-Kin1p transformants grew well and were motile. Although we could not detect any GFP autofluorescence in live transformant cells, we detected the GFP-Kin1p signal in fixed cells by immunofluorescence using polyclonal anti-GFP antibodies. In WT nondividing (Figure 7A) and dividing cells (Figure 7M), anti-GFP antibodies produced only a weak background staining in the cell body, whereas cilia were not stained. In cells rescued by a GFP-KIN1 fragment, a weak GFP signal was detected in cilia, and there was an increase of signal in the cell body relative to negative control cells (Figure 7G). Most locomotory cilia were weakly labeled, except a few cilia, which were labeled more strongly and were generally shorter and therefore could be immature cilia in the process of their assembly (Figure 7, G and H boxed areas). Strong GFP labeling was observed in oral cilia in the developing oral apparatus in dividing cells (Figure 7O, arrows), and only weak labeling was detected in mature oral cilia of nondividing cells (Figure 7G, arrows). Thus, it appears that Kin1p preferentially accumulates in locomotory and oral cilia, which undergo active assembly. To test this hypothesis, we analyzed the localization of GFP-Kin1p in nongrowing cells (incubated in a starvation medium overnight), which were deciliated and allowed to regenerated cilia. These cells were double labeled using anti-GFP antibodies and the TAP952 monoclonal antibodies, which recognize monoglycylated tubulins (Bre et al., 1996).
Figure 7.
Wild-type (A–F, M, and N) and GFP-kin1p rescued (G–L, O, and P) cells were isolated from exponentially growing cultures and processed for confocal analysis using anti-GFP antibodies and TAP952 anti-tubulin antibodies. GFP and tubulin were detected using secondary antibodies coupled to Cy3 and FITC, respectively. For individual cells corresponding images of GFP (A, C, E, G, I, K, M, and O) and tubulin (B, D, F, H, J, L, N, and P) are shown. (A and B) Negative control cell. Some background staining with GFP antibodies is present in the cell body (A). (C–F) higher magnifications of the boxes shown in panels A and B. (G and H) Nondividing cell expressing GFP-Kin1p. Note weak staining of cilia by anti-GFP antibodies. Some cilia shown in boxed areas are labeled more strongly by anti-GFP antibodies (G). These cilia are shorter and label more uniformly by the TAP952 antibodies (H). (I–L) Higher magnifications of the boxes shown in G and H. Arrowheads show shorter immature cilia, which label uniformly with TAP952 antibodies and also show more of GFP signal. Stars indicate mature cilia in which the TAP952 labeling is restricted to ciliary tips, and there is relatively less GFP signal. (M and N) Negative control dividing cell. Two oral apparatuses are present. (O and P) Dividing cell expressing GFP-Kin1p. The oral cilia are labeled heavily by anti-GFP antibodies (O) and uniformly by TAP952 antibodies (P). Note relatively weak staining for GFP in the oral apparatus of a nondividing cell (G). Bars: A and M, 15 μm; C, 1 μm. OA, oral apparatus; LM, longitudinal microtubule bundle.
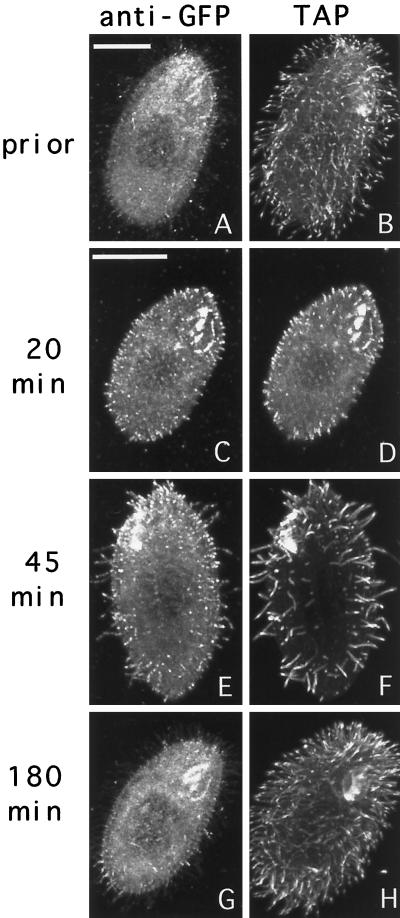
In starved cells before deciliation, the TAP952 antibody primarily labeled the tips of cilia (Figure 8, B). However, in regenerating cells, newly assembled cilia were labeled more uniformly by TAP952 (Figure 8, D and F). Thus, the uniform labeling with the TAP952 antibody can be used as a marker for newly assembled cilia. In starved cells before deciliation, the pattern of distribution of GFP-Kin1p was similar to the pattern seen in vegetatively growing cells, with most of the GFP signal present in cilia and some staining of the cell body (Figure 8A). Twenty minutes after deciliation, short cilia were already present, and virtually all cilia were labeled heavily by anti-GFP and uniformly by TAP952 antibodies (Figure 8, C and D). At 45 min most cilia were also stained brightly by anti-GFP antibodies and uniformly by the TAP952 antibodies (Figure 8, E and F). Negative control cells did not show any ciliary labeling by anti-GFP antibodies at 45 min (our unpublished results). By 180 min, most cilia in cells expressing GFP-Kin1p were already fully assembled, as indicated by the pattern of labeling of TAP952 (mainly tips of cilia), and there was a dramatic decrease in the staining intensity of cilia by anti-GFP antibodies (Figure 8, G and H). Thus, in cilia regenerating starved cells, Kin1p preferentially accumulates in cilia that actively assemble, and its abundance decreases dramatically after ciliary assembly is completed.
