A Defect in Menadione Biosynthesis Induces Global Changes in Gene Expression in Staphylococcus aureus (original) (raw)
Abstract
Both the high-resolution two-dimensional protein gel electrophoresis technique and full-genome DNA microarrays were used for identification of Staphylococcus aureus genes whose expression was changed by a mutation in menD. Because the electron transport chain is interrupted, the mutant should be unable to use oxygen and nitrate as terminal electron acceptors. Consistent with this, a mutation in menD was found to cause a gene expression pattern typically detected under anaerobic conditions in wild-type cells: proteins involved in glycolytic as well as in fermentation pathways were upregulated, whereas tricarboxylic acid (TCA) cycle enzymes were significantly downregulated. Moreover, the expression of genes encoding enzymes for nitrate respiration and the arginine deiminase pathway was strongly increased in the mutant strain. These results indicate that the menD mutant, just as the site-directed S. aureus hemB mutant, generates ATP from glucose or fructose mainly by substrate phosphorylation and might be defective in utilizing a variety of carbon sources, including TCA cycle intermediates and compounds that generate ATP only via electron transport phosphorylation. Of particular interest is that there are also differences in the gene expression patterns between hemB and menD mutants. While some anaerobically active enzymes were present in equal amounts in both strains (Ldh1, SACOL2535), other classically anaerobic enzymes seem to be present in higher amounts either in the hemB mutant (e.g., PflB, Ald1, IlvA1) or in the menD mutant (arc operon). Only genes involved in nitrate respiration and the ald1 operon seem to be additionally regulated by a depletion of oxygen in the hemB and/or menD mutant.
Staphylococcus aureus has become one of the most feared pathogens in the hospital due to its ability to cause severe life-threatening infections (29). A large number of surface-associated and extracellular proteins are considered to play an important role in the virulence of this pathogen. During recent decades, S. aureus infections have become ever more difficult to treat because of the emerging resistance of this microorganism to various antimicrobial agents. The development of new therapeutic strategies to overcome S. aureus infections is thus the ambitious goal for the near future. For that purpose, a more comprehensive understanding of the physiological processes that facilitate the survival and replication of S. aureus within the host is urgently needed. Studying these phenomena may provide completely new targets for antimicrobial compounds.
Besides strains that possess genetically defined resistance determinants, physiological variants such as biofilm-forming strains and small-colony variants (SCVs) are difficult to treat with the conventional therapeutic approach (3, 7). Consequently, physiological characterization of these variants might provide some hints as to the mechanisms by which these variants are protected from host defenses and exposure to antibiotics. SCVs are characterized by a low growth rate, an atypical colony morphology, and an unusual bacterial physiology. Clinical S. aureus SCVs are often auxotrophic for hemin and menadione, which are used in cytochrome and menaquinone biosynthesis, respectively. Consequently, the electron transport chain should be interrupted in these strains (for a review, see reference 38). Because most of the clinical SCVs are genetically undefined and exhibit a high rate of reversion to the normal phenotype, mutants with defects in the electron transport chain were created in different S. aureus strains with defined genetic backgrounds (8325-4, COL) by deleting genes involved in the biosynthesis of heme (hemB, coding for porphobilinogen synthase) and menadione (menD, coding for 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase [SHCHC]) (27, 51, 52). These mutants show all the features typically found for SCVs recovered from clinical specimens (23, 37, 38, 43, 49-51).
As components of the respiratory chain, heme and menaquinone are important for the electron transport to both oxygen and nitrate (see Fig. S1 in the supplemental material). Previous studies on global gene expression analyses of a hemB mutant showed that enzymes of glycolytic and related pathways as well as of fermentation pathways are highly upregulated, while enzymes of the TCA cycle are downregulated by a defect in heme biosynthesis (27, 42). Consistent with this, a biochemical study showed that a hemB mutant is deficient in utilizing carboxylic acids or amino acids (52). These results strongly indicate that ATP is provided only by substrate phosphorylation. The only way to regenerate NADH might be the reduction of pyruvate to lactate (27). The phenotype of a menD mutant is very similar to that of a hemB mutant (45, 52). However, defects in carbon metabolism are remarkably more pronounced than those observed for the hemB mutant (52). This might be due to the fact that menaquinones are additionally required for the activity of some enzymes involved in central metabolic pathways such as malate:quinol oxidoreductase (Mqo2) and dihydroorotate dehydrogenase (PyrD). A menD mutant should be unable to convert oxaloacetate to malate and consequently to fumarate due to the loss of Mqo2 activity. This pathway should be active in the hemB mutant. Nevertheless, malate could be produced from pyruvate by the activity of the malate enzyme (SACOL1749). Finally, menaquinones could have regulatory functions like those described for ubiquinones in Escherichia coli (18, 32, 33).
In the present study, global gene expression of a menD mutant was analyzed by using both a transcriptomic and a proteomic approach. The expression profile of the menD mutant was first compared with that of the wild type. Subsequently, the gene expression profile of a hemB mutant was included in this study in order to find differences in gene expression between the menD and the hemB mutants. Finally, both the menD mutant and the hemB mutant were exposed to anaerobic conditions in order to find proteins induced by the lack of oxygen in these mutants.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. aureus wild-type strain COL (44) and its isogenic hemB (hemB::ermB) and menD (menD::ermC) mutants were used in this study. The hemB mutant as well the menD mutant was constructed by allelic replacement as previously described (2, 51, 52). For overnight cultures, 5 ml of tryptic soy broth (TSB) (Oxoid, Wesel, Germany) was inoculated with a single colony and cultivated at 37°C under vigorous agitation. For selective pressure, erythromycin was added to the overnight culture of the mutant strains to a final concentration of 2.5 μg/ml. For protein and RNA preparation, 100 ml of TSB was inoculated with exponentially growing cells of the respective S. aureus strain to an initial optical density at 540 nm (OD540) of 0.05, and the cultures were cultivated in 500-ml Erlenmeyer flasks under vigorous agitation at 37°C. For anaerobic experiments, 50-ml BD Falcon tubes (BD) completely filled with TSB were inoculated with exponentially growing cells of the respective S. aureus strains to initial OD540s of 0.05 and the cultures were grown under gentle agitation at 37°C. Anaerobic conditions were verified by using 0.001% resazurin as a redox indicator (also see reference 17).
Preparation of cytoplasmic proteins for preparative two-dimensional (2D) gel electrophoresis.
Cells of 50-ml cultures were harvested on ice and centrifuged for 10 min at 7,000 × g and 4°C. Cells were washed twice with ice-cold Tris-EDTA buffer and resuspended in 1 ml of lysis buffer (10 mM Tris, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, pH 7.5). For mechanical disruption, the cell suspension was replaced to screw-cap microtubes (Sarstedt, Germany) containing 500 μl of glass beads (diameter, 0.10 to 0.11 mm; Sartorius, Goettingen, Germany). Cells were disrupted by homogenization using a Ribolyser (Thermo Electron Corporation) at 6.5 m/s for 35 s. The lysate was centrifuged for 25 min at 21,000 × g (4°C). In order to remove membrane fragments and insoluble proteins, the centrifugation step was repeated for 45 min at 21,000 × g (4°C). The protein concentration was determined using Roti-Nanoquant (Roth, Germany), and the protein solution was stored at −20°C.
Analytical and preparative 2D polyacrylamide gel electrophoresis.
2D polyacrylamide gel electrophoresis was performed using the immobilized pH gradient technique described previously (8). In the first dimension, the protein samples (300 μg) were separated on immobilized pH gradient strips (GE Healthcare, Little Chalfont, United Kingdom) with a pH range of 4 to 7. The proteins separated on 2D gels were stained with colloidal Coomassie brilliant blue (9). The stained gels were scanned with a light scanner with an integrated transparency unit (Quatographic, Braunschweig, Germany).
Protein identification.
For identification of proteins by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS), Coomassie brilliant blue-stained protein spots were cut from gels by use of a spot cutter (Proteome Work) with a picker head of 2 mm and transferred into 96-well microtiter plates. Digestion with trypsin and subsequent spotting of peptide solutions onto the MALDI targets were performed automatically in an Ettan spot handling workstation (GE Healthcare, Little Chalfont, United Kingdom) by use of a modified standard protocol (13). MALDI-TOF MS analyses of spotted peptide solutions were carried out on a proteome analyzer 4700/4800 (Applied Biosystems, Foster City, CA). The spectra were recorded in a reflector mode in a mass range from 900 to 3,700 Da. Automatic or manual calibration was performed as described previously (13). After calibration, the peak lists were created using the “peak-to-mascot” script of the 4700 Explorer software with the following settings: mass range from 900 to 3,700 Da, peak density of 15 peaks per range of 200 Da, minimal area of 100 and maximal 60 peaks per protein spot, and minimal signal-to-noise (S/N) ratio of 15. Database searches were performed using the GPS explorer software version 3.6 (build 329) with the organism-specific databases.
Peptide mixtures that at least twice gave a Mowse score of at least 64 and a sequence coverage of at least 30% were regarded as positive identifications (IDs). Proteins that failed to exceed the 30% sequence coverage cutoff were subjected to MALDI tandem MS (13). MALDI-TOF analysis was performed for the three highest peaks of the TOF spectrum. For one main spectrum, 25 subspectra with 125 shots per subspectrum were accumulated using a random search pattern. The internal calibration was automatically performed as one-point calibration if the monoisotopic arginine (M+H)+ m/z at 175.119 or lysine (M+H)+ m/z at 147.107 reached an S/N ratio of at least 5. The peak lists were created using the “peak-to-mascot” script of the 4700 Explorer software with the following settings: mass range from 60 Da to a mass that was 20 Da lower than the precursor mass, peak density of 15 peaks per 200 Da, minimal area of 100 and maximal 65 peaks per precursor, and a minimum S/N ratio of 10. Database searches were performed using the GPS explorer software version 3.6 (build 329) with the organism-specific databases.
