Detection and Molecular Characterization of 9000-Year-Old Mycobacterium tuberculosis from a Neolithic Settlement in the Eastern Mediterranean (original) (raw)
Abstract
Background
Mycobacterium tuberculosis is the principal etiologic agent of human tuberculosis. It has no environmental reservoir and is believed to have co-evolved with its host over millennia. This is supported by skeletal evidence of the disease in early humans, and inferred from M. tuberculosis genomic analysis. Direct examination of ancient human remains for M. tuberculosis biomarkers should aid our understanding of the nature of prehistoric tuberculosis and the host/pathogen relationship.
Methodology/Principal Findings
We used conventional PCR to examine bone samples with typical tuberculosis lesions from a woman and infant, who were buried together in the now submerged site of Atlit-Yam in the Eastern Mediterranean, dating from 9250-8160 years ago. Rigorous precautions were taken to prevent contamination, and independent centers were used to confirm authenticity of findings. DNA from five M tuberculosis genetic loci was detected and had characteristics consistent with extant genetic lineages. High performance liquid chromatography was used as an independent method of verification and it directly detected mycolic acid lipid biomarkers, specific for the M. tuberculosis complex.
Conclusions/Significance
Human tuberculosis was confirmed by morphological and molecular methods in a population living in one of the first villages with evidence of agriculture and animal domestication. The widespread use of animals was not a source of infection but may have supported a denser human population that facilitated transmission of the tubercle bacillus. The similarity of the M. tuberculosis genetic signature with those of today gives support to the theory of a long-term co-existence of host and pathogen.
Introduction
Tuberculosis is a major global cause of death and disease and around 2 billion people, about one third of the world's total population, are believed to be infected with tubercle bacilli [1]. However, only around 10% of infected persons become ill with active disease, and this high level of latent infection is an indication of long-term co-existence of human host and bacterial pathogen [2]. Tuberculosis is caused by a group of closely related bacterial species termed the Mycobacterium tuberculosis complex. Today the principal cause of human tuberculosis is Mycobacterium tuberculosis. Mycobacterium bovis has a wider host range and is the main cause of tuberculosis in other animal species. Humans become infected by M. bovis, usually via milk, milk products or meat from an infected animal. It is estimated that in the pre-antibiotic era M. bovis was responsible for about 6% of tuberculosis deaths in humans [3], [4]. Other members of the M. tuberculosis complex include the human pathogens Mycobacterium canettii, Mycobacterium africanum, and species usually associated with animal infections, such as Mycobacterium microti, Mycobacterium caprae and Mycobacterium pinnipedii.
Tuberculosis can cause characteristic skeletal changes, such as collapse of the vertebrae (Pott's disease), periosteal reactive lesions, and osteomyelitis [5]. Such paleopathological changes have been reported in pre-dynastic (3500-2650 BC) Egypt [6], and Neolithic (3200-2300 BC) Sweden culturally associated with the earliest cattle breeders [7]. These are the oldest cases of human tuberculosis confirmed by ancient DNA. Older cases recognized by skeletal changes alone were found in Neolithic Italy at the beginning of the fourth millennium BC [8], [9].
Erosive lesions suggestive of tuberculosis have been found on fossil fauna from the Natural Trap Cave in Wyoming, dated from the 17,000 to 20,000 year level [10] and tuberculosis in one specimen was confirmed by biomolecular methods [11]. Initially it was believed that humans acquired tuberculosis from animals, especially after domestication [12]–[14]. Whole genome sequencing has since revealed that the M. tuberculosis complex has accumulated deletions over time, which can be used to distinguish individual species and lineages [15] and earlier ideas about the evolution of the M. tuberculosis complex have been revised [16], [17]. An intriguing indication of the antiquity of the disease is the finding of non-specific morphological changes consistent with tuberculosis in a fossil Homo erectus dating from the middle Pleistocene (490–510,000 years BP) from Turkey [18].
The emergence of human infectious diseases has been linked to changes in human ecology and to interactions between populations [19]. The change from a gatherer-hunter lifestyle to settled farming communities appears to coincide with the appearance of diseases such as smallpox, measles, malaria, schistosomiasis, and tuberculosis [20]. Our aim was to investigate this stage of human history by the use of molecular methods to examine human remains that pre-date the earliest verified cases of tuberculosis, but with paleopathology consistent with this disease. A further aim was to elucidate the molecular characteristics of the causative organism. It is believed that the denser, settled populations associated with agriculture and animal domestication enabled human pathogens such as M. tuberculosis to be maintained indefinitely [21]. Therefore, we examined one of the earliest villages with evidence of both animal domestication and agriculture, Atlit-Yam [22], for the presence of tuberculosis in human remains with characteristic lesions. M. tuberculosis was confirmed in the skeletal remains of a woman and child, using both ancient DNA and bacterial cell-wall specific lipid markers. Deletion analysis indicates that the modern M. tuberculosis lineage characterized by the TbD1 deletion existed 9000 years ago.