Figure 8.
Cells expressing GFP-Kin1p were starved for 24 h, deciliated, and allowed to regenerate cilia. At various times, samples of cells were processed for immunofluorescence using anti-GFP antibodies plus secondary antibodies coupled to Cy3 and TAP952 anti-tubulin antibodies plus secondary antibodies coupled to FITC. Pairs of corresponding GFP (A, C, E, and G) and tubulin (B, D, F, and H) images are shown for individual cells. Bars, 15 μm.
Strikingly, in vegetatively growing cells individual locomotory cilia, which were labeled strongly with anti-GFP, antibodies were also labeled uniformly by the TAP952. In most cases, scattered single growing cilia (more uniform TAP952 labeling) were strongly labeled by anti-GFP antibodies and were immediately adjacent to mature cilia (tip labeling by TAP952), which were only very weakly labeled by anti-GFP antibodies (Figure 7, I–L). Negative control cells lacking GFP-Kin1p showed no anti-GFP labeling in both mature and growing cilia (Figure 7, C–F). Newly developed oral cilia, which were labeled heavily by anti-GFP antibodies, were also uniformly labeled by TAP952 antibodies (Figure 7, O and P). Thus, Kin1p is preferentially targeted to a subset of cilia that undergo active assembly in both vegetatively growing and cilia regenerating cells. The targeting mechanism appears to operate at the resolution level of a single cilium.
Loss of Viability of Double Knockout Cells Is Caused by Inability to Phagocytose
In Tetrahymena cells a subset of specialized cilia is organized into four oral membranelles that surround the oral cavity. Coordinated beating of oral cilia is required for directing food particles into the phagocytic vacuoles formed at the bottom of the oral cavity. At 41–60 h, (Figure 3, G and H) most ΔKIN1ΔKIN2 cells appeared to lack any oral membranelles, which were easily identified in WT cells (Figure 3A) or in ΔKIN1ΔKIN2 cells at earlier time points (Figure 3, C–F). Microscopic observations of live ΔKIN1ΔKIN2 cells showed absence of any food vacuoles inside the cell body. Thus, the ΔKIN1ΔKIN2 cells lose their ability to phagocytose, likely because they lack oral cilia. This observation raises the possibility that ΔKIN1ΔKIN2 cells die on the standard medium (SPP) because they are unable to feed. Although Tetrahymena cells require phagocytosis to grow on the SPP medium used so far in this study, mutants of Tetrahymena that lack a functional oral apparatus, can be grown in a modified medium (MEPP). Presumably, this medium stimulates alternative routes for nutrient uptake, such as micropinocytosis (Orias and Rasmussen, 1976). Strikingly, unlike in SPP medium, in MEPP most ΔKIN1ΔKIN2 cells remained viable and divided with a doubling time of ∼8.5 h. Thus, the cause of lethality of double knockout cells in SPP is the inability to perform phagocytosis, resulting from the loss of oral cilia. Although ΔKIN1ΔKIN2 cells continued to grow and divide in the MEPP medium, they remained completely paralyzed, and many remained multinucleated (Figures 3I and 5).
