Gene expression changes in the course of normal brain aging are sexually dimorphic (original) (raw)
Abstract
Gene expression profiles were assessed in the hippocampus, entorhinal cortex, superior-frontal gyrus, and postcentral gyrus across the lifespan of 55 cognitively intact individuals aged 20–99 years. Perspectives on global gene changes that are associated with brain aging emerged, revealing two overarching concepts. First, different regions of the forebrain exhibited substantially different gene profile changes with age. For example, comparing equally powered groups, 5,029 probe sets were significantly altered with age in the superior-frontal gyrus, compared with 1,110 in the entorhinal cortex. Prominent change occurred in the sixth to seventh decades across cortical regions, suggesting that this period is a critical transition point in brain aging, particularly in males. Second, clear gender differences in brain aging were evident, suggesting that the brain undergoes sexually dimorphic changes in gene expression not only in development but also in later life. Globally across all brain regions, males showed more gene change than females. Further, Gene Ontology analysis revealed that different categories of genes were predominantly affected in males vs. females. Notably, the male brain was characterized by global decreased catabolic and anabolic capacity with aging, with down-regulated genes heavily enriched in energy production and protein synthesis/transport categories. Increased immune activation was a prominent feature of aging in both sexes, with proportionally greater activation in the female brain. These data open opportunities to explore age-dependent changes in gene expression that set the balance between neurodegeneration and compensatory mechanisms in the brain and suggest that this balance is set differently in males and females, an intriguing idea.
Keywords: entorhinal cortex, hippocampus, microarray, sex differences, superior frontal gyrus
Aging is associated with mild changes in cognitive capacity even in cognitively intact humans, including declines in memory and executive function that are associated with the hippocampus (HC) and frontal cortex. Paradoxically, these age-related changes in cognitive function are not well accounted for by corresponding age-related neuron loss and synaptic change in cortical or temporal structures. For example, despite reductions in cortical thickness, unbiased stereological assessment reveals that overall neuronal number in the human brain declines <10% over the age range of 20–90 years (1), and cortical neuron and synapse numbers are relatively maintained. Although the hilus of the HC does appear to undergo mild age-related neuron loss, other hippocampal subregions show increased dendritic and synaptic complexity with increasing age (2, 3), and similar synaptic remodeling is apparent in the frontal and temporal cortex (4).
Alterations at the gene expression level may help account for mild cognitive decline in aging even in the absence of gross histopathologic changes. Changes in gene expression can have robust effects on brain function from the cellular to the behavioral level, with potential critical effects on cognitive function if they occur in frontal and temporal areas. Although a number of studies have investigated gene expression changes in conditions of profound cognitive decline such as Alzheimer's disease (AD), little is known about the genomic changes that occur in the brains of cognitively intact humans across the normal adult lifespan. Currently, two microarray studies have assessed gene expression profiles across aging, both of which were limited to the frontal/prefrontal cortex (5, 6). How gene expression profiles change across the human adult lifespan in other brain regions is unknown. For example, of particular interest are temporal cortical regions such as the HC and entorhinal cortex (EC), key regions affected in AD and mild cognitive impairment (MCI). Understanding the aging-related changes in gene profiles that occur in temporal and prefrontal brain regions may help elucidate the vulnerability of these regions to functional decline in normal aging and in age-related neurodegenerative diseases. In addition, it is likely that other brain regions undergo changes in gene expression across the lifespan, including less vulnerable brain regions such as the postcentral gyrus (PCG) (somatosensory cortex), that is virtually devoid of pathology in aging and AD. Global examination of how aging alters gene expression over the lifespan, in both cognitive and noncognitive brain regions, may reveal that there are common themes of gene change in aging that occur globally across the brain, an idea that has not yet been addressed.
A second issue that has not been fully explored by genomic profile analysis is that aging may impact the brain differently in men and women. Differential neuroanatomical aging has been documented between the sexes, with men generally exhibiting larger age-related brain atrophy and cerebrospinal fluid increases than women over the entire lifespan (7, 8), particularly in the frontal and temporal lobes (9, 10). However, previous gene chip studies in humans have grouped males and females in the analyses, so that possible gender-related alterations may have been obscured. In adult mice, thousands of genes show sexual dimorphism in tissues as diverse as liver, fat, muscle, and brain (11). The human brain is unlikely to be an exception. Moreover, if male and female brains undergo unique profiles in development, as is generally accepted, might they not also exhibit sexually dimorphic responses in aging?
Recent advents in high-throughput gene expression technology have made possible the simultaneous assessment of activity of all genes in the genome. This technology offers enormous power to assess global changes in gene activity and is ideally suited for gaining broad insight into genomewide effects of aging in different brain regions and potentially unique patterns of brain aging between males and females. The present study addresses these gaps in knowledge by assessing gene expression profiles in the HC, EC, superior frontal gyrus (SFG), and PCG across the lifespan of intact males and females, aged 20–99 years.
Results
Brain Regions Show Differential Responsiveness to Aging Across the Lifespan.
