GERp95, a Membrane-associated Protein that Belongs to a Family of Proteins Involved in Stem Cell Differentiation (original) (raw)
Abstract
A panel of mAbs was elicited against intracellular membrane fractions from rat pancreas. One of the antibodies reacted with a 95-kDa protein that localizes primarily to the Golgi complex or the endoplasmic reticulum (ER), depending on cell type. The corresponding cDNA was cloned and sequenced and found to encode a protein of 97.6 kDa that we call GERp95 (Golgi ER protein 95 kDa). The protein copurifies with intracellular membranes but does not contain hydrophobic regions that could function as signal peptides or transmembrane domains. Biochemical analysis suggests that GERp95 is a cytoplasmically exposed peripheral membrane protein that exists in a protease-resistant complex. GERp95 belongs to a family of highly conserved proteins in metazoans and Schizosaccharomyces pombe. It has recently been determined that plant and Drosophila homologues of GERp95 are important for controlling the differentiation of stem cells (Bohmert et al., 1998; Cox et al., 1998; Moussian et al., 1998). In Caenorhabditis elegans, there are at least 20 members of this protein family. To this end, we have used RNA interference to show that the GERp95 orthologue in C. elegans is important for maturation of germ-line stem cells in the gonad. GERp95 and related proteins are an emerging new family of proteins that have important roles in metazoan development. The present study suggests that these proteins may exert their effects on cell differentiation from the level of intracellular membranes.
INTRODUCTION
The cytoplasm of eukaryotic cells is partitioned into more than a dozen membrane-bound organelles. Compartmentalization serves to increase the efficiencies of cellular processes by controlling the spatial and temporal interactions of proteins, nucleic acids and lipids. The endoplasmic reticulum (ER) and Golgi complex play central roles in the biogenesis and operational fidelity of eukaryotic cells by orchestrating the synthesis and movement of proteins and lipids (Hurtley and Helenius, 1989; Narula et al., 1992; Sitia and Meldolesi, 1992; Bergeron et al., 1994; Hammond and Helenius, 1995; Farquhar and Hauri, 1997; Hauri and Schweizer, 1997; Farquhar and Palade, 1998). It is now clear that these two organelles are directly involved in processes that affect cellular differentiation because mistargeting and/or altered expression of resident proteins of the ER and Golgi complex can have profound effects on cell growth, morphology, and tumorigenicity. Moreover, cellular defects at the ER/Golgi level underlie the pathophysiology of many human diseases such as familial hypercholesterolemia, polycystic kidney disease, Tangier disease, cystic fibrosis, mucopolysaccharidosis types I, VI, and VII, progeroid syndrome, and many others (for review, see Brooks, 1997; Gonatas, 1997).
We are interested in identifying novel ER- and Golgi-associated proteins that are involved in the function of these organelles. The approach we adopted was to generate a panel of mAbs against ER/Golgi membrane fractions so that they may be used to identify and then immunoaffinity purify their cognate antigens for sequence analysis. In the recent past, this technique has facilitated the characterization of ER- and Golgi-associated proteins that otherwise may not have been discovered by conventional biochemical or genetic approaches (Saraste et al., 1987; Schweizer et al., 1988; Fritzler et al., 1995; Erlich et al., 1996; Griffith et al., 1997).
In the present study, we report the purification and characterization of a 95-kDa protein that exhibits cell-dependent differential localization. The protein was named GERp95 (Golgi ER protein 95 kDa) to reflect the fact that it localizes to the Golgi and/or ER. In epithelioid NRK cells and pancreas acinar cells, GERp95 was concentrated in the Golgi complex, whereas in all other cell types examined it was found to be associated with the ER. The cDNA encoding this protein was isolated and sequenced, revealing that GERp95 belongs to a family of highly conserved proteins found in multicellular eukaryotes and Schizosaccharomyces pombe. Recently GERp95 homologues have been implicated in various aspects of stem cell differentiation and tissue development (Bohmert et al., 1998; Cox et al., 1998; Moussian et al., 1998; Lynn et al., 1999; Schmidt et al., 1999). In the present study, we provide evidence that suggests a role for the GERp95 Caenorhabditis elegans orthologue in germ-line stem cell maturation.
MATERIALS AND METHODS
Reagents
Reagents and supplies were from the following sources. Protein A-Sepharose was purchased from Pharmacia (Alameda, CA). BioMag goat anti-mouse IgG (Fc-specific) coated magnetic beads were purchased from PerSeptive Diagnostics (Cambridge, MA). Fibronectin, PMSF, SDS, and BSA were purchased from Sigma (St. Louis, MO). Promix [35S] methionine/cysteine (1000 Ci/mM), translation grade [35S] methionine (1000 Ci/mM), and [14C]-labeled protein standards were purchased from Amersham (Arlington Heights, IL). Texas Red-conjugated goat anti-mouse IgG and FITC-conjugated donkey anti-rabbit IgG (each double-labeling grade) were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA). Goat anti-mouse IgG and anti-rabbit IgG conjugated to horseradish peroxidase were purchased from Bio-Rad (Richmond, CA). Optimem serum-free media, FBS, and DMEM hi-glucose were purchased from Life Technologies (Gaithersburg, MD). MEM lacking cysteine/methionine was purchased from ICN Biomedicals (Irvine, CA). Chymotrypsin, trypsin, aprotinin, tunicamycin, Pefabloc, and Pwo polymerase were purchased from Boehringer Mannheim (Laval, Quebec). Reagents for coupled transcription/translation were purchased from Promega (Madison, WI). Rabbit antiserum to α-mannosidase II (Man II) was a gift from Drs. Marilyn Farquhar (University of California, San Diego, CA) and Kelley Moremen (University of Georgia, Athens, GA). Rabbit antibodies to calnexin, BiP, and the constitutive form of HSP70 were purchased from Stressgen (Victoria, British Columbia, Canada). Antiserum to the β-subunit of glucosidase II (Arendt and Ostergaard, 1997) was provided by Dr. Hanne Ostergaard (University of Alberta, Alberta, Canada). Rabbit antibodies to ERp72 were provided by Dr. Paul Kim (Harvard Medical School, Boston, MA). BHK-21, Clone 9, REF-52, NRK52E and NRK49F, and COS cells were obtained from the American Type Culture Collection (Rockville, MD). An expression vector containing the Z0–3 cDNA (Haskins et al., 1998) was obtained from Dr. Bruce Stevenson (University of Alberta). Canine pancreatic microsomes were provided by Dr. Chris Nicchitta (Duke University, Durham, NC). The Indiana strain of vesicular stomatitis virus (VSV), a cDNA encoding VSV G protein for use in in vitro transcription/translation and antibodies to VSV were gifts from Dr. Carolyn Machamer (The Johns Hopkins University, Baltimore, MD).
mAb Production
Intracellular membrane fractions were isolated from the pancreas of Sprague Dawley rats essentially as described (Saraste et al., 1987). The B2 fraction, which contains membranes with a density of 1.17 g/cm3 and is enriched in _cis_-Golgi elements, was subjected to phase separation using Triton X-114 as described (Bordier, 1981). The detergent phase was used to immunize mice for hybridoma production (Harlow and Lane, 1988). Hybridoma supernatants were screened by indirect immunofluorescence on various cell types. One of the hybridomas. LCH-7, an IgG1, was selected for further study.
Metabolic Labeling and Radioimmunoprecipitation
Confluent dishes of cells were washed once with PBS, incubated in MEM minus cysteine and methionine/5% dialyzed FBS for 15 min at 37°C. Cells were labeled for various time periods with 500-1000 μCi [35S] Promix/ml in the same media. Where indicated, cells were treated with tunicamycin (3 μg/ml) before and during the labeling period. Radiolabeled cells were washed three times with ice-cold PBS, lysed on ice in 1% NP-40, 150 mM NaCl, 2 mM EDTA, 50 mM Tris-HCl, pH 8.0, containing 100 μg/ml the protease inhibitor Pefabloc. Lysates were centrifuged at 14,000 × g for 5 min at 4°C before immunoprecipitation with antibodies and protein A-Sepharose. Immune complexes prepared using LCH-7 were washed twice with 1% Triton X-100, 500 mM NaCl, 50 mM Tris-HCl, pH 7.4, once with 0.2% Triton X-100, 1.0 M NaCl, 50 mM Tris-HCl, pH 7.4, and once with water. When using rabbit anti-GERp95, samples were washed three times with RIPA buffer (1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, 50 mM Tris-HCl, pH 8.0) and once with water. Samples were heated at 95°C in 2× SDS-gel sample buffer for 5 min before loading onto gels.
SDS-PAGE and Autoradiography
Proteins were separated on 8 or 10% polyacrylamide gels before fixation in isopropanol:water:acetic acid (25:65:10) for 30 min. Gels were then soaked in 1.0 M sodium salicylate/0.01% 2-mercaptoethanol for 20 min before drying and exposure to Kodak XAR film at −80°C.
Immunoblotting
Proteins were transferred from polyacrylamide gels to PVDF membranes using a semidry transfer apparatus (Tyler Instruments, Edmonton, AB) according to manufacturer’s instructions. Membranes were blocked in TBS containing 0.05% Tween 20 and 4% skim milk. Primary and secondary antibody incubations were done in the same solution. Membranes were washed three to four times (10 min each) after each antibody in TBS, 0.05% Tween 20. Blots were then developed using ECL reagents from Amersham Canada (Oakville, ON) and exposed to Fuji RX film.
Immunofluorescence Microscopy
Cells were grown on 12-mm glass coverslips, fixed, and permeabilized with methanol at −20°C or with 4% paraformaldehyde and 0.05% saponin and processed for indirect immunofluorescence as described (Kuismanen and Saraste, 1989; Hobman et al., 1992). When rabbit anti-GERp95 antibodies were used, cells were fixed with freshly prepared ethanol:acetic acid (19:1) at −20°C for 6 min before processing. Rat pancreas obtained from Dr. Teresa Krukoff (Department of Cell Biology, University of Alberta) was perfused in situ with 4% paraformaldehyde, and frozen thin sections (0.3 mm) were prepared and processed for indirect immunofluorescence essentially as described (Velasco et al., 1993).