The combined MS and MS/MS peak lists were searched against an S. aureus COL and S. aureus N315 protein database with the Mascot search engine version 2.1.0.4 (Matrix Science, London, United Kingdom). Search parameters were as follows: trypsin digestion with one missed cleavage permitted, variable modifications (oxidation of methionine and cabamidomethylation of cysteine), mass tolerance for MS data of 50 ppm, and mass tolerance for precursor ions of 0.6 Da. Search results that yielded Mowse scores of at least 64 (P < 0.05) for MS data and 15 (P < 0.05) for MS/MS data and a sequence coverage of at least 20% were regarded as positive IDs.
Protein quantitation approaches.
The 2D gel images were analyzed with the software Delta2D (Decodon GmbH, Greifswald, Germany). Three different data sets were included in order to screen for differences in the accumulations of cytoplasmic proteins identified on 2D gels in different strains and under aerobic and anaerobic growth conditions.
RNA preparation.
Total RNA was isolated using the acid phenol method (17, 19) with some modifications. Briefly, 40-ml portions of aerobic and anaerobic cultures in the exponential growth phase (OD540 of 0.5) and 20 ml from aerobic and anaerobic cultures in the stationary growth phase (Fig. 1A) were treated with 10 ml of ice-cold killing buffer (20 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 20 mM NaN3). The cells were immediately separated from the supernatant by centrifugation (for 5 min at 7,155 × g and 4°C), washed with ice-cold killing buffer, and resuspended in lysis buffer (3 mM EDTA, 200 mM NaCl). For mechanical disruption, the cell suspension was transferred into screw-top tubes containing 500 μl of glass beads (diameter, 0.1 to 0.11 mm; Sartorius, Goettingen, Germany) and 500 μl of water-saturated phenol-chloroform-isoamyl alcohol (25:24:1 [vol/vol/vol]); the cells were then disrupted by homogenization using a Ribolyser (Thermo Electron Corporation) for 30 s at 6.5 m/s. Afterwards, the resulting RNA solution was extracted once with water-saturated phenol-chloroform-isoamyl alcohol (25:24:1 [vol/vol/vol]) and twice with chloroform-isoamyl alcohol (24:1 [vol/vol]). RNA was precipitated by using 70% ethanol and resuspended in deionized water. The quality of RNA was ensured by gel electrophoresis and by analysis with a bioanalyzer (Agilent Technologies, Palo Alto, CA).
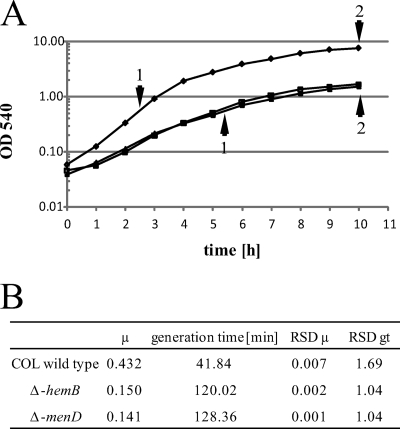
FIG. 1.
Growth curves (A) and growth parameters (B) for S. aureus COL (⧫), its isogenic menD mutant (▴), and the hemB mutant (▪). Sampling points for transcriptional and proteomic analyses are indicated by arrowheads on the growth curves. Abbreviations: RSD, relative standard deviation; μ, growth rate; gt, generation time.
Northern blot analyses.
Digoxigenin-labeled RNA probes were prepared by in vitro transcription with T7 RNA polymerase and appropriate PCR fragments as templates (17, 19). The PCR fragments were generated by using chromosomal DNA of S. aureus COL isolated with the chromosomal DNA isolation kit (Promega, Madison, WI) according to the manufacturer's recommendations and the respective oligonucleotides (see Table S1 in the supplemental material). Northern blot analyses were carried out as previously described (17). The digoxigenin-labeled RNA marker I (Roche, Indianapolis, IN) was used to calculate the sizes of the transcripts. The hybridization signals were detected using a Lumi-Imager (Roche Diagnostics, Mannheim, Germany) and analyzed using the software package Lumi-Analyst (Roche Diagnostics, Mannheim, Germany).
Microarray analyses.
The DNA microarrays used in this study (S. aureus N315; sciTRACER) were purchased from Scienion (Berlin, Germany) and contain 2,338 PCR products of 200 to 500 bp in length covering about 90% of the S. aureus N315 genes (NCBI NC_002745). PCR products were designed by the manufacturer (Scienion AG, Berlin, Germany), were optimized for similar thermodynamic hybridization parameters, and displayed no significant cross-hybridization with other open reading frames of the N315 genome. Each PCR product was spotted in duplicate on glass slides.
For DNA microarray analysis, RNAs of two independent experiments were prepared. Samples were taken at the exponential growth phase (OD540 = 0.5) and at the stationary phase.
For cDNA synthesis, 10 μg of total RNA was mixed with 0.5 μg of random hexamers (GE Healthcare, Little Chalfont, United Kingdom) in a total volume of 15 μl and denatured at 70°C for 2 min. The denatured RNA was then added to the labeling mixture (8 μl of 5× first-strand buffer [Invitrogen, Karlsruhe, Germany], 4 μl of Cy3-dUTP or Cy5-dUTP [GE Healthcare, Little Chalfont, United Kingdom], 4 μl of 0.1 M dithiothreitol [Invitrogen, Karlsruhe, Germany], 1 μl of RNasin [40 U/μl; Promega, Madison, WI], and 4 μl of deoxynucleoside triphosphate mix [dATP, dCTP, and dGTP at a 5 mM concentration and dTTP at 2 mM; GE Healthcare, Little Chalfont, United Kingdom]) and incubated for 25 min at 42°C after the addition of 2 μl of Superscript II (200 U/μl; Invitrogen, Karlsruhe, Germany). This first incubation was followed by a second incubation at 42°C for 35 min in the presence of 2 μl of freshly added Superscript II. The reaction was stopped by the addition of 5 μl of EDTA (0.5 M), and the RNA was hydrolyzed by the addition of 10 μl of NaOH (1 M) and incubation at 65°C for 15 min. The solution was neutralized with 25 μl of 1 M Tris-HCl (pH 7.5). Purification of labeled cDNA was done with a CyScribe purification kit (GE Healthcare, Little Chalfont, United Kingdom) according to the manufacturer's instruction. The purified cDNA was then concentrated in a SpeedVac, and the concentrate (ca. 2 μl) was dissolved in 40 μl of hybridization buffer, provided by Scienion (Berlin, Germany). Prior to the hybridization, the labeled cDNA hybridization buffer mixture was incubated for 2 min at 65°C. The slides were hybridized overnight (16 h) in a water bath at 42°C. After hybridization and three washing steps (0.03% sodium dodecyl sulfate and 2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate] for 5 min at room temperature, 0.2× SSC for 5 min at 42°C, and 0.06× SSC for 1 min at room temperature), the slides were dried by centrifugation.
Statistical analysis.
Slides were scanned using a ScanArray scanner (PE Biosystems, Weiterstadt, Germany). The obtained images were quantified with the ScanArray Express software, version 3.0 (PE Biosystems, Weiterstadt, Germany), using the adaptive threshold as the quantitation method. The measured fluorescence signals were subjected to local background subtraction followed by LOWESS subgrid normalization (ArrayInformatics software; Perkin Elmer). To identify genes with significantly altered transcription levels, a series of statistical analyses (filtering) were performed; cutoff values for ratio (log2, ratio Cy3/Cy5) of transcription levels of ≥1 and ≤−1 were used to filter genes with changes in transcript levels greater than ±1 (log2). Genes with a relative standard deviation of ≥0.30 across all changes of transcription level (ratio of Cy3 to Cy5 per gene) were excluded. Finally, to test if genes were differentially transcribed in wild-type strain COL versus the menD mutant, in wild-type strain COL versus the hemB mutant, and in the hemB mutant versus the menD mutant, we used the Student t test. Only those genes with significantly different mRNA levels (α ≤ 0.05) were selected.
Data visualization.
Quantitative transcriptomic and proteomic data were visualized by using the open source tool “ManyEyes” provided by IBM (http://services.alphaworks.ibm.com/manyeyes/home) (48). The transcriptome and proteome data are provided at http://microbio1.biologie.uni-greifswald.de/csp/bio/login.csp. For that purpose, all genes that showed changes of the transcript or of the protein amount in their respective experiments were summarized in a table together with their induction ratios. The genes were classified into functional groups according to TIGR and KEGG databases. This table was then used to create tree maps representing all the genes in their respective functional group. A special color code was used to visualize the induction ratios for each gene obtained by the proteomic and transcriptomic approaches.
Quantification of extracellular metabolites by 1H nuclear magnetic resonance.
Extracellular metabolites, i.e., glucose, lactate, acetate, ethanol, 2,3-butanediol, formate, and ornithine, were measured and quantified in the supernatants of the wild type, the hemB mutant, and the menD mutant grown in TSB medium under aerobic and anaerobic conditions (with the latter only for the wild type) by 1H nuclear magnetic resonance as described previously (22).
RESULTS AND DISCUSSION
Effects of a mutation in the menadione biosynthesis gene menD on global gene expression in S. aureus.
menD mutants are described as slow-growing subpopulations showing a restricted carbon metabolism (52). Accordingly, we found a clearly decreased growth rate for the menD mutant grown in TSB medium compared to that for the wild-type COL. In addition, the menD mutant reached cell densities conspicuously lower than those seen for the wild type. This growth phenotype is very similar to that found for a hemB mutant (Fig. 1) (27, 42).