Gene chips (Affymetrix U133 plus 2.0) were run on 174 samples from the HC, SFG, EC, and PCG from 55 individuals across four age categories (20–39, 40–59, 60–79, and 80–99 years) (supporting information (SI) Table S1). Clustering analysis of the individual cases revealed that samples within a group were correlated (Fig. S1). Reliably present probe sets (>50% present call across chips) were processed for differential expression by using GeneSpring software (Agilent Technologies), based on GC Robust Multi-array Average (GC-RMA) summarized expression values and a relatively stringent statistical cut-off (P < 0.01, cross-gene error model estimate of variability). To further increase stringency while minimizing loss of true positives, GC-RMA-based significant probe sets were validated by reanalysis of the chips by using an independent algorithm-based approach (Probe Logarithmic Intensity Error, PLIER) applying the same stringent criteria as for GC-RMA. Only probe sets validated by the two approaches were considered for further analysis to balance the risk of type I and type II errors inherent in the analysis of gene chip data (for details, see SI Text). Equally powered groups were used across all comparisons (Table S2). For an initial global view of the relationship between brain regions and age, unsupervised hierarchical clustering of reliably present probe sets was used to group expression profiles of the four brain regions (HC, SFG, EC, and PCG) at each age category (20–39, 40–59, 60–79, and 80–99 years).
The SFG and PCG show the largest number of aging-related changes, whereas the EC shows the fewest.
Clustering results suggested that each brain region has a distinct “signature” gene expression change with aging (Fig. S2). Age was a more dominant clustering factor than region in the SFG and PCG, suggesting that greater aging-related changes may occur in these regions than in the HC and EC. In contrast, groups clustered first by region then by age in the HC and EC, suggesting that the HC and EC undergo gene profile changes with age that are unique from how aging affects other brain regions. In all regions, the youngest age groups (20–39 and 40–59 years) clustered and the two older cohorts (60–79 and 80–99 years) clustered, suggesting that cases could be broadly grouped into a younger cohort of individuals (20–59 years) and an aged cohort (60–99 years).
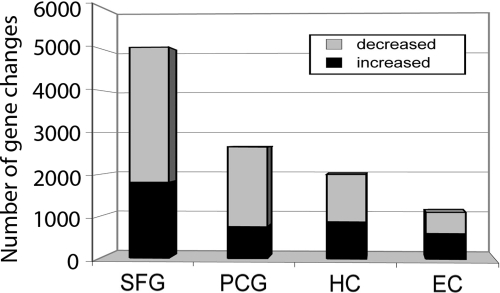
Based on these clustering results, gene expression was first compared between young (20–59 years) and aged (60–99 years) cases independently in each brain region, for an initial broad view of how different brain regions respond to aging. Numbers of differentially expressed genes were used as an index to estimate the magnitude of age-related change, using equally powered comparisons across regions (Table S2), revealing surprising heterogeneity in the responses of different brain regions to aging (Fig. 1). Notably, the SFG showed the largest number of differentially expressed genes between the two age groups (5,029 probe sets), followed by the PCG (2,653 probe sets), HC (2,003 probe sets), and EC (1,110 probe sets). The majority of genes in the SFG and PCG (64% and 72%, respectively) showed down-regulated expression in aging, whereas the distribution of up-regulated and down-regulated genes was nearly balanced in the HC and EC. Interestingly, the EC (in the temporal area, phylogenetically older than the SFG and PCG, and particularly vulnerable to decline in MCI and AD), exhibited the fewest gene changes with increasing age, undergoing nearly five times fewer gene response than the SFG.
Fig. 1.
Differential gene responsiveness to aging across the brain in cognitively normal controls. The SFG shows the greatest number of gene changes with aging, followed by the PCG and HC. The EC showed the fewest responsive genes.
The sixth and seventh decades are a period of robust gene change.
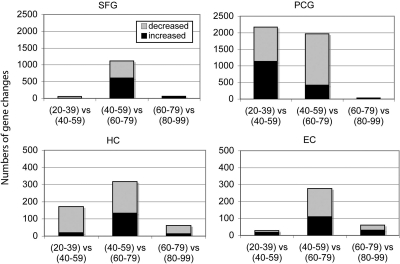
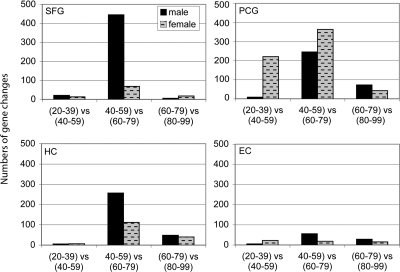
Characterizing the time course of change across aging in further detail, patterns of gene changes over 20-year increments were compared to determine whether specific windows of time were characterized by more extensive gene change and how they compared across different brain regions. For this analysis, gene profiles in the following age groups were compared: (20–39 vs. 40–59), (40–59 vs. 60–79), and (60–79 vs. 80–99), using equally powered groups for comparison (Table S2). Paralleling the initial results, the four brain regions continued to show unique patterns of response across aging. Notably, the greatest number of gene changes was observed in the shift from ages 40–59 to 60–79, a pattern that was observed in all brain regions (Fig. 2). These results suggest that ages 60–79 represents a period of dynamic change in gene activity for cortical regions of the brain, in particular the SFG and PCG. Interestingly, in addition to a large increase in gene change during the sixth and seventh decades, the PCG showed gene responses earlier in the fourth and fifth decades, a time period where the other brain regions showed relatively little gene change. These results indicate that aging does not affect all brain regions to a similar degree, and that the sixth and seventh decades of life may be a critical transition phase for the brain, particularly for cortical regions.