Cell Division Phenotype in Double Knockout Cells Is Most Likely Induced by Cell Paralysis
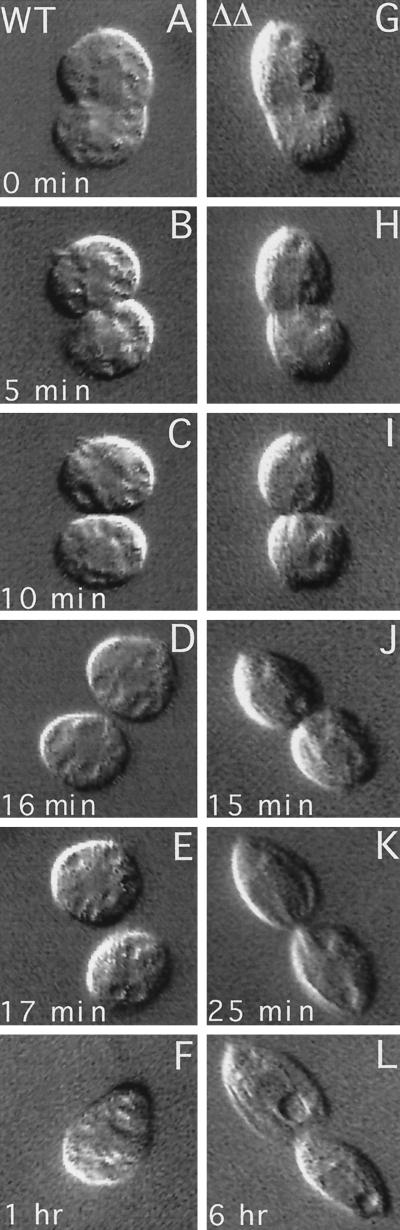
To assess how the absence of Kin1p and Kin2p affects the course of cytokinesis, we analyzed live WT and ΔKIN1ΔKIN2 cells during cell division using video microscopy. In T. thermophila, initially the cleavage furrow is formed asymmetrically on one side of the cell. We isolated early dividers having a unilateral cleavage furrow from WT and ΔKIN1ΔKIN2 populations and analyzed the course of cytokinesis (Figure 9). It took ∼20 min for WT cells to complete cell division starting from the unilateral cleavage stage (Figure 9, A–F). Mutant cells showed a nearly normal cleavage furrow ingression (Figure 9, G–K), but many failed to separate at the final stage of cytokinesis. Figure 9 shows a ΔKIN1ΔKIN2 cell that had almost a complete cleavage furrow ingression at 25 min (Figure 9K) but failed to separate completely and after 6 h showed signs of cortical integration, with the cytoplasmic content of the posterior cell being absorbed by the anterior cell (Figure 9L).
Figure 9.
Comparison of cleavage furrow progression in WT (A–F) and ΔKIN1ΔKIN2 (G–L) cells. Single cells were isolated and analyzed using differential interference contrast video microscopy.
As already mentioned, the temporal analysis of phenotypic traits indicated that the cell division arrests occur after cells become paralyzed. Furthermore, we did not see any Kin1p in association with the cleavage furrow (Figure 7O). All these observations taken raised a possibility that the arrests at cytokinesis in ΔKIN1ΔKIN2 cells are caused by lack of ciliary motility. Thus, Tetrahymena cells may require cell locomotion to complete cytokinesis. Dividing WT cells are generally less motile compared with nondividing cells and tend to sit at the bottom of a culture dish. However, a video analysis of a number of WT cells revealed that within ∼2 min before cell separation the posterior daughter cell often rotates along its longitudinal axis, and virtually all daughter cells briefly pull apart immediately before their final separation (our unpublished results). It is likely that the rotations create a strain within the membranous channel connecting the daughter cells, which facilitate its breakage when the cells pull apart. The detailed analysis of the motile activity of dividing cells will be published elsewhere (Brown, Hardin, and Gaertig, unpublished data). The dividing double knockout cells remain completely paralyzed in the course of cell division. Thus, it appears that the arrests in cytokinesis frequently seen in ΔKIN1ΔKIN2 cells are induced by cell paralysis.
DISCUSSION
To investigate the function of kinesin-II in vivo, we constructed strains of T. thermophila, which lack the kinesin-II-encoding genes KIN1 and KIN2. Cells lacking either KIN1 or KIN2 exhibited several subtle phenotypes. Some of these, such as slow growth and temperature sensitivity were observed in cells lacking either of the two genes. However, only cells lacking KIN1 showed reduced cell motility and impaired assembly of cilia. The reduction in growth rate was more severe in the KIN2 null cells. Thus, although the phenotypes caused by deletion of either gene partly overlap, KIN1 appears to be more important for ciliary functions, whereas KIN2 is more important for cell multiplication.