DNA arrays and 2D gel analysis were used to get an overview on global changes in gene expression caused by a mutation in menD in S. aureus COL. Total RNA and cytoplasmic proteins were prepared from aerobically grown cells of the wild-type COL and the respective menD mutant at the exponential and stationary growth phases (Fig. 1). By comparing the gene transcription of the wild type with that of an isogenic menD mutant, differently transcribed genes could be identified. Altogether, at the exponential phase the transcription of 210 genes was changed. Among these, 104 genes showed an increased transcription rate in the mutant. At the stationary phase, 291 genes revealed changes in the transcript level between the wild type and the mutant. Additionally, we analyzed the cytoplasmic proteome of the wild type and the menD mutant. At a pI range of 4 to 7, most of the metabolic enzymes can be visualized on 2D gels (28), enabling us to compare the amounts of these proteins in both strains. Of the genes whose transcription was affected by a mutation in menD, 135 proteins were identified on the gels. In accordance with the transcriptional analyses, the amounts of 79 proteins changed in the mutant. In contrast, changes in the transcript levels of 56 genes did not result in alterations in protein amount. Moreover, the amounts of 88 proteins differed between the wild type and the mutant, while the respective transcript levels remained unchanged.
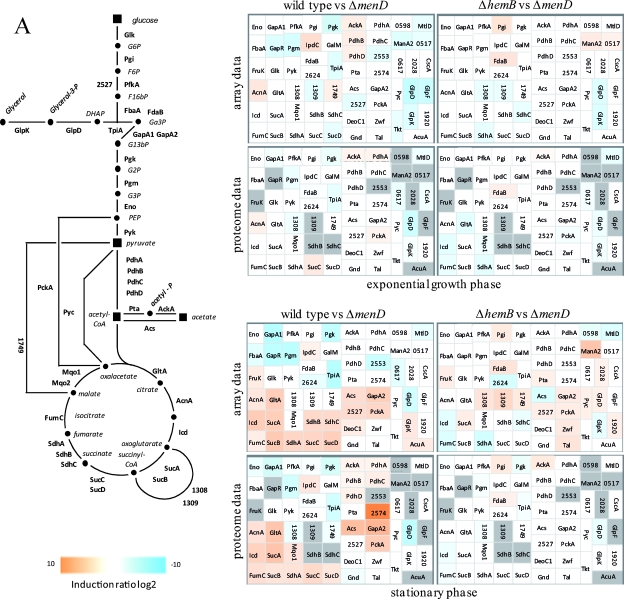
As observed with the hemB mutant (27, 42), the deletion of menD led to an increased expression of genes involved in glycolysis, fermentation, and anaerobic respiration (Fig. 2 and 3A; also see Table S2 in the supplemental material). Like heme, menadione is required for aerobic and anaerobic electron transport phosphorylation. Accordingly, the menD mutant also should be unable to use oxygen and nitrate as terminal electron acceptors (40). The higher glycolytic rate may compensate for the loss of electron transport phosphorylation. Simultaneously, the activity of the pyruvate dehydrogenase complex and of the TCA cycle as the main producer of NADH is downregulated, as indicated by reduced amounts of the respective enzymes (Fig. 2A and 3A).
FIG. 2.
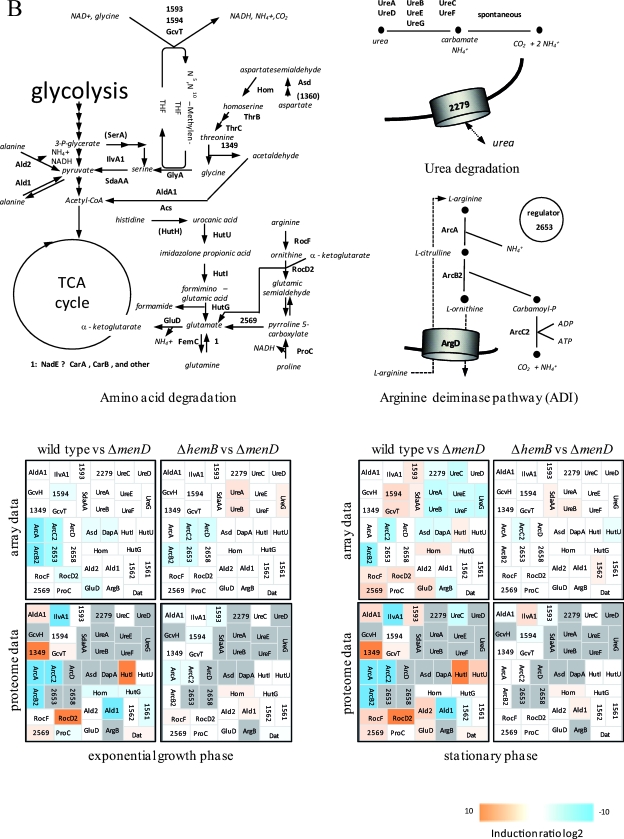
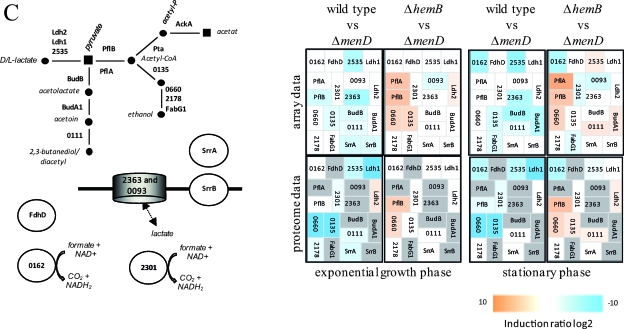
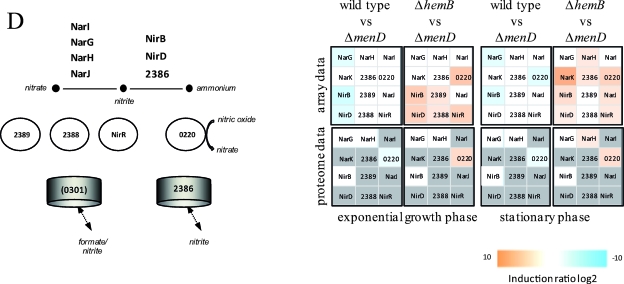
Visualization of transcriptomic (DNA microarrays) and proteomic (2D gels) data for selected genes/proteins of S. aureus COL (wild type) compared to its isogenic menD mutant (Δ_menD_) and for the hemB mutant (Δ_hemB_) compared to the menD mutant (Δ_menD_) grown in TSB medium at the exponential growth phase and at early stationary phase. (A) Carbohydrate metabolism. (B) Amino acid metabolism. (C) Fermentation. (D) Nitrate/nitrite respiration. Induction ratios were given as logarithms to the base 2. Boxes ranging in color from light orange to dark orange indicate transcripts or proteins that were significantly downregulated in the menD mutant, and boxes ranging from light blue to dark blue indicate transcripts or proteins that were significantly upregulated in the menD mutant. White boxes indicate transcripts or proteins with no changes between the menD mutant and the wild type or the hemB mutant. Gray boxes represent proteins that were not identified on the gels. The IDs, symbols, and functions are based on TIGR annotation for S. aureus COL (http://www.tigr.org). Glycolysis: Eno, enolase; FbaA, fructose-bisphosphate aldolase; FdaB, fructose-bisphosphate aldolase (class I); FruK, 1-phosphofructokinase; GalM, aldose 1-epimerase; GapA1, glyceraldehyde-3-phosphate dehydrogenase; GapR, glycolytic operon regulator; Glk, glucokinase; IpdC, pyruvate decarboxylase; PfkA, 6-phosphofructokinase; Pgi, glucose-6-phosphate isomerase A; Pgk, phosphoglycerate kinase; Pgm, phosphoglycerate mutase; Pyk, pyruvate kinase; TpiA, triosephosphate isomerase; 2624, acetyl coenzyme A (acetyl-CoA) synthetase. TCA cycle: AcnA, aconitate hydratase; FumC, fumarate hydratase; GltA, citrate synthase; Icd, isocitrate dehyrogenase; Mqo1, malate:quinone oxidoreductase; SdhA, succinate dehydrogenase flavoprotein subunit; SdhB, succinate dehydrogenase iron-sulfur protein subunit; SdhC, succinate dehydrogenase cytochrome _b_-558; SucA, 2-oxoglutarate dehydrogenase E1; SucB, dihydrolipoamide succinyltransferase; SucCD, succinyl-CoA synthetase beta/alpha chain; 1308/1309, 2-oxoacid ferredoxin oxidoreductase, alpha/beta subunit; 1749, malate dehydrogenase homolog. Pyruvate metabolism: AckA, acetate kinase; PdhABCD, pyruvate dehydrogenase E1; Pta, phosphotransacetylase; 2553, putative pyruvate oxidase; 2574, putative d-lactate dehydrogenase. Gluconeogenesis: Acs, acetyl-CoA synthetase; GapA2, glyceraldehyde 3-phosphate dehydrogenase 2; PckA, phosphoenolpyruvate carboxykinase; Pyc, pyruvate carboxylase; 2527, fructose-bisphosphatase. Pentose phosphate pathway: DeoC1, deoxyribose-phosphate aldolase; Gnd, phosphogluconate dehydrogenase; Tal, transaldolase; Zwf, glucose-6-phosphate 1-dehydrogenase. Sugar usage: CscA, sucrose-6-phosphate hydrolase; ManA2, mannose-6-phosphate isomerase; MtlD, mannitol-1-phosphate 5-dehydrogenase; 0517, trehalose-6-phosphate hydrolase; 0598, l-ribulokinase; 0617, putative hexulose-6-phosphate synthase; 2028, putative fructokinase. Glycerol degradation: GlpD, glycerol-3-phosphate dehydrogenase; GlpF, glycerol uptake facilitator; GlpK, glycerol kinase; 1920, putative glycerate dehydrogenase. Acetoin usage: AcuA, acetoin utilization protein. Amino acid degradation: Ald1, alanine dehydrogenase; Ald2, alanine dehydrogenase; AldA1, aldehyde dehydrogenase; ArgB, _N_-acetylglutamate 5-phosphotransferase; Asd, aspartate semialdehyde dehydrogenase; DapA, dihydrodipicolinate synthase; Dat, d-alanine aminotransferase; GcvH, glycine cleavage system protein H homolog; GcvT, aminomethyltransferase; GluD, NAD-specific glutamate dehydrogenase; Hom, homoserine dehydrogenase; HutG, formiminoglutamase; HutI, imidazolonepropionase; HutU, urocanate hydratase; IlvA1, threonine deaminase IlvA homolog; ProC, pyrroline-5-carboxylate reductase; RocD2, ornithine aminotransferase; RocF, arginase; SdaAA, l-serine dehydratase; 1349, threonine aldolase; 1562, branched-chain alpha-keto acid dehydrogenase E1; 1561, branched-chain alpha-keto acid dehydrogenase E1; 1593, glycine dehydrogenase subunit 2 homolog; 1594, glycine dehydrogenase subunit 1; 2569, 1-pyrroline-5-carboxylate dehydrogenase. Urea degradation: UreA, UreB, UreC, UreD, UreE, UreG, and UreF, urease subunits and urease accessory proteins; 2279, urea transporter. Arginine deiminase pathway (ADI): ArcA, arginine deiminase; ArcB2, ornithine transcarbamoylase; ArcC2, carbamate kinase; ArcD, arginine/ornithine antiporter; 2653, ArcR protein; 2658, arginine repressor. Fermentation: BudB, acetolactate synthase large subunit; BudA1, alpha-acetolactate decarboxylase; FabG1, short-chain dehydrogenase; FdhD, formate dehydrogenase accessory protein; Ldh1, l-lactate dehydrogenase; Ldh2, l-lactate dehydrogenase; PflA, formate acetyltransferase activating enzyme; PflB, formate acetyltransferase; SrrA, staphylococcal respiratory response protein SrrA (response regulator); SrrB, staphylococcal respiratory response protein SrrB (sensor kinase); 0093, lactate transporter; 0111, acetoinreductase; 0135, alcohol dehydrogenase/acetaldehyde dehydrogenase; 0162, NAD-dependent formate dehydrogenase; 0660, alcohol dehydrogenase I; 2178, putative alcohol dehydrogenase (zinc containing); 2301, formate dehydrogenase homolog; 2363, l-lactate permease; 2535, d-specific d-2-hydroxyacid dehydrogenase. Nitrate/nitrite respiration: NarG, respiratory nitrate reductase alpha chain; NarH, nitrate reductase beta chain; NarI, nitrate reductase gamma chain; NarJ, nitrate reductase delta chain; NirB, nitrite reductase; NirD, nitrite reductase [NAD(P)H] (small subunit); NirR, transcriptional regulator; 0220, flavohemoprotein Hmp; 0301, formate/nitrite transporter; 2386, nitrite extrusion protein; 2388, two-component sensor histidine kinase NreB; 2389, regulator of two-component regulatory NreC.