Protein Purification and Sequencing
Four confluent 1800 cm2 rollerbottle cultures of NRK52E and BHK-21 cells were harvested by trypsinization. Cells were washed in growth media containing 5% calf serum and twice with cold PBS, before resuspension in PBS containing PMSF (400 μM). Triton X-100 was added to a final concentration of 1%, and cells were rocked at 4°C for 15 min to ensure efficient lysis. Lysates were subjected to centrifugation at 16,000 × g for 10 min at 4°C, and the resulting supernatants were incubated with LCH-7 for 4 h at 4°C on a rocking device after which BioMag goat anti-mouse IgG (Fc-specific) beads were added and incubation was continued for another 12 h. The magnetic beads were washed three times with 1% Triton X-100, 500 mM NaCl, 50 mM Tris-HCl, pH 7.4, three times with 0.2% Triton X-100, 1.0 M NaCl, 50 mM Tris-HCl, pH 7.4, and once with water. Proteins were eluted from the beads in nonreducing SDS sample at 95°C for 5 min. Eluates were reduced with 2-mercaptoethanol before SDS-PAGE on 10% gels and transfer to PVDF membranes. The membranes were washed three times with water followed by staining for 5 min with 0.1% amido black prepared in 45% methanol:10% acetic acid. Blots were destained with 50% methanol:10% acetic acid, three washes, 10 min each, followed by five washes in water. After air drying, the section of the blot containing the 95-kDa protein was excised and processed for internal sequencing at the Protein/DNA Technology Center at Rockefeller University (Fernandez et al., 1994).
Library Screening and cDNA Cloning and Sequencing
One of the internal peptide sequences obtained from GERp95, VQVHQDTLRT, was screened against the EST data banks using the program tblastn (Altschul et al., 1990). A rat EST that matched the sequence of the peptide was identified. A forward primer, 5′-GAA GGA AGC CAT ACC TCT G-3′, and a reverse primer, 5′-CAA CCA AAA GTA CAT GGT-3′, were used in PCR to amplify a 102-bp region of the rat EST from a rat N1S1 cDNA library prepared in λZAP (Wozniak et al., 1989). The 102-bp cDNA fragment was purified and subjected to a second round of amplification using the same primers with 32Pα-dATP substituted for dATP. The 32P-labeled cDNA product was used to screen the rat N1S1 cDNA library essentially as described (Sambrook et al., 1989). The 5′ 350 bp of the cDNA was obtained from a rat liver cDNA library made in λZAPII. Isolated clones were subjected to in vitro excision using VCSM13 helper phage, and both strands of the inserts were sequenced using the Department of Biochemistry DNA Sequencing Facility (University of Alberta).
Northern Blot Analysis
Total RNA was extracted from cultured cells and mouse tissues essentially as described (Chirgwin et al., 1979). Typically, 50 μg of RNA were separated on 1% agarose gels containing 6% formaldehyde and transferred to Hybond-N (Amersham) membranes by capillary blotting. Northern blots with poly(A)+ RNA prepared from human endocrine tissues and cancer cell lines were purchased from Clontech (Palo Alto, CA). Blots were hybridized with 32P-labeled cDNA probes corresponding to the 5′ end of the GERp95 cDNA, washed, and subjected to autoradiography as described (Sambrook et al., 1989).
Expression of Recombinant GERp95 and Polyclonal Antibody Production
Amino acids 197 to 430 of GERp95 were expressed as a GST fusion protein in Escherichia coli BL21 using the vector PGEX-3X (Pharmacia). Expression was induced with 0.1 mM isopropyl-β-d-thiogalactoside for 3 h at 37°C after which inclusion bodies containing the GST–GERp95 fusion protein were isolated (Harlow and Lane, 1988). The inclusion bodies were solubilized in SDS gel sample buffer and separated on 10% acrylamide gels. The gels were stained with ice-cold 250 mM KCl to visualize proteins, and the GST–GERp95 fusion protein was excised and electroeluted into dialysis membrane (14-kDa exclusion size) for 48 h at 20 mA in 40 mM Tris-acetate, pH 8.4, 0.02% SDS. The purified protein was dialyzed against 200 mM sodium bicarbonate/0.01% SDS before it was injected into rabbits (500 μg per animal).
Transcription/Translation
35S-labeled GERp95 was synthesized in vitro using a T7 polymerase/rabbit reticulocyte-coupled kit according to the manufacturer’s specifications. Where indicated, some reactions contained canine pancreatic microsomes. After translation, samples were adjusted to 10 mM CaCl2 and held on ice for 10 minutes. Samples were then incubated with a mixture of trypsin and chymotrypsin in the presence or absence of Triton X-100 (1%) on ice for 60 min. Reactions were terminated by the addition of aprotinin. VSV G protein was used as a positive control to verify the translocational activity of the microsomes.
Cell Fractionation and Membrane Extractions
NRK52E and NRK49F cells grown to confluence in 1800 cm2 rollerbottles were harvested by trypsinization and washed sequentially with ice-cold PBS containing 5%FBS and HME (10 mM HEPES, 250 mM mannitol, 0.5 mM EDTA, pH 7.4). Cells were resuspended in five volumes of HME containing 0.1 mM PMSF and homogenized with a ball-bearing style homogenizer (Balch et al., 1984). Homogenates were centrifuged at 1500 × g for 10 min at 4°C to pellet nuclei and unbroken cells. The postnuclear supernatant was centrifuged at 10,000 × g for 10 min at 4°C, and the resulting supernatant was collected and underlaid with a 20% sucrose/PBS cushion. Microsomes were collected by centrifugation at 100,000 × g average for 60 min at 4°C, resuspended in HME using a ground-glass tissue grinder, then used immediately for assays, or they were aliquotted, snap-frozen with liquid nitrogen, and stored at −80°C for further use.
For protease protection assays, microsomes (15–25 μl) were adjusted to 10 mM CaCl2 and incubated at 0°C for 40 min with varying amounts of trypsin and chymotrypsin (up to 40 μg each) with or without 1% Triton X-100 present. After 40 min, 40 μg of aprotinin were added to each sample, and incubation was continued for 5 min on ice. Samples were then analyzed by SDS-PAGE and immunoblotting.
Alkaline and high salt extraction of microsomes or 10,000 × g centrifugation of cell supernatants was performed as described (Suomalainen et al., 1990). Briefly, cell fractions (50–100 μl) were incubated with 10–20 volumes of HME, 50 mM sodium carbonate, pH 11.5, or 0.5–1.0 M KCl at 0°C for 45 min. Samples were underlaid with 0.1 volumes of 0.2 M sucrose, 30 mM HEPES, pH 11.5, 150 mM potassium acetate, 2.5 mM magnesium acetate, 1 mM DTT, and centrifuged at 50,000 rpm (100,000 × g average) for 30 min in a TLA 120.2 centrifuge at 4°C. For HME and KCl extractions, the 0.2 M sucrose cushions were prepared in PBS. The supernatants were then precipitated with trichloroacetic acid. Supernatant and pellet fractions were resuspended in 2× SDS gel sample buffer followed by SDS-PAGE and immunoblotting.
Rat liver membrane fractions were prepared as described (Cui et al., 1993).
Expression of GERp95 in Cultured Cells
For expression in NRK52E and NRK49F cells, it was necessary to use an alphavirus-mediated gene expression system. The cDNA for GERp95 was modified by PCR to include a 10 amino acid epitope tag (TDIEMNRLGK) from the C terminus of VSV G protein. Rabbit antibodies to the VSV G tag were used to differentiate between exogenous GERp95 and endogenous GERp95 in transfected cells. The GERp95 cDNA was excised from pBluescript (Stratagene, La Jolla, CA) with AvrII and _Apa_I and ligated into the Sindbis virus replicon vector pSinRep5 (Invitrogen, San Diego, CA) between the _Xba_I and _Apa_I sites. The resulting pSinRep5-GERp95 plasmid was linearized with _Not_I, and capped RNA was synthesized using an SP6 MessageMachine Kit from Ambion (Austin, TX). The plasmid DHBB that encodes the structural proteins of Sindbis virus was linearized with _Xho_I, and capped RNA was synthesized as above. BHK21 cells (5 × 106) were electroporated twice with equal molar amounts of DHBB and SinRep5-GERp95 RNAs (>5 μg each) using settings of 1.5 kV, 25 μF, and infinite resistance. Recombinant Sindbis viruses were harvested 24 h after electroporation. Cells grown on coverslips were infected with viruses encoding GERp95 and examined by indirect immunofluorescence after 16 h. COS cells were transfected with pCMV5-GERp95 using the calcium phosphate method and examined after 24–40 h (Sambrook et al., 1989).
RNA Interference
PCR was used to amplify the 5′ end of the C. elegans orthologue T07D3 (GenBank no. AF016682) on chromosome II from Bristol strain N2 genomic DNA. Sense and antisense RNA was synthesized in vitro using the Ambion Megascript kit. Equal volumes of sense and antisense RNA were combined with an equal volume of 3× injection buffer (20 mM potassium phosphate, 3 mM potassium citrate, 2% polyethylene glycol 6000, pH 7.5) and heated at 68°C for 10 min and then 37°C for 30 min to facilitate formation of double-strand RNA (Fire et al., 1998). RNA was injected into the syncytial cytoplasm of adult hermaphrodite gonads, and the F1 progeny were collected beginning 7 h after injection. Animals were examined by differential interference contrast microscopy. For each RNA sample, a minimum of four separate experiments were conducted using 10 injected animals per experiment. Typically, >80% of the F1 progeny showed the same RNA-specific phenotypes.