Materials and Methods
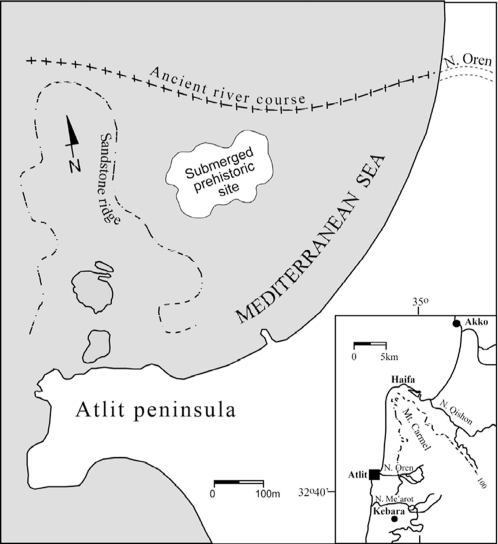
The site of Atlit-Yam is now located 300–500 m offshore, (34°56′ E, 32°42.5′ N), 8–12 m below sea level in the North Bay of Atlit, 10 km south of Haifa (Figure 1). Calibrated radiocarbon dates range from 9250-8160 years BP [23], indicating a date during the last phase of the Pre-Pottery Neolithic C period, when human society accomplished a full shift from hunting and gathering to farming, fishing and animal husbandry. The rich finds included botanical remains, tools, animal and human bones. The many animal remains that were excavated included goat (44%), cattle (43%), pig (9%), gazelle and deer (3.3%).
Figure 1. Map of Atlit-Yam site in the North Bay of Atlit, 10 km south of Haifa (34°56′ E, 32°42.5′ N). Inset shows general geographical location.
Human skeletons, which were embedded in dark clay, were carefully excavated and soaked in fresh-water tanks to dissolve the salts. The skeletal remains [24], [25] were generally well-preserved (Supporting Figure S1A) and some showed paleopathological features consistent with a diagnosis of tuberculosis. Samples were taken for molecular examination from the skeletal remains of a woman buried together with an infant (Supporting Figure S1B). We analyzed the ribs, arm bones (adult) and long bones (infant). The work was done in separate centers to provide verification of data, and stringent precautions were taken against contamination (Supporting Materials and Methods S1). Separate areas and pipettes were used for extraction, PCR set-up and product analysis. Filter tips were used routinely and surfaces and equipment were cleaned before each assay. DNA extracts were prepared and, using the polymerase chain reaction (PCR), both multi-copy and single copy target loci were amplified (Table 1) and sequenced to confirm their identity. Screening PCRs detected the M. tuberculosis complex and nested or hemi-nested PCR was used to increase the likelihood of detection (Supporting Materiasls and Methods S1). A single copy conserved membrane protein locus (CMP) found in the M. tuberculosis complex was examined by PCR to assess the feasibility of seeking further single-copy loci. PCR target regions, based on specific deletions, were used to distinguish between M. tuberculosis and M. bovis. Extracts were analyzed further by reverse dot-blot hybridization of the M. tuberculosis complex-specific Direct Repeat (DR) region, a procedure known as spoligotyping [26]. In addition, samples from both the infant and adult were analyzed by high performance liquid chromatography (HPLC) for mycobacterial cell wall mycolic acids [27], [28] (Supporting Tables S1, S2 and S3 and Supporting Figure S2 A and B). Long chain fatty acids were converted to pyrenebutyric acid-pentafluorobenzyl mycolates, and reverse phase HPLC examined for profiles similar to standard M. tuberculosis. Further normal and reverse phase HPLC was performed to give detailed profiles for each sample. These were used to determine the percentage ratios and absolute amounts of mycolic acids extracted from bone samples.
Table 1. Primer sequences and PCR details.1.
| Locus | Primers (5′ - 3′) | MgCl2 (mM) | Annealing temp. (°C) | Product (bp) |
|---|---|---|---|---|
| IS_6110_ | P1: CTCGTCCAGCGCCGCTTCGG | |||
| Outer | P2: CCTGCGAGCGTAGGCGTCGG | 1.5 | 68 | 123 |
| IS_6110_ | IS3: TTCGGACCACCAGCACCTAA | |||
| Nested | IS4: TCGGTGACAAAGGCCACGTA | 1.5 | 58 | 92 |
| IS_1081_ | F2: CTGCTCTCGACGTTCATCGCCG | |||
| Outer | R2: GGCACGGGTGTCGAAATCACG | 1.5 | 58 | 135 |
| IS_1081_ | F2: CTGCTCTCGACGTTCATCGCCG | |||
| Hemi-nested | R3: TGGCGGTAGCCGTTGCGC | 2.0 | 58 | 113 |
| TbD1 | TbD1a: CTAACGGGTGCAGGGGATTTC | |||
| Flanking outer | TbD1b: CCAAGGTTACGGTCACGCTGGC | 1.5 | 60 | 128 |
| TbD1 | TbD1c: GCAGGGGATTTCAGTGACTG | |||
| Flanking inner | TbD1d: GCTGGCCAGCTGCTCGCCG | 1.5 | 58 | 103 |
| CMP | F2: TCGGTCAGCAAGACGTTGAAG | |||
| R: ACTTCAGTGCTGGTTCGTGG | 2.0 | 58 | 105 | |
| RD2 | BV1: ATCTTGCGGCCCAATGAATC | |||
| Outer | BV2: CAACGTCTTGCTGACCGACA | 1.5 | 58 | 124 |
| RD2 | BV3: ATGAATCGGCCGCGTTCG | |||
| Nested | BV4: GACCGACATCGGTGCCGCG | 1.5 | 58 | 99 |
| DR | DRa: GGTTTTGGGTGTGACGAC 2 | Not applicable | ||
| DRb: CCGAGAGGGGACGGAAAC | 3.0 | 55 |
Results
Paleopathology
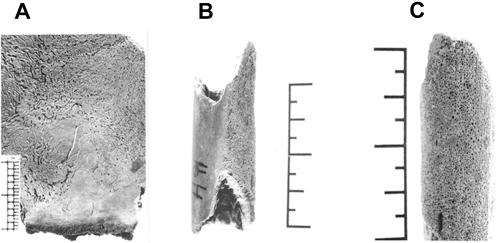
The infant, though small in size, was estimated to be about 12 months old, based on crown development and long bone dimensions. On the inner aspect of the infant cranial bones were serpentine engravings (serpens endocrania symmetrica, SES; Figure 2A), a reliable diagnostic criterion for intra-thoracic inflammation [29] and associated with tuberculosis. The infant tubular bones also demonstrated lesions identified as hypertrophic osteoarthropathy (HOA), highly suggestive of tuberculosis [30] and characterized by the formation of an expanded shell of periosteal reactive bone (Figure 2B and C). The woman was estimated to be around 25 years old, based on dental attrition, epiphyseal ring ankylosis and symphysis pubis. There was a slight periosteal reaction affecting the distal diaphysis of the one tibia available for examination, a bony change consistent with HOA [5], [31], [32]. However, the changes were not so marked as to be diagnostic.
