Transforming Growth Factor β-Induced Smad1/5 Phosphorylation in Epithelial Cells Is Mediated by Novel Receptor Complexes and Is Essential for Anchorage-Independent Growth (original) (raw)
Abstract
Transforming growth factor β (TGF-β) signals predominantly through a receptor complex comprising ALK5 and TβRII to activate receptor-regulated Smads (R-Smads) Smad2 and Smad3. In endothelial cells, however, TGF-β can additionally activate Smad1 and Smad5. Here, we report that TGF-β also strongly induces phosphorylation of Smad1/5 in many different normal epithelial cells, epithelium-derived tumor cells, and fibroblasts. We demonstrate that TβRII and ALK5, as well as ALK2 and/or ALK3, are required for TGF-β-induced Smad1/5 phosphorylation. We show that the simultaneous activation of the R-Smads Smad2/3 and Smad1/5 by TGF-β results in the formation of mixed R-Smad complexes, containing, for example, phosphorylated Smad1 and Smad2. The prevalence of these mixed R-Smad complexes explains why TGF-β-induced Smad1/5 phosphorylation does not result in transcriptional activation via bone morphogenetic protein (BMP)-responsive elements, which bind activated Smad1/5-Smad4 complexes that are induced by BMP stimulation. Thus, TGF-β induces two parallel pathways: one signaling via Smad2-Smad4 or Smad3-Smad4 complexes and the other signaling via mixed R-Smad complexes. Finally, we assess the function of the novel arm of TGF-β signaling and show that TGF-β-induced Smad1/5 activation is not required for the growth-inhibitory effects of TGF-β but is specifically required for TGF-β-induced anchorage-independent growth.
Ligands of the transforming growth factor β (TGF-β) superfamily control numerous cellular processes, such as proliferation, apoptosis, differentiation, adhesion, and mobility. As a result, they play essential roles in organisms undergoing early development and in adult organisms, in both healthy and diseased states (28). This ligand superfamily can be divided into three major subgroups: the TGF-βs, the activins and Nodals, and the bone morphogenetic proteins (BMPs)/growth and differentiation factors (40). Signal transduction is mediated by receptor complexes comprising two type II receptors and two type I receptors, both of which are serine/threonine kinases (12). There are five type II receptors in the human genome and seven type I receptors which are named activin receptor-like kinases 1 to 7 (ALK1 to ALK7) (12). It has been very difficult to precisely define which ligands bind which type II-type I receptor complexes, and in fact, recent evidence suggests that multiple combinations can occur. Some type I receptors, such as ALK5, appear to act predominantly with one type II receptor and to bind one class of ligand, in this case, TβRII and the TGF-βs, respectively. However, other type I receptors are more promiscuous, for example, ALK2, which acts with a number of different type II receptors and appears to be able to mediate signals from all the subgroups of ligands in different cellular contexts (12).
Binding of ligand induces formation of a type II-type I receptor complex in which the constitutively active type II receptor phosphorylates and activates the type I receptor. The signal is then transduced to the nucleus predominantly by members of the Smad family. This is achieved through the phosphorylation of specific receptor-regulated Smads (R-Smads) by an activated type I receptor at two serine residues in an S-M/V-S motif at the extreme C terminus of the R-Smad. There are five different R-Smads: Smad1, Smad2, Smad3, Smad5, and Smad8. Which R-Smads are phosphorylated by which type I receptor is determined by the sequence of the so-called L45 loop in the type I receptor and the L3 loop in the C-terminal Mad homology 2 domain of the R-Smad (5, 12, 33). ALK1, ALK2, ALK3, and ALK6 bind and phosphorylate Smad1, Smad5, and Smad8, whereas ALK4, ALK5, and ALK7 bind and phosphorylate Smad2 and Smad3. This phosphorylation promotes formation of both homomeric Smad complexes and heteromeric complexes with the common mediator Smad, Smad4. These activated Smad complexes accumulate in the nucleus, where they are directly involved in the regulation of target genes (12). Different R-Smad-Smad4 complexes recognize distinct promoter elements (37). Smad3-Smad4 complexes bind repeats of the sequence AGAC or its complement GTCT, which are known as Smad-binding elements. In contrast, the BMP-regulated R-Smads preferentially bind to GC-rich sequences, which are commonly found next to a Smad-binding element, allowing the Smad4 in a Smad1/5-Smad4 complex to also contact DNA. This occurs in conjunction with the transcriptional regulator Schnurri (35, 45). The affinities of the Smads for all these binding sites are relatively weak, and thus multimers of the Smad binding sites are required for efficient ligand-induced activation. In contrast to Smad1, Smad3, Smad4, and Smad5, Smad2 lacks any inherent DNA-binding activity, and thus Smad2-Smad4 complexes are recruited to DNA via other transcription factors, for example, FoxH1 (4).
The original view of the TGF-β superfamily signaling pathways was that there were essentially two branches: a TGF-β/Nodal/activin branch that signals through activation of Smad2/3, and a BMP/growth and differentiation factor branch that signals through Smad1, Smad5, and Smad8 (40). However, the concept that one ligand subfamily activates one class of R-Smads was revised when it became clear that in endothelial cells, TGF-β activates both Smad2/3 and Smad1/5 through a heteromeric receptor complex comprising TβRII, ALK1, and ALK5 (15, 16). In these cells, ALK5 promotes a TGF-β-dependent recruitment of ALK1 into a receptor complex, and for maximal ALK1 activation, ALK5 kinase activity is required. Furthermore, TGF-β signaling through ALK1 is promoted by the coreceptor endoglin (22). ALK1 was shown to directly antagonize ALK5 signaling and induce biological responses opposite those of ALK5. Thus, the biological responses of endothelial cells are determined by the fine balance between ALK5 and ALK1 signaling.
Since ALK1 is an endothelial cell-specific receptor (41), the phenomenon of dual signaling by TGF-β might be expected to be specific to endothelial cells. One exception to this rule has been reported, namely, TGF-β3-induced Smad1 phosphorylation was observed in Hs578T human breast cancer cells, but the mechanism underlying this was not investigated (25). To determine how widespread this dual-signaling phenomenon is, we investigated the ability of TGF-β to induce simultaneous phosphorylation of Smad1/5 and Smad2/3 in a panel of normal epithelial cells, fibroblasts, and epithelium-derived tumor cell lines of different tissue origins. Unexpectedly, we observe TGF-β-induced Smad1/5 phosphorylation in many different cell lines, both transformed and untransformed, and conclude that it is an extremely widespread event. We demonstrate that this dual signaling requires TβRII, ALK5, and, in addition, ALK2 and/or ALK3. We show that coactivation of phosphorylated Smad1/5 and Smad2/3 by TGF-β results in the production of novel mixed R-Smad complexes that we hypothesize are responsible for transducing the signal to the nucleus. We did not find that TGF-β-induced Smad1/5 signaling antagonized Smad2/3 signaling, in contrast to results obtained with endothelial cells. Instead, we demonstrate that TGF-β-induced phosphorylation of Smad1/5 is critically important for a subset of TGF-β responses. It is not required for TGF-β-induced growth arrest or induction of epithelial-to-mesenchymal transition (EMT) but is essential for the ability of TGF-β to promote anchorage-independent growth.
MATERIALS AND METHODS
Plasmids.
The following plasmids were as previously described: (CAGA)12-Luc (9), c-JunSBR6-Luc (23), BRE-Luc (21), the EF-LacZ control plasmid (1), and the TK-Renilla plasmid (Promega) (24). Plasmids expressing green fluorescent protein (GFP)-ALK5 and FLAG-ALK2 were generated using standard PCR methods, and the open reading frames were inserted into PAGFPC1 (39) and EF-Plink (18), respectively. All constructs were verified by sequencing. The plasmids expressing hemagglutinin (HA)-ALK5 and TβRII were gifts from Anita Roberts, and plasmids expressing HA-ALK1 and HA-ALK3 were a gift from Peter ten Dijke.
Cell culture and treatments.