RESULTS
mAb LCH-7 Recognizes a Golgi- or ER-Associated Antigen
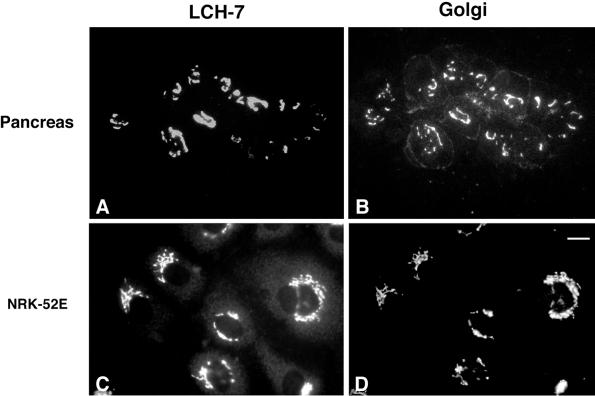
The objective of this study was to identify novel proteins of the ER and Golgi complex using mAbs prepared against intracellular membrane fractions. Pancreas membranes were used as the immunogen because acinar cells, which make up >90% of the organ tissue, are highly specialized for secretion and have a poorly developed endocytic system. Consequently, most of the acinar cell membranes are ER- and Golgi-derived (Bolender, 1974). Mice were immunized with an intracellular membrane fraction (density = 1.17 g/cm3) that consists primarily of smooth vesicles and is enriched in _cis_-Golgi elements (Saraste et al., 1987). A panel of hybridoma supernatants was screened by indirect immunofluorescence using various cell types. One of the hybridomas, LCH-7, secreted an antibody that reacted with the Golgi complex of pancreas (Figure 1, A and B, arrowheads). LCH-7 staining overlapped with Man II in the Golgi region; however, unlike Man II (Velasco et al., 1993), the LCH-7 antigen was not detected on the plasma membrane of acinar cells. We next determined whether the Golgi antigen recognized by LCH-7 was present in cells other than pancreas. NRK52E cells, which are an epithelial-type cell line derived from rat kidney, also exhibited Golgi staining for LCH-7 (Figure 1, C and D).
Figure 1.
LCH-7 recognizes an antigen in the Golgi complex of pancreas acinar and epithelial NRK cells. Rat pancreas sections and NRK cells were fixed and processed for double-label indirect immunofluorescence as described. Samples were incubated with the mAb LCH-7 (A and C) and the Golgi marker rabbit anti-Man II (B and D). Colocalization of LCH-7 and Man II can be seen in the Golgi of pancreas acinar and NRK52E cells (A–D). A limited amount of staining for Man II is also observed on the plasma membrane of the acinar cells (B). Bar in D, ∼10 μm.
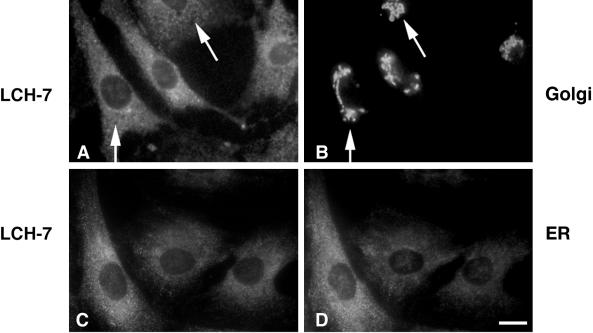
In contrast to NRK52E and pancreas acinar cells, LCH-7 did not stain the Golgi of the fibroblast NRK49F cells (Figure 2, A and B, arrows). LCH-7 recognized weakly staining cytoplasmic structures in NRK49F cells. Staining of other cultured cells such as BHK-21, CHO, clone 9, and REF-52 with LCH-7 revealed similar punctate cytoplasmic structures, but no Golgi staining (our unpublished results). The punctate cytoplasmic LCH-7 staining pattern in these cells is reminiscent of low-abundance ER proteins that do not display the classic continuous reticular pattern of ER membrane proteins (Hauri and Schweizer, 1997). When NRK49F cells were costained with LCH-7 and an antibody to the lumenal ER protein ERp72 (Urade et al., 1993), the staining patterns were very similar but not identical (Figure 2, C and D). Similar results were obtained using BHK cells (our unpublished results). Fractionation of rat liver membranes and expression of GERp95 in transfected cells confirmed that the highest concentrations of GERp95 are present in ER membranes (see Figures 7 and 8). These results indicate that the antigen recognized by LCH-7 is confined to ER-associated structures in NRK49F cells. Attempts to localize this antigen to the ER in NRK49F and BHK-21 cells by immunoelectron microscopy were unsuccessful, possibly because of poor reactivity of the antibody on fixed specimens.
Figure 2.
LCH-7 stains ER-associated structures in fibroblastic NRK cells. Cells were processed for double-label indirect immunofluorescence by staining with LCH-7 (A and C) and rabbit anti-Man II to label Golgi (B) or rabbit anti-ERp72 to label ER (D). No colocalization between LCH-7 and anti-Man II is evident (A and B, arrowheads), but some overlap is seen with the anti-ER antibody (C and D). Bar, ∼10 μm.
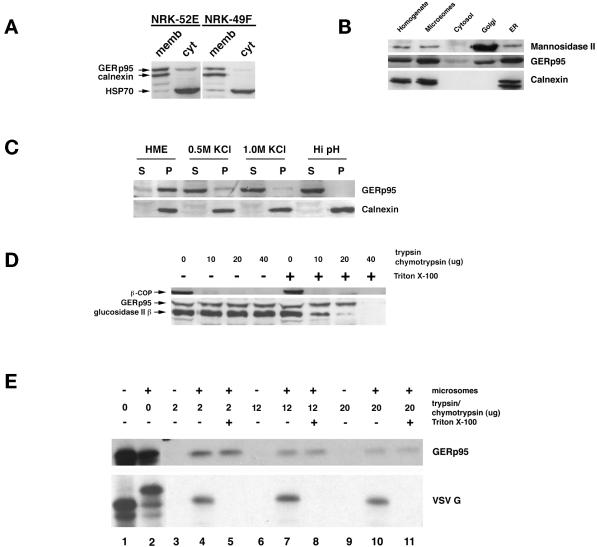
Figure 7.
Interaction of GERp95 with membranes in vivo and in vitro. (A) Crude microsomes and cytosol were prepared from NRK52E and NRK49F cells, and equivalent proportions normalized to starting volumes were separated by SDS-PAGE and transferred to PVDF membranes. The membranes were probed with rabbit antisera to GERp95, calnexin, and rat anti-HSP70. The majority of GERp95 and calnexin are found in the membrane fractions, whereas HSP70 is in the soluble fraction. (B) Fifty micrograms (protein) of rat liver fractions were subjected to SDS-PAGE and immunoblotting with antibodies to GERp95, and calnexin (ER marker) and Man II (Golgi). The highest concentrations of GERp95 are found in rough ER fractions. (C) NRK52E microsomes were extracted with HME, 0.5 M KCl, 1.0 M KCl, or Hi pH buffer (0.1 M sodium carbonate, pH 11.5), and then centrifuged at 100,000 × g for 60 min to obtain pellet (P) and soluble (S) fractions. Alkaline treatment and KCl washing resulted in complete extraction of GERp95 from membranes. (D) Microsomes were incubated at 0°C with increasing amounts of trypsin/chymotrypsin with or without 1% Triton X-100 followed by SDS-PAGE and immunoblotting with rabbit antiserum to GERp95, β-COP, and glucosidase II β-subunit. In the absence of detergent, GERp95 and glucosidase II β are insensitive to protease, but are completely digested when Triton X-100 is included. In contrast, β-COP, a peripheral membrane protein on the cytosolic side of membranes is completely degraded by the proteases even in the absence of detergent. (E) 35S-labeled GERp95 was synthesized in vitro in the presence or absence of canine pancreatic microsomes. Samples were subjected to digestion with varying amounts of trypsin/chymotrypsin in the presence or absence of Triton X-100 before SDS-PAGE and autoradiography. VSV G protein was used as a positive control to show translocational activity of the microsomes. VSV G is only protected from protease when microsomes are present and detergent is absent.
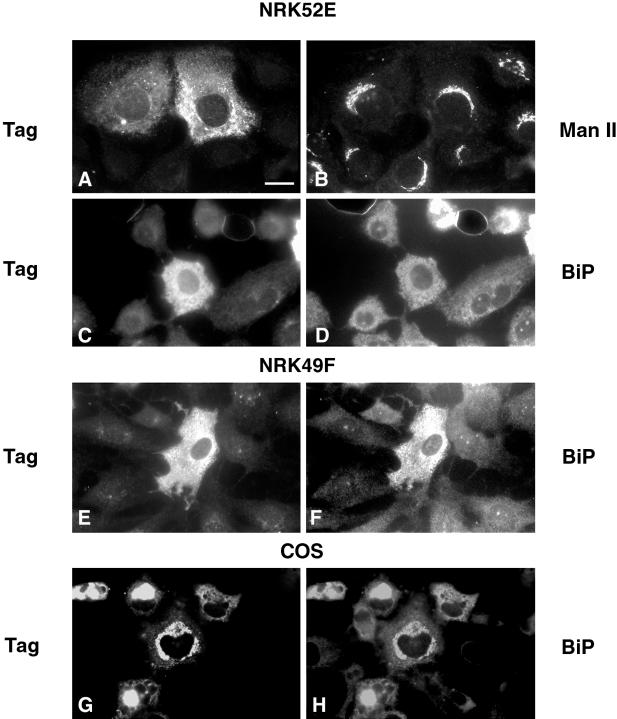
Figure 8.
Expression of liver-derived GERp95 cDNA in NRK and COS cells. Cells were infected with recombinant Sindbis viruses encoding liver-derived GERp95 cDNA, which includes an epitope tag. Sixteen hours after infection, cells were processed for double-label indirect immunofluorescence using rabbit anti-tag (A, C, E, and G), mouse anti-Man II (B), and mouse anti-BiP (D, F, and H). COS cells were transfected with a mammalian expression vector encoding epitope-tagged GERp95 and processed for indirect immunofluorescence 24–40 h later. Bar, 10 μm.
Purification of a 95-kDa Protein and Cloning of Its Corresponding cDNA
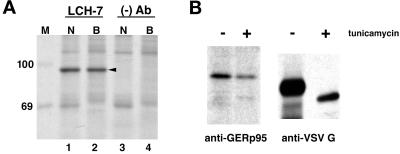
NRK52E and BHK-21 cells were biosynthetically labeled with [35S]methionine/cysteine, lysed, and immunoprecipitated with LCH-7 antibody followed by SDS-PAGE and fluorography. In both cell types, LCH-7 immunoprecipitated a protein with an apparent molecular mass of ∼95 kDa (Figure 3A, lanes 1 and 2). Similar results were obtained using NRK49F cells and primary rat hepatocytes (our unpublished results). The 95-kDa protein was immunoaffinity-purified using LCH-7 from both NRK52E and BHK-21 cells and prepared for microsequencing. Three internal peptides generated by lys-C digestion were sequenced (Fernandez et al., 1994). The peptide sequences were used to screen protein and nucleic acid data bases with the BLAST algorithm (Altschul et al., 1990). One of the peptides matched the sequence of a rat EST.