FIG. 3.
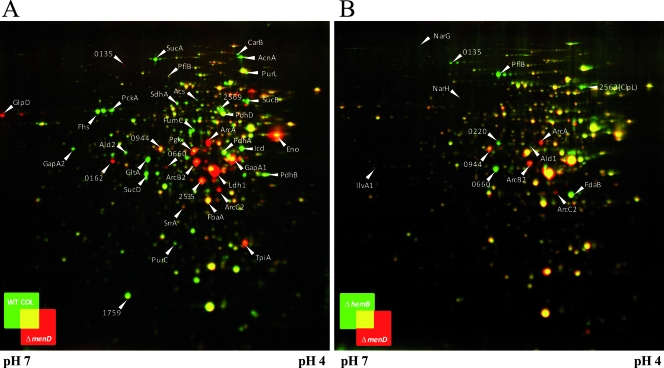
Cytoplasmic protein pattern of S. aureus COL and its isogenic hemB and menD mutants. The 2D gel images of cytoplasmic proteins of the menD mutant and the wild type (A) and of the menD mutant and the hemB mutant (B) were overlaid using Delta2D software (Decodon GmbH, Greifswald, Germany). Cytoplasmic proteins were prepared from cells grown in TSB under aerobic conditions at the exponential and early stationary phases. Portions (300 μg) of proteins were separated on 2D gels by using immobilized pH gradient strips with a pH range of 4 to 7. Proteins were stained with colloidal Coomassie brilliant blue. Proteins whose amounts were increased in the wild-type COL or the hemB mutant are indicated in green, and those proteins that were accumulated in the menD mutant appear in red. Spots of interest are labeled with the respective protein IDs according to the annotation of S. aureus COL (http://www.tigr.org).
Besides being involved in the generation of ATP, the electron transport chain is also involved in the oxidation of NADH. The upregulation of fermentative enzymes indicates that the menD mutant utilizes fermentation pathways for NAD regeneration. According to the gene expression data, production of lactate and 2,3-butanediol could be postulated. Consistent with this, a gene coding for a putative lactate permease (SACOL2363) was among the strongly upregulated genes, too. Although the amounts of PflB, SACOL0660 (Adh1), and SACOL0135 (AdhE) were also slightly increased, the production of ethanol and acetate should be restricted, as the pyruvate dehydrogenase is possibly inhibited by NADH and the pyruvate formate lyase was not activated when the cells were grown aerobically (27). Moreover, the arginine deiminase pathway and also enzymes involved in anaerobic respiration are highly expressed in the mutant. Besides generating ATP, an active arginine deiminase pathway might protect the cells against the damaging effects of an acidic environment by releasing ammonia. The same could be true for urea metabolism, which is also upregulated in the mutant (Fig. 2B; also see Table S2 in the supplemental material).
In the menD mutant, the mRNA levels of some genes with regulatory functions in metabolic adaptation processes were increased. Among them was gapR, whose homologous gene product in B. subtilis, CggR, is known to regulate the transcription of the gap operon (10, 16, 30). Moreover, the transcription of srrAB and the SACOL2653 gene (arcR) was upregulated. The two-component system SrrAB as well as the DNA binding protein ArcR is involved in anaerobic gene regulation and thus activates the fermentation metabolism in S. aureus in the absence of oxygen (31, 46). While SrrA/B might have a more global function in anaerobic gene regulation by affecting the expression of several genes involved in fermentation processes and the TCA cycle, the ArcR protein specifically regulates the expression of the arc operon in S. aureus by binding to a consensus sequence in front of the operon under anaerobic conditions and thereby activating gene transcription (31, 46). The mechanism by which SrrAB regulates the transcription of its target genes under both aerobic and anaerobic conditions is only poorly understood so far.
Likewise, differences in the mRNA levels of genes coding for components of the electron transport chain could be found between the menD mutant and the wild type (see Table S1 and Fig. S2 in the supplemental material). Transcription of two NADH dehydrogenases was increased in the mutant: type II NADH dehydrogenase encoded by the SACOL0944 gene and a subunit of a putative membrane-associated NADH dehydrogenase encoded by nuoF. The two terminal menaquinone oxidases that exist in S. aureus, QoxABCD and CydAB, seem to be differentially regulated. While the cydAB operon was strongly downregulated in the mutant, no influence on the transcript level of qoxABCD could be observed.
A further group of genes consisting of glpF, glpK, glpD, mltD, manA2, and SACOL0517, whose transcription is upregulated by a deletion of menD in exponentially growing cells, is involved in the utilization of alternative carbon sources such as glycerol, mannitol, mannose, and trehalose (see Table S2 in the supplemental material). Additionally, we found an increased transcription of genes whose gene products might play a role in the uptake of trehalose (SACOL0516), mannitol (SACOL2146), and fructose (SACOL2663) and also of glucose (ptsI, SACOL0175, SACOL2552) (see Table S2 in the supplemental material). Very recently, it was shown that the menD mutant is able to grow on glucose, trehalose, and mannose, although with growth rates that were very low compared to those of the wild type. However, on mannitol the growth of the mutant was almost completely restricted. The metabolism of mannitol might be more impaired than that of mannose, possibly by the inhibition of the mannitol-1-phosphate 5-dehydrogenase MltD activity in the presence of high NADH levels (52).
Also of particular interest were virulence genes whose expression was changed in a _menD_-deficient strain. It is interesting that the transcription levels of most of the genes involved in capsule synthesis were upregulated in the mutant. As found for the hemB mutant, virulence genes whose expression is induced at higher ODs in the wild-type COL failed to be induced in the menD mutant during growth in TSB medium. Among them are geh, hlY, and hlb and the SACOL0317 (lip) and SACOL2418 (sbi) genes (see Table S2 in the supplemental material). In accordance to this observation, the transcription of virulence-associated regulatory genes such as agrA and agrC2, rot, saeR and saeS, and sarS was downregulated in the mutant strain. However, there are also virulence genes showing a higher mRNA level in the mutant, namely, clf and aur and the SACOL1522 and SACOL2006 genes. The regulatory gene sarU was among the upregulated genes, too (see Table S2 in the supplemental material). The increased expression of genes encoding surface-associated proteins (e.g., Clf) and the reduced amount of toxins such as HlY should benefit internalization by human cells and the intracellular persistence of the menD mutant, as previously described for hemB mutant strains (38, 41, 47, 51).