All cell lines were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum with a few exceptions. NMuMG cells were additionally supplemented with 10 mg/ml insulin, Colo-357 and LUDLU-1 cells were cultured in RPMI medium with 10% fetal calf serum, and bEnd5 endothelioma cells (36), MCF-10A cells (8), enhanced GFP (EGFP)-Smad2-containing HaCaT cells (39), and EGFP-Smad3-containing HaCaT cells (30) were maintained as previously described. Cells were induced at the indicated times with 2 ng/ml TGF-β1 (PeproTech), 200 ng/ml BMP2 (R&D Systems), 20 ng/ml BMP4 (R&D Systems), or 200 ng/ml BMP7 (R&D Systems). The ALK2/3/6 inhibitor dorsomorphin, which was obtained from Paul Yu (46), and SB-431542 (Tocris) were used at a final concentration of 10 μM (19). Recombinant Noggin was a gift from Senyon Choe and was used at a 300-ng/ml final concentration.
Transfections, luciferase assays, and reverse transcription-PCR (RT-PCR).
Small interfering RNA (siRNA) transfections in EpH4 cells and EpH4 cells that have been transformed by stable expression of oncogenic Ras (EpRas cells) were performed in six-well plates. Cells were seeded at 5 × 104 cells per well the day prior to transfection. Each well was transfected with a 75 nM final concentration of siRNA by using Dharmafect reagent 2. Cells were incubated for 48 h and then replated into six-well plates. After 24 h, cells were treated with growth factors. For the soft agar assay, siRNA-treated EpRas cells were split after 48 h and seeded into the soft agar. When two siRNA pools were combined, 50 nM of the Smad1 siRNA oligonucleotide and 25 nM of the Smad5 oligonucleotide were transfected (see Fig. 7).
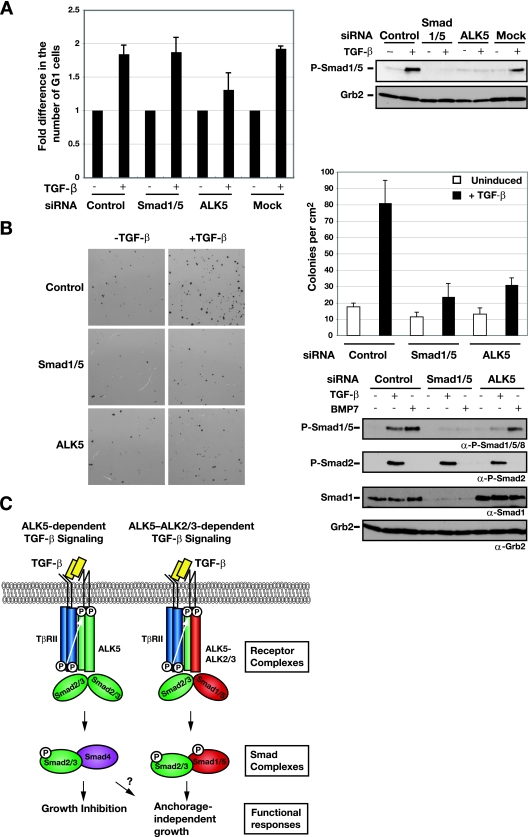
FIG. 7.
Knockdown of Smad1/5 has no effect on TGF-β-induced growth arrest, but Smad1/5 is required for anchorage-independent growth in soft agar. (A) Knockdown of Smad1/5 has no effect on TGF-β-induced growth arrest. EpH4 cells were transfected with siRNA SMARTpools against Smad1/5, ALK5 siRNA duplexes, or a control siRNA oligonucleotide. After 48 h, cells were either left uninduced (−) or stimulated with TGF-β1 (+) for 20 h. Samples were collected for FACS analysis to determine the percentages of cells in the G1, S, and G2/M phases. The percentage of cells in G1 was normalized to the percentage of cells in G1 under unstimulated conditions (no TGF-β). The values shown are the averages from three independent experiments. For the control siRNA samples, the average percentage of cells in G1 in the absence of TGF-β was 24.5%, and that in the presence of TGF-β was 44.8%. Samples were also analyzed to confirm depletion of TGF-β-induced phosphorylation of Smad1/5. After 72 h incubation, cells were induced with TGF-β1 for 45 min and cell lysates were analyzed by Western blotting using antibodies against phosphorylated Smad1/5 (P-Smad1/5) and Grb2 (right panel). (B) Activation of Smad1/5 in combination with Smad2/3 by TGF-β is required for the growth of EpRas cells in soft agar in response to TGF-β. EpRas cells were transfected with siRNA SMARTpools against Smad1/5, an ALK5 siRNA duplex, or a control siRNA oligonucleotide, as indicated. After 48 h, the cells were assayed for their ability to grow in soft agar in the absence or presence of 2 ng/ml TGF-β as described in Materials and Methods. After 12 days, the number of colonies was assessed by staining with thiazolyl blue tetrazolium bromide (left panel). Each field is equal to 1 cm2. The means and standard deviations (error bars) for three replicate wells from a representative experiment are shown (upper-right panel). Confirmation of depletion of TGF-β-induced phosphorylation of Smad1/5 Western blot analysis as described for panel A is also shown (lower-right panel). α, anti. (C) Schematic illustration of the signaling events downstream of TGF-β stimulation in epithelial cells. Different sets of receptor complexes are formed to phosphorylate the R-Smads, which then form complexes in a variety of combinations. The complexes accumulate in the nucleus, where they are involved in transcriptional activation and repression of a plethora of gene promoters. See the text for a discussion. For simplicity, Smad complexes are portrayed as dimers.
Both MDA-MB-231 and Colo-357 cells were transfected with INTERFERin (Polyplus-transfection) using siRNA SMARTpools at concentrations of 1 nM and 5 nM, respectively. When siRNA SMARTpools were combined, 1 nM of each SMARTpool was used (see Fig. 1B and 4A). When siRNA oligonucleotides were combined together in Colo-357 cells, siRNAs were transfected using 5 nM of each (see Fig. 4B). Samples for Western blotting were taken 72 h posttransfection after treatment with TGF-β, BMP4, or BMP7. See Table S1 in the supplemental material for a comprehensive list of the siRNA oligonucleotides used. Plasmid transfections were performed using either Lipofectamine reagent (Invitrogen) for NIH 3T3, C2C12, and MDA-MB-231 cells or Lipofectamine2000 reagent (Invitrogen) for EpH4 and EpRas cells according to the manufacturer's protocol. For luciferase assays with EpH4 and EpRas cells, cells were transfected in six-well plates and the next day seeded into 12-well plates. For all other cells, transfection was performed in the 12-well format. After overnight incubation, cells were induced with TGF-β for 8 h and assayed for luciferase activity by using either a dual-luciferase reporter system (Promega) or as previously described (13). All experiments were performed in triplicate.
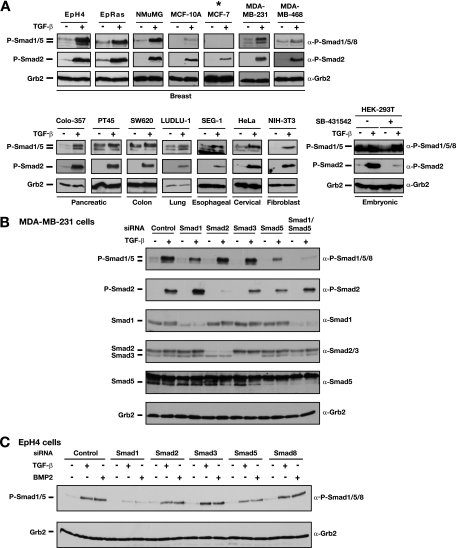
FIG. 1.
TGF-β induces phosphorylation of Smad1 and Smad5 in epithelial cells. α, anti. (A) A panel of cell lines including normal and cancerous breast cell lines and various human cancer cell lines and fibroblasts were tested for the ability of TGF-β to induce phosphorylation of Smad1/Smad5. Cells were either left untreated (−) or stimulated with 2 ng/ml TGF-β1 (+) for 1 h as indicated. An asterisk indicates the cell line in which TGF-β did not induce Smad1/5 phosphorylation. The lower right panel shows results for HEK-293T cells that were not treated with SB-431542 or treated with 10 μM SB-431542 overnight. The untreated cells were treated with or without TGF-β for 1 h. Of the cells treated with SB-431542, those to be induced with TGF-β were washed twice with phosphate-buffered saline and then stimulated with TGF-β for 1 h. Whole-cell extracts were analyzed by Western blotting using antibodies against phosphorylated Smad1/5/8 (P-Smad1/5/8), P-Smad2, and Grb2 as a loading control. (B) Smad1 and Smad5 are phosphorylated in response to TGF-β in MDA-MB-231 cells. MDA-MB-231 cells were transfected with siRNA SMARTpools against the R-Smads or a control siRNA oligonucleotide as indicated. At 72 h posttransfection, cells were left untreated (−) or treated with TGF-β1 for 45 min (+), and whole-cell extracts were prepared and analyzed by Western blotting using antibodies against Smad1, Smad2/3, Smad5, P-Smad1/5/8, P-Smad2, and Grb2. Note that P-Smad1/5 is a doublet. The siRNA knockdown results suggest that the upper band is a mixture of Smad1 and Smad5 and the lower band is predominantly an isoform of Smad5. (C) EpH4 cells were transfected with siRNA SMARTpools against the R-Smads or a control siRNA oligonucleotide as indicated. Cells were either left uninduced or treated with TGF-β1 (2 ng/ml) or BMP2 (200 ng/ml) for 45 min as indicated. Cell lysates were analyzed as described for panel A.