Figure 2. Paleopathological lesions on Neolithic infant bones.
A. Endocranial surface of the infant showing marked engravings (serpens endocrania symmetrica, SES), which indicate chronic respiratory malfunction, and are usually associated with tuberculosis. B. Fragment of long bone of the infant. Note the intensive bone remodeling (hypertrophic osteoarthropathy, HOA) at the surface on the right side. C. Higher magnification of the HOA on the infant bone.
Mycobacterium tuberculosis complex and M. tuberculosis DNA
M. tuberculosis complex DNA was detected in the bones of woman and infant (Table 2). Positive results with the multi-copy IS_6110_ [33], [34] and IS_1081_ [35] PCRs were obtained with the rib sample from the woman and infant long bone sample, and confirmed by sequencing. An IS_6110_ 123 bp product from the woman (right rib) and a 92 bp nested IS_6110_ product from the infant were obtained, identical to those in the NCBI database. Additionally, a 104 bp sequence identical to the relevant NCBI sequence in the IS_1081_ product was obtained from the infant.
Table 2. Summary of PCR results.
| PCR locus | Woman | Infant | ||
|---|---|---|---|---|
| 1st stage PCR | 2nd stage PCR | 1st stage PCR | 2nd stage PCR | |
| IS_6110_ | Positive | Positive | Positive1 | Negative |
| IS_1081_ | Positive | Positive | Positive1 | Positive |
| Flanking TbD1 | Positive | Negative | Positive1 | Positive |
| RD2 | Negative | Negative | Negative | Negative |
| CMP | Positive | Not done | Positive1 | Not done |
The single copy TbD1 flanking PCR was positive from the infant sample and a complete DNA sequence for the 128 bp amplicon with the outer primers was obtained (Supporting Figure S3A and B). The consensus sequence was identical to that in the NCBI database. The strong signal indicates the excellent preservation at this locus of the M. tuberculosis DNA template. Nested PCR was also successful. Weak positives were obtained with the outer primers from the female sample. These findings are evidence that the infecting organism was M. tuberculosis from a lineage in which the TbD1 deletion had occurred [16]. Results from the infant for the single copy CMP PCR were faint and a partial sequence was obtained (Supporting Figure S3C) with some mismatched bases compared with the database, attributed to poor DNA preservation. No demonstrable amplicons of human nuclear microsatellite DNA were obtained from the bone samples. Spoligotyping provided additional evidence for M. tuberculosis complex DNA for both the adult and infant specimens (Supporting Figure S4 A–D), although there were several faint or dubious positives and inconsistencies between replicates, as might be expected of ancient specimens.
Lipid biomarkers of M. tuberculosis
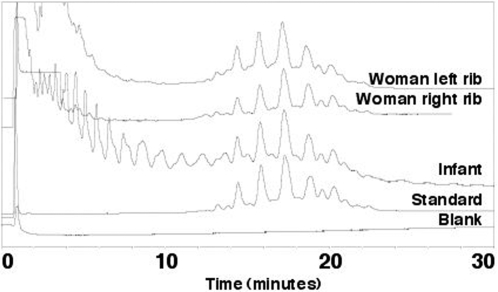
Modifying an established protocol [28], [36], long-chain fatty acids were extracted as pentafluorobenzyl (PFB) esters, and fractions corresponding to PFB mycolates were obtained (Supporting Tables S4, S5). After treatment with pyrenebutyric acid (PBA) these fractions produced PBA-PFB mycolates, which, after reverse phase HPLC, gave profiles closely similar to those produced by the M. tuberculosis complex, as indicated by a standard M. tuberculosis strain (Figure 3). Further normal and reverse phase HPLC gave detailed profiles for each sample, reinforcing the close identity with M. tuberculosis (Supporting Figure S2 C and D).