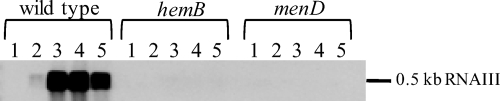
RNAIII is the major regulator of virulence genes in S. aureus and activates the transcription of various extracellular proteins and toxins, while the transcription of genes encoding surface-associated virulence factors is repressed by RNAIII. Since RNAIII is not represented on the DNA array used in the present study, Northern blot experiments were performed to evaluate RNAIII levels in the wild type compared to those in the menD mutant. As expected, RNAIII was not transcribed in the menD mutant (Fig. 4), possibly due to the conspicuously low ODs reached in the menD mutant culture and hence the resulting low concentration of the quorum-sensing peptide (24, 27). Moreover, SrrAB, which was enhanced in the menD mutant, could be involved in the repression of RNAIII as well (36). The loss of RNAIII was shown to affect virulence gene expression in S. aureus dramatically (11, 54) and might be an explanation for the reduced virulence potential observed for a hemB and a menD mutant in the Caenorhabditis elegans infection model (45).
FIG. 4.
Northern blot analyses of RNAIII. S. aureus COL wild type and its isogenic hemB and menD mutants were grown in TSB at 37°C. RNA was isolated at different growth stages, indicated as follows above the gels: 1, exponential growth phase; 2, late exponential growth phase; 3, transient phase; 4, early stationary phase; and 5, middle stationary phase. Portions (4 μg) of RNA from each sampling point were separated on denaturing agarose gels containing formaldehyde. Afterwards, the RNA was blotted onto positively charged nylon membranes and hybridized with an RNAIII-specific RNA probe.
In bacteria, the earliest response to the reduction of growth is the induction of the stringent response (21, 35, 53). In particular, genes involved in translation and amino acid biosynthesis are targets of a negative stringent control (1, 4, 12, 14, 26, 34). Consistent with the decreased growth rate of the menD mutant grown in TSB medium, a lower translational capacity was observed for the mutant, as indicated by a reduced transcriptional activity of genes encoding ribosomal proteins, aminoacyl tRNA synthetases, and translational factors.
Interestingly, among the differently expressed genes, we found 153 genes whose function has not been characterized so far (see Table S2 in the supplemental material). While the expression of 84 of these genes was found to be upregulated in the menD mutant, 69 genes showed a decreased expression rate. Interestingly, the expression of 44 of the up- and downregulated genes was similarly affected by a mutation in hemB (also see references 27 and 42). Elucidation of the function of these particular genes is a challenging task for future studies and might provide completely new insights into S. aureus physiology and virulence.
Detailed comparison of the gene expression profile of a menD mutant with that of a hemB mutant.
Apparently, most of the genes whose expressions were up- or downregulated in the menD mutant compared to the wild type were also found with changed transcript and protein levels in the hemB mutant and represent a transcriptome/proteome signature typical for electron transport deficiency in S. aureus (27, 42). It is noteworthy that this transcriptome/proteome signature was also found for S. aureus cells exposed to sublethal concentrations of nitric oxide known to inhibit aerobic and anaerobic respiration by reversible binding to cytochromes (22, 39). A recently published phenotypic microarray profiling study indicated that the phenotype of a menD mutant was very similar to that of a hemB mutant; however, the carbon metabolism was more profoundly affected in the menD mutant (52). In particular, the metabolism of mannitol, maltotriose, and dextrin was even more impaired by the loss of menadione. Moreover, it was shown that a defect in menadione biosynthesis also decreased heme biosynthesis (5). These differences might be explained by the fact that in addition to having a function in the electron transport chain, menaquinones are also found in enzymes of central metabolic pathways. Consequently, the activity of these enzymes should be affected by the loss of menadione biosynthesis, and these pathways should be impaired in this strain. Moreover, menaquinones could have regulatory functions as described for quinone electron carriers in E. coli. Here, quinones have been shown to efficiently oxidize cytoplasmically located cysteine residues of the sensor kinase ArcB and thereby inhibit its kinase activity (32). It is also possible that in S. aureus, quinones could be involved in redox signaling by oxidizing cysteine residues of a protein(s) that has regulatory functions. Hence, we compared the gene expression profile of the menD mutant with that of a hemB mutant in order to identify differently regulated genes between both mutants in more detail, thereby explaining the different phenotypes. Altogether, 346 genes were found to be differently expressed: 71 were expressed at the exponential phase and 303 were expressed at the stationary phase. Of the 346 genes, 189 showed a higher expression level in the hemB mutant than in the menD mutant (see Table S2 in the supplemental material). Strikingly, compared to the wild type, the expression levels of a small number of these genes, 83 of 346, were exclusively affected either in the hemB or in the menD mutant. The expression of 41 genes was up- or downregulated both by a mutation in hemB as well as by one in menD but on a different scale. Interestingly, the most significant differences in gene expression between both mutants were observed for genes of fermentation pathways and anaerobic respiration (Fig. 2C and D and 3B; also see Table S2 in the supplemental material). Thus, the transcription of pflA and pflB and of the SACOL0135 (adhE) and SACOL0660 (adh1) genes was strongly decreased in the menD mutant, indicating that acetate, formate, and ethanol formation should be strongly impaired by a deletion in menD. While pflB belongs to the genes whose transcription almost completely failed to be upregulated in the menD mutant, the transcription of the SACOL0660 (adh1) gene was considerably induced in both the hemB and menD mutants but to a higher extent in the hemB mutant (Fig. 2 and 5; also see Table S2 in the supplemental material). The same phenomenon was observed for genes involved in nitrate/nitrite respiration and ATP synthesis; the SACOL2563 gene, encoding the chaperone ClpL; nrdD, encoding an anaerobic ribonucleoside triphosphate reductase; and the cydAB operon as well as ald1 and ilv1 (Fig. 2, 3B, and 5; also see Table S2 in the supplemental material). In contrast, genes coding for components of the arginine deiminase pathway were more highly expressed in the menadione-deficient strain (Fig. 2 and 3B; also see Table S2 in the supplemental material).
FIG. 5.
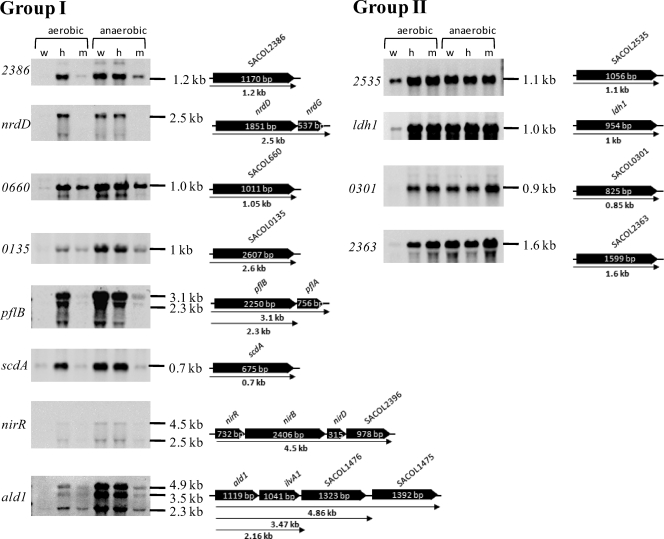
Northern blot analyses of genes whose transcription was induced under anaerobic conditions. Group I consists of genes whose transcriptions were induced in the hemB mutant but failed to be induced in the menD mutant. Group II consists of genes whose transcriptions were induced to similar levels in the hemB and menD mutants. Total RNA was isolated from S. aureus COL wild type (w) and its isogenic hemB (h) and menD (m) mutants grown under aerobic and anaerobic conditions to an OD540 of 0.5. Portions (5 μg [ldh, pflB, SACOL0660] or 10 μg [other genes] of RNA from each sampling point were separated on denaturing agarose gels containing formaldehyde. Afterwards, the RNA was blotted onto positively charged nylon membranes and hybridized with gene-specific RNA probes. Schematic representations of gene loci based on the sequence of S. aureus COL are shown (locus names are shown in italics and without the “SACOL” prefix). ald1, alanine dehydrogenase; ldh1, l-lactate dehydrogenase; nirR, transcriptional regulator NirR; nrdD, anaerobic ribonucleoside-triphosphate reductase; pflB, formate acetyltransferase; scdA, isochorismatase family protein ScdA; srrA, staphylococcal respiratory response protein SrrA, response regulator; SACOL0135, alcohol dehydrogenase/acetaldehyde dehydrogenase; SACOL0301, formate/nitrite transporter; SACOL0660, alcohol dehydrogenase I; SACOL2363, l-lactate permease; SACOL2386, nitrate/nitrite extrusion protein; SACOL2535, d-specific d-2-hydroxyacid dehydrogenase (d-lactate dehydrogenase).
Apparently, the expression of these genes partially depends on the presence of menadione, which supports the hypothesis that menaquinone could also have regulatory functions in S. aureus and might play a role in anaerobic gene regulation. However, there are also anaerobically regulated genes whose expression was similarly increased in the menD mutant and the hemB mutant (Fig. 5). Among these are ldh1 (encoding an l-lactate dehydrogenase), the SACOL2535 gene (d-lactate dehydrogenase), and the SACOL2363 gene (lactate permease), which are also known to be strongly induced under anaerobic conditions.
In order to strengthen the expression data for genes involved in fermentation pathways, we performed additional experiments on the formation of the particular fermentation products as well as on the consumption of glucose (see Fig. S3 in the supplemental material). With respect to lactate and acetate formation, aerobically grown hemB and menD mutant cells behaved like wild-type cells cultivated under anaerobic conditions. Hence, lactate but not acetate was detected in the supernatant. By contrast, neither the accumulation of 2,3-butanediol nor the accumulation of formate and ethanol was evident for the mutants. This might be explained either by the missing accumulation of the enzymes involved in these pathways (e.g., PflB in the menD mutant) or by the failed activation of these proteins, as already shown for PflB in the hemB mutant (27). Interestingly, the observed higher expression of the arc operon only in the menD mutant was accompanied by an increased formation of ornithine.