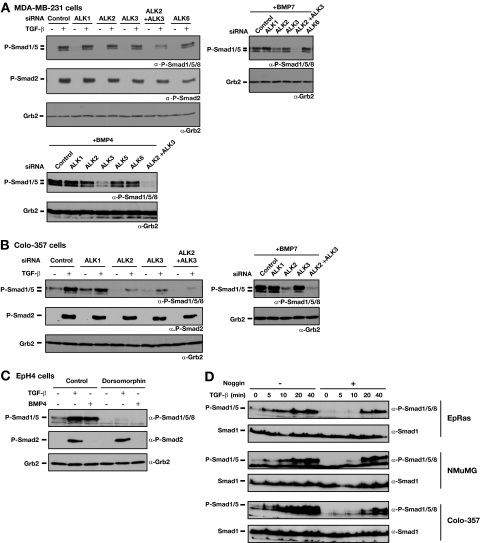
FIG. 4.
ALK2 and ALK3 are required for TGF-β-induced phosphorylation of Smad1/5. α, anti. (A) Effect of knockdown of ALK2 and ALK3 on Smad1/5 phosphorylation in MDA-MB-231 cells. Cells were transfected with siRNA SMARTpools against the ALKs for 72 h. Cells were either left uninduced or stimulated with 2 ng/ml TGF-β1 (upper-left panel), 200 ng/ml BMP7 (upper-right panel), or 20 ng/ml BMP4 (lower panel) for 45 min. Whole-cell extracts were analyzed by Western blotting using antibodies against phosphorylated Smad1/5/8 (P-Smad1/5/8), P-Smad2, and Grb2 as a loading control. (B) Colo-357 cells were transfected with the indicated siRNA SMARTpools for 72 h. Cells were either left uninduced or stimulated with 2 ng/ml TGF-β1 (left panel) or 200 ng/ml BMP7 (right panel) for 45 min. Whole-cell extracts were analyzed by Western blotting using antibodies against P-Smad1/5/8, P-Smad2, and Grb2 as a loading control. (C) Inhibition of ALK2, ALK3, and ALK6 activity abolishes phosphorylation of Smad1/5 by TGF-β. EpH4 cells were incubated with dorsomorphin (10 μM), a selective inhibitor of ALK2, ALK3, and ALK6, for 1 h prior to stimulation with TGF-β1 or BMP4 for 45 min. Cell lysates were analyzed by Western blotting as described above. (D) Phosphorylation of Smad1/5 in response to TGF-β is direct and independent of BMP signaling. EpRas, NMuMG, and Colo-357 cells were treated with or without 300 ng/ml Noggin for 2 h and then treated with TGF-β for the times indicated. Whole-cell extracts were analyzed by Western blotting using antibodies against phospho-Smad1/5/8 and Smad1.
RT-PCR was performed as previously described (23). The sequences of the primers used for analysis of ALK mRNAs are given in Table S2 in the supplemental material.
Cell lysis, Western blotting, and immunoprecipitations (IPs).
Whole-cell extracts were prepared using radioimmunoprecipitation assay buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1 mM dithiothreitol, 25 mM NaF, 25 mM Na β-glycerophosphate, and protease inhibitors). Western blotting was performed using standard procedures. The following antibodies were used: Smad1 (Zymed); Smad5 (Upstate); Smad2/3 and Grb2 (BD Biosciences); Smad4 (B8; Santa Cruz); ALK5 (V22; Santa Cruz); phospho-Smad2, phospho-Smad1/5/8 (catalog no. 9511), and phospho-Smad1/3/5 (catalog no. 9514) (Cell Signaling Technology); TβRII (06-318; Upstate); FLAG and FLAG-horseradish peroxidase (Sigma); and HA, HA-horseradish peroxidase, and GFP (Roche) antibodies.
For IPs of endogenous proteins, EpH4 cells were either left uninduced or induced with 2 ng/ml of TGF-β for 45 min, and whole-cell extracts were prepared using IP buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM EDTA, 0.1% Triton X-100, 1 mM dithiothreitol, 25 mM NaF, 25 mM Na β-glycerophosphate, and protease inhibitors). Lysates were precleared with protein A beads and then incubated with 1.25 μg of the corresponding antibody coupled to bovine serum albumin-blocked protein A beads. Beads were washed three times with IP buffer and immunoprecipitates were fractionated on 15% SDS-polyacrylamide gels and Western blotted. IP of receptors was performed with anti-FLAG beads (Sigma) or anti-GFP antibody (Roche) coupled to protein G beads in lysis buffer (100 mM NaCl, 50 mM Tris-HCl, pH 7.5, 0.1% NP-40, 25 mM NaF, 25 mM Na β-glycerophosphate, and protease inhibitors). Immunoprecipitates were fractionated on 15% SDS-polyacrylamide gels and Western blotted.
Cell cycle analysis and soft agar assay.
The growth inhibition assays with EpH4 cells were performed essentially as previously described (34). Briefly, the cells were transfected by siRNA, grown to confluence over 48 h, trypsin released, and replated at a low density in the presence or absence of 2 ng/ml TGF-β. Samples for fluorescence-activated cell sorting (FACS) analysis were collected 20 to 24 h later.
For soft agar assays, a concentrated bottom layer comprising 2 ml of growth medium containing 0.6% agarose (Sigma) was poured into a six-well dish. A total of 5 × 103 EpRas cells was resuspended in 2 ml of medium containing 0.3% agarose and overlaid on the hardened bottom layer. After 12 days of incubation, colonies were visualized by staining with 0.5 mg/ml thiazolyl blue tetrazolium bromide (Sigma) and scanned on a Umax PowerLook 1120 scanner to visualize the colonies. Colonies (≥100 μm) were counted from three different fields (1 cm2) within each triplicate well.
RESULTS
TGF-β induces phosphorylation of Smad1 and Smad5 in epithelial cells.
It has previously been shown that TGF-β induces phosphorylation of Smad1/5 in addition to Smad2/3 in endothelial cells (15, 16). To investigate whether this signaling is restricted to endothelial cells, we tested a panel of epithelial cell lines for their ability to phosphorylate Smad1/5 in response to TGF-β. This panel included normal human and mouse mammary epithelial lines (NMuMG, EpH4, and MCF-10A), transformed mammary cell lines (MCF-7, MDA-MB-231, and MDA-MB-468), human pancreatic adenocarcinomas (Colo-357 and PT45), carcinomas from a range of other tissues, and fibroblasts (Fig. 1A). Cells were treated with TGF-β for 1 h and then analyzed with a phospho-Smad1/5/8-specific antibody which we proved does not recognize phosphorylated forms of Smad2 or Smad3 (Fig. 1B; see Fig. S1 in the supplemental material). Interestingly, in addition to inducing Smad2 phosphorylation in these cells, TGF-β also induced a rapid phosphorylation of Smad1/5/8 in 13 out of the 14 cell lines (Fig. 1A). We also observed a robust TGF-β-induced Smad1/5/8 phosphorylation in the untransformed keratinocyte cell line HaCaT (see Fig. S1 in the supplemental material). The levels of Smad1/5/8 phosphorylation in the different cell lines were variable, with the strongest inductions observed for mammary epithelial lines, for the Colo-357 pancreatic adenocarcinoma, and for the NIH 3T3 fibroblasts (Fig. 1A). For some cells, we observed high basal levels of phospho-Smad1/5/8, which result from a combination of autocrine BMP signaling (see below) and autocrine TGF-β signaling. In fact, in some cases, this masked the induction of phospho-Smad1/5/8 by TGF-β, in particular, in HEK-293T cells. These cells show little induction of phospho-Smad1/5/8 when TGF-β is simply added to the growth medium. However, they exhibit a strong response if the basal phospho-Smad1/5/8 levels are first reduced by incubation of the cells with the ALK5 kinase inhibitor SB-431542 (19) prior to washout of the inhibitor and TGF-β stimulation (Fig. 1A, bottom-right panel).