Figure 3. Detection of Mycobacterium tuberculosis mycolic acid pyrenebutyric acid-pentafluorobenzyl (PBA-PFB) derivatives by reverse phase fluorescence high performance liquid chromatography (HPLC), from the Neolithic woman and infant.
The characteristic tight envelopes of peaks are the total mixture of homologues for the different α-, methoxy- and ketomycolates. The Y-axis in the profiles represents absorbance; absolute values of the mycolates detected are shown in Supporting Table 5.
Discussion
It is believed that inundation of the Atlit-Yam site occurred shortly after abandonment [24] and thereafter the environment remained unchanged for 9,000 years. The Atlit-Yam site was located within marshland; the graves were encased in clay, eventually covered by thick layer of sand and later by salt water, thus providing anaerobic conditions that retard degradation. The excellent preservation of the skeletal remains is consistent with the excellent physical state of the organic artifacts (wooden bowls, reed mats) that were found on the site. The paleopathogical lesions of SES in the infant cranium and HOA in the infant long bones and possibly also the tibia of the woman buried with the infant, presumed to be the mother, suggest that both suffered from, and died of, tuberculosis.
Anaerobic conditions are also conducive for DNA preservation [37] and DNA analysis supports the paleopathological diagnosis of tuberculosis. Overall, the PCR data provide strong evidence of the M. tuberculosis complex as specific DNA was detected in five different genetic loci, including the TbD1 locus with a deletion that is specific for a broadly-defined modern lineage of M. tuberculosis. Failure to detect human DNA (data not included) may reflect the greater stability of the GC-rich mycobacterial DNA, which additionally benefits from the robust hydrophobic bacterial cell wall [28], [38], [39].
Direct detection of cell wall mycolic acids specific for the Mycobacterium tuberculosis complex, without any amplification step, provides independent, robust confirmation of the presence of tuberculosis. The quantity of mycolic acids appeared lower in the infant sample (Supporting Table S5), in contrast to the DNA studies where the infant gave better results. However, the mycolate analyses were carried out on three combined rib samples from the baby, not all of which had been tested for MTB DNA. These extremely hydrophobic high molecular weight molecules are more stable than DNA and have been used previously to confirm diagnoses of ancient tuberculosis [27], [28].
We conclude that both individuals in our study were infected with M. tuberculosis, and that our findings are supported when we consider the nature of the site, the stringent precautions taken to prevent cross-contamination and verification by the specific lipid biomarkers. Furthermore, we believe that this is the earliest report of the disease in humans that has been confirmed by molecular means. The infant is likely to have had disseminated primary tuberculosis: - the only DNA sequences for single copy loci were obtained from the infant material, which suggests a higher bacterial load during life. In infants less than a year old the present risk of developing active disease on infection with M. tuberculosis is as high as 43% [40] due to the inadequacy of their immune system. This compares with 5–10% in adults, 15% in adolescents, and 24% in children aged 1–5 years.
The size of the infant's bones, and the extent of the bony changes, suggest a case of acquired neonatal tuberculosis, in which an adult suffering from contagious pulmonary tuberculosis infects an infant shortly after birth. Childhood tuberculosis is closely linked to adult disease and is usually a sentinel event in the community, demonstrating recent transmission. Infant and maternal mortality rates from untreated tuberculosis in recent times was between 30% and 40% [41], so it is unsurprising for both mother and child to succumb and be buried together.
Spoligotyping should be a useful method of examining DNA from archaeological material as even fragmented DNA gives results, due to the increased sensitivity from the combination of amplification and hybridization [42]. M. tuberculosis complex DNA from the lesion of a 17,000-old extinct Pleistocene bison [11] yielded spoligotyping patterns most similar to Mycobacterium africanum or M. tuberculosis [43], and distinct from present day M. bovis. Zink et al. [44] obtained spoligotypes from ancient Egyptian human bone and soft tissue samples, dating back to about 4000 years. Of their 12 positive samples, spoligotyping indicated M. tuberculosis or, in some older Middle Kingdom samples, M. africanum patterns, but not those of M. bovis.
We carried out spoligotyping on specimens from the woman and infant but replicated typing gave inconsistent results, which suggests there may be poor DNA preservation of some of the single-copy spacer regions. The observed patterns do not match any in the International Data Base spoldb4: www.pasteur-guadeloupe.fr/tb/spoldb4 but appear similar to an ancestral pattern. Results need to be interpreted with caution, as spoldb4 is based on data obtained from cultured organisms and spoligotyping has not been validated for application to DNA extracts prepared from degraded or archival specimens. The spoligotyping technique is based on modern M. tuberculosis strains from around the world. As the main variation in types is caused by unidirectional deletions, all ancestral strains are likely to produce a near-complete profile of the DR region and therefore to resemble each other. This may explain why the spoligotypes from the Atlit Yam skeletons resemble those of the Pleistocene bison [11].
Deletion analysis is a more robust method of examining ancient material [16], [45], and based on the TbD1 deletion, the genetic lineage resembles modern lineages of M. tuberculosis [16], [43], [51].