Figure 3.
Immunoprecipiation of GERp95 from cultured cells. (A) NRK52E (N) and BHK-21 (B) cells were biosynthetically labeled with 35S cysteine/methionine for 6 h before lysis and immunoprecipitation with LCH-7 or a mAb to CD8 ((−) Ab). Samples were subjected to SDS-PAGE on 8% gels followed by fluorography. A 95-kDa protein, GERp95 (arrowhead), is immunoprecipitated from both cell types with the LCH-7 antibody (lanes 1 and 2) but not the negative control Ab (lanes 3 and 4). 14C-labeled protein standards (kDa) are shown lane M. (B) NRK52E cells were biosynthetically labeled with 35S as above in the presence and absence of tunicamycin (3 μg/ml) before lysis and immunoprecipitation with LCH-7. Cells were infected with VSV and radiolabeled using the same conditions, and the VSV G protein was immunoprecipitated and subjected to SDS-PAGE and fluorography. The mobility of VSV G is increased in the presence of tunicamycin attributable to inhibition of N-linked glycosylation.
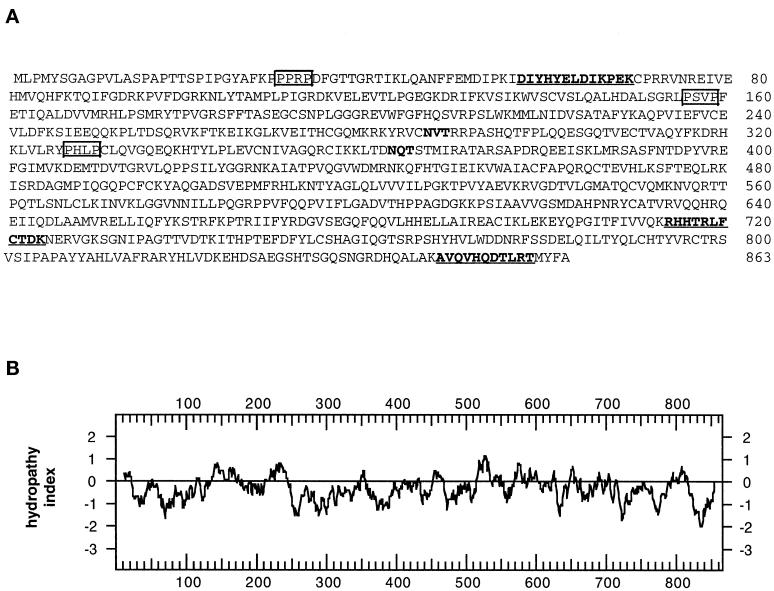
A 32P-labeled 102-bp cDNA probe was produced by PCR from a rat N1S1 hepatoma cDNA library using primers derived from the rat EST sequence. The probe was used to screen rat hepatoma and liver cDNA libraries from which we obtained a series of overlapping cDNA clones that comprised a total of 2743 nucleotides. The overlapping clones contained a single ORF that encodes a protein of 863 amino acids with a pI of 9.32 (Figure 4A). If translation occurs from the first in-frame methionine, the cDNA encodes a protein with a predicted molecular mass of 97,597 Da, which is in good agreement with the apparent molecular mass of ∼95 kDa (Figure 3A). All three of the peptides obtained from microsequencing of the purified protein were found in the translated cDNA sequence and were preceded by lysine residues as would be expected for lys-C–generated peptide fragments (Figure 4A, bold and underlined). Hydropathy analysis (Kyte and Doolittle, 1982) indicated that the protein does not contain hydrophobic regions that could function as signal peptides or transmembrane domains (Figure 4B). Accordingly, this suggested that GERp95 kDa is cytosolic and that the two consensus sites for addition of asparagine-linked carbohydrates (Figure 4A) are not used. This was confirmed by the experiment shown in Figure 3B. Treatment of the cells with tunicamycin did not affect the relative mobility of the 95-kDa protein by SDS-PAGE. To demonstrate that tunicamycin was inhibiting N-linked glycosylation in NRK52E, cells were infected with VSV, followed by biosynthetic labeling in the presence and absence of tunicamycin. As expected, the mobility of VSV G protein was increased in the presence of tunicamycin (Figure 3B).
Figure 4.
Predicted amino acid sequence and hydropathy analysis of GERp95. (A) Predicted amino acid sequence of GERp95. Internal peptide sequences obtained by microsequencing immunoaffinity-purified protein are underlined and shown in bold. Two consensus sites for N-linked glycosylation are highlighted in bold. Potential high-affinity SH3-binding sites (PxxP) are boxed. (B) Hydropathy analysis of GERp95 according to the algorithm of Kyte and Doolittle (1982) using a window size of 19 amino acids for scanning. (C) Sequence alignment of rat GERp95 and related proteins from C. elegans, A. thaliana, and S. pombe. Identical and conserved amino acids are indicated by shading in black and gray, respectively. The genomes of C. elegans and A. thaliana each encode multiple GERp95-related proteins, but only the closest relatives are shown in this figure.
Analysis of the GERp95 sequence with the Procite algorithm revealed the presence of numerous potential sites of phosphorylation for casein kinase II, cCMP, and protein kinase C (PKC), but metabolic labeling with 32P Pi revealed that the protein was not stably phosphorylated in NRK cells (our unpublished results). The amino terminal 123 amino acids of GERp95 is proline rich (14%) and contains three potential high-affinity SH3-binding sites represented by the sequence PxxP (Figure 4A). Finally, a glutamine-rich domain (11.2%) is located between amino acids 549 and 682.
A search of the nonredundant protein/nucleic acid data bases showed that GERp95 homologues were present in numerous animal and plant species. No homologues were found in the Saccharomyces cerevisiae or bacterial genome databases. GERp95 is 93.5% identical to a rabbit protein named eIF2C protein (Zou et al., 1998). EIF2C is involved in eukaryotic peptide chain initiation (Chakravarty et al., 1985); however, in the above paper (Zou et al., 1998), it was not shown that the cDNA reported actually encodes a protein with this activity. Over the continuous 813 amino acid stretch, the two proteins are 99.5% identical, indicating that they are orthologues. The published sequence of eIF2C is only 813 amino acids long and is missing the amino terminal 50 amino acids of GERp95. We have constructed deletion mutants of GERp95, which were expressed in vitro and in vivo, and the results indicate that GERp95 uses the start site reported in our sequence (our unpublished results).
The plant Arabidopsis thaliana contains at least four GERp95 homologues (Bohmert et al., 1998; Moussian et al., 1998; Lynn et al., 1999). The most well characterized of these proteins, AGO1 and ZLL, share 65% identity with each other, and 37.2 and 37.8% identity, respectively, with GERp95 at the amino acid level. Mutations in AGO1 and ZLL genes result in specific defects in plant architecture during early development, but their molecular functions remain unknown. Two homologues of GERp95, Piwi and Sting, have been described in Drosophila (Cox et al., 1998; Schmidt et al., 1999). The C. elegans genome contains at least 20 genes that encode proteins related to AGO1, ZLL, Piwi, Sting, and GERp95. The most closely related homologue in C. elegans is 64.8% identical to rat GERp95 at the amino acid level (Figure 4C). Finally, a hypothetical protein of unknown function encoded by a gene on S. pombe chromosome III shares 32.4% identity with GERp95. Sequence conservation within this family is greatest in the C terminal two-thirds of the proteins, whereas the amino terminal regions share much less homology (Figure 4C).
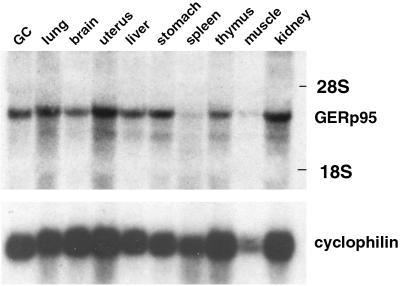
EST database searches revealed that GERp95 mRNAs are expressed in a wide variety of mammalian tissues and cell types such as placenta, fetal liver and spleen, pregnant uterus, colon and synovial carcinomas, and melanocytes. We probed total RNA isolated from various mouse tissues with a 32P-labeled cDNA probe derived from the 5′ end of the GERp95 coding region. GERp95 mRNAs (∼3 kb) are expressed at moderate levels in most tissues except for spleen (Figure 5). Similar results were obtained using RNA prepared from NRK52E, NRK49F, and BHK21 cells and poly(A)+ RNA isolated from human tissues and cell lines (our unpublished results). There were no apparent differences between GERp95-specific RNAs isolated from fibroblastic and epithelial NRK cells.
Figure 5.
Expression of GERp95-specific RNA in mouse tissues. A membrane containing 50 μg of total RNA isolated from mouse tissues was hybridized with a 32P-labeled cDNA probe derived from the 5′ coding region of the rat GERp95 cDNA. The blot was stripped and probed with a radiolabeled cyclophilin cDNA to show relative loading and integrity of RNA. The positions the 28S and 18S rRNAs are indicated.