We noted that the phospho-Smad1/5/8 antibody frequently recognized a doublet of bands. To identify them, we performed siRNA knockdown of all the individual Smad proteins. Knockdown of R-Smads in MDA-MB-231 and Colo-357 cells revealed that the upper band is comprised mainly of Smad1 with some Smad5, whereas the lower band is an isoform of Smad5 (Fig. 1B; see Fig. S2A in the supplemental material). Knockdown of Smad2 or Smad3 had no effect on either of these bands (Fig. 1B). In EpH4 cells, both TGF-β and BMP2 treatments resulted predominantly in phosphorylation of Smad1 (Fig. 1C; see Fig. S2B in the supplemental material). Taken together, these data confirm that both Smad1 and Smad5 are phosphorylated in response to TGF-β in a wide range of cells of different tissue origins.
ALK5 and TβRII are required for Smad1/5 phosphorylation in response to TGF-β.
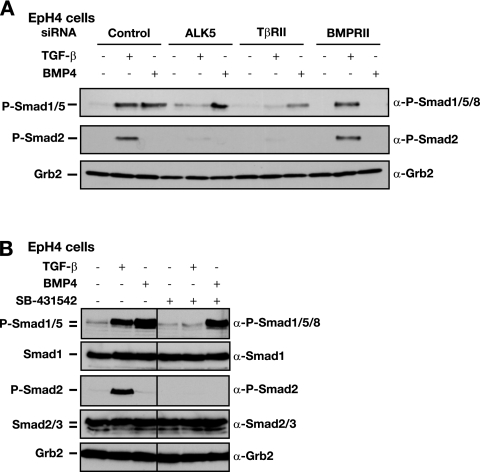
We next investigated whether the canonical TGF-β receptors were required for the activation of Smad1/5 in response to TGF-β in epithelial cells. siRNA knockdown of either ALK5 or TβRII protein (see Fig. S3A in the supplemental material) abrogated phosphorylation of Smad1/5 in response to TGF-β, whereas BMP4-induced Smad1/5 phosphorylation was relatively unaffected (Fig. 2A). Phosphorylation of Smad2 was also abolished in ALK5- and TβRII-depleted cells, as expected (Fig. 2A). As a negative control, knockdown of the BMP type II receptor (BMPRII) had no effect on TGF-β-induced Smad1/5 or Smad2 phosphorylation, whereas BMP4-induced Smad1/5 phosphorylation was abolished (Fig. 2A). These results indicate that both ALK5 and TβRII are required for phosphorylation of Smad1/5 in response to TGF-β. Moreover, they also indicate that the effect is not due to TGF-β induction of BMP expression. To examine whether ALK5 kinase activity was also involved in TGF-β induction of Smad1/5 phosphorylation, EpH4 cells were preincubated with the ALK5 kinase inhibitor SB-431542 before stimulation with TGF-β. This treatment completely abolished TGF-β-induced Smad1/5 phosphorylation but had no effect on BMP4-induced Smad1/5 phosphorylation (Fig. 2B). These data show that TGF-β-induced activation of Smad1/5 in epithelial cells is critically dependent on TβRII and ALK5 and requires the kinase activity of ALK5.
FIG. 2.
ALK5 and TβRII are required for the activation of Smad1/5 by TGF-β. α, anti. (A) Depletion of ALK5 and TβRII by siRNA silencing abolishes TGF-β-induced Smad1/5 phosphorylation. EpH4 cells were transfected with siRNA SMARTpools against TβRII, BMPRII, an siRNA duplex against ALK5, or a control siRNA oligonucleotide. After 72 h, cells were either left uninduced or stimulated with TGF-β1 or BMP4 for 45 min as indicated. Whole-cell extracts were analyzed by Western blotting using antibodies against phosphorylated Smad1/5/8 (P-Smad1/5/8), P-Smad2, and Grb2 as a control for protein loading. (B) ALK5 kinase activity is required for TGF-β-induced Smad1/5 phosphorylation. EpH4 cells were either left untreated or treated with the ALK5 inhibitor SB-431542 (10 μM) 15 min prior to stimulation with TGF-β or BMP4 for 45 min. Cell lysates were analyzed by Western blotting using antibodies against P-Smad1/5/8, Smad1, P-Smad2, Smad2/3, and Grb2 as a loading control.
Differential regulation of Smad1/5 phosphorylation and Smad2/3 phosphorylation in response to TGF-β.
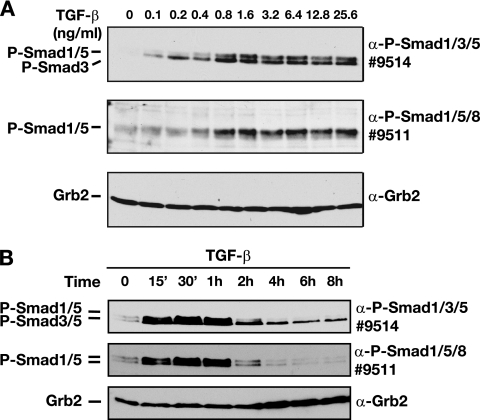
We further characterized this novel arm of TGF-β/Smad signaling in EpH4 cells by comparing its TGF-β dose dependency and kinetics of signaling with those of the canonical branch of the TGF-β/Smad pathway mediated by phosphorylation of Smad3 by using an antibody that detects both phosphorylated Smad1/5 and phosphorylated Smad3 (see Fig. S2C in the supplemental material). TGF-β readily induced detectable Smad3 phosphorylation when used at a concentration of 0.1 ng/ml and reached a plateau at 0.8 ng/ml of TGF-β (Fig. 3A, top panel). In contrast, Smad1/5 phosphorylation was first detected at 0.4 ng/ml and reached a maximum at 0.8 ng/ml of TGF-β (Fig. 3A). Thus, the activation of Smad1/5 by TGF-β requires concentrations of ligand higher than those required for phosphorylation of Smad3.
FIG. 3.
Differential regulation of TGF-β-induced phosphorylated Smad1/5 (P-Smad1/5) and P-Smad2/3. α, anti. (A) High doses of ligand are required for TGF-β-induced Smad1/5 phosphorylation. EpH4 cells were stimulated with increasing concentrations of TGF-β1 for 1 h as indicated. Whole-cell extracts were analyzed by Western blotting using antibodies against P-Smad1/5/8 (antibody 9511), P-Smad1/3/5 (antibody 9514), and Grb2 as a loading control. Note that the Cell Signaling Technology antibody 9514 recognizes phosphorylated Smad3 in addition to phosphorylated Smad1/5. See Fig. S2C in the supplemental material for data on which the assignment of the different bands to the different Smads is based. (B) TGF-β-induced Smad1/5 phosphorylation is transient and disappears between 2 and 4 h after stimulation, whereas Smad3 phosphorylation is readily detectable for at least 8 h. EpH4 cells were stimulated with TGF-β1 (2 ng/ml) for different time periods at 37°C before lysis. Cell lysates were fractionated by SDS-polyacrylamide gel electrophoresis and analyzed by Western blotting with antibodies against P-Smad1/3/5, P-Smad1/5/8, and Grb2.
Next, we examined the kinetics of TGF-β-induced phosphorylation of Smad1/5 in comparison to the kinetics of TGF-β-induced phosphorylation of Smad3. TGF-β (2 ng/ml) rapidly induced phosphorylation of both Smad1/5 and Smad3, which plateaued after 30 min (Fig. 3B). Phospho-Smad3 levels declined after 4 h but did not decrease to zero even after 8 h (Fig. 3B), consistent with the duration of signaling previously observed for the human keratinocyte cell line HaCaT (20). In contrast, TGF-β-induced Smad1/5 phosphorylation was dramatically reduced by 2 h and absent 4 h poststimulation (Fig. 3B). Thus, the TGF-β-induced phospho-Smad1/5 is more transient than the TGF-β-induced phospho-Smad3.
ALK2 and ALK3 are required for TGF-β-induced Smad1/5 phosphorylation.