Three lines of evidence indicate that KIN1 and KIN2 arose relatively recently as a result of the duplication of a common ancestor kinesin-II gene: 1) sequences of KIN1 and KIN2 are more similar to each other than to any other known kinesins-II; 2) positions of two of three introns are conserved; and 3) the two genes are genetically linked and thus are located not far from each other on the same micronuclear chromosome. To address the possibility of overlapping functions between KIN1 and KIN2, we created cells lacking all copies of both genes, using a novel approach based on construction of heterokaryon strains lacking specific genes in the germ line micronucleus and having normal gene copies in the somatic macronucleus (Hai and Gorovsky, 1997). The heterokaryons are phenotypically normal, because only the macronuclear genes are expressed during the vegetative life of Tetrahymena. However, when two heterokaryons mate, they form new MACs from the micronuclei, which lack all functional copies of the targeted genes. This approach effectively produces an inducible gene knockout. Double knockout cells showed two major defects: 1) extreme shortening of locomotory and oral cilia and 2) frequent failure in cytokinesis. Double knockout cells were not viable on a standard medium but could grow on a modified medium on which Tetrahymena cells do not require phagocytosis for their survival (Orias and Rasmussen, 1976). Thus, the lethality observed on the standard medium is most likely caused by the loss of oral cilia, which are essential for phagocytosis in Tetrahymena. Consequently, Tetrahymena cells do not require KIN1 and KIN2 for their survival under conditions in which cells are not dependent on phagocytosis.
The most dramatic phenotypic change in double knockout Tetrahymena cells is the almost complete loss of cilia. Similarly, mutation of the Chlamydomonas kinesin-II FLA10 leads to complete resorbtion of existing flagella (Lux and Dutcher, 1991; Walther et al., 1994). Knockout of the kinesin-II gene in mouse, KIF3B, caused death of embryos before midgestation. However, the nodal cells in the KIF3B-deficient embryos completely failed to assemble cilia (Nonaka et al., 1998). This phenotype is essentially identical to the phenotype caused by elimination of both Kin1p and Kin2p in Tetrahymena. Thus, kinesin-II genes function universally in ciliary assembly.
We show that in addition to its role in the assembly of new cilia, kinesin-II is also essential for ciliary maintenance. In wild-type Tetrahymena, most if not all locomotory cilia are never resorbed during the vegetative life cycle. Strikingly, we found that after induction of the knockout phenotype, mutant cells not only failed to assemble new cilia but also resorbed all of their preexisting cilia. Moreover, an epitope-tagged Kin1p was found along the full length of cilia in nongrowing (starved) cells, consistent with the involvement of kinesin-II in ciliary maintenance. These data are consistent with a high level of turnover of axonemal subunits reported for morphostatic cilia and flagella (Rosenbaum and Child, 1967; Nelsen, 1975; Stephens, 1997) and suggest that the subunit turnover is driven by kinesin-II and possibly other molecular motors.
Although kinesin-II is required for both assembly and maintenance of cilia, we found that epitope-tagged motor proteins preferentially accumulate in cilia that undergo assembly. This phenomenon was observed in regenerating cilia and in a subset of oral and locomotory cilia that assemble in vegetative cells. Tetrahymena cells appear to use a mechanism that preferentially directs kinesin-II to the newly assembled cilia or causes its preferential retention by assembling cilia. Because in Tetrahymena new cilia are formed immediately adjacent to the preexisting cilia, the proposed targeting and retention mechanism must operate at the resolution level of a single cilium. Interestingly, immunofluorescence studies in Chlamydomonas showed that the kinesin-II epitopes were more concentrated near the basal bodies and in the proximal part of the axoneme (Vashishtha et al., 1996; Cole et al., 1998). Thus, initially, kinesin-II may be targeted to the basal bodies and later to move to the adjacent axoneme.