Fig. 2.
Brain regions show different patterns of change across the lifespan. Patterns of gene changes over 20-year increments were compared, with males and females combined. A notably large increase in gene change was observed in the shift from 40–59 to 60–79 years, occurring in all brain regions. The PCG additionally shows gene responses earlier in the fourth and fifth decades, a time period when other brain regions show relatively little gene change.
The Brain Undergoes Sexually Dimorphic Responses Across Aging.
Taking advantage of the power provided by the relatively large sample size across the aging time-course, it was possible to separately analyze gene changes in men and women, thereby addressing the novel idea that gender-specific differences in brain aging may exist at the level of gene activity.
The male brain undergoes more global gene change than the female brain during aging.
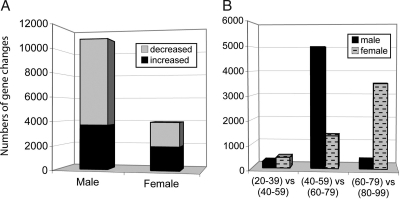
To obtain a global view of potential sex differences in brain aging, gene expression was compared between young (20–59 years) and aged (60–99 years) cases globally across the brain, independently for each sex, by using equally powered comparisons (Table S2). Marked differences emerged between men and women in the pattern of gene change occurring with aging (Fig. 3A). Globally, males showed more than three times as many gene changes across the brain than did females (≈10,891 vs. 3,491 probe sets, respectively) (Fig. 3A). Moreover, when global brain aging changes were analyzed across 20-year increments (Fig. 3B), the gender-specific analysis revealed that males underwent substantial gene change in the transition to the sixth and seventh decades of life (5,002 probe sets), whereas females showed relatively few gene changes (1,353 probe sets) during this age range. In contrast, females showed progressively more gene changes with increasing age, with the largest numbers of genes responding in the eighth and ninth decades of life. Interestingly, whereas males showed a large gene response in the sixth and seventh decades, few genes showed altered expression in the subsequent decades. These results suggest that in the male brain gene expression generally remains stable after age 80, whereas in the female brain genes continue to undergo age-related change into the eighth and ninth decades of life.
Fig. 3.
The male brain undergoes more global gene change than the female brain during aging [(20–59 yrs) vs. (60–99 yrs)]. (A) Gene change was assessed independently for males and females. Relative to females, males showed more than three times as many gene changes globally across the brain and more down-regulated than up-regulated genes. (B) Patterns of global gene changes over 20-year increments reveal that males underwent substantial gene change in the transition to the sixth and seventh decades of life, whereas females showed relatively few gene changes at this age range. In contrast, whereas gene expression appears to be stable after age 80 in the male brain, females show more gene changes with increasing age, with the largest numbers of genes responding across the brain in the eighth and ninth decades.
Region-specific sexually dimorphic patterns of aging.
Striking sex-specific patterns of gene change were further apparent when each brain region was assessed independently for males and females. Sex differences were observed at a number of levels, including differences in overall aging [e.g., when comparing the gene profiles of the young (20–59 years) and aged (60–99 years) cohorts] (Fig. 4) and at specific time windows across the lifespan (Fig. 5).
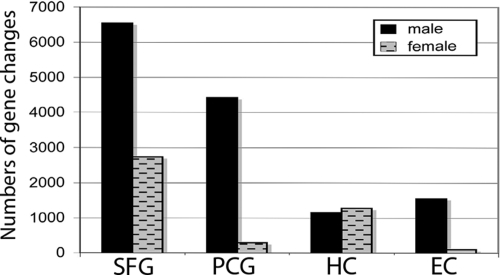
Fig. 4.
In aging, males show more gene changes than females in all brain regions except the HC. Numbers of genes showing differential expression between young (20–59 years) and aged (60–99 years) cases was assessed independently for males and females in each brain region.
Fig. 5.
Gene expression profiles across the lifespan undergo sexually dimorphic and region-specific changes. Patterns of global gene changes over 20-year increments were compared independently for males and females in each brain region. Pronounced sexually dimorphic patterns are apparent in the SFG and PCG, whereas the HC and EC show less dramatic differences between males and females.
The SFG, PCG, and EC all showed striking sexual dimorphism in the numbers of genes showing aging-related changes, with males consistently showing more gene change in the SFG, PCG, and EC (Fig. 4). Strikingly, in males, all brain regions show a marked increase in gene response in the transition to the seventh and eighth decades, which was most notable in the SFG (Fig. 5). Although females tended to show a slight increase in the numbers of genes changing in the seventh and eighth decades as well, the tendency was less pronounced than in males in all brain regions with the exception of the PCG (Fig. 5). A very different sexually dimorphic pattern was observed in the PCG, where it was the female PCG that showed more gene change than males in both the transitions to the fourth and fifth decades, as well as the sixth and seventh decades. Interestingly, the change in gene expression in the responding genes in the female PCG appearing to be a transitory effect, as most of the genes that spike in the fourth and fifth decades return to near baseline expression in the sixth and seventh decades (Fig. S3), such that the female PCG shows little change in gene profile overall in aging (young vs. aged) (Fig. 4). In contrast, whereas the female PCG showed little change in gene profile across aging, the male PCG showed substantial change in aging overall (Fig. 4), with a prominent spike in the seventh and eighth decades (Fig. 5). Finally, in the HC, whereas males showed more gene change during the transition to the seventh and eighth decades than females (Fig. 5), overall in aging the gene change was similar between the sexes (Fig. 4). Taken together, these results indicate that aging does not affect all brain regions to a similar degree, that males show greater numbers of gene changes with aging in several brain regions, and that the HC shows minimal sexually dimorphic changes in aging, unlike the SFG, PCG, and EC.