The observation that the dose dependency and the kinetics of Smad1/5 phosphorylation were distinct from Smad3 phosphorylation, coupled with the fact that ALK5 alone is unable to phosphorylate Smad1/5 (5) indicated that a receptor complex distinct from the canonical TβRII-ALK5 complex must be responsible for the TGF-β-induced Smad1/5 phosphorylation. In endothelial cells, ALK1 forms a complex with ALK5 and TβRII to mediate TGF-β-induced Smad1/5 phosphorylation (15), but we reasoned that ALK1 could not be responsible in epithelial cells or fibroblasts, as it is specific to endothelial cells (41). Investigation of the expression profiles of all seven TGF-β superfamily type I receptors (ALK1 to ALK7) in a panel of cell lines by RT-PCR revealed that the cell lines that induce Smad1/5 phosphorylation in response to TGF-β express ALK2, ALK3, ALK4, and ALK5 mRNA (see Fig. S4A in the supplemental material). In addition, EpRas cells express low levels of ALK1 mRNA, but this expression was completely absent in the other responsive cells lines. We deduced that, of these receptors, the most likely candidates responsible for TGF-β-induced Smad1/5 phosphorylation would be ALK2 and ALK3. These two receptors have been shown to bind TGF-β in conjunction with TβRII (10, 42), are capable of phosphorylating Smad1/5 (12), and are expressed in all the responsive cell lines tested. We therefore investigated the involvement of these receptors in TGF-β-induced Smad1/5 phosphorylation in several different mouse and human cell lines.
Knockdown of either ALK2 or ALK3 in MDA-MB-231 cells had little effect, but knockdown of ALK2 and ALK3 together substantially reduced TGF-β induction of Smad1/5 phosphorylation (Fig. 4A), suggesting that they act redundantly. For a control for the efficiency of the knockdown, we investigated the effect of the ALK2 and ALK3 siRNAs on BMP7 and BMP4 induction. BMP7 preferentially binds ALK2, whereas BMP2 and BMP4 have higher affinity for ALK3 (26, 43). Consistent with this, BMP7-induced Smad1/5 phosphorylation in ALK2-depleted cells was reduced, whereas it was only marginally affected by knockdown of ALK3 (Fig. 4A). Knockdown of ALK3, in contrast, resulted in a dramatic reduction of BMP4-dependent Smad1/5 phosphorylation, whereas ALK2 knockdown had relatively little effect (Fig. 4A). In both cases, knocking down both ALK2 and ALK3 abolished BMP-induced Smad1/5 phosphorylation (Fig. 4A). Importantly, we could also show that, in human cells, the siRNA SMARTpools against ALK1, ALK2, ALK3, and ALK5 were efficient at knocking down the cognate receptors and had no effect on expression of other ALKs (see Fig. S3B and C in the supplemental material). For Colo-357 cells, siRNA knockdown experiments also indicated that both ALK2 and ALK3 were involved in TGF-β-induced Smad1/5 phosphorylation, but in this case, knockdown of either was sufficient to substantially reduce TGF-β-induced Smad1/5 phosphorylation (Fig. 4B). This is probably because the level of the other ALK is not sufficiently high to be able to compensate. In EpH4 cells, TGF-β-induced Smad1/5 phosphorylation appears to be predominantly mediated via ALK2, as the knockdown of ALK2 had a dramatic effect (see Fig. S4B in the supplemental material), whereas knockdown of ALK3 in these cells had only a minor effect (data not shown). Thus, we conclude that both ALK2 and ALK3 are involved in TGF-β-induced Smad1/5 phosphorylation, but the relative contribution of each receptor is dependent on cell type.
To consolidate the data acquired using siRNA oligonucleotides against ALK2 and ALK3, we used a small molecule inhibitor of BMP signaling, dorsomorphin, which has recently been shown to inhibit ALK2, ALK3, and ALK6 activity (46). Pretreatment of EpH4 cells with dorsomorphin for 1 h completely inhibited both TGF-β- and BMP-induced phosphorylation of Smad1/5 (Fig. 4C). In contrast, TGF-β-induced phosphorylation of Smad2 was unaffected by the inhibitor, indicating that it had no effect on ALK5 kinase activity. This confirms the requirement of ALK2 and ALK3 for TGF-β-induced Smad1/5 phosphorylation.
Since ALK2 and ALK3 are more commonly thought of as BMP receptors (12), it was imperative to prove that the TGF-β-induced Smad1/5 phosphorylation that we observed was not mediated via regulated BMP expression. We therefore treated three different cell lines (EpRas, NMuMG, and Colo-357) with TGF-β for different times in the presence or absence of the BMP inhibitor Noggin (Fig. 4D). In all cases, Noggin clearly inhibited endogenous BMP signaling, as it abolished the basal phospho-Smad1/5 levels. However, TGF-β was still able to strongly induce phosphorylation of Smad1/5. This could be visualized as early as after 10 min in NMuMG cells and was robust by 20 min in all three cell lines (Fig. 4D). Thus, we conclude that TGF-β induces phosphorylation of Smad1/5 directly and this is not mediated via BMP.
ALK2 and ALK3 can form a heteromeric complex with ALK5.
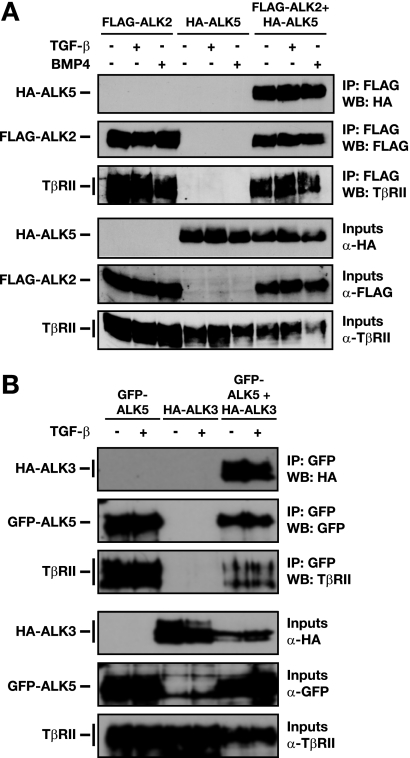
The dual requirement of two distinct classes of type I receptors suggests that both ALK5 and the lower-affinity TGF-β receptors ALK2 and ALK3 may function in concert to activate Smad1/5. Previous evidence suggests that TGF-β signals through a heterotetrameric receptor complex, which comprises homodimers of TβRII and ALK5. In addition, ALK1, ALK5, and TβRII are proposed to form a heterotetrameric complex which is responsible for Smad1/5 activation by TGF-β in endothelial cells (15). To investigate whether ALK5 forms an analogous complex with either ALK2 or ALK3 and TβRII, we overexpressed epitope-tagged versions of the ALKs and TβRII in NIH 3T3 cells and performed IPs. IP of FLAG-ALK2 could coprecipitate ALK5 and TβRII (Fig. 5A). A similar result was observed for tagged ALK3, ALK5, and TβRII (Fig. 5B). Surprisingly, the complexes can form in the absence of ligand, as stimulation with either TGF-β or BMP had no effect. This is most likely due to the overexpression of the receptors. Taken together with the loss-of-function data and the fact that TGF-β acts in a direct manner, these data suggest that TGF-β-induced Smad1/5 phosphorylation could be mediated by heteromeric complexes comprising TβRII, ALK5, and either ALK2 or ALK3.
FIG. 5.
ALK2 and ALK3 form a heteromeric complex with ALK5. α, anti. (A and B) Interaction of ALK2 or ALK3 with ALK5 was assayed by IP with overexpressed tagged ALKs and Western blot (WB) analysis. NIH 3T3 cells were transfected with either FLAG-tagged ALK2 (A) or HA-ALK3 (B) and/or HA-ALK5 (A) or GFP-ALK5 (B) together with TβRII. After 48 h, cells were treated with TGF-β1 or BMP4 for 1 h, as indicated. Whole-cell extracts were analyzed by Western blotting using antibodies against HA, FLAG, GFP, or TβRII either directly (input) or after IP with anti-FLAG beads (A) or anti-GFP antibody and protein G beads (B).
TGF-β-induced Smad1/5 phosphorylation does not regulate transcription of a BMP-responsive reporter.