The molecular nature of kinesin-II involvement in ciliogenesis and ciliary maintenance is not well understood. In Chlamydomonas, FLA10 activity is required to maintain intraflagellar transport, the motility of raft particles detected beneath the flagellar membrane (Kozminski et al., 1995). Induction of the fla10 mutant phenotype caused loss of rafts and the loss of two types of 16S protein complexes from flagella, suggesting that 16S complexes are both components of rafts and the FLA10 cargo (Piperno and Mead, 1997; Cole et al., 1998). One of the components of the 16S complex in Chlamydomonas is a protein homologous to the OSM-6 protein of C. elegans, which plays an essential role in the function of chemosensory ciliary neurons (Cole et al., 1998; Collet et al., 1998). Recently, fluorescent OSM-6-GFP protein was observed to move inside the chemosensory cilia of living C. elegans worms at the same rate as the rate of movement of the KAP subunit of the kinesin-II complex (Orozco et al., 1999). The 16S complexes are most likely components of rafts seen in live cells by differential interference contrast microscopy and are proposed to function in transport of flagellar subunits (Rosenbaum et al., 1999). Consistent with this hypothesis, induction of the fla10 phenotype blocked transport of an inner dynein arm polypeptide but allowed for transport of an outer dynein arm component (Piperno et al., 1996), suggesting that the inner but not outer arm components are one of the cargoes of kinesin-II. It seems unlikely, however, that a failure to transport inner dynein arm components by kinesin-II would result in a complete block in axonemal assembly, because numerous mutants lacking specific axonemal components (dynein arms and radial spokes) have been described that still assemble axonemes (Dutcher, 1995). It is likely that kinesin-II in addition to its role in the transport of dynein arm components is also involved in the transport of some structural components required for the initial elongation of the axoneme such as microtubule subunits or ciliary membranes. Interestingly, longitudinal sections of basal bodies on ΔKIN1ΔKIN2 cells showed that many of the very short axonemal fragments are covered by a bulge of ciliary membrane, suggesting that membranes are properly delivered to the growing axoneme. It appears that kinesin-II is involved in the delivery of a basic structural component of cilia that is essential for elongation of axonemal microtubules, such as tubulin dimers or oligomers.
The most unexpected result of our study was the frequent failure of double knockout cells to complete cytokinesis. Some previous studies suggested that kinesin-II plays a direct role in cell division. Immunofluorescent studies in sea urchin embryos showed an accumulation of kinesin-II proteins in the interzone of the mitotic spindle during anaphase before the formation of the cleavage furrow (Henson et al., 1995). Also, the fla10 mutation results in synthetic defects in the cell cycle when combined with another mutation (Lux and Dutcher, 1991), and FLA10 protein transiently associates with the centrioles in Chlamydomonas (Vashishtha et al., 1996). However, despite dramatic cytokinesis defects in our kinesin-II mutants, we did not find any evidence that would support direct involvement of kinesin-II in cytokinesis. First, immunolocalization studies showed that epitope-tagged Kin1p is highly concentrated in cilia. Although some Kin1p was detected in the cell body by immunofluorescence, this pool did not show any clear association with the cleavage furrow or changes in the distribution during the cell cycle. More likely, the cell body pool of Kin1p represents the newly synthesized motor subunits before their delivery to cilia. Furthermore, the double KIN mutants appear not to be affected in the initiation and ingression of the cleavage furrow until the final stage of cytokinesis when the daughter cells break the cytoplasmic connection. At that time in wild-type cells we found that the posterior daughter cells often rotate along their longitudinal axis, and both daughters pull apart. These observations suggest that Tetrahymena cells use mechanical force generated by ciliary beating to culminate cytokinesis. This hypothesis is consistent with the earlier onset of the cell paralysis phenotype compared with the cell division arrest phenotype (Figure 6D). Although cell locomotion is not absolutely required for cell division, it appears to be an evolutionary adaptation to support the unusually high rate of culture growth of Tetrahymena.
Supplementary Material
View video 9a (associated with Figure 9)
View video 9b (associated with Figure 9)
ACKNOWLEDGMENTS
We are grateful to Dr. Donna Cassidy-Hanley for assistance with the use of the biolistic gun. We are most grateful to University of Georgia Athens undergraduates Curtis McNiff, Brett Margolias, Wood Pope, and Clyde Hardin for contribution in analyses of knockout phenotypes and Naishaj Shah for construction of the GFP-KIN1 plasmid. We thank Dr. Mark Farmer (Center for Advanced Ultrastructural Research, University of Georgia) for help with the use of the video imaging system and for suggestions regarding capture of DAPI images on the confocal microscope. We thank Jian Zhao Shen for assistance in preparation of thin sections for electron microscopy and Yan Gao for assistance with Western blots. We also thank Dr. Mitchell Bernstein (Albert Einstein College of Medicine) for providing the sequence of the cloned KIN5 gene fragment, Dr. Nicolette Levilliers (University Paris-Sud, Paris, France) for providing the TAP952 and AXO49 antibodies, Dr. Marty Gorovsky (University of Rochester, Rochester, NY) for the SG antibody, Dr. David Allis (University of Virginia, Charlottesville, VA) for the anti-histone hv1 antibodies, Dr. Karl Saxe (Emory University, Atlanta, GA) for providing the bsr plasmid, Dr. Keith Kozminski for providing the Myc construct, and Dr. Aaron Turkewitz for the GFP construct. We also thank Dr. Marty Gorovsky and Dr. Joseph Frankel (University of Iowa, Iowa City, IA) for reading the manuscript and for helpful suggestions. The biolistic bombardment experiment for the disruption of the KIN1 gene was performed at the biolistic facility of the Plant Science Center at Cornell University. This work was supported by US Public Health Service grant GM-54017 from the National Institutes of Health to J.G.