Suggestions that human tuberculosis arose from M. bovis in hunted or domesticated animals have been revised since comparative genomic studies demonstrate that M. bovis represents a later lineage [16], [17]. Members of the M. tuberculosis complex are genetically very similar and were believed to be the result of a clonal expansion following an evolutionary bottleneck 20,000–35,000 years ago [16], [46], [47]. However, further genomic studies of the M. tuberculosis complex indicate a more ancient origin of this group of closely related species than had previously been believed, and that possibly an early progenitor, perhaps similar to M. canettii, was present in East Africa as early as 3 million years ago [48], [49]. The observation of non-specific lesions consistent with tuberculosis found in a 500,000 year-old skeleton of Homo erectus [18] may also indicate the long-term co-existence of host and pathogen, although the diagnosis in this particular case has been questioned. However, M. tuberculosis appears to have undergone long-term co-evolution with its human host prior to the evolutionary bottleneck and well before the development of agriculture and domestication, comparable to other long-term human pathogens such as Helicobacter pylori [50], [51].
The present study of a population from 9250-8160 years ago, around the time of the first great transition from hunter-gatherers to a settled agriculture-based lifestyle [19], helps us to understand the nature of tuberculosis within the Middle East. Could the presence of cattle be pertinent? Atlit-Yam is among the very few Pre-Pottery Neolithic sites where domesticated cattle have been found. Furthermore, it is the only Neolithic site where there were quantities of bovine bones, indicating that cattle were a major dietary component [23]. We suggest that in the absence of detectable M. bovis, the cattle may be important by supporting a larger and denser human population, thus indirectly encouraging the conditions for the long-term maintenance and transmission of M. tuberculosis [21].
Supporting Information
Materials and Methods S1
Text
(0.05 MB DOC)
Table S1
The solvent sequence used for the silica gel normal phase cartridge fractionation of long-chain compounds
(0.03 MB DOC)
Table S2
The solvent sequence used for the reverse phase cartridge purification of PBA-PFB mycolates
(0.02 MB DOC)
Table S3
Conditions for HPLC analysis of PBA-PFB mycolates
(0.02 MB DOC)
Table S4
Percentage ratios of alpha-, methoxy- and ketomycolates in archaeological samples and M. tuberculosis standard determined in normal phase HPLC
(0.02 MB DOC)
Table S5
Absolute amounts of mycolic acids extracted from bone samples
(0.02 MB DOC)
Figure S1
Atlit-Yam burials. A. An example of human remains with excellent preservation. B. Partial excavation of the burial site with the adult female and infant skeleton (arrow).
(2.46 MB TIF)
Figure S2
High Performance Liquid Chromatography (HPLC) methodology. A. Representative structures of the mycolic acids from M. tuberculosis. Natural mixtures of mycolates express a range of homologous components with varying chain lengths. B. Strategy for the release and derivatization of mycolic acids for fluorescence HPLC. (a) Hydrolysis with KOH/ methanol/toluene to release mycolic acids. (b) Phase-transfer catalyzed esterification of mycolic acids with pentafluorobenzyl bromide (PFB) to give PFB mycolates. (c) Esterification of PFB mycolates, by reaction with pyrenebutyric acid (PBA), to produce PBA-PFB mycolates, catalyzed by dicyclohexylcarbodiimide and pyrrolidinopyridine. R- represents the remainder of the mycolate molecule. C. Normal phase HPLC of PBA-PFB mycolates from bone samples and standard M. tuberculosis. D. Reverse phase HPLC of individual α -, methoxy- and ketomycolic acid PBA-PFB derivatives from bone samples and standard M. tuberculosis. The number of carbons in the individual underivatized mycolic acids is shown.
(0.83 MB TIF)
Figure S3
DNA sequence data. A. M. tuberculosis TbD1 flanking region (128 bp), obtained from the infant sample (5′-3′ strand). B. M. tuberculosis TbD1 flanking region (128 bp), obtained from the infant sample (3′-5′ strand). C. M. tuberculosis conserved membrane protein (CMP) region, obtained from the infant sample (5′-3′ strand only).
(12.09 MB TIF)
Figure S4
Repeated spoligotypes from the Atlit-Yam samples and controls. Each set of spoligotyping data (A–C) represents, from top to bottom, M. tuberculosis, M. bovis (BCG) controls, Atlit Yam female and Atlit Yam infant. D. Diagram of spoligotyping data, including M. africanum spoligotypes (Donoghue et al 2004), and a consensus pattern based on one or more positive results.
(5.61 MB TIF)
Acknowledgments
We thank Kim Vernon for laboratory assistance in Jerusalem. G. Michael Taylor (UCL) supplied primers for the RD2 and CMP regions, and kindly prepared the PCR products for sequencing. He and Professor Robin Weiss, Wohl Virion Centre, Division of Infection and Immunity, UCL are thanked for their constructive criticism.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The CARE, MAFCAF and Dan David Foundation supported the archaeological and anthropological work. A grant from The Leverhulme Trust (F/125/AK) to AMG and DEM is acknowledged; DEM is a Leverhulme Emeritus Fellow. MS is funded by the Deutsche Forschungsgemeinschaft (DFG) for a trilateral project between German, Israeli and Palestinian researchers. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.World Health Organisation. Tuberculosis. Fact sheet 104. 2008. Revised April 2008. Available: http://www.who.int/tb/publications/2008/factsheet_april08.pdf Accessed 2008 Jun 2.