Males and females show different themes of global gene change with aging.
Previous gene chip studies of brain aging and AD have typically focused on the number of genes altered in a particular age group or condition, as above, or have emphasized changes in a few genes that may be particularly salient to the aging process or disease state. Putting the various genes that exhibit significant alterations into a broader, functional context, however, may have several benefits. In particular, to the extent that alterations in multiple genes subserving particular biochemical or physiological pathways are internally consistent, greater confidence in the individual results may be provided (12). Curated databases such as Gene Ontology (GO) (13) can be applied to gene lists to assign significant genes to functional categories, providing insight into alterations in classes of genes or signaling pathways. In addition to assigning genes to functional groups within various ontologies, a P value can be calculated for each enriched category as a measure of overrepresentation, providing a “rank ordering” of significant pathways or functional categories.
Accordingly, GO categorization was next applied to functionally characterize the sexually dimorphic responses that occur in the brain across the lifespan. Global gene responses across the brain were characterized to identify themes in aging that occur across the brain and to determine how these themes compare in males and females. To identify classes of genes that show aging-related changes globally in the brain, GO categorization was applied to the significant gene lists from the comparison of young (20–59 years) vs. aged (60–99 years) groups across brain regions for males (Table S3) and females (Table S4). These identified genes were first parsed into lists of decreasing or increasing genes for each sex, followed by GO categorization and ranking of GO category enrichment based on P value. To identify the most biologically meaningful categories, GO categories were restricted by the following criteria: a minimum enrichment of P < 0.01, consisting of no more than 20% of the probe sets on the gene chip and containing at minimum eight probe sets.
Down-regulated genes show sexually dimorphic patterns in aging.
As shown in Fig. 3A, >3-fold more gene changes occur across the male brain during aging than in the female brain. These genes can be separated into those undergoing decreased or increased expression. Notably, a greater percentage of these genes showed down-regulated expression in males than in females. Down-regulated genes constituted 66% of the gene list in males (7,202 genes, of which 4,303 had GO annotations) and 50% of the gene list in females (1,994 genes, 1,015 GO-annotated). Further, GO categorization of these genes revealed that different functional categories of genes were affected by aging in male versus female brains.
In males, there appeared to be a general decreased catabolic and anabolic capacity with aging, with down-regulated genes showing heavy enrichment in categories related to energy production, RNA processing, and protein synthesis/transport. For example, enriched categories related to energy production included down-regulation of genes involved in electron transport, oxidative phosphorylation, ribonucleotide metabolism, ATP metabolism/biosynthesis, and mitochondrial transport (Table S5). Enrichment of these categories selectively in males suggests that a predominant theme in aging in the male brain is a decreased capacity for energy production, whereas this does not appear to be a major theme in aging of the female brain (Table S6). In addition to a broad decrease in genes related to energy production, another principal theme of aging in the male brain is related to a general decreased capacity for protein synthesis and transport. Specifically, highly enriched categories included ribosome-related processes (rRNA processing, ribosome biogenesis), mRNA processing, translation initiation and elongation, and general protein processing (folding, localization, and transport, including retrograde transport of proteins from Golgi to the endoplasmic reticulum) (Table S5). These results suggest that, in parallel with a decreased capacity for energy production, the male brain shows decreased capacity for protein generation, suggesting broad decreased catabolic and anabolic capacity with aging.
Whereas down-regulated genes in the male brain showed a broad enrichment of energy-related categories, this was not seen in the female brain. In contrast, down-regulated genes in the female brain showed unique enrichment in categories of neuronal morphogenesis (axon guidance and neurite morphogenesis) and intracellular signaling and signal transduction (Table S6). Like males, females showed some decline in anabolic capacity (specifically related to biopolymer metabolism and protein ubiquination), but to a much lesser degree than in males, where decreased capacity for protein generation appeared on a multitude of levels. Interestingly, both sexes showed a similar down-regulation of genes involved in transmission of nerve impulse/synaptic transmission in aging.
These results suggest that different main themes of change predominate in male versus female brains in the aging process. Overall in aging, the female brain showed fewer gene changes than males, and the male brain in particular may have a decreased capacity for anabolic and catabolic function with increasing age.
Age-related activation of immune- and inflammation-related genes in both male and female brains.
Of the genes identified as significantly different in young versus aged cohorts, up-regulated genes constituted 34% of the gene list in males (3,689 probe sets) and 50% of the gene list in females (1,947 probe sets). In contrast to the striking differential enrichment between males and females in categories of genes undergoing down-regulation, there were many similar categories of genes showing increased expression with age across the brain in both sexes.