Footnotes
V⃞
Online version of this article contains video material for Figure 9. Online version available at www.molbiolcell.org.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Sequence homology searches done using BLAST. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bre M-H, et al. Axonemal tubulin polyglycylation probed with two monoclonal antibodies: widespread evolutionary distribution, appearance during spermatozoan maturation and possible function in motility. J Cell Sci. 1996;109:727–738. doi: 10.1242/jcs.109.4.727. [DOI] [PubMed] [Google Scholar]
- Callen A-M, et al. Isolation and characterization of libraries of monoclonal antibodies directed against various forms of tubulin in Paramecium. Biol Cell. 1994;81:95–119. doi: 10.1016/s0248-4900(94)80002-2. [DOI] [PubMed] [Google Scholar]
- Calzone FJ, Gorovsky MA. Cilia regeneration in Tetrahymena. Exp Cell Res. 1982;140:474–476. doi: 10.1016/0014-4827(82)90144-6. [DOI] [PubMed] [Google Scholar]
- Cassidy-Hanley D, Bowen J, Lee J, Cole ES, VerPlank LA, Gaertig J, Gorovsky MA, Bruns PJ. Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics. 1997;146:135–147. doi: 10.1093/genetics/146.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole DG, Chinn SW, Wedaman KP, Hall K, Vuong T, Scholey JM. Novel heterotrimeric kinesin-related protein purified from sea urchin eggs. Nature. 1993;366:268–270. doi: 10.1038/366268a0. [DOI] [PubMed] [Google Scholar]
- Cole DG, Diener DR, Himelblau AL, Beech PL, Fuster JC, Rosenbaum JL. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol. 1998;18:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collet J, Spike CA, Lundquist EA, Shaw JE, Herman RK. Analysis of osm-6, a gene that affects sensory cilium structure and sensory neuron function in Caenorhabditis elegans. Genetics. 1998;148:187–200. doi: 10.1093/genetics/148.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dippell RV. The development of basal bodies in Paramecium. Proc Natl Acad Sci USA. 1968;61:461–468. doi: 10.1073/pnas.61.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher SK. Flagellar assembly in two hundred and fifty easy-to-follow steps. Trends Genet. 1995;11:398–404. doi: 10.1016/s0168-9525(00)89123-4. [DOI] [PubMed] [Google Scholar]
- Frohman MA. RACE: rapid amplification of cDNA ends. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols. San Diego: Academic Press; 1990. pp. 28–38. [Google Scholar]
- Gaertig J, Cruz MA, Bowen J, Gu L, Pennock DG, Gorovsky MA. Acetylation of lysine 40 in alpha-tubulin is not essential in Tetrahymena thermophila. J Cell Biol. 1995;129:1301–1310. doi: 10.1083/jcb.129.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertig J, Gu L, Hai B, Gorovsky MA. High frequency vector-mediated transformation and gene replacement in Tetrahymena. Nucleic Acids Res. 1994a;22:5391–5398. doi: 10.1093/nar/22.24.5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertig J, Thatcher TH, Gu L, Gorovsky MA. Electroporation-mediated replacement of a positively and negatively selectable β-tubulin gene in Tetrahymena thermophila. Proc Natl Acad Sci USA. 1994b;91:4549–4553. doi: 10.1073/pnas.91.10.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorovsky MA. Macro- and micronuclei of Tetrahymena pyriformis: a model system for studying the structure and function of eukaryotic nuclei. J Protozool. 1973;20:19–25. doi: 10.1111/j.1550-7408.1973.tb05995.x. [DOI] [PubMed] [Google Scholar]
- Guttman SD, Gorovsky MA. Cilia regeneration in starved Tetrahymena: an inducible system for studying gene expression and organelle biogenesis. Cell. 1979;17:307–317. doi: 10.1016/0092-8674(79)90156-9. [DOI] [PubMed] [Google Scholar]
- Haddad A, Turkewitz AP. Analysis of exocytosis mutants indicates close coupling between regulated secretion and transcription activation in Tetrahymena. Proc Natl Acad Sci USA. 1997;94:10675–10680. doi: 10.1073/pnas.94.20.10675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai B, Gorovsky MA. Germ-line knockout heterokaryons of an essential alpha-tubulin gene enable high-frequency gene replacement and a test of gene transfer from somatic to germ-line nuclei in Tetrahymena thermophila. Proc Natl Acad Sci USA. 1997;94:1310–1315. doi: 10.1073/pnas.94.4.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson JH, Cole DG, Terasaki M, Rashid D, Scholey JM. Immunolocalization of the heterotrimeric kinesin-related protein KRP(85/95) in the mitotic apparatus of sea urchin embryos. Dev Biol. 1995;171:182–194. doi: 10.1006/dbio.1995.1270. [DOI] [PubMed] [Google Scholar]