- 2.Hirsh AE, Tsolaki AG, DeRiemer K, Feldman MW, Small PM. Stable association between strains of Mycobacterium tuberculosis and their human host populations. Proc Natl Acad Sci USA. 2004;101:4871–4876. doi: 10.1073/pnas.0305627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hardie RM, Watson JM. Mycobacterium bovis in England and Wales: past, present and future. Epidemiol Infect. 1992;109:23–33. [PMC free article] [PubMed] [Google Scholar]
- 4.O'Reilly LM, Daborn CJ. The epidemiology of Mycobacterium bovis infections in animals and man: a review. Tuber Lung Dis 76 Suppl. 1995;1:1–46. doi: 10.1016/0962-8479(95)90591-x. [DOI] [PubMed] [Google Scholar]
- 5.Ortner DJ, Putschar WG. Identification of pathological conditions on human skeletal remains. Washington DC: Smithsonian Institution Press; 1981. [Google Scholar]
- 6.Zink A, Haas CJ, Reischl U, Szeimies U, Nerlich AG. Molecular analysis of skeletal tuberculosis in an ancient Egyptian population. J Med Microbiol. 2001;50:355–366. doi: 10.1099/0022-1317-50-4-355. [DOI] [PubMed] [Google Scholar]
- 7.Nuorala E, Götherström A, Ahlström T, Donoghue HD, Spigelman M, et al. MTB complex DNA in a Scandinavian Neolithic passage grave (Theses and Papers in Scientific Archaeology 6, Archaeological Research Laboratory, Stockholm University), paper I 2004 [Google Scholar]
- 8.Formicola V, Milanesi Q, Scarsini C. Evidence of spinal tuberculosis at the beginning of the fourth millennium BC from Arene Candide cave (Liguria, Italy). Am J Phys Anthropol. 1987;72:1–6. doi: 10.1002/ajpa.1330720102. [DOI] [PubMed] [Google Scholar]
- 9.Canci A, Minozzi S, Borgognini Tarli SM. New evidence of tuberculous spondylitis from Neolithic Liguria (Italy). Int J Osteoarchaeol. 1996;6:497–501. [Google Scholar]
- 10.Rothschild BM, Martin LD. Frequency of pathology in a large natural sample from Natural Trap Cave with special remarks on erosive disease in the Pleistocene. Reumatismo. 2003;55:58–65. doi: 10.4081/reumatismo.2003.58. [DOI] [PubMed] [Google Scholar]
- 11.Rothschild BM, Martin LD, Lev G, Bercovier H, Bar-Gal GK, et al. Mycobacterium tuberculosis complex DNA from an extinct bison dated 17,000 years before the present. Clin Infect Dis. 2001;33:305–311. doi: 10.1086/321886. [DOI] [PubMed] [Google Scholar]
- 12.Steinbock RT. Paleopathological diagnosis and interpretation: Bone disease in ancient human populations. Springfield, Illinois: Charles Thomas; 1976. [Google Scholar]
- 13.Manchester K. Tuberculosis and leprosy in antiquity: an interpretation. Med History. 1984;28:162–173. doi: 10.1017/s0025727300035705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark GA, Kelley MA, Grange JM, Hill MC. The evolution of mycobacterial disease in human populations. A reevaluation. Curr Anthropol. 1987;28:45–62. doi: 10.1086/203490. [DOI] [PubMed] [Google Scholar]
- 15.Parsons LM, Brosch R, Cole ST, Somoskövi Á, Loder A, et al. Rapid and simple approach for identification of Mycobacterium tuberculosis complex isolates by PCR-based genomic deletion analysis. J Clin Microbiol. 2002;40:2339–2345. doi: 10.1128/JCM.40.7.2339-2345.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brosch R, Gordon SV, Marmiesse M, Brodin P, Buchreiser C, et al. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc Natl Acad Sci USA. 2002;99:3684–3689. doi: 10.1073/pnas.052548299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mostowy S, Cousins D, Brinkman J, Aranaz A, Behr MA. Genomic deletions suggest a phylogeny for the Mycobacterium tuberculosis complex. J Infect Dis. 2002;186:74–80. doi: 10.1086/341068. [DOI] [PubMed] [Google Scholar]
- 18.Kappelman J, Alçiçek MC, Kazanci N, Schultz M, Ozkul M, et al. First Homo erectus from Turkey and implications for migrations into temperate Eurasia. Am J Phys Anthropol. 2008;135:110–116. doi: 10.1002/ajpa.20739. [DOI] [PubMed] [Google Scholar]
- 19.McMichael AJ. Environmental and social influences on emerging infectious diseases: past, present and future. Phil Trans R Soc Lond B. 2004;359:1049–1058. doi: 10.1098/rstb.2004.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen MN, Armelagos GJ. Paleopathology at the origins of agriculture: editors' summation. In: Cohen MN, Armelagos GJ, editors. Paleopathology at the origins of agriculture. New York: Academic Press; 1984. pp. 585–601. [Google Scholar]
- 21.Weiss RA, McMichael AJ. Social and environmental risk factors in the emergence of infectious diseases. Nature Med. 2004;10(Suppl):S70–S76. doi: 10.1038/nm1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galili E, Weinstein-Evron M, Hershkovitz I, Gopher A, Kislev M, et al. Atlit-Yam: A prehistoric site on the sea floor off the Israeli coast. J Field Archaeol. 1993;20:133–157. [Google Scholar]