Notably, inflammation and immune function genes emerged as the top enriched category for both sexes, including genes involved in classical complement signaling, toll-like receptor (TLR) signaling, antigen processing via MHCI and MHCII, IFκ-B/NF-κB signaling, and macrophage activation (Tables S7 and S8). Across the brain, up-regulation of genes related to inflammation and immune components appeared to be proportionally greater for females, making up 10% of up-regulated genes for females and 5.3% for males. Of the individual brain regions, the HC and EC showed the most significant enrichment of inflammation and immune-related genes in both males and females, with 17% of gene changes in the aging HC relating to inflammation/immune function. In addition to the EC and HC, females but not males showed additional highly significant enrichment of immune-related genes in the PCG. Aging-dependent changes in immune- and inflammation-related genes in the HC were validated by RT-PCR, focusing on a subset of key factors including complement component C3, CD14, TLR 2, TLR 4, TLR 7, and TOLLIP, an inhibitor of the Toll-like signaling pathway that may be particularly important in controlling innate inflammatory mechanisms. The RT-PCR data paralleled the microarray results for these genes and showed comparable age-related increases in C3, CD14, TLR2, TLR4, and TLR7 and decreased expression of TOLLIP (Fig. S4). These data suggest that immune activation occurs in the HC with normal aging and is accompanied by decreased compensatory inhibition, particularly of the innate immune system (e.g., decreased TOLLIP expression). Further, whereas immune activation occurs across the brain in both males and females during aging, the female brain shows a proportionally greater immune representation (relative to all aging up-regulated genes), suggesting that this is a more prominent component of aging in the female brain.
Up-regulated gene categories show overlapping and sexual dimorphic enrichment.
In addition to a marked increase in immune components, several other categories of up-regulated genes showed enrichment across the brain in aging. Males and females showed similar up-regulation across the brain of genes involved in cell death (≈6.5% of list), angiogenesis/blood vessel development (1.1% of list), and oxygen/gas transport, among others (Tables S7 and S8). Finally, although many categories of up-regulated genes were common to both the male and female brain, several categories showed sex-specific enrichment. For example, females (but not males) showed significant up-regulation of genes involved in integrin-mediated signaling (1% of list) and hemostasis (1.7% of list) (Table S4). In contrast, up-regulated genes in males (but not females) showed significant enrichment in RNA catabolism (Table S7).
Sex differences in age-related gene profiles reflect gender-specific signatures and differential longevity.
Sex-differences in gene profiles are likely a reflection of both sex-specific expression patterns and differential longevity trends between males and females. Women live on average 5–10 years longer than men, suggesting that some of the sex differences in brain aging may be caused by gene responses being shifted on the aging curve for women relative to men. Such a shift would predict that the types of changes that occur in males in the transition to the seventh and eighth decades would occur later in females, for example, in the transition to the ninth and 10th decades. To assess this contribution to the gender differences, gene changes were compared between males and females in these two timeframes. The significant gene list from males in the transition to the seventh and eighth decades (5,002 probe sets) were compared with the significant genes changing in females in the ninth and 10th decades (3,493 probe sets), to identify the degree of overlap. A total of 873 genes were shared in common, representing only 17% of genes in the male list and 25% of the genes in the female list, indicating that >75% of the genes changing in males and females in these age ranges are unique to one gender or the other. To evaluate whether the categories of gene change were similar, even if the genes themselves showed little overlap, GO categories of gene change occurring in these time frames were compared. GO analysis revealed that different patterns of gene change occur in males and females in these timeframes, with males uniquely showing enrichment in genes related to oxidative phosphorylation/energy production as a top enriched category. Clearly in the data set, there are a number of genes that are not showing delayed expression changes in females and represent true gender differences in aging. However, some highly enriched categories of gene function were similar between males and females, in particular related to protein transport and targeting, suggesting that these categories of genes are shifted in the age profile with females showing these changes later than males. Taken together, these data suggest that although some of the gender differences in aging may be related to longevity a majority of gene changes remain relatively unique to each gender.