- Johnson KA, Rosenbaum JL. Polarity of flagellar assembly in Chlamydomonas. J Cell Biol. 1992;119:1605–1611. doi: 10.1083/jcb.119.6.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneller DG, Cohen FE, Langridge R. Improvements in protein secondary structure prediction by an enhanced neural network. J Mol Biol. 1990;214:171–182. doi: 10.1016/0022-2836(90)90154-E. [DOI] [PubMed] [Google Scholar]
- Kondo S, Sato Yoshitake R, Noda Y, Aizawa H, Nakata T, Matsuura Y, Hirokawa N. KIF3A is a new microtubule-based anterograde motor in the nerve axon. J Cell Biol. 1994;125:1095–1107. doi: 10.1083/jcb.125.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski KG, Beech PL, Rosenbaum JL. The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J Cell Biol. 1995;131:1517–1527. doi: 10.1083/jcb.131.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski KG, Johnson KA, Forscher P, Rosenbaum JL. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc Natl Acad Sci USA. 1993;90:5519–5523. doi: 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Lux F, III, Dutcher SK. Genetic interactions at the FLA10 locus: suppressors and synthetic phenotypes that affect the cell cycle and flagellar function in Chlamydomonas reinhardtii. Genetics. 1991;128:549–561. doi: 10.1093/genetics/128.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marszalek JR, Ruiz-Lozano P, Roberts E, Chien KR, Goldstein LS. Situs inversus and embryonic ciliary morphogenesis defects in mouse mutants lacking the KIF3A subunit of kinesin-II. Proc Natl Acad Sci USA. 1999;27:5043–5048. doi: 10.1073/pnas.96.9.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RL, Scholey JM. Heterotrimeric kinesin-II is required for the assembly of motile 9 + 2 ciliary axonemes on sea urchin embryos. J Cell Biol. 1997;138:1009–1022. doi: 10.1083/jcb.138.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muresan V, Abramson T, Lyass A, Winter D, Porro E, Hong F, Chamberlin NL, Schnapp BJ. KIF3C is a novel neuronal kinesin that interacts with KIF3A and associates with membrane vesicles. Mol Biol Cell. 1998;9:637–652. doi: 10.1091/mbc.9.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelsen EM. Regulation of tubulin during ciliary regeneration of nongrowing Tetrahymena. Exp Cell Res. 1975;94:152–158. doi: 10.1016/0014-4827(75)90542-x. [DOI] [PubMed] [Google Scholar]
- Nonaka S, Tanaka Y, Okada Y, Takeda S, Harada A, Kanai Y, Kido M, Hirokawa N. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 1998;95:829–837. doi: 10.1016/s0092-8674(00)81705-5. [DOI] [PubMed] [Google Scholar]
- Orias E, Bruns PJ. Induction and isolation of mutants in Tetrahymena. In: Prescott DM, editor. Methods in Cell Biology. New York: Academic Press; 1976. pp. 247–282. [PubMed] [Google Scholar]
- Orias E, Rasmussen L. Dual capacity for nutrient uptake in Tetrahymena. IV. Growth without food vacuoles. Exp Cell Res. 1976;102:127–137. doi: 10.1016/0014-4827(76)90307-4. [DOI] [PubMed] [Google Scholar]
- Orozco JT, Wedaman KP, Signor D, Brown H, Rose L, Scholey JM. Movement of motor and cargo along cilia. Nature. 1999;398:674. doi: 10.1038/19448. [DOI] [PubMed] [Google Scholar]