Production of Antibodies to GERp95
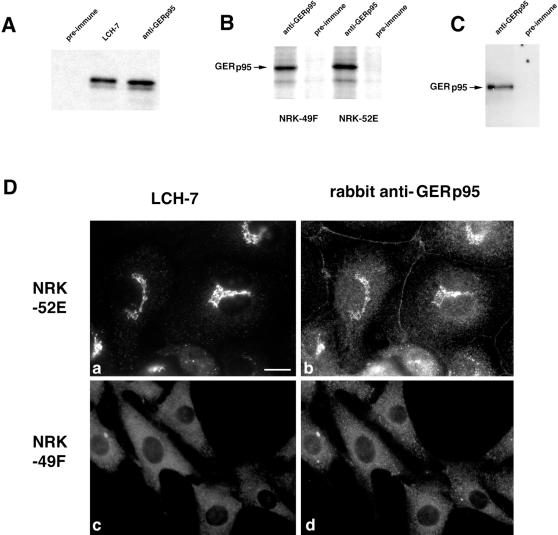
It was necessary to develop a polyclonal antibody to GERp95 that could be used for subcellular fractionation and immunoblot analysis. Amino acids 197 to 430 of GERp95 were expressed as a GST–fusion protein in E. coli, purified, and injected into rabbits. To demonstrate that the rabbit antibody recognized the same protein as LCH-7, radiolabeled GERp95 was synthesized using a coupled transcription/translation system, and the products were subjected to immunoprecipitation using LCH-7, rabbit anti-GERp95, or rabbit preimmune serum. Similar to LCH-7, rabbit anti-GERp95 precipitated a ∼95 kDa doublet, whereas preimmune serum did not (Figure 6A). The polyclonal antiserum immunoprecipitated a 95-kDa protein from radiolabeled NRK52E and 49F cells (Figure 6B) and also worked well for immunoblotting (Figure 6C). In addition, the signal from rabbit anti-GERp95 overlapped with that of LCH-7 in the Golgi region of NRK52E cells by indirect immunofluorescence (Figure 6D, a and b). As with LCH-7, rabbit anti-GERp95 did not react with the Golgi complex of NRK49F cells (Figure 6D, c and d). Instead, rabbit anti-GERp95 and LCH-7 staining were confined to punctate cytoplasmic elements in these cells. A variable amount of nonspecific cell border staining was observed in NRK52E cells when the polyclonal antibody was used for staining, but this was not the case for NRK49F cells (Figure 6D, a and b). These results indicate that the rabbit anti-GERp95 antibody recognizes the same protein as LCH-7.
Figure 6.
Characterization of a polyclonal antibody to recombinant GERp95. (A) GERp95 was expressed in vitro using a coupled transcription/translation system containing 35S methionine. The samples were subjected to immunoprecipitation with LCH-7, rabbit preimmune, or anti-GERp95 sera. Samples were subjected to SDS-PAGE on 8% gels followed by fluorography. Both LCH-7 and rabbit anti-GERp95 precipitate a major band of ∼95 kDa. (B) NRK49F and NRK52E cells were biosynthetically labeled with 35S cysteine/methionine for 4 h before lysis and immunoprecipitation with rabbit preimmune or anti-GERp95 serum. Rabbit anti-GERp95 serum specifically immunoprecipitates a protein with an apparent molecular weight of ∼95 kDa from both NRK49F and NRK52E cells. (C) NRK52E lysates were separated by SDS-PAGE, transferred to PVDF membrane, and probed with rabbit preimmune or anti-GERp95 serum. (D) NRK52E (a and b) and NRK49F (c and d) cells fixed with acid-alcohol were incubated with LCH-7 and rabbit anti-GERp95 serum. Both LCH-7 and rabbit anti-GERp95 stain the Golgi complex in NRK52E cells (a and b) but not in NRK49F cells (c and d). Bar, 10 μm.
GERp95 Is a Peripheral Membrane Protein
To determine what proportion of GERp95 was membrane-associated in NRK52E and NRK49F cells, crude membrane and cytosolic fractions were prepared for use in immunoblotting. In both epithelial- and fibroblast-type NRK cells, most GERp95 was found to copurify with membranes (Figure 7A). A small fraction of GERp95 partitioned into the cytosol of both cell types, whereas calnexin, an integral membrane protein of the ER (Wada et al., 1991), was found exclusively in the membrane fraction (Figure 7A). The constitutive form of HSP70 (Welch, 1991) was found mainly in the soluble fraction of both cell types (Figure 7A). GERp95 was also concentrated in membrane fractions prepared from rat liver hepatocytes, particularly in membranes of the rough ER (Figure 7B). Considerably less GERp95 was found in Golgi-enriched fractions of hepatocytes, which is consistent with indirect immunofluorescent staining of these cells with LCH-7.
We predicted that GERp95 was bound to the cytosolic side of membranes on the basis of the following observations. 1) Hydropathy analysis indicated that it does not contain a signal peptide that could mediate translocation across the ER (Figure 4B). 2) GERp95 contains two N-linked glycosylation sites that are not used (Figure 3B). We tested our prediction by extraction analysis of membranes isolated from NRK cells. Membranes were extracted with HME (physiological buffer), sodium carbonate, pH 11.5, or high salt (0.5–1.0 M KCl) and separated into membrane pellets and soluble fractions by centrifugation at 100,000 × g (Suomalainen et al., 1990). Alkaline treatment resulted in complete stripping of GERp95 from microsomes and its subsequent partitioning into the soluble phase, whereas calnexin, an integral membrane protein, remained in association with the membrane fraction (Figure 7C). Extraction with 0.5 and 1.0 M KCl, but not HME, also resulted in dissociation of the bulk of GERp95 from membranes (Figure 7C).
As a second means to verify the membrane orientation of GERp95, the microsomes were subjected to protease digestion with or without Triton X-100 present. The glucosidase II β-subunit, which is a lumenal ER protein (Arendt and Ostergaard, 1997), was used as a control for microsome integrity. Proteolysis of this 80-kDa protein was observed only when detergent was included in the assay, whereas β-COP, a peripheral membrane protein located on the cytosolic side of microsomes (Duden et al., 1991), was sensitive to protease in the absence of detergent (Figure 7D). Unexpectedly, GERp95 was resistant to trypsin/chymotrypsin in the absence of detergent and was digested only at the highest concentrations of protease when detergent was included (Figure 7D). In conditions where >80% of glucosidase II was proteolyzed (e.g., addition of 20 μg of each protease/detergent), the amount of intact GERp95 was not affected. This is not simply the result of this protein being inherently resistant to trypsin/chymotrypsin, because in vitro synthesized GERp95 is very sensitive to proteolysis (Figure 7E, lanes 3, 6, and 9). GERp95 that was synthesized in the presence of canine pancreatic microsomes was partially resistant to protease whether or not Triton X-100 was included (Figure 7E, lanes 4, 5, 7, 8, 10, and 11). As a positive control to show integrity of the microsomes, VSV G was used. VSV G is a type I glycoprotein of which all but 29 amino acids are translocated in microsomes (Katz et al., 1977). Consequently, G protein synthesized in the presence of microsomes was sensitive to protease when detergent was included during the incubation with protease, but not in its absence (Figure 7E, compare lanes 5, 8, and 11 with 4, 7, and 10). Unlike VSV G protein, inclusion of microsomes in the translation reactions did not alter the mobility of GERp95 attributable to glycosylation and/or signal peptide cleavage (lanes 1 and 2). Together these results are consistent with the possibility that GERp95 associates with the cytoplasmic side of membranes and is incorporated into a protease-resistant complex. The fact that not all of the GERp95 becomes resistant to protease in vitro could mean that components needed to assemble this complex are limiting in the rabbit reticulocyte lysate or microsome preparations.
Expression of GERp95 in Cultured Cells
As a first step toward understanding how differential distribution of GERp95 in fibroblast and epithelial-type NRK cells occurs, we constructed recombinant Sindbis viruses containing cDNAs encoding liver-derived epitope-tagged GERp95 that were then used to infect NRK and BHK cells. It was necessary to use this type of expression system because transient transfection of NRK cells was inefficient, and attempts to make stable cell lines expressing the GERp95 cDNA were unsuccessful. NRK49F and NRK52E cells were infected with recombinant Sindbis viruses and processed for indirect immunofluorescence. The exogenously expressed GERp95 did not overlap with the Golgi marker Man II in NRK52E cells (Figure 8A and B); however, the Sindbis virus-expressed GERp95 did overlap with the ER marker BiP in both types of NRK and COS cells (Figure 8, C–H). The experiments were also conducted with infected BHK cells, and the results were identical (our unpublished results). A significant number of cells that were expressing exogenous GERp95 appeared to contain elevated levels of BiP (Figure 8, F and H). Overexpression of another peripheral membrane protein Z0–3 (Haskins et al., 1998) did not result in its association with the ER in transfected cells (our unpublished results). These results indicate that the same GERp95 protein expressed in different cell types exhibits the same intracellular localization (ER) and suggest that cell-dependent differential localization is due to the presence of multiple GERp95 isoforms with different targeting information. The possibility that GERp95 becomes incorporated into different protein complexes in different cell types cannot be ruled out at this point.
A GERp95 Homologue Is Important for Germ Cell Development in C. elegans
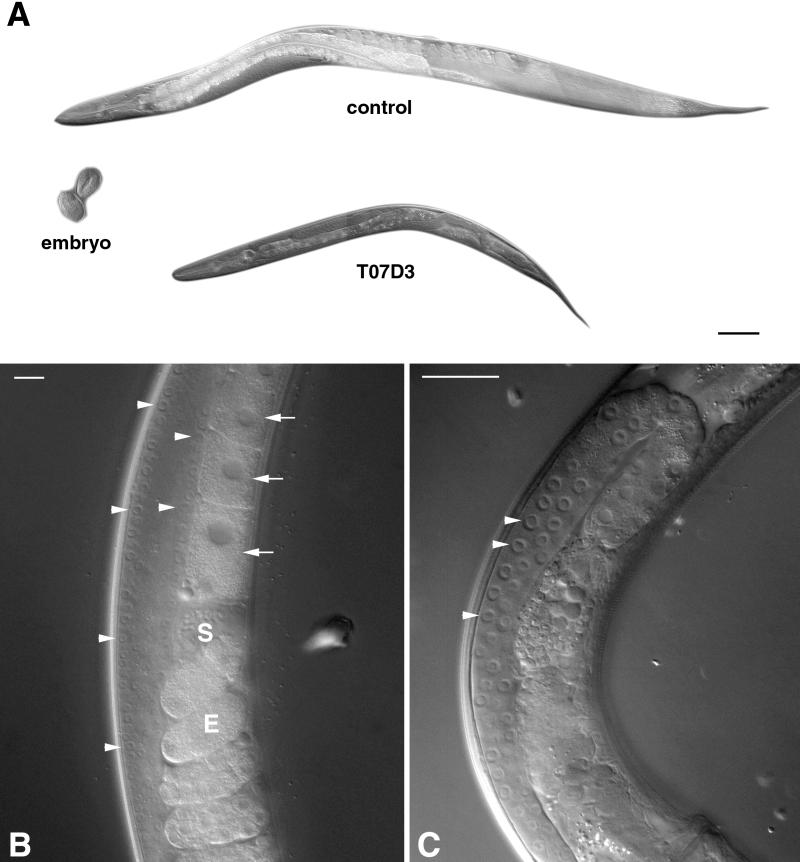
To ascertain whether GERp95-related proteins are important for development in animals, we used RNA interference to obtain the probable null phenotype for the C. elegans GERp95 orthologue, which shares 64.8% identity to rat GERp95 at the amino acid level. Injection of double-stranded RNA into germ-line tissues often results in the production of F1 progeny that are phenocopied for the null phenotype in that gene and thus allows rapid analysis of gene function in worms and Drosophila ( Sluder et al., 1997; Cox et al., 1998; Fire et al., 1998; Kennerdell and Carthew, 1998; Hobert et al., 1999). Double-stranded uncapped RNA from the 5′ coding region of the GERp95 orthologue on chromosome II was synthesized in vitro. To minimize the possibility of affecting the function of more than one gene, we avoided using a sequence from the 3′ end because this region is highly conserved throughout the gene family. The F1 progeny of injected hermaphrodites were viable and appeared otherwise normal except that they were much smaller than control-injected worms (Figure 9A). On average, the T07D3-injected worms were 50% smaller than control worms of the same age when measured for diameter and length. On closer examination of the animals, it was evident that there was a defect in the germ cells of the proximal gonad. Oocytes were formed normally and sperm was visible in the proximal gonads of control-injected worms (Figure 9B), but the gonads of T07D3-injected animals showed defects. Specifically, oocytes and sperm were never seen in the proximal gonads (Figure 9C). Consequently, these animals did not produce fertilized eggs or progeny, although they had germ-line stem cells in the mitotic zones of their distal gonads. Germ-line stem cells normally go through meiosis as they migrate toward the proximal gonad and develop into oocytes and sperm. There was no evidence of meiosis in the gonads of T07D3-injected worms as assayed by DAPI staining of fixed animals (our unpublished results). The germ-line stem cells, although normal in appearance, were fewer in number in the affected animals (Figure 9C). It remains to be determined whether this was simply due to the smaller size of the gonads in these animals or whether self-renewal of germ-line stem cells was affected.