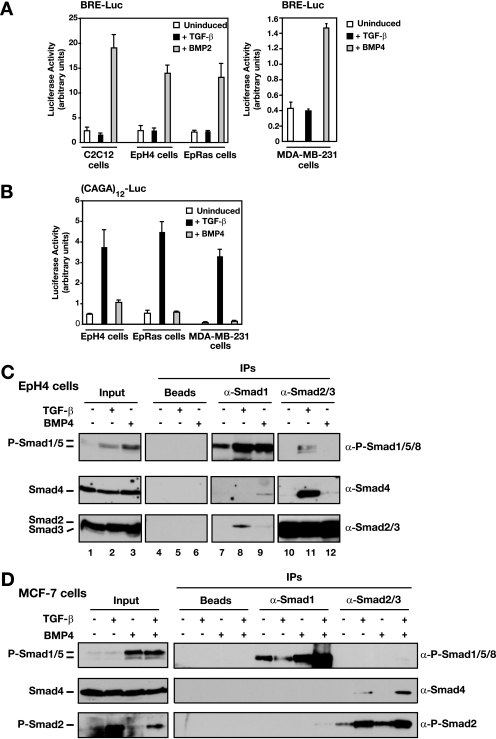
Having demonstrated that TGF-β induces Smad1/5 phosphorylation in epithelial cells and fibroblasts, we next investigated the downstream transcriptional response mediated by the phosphorylated Smads. The BRE-Luc reporter construct consists of two repeats of a BMP response element (BRE) from the Id1 promoter driving luciferase and is used as a transcriptional readout of BMP-induced signaling (21). The consensus element in this reporter has been shown to bind an activated Smad1-Smad4 complex with the transcriptional regulator Schnurri (35, 45). As a positive control, EpH4, EpRas, and MDA-MB-231 cells and the myoblast cell line C2C12 were treated with BMP2 or BMP4, which strongly induced transcription of the BRE-Luc reporter in all four cell lines (Fig. 6A). TGF-β is as potent as BMP in stimulating phosphorylation of Smad1/5 in EpH4 cells (Fig. 1C and 4C) and therefore might be expected to elicit an equivalent transcriptional response of the BRE-Luc reporter. Strikingly, however, TGF-β failed to induce transcription of the BRE-Luc reporter (Fig. 6A). In contrast, TGF-β readily induced transcription of the Smad3-dependent reporter (CAGA)12-Luc (9) in all the cell lines tested, whereas BMP stimulation did not (Fig. 6B). Thus, TGF-β-induced phosphorylation of Smad1/5 cannot initiate transcription via BREs in epithelial cells.
FIG. 6.
TGF-β-induced phospho-Smad1/5 (P-Smad1/5) fails to activate transcription from a BRE because it forms mixed R-Smad complexes with Smad2/3. α, anti. (A and B) Luciferase reporter assays with EpH4, EpRas, MDA-MB-231, and C2C12 cells. Cells were transfected with either BRE-Luc (A) or (CAGA)12-Luc (B) reporter and induced with TGF-β1 or BMP4 or BMP2 for 8 h as indicated. Luciferase activity was assayed and normalized. The data are the means and standard deviations for three independent experiments. (C) Smad1 forms complexes with Smad2 and Smad3 after stimulation with TGF-β and with Smad4 after stimulation with BMP4. Interaction of Smad1/5 with Smad2/3 was assayed by IP with anti-Smad antibodies and Western blot analysis. EpH4 cells were either left untreated or stimulated with TGF-β1 (2 ng/ml) or BMP4 (20 ng/ml) for 45 min before lysis. Whole-cell extracts were prepared, and equal amounts of protein were immunoprecipitated with antibodies against Smad1 or Smad2/3 or with beads alone. The IP reactions were analyzed by Western blotting with antibodies against Smad2/3, Smad4, and P-Smad1/5/8. As controls, 20% inputs are also shown. (D) Mixed R-Smad complexes are not formed by dual phosphorylation of Smad1/5 and Smad2/3 after costimulation with TGF-β and BMP4. Interactions of Smad1/5 with Smad2, Smad3, and Smad4 were assayed by IP with anti-Smad antibodies and Western blot analysis. MCF-7 cells were either untreated or stimulated with TGF-β1 (2 ng/ml) and/or BMP4 (20 ng/ml) for 45 min before lysis. Whole-cell extracts were prepared, and equal amounts of protein were immunoprecipitated with antibodies against Smad1 and Smad2/3. The IP reactions were analyzed by Western blotting with antibodies against Smad4, P-Smad2, and P-Smad1/5/8. As controls, inputs are also shown on the left of the panel.
Smad1/5 and Smad2/3 form mixed R-Smad complexes in response to TGF-β.
The induction of transcription via the BRE requires binding of phosphorylated Smad1/5 complexed with Smad4. Given the failure of TGF-β-induced Smad1/5 to activate transcription via the BRE and the fact that, in these cells, TGF-β simultaneously induces both phosphorylated Smad1/5 and Smad2/3, we investigated whether mixed R-Smad complexes were formed in response to TGF-β in preference to Smad1/5-Smad4 complexes. Endogenous R-Smads, Smad1 and Smad2/3, were therefore immunoprecipitated from EpH4 whole-cell extracts, and the immunoprecipitates were Western blotted for phospho-Smad1/5, Smad4, or Smad2/3. Smad2/3 clearly coprecipitated with phospho-Smad1/5 upon TGF-β treatment but not in response to BMP4 (Fig. 6C, upper panel, compare lanes 11 and 12), and conversely, Smad1 coprecipitated with Smad2 in response to TGF-β but not BMP4 (Fig. 6C, lower panel, compare lanes 8 and 9). As previously reported, Smad2/3 coprecipitated with Smad4 in response to TGF-β but not BMP4 (Fig. 6C, middle panel, compare lanes 11 and 12). Surprisingly, Smad1 coimmunoprecipitated with Smad4 only after stimulation with BMP4 but not TGF-β (Fig. 6C, middle panel, compare lanes 8 and 9). These data indicate that stimulation of epithelial cells with TGF-β results in the formation of novel endogenous mixed R-Smad complexes. Similar mixed R-Smad complexes were also readily detected in TGF-β-induced NMuMG cells, in which TGF-β also failed to induce BRE-Luc activity (see Fig. S5 in the supplemental material).
It was important to confirm that these activated mixed R-Smad complexes were formed as a result of the different classes of R-Smads being simultaneously phosphorylated by what we presume to be a heteromeric receptor complex and were not simply formed as a result of extract preparation. We therefore tested whether they would form when phosphorylated Smad1/5 and phosphorylated Smad2/3 were induced independently by costimulation of cells with TGF-β and BMP4. To do this, we used MCF-7 cells, which do not induce phosphorylation of Smad1/5 in response to TGF-β but do in response to BMP4 (Fig. 1A and 6D). In this case, we could not detect formation of complexes of phosphorylated Smad1/5 with Smad2 or Smad3 when cells were coactivated with TGF-β and BMP4 (Fig. 6D). We thus conclude that coactivation of Smad2/3 and Smad1/5 by a putative heteromeric receptor complex in response to TGF-β is required for mixed R-Smad complex formation.
Smad1/5 activation is not involved in TGF-β-induced growth arrest but is essential for TGF-β-induced anchorage-independent growth.
Having demonstrated that the ability of TGF-β to induce phosphorylation of Smad1/5 in addition to Smad2/3 results in a novel class of activated Smad complexes, we investigated the functional consequences of this branch of TGF-β signaling. We used the mouse mammary epithelial system with which we observed the strongest Smad1/5 response. The parental cells, EpH4 cells, are nontransformed mouse mammary gland epithelial cells, which are nontumorigenic and undergo growth inhibition and apoptosis in response to TGF-β (31, 32). However, EpRas cells undergo an EMT in response to TGF-β and form rapidly growing tumors in mice (31, 32).
First, we examined the function of Smad1/5 phosphorylation in TGF-β-induced growth arrest as determined by the ability of cells to progress through G1/S in the absence or presence of TGF-β (32, 34). EpH4 cells were transiently transfected with siRNA oligonucleotides against Smad1/5 or ALK5, as a positive control, and the cell cycle distribution was assayed. In control samples, the addition of TGF-β prevented EpH4 cells from entering S phase, and thus, a much higher percentage of cells remained in G1 (Fig. 7A). This effect of TGF-β was eliminated in ALK5-depleted cells (Fig. 7A). In contrast, knockdown of Smad1/5 had no obvious effect, despite efficient reduction of phospho-Smad1/5 levels (Fig. 7A, right panel). Thus, TGF-β-induced Smad1/5 phosphorylation is not involved in TGF-β-induced growth inhibition.