- Pazour GJ, Wilkerson CG, Witman GB. A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT) J Cell Biol. 1998;141:979–992. doi: 10.1083/jcb.141.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Mead K. Transport of a novel complex in the cytoplasmic matrix of Chlamydomonas flagella. Proc Natl Acad Sci USA. 1997;94:4457–4462. doi: 10.1073/pnas.94.9.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Mead K, Henderson S. Inner dynein arms but not outer dynein arms require the activity of kinesin homologue protein KHP1(FLA10) to reach the distal part of flagella in Chlamydomonas. J Cell Biol. 1996;133:371–379. doi: 10.1083/jcb.133.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Siuda E, Henderson S, Segil M, Vaananen H, Sassaroli M. Distinct mutants of retrograde intraflagellar transport (IFT) share similar morphological and molecular defects. J Cell Biol. 1998;143:1591–601. doi: 10.1083/jcb.143.6.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter ME, Bower R, Knott JA, Byrd P, Dentler W. Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas. Mol Biol Cell. 1999;10:693–712. doi: 10.1091/mbc.10.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SL, Tint IS, Fanapour PC, Gelfand VI. Regulated bidirectional motility of melanophore pigment granules along microtubules in vitro. Proc Natl Acad Sci USA. 1997;94:3720–3725. doi: 10.1073/pnas.94.8.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JL, Child FM. Flagellar regeneration in protozoan flagellates. J Cell Biol. 1967;34:345–364. doi: 10.1083/jcb.34.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JL, Cole DG, Diener DR. Intraflagellar transport: the eyes have it. J Cell Biol. 1999;144:385–388. doi: 10.1083/jcb.144.3.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JC, DeVit MJ, Russell JA, Smith FD, Harpending PR, Roy MK, Johnston SA. An improved, helium-driven biolistic device. Technique. 1991;3:3–16. [Google Scholar]
- Signor D, Wedaman KP, Rose LS, Scholey JM. Two heteromeric kinesin complexes in chemosensory neurons and sensory cilia of Caenorhabditis elegans. Mol Biol Cell. 1999;10:345–360. doi: 10.1091/mbc.10.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stargell LA, Bowen J, Dadd CA, Dedon PC, Davis M, Cook RG, Allis CD, Gorovsky MA. Temporal and spatial association of histone H2A variant hv1 with transcriptionally competent chromatin during nuclear development in Tetrahymena thermophila. Genes & Dev. 1993;7:2641–2651. doi: 10.1101/gad.7.12b.2641. [DOI] [PubMed] [Google Scholar]
- Stephens RE. Synthesis and turnover of embryonic sea urchin ciliary proteins during selective inhibition of tubulin synthesis and assembly. Mol Biol Cell. 1997;8:2187–2198. doi: 10.1091/mbc.8.11.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutoh K. A transformation vector for Dictyostelium discoideum with a new selectable marker bsr. Plasmid. 1993;30:150–154. doi: 10.1006/plas.1993.1042. [DOI] [PubMed] [Google Scholar]
- Takeda S, Yonekawa Y, Tanaka Y, Okada Y, Nonaka S, Hirokawa N. Left-right asymmetry and kinesin superfamily protein KIF3A: new insights in determination of laterality and mesoderm induction by kif3A−/− mice analysis. J Cell Biol. 1999;145:826–835. doi: 10.1083/jcb.145.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashishtha M, Walther Z, Hall JL. The kinesin-homologous protein encoded by the Chlamydomonas FLA10 gene is associated with basal bodies and centrioles. J Cell Sci. 1996;109:541–549. doi: 10.1242/jcs.109.3.541. [DOI] [PubMed] [Google Scholar]
- Walther Z, Vashistha M, Hall JL. The Chlamydomonas FLA10 gene encodes a novel kinesin-homologous protein. J Cell Biol. 1994;126:175–188. doi: 10.1083/jcb.126.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H, Nakata T, Okada Y, Hirokawa N. KIF3A/B: a heterodimeric kinesin suparfamily protein that works as a microtubule plus end-directed motor for membrane organelle transport. J Cell Biol. 1995;130:1387–1399. doi: 10.1083/jcb.130.6.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H, Nakata T, Okada Y, Hirokawa N. Cloning and characterization of KAP3: a novel kinesin superfamily-associated protein of KIF3A/3B. Proc Natl Acad Sci USA. 1996;93:8443–8448. doi: 10.1073/pnas.93.16.8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JT, Layman RA, Goldstein LSB. A three-domain structure of kinesin heavy chain revealed by DNA sequence and microtubule binding assays. Cell. 1989;56:879–889. doi: 10.1016/0092-8674(89)90692-2. [DOI] [PubMed] [Google Scholar]
- Yang Z, Goldstein LSB. Characterization of the KIF3C neural kinesin-like motor from mouse. Mol Biol Cell. 1998;9:249–261. doi: 10.1091/mbc.9.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
View video 9a (associated with Figure 9)
View video 9b (associated with Figure 9)