Discussion
Gene expression patterns are dynamic throughout the course of life. Here, we report that changes in gene profiles across the lifespan can be gender- and region-specific in the human brain. A priori, it might be expected that the HC and EC would show the greatest gene profile alterations across the lifespan, as these brain regions are particularly vulnerable to degeneration in age-related disease, whereas least affected would be cortical regions such as the somatosensory cortex. Contrary to our expectations, the HC and EC show relatively stable gene expression profiles across normal aging, whereas the SFG and PCG regions are the most changed across the lifespan. Notably, the SFG shows extensive changes in gene profile, changes that are particularly prominent in the sixth and seventh decades in males. In general, in the male brain, the sixth and seventh decades appear to be a period of prominent aging across several brain regions, with subsequent gene expression generally remaining stable after age 80. In contrast, in the female brain, changes in gene profiles tend to be more progressive with increasing age, depending on the brain region, and genes continue to change into the eighth and ninth decades of life. Interestingly, these trends are consistent with the gender differences observed in the age-related risk of dementia (14, 15). Whereas females show nearly linearly increasing prevalence of dementia with increasing age from 77 to >95 years, the prevalence for men does not change after ≈85 years of age (15), suggesting that the female brain shows increased vulnerability to decline with advanced age, more so than the male brain. Thus, our data reveal that, across the lifespan, there are certain timeframes that are characterized by greater gene change, depending on brain region and gender, such that the gene profile does not necessarily show progressively more change with increasing age. The observation that aging affects the SFG, PCG, HC, and EC brain regions differently is in agreement with a previous study demonstrating that the cortex undergoes more gene expression changes with age than the cerebellum (16). In addition, the observation that the SFG undergoes extensive age-related change in gene profile is in agreement with a previous study documenting that expression of ≈7.5% of interrogated probesets in two subregions of the prefrontal cortex show age-related change across the lifespan (6). Notably, the aging-related genes identified in the SFG in the current study show a 75%–81% concordance with the genes identified in the two previous microarray studies investigating the frontal/prefrontal cortex, despite differences in the precise anatomical regions used (e.g., BA10 vs. BA9) or tissue composition (pure gray matter vs. gray/white matter mixed) (5, 6). The gene changes in the SFG are likely related to the evolving capacities for reasoning, attentional processes, and executive function that occur across the lifespan and suggest that the SFG is continually reprogramming throughout life.
The overall patterns of GO categories affected in aging that we document globally across the brain are generally in agreement with reports on the prefrontal cortex, the only region previously assessed for aging-related changes in human by using microarrays (5, 6). These studies reported that across the lifespan the greatest changes in the prefrontal cortex occurred in pathways related to synaptic plasticity, signal transduction, mitochondrial function, and potential cellular defense mechanisms (e.g., induction of stress responses, antioxidant responses, and DNA repair genes) (5, 6). By looking at categories of change across multiple brain regions, our results expand current knowledge of aging-related gene change that has until now been limited to the prefrontal region. In addition, our study reveals that different categories of gene change dominate in the male and female brain, underscoring the observation that the human brain undergoes sexually dimorphic changes in gene profiles during cognitively normal aging.
Our study reveals that, globally across the brain in aging, the majority of genes show decreased expression, a pattern that was more prominent in the male brain. Notably, the major categories of genes showing down-regulated expression in males were related to protein processing and energy generation, suggesting that broad decreased catabolic and anabolic capacity occurs with aging, particularly in the male brain. Previous microarray studies in rodents have consistently reported altered protein turnover (increased degradation) in aging (17–19), a theme of brain aging that has not previously emerged in human microarray studies of the prefrontal cortex alone (6). Consistent with the present findings, metabolic decline has been documented in human imaging studies (20, 21). Moreover, gender-dependent changes in cerebral blood flow and brain activity with aging (10, 22), task performance (23–25), and memory (26, 27) have been suggested in several studies.
Although the majority of genes that were altered globally across the brain showed decreased expression in aging, there were classes of genes that were up-regulated. Consistent with the emerging idea that increased inflammation is a feature of aging across tissues and organisms (e.g., for reviews in humans, see refs. 28 and 29), the most significant categories of up-regulated genes in both males and females were related to immune activation and inflammation. Inflammation has multiple complex roles, with the capacity to be neuroprotective under certain circumstances, while tending to be detrimental to neuronal health with prolonged or extensive inflammation. In addition to immune activation, other classes of genes that are up-regulated globally across the brain in aging include genes controlling angiogenesis, cell death, certain families of signal transduction, and cell growth responses. Although many gene changes are likely contributing to age-related declines in function, some gene changes may reflect compensatory responses that slow or counteract aging-related changes.
The mechanisms responsible for the sexually dimorphic patterns in gene expression profiles in brain aging are undoubtedly multifaceted. Gender differences are probably a composite of hormonal elements, environmental factors, and overall lifestyle components that affect overall health. Sex differences are not only determined by gender-related differences in sex steroid levels, but also by gender-specific tissue and cellular characteristics that mediate sex-specific responses to a variety of stimuli. It is clear that this is the case in vascular biology (30) and is likely to be true for other organ systems, including the brain. Such gender-specific tissue and cellular differences likely become amplified with age, in conjunction with environmental exposure and overall lifestyle. In addition, differences in longevity between men and women may contribute to some of the gender differences in brain aging, with women shifted on the aging curve relative to men. Our data reveal that changes in some genes appear to be related to the longevity, particularly those related to protein transport and targeting, whereas other gene changes remain relatively unique to each gender, with profound differences in gene profile that are gender- and region-dependent. Age-dependent changes in gene expression set the balance between neurodegeneration and compensatory mechanisms in the brain and suggest that this balance is set differently in males and females.
Clearly, molecular changes observed in various brain regions in the present study are the products of many cell types, as the tissue used in this study was heterogeneous for neuronal and glial elements. Although the present study did not determine the cellular sources of the observed gene changes, a previous report has profiled gene changes with age in white versus gray matter in the human prefrontal cortex (6). It was suggested that glial-enriched gene groups tended to be up-regulated with age and were largely associated with cellular defenses (immune system, complement activation), whereas neuronal-enriched genes were largely down-regulated with age and were related to synaptic function and signal transduction, including ion channels, transmembrane receptors, calcium regulation, and synaptic structure (6). Although this earlier study sheds light on cellular sources of gene transcripts, it is increasingly recognized that neurons and glia can express many of the same genes, including those traditionally thought to be purely glial. For example, it has been shown that mRNAs for major classical complement pathway proteins are more clearly expressed by neurons than glia (31).