We next investigated the contribution of Smad1/5 signaling in TGF-β responses in EpRas cells. First, we tested the effect of Smad1/5 knockdown on the ability of these cells to undergo TGF-β-induced EMT (32). However, no apparent differences were observed in either cell morphology or mesenchymal marker expression between Smad1/5-depleted cells and the wild-type cells (data not shown). EpRas cells have also been shown to form rapidly growing tumors in mice, and this is dependent on continuous TGF-β signaling (32). We therefore tested the ability of EpRas cells to grow in soft agar in response to TGF-β and investigated the effect of knockdown of Smad1/5 by using SMARTpools against Smad1 and Smad5. Indeed, EpRas cells formed colonies in soft agar, and the colonies were markedly increased upon the addition of TGF-β (control samples; Fig. 7B). Strikingly, knockdown of Smad1/5 dramatically reduced this effect to a level comparable with that seen with the reduction of ALK5 (Fig. 7B). However, it had little effect on the number of colonies in the absence of ligand, indicating that the siRNAs were not generally detrimental to cell viability. Knockdown of ALK5 in the particular experiment shown in Fig. 7B was not complete. It was sufficient to abolish Smad1/5 phosphorylation but was not effective in preventing phosphorylation of Smad2 (Fig. 7B, right panel). The fact that this was still sufficient to strongly reduce the number of TGF-β-induced colonies supports a specific role of Smad1/5 phosphorylation in TGF-β-induced anchorage-independent growth. EpRas cells express levels of Smad1 that are considerably higher than those of Smad5 (data not shown). We therefore also confirmed that knockdown of Smad1 by two individual siRNA oligonucleotides inhibited TGF-β-induced anchorage-independent growth (see Fig. S6 in the supplemental material). In addition, we tried to mimic the effect of Smad1/5 knockdown on TGF-β-induced anchorage-independent growth by knocking down ALK2, ALK3, and also ALK1, which is expressed at low levels in these cells. However, we could never achieve a sufficiently high efficiency of knockdown of these receptor kinases to effectively reduce levels of phospho-Smad1/5 in these cells.
Since depletion of Smad1/5 would also be expected to affect autocrine BMP signaling, we wanted to eliminate any contribution of BMP signaling in the TGF-β-induced anchorage-independent growth assay. We therefore treated cells with or without the BMP inhibitor Noggin. Identical profiles were seen for cells treated with and without Noggin, thus eliminating a role for BMP-induced regulation of Smad1/5 in this assay (data not shown).
Taken together, these data indicate that TGF-β-induced Smad1/5 phosphorylation is not involved in TGF-β-induced growth inhibition but is required for the transformation of EpRas cells in response to TGF-β in vitro.
DISCUSSION
To date, the induction of Smad1/5 phosphorylation by TGF-β has been thought to be predominantly specific to endothelial cells, primarily due to the expression of ALK1 in these cells (15, 16). Here, we have shown that Smad1/5 phosphorylation is not restricted to endothelial cells. We have demonstrated that TGF-β induces phosphorylation of both Smad1 and Smad5 in addition to phosphorylation of Smad2/3 in a number of normal epithelial cell lines, fibroblasts, and cell lines of tumor origin. In agreement with our data, a very recent study has also reported TGF-β-induced Smad1/5 phosphorylation in HaCaT cells (2).
TβRII, ALK5, and either ALK2 or ALK3 are required for TGF-β-induced Smad1/5 phosphorylation.
Our data indicate that the rapid and direct TGF-β induction of Smad1/5 phosphorylation is critically dependent on ALK5 and TβRII. It requires not only the expression of ALK5 but also its kinase activity. Previous work has shown that the substrate specificity of type I receptors, including ALK5, is tightly restricted (5, 6, 11, 33). Importantly, the L45 loop sequence of ALK5 is compatible with the L3 loop configuration of Smad2 and Smad3 but not of Smad1 or Smad5 (5). Thus, ALK5 alone cannot activate Smad1/5, and a second type I receptor activity must be required for the phosphorylation of Smad1/5. ALK1 mRNA is not generally expressed in epithelial cell lines at detectable levels, and knockdown of this receptor had no effect on Smad1/5 phosphorylation, excluding a functional role for ALK1 in these cells.
Instead, the evidence provided here supports a role for ALK2 and/or ALK3 in the TGF-β-induced Smad1/5 phosphorylation in epithelial cells and tumor cells. Both of these receptors can bind TGF-β in concert with TβRII (10, 42). Moreover, ALK2 has previously been proposed to be a TGF-β receptor (29), and sequence analysis indicates that it is in the same subgroup of type I receptors as ALK1 (6), making it an excellent candidate for mediating TGF-β-induced Smad1/5 phosphorylation. We found that knockdown of ALK2 in EpH4 cells was indeed sufficient to virtually abolish TGF-β-induced Smad1/5 phosphorylation (see Fig. S4 in the supplemental material). However, for Colo-357 and MDA-MB-231 cells, it was necessary to additionally knock down ALK3 to eliminate Smad1/5 phosphorylation in response to TGF-β. This suggests that in these two cell lines, ALK2 and ALK3 act redundantly. Furthermore, we have confirmed the involvement of ALK2 and ALK3 by using the recently characterized ALK2/ALK3/ALK6 inhibitor dorsomorphin, which completely abolishes Smad1/5 phosphorylation in response to TGF-β but has no effect on TGF-β-induced Smad2/3 phosphorylation.
Given the requirement of TβRII and ALK5 with ALK2 and/or ALK3, the demonstration that TGF-β induces Smad1/5 rapidly and directly, and given the fact that TGF-β superfamily receptor complexes are thought to comprise two type II receptors and two type I receptors, we propose that TGF-β induction of Smad1/5 phosphorylation may be mediated by a heterotetrameric receptor complex comprising TβRII and ALK5 with either ALK2 or ALK3 (Fig. 7C). This would be analogous to the heteromeric ALK1/ALK5/TβRII receptor complex proposed for endothelial cells (15). However, the final proof of the existence of such heteromeric complexes awaits the development of antibodies that detect endogenous ALK2 and ALK3. Although our data allow us to define what receptors are required for TGF-β-induced Smad1/5 activation, it is not clear whether they are sufficient. Attempts to reconstitute a TGF-β induction of Smad1/5 phosphorylation in the nonresponsive MCF-7 cells by overexpressing ALK5 with ALK2 or ALK3 were not successful, leaving open the possibility that other components are additionally required. It is not clear how ALK2 and ALK3 are activated in response to TGF-β. This may be achieved by phosphorylation mediated by TβRII, or possibly by ALK5, which would explain the requirement for the kinase activity of ALK5. However, in preliminary experiments, we have been unable to detect any increase in ALK2 activity in IP kinase assays as a result of coexpression of activated ALK5. Further work is required to clarify this issue.
Our demonstration that ALK2 and ALK3 are directly involved in TGF-β signaling in addition to their previously characterized roles in BMP6/7 and BMP2/4 signaling, respectively (26, 43), suggests that these receptors can bind different ligands in different contexts. This concept has already been shown for ALK1, which has been shown to act downstream of both TGF-β and BMP9/10 (7, 15, 16, 38). We propose that, in the case of ALK2 and ALK3, they bind BMPs when complexed with BMP type II receptors such as BMPRII, ActRII, and ActRIIB (12) but bind TGF-β in a heteromeric complex with TβRII and ALK5.
Recent structural data have demonstrated that a molecule of ALK2 or ALK3 would not be able to directly replace a molecule of ALK5 in the high-affinity, cooperative TβRII/ALK5/TGF-β complex (17). This indicates that a mixed complex of type I receptors would have to have a distinct architecture from the canonical TGF-β receptor complex. Indeed, such complexes may be more similar to BMP receptor complexes. In experiments designed to prove that all three receptors were present in the same complex by serial IPs, we found that the receptor complexes fell apart as they were purified (our unpublished data). This suggests that it is the avidity from membrane localization and/or the presence of ligand that promotes assembly of the receptor complexes, as is the case for the BMP receptors, which do not actually interact directly with each other (17). Given these structural considerations, a mixed type I receptor complex is likely to have a lower affinity for TGF-β than the canonical ALK5-TβR complex. Therefore, high concentrations of TGF-β and/or high cell surface expression levels of ALK5 would be expected to be important for the formation of TGF-β-induced ALK5-ALK2/3 complexes and, hence, TGF-β-induced Smad1/5 phosphorylation. Indeed, with EpH4 cells, we have demonstrated that the dose of TGF-β required for Smad1/5 phosphorylation is higher than that required for Smad3 phosphorylation. We have also shown that high ALK5 levels are required for Smad1/5 phosphorylation. Partial knockdown of ALK5 in EpRas cells resulted in complete abrogation of Smad1/5 phosphorylation, whereas activation of Smad2/3 was maintained (Fig. 7B). It is tempting to speculate that our observation that low levels of TGF-β signaling can induce only the canonical arm of TGF-β/Smad signaling, while higher doses of TGF-β additionally induce Smad1/5 phosphorylation, might explain dose-dependent responses to TGF-β (27).