- 23.Galili E, Rosen B, Gopher A, Horwitz LK. The emergence and dispersion of the Eastern Mediterranean fishing village: evidence from submerged Neolithic settlements off the Carmel coast, Israel. J Medit Archaeol. 2002;15:167–198. [Google Scholar]
- 24.Hershkovitz I, Galili E. 8000 year-old human remains on the sea floor near Atlit, Israel. Hum Evol. 1990;5:319–358. [Google Scholar]
- 25.Galili E, Eshed V, Gopher A, Hershkovitz I. Burial practices at the submerged pre-pottery Neolithic C site of Atlit-Yam, Northern Coast of Israel. BASOR. 2005;339:1–19. [Google Scholar]
- 26.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, et al. Simultaneous detection and strain identification of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Donoghue HD, Spigelman J, Zias J, Gernaey-Child AM, Minnikin DE. Mycobacterium tuberculosis complex DNA in calcified pleura from remains 1400 years old. Lett Appl Microbiol. 1998;27:265–269. [PubMed] [Google Scholar]
- 28.Gernaey AM, Minnikin DE, Copley MS, Dixon RA, Middleton JC, et al. Mycolic acids and ancient DNA confirm an osteological diagnosis of tuberculosis. Tuberculosis (Edinb) 2001;81:259–265. doi: 10.1054/tube.2001.0295. [DOI] [PubMed] [Google Scholar]
- 29.Hershkovitz I, Greenwald CM, Latimer B, Jellema LM, Wish-Baratz S, et al. Serpens endocrania symmetrica (SES): a new term and a possible clue for identifying intrathoracic disease in skeletal populations. Am J Phys Anthropol. 2002;118:201–216. doi: 10.1002/ajpa.10077. [DOI] [PubMed] [Google Scholar]
- 30.Mensforth RP, Lovejoy CO, Lallo JW, Armelagos GJ. The role of constitutional factors, diet, and infectious disease in the etiology of porotic hyperostosis and periosteal reactions in prehistoric infants and children. Med Anthropol. 1978;2:1–59. doi: 10.1080/01459740.1978.9986939. [DOI] [PubMed] [Google Scholar]
- 31.Rothschild BM, Rothschild C. Recognition of hypertrophic osteoarthropathy in skeletal remains. J Rheumatol. 1998;25:2221–2227. [PubMed] [Google Scholar]
- 32.Mays S, Taylor GM. Osteological and biomolecular study of two possible cases of Hypertrophic Osteoarthropathy from Mediaeval England. Am J Phys Anthropol. 2002;29:1267–1276. [Google Scholar]
- 33.Eisenach KD, Cave MD, Bates JH, Crawford JT. Polymerase chain reaction amplification of a repetitive DNA sequence specific for Mycobacterium tuberculosis. J Infect Dis. 1990;161:977–981. doi: 10.1093/infdis/161.5.977. [DOI] [PubMed] [Google Scholar]
- 34.Taylor GM, Crossey M, Saldanha J, Waldron T. Mycobacterium tuberculosis identified in mediaeval human skeletal remains using polymerase chain reaction. J Archaeol Sci. 1996;23:789–798. [Google Scholar]
- 35.Taylor GM, Stewart GR, Cooke M, Chaplin S, Ladva S, et al. Koch's Bacillus – a look at the first isolate of Mycobacterium tuberculosis from a modern perspective. Microbiology. 2003;149:3213–3220. doi: 10.1099/mic.0.26654-0. [DOI] [PubMed] [Google Scholar]
- 36.Minnikin DE, Bolton DE, Dobson G, Mallet AI. Mass spectrometric analysis of multimethyl branched fatty acids and phthiocerols from clinically-significant mycobacteria. Proc Jap Soc Med Mass Spectrom. 1987;12:23–32. [Google Scholar]
- 37.Drancourt M, Raoult D. Palaeomicrobiology: current issues and perspectives. Nat Rev Microbiol. 2005;3:23–35. doi: 10.1038/nrmicro1063. [DOI] [PubMed] [Google Scholar]
- 38.Minnikin DE, Kremer L, Dover LG, Besra GS. The methyl-branched fortifications of Mycobacterium tuberculosis. Chem Biol. 2002;9:545–553. doi: 10.1016/s1074-5521(02)00142-4. [DOI] [PubMed] [Google Scholar]
- 39.Spigelman M, Donoghue HD. Palaeobacteriology with special reference to pathogenic mycobacteria. In: Greenblatt CL, Spigelman M, editors. Emerging pathogens: archaeology, ecology and evolution of infectious disease. Oxford: Oxford University Press; 2003. pp. 175–188. [Google Scholar]
- 40.Walls T, Shingadia D. Global epidemiology of pediatric tuberculosis. J Infection. 2004;48:13–22. doi: 10.1016/s0163-4453(03)00121-x. [DOI] [PubMed] [Google Scholar]
- 41.Starke JR. Pediatric tuberculosis: time for a new approach. Tuberculosis (Edinb) 2003;83:208–212. doi: 10.1016/s1472-9792(02)00088-4. [DOI] [PubMed] [Google Scholar]
- 42.Donoghue HD, Spigelman M, Greenblatt CL, Lev-Maor G, Kahila Bar-Gal G, et al. Tuberculosis: from prehistory to Robert Koch, as revealed by ancient DNA. Lancet Infect Dis. 2004;4:584–592. doi: 10.1016/S1473-3099(04)01133-8. [DOI] [PubMed] [Google Scholar]