Influence of oxygen on the cytoplasmic protein pattern of electron transport-deficient mutants.
The recently described increased expression of genes involved in anaerobic respiration and fermentation pathways in a hemB mutant under aerobic conditions (17, 27, 42) was the first evidence that the oxygen concentration per se might not be crucial for the regulation of most of these genes. It was speculated that the redox balance (NAD/NADH ratio) in the cell, the membrane potential, and/or components of the respiratory chain are involved in the signaling of anaerobic gene regulation. The results concerning anaerobic gene expression obtained with the menD mutant emphasize this observation.
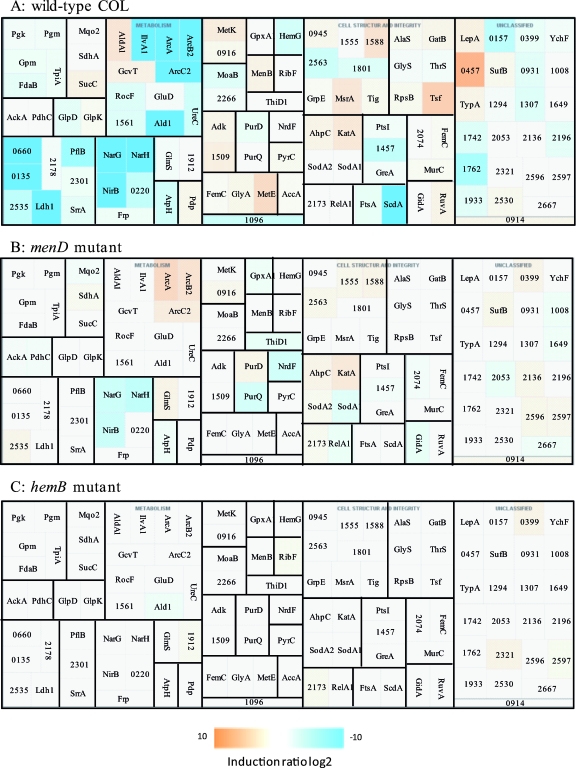
Next, we wanted to identify genes whose expression would be changed in the hemB and menD mutants under oxygen depletion conditions. An Fnr protein as described in other bacteria does not exist in S. aureus. The only known regulatory system in S. aureus that directly senses oxygen is the two-component system NreBC. The sensor NreB is a cytoplasmic protein, and its activity depends on iron sulfur reconstitution and oxygen availability. It was demonstrated that NreB is a direct oxygen sensor and activates the response regulator NreC (25). In Staphylococcus carnosus, the two-component system positively controls the nitrate reductase and the nitrite reductase operon and the putative nitrate transporter (15). The expression of these genes should be dependent on the oxygen concentration. The cytoplasmic protein patterns of the wild type and its isogenic hemB and menD mutants grown under aerobic and anaerobic conditions were compared. While in the wild type under anaerobic conditions at least 55 proteins accumulated, among them proteins of the arginine deiminase pathway (ArcA, ArcB2, ArcC2), Ald1, IlvA1, Ldh1, NarG, NarH, NirB, PflB, SACOL0660, SACOL0135, ScdA, and some enzymes of the glycolytic pathway, the amounts of most these proteins remained unchanged in the hemB mutant as well as in the menD mutant after a shift to anaerobic conditions (Fig. 5 and 6). This confirms our observation that most of the proteins whose amounts were affected under anaerobic conditions in the wild type are already present at elevated or decreased levels in the hemB mutant and/or in the menD mutant. However, for the menD mutant, we found a slightly increased accumulation of proteins involved in the reduction of nitrate (NarG, NarH, NirB) in cells grown without oxygen. Looking at the transcriptional level, a considerably oxygen-dependent induction of the transcription of the nir operon was observed only for the wild type. In the hemB mutant, the transcription of the operon was already upregulated under aerobic conditions, while in the menD mutant, almost no specific transcript could be detected under the same conditions. For both mutants, only a very slight increase of the transcript level under anaerobic conditions was found. These results indicate that the expression of these genes needs an active menaquinone system, and the oxygen-dependent expression is possibly activated via the presence of reduced menaquinone. However, it remains unclear which role NreBC may play for the activation of these genes under experimental conditions. In contrast, the expression of ald1 was additionally induced under these conditions only in the hemB mutant (Fig. 6).
FIG. 6.
Visualization of proteomic (2D gels) data for selected genes/proteins of S. aureus COL (wild type) and its isogenic menD mutant and hemB mutant grown in TSB medium under aerobic and anaerobic conditions at the exponential growth phase. (A) Wild type. (B) menD mutant. (C) hemB mutant. Induction ratios were given as logarithm to the base 2. Boxes ranging in color from light orange to dark orange indicate proteins whose amounts were decreased under anaerobic conditions, and boxes ranging from light blue to dark blue indicate proteins whose amounts were increased under anaerobic conditions. White boxes indicate proteins whose amounts did not change. The IDs, symbols, and functions are based on TIGR annotation for S. aureus COL (http://www.tigr.org). AccA, acetyl-CoA carboxylase, carboxyl transferase, alpha subunit; AckA, acetate kinase; Adk, adenylate kinase; AhpC, alkyl hydroperoxide reductase C subunit; AlaS, alanyl-tRNA synthetase; Ald1, alanine dehydrogenase; AldA1, aldehyde dehydrogenase; ArcA, arginine deiminase; ArcB2, ornithine carbamoyltransferase; ArcC2, carbamate kinase; AtpH, ATP synthase F1, delta subunit; FdaB, fructose-bisphosphate aldolase, class I; FemC, glutamine synthetase; Frp, NAD(P)H-flavin oxidoreductase; FtsA, cell division protein; GatB, glutamyl-tRNA (Gln) amidotransferase, B subunit; GcvT, glycine cleavage system T protein; GidA, glucose-inhibited division protein A; GlmS, glucosamine-fructose-6-phosphate aminotransferase (isomerizing); GlpD, aerobic glycerol-3-phosphate dehydrogenase; GlpK, glycerol kinase; GluD, glutamate dehydrogenase, NAD-specific; GlyA, serine hydroxymethyltransferase; GlyS, glycyl-tRNA synthetase; Gpm, phosphoglycerate mutase; GpxA1, glutathione peroxidase; GreA, transcription elongation factor; GrpE, heat shock protein; HemG, protoporphyrinogen oxidase; IlvA1, threonine dehydratase, catabolic; KatA, catalase; Ldh1, l-lactate dehydrogenase; LepA, GTP-binding protein; MenB, naphthoate synthase; MetE, 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase; MetK, _S_-adenosylmethionine synthetase; MoaB, molybdenum cofactor biosynthesis protein B; MsrA, peptide methionine sulfoxide reductase; MurC, UDP-_N_-acetylmuramate-alanine ligase; NarG, respiratory nitrate reductase, alpha subunit; NarH, respiratory nitrate reductase beta subunit; NirB, nitrite reductase [NAD(P)H] large subunit; NrdF, ribonucleoside-diphosphate reductase 2 beta subunit; PdhC, pyruvate dehydrogenase complex E2 component dihydrolipoamide acetyltransferase; Pdp, pyrimidine-nucleoside phosphorylase; PflB, formate acetyltransferase; Pgk, phosphoglycerate kinase; Pgm, phosphoglycerate mutase, 2,3-bisphosphoglycerate independent; PtsI, phosphoenolpyruvate-protein phosphotransferase; PurD, phosphoribosylamine-glycine ligase; PurQ, phosphoribosylformylglycinamidine synthase I; PyrC, dihydroorotase; RelA1, GTP pyrophosphokinase; RibF, riboflavin biosynthesis protein; RocF, arginase; RpsB, ribosomal protein S2; RuvA, Holliday junction DNA helicase; SACOL0135, alcohol dehydrogenase, iron containing; SACOL0157, hypothetical protein; SACOL0220, flavohemoprotein, putative; SACOL0399, oxidoreductase, putative; SACOL0457, hypothetical protein; SACOL0660, alcohol dehydrogenase, zinc containing; SACOL0914, FeS assembly ATPase SufC; SACOL0916, cysteine desulfurase, SufS subfamily; SACOL0931, hydrolase, haloacid dehalogenase-like family; SACOL0945, cytosol aminopeptidase; SACOL1008, hypothetical protein; SACOL1096, TrkA potassium uptake family protein; SACOL1294, metallo-β-lactamase family protein; SACOL1307, hypothetical protein; SACOL1457, PTS system, IIA component; SACOL1509, nucleoside diphosphate kinase; SACOL1555, peptidase, M20/M25/M40 family; SACOL1561, 2-oxoisovalerate dehydrogenase, E1 component beta subunit; SACOL1588, proline dipeptidase; SACOL1649, hypothetical protein; SACOL1724, MutT/nudix family protein; SACOL1762, thiol peroxidase, putative; SACOL1801, peptidase, M20/M25/M40 family; SACOL1912, glucosamine-6-phosphate isomerase, putative; SACOL1933, ThiJ/PfpI family protein; SACOL2053, S1 RNA binding domain protein; SACOL2074, d-alanine-d-alanine ligase; SACOL2136, hypothetical protein; SACOL2173, alkaline shock protein 23; SACOL2178, alcohol dehydrogenase, zinc containing; SACOL2196, hypothetical protein; SACOL2266, molybdopterin biosynthesis protein MoeA, putative; SACOL2301, formate dehydrogenase, alpha subunit, putative; SACOL2321, oxidoreductase, short-chain dehydrogenase/reductase family; SACOL2530, hypothetical protein; SACOL2535, d-isomer-specific 2-hydroxyacid dehydrogenase family protein; SACOL2563, ATP-dependent Clp protease, putative; SACOL2596, hypothetical protein; SACOL2597, hydrolase, alpha/beta hydrolase fold family; SACOL2667, isochorismatase family protein; ScdA, ScdA protein; SdhA, succinate dehydrogenase, flavoprotein subunit; SodA1, superoxide dismutase; SodA2, superoxide dismutase; SrrA, DNA binding response regulator SrrA; SucC, succinyl-CoA synthase, beta subunit; SufB, FeS assembly protein; ThiD1, phosphomethylpyrimidine kinase; ThrS, threonyl-tRNA synthetase; Tig, trigger factor; TpiA, triosephosphate isomerase; Tsf, translation elongation factor Ts; TypA, GTP-binding protein; UreC, urease, alpha subunit; YchF, hypothetical protein.