Figure 9.
The GERp95 orthologue in C. elegans is important for oocyte development. Gonads of young hermaphrodite worms were injected with double-stranded RNA synthesized from empty pBluescript vector (control) or a C. elegans GERp95-like gene (T07D3) on chromosome II. A shows the progeny of injected worms that are now adult hermaphrodites of the same age. Two wild-type embryos are shown for comparison. Bar, ∼100 μm. Higher magnifications of the gonad regions of injected worms are shown in B and C. In control worms (B), germ-line stem cells in the mitotic proliferation zone of the distal gonad are indicated by arrowheads. Brick-shaped oocytes with large nuclei are indicated by arrows. Sperm (S) and eggs (E) are also visible in the proximal gonads of control worms but not in progeny of T07D3-injected animals (C). In the progeny of T07D3-injected worms, normal-looking germ-line stem cells are present in the distal gonad (arrowheads), but oocytes and sperm are absent from the proximal gonad. Bars in B and C, ∼25 μm.
To show the specificity of the technique, animals were injected with doubled-stranded RNAs from genes that have been shown to have defects either early or late in C. elegans development. Injection of apx-1 RNA resulted in F1 progeny that arrested embryonically (Mango et al., 1994), whereas animals injected with unc-119 produce progeny of normal size and morphology, but with neurological abnormalities (Maduro and Pilgrim, 1995) (our unpublished results). Interestingly, injection of double-strand RNA corresponding to the analogous 5′ region of two other C. elegans GERp95 homologues did not produce the same phenotype as the T07D3 (Pilgrim, Cikaluk, Hansen, and Hobman, unpublished observations). This suggests that the GERp95 orthologue is important for differentiation of germ-line stem cells into sperm and oocytes in C. elegans.
DISCUSSION
GERp95 Belongs to a Highly Conserved Family of Proteins
In this article we report the identification and characterization of GERp95, a membrane-associated protein that belongs to a family of highly conserved proteins in multicellular organisms and S. pombe. Sequence conservation is greatest in the C terminal two-thirds of this protein family (Figure 4C), indicating that this region may define the general functionality of these proteins. In contrast, the amino terminal regions are not well conserved and may be involved in isoform-specific functions. Protein database searches did not turn up any matches with known functional domains, such as kinase or nucleoside-binding domains, etc.
Articles describing mutations in GERp95 homologues from A. thaliana and Drosophila have recently been published (Bohmert et al., 1998; Cox et al., 1998; Moussian et al., 1998; Lynn et al., 1999; Schmidt et al., 1999). The functions of these proteins are discussed below. The studies listed above were limited to genetic and morphological analysis of mutant plants/animals, and nothing is known about the intracellular localization of these proteins, including whether they are membrane-associated. In C. elegans, there are at least 20 homologues of GERp95. T07D3 shares the highest degree of identity (64.8%) to GERp95 and is assumed to be the worm orthologue.
Differential Localization of GERp95
One of the most intriguing properties of GERp95 is that it is differentially localized to the Golgi or ER depending on cell type. Our data is most consistent with the possibility that GERp95 exists in a protease-resistant complex on the cytoplasmic side of intracellular membranes. In certain types of epithelial cells (NRK52E and pancreatic acinar cells), GERp95 is found primarily on Golgi membranes, whereas in the six other cell types examined (NRK49F, BHK-21, Clone 9, CHO, REF52, and rat hepatocytes), two of which are epithelial, the protein is confined to ER-associated structures. We found no evidence to suggest that GERp95 cycles between the ER and Golgi complex (our unpublished results). Furthermore, unlike coatomer (Klausner et al., 1992), brefeldin A does not cause rapid dissociation of GERp95 from Golgi membranes in NRK52E cells.
One obvious possibility to account for the differential localization of GERp95 is that there are cell-specific isoforms of this protein. Presumably, GERp95 in pancreas acinar and NRK52E cells contains a Golgi localization signal, whereas in NRK49F and other cell types (e.g., hepatocytes), it harbors an ER-targeting motif. The fact that a liver-specific GERp95 localizes to the ER in NRK52E cells is consistent with this scenario. Our preliminary work indicates that GERp95 is not stably phosphorylated or palmitoylated, nor are there any potential myristoylation sites within the protein (our unpublished results). Therefore it seems unlikely that these types of posttranslational modifications could mediate the membrane association of this protein.
Differential targeting of protein isoforms can be accomplished by various mechanisms. For example, there are two isoforms of glutamic acid decarboxylase, one of which is associated with the Golgi (Solimena et al., 1993). The two isoforms are encoded by separate genes, but they differ significantly only in their amino terminal regions (Erlander and Tobin, 1991). Second, there are at least 11 different isoforms of PKC, most of which are encoded by separate genes, that localize to various intracellular structures including the ER and Golgi complex (Goodnight et al., 1995). Alternative translation or splicing of mRNA from the same gene is another means by which cells generate protein isoforms that are targeted to different intracellular locations. For instance, alternative initiation of translation in the syntaxin 5 mRNA results in two forms of the protein, one of which is localized to the ER, the other to the Golgi (Hui et al., 1997). Alternative splicing of the mRNA for the heterotrimeric G protein subunit Gαi2 results in products that are targeted to the plasma membrane or the Golgi complex (Montmayeur and Borelli, 1994). RT-PCR was used to prepare GERp95-specific cDNA from NRK52E cells for sequencing. NRK52E and liver GERp95 cDNA sequences were identical over 2589 nucleotides of coding region (our unpublished results). It is possible, however, that the NRK52E mRNA differs from liver mRNA in the 5′end. These differences would not have been detected by our RT-PCR analysis and can only be examined by isolating GERp95-specific cDNAs from an NRK52E cDNA library.
Functions of GERp95 and Related Proteins
The molecular functions of GERp95 and related proteins are unknown at this point, but mutations in the genes that encode these proteins produce drastic phenotypes in plants and animals, indicating that they have important roles in development. ZLL and AGO1 are GERp95 homologues in A. thaliana that were identified by screening for leaf development mutants. They share 37.2 and 37.8% identity, respectively, with GERp95 and have overlapping functions in maintaining stem cells in an undifferentiated state during the transition from embryo-specific development to repetitive organ formation (Bohmert et al., 1998; Moussian et al., 1998; Lynn et al., 1999). AGO1 may have additional roles in development because mutations in this gene result in pleiotropic defects, not all of which can be attributed to abnormal stem cell division. Presumptive null mutations in a GERp95 homologue of Drosophila, Sting, results in male sterility and lethal maternal effects (Schmidt et al., 1999). A second Drosophila homologue, Piwi, is required for germ-line stem cell renewal (Cox et al., 1998). Piwi is required for asymmetric division of germ-line stem cells, and decreased expression of this protein causes a reduction in the proliferation of these cells attributable to differentiation of daughter cells. Drosophila Piwi and Sting proteins are 23.4 and 22.4% identical, respectively, to rat GERp95 and are therefore paralogues of GERp95 rather than orthologues. RNA interference in C. elegans with a Piwi homologue termed PRG-1 indicated that this protein is required for germ-line stem cell maintenance in worms too (Cox et al., 1998). The data from this study suggest a role for the GERp95 orthologue T07D3 in germ-line stem cell maturation. Affected animals do not produce progeny, presumably because of failure of germ-line stem cells to differentiate into oocytes and sperm. In contrast, PRG-1–injected animals do produce germ cells and progeny, albeit at reduced levels (Cox et al., 1998). A potentially important difference between GERp95 (and its C. elegans orthologue T07D3) and Sting, Piwi, AGO1, and ZLL, is that the latter four proteins have nuclear localization signals, whereas GERp95 does not. We have also shown by indirect immunofluorescence and subcellular fractionation that GERp95 is associated with cytoplasmic membranes. The intracellular localizations of Sting, Piwi, AGO1, and ZLL have not been investigated.