- 43.Huard RC, Fabre M, de Haas P, Lazzarini LC, van Soolingen D, et al. Novel genetic polymorphisms that further delineate the phylogeny of the Mycobacterium tuberculosis complex. J Bacteriol. 2006;188:4271–4287. doi: 10.1128/JB.01783-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zink AR, Sola C, Reischl U, Grabner W, Rastogi N, et al. Characterization of Mycobacterium tuberculosis complex DNAs from Egyptian mummies by spoligotyping. J Clin Microbiol. 2003;41:359–367. doi: 10.1128/JCM.41.1.359-367.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor GM, Young DB, Mays SA. Genotypic analysis of the earliest known prehistoric case of tuberculosis in Britain. J Clin Microbiol. 2005;43:2236–2240. doi: 10.1128/JCM.43.5.2236-2240.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sreevatsan S, Pan X, Stockbauer KE, Connell ND, Kreiswirth BN, et al. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci U S A. 1997;94:9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hughes AL, Friedman R, Murray M. Genomewide pattern of synonymous nucleotide substitution in two complete genomes of Mycobacterium tuberculosis. Emerg Infect Dis. 2002;8:1342–1346. doi: 10.3201/eid0811.020064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gutierrez MC, Brisse S, Brosch R, Fabre M, Omaïs B, et al. Ancient origin and gene mosaicism of the progenitor of Mycobacterium tuberculosis. PLoS Pathog. 2005;1:55–61. doi: 10.1371/journal.ppat.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Daniel TM. The history of tuberculosis. Resp Med. 2006;100:1862–1870. doi: 10.1016/j.rmed.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 50.Pearce-Duvet JMC. The origin of human pathogens: evaluating the role of agriculture and domestic animals in the evolution of human disease. Biol Rev Camb Philos Soc. 2006;81:369–382. doi: 10.1017/S1464793106007020. [DOI] [PubMed] [Google Scholar]
- 51.Wirth T, Hildebrand F, Allix-Béguec C, Wölbeling F, Kubica T, et al. Origin, Spread and Demography of the Mycobacterium tuberculosis Complex. PLoS Pathogens. 2008 doi: 10.1371/journal.ppat.1000160. 4(9): e1000160 doi:10.1371/journal.ppat.1000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials and Methods S1
Text
(0.05 MB DOC)
Table S1
The solvent sequence used for the silica gel normal phase cartridge fractionation of long-chain compounds
(0.03 MB DOC)
Table S2
The solvent sequence used for the reverse phase cartridge purification of PBA-PFB mycolates
(0.02 MB DOC)
Table S3
Conditions for HPLC analysis of PBA-PFB mycolates
(0.02 MB DOC)
Table S4
Percentage ratios of alpha-, methoxy- and ketomycolates in archaeological samples and M. tuberculosis standard determined in normal phase HPLC
(0.02 MB DOC)
Table S5
Absolute amounts of mycolic acids extracted from bone samples
(0.02 MB DOC)
Figure S1
Atlit-Yam burials. A. An example of human remains with excellent preservation. B. Partial excavation of the burial site with the adult female and infant skeleton (arrow).
(2.46 MB TIF)
Figure S2
High Performance Liquid Chromatography (HPLC) methodology. A. Representative structures of the mycolic acids from M. tuberculosis. Natural mixtures of mycolates express a range of homologous components with varying chain lengths. B. Strategy for the release and derivatization of mycolic acids for fluorescence HPLC. (a) Hydrolysis with KOH/ methanol/toluene to release mycolic acids. (b) Phase-transfer catalyzed esterification of mycolic acids with pentafluorobenzyl bromide (PFB) to give PFB mycolates. (c) Esterification of PFB mycolates, by reaction with pyrenebutyric acid (PBA), to produce PBA-PFB mycolates, catalyzed by dicyclohexylcarbodiimide and pyrrolidinopyridine. R- represents the remainder of the mycolate molecule. C. Normal phase HPLC of PBA-PFB mycolates from bone samples and standard M. tuberculosis. D. Reverse phase HPLC of individual α -, methoxy- and ketomycolic acid PBA-PFB derivatives from bone samples and standard M. tuberculosis. The number of carbons in the individual underivatized mycolic acids is shown.
(0.83 MB TIF)
Figure S3
DNA sequence data. A. M. tuberculosis TbD1 flanking region (128 bp), obtained from the infant sample (5′-3′ strand). B. M. tuberculosis TbD1 flanking region (128 bp), obtained from the infant sample (3′-5′ strand). C. M. tuberculosis conserved membrane protein (CMP) region, obtained from the infant sample (5′-3′ strand only).
(12.09 MB TIF)
Figure S4
Repeated spoligotypes from the Atlit-Yam samples and controls. Each set of spoligotyping data (A–C) represents, from top to bottom, M. tuberculosis, M. bovis (BCG) controls, Atlit Yam female and Atlit Yam infant. D. Diagram of spoligotyping data, including M. africanum spoligotypes (Donoghue et al 2004), and a consensus pattern based on one or more positive results.
(5.61 MB TIF)