The results presented here are, of course, subject to the limitations of microarray-based approaches. These include false discovery resulting from multiple statistical tests of significance, although this source of error was limited by the use of two different analytical methods to validate the gene lists, as well as conventional validation by RT-PCR. The obvious caveats notwithstanding, these data illustrate the rich gene expression changes that occur in the brain across the lifespan and will lead to insights into the regulatory mechanisms underlying the general and gender-specific changes that occur with age. If, as is commonly thought, gene expression changes help dictate sexual dimorphism from embryogenesis to adulthood, then it should be reasonable to consider that sexually dimorphic gene expression profiles may continue to be influential from adulthood to senescence.
Materials and Methods
Frozen unfixed tissue was obtained from 58 normal controls aged 20–99 years and categorized into four age ranges: 20–39, 40–59, 60–79, and 80–99 years (Table S9). Brain regions used in the study were the SFG, PCG, HC, and EC, and cases were preferentially selected where tissue from at least two brain regions were available from the same individual brain. Total RNA was extracted and purified, and quality was verified by using the Agilent BioAnalyzer (Agilent Technologies) (SI Text). A total of 177 microarrays were processed at the University of California Irvine DNA and Protein MicroArray Facility (UCI Core), using Affymetrix Hg-U133plus 2.0 microarrays, using a robotic system and following manufacturer's recommendations (SI Text). Microarrays were assessed for data quality by using Affymetrix Expression Console software, following sample and chip quality guidelines from Affymetrix. GeneSpring software was used for statistical analysis of GC-RMA and PLIER expression values (P < 0.01, cross-gene error model) and for data mining based on GO. Age-related gene expression changes in the HC were validated for a subset of genes with quantitative RT-PCR, using a subset of the young (20–59 years, n = 10) and aged (60–99 years, n = 10) cases used in the microarray analysis. Statistical significance was determined by using Statview. See SI Text for further details.
Supplementary Material
Supporting Information
Acknowledgments.
We thank Lana Bordcosh and J. Denis Heck at the UCI Core for microarray expertise, and the tissue repositories at the Institute for Brain Aging at the University of California at Irvine, the University of Rochester, Johns Hopkins University, the National Institute of Child Health and Human Development, Brain and Tissue Bank for Developmental Disorders at the University of Maryland, the University of Pennsylvania, the University of Southern California, the Sun Health Research Institute Brain Donation Program, and the Arizona Biomedical Research Commission for contributing tissue for this study. Funding for this work was provided by National Institute on Aging Grants R01 AG023173, P50 AG16573, and AG00538 (to C.W.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE11882).
References
- 1.Pakkenberg B, et al. Aging and the human neocortex. Exp Gerontol. 2003;38:95–99. doi: 10.1016/s0531-5565(02)00151-1. [DOI] [PubMed] [Google Scholar]
- 2.Flood DG, Buell SJ, Horwitz GJ, Coleman PD. Dendritic extent in human dentate gyrus granule cells in normal aging and senile dementia. Brain Res. 1987;402:205–216. doi: 10.1016/0006-8993(87)90027-8. [DOI] [PubMed] [Google Scholar]
- 3.Flood DG, Guarnaccia M, Coleman PD. Dendritic extent in human CA2–3 hippocampal pyramidal neurons in normal aging and senile dementia. Brain Res. 1987;409:88–96. doi: 10.1016/0006-8993(87)90744-x. [DOI] [PubMed] [Google Scholar]
- 4.Bertoni-Freddari C, et al. Alterations of synaptic turnover rate in aging may trigger senile plaque formation and neurodegeneration. Ann NY Acad Sci. 2007;1096:128–137. doi: 10.1196/annals.1397.078. [DOI] [PubMed] [Google Scholar]
- 5.Lu T, et al. Gene regulation and DNA damage in the aging human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 6.Erraji-Benchekroun L, et al. Molecular aging in human prefrontal cortex is selective and continuous throughout adult life. Biol Psychiatry. 2005;57:549–558. doi: 10.1016/j.biopsych.2004.10.034. [DOI] [PubMed] [Google Scholar]
- 7.Coffey CE, et al. Sex differences in brain aging: A quantitative magnetic resonance imaging study. Arch Neurol. 1998;55:169–179. doi: 10.1001/archneur.55.2.169. [DOI] [PubMed] [Google Scholar]
- 8.Gur RC, et al. Sex differences in brain gray and white matter in healthy young adults: Correlations with cognitive performance. J Neurosci. 1999;19:4065–4072. doi: 10.1523/JNEUROSCI.19-10-04065.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gur RC, Gunning-Dixon FM, Turetsky BI, Bilker WB, Gur RE. Brain region and sex differences in age association with brain volume: A quantitative MRI study of healthy young adults. Am J Geriatr Psychiatry. 2002;10:72–80. [PubMed] [Google Scholar]