Recently the sequence of a rabbit cDNA encoding a protein (eIF2C) with 93.5% overall identity to rat GERp95 was reported (Zou et al., 1998). Our results suggest that the published sequence of eIF2C is missing the 5′ end of the cDNA. This protein, which is clearly the rabbit orthologue of GERp95, has been suggested to play an important role in eukaryotic peptide chain initiation; however, this activity was never demonstrated. Furthermore, it seems unlikely that GERp95 and related proteins would have general roles in protein translation, given the fact that probable null mutations in a number of A. thaliana and Drosophila homologues produce very specific developmental phenotypes but are not lethal (Bohmert et al., 1998; Cox et al., 1998; Moussian et al., 1998; Lynn et al., 1999). We have also tested two different polyclonal GERp95 antibodies to determine whether they could inhibit translation in vitro, and they did not (our unpublished results). Together, these data are not consistent with GERp95 and related proteins having general roles in protein translation.
The fission yeast S. pombe encodes a hypothetical protein that shares 32.4% amino acid identity with rat GERp95. It is thought that S. pombe diverged from other ascomycete yeasts very early, and it is considered to be much more similar to mammalian cells than S. cerevisiae (Moreno et al., 1991). Accordingly, fission yeast such as S. pombe are highly polarized structures, and unlike budding yeast, the control of growth occurs at different times and places within the cell (Nurse, 1994; Mata and Nurse, 1998). The function of this yeast protein is unknown, but it is highly probable that it has functions unrelated to cell differentiation.
GERp95 and many of its homologues contain potential high-affinity SH3-binding sites (PxxP). SH3 domains are found on a wide variety of proteins, including cytoskeletal components and proteins involved in signal transduction (Mayer and Eck, 1995). For example, the cytoskeletal-associated protein spectrin contains a centrally located SH3 domain (De Matteis and Morrow, 1998). There are more than one dozen isoforms of spectrin, some of which are associated with the Golgi complex and/or ER (Zagon et al., 1986; Beck et al., 1994; Burkhardt, 1998). These observations come into perspective when one considers some of the parameters that affect differentiation of stem cells. Asymmetric division of germ-line stem cells is thought to be controlled in part by a cytoplasmic organelle called the fibrous body-membranous organelle or spectrosome/fusome (Deng and Lin, 1997; L’Hernault, 1997). To our knowledge, the ultrastructural features of these organelles, which are found in germ-line stem cells and cytoblasts, have not been described, but it is thought that they are derived from the ER and/or Golgi. They are enriched in protein such as spectrin, ankyrin, and dynein, which may facilitate the interaction of these organelles with cytoskeletal networks. Given the fact that they sequester many of the same types of cytoplasmic proteins that the ER and Golgi do, it is tempting to speculate that GERp95 may also associate with these structures in stem cells. If this is true, it would imply that GERp95 has different functions in differentiated cells than it does in germ-line stem cells. Interestingly, mutations in ER/Golgi proteins that function in protein translocation and protein trafficking, such as Sec61p and presenilins, respectively, cause specific defects in oocyte and sperm formation, respectively, in C. elegans (Iwasaki et al., 1996; Arduengo et al., 1998).
In summary, GERp95 and related proteins represent a diverse family of proteins that may perform similar functions at different intracellular localizations. Analysis of the 20 GERp95-related proteins in C. elegans with the PSORT II program suggests that members of this family may have diverse intracellular localization, including the nucleus, mitochondria, cytoplasm, ER, and plasma membrane. In this respect, the GERp95 family may be analogous to PKC isoforms that localize to many different subcellular compartments (Goodnight et al., 1995). Induction of cultured cells to undergo morphological changes can result in the concomitant recruitment of PKC-α to the ER and translocation of PKC-γ to the Golgi (Goodnight et al., 1995). Some isoforms of PKC are constitutively associated with specific organelles, whereas in other cases, similar to GERp95, different intracellular locations for the same PKC isozyme have been described in different cell types (Germano et al., 1994; Goodnight et al., 1995). RACKs, the receptors that mediate localization of PKC, have been shown to interact with other proteins, including integrins, and may therefore provide the link for signal transduction from intracellular membranes where PKC isoforms localize to the cell surface or nucleus, thereby mediating changes in cell morphology and physiology (Liliental and Chang, 1998). We hypothesize that the localization of GERp95 to distinct intracellular membranes has functional consequences, similar to PKC. The identification of proteins that interact with members of the GERp95 family may provide clues as to how they function and how differential localization occurs. We are currently engaged in mapping targeting signals within GERp95 and identifying potential interacting proteins.
ACKNOWLEDGMENTS
We are grateful to Drs. Chris Nicchitta, Marilyn Farquhar, Teresa Krukoff, Kelly Moremen, Hanne Ostergaard, Bruce Stevenson, Carolyn Machamer, Paul Kim, and Jean Vance for their generous gifts of reagents, and to Drs. Marita Hobman, Paul Melançon, and Bruce Stevenson for critical reading of this manuscript. We thank LeighAnn Giebelhaus and Honey Chan for technical assistance and preparation of the electron micrographs, respectively. This work was supported by grants from the Alberta Heritage Foundation for Medical Research and the Medical Research Council of Canada awarded to T.C.H.
Abbreviations used:
ER
endoplasmic reticulum
GERp95
Golgi ER protein 95 kDa
Man II
mannosidase II
VSV
vesicular stomatitis virus
REFERENCES
- Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arduengo PM, Appleberry OK, Chuang P, L’Hernault SW. The presenilin protein family member SPE-4 localizes to an ER/Golgi derived organelle and is required for proper cytoplasmic partitioning during Caenorhabditis elegans spermatogenesis. J Cell Sci. 1998;111:3645–54. doi: 10.1242/jcs.111.24.3645. [DOI] [PubMed] [Google Scholar]
- Arendt CW, Ostergaard HL. Identification of the CD45-associated 116-kDa and 80-kDa proteins as the alpha- and beta-subunits of the alpha-glucosidase II. J Biol Chem. 1997;272:13117–13125. doi: 10.1074/jbc.272.20.13117. [DOI] [PubMed] [Google Scholar]
- Balch WE, Dunphy WG, Braell WA, Rothman JE. Reconstitution of the transport of protein between successive compartments of the Golgi measured by the coupled incorporation of N-acetylglucosamine. Cell. 1984;39:405–416. doi: 10.1016/0092-8674(84)90019-9. [DOI] [PubMed] [Google Scholar]
- Beck KA, Buchanan J, Malhotra V, Nelson WJ. Golgi spectrin: identification of an erythroid β-spectrin homolog associated with the Golgi complex. J Cell Biol. 1994;127:707–723. doi: 10.1083/jcb.127.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron JJM, Brenner MB, Thomas DY, Williams DB. Calnexin: a membrane-bound chaperone of the endoplasmic reticulum. Trends Biochem Sci. 1994;19:124–128. doi: 10.1016/0968-0004(94)90205-4. [DOI] [PubMed] [Google Scholar]
- Bohmert K, Camus I, Bellini C, Bouchez D, Caboche M, Benning C. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 1998;17:170–180. doi: 10.1093/emboj/17.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolender RP. Stereological analysis of the guinea pig pancreas. J Cell Biol. 1974;61:269–287. doi: 10.1083/jcb.61.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- Brooks D. Protein processing: a role in the pathophysiology of genetic disease. FEBS Lett. 1997;409:115–120. doi: 10.1016/s0014-5793(97)00423-7. [DOI] [PubMed] [Google Scholar]
- Burkhardt JK. The role of microtubule-based motor proteins in maintaining the structure and function of the Golgi complex. Biochim Biophys Acta. 1998;1404:113–126. doi: 10.1016/s0167-4889(98)00052-4. [DOI] [PubMed] [Google Scholar]
- Chakravarty I, Bagchi MK, Roy R, Banerjee AC, Gupta NK. Protein synthesis in rabbit reticulocytes. Purification and properties of an Mr 80,000 polypeptide (CoeIF-2A80) with CoeIF-2A activity. J Biol Chem. 1985;260:6945–6949. [PubMed] [Google Scholar]
- Chirgwin J, Przybyla A, MacDonald R, Rutter W. Isolation of biologically active RNA from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Z, Vance JE, Chen MH, Voelker DR, Vance DE. Cloning and expression of a novel phosphatidylethanolamine N-methyltransferase. J Biol Chem. 1993;268:16655–16663. [PubMed] [Google Scholar]
- De Matteis MA, Morrow JS. The role of ankyrin and spectrin in membrane transport and domain formation. Curr Opin Cell Biol. 1998;10:542–9. doi: 10.1016/s0955-0674(98)80071-9. [DOI] [PubMed] [Google Scholar]
- Deng W, Lin H. Spectrosomes and fusomes anchor mitotic spindles during asymmetric germ cell divisions and facilitate the formation of a polarized microtubule array for oocyte specification in Drosophila. Dev Biol. 1997;189:79–94. doi: 10.1006/dbio.1997.8669. [DOI] [PubMed] [Google Scholar]
- Duden R, Griffiths G, Frank R, Argos P, Kreis TE. β-COP, a 110 kDa protein associated with nonclathrin-coated vesicles and the Golgi complex, shows homology to β-adaptin. Cell. 1991;64:649–665. doi: 10.1016/0092-8674(91)90248-w. [DOI] [PubMed] [Google Scholar]
- Erlander M, Tobin A. Two genes encode distinct glutamate decarboxylases. Neuron. 1991;7:91–100. doi: 10.1016/0896-6273(91)90077-d. [DOI] [PubMed] [Google Scholar]
- Erlich R, Gleeson PA, Campbell P, Dietzsch E, Toh B-H. Molecular characterization of trans-Golgi p230. J Biol Chem. 1996;271:8328–8337. doi: 10.1074/jbc.271.14.8328. [DOI] [PubMed] [Google Scholar]
- Farquhar M, Hauri H-P. Protein sorting and vesicular traffic in the Golgi apparatus. In: Berger E, Roth J, editors. The Golgi Apparatus. Basel: Birkhauser Verlag; 1997. pp. 63–128. [Google Scholar]