Conclusions.
A defect in menadione biosynthesis induces an expression pattern very similar to that found for mutants with a defect in heme biosynthesis and for wild-type cells grown under anaerobic conditions (17, 27, 42). This was not very surprising and confirmed our previous knowledge on the physiology of electron transport-deficient mutants. However, comparing the gene expression pattern of the menD mutant with that of a hemB mutant, the most prominent differences in gene expression were found for genes involved in nitrate respiration and fermentation pathways. The expression of some of these genes showed a lower induction ratio in the absence of active menaquinone and could not be further induced under anaerobic conditions. The quinone pool, which acts both as a collector of electrons from various dehydrogenases and as an electron donor to reductases and oxidases, is a central component of the electron transport chain. The redox state of quinones will change rapidly under conditions that affect the electron flow in the electron transport chain, for example, a shift from aerobic to anaerobic conditions and vice versa, but also in mutants with defects in cytochrome biosynthesis (such as hemB mutants). As the electrons cannot flow from quinones toward O2 via cytochrome oxidases, the quinones should be present in the reduced state and are not able to accept electrons any longer. In E. coli, oxidized cysteine residues of sensor kinase ArcB switch to the reduced form under anaerobic conditions by the uptake of electrons from the reduced quinones, resulting in an increased kinase activity. This mechanism allows a rapid response of bacterial cells to changes in oxygen concentrations in the environment (18, 32, 33). The present data provide evidence that a very similar mechanism of anaerobic gene regulation might exist in S. aureus. In the absence of oxygen, menaquinone could transfer electrons to a sensor kinase which is active in its reduced form. Among the sensor kinases encoded by the genome of S. aureus are several proteins which possess cysteine residues within the cytoplasmic domain. Among these are SrrB (three cysteine residues) and VicK (one cysteine residue). The most promising candidate might be SrrB, which is already known to play a role in anaerobic gene regulation. Interestingly, the mechanism by which the kinase activity of SrrB is induced is completely unknown, and it would be interesting to analyze whether the cysteine residues are important for the function of the protein.
Obviously, at least three mechanisms might exist in S. aureus that enable the bacterium to adapt to the consequences of low oxygen concentrations with expression of genes involved in anaerobic respiration and fermentation pathways. The first mechanism, which is active in electron transport-deficient mutants and under anaerobic conditions, might react to changes in the redox balance within the cells. Interestingly, an inverted repeat found in front of some of these genes showed strong similarities to possible Rex binding sites in B. subtilis and Streptomyces (17). Rex is a repressor and could bind either NAD or NADH. However, the protein is active, binds to DNA, and represses the expression of its target genes only in association with NAD (6, 20). A second mechanism might be dependent on the menaquinone system and consequently induce gene expression in the hemB mutant and under anaerobic conditions in the wild type but never in the menD mutant. The regulatory components of this mechanism have yet to be elucidated. The third mechanism includes proteins such as NreBC that could directly measure the oxygen concentration. This mechanism should be active in the wild type as well as in electron transport-deficient mutants but exclusively under anaerobic conditions. Obviously, most of the genes are regulated by more than one mechanism.
Supplementary Material
[Supplemental material]
Acknowledgments
We thank Dirk Albrecht for support in protein identification and Thomas Meier for excellent technical assistance. Furthermore, Decodon GmbH (Greifswald, Germany) is acknowledged for providing Delta2D software.
The study was supported by research grants from the BMBF (031U107A/-207A; 031U213B), the Deutsche Forschungsgemeinschaft (SFB/TRR34/1-2006, GK212/3-00), EU (Staphdynamics) to M.H. and S.E., and the Deutsche Forschungsgemeinschaft (EI247/7-1) to C.V.E.
Footnotes
▿
Published ahead of print on 1 August 2008.
REFERENCES
- 1.Anderson, K. L., C. Roberts, T. Disz, V. Vonstein, K. Hwang, R. Overbeek, P. D. Olson, S. J. Projan, and P. M. Dunman. 2006. Characterization of the Staphylococcus aureus heat shock, cold shock, stringent, and SOS responses and their effects on log-phase mRNA turnover. J. Bacteriol. 1886739-6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bates, D. M., C. von Eiff, P. J. McNamara, G. Peters, M. R. Yeaman, A. S. Bayer, and R. A. Proctor. 2003. Staphylococcus aureus menD and hemB mutants are as infective as the parent strains, but the menadione biosynthetic mutant persists within the kidney. J. Infect. Dis. 1871654-1661. [DOI] [PubMed] [Google Scholar]
- 3.Baumert, N., C. von Eiff, F. Schaaff, G. Peters, R. A. Proctor, and H. G. Sahl. 2002. Physiology and antibiotic susceptibility of Staphylococcus aureus small colony variants. Microb. Drug Resist. 8253-260. [DOI] [PubMed] [Google Scholar]
- 4.Bernhardt, J., J. Weibezahn, C. Scharf, and M. Hecker. 2003. Bacillus subtilis during feast and famine: visualization of the overall regulation of protein synthesis during glucose starvation by proteome analysis. Genome Res. 13224-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bochner, B. R., P. Gadzinski, and E. Panomitros. 2001. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 111246-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brekasis, D., and M. S. Paget. 2003. A novel sensor of NADH/NAD+ redox poise in Streptomyces coelicolor A3(2). EMBO J. 224856-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brouillette, E., A. Martinez, B. J. Boyll, N. E. Allen, and F. Malouin. 2004. Persistence of a Staphylococcus aureus small-colony variant under antibiotic pressure in vivo. FEMS Immunol. Med. Microbiol. 4135-41. [DOI] [PubMed] [Google Scholar]
- 8.Büttner, K., J. Bernhardt, C. Scharf, R. Schmid, U. Mäder, C. Eymann, H. Antelmann, A. Völker, U. Völker, and M. Hecker. 2001. A comprehensive two-dimensional map of cytosolic proteins of Bacillus subtilis. Electrophoresis 222908-2935. [DOI] [PubMed] [Google Scholar]
- 9.Candiano, G., M. Bruschi, L. Musante, L. Santucci, G. M. Ghiggeri, B. Carnemolla, P. Orecchia, L. Zardi, and P. G. Righetti. 2004. Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 251327-1333. [DOI] [PubMed] [Google Scholar]
- 10.Doan, T., and S. Aymerich. 2003. Regulation of the central glycolytic genes in Bacillus subtilis: binding of the repressor CggR to its single DNA target sequence is modulated by fructose-1,6-bisphosphate. Mol. Microbiol. 471709-1721. [DOI] [PubMed] [Google Scholar]
- 11.Dunman, P. M., E. Murphy, S. Haney, D. Palacios, G. Tucker-Kellogg, S. Wu, E. L. Brown, R. J. Zagursky, D. Shlaes, and S. J. Projan. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 1837341-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durfee, T., A. M. Hansen, H. Zhi, F. R. Blattner, and D. J. Jin. 2008. Transcription profiling of the stringent response in Escherichia coli. J. Bacteriol. 1901084-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eymann, C., A. Dreisbach, D. Albrecht, J. Bernhardt, D. Becher, S. Gentner, T. Tam Le, K. Büttner, G. Buurman, C. Scharf, S. Venz, U. Völker, and M. Hecker. 2004. A comprehensive proteome map of growing Bacillus subtilis cells. Proteomics 42849-2876. [DOI] [PubMed] [Google Scholar]
- 14.Eymann, C., G. Homuth, C. Scharf, and M. Hecker. 2002. Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis. J. Bacteriol. 1842500-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fedtke, I., A. Kamps, B. Krismer, and F. Götz. 2002. The nitrate reductase and nitrite reductase operons and the narT gene of Staphylococcus carnosus are positively controlled by the novel two-component system NreBC. J. Bacteriol. 1846624-6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fillinger, S., S. Boschi-Muller, S. Azza, E. Dervyn, G. Branlant, and S. Aymerich. 2000. Two glyceraldehyde-3-phosphate dehydrogenases with opposite physiological roles in a nonphotosynthetic bacterium. J. Biol. Chem. 27514031-14037. [DOI] [PubMed] [Google Scholar]
- 17.Fuchs, S., J. Pané-Farré, C. Kohler, M. Hecker, and S. Engelmann. 2007. Anaerobic gene expression in Staphylococcus aureus. J. Bacteriol. 1894275-4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Georgellis, D., O. Kwon, and E. C. Lin. 2001. Quinones as the redox signal for the arc two-component system of bacteria. Science 2922314-2316. [DOI] [PubMed] [Google Scholar]
- 19.Gertz, S., S. Engelmann, R. Schmid, K. Ohlsen, J. Hacker, and M. Hecker. 1999. Regulation of sigmaB-dependent transcription of sigB and asp23 in two different Staphylococcus aureus strains. Mol. Gen. Genet. 261558-566. [DOI] [PubMed] [Google Scholar]