- 10.Murphy DG, et al. Sex differences in human brain morphometry and metabolism: An in vivo quantitative magnetic resonance imaging and positron emission tomography study on the effect of aging. Arch Gen Psychiatry. 1996;53:585–594. doi: 10.1001/archpsyc.1996.01830070031007. [DOI] [PubMed] [Google Scholar]
- 11.Yang X, et al. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16:995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blalock EM, et al. Harnessing the power of gene microarrays for the study of brain aging and Alzheimer's disease: Statistical reliability and functional correlation. Aging Res Rev. 2005;4:481–512. doi: 10.1016/j.arr.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Harris MA, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corrada MM, Brookmeyer R, Berlau D, Paganini-Hill A, Kawas CH. Prevalence of dementia after age 90: Results from the 90+ study. Neurology. 2008;71(5):337–343. doi: 10.1212/01.wnl.0000310773.65918.cd. [DOI] [PubMed] [Google Scholar]
- 15.von Strauss E, Viitanen M, De Ronchi D, Winblad B, Fratiglioni L. Aging and the occurrence of dementia: Findings from a population-based cohort with a large sample of nonagenarians. Arch Neurol. 1999;56:587–592. doi: 10.1001/archneur.56.5.587. [DOI] [PubMed] [Google Scholar]
- 16.Fraser HB, Khaitovich P, Plotkin JB, Paabo S, Eisen MB. Aging and gene expression in the primate brain. PLoS Biol. 2005;3:e274. doi: 10.1371/journal.pbio.0030274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blalock EM, et al. Gene microarrays in hippocampal aging: Statistical profiling identifies novel processes correlated with cognitive impairment. J Neurosci. 2003;23:3807–3819. doi: 10.1523/JNEUROSCI.23-09-03807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang CH, Tsien JZ, Schultz PG, Hu Y. The effects of aging on gene expression in the hypothalamus and cortex of mice. Proc Natl Acad Sci USA. 2001;98:1930–1934. doi: 10.1073/pnas.98.4.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee CK, Weindruch R, Prolla TA. Gene-expression profile of the ageing brain in mice. Nat Genet. 2000;25:294–297. doi: 10.1038/77046. [DOI] [PubMed] [Google Scholar]
- 20.Loessner A, et al. Regional cerebral function determined by FDG-PET in healthy volunteers: Normal patterns and changes with age. J Nucl Med. 1995;36:1141–1149. [PubMed] [Google Scholar]
- 21.Martin AJ, Friston KJ, Colebatch JG, Frackowiak RS. Decreases in regional cerebral blood flow with normal aging. J Cereb Blood Flow Metab. 1991;11:684–689. doi: 10.1038/jcbfm.1991.121. [DOI] [PubMed] [Google Scholar]
- 22.Kawachi T, et al. Gender differences in cerebral glucose metabolism: A PET study. J Neurol Sci. 2002;199:79–83. doi: 10.1016/s0022-510x(02)00112-0. [DOI] [PubMed] [Google Scholar]
- 23.Bell EC, Willson MC, Wilman AH, Dave S, Silverstone PH. Males and females differ in brain activation during cognitive tasks. NeuroImage. 2006;30:529–538. doi: 10.1016/j.neuroimage.2005.09.049. [DOI] [PubMed] [Google Scholar]
- 24.Boghi A, et al. The effect of gender on planning: An fMRI study using the Tower of London task. NeuroImage. 2006;33:999–1010. doi: 10.1016/j.neuroimage.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 25.Li CS, Huang C, Constable RT, Sinha R. Gender differences in the neural correlates of response inhibition during a stop signal task. NeuroImage. 2006;32:1918–1929. doi: 10.1016/j.neuroimage.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 26.Cahill L, Uncapher M, Kilpatrick L, Alkire MT, Turner J. Sex-related hemispheric lateralization of amygdala function in emotionally influenced memory: An FMRI investigation. Learn Mem. 2004;11:261–266. doi: 10.1101/lm.70504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piefke M, Weiss PH, Markowitsch HJ, Fink GR. Gender differences in the functional neuroanatomy of emotional episodic autobiographical memory. Hum Brain Mapp. 2005;24:313–324. doi: 10.1002/hbm.20092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vasto S, et al. Inflammatory networks in ageing, Age-related diseases and longevity. Mech Aging Dev. 2007;128:83–91. doi: 10.1016/j.mad.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 29.Chung HY, Sung B, Jung KJ, Zou Y, Yu BP. The molecular inflammatory process in aging. Antioxid Redox Signal. 2006;8:572–581. doi: 10.1089/ars.2006.8.572. [DOI] [PubMed] [Google Scholar]
- 30.Ng MK. New perspectives on Mars and Venus: Unraveling the role of androgens in gender differences in cardiovascular biology and disease. Heart Lung Circ. 2007;16:185–192. doi: 10.1016/j.hlc.2007.02.108. [DOI] [PubMed] [Google Scholar]
- 31.Shen Y, Li R, McGeer EG, McGeer PL. Neuronal expression of mRNAs for complement proteins of the classical pathway in Alzheimer brain. Brain Res. 1997;769:391–395. doi: 10.1016/s0006-8993(97)00850-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information