- Farquhar M, Palade G. The Golgi apparatus: 100 years of progress and controversy. Trends Cell Biol. 1998;8:2–10. doi: 10.1016/S0962-8924(97)01187-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez J, Andrews L, Mische S. An improved procedure for enzymatic digestion of polyvinylidene difluoride-bound proteins for internal sequence analysis. Anal Biochem. 1994;214:112–117. doi: 10.1006/abio.1994.1148. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fritzler M, Lung C, Hamel J, Grifitth K, Chan E. Molecular characterization of Golgin-245, a novel Golgi complex protein containing a granin signature. J Biol Chem. 1995;270:31262–31268. doi: 10.1074/jbc.270.52.31262. [DOI] [PubMed] [Google Scholar]
- Germano P, Gomez J, Kazanietz M, Blumberg P, Rivera J. Phosphorylation of the g chain of the high affinity receptor for immunoglobulin E by receptor-associated protein kinase C-δ. J Biol Chem. 1994;269:23102–23107. [PubMed] [Google Scholar]
- Gonatas N. The Golgi apparatus in disease. In: Berger E, Roth J, editors. The Golgi Apparatus. Basel: Birkhauser Verlag; 1997. pp. 247–273. [Google Scholar]
- Goodnight J, Mischak H, Kolch W, Mushinski J. Immunocytochemical localization of eight protein kinase C isozymes overexpressed in NIH 3T3 fibroblasts. J Biol Chem. 1995;270:9991–10001. doi: 10.1074/jbc.270.17.9991. [DOI] [PubMed] [Google Scholar]
- Griffith K, Chan E, Lung C, Hamel J, Guo X, Miyachi K, Fritzler M. Molecular cloning of a novel 97-kDa Golgi complex autoantigen associated with Sjorgren’s syndrome. Arthritis Rheum. 1997;40:1963–1702. doi: 10.1002/art.1780400920. [DOI] [PubMed] [Google Scholar]
- Hammond C, Helenius A. Quality control in the secretory pathway. Curr Opin Cell Biol. 1995;7:523–529. doi: 10.1016/0955-0674(95)80009-3. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Haskins J, Gu L, Wittchen E, Hibbard J, Stevenson B. ZO-3, a novel member of the MAGUK protein family found at the tight junction, interacts with ZO-1 and occludin. J Cell Biol. 1998;141:199–208. doi: 10.1083/jcb.141.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauri H-P, Schweizer A. The ER-Golgi membrane system: compartmental organization and protein traffic. In: Hoffmann J, Jamieson J, editors. Handbook of Physiology. New York: Oxford University Press; 1997. pp. 605–642. [Google Scholar]
- Hobert O, Moerman DG, Clark KA, Beckerle MC, Ruvkun G. A conserved LIM protein that affects muscular adherens junction integrity and mechanosensory function in Caenorhabditis elegans. J Cell Biol. 1999;144:45–57. doi: 10.1083/jcb.144.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobman TC, Woodward L, Farquhar MG. The rubella virus E1 glycoprotein is arrested in a novel postER, preGolgi compartment. J Cell Biol. 1992;118:795–811. doi: 10.1083/jcb.118.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui N, Nakamura N, Sonnichsen B, Shima D, Nilsson T, Warren G. An isoform of the Golgi t-SNARE, Syntaxin 5, with an endoplasmic reticulum retrieval signal. Mol Biol Cell. 1997;8:1777–1787. doi: 10.1091/mbc.8.9.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtley S, Helenius A. Protein oligomerization in the endoplasmic reticulum. Annu Rev Cell Biol. 1989;5:277–307. doi: 10.1146/annurev.cb.05.110189.001425. [DOI] [PubMed] [Google Scholar]
- Iwasaki K, McCarter J, Francis R, Schedl T. emo-1, a Caenorhabditis elegans Sec61p gamma homologue, is required for oocyte development and ovulation. J Cell Biol. 1996;134:699–714. doi: 10.1083/jcb.134.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz FN, Rothman JE, Knipe DM, Lodish HF. Membrane assembly: synthesis and intracellular processing of the vesicular stomatitis viral glycoprotein. J Supramol Struct. 1977;7:353–70. doi: 10.1002/jss.400070308. [DOI] [PubMed] [Google Scholar]
- Kennerdell JR, Carthew RW. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell. 1998;95:1017–1026. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- Klausner RD, Donaldson JG, Lippincott-Schwartz J. Brefeldin A: insights in the control of membrane traffic and organelle structure. J Cell Biol. 1992;116:1071–1080. doi: 10.1083/jcb.116.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuismanen E, Saraste J. Low temperature-induced transport blocks as tools to manipulate membrane traffic. In: Tartakoff A, editor. Methods in Cell Biology. San Diego: Academic Press; 1989. pp. 257–277. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle R. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- L’Hernault WW. Spermatogenesis. In: Riddle D, Blumenthal T, Meyer B, Priess J, editors. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1997. pp. 274–294. [PubMed] [Google Scholar]
- Liliental J, Chang D. Rack1, a receptor for activated protein kinase C, interacts with integrin β subunit. J Biol Chem. 1998;273:2379–2383. doi: 10.1074/jbc.273.4.2379. [DOI] [PubMed] [Google Scholar]
- Lynn K, Fernandez A, Aida M, Sedbrook J, Tasaka M, Masson P, Barton MK. The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development. 1999;126:469–481. doi: 10.1242/dev.126.3.469. [DOI] [PubMed] [Google Scholar]
- Maduro M, Pilgrim D. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics. 1995;141:977–988. doi: 10.1093/genetics/141.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mango S, Thorpe C, Martin P, Chamberlain S, Bowerman B. Two maternal genes, apx-1 and pie-1, are required to distinguish the fates of equivalent blastomeres in the early Caenorhabditis elegans embryo. Development. 1994;120:2305–2315. doi: 10.1242/dev.120.8.2305. [DOI] [PubMed] [Google Scholar]
- Mata J, Nurse P. Discovering the poles in yeast. Trends Cell Biol. 1998;8:163–167. doi: 10.1016/s0962-8924(98)01224-0. [DOI] [PubMed] [Google Scholar]
- Mayer BJ, Eck MJ. SH3 domains. Minding your p’s and q’s. Curr Biol. 1995;5:364–367. doi: 10.1016/s0960-9822(95)00073-x. [DOI] [PubMed] [Google Scholar]
- Montmayeur J-P, Borelli E. Targeting of Gα12 to the Golgi by alternative spliced carboxyl-terminal region. Science. 1994;263:95–98. doi: 10.1126/science.8272874. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Moussian B, Schoof H, Haecker A, Juergens G, Laux T. Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBO J. 1998;17:1799–1809. doi: 10.1093/emboj/17.6.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narula N, McMorrow I, Plopper G, Doherty J, Matlin K, Burke B, Stow J. Identification of a 200-kDa, brefeldin-sensitive protein on Golgi membranes. J Cell Biol. 1992;117:27–38. doi: 10.1083/jcb.117.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P. Fission yeast morphogenesis-posing the problems. Mol Biol Cell. 1994;5:613–616. doi: 10.1091/mbc.5.6.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Saraste J, Palade GE, Farquhar MG. Antibodies to rat pancreas Golgi subfractions: identification of a 58 kDa cis-Golgi protein. J Cell Biol. 1987;105:2021–2029. doi: 10.1083/jcb.105.5.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Palumbo G, Bozzetti MP, Tritto P, Pimpinelli S, Schafer U. Genetic and molecular characterization of sting, a gene involved in crystal formation and meiotic drive in the male germ line of Drosophila melanogaster. Genetics. 1999;151:749–760. doi: 10.1093/genetics/151.2.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer A, Fransen JAM, Bächi T, Ginsel L, Hauri H-P. Identification, by a mAb, of a 53-kDa protein associated with a tubulo-vesicular compartment at the cis-side of the Golgi apparatus. J Cell Biol. 1988;107:1643–1653. doi: 10.1083/jcb.107.5.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitia R, Meldolesi J. Endoplasmic reticulum: a dynamic patchwork of specialized subregions. Mol Biol Cell. 1992;3:1067–1072. doi: 10.1091/mbc.3.10.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluder A, Lindblom T, Ruvkun G. The Caenorhabditis elegans orphan nuclear hormone receptor gene nhr-2 functions in early embryonic development. Dev Biol. 1997;184:303–319. doi: 10.1006/dbio.1997.8544. [DOI] [PubMed] [Google Scholar]
- Solimena M, Aggujaro D, Muntzel C, Dirkx R, Butler M, De Camilli P, Hayday A. Association of GAD-65, but not GAD-67, with the Golgi complex of transfected Chinese hamster ovary cells mediated by the N-terminal region. Proc Natl Acad Sci USA. 1993;90:3073–3077. doi: 10.1073/pnas.90.7.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen M, Garoff H, Baron MD. The E2 signal sequence of Rubella virus remains part of the capsid protein and confers membrane association in vitro. J Virol. 1990;64:5500–5509. doi: 10.1128/jvi.64.11.5500-5509.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urade R, Takenaka Y, Kito M. Protein degradation by ERp72 from rat and mouse liver endoplasmic reticulum. J Biol Chem. 1993;268:22004–22009. [PubMed] [Google Scholar]
- Velasco A, Hendricks L, Moremen KW, Tulsiani DRP, Touster O, Farquhar MG. Cell type-dependent variations in the subcellular distribution of α-mannosidase I and II. J Cell Biol. 1993;122:39–51. doi: 10.1083/jcb.122.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada I, Rindress D, Cameron PH, Ou W-J, Doherty JJ, Louvard D, Bell AW, Dignard D, Thomas DY, Bergeron JJM. SSRa and associated calnexin are major calcium binding proteins of the endoplasmic reticulum membrane. J Biol Chem. 1991;266:19599–19610. [PubMed] [Google Scholar]
- Welch WJ. The role of heat-shock proteins as molecular chaperones. Curr Opin Cell Biol. 1991;3:1033–1038. doi: 10.1016/0955-0674(91)90125-i. [DOI] [PubMed] [Google Scholar]
- Wozniak RW, Bartnik E, Blobel G. Primary structure analysis of an integral membrane glycoprotein of the nuclear pore. J Cell Biol. 1989;108:2083–2092. doi: 10.1083/jcb.108.6.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagon IS, Higbee R, Riederer BM, Goodman SR. Spectrin subtypes in mammalian brain: an immunoelectron microscopic study. J Neurosci. 1986;6:2977–2986. doi: 10.1523/JNEUROSCI.06-10-02977.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou C, Zhang Z, Wu S, Osterman J. Molecular cloning and characterization of a rabbit eIF2C protein. Gene. 1998;211:187–194. doi: 10.1016/s0378-1119(98)00107-3. [DOI] [PubMed] [Google Scholar]