- 20.Gyan, S., Y. Shiohira, I. Sato, M. Takeuchi, and T. Sato. 2006. Regulatory loop between redox sensing of the NADH/NAD+ ratio by Rex (YdiH) and oxidation of NADH by NADH dehydrogenase Ndh in Bacillus subtilis. J. Bacteriol. 1887062-7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hecker, M., A. Richter, A. Schroeter, L. Wölfel, and F. Mach. 1987. Synthesis of heat shock proteins following amino acid or oxygen limitation in _Bacillus subtilis relA_+ and relA strains. Z. Naturforsch. 42941-947. [PubMed] [Google Scholar]
- 22.Hochgräfe, F., C. Wolf, S. Fuchs, S. Engelmann, and M. Hecker. 2008. Nitric oxide stress induces different responses but mediates comparable protein thiol protection in Bacillus subtilis and Staphylococcus aureus. J. Bacteriol. 1904997-5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kahl, B., M. Herrmann, A. S. Everding, H. G. Koch, K. Becker, E. Harms, R. A. Proctor, and G. Peters. 1998. Persistent infection with small colony variant strains of Staphylococcus aureus in patients with cystic fibrosis. J. Infect. Dis. 1771023-1029. [DOI] [PubMed] [Google Scholar]
- 24.Kahl, B. C., G. Belling, P. Becker, I. Chatterjee, K. Wardecki, K. Hilgert, A. L. Cheung, G. Peters, and M. Herrmann. 2005. Thymidine-dependent Staphylococcus aureus small-colony variants are associated with extensive alterations in regulator and virulence gene expression profiles. Infect. Immun. 734119-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamps, A., S. Achebach, I. Fedtke, G. Unden, and F. Götz. 2004. Staphylococcal NreB: an O2-sensing histidine protein kinase with an O2-labile iron-sulphur cluster of the FNR type. Mol. Microbiol. 52713-723. [DOI] [PubMed] [Google Scholar]
- 26.Koburger, T., J. Weibezahn, J. Bernhardt, G. Homuth, and M. Hecker. 2005. Genome-wide mRNA profiling in glucose starved Bacillus subtilis cells. Mol. Genet. Genomics 2741-12. [DOI] [PubMed] [Google Scholar]
- 27.Kohler, C., C. von Eiff, G. Peters, R. A. Proctor, M. Hecker, and S. Engelmann. 2003. Physiological characterization of a heme-deficient mutant of Staphylococcus aureus by a proteomic approach. J. Bacteriol. 1856928-6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohler, C., S. Wolff, D. Albrecht, S. Fuchs, D. Becher, K. Büttner, S. Engelmann, and M. Hecker. 2005. Proteome analyses of Staphylococcus aureus in growing and non-growing cells: a physiological approach. Int. J. Med. Microbiol. 295547-565. [DOI] [PubMed] [Google Scholar]
- 29.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339520-532. [DOI] [PubMed] [Google Scholar]
- 30.Ludwig, H., G. Homuth, M. Schmalisch, F. M. Dyka, M. Hecker, and J. Stülke. 2001. Transcription of glycolytic genes and operons in Bacillus subtilis: evidence for the presence of multiple levels of control of the gapA operon. Mol. Microbiol. 41409-422. [DOI] [PubMed] [Google Scholar]
- 31.Makhlin, J., T. Kofman, I. Borovok, C. Kohler, S. Engelmann, G. Cohen, and Y. Aharonowitz. 2007. Staphylococcus aureus ArcR controls expression of the arginine deiminase operon. J. Bacteriol. 1895976-5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malpica, R., B. Franco, C. Rodriguez, O. Kwon, and D. Georgellis. 2004. Identification of a quinone-sensitive redox switch in the ArcB sensor kinase. Proc. Natl. Acad. Sci. USA 10113318-13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malpica, R., G. R. Sandoval, C. Rodriguez, B. Franco, and D. Georgellis. 2006. Signaling by the arc two-component system provides a link between the redox state of the quinone pool and gene expression. Antioxid. Redox Signal. 8781-795. [DOI] [PubMed] [Google Scholar]
- 34.Nascimento, M. M., J. A. Lemos, J. Abranches, V. K. Lin, and R. A. Burne. 2008. Role of RelA of Streptococcus mutans in global control of gene expression. J. Bacteriol. 19028-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishino, T., J. Gallant, P. Shalit, L. Palmer, and T. Wehr. 1979. Regulatory nucleotides involved in the Rel function of Bacillus subtilis. J. Bacteriol. 140671-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pragman, A. A., Y. Ji, and P. M. Schlievert. 2007. Repression of Staphylococcus aureus SrrAB using inducible antisense srrA alters growth and virulence factor transcript levels. Biochemistry 46314-321. [DOI] [PubMed] [Google Scholar]
- 37.Proctor, R. A., P. van Langevelde, M. Kristjansson, J. N. Maslow, and R. D. Arbeit. 1995. Persistent and relapsing infections associated with small-colony variants of Staphylococcus aureus. Clin. Infect. Dis. 2095-102. [DOI] [PubMed] [Google Scholar]
- 38.Proctor, R. A., C. von Eiff, B. C. Kahl, K. Becker, P. McNamara, M. Herrmann, and G. Peters. 2006. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 4295-305. [DOI] [PubMed] [Google Scholar]
- 39.Richardson, A. R., P. M. Dunman, and F. C. Fang. 2006. The nitrosative stress response of Staphylococcus aureus is required for resistance to innate immunity. Mol. Microbiol. 61927-939. [DOI] [PubMed] [Google Scholar]
- 40.Sasarman, A., P. Purvis, and V. Portelance. 1974. Role of menaquinone in nitrate respiration in Staphylococcus aureus. J. Bacteriol. 117911-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schröder, A., R. Kland, A. Peschel, C. von Eiff, and M. Aepfelbacher. 2006. Live cell imaging of phagosome maturation in Staphylococcus aureus infected human endothelial cells: small colony variants are able to survive in lysosomes. Med. Microbiol. Immunol. 195185-194. [DOI] [PubMed] [Google Scholar]
- 42.Seggewiss, J., K. Becker, O. Kotte, M. Eisenacher, M. R. Yazdi, A. Fischer, P. McNamara, N. Al Laham, R. Proctor, G. Peters, M. Heinemann, and C. von Eiff. 2006. Reporter metabolite analysis of transcriptional profiles of a Staphylococcus aureus strain with normal phenotype and its isogenic hemB mutant displaying the small-colony-variant phenotype. J. Bacteriol. 1887765-7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seifert, H., C. von Eiff, and G. Fatkenheuer. 1999. Fatal case due to methicillin-resistant Staphylococcus aureus small colony variants in an AIDS patient. Emerg. Infect. Dis. 5450-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shafer, W. M., and J. J. Iandolo. 1979. Genetics of staphylococcal enterotoxin B in methicillin-resistant isolates of Staphylococcus aureus. Infect. Immun. 25902-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sifri, C. D., A. Baresch-Bernal, S. B. Calderwood, and C. von Eiff. 2006. Virulence of Staphylococcus aureus small colony variants in the Caenorhabditis elegans infection model. Infect. Immun. 741091-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Throup, J. P., F. Zappacosta, R. D. Lunsford, R. S. Annan, S. A. Carr, J. T. Lonsdale, A. P. Bryant, D. McDevitt, M. Rosenberg, and M. K. Burnham. 2001. The srhSR gene pair from Staphylococcus aureus: genomic and proteomic approaches to the identification and characterization of gene function. Biochemistry 4010392-10401. [DOI] [PubMed] [Google Scholar]
- 47.Vaudaux, P., P. Francois, C. Bisognano, W. L. Kelley, D. P. Lew, J. Schrenzel, R. A. Proctor, P. J. McNamara, G. Peters, and C. von Eiff. 2002. Increased expression of clumping factor and fibronectin-binding proteins by hemB mutants of Staphylococcus aureus expressing small colony variant phenotypes. Infect. Immun. 705428-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Viegas, F. B., M. Wattenberg, F. van Ham, J. Kriss, and M. McKeon. 2007. ManyEyes: a site for visualization at Internet scale. IEEE Trans. Vis. Comput. Graph. 131121-1128. [DOI] [PubMed] [Google Scholar]
- 49.von Eiff, C., K. Becker, D. Metze, G. Lubritz, J. Hockmann, T. Schwarz, and G. Peters. 2001. Intracellular persistence of Staphylococcus aureus small-colony variants within keratinocytes: a cause for antibiotic treatment failure in a patient with Darier's disease. Clin. Infect. Dis. 321643-1647. [DOI] [PubMed] [Google Scholar]
- 50.von Eiff, C., D. Bettin, R. A. Proctor, B. Rolauffs, N. Lindner, W. Winkelmann, and G. Peters. 1997. Recovery of small colony variants of Staphylococcus aureus following gentamicin bead placement for osteomyelitis. Clin. Infect. Dis. 251250-1251. [DOI] [PubMed] [Google Scholar]
- 51.von Eiff, C., C. Heilmann, R. A. Proctor, C. Woltz, G. Peters, and F. Gotz. 1997. A site-directed Staphylococcus aureus hemB mutant is a small-colony variant which persists intracellularly. J. Bacteriol. 1794706-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.von Eiff, C., P. McNamara, K. Becker, D. Bates, X. H. Lei, M. Ziman, B. R. Bochner, G. Peters, and R. A. Proctor. 2006. Phenotype microarray profiling of Staphylococcus aureus menD and hemB mutants with the small-colony-variant phenotype. J. Bacteriol. 188687-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wendrich, T. M., and M. A. Marahiel. 1997. Cloning and characterization of a relA/spoT homologue from Bacillus subtilis. Mol. Microbiol. 2665-79. [DOI] [PubMed] [Google Scholar]
- 54.Ziebandt, A. K., D. Becher, K. Ohlsen, J. Hacker, M. Hecker, and S. Engelmann. 2004. The influence of agr and sigmaB in growth phase dependent regulation of virulence factors in Staphylococcus aureus. Proteomics 43034-3047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]