Formation of mixed R-Smad complexes.
TGF-β stimulation of epithelial cells has previously been shown to induce Smad2-Smad4 and Smad3-Smad4 complexes. Our data now indicate that activation of ALK5-ALK2/3 receptor complexes results in the simultaneous phosphorylation of Smad1/5 and Smad2/3 and the subsequent formation of novel mixed R-Smad complexes. This is supported by strong evidence for endogenous complexes containing activated Smad2/3 and Smad1 in response to TGF-β but not BMP. In addition, when we performed IPs with antibodies that are specific for Smad2 or Smad3, we could detect complexes containing activated Smad1/Smad2 and Smad1/Smad3 in response to TGF-β in EpH4 cells (our unpublished data). The exact stoichiometry and precise combinations of the mixed R-Smad complexes have not yet been determined. We conclude that mixed R-Smad complexes form via simultaneous activation at the cell surface, as we could not detect them in MCF-7 cells costimulated with TGF-β (which induces only phosphorylated Smad2/3 in these cells) and BMP4 (which induces phosphorylation of Smad1/5). Previously characterized Smad1-Smad4, Smad2-Smad4, and Smad3-Smad4 complexes have been shown to bind with different affinities to distinct promoter elements and thus elicit different transcriptional responses (37). We suggest that these mixed activated Smad1/5-Smad2/3 complexes are also distinct entities, binding unique promoter elements, and are responsible for transducing this novel branch of TGF-β signaling to the nucleus. The absence of BRE-Luc activity upon TGF-β stimulation in cell lines in which TGF-β induces strong phosphorylation of Smad1/5 provides evidence for this hypothesis. The levels of Smad1/5 phosphorylation upon TGF-β and BMP stimulation are equivalent, but the transcriptional readouts in response to ligand are dramatically different. We do not think that the inability of TGF-β-induced phospho-Smad1/5 to initiate transcription of BRE-Luc is due to the transient phospho-Smad1/5 signal, as 1 to 2 h of TGF-β signaling is sufficient for a measurable induction of a variety of reporters (20). It is more likely due to the fact that the majority of activated Smad1 or Smad5 is complexed with Smad2/3 and therefore cannot bind effectively to the BRE in a complex with Smad4 (3). Furthermore, the idea that the mixed R-Smad complexes might target a unique set of genes is supported by the observation that, in endothelial cells, TGF-β regulates a greater number of genes than constitutively active ALK1 or ALK5 (44).
Function of TGF-β-induced Smad1/5 phosphorylation.
The study of the cellular function of TGF-β-induced phospho-Smad1/5 in endothelial cells concluded that ALK1 activity antagonizes Smad2/3 signaling via ALK5, as demonstrated by decreased (CAGA)12-Luc activity upon expression of ALK1 in endothelial cells (15). This antagonism was thought to be exerted at the Smad level (15). Furthermore, ALK1 activity was proposed to promote proliferation and migration of endothelial cells, whereas ALK5 has been shown to inhibit both processes (15, 16). Thus, for endothelial cells, TGF-β was thought to activate two independent Smad signaling pathways which have opposing effects.
We have shown, in contrast to these findings, that Smad2/3 signaling is not inhibited by Smad1/5 activation in epithelial cells. We found that knockdown of Smad1 and Smad5 did not significantly affect the transcriptional activation of the (CAGA)12-Luc reporter (our unpublished data). More importantly, knockdown of Smad1/5 had no effect on the antiproliferative effects of TGF-β in EpH4 cells or TGF-β-induced EMT in EpRas cells, suggesting that Smad1/5 activation did not cause a general inhibition of Smad2/3 signaling. In addition, we have shown that Smad1/5 is required for the transforming effects of TGF-β in EpRas cells in soft agar, indicating that Smad1/5 signaling has specific biological functions downstream of TGF-β. These functional data are consistent with the proposed model, whereby ALK5-induced Smad2/3 signaling and ALK5-ALK2/3-induced dual Smad1/5-Smad2/3 signaling each carry out a subset of functions (Fig. 7C). We are currently investigating this in more detail, determining which TGF-β target genes require phosphorylated Smad1/5 for their regulation.
Finally, our analysis shows that signaling by TGF-β is much more complex than previously appreciated. The precise TGF-β response is dependent on the nature of the activated receptor complexes, which have distinct dose requirements and exhibit independent regulation. However, many more questions remain to be answered. Does cooperation between the ALK5 and ALK2/3-ALK5 signaling pathways exist? What other components, if any, are required for ALK2/3-ALK5 signaling? What are the genes regulated by the mixed R-Smad complexes? In addition, our in vitro analysis suggests that the ability of TGF-β to induce Smad1/5 phosphorylation is important for transformation. It will be important to address whether this is relevant for tumor progression in vivo.
Supplementary Material
[Supplemental material]
Acknowledgments
We thank Senyon Choe for recombinant Noggin, Holger Gerhardt for endothelioma cells, Anita Roberts for the HA-tagged ALK5 and TβRII plasmids, and Peter ten Dijke for ALK1, ALK2, and ALK3 expression plasmids and the BRE-Luc construct. We thank Paul Yu for generously providing dorsomorphin and data prior to publication. We are grateful to the Cancer Research UK FACS laboratory for help with FACS analyses and to the Cancer Research UK Cell Services for providing numerous cell lines. We thank Karel Dorey, Mike Howell, Laurence Levy, Erik Sahai, and Bernhard Schmierer for helpful discussions and advice and/or useful comments on the manuscript.
This work was supported by Cancer Research UK and by a European Union Research Training Network grant (MRTN-CT-2004-005428).
Footnotes
▿
Published ahead of print on 15 September 2008.
REFERENCES
- 1.Bardwell, V. J., and R. Treisman. 1994. The POZ domain: a conserved protein-protein interaction motif. Genes Dev. 81664-1677. [DOI] [PubMed] [Google Scholar]
- 2.Bharathy, S., W. Xie, J. M. Yingling, and M. Reiss. 2008. Cancer-associated transforming growth factor β type II receptor gene mutant causes activation of bone morphogenic protein-Smads and invasive phenotype. Cancer Res. 681656-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byfield, S. D., and A. B. Roberts. 2004. Lateral signaling enhances TGF-β response complexity. Trends Cell Biol. 14107-111. [DOI] [PubMed] [Google Scholar]
- 4.Chen, X., M. J. Rubock, and M. Whitman. 1996. A transcriptional partner for MAD proteins in TGF-β signalling. Nature 383691-696. [DOI] [PubMed] [Google Scholar]
- 5.Chen, Y. G., A. Hata, R. S. Lo, D. Wotton, Y. Shi, N. Pavletich, and J. Massagué. 1998. Determinants of specificity in TGF-β signal transduction. Genes Dev. 122144-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, Y. G., and J. Massague. 1999. Smad1 recognition and activation by the ALK1 group of transforming growth factor-β family receptors. J. Biol. Chem. 2743672-3677. [DOI] [PubMed] [Google Scholar]
- 7.David, L., C. Mallet, S. Mazerbourg, J. J. Feige, and S. Bailly. 2007. Identification of BMP9 and BMP10 as functional activators of the orphan activin receptor-like kinase 1 (ALK1) in endothelial cells. Blood 1091953-1961. [DOI] [PubMed] [Google Scholar]
- 8.Debnath, J., S. K. Muthuswamy, and J. S. Brugge. 2003. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30256-268. [DOI] [PubMed] [Google Scholar]
- 9.Dennler, S., S. Itoh, D. Vivien, P. ten Dijke, S. Huet, and J. M. Gauthier. 1998. Direct binding of Smad3 and Smad4 to critical TGF β-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 173091-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ebner, R., R. H. Chen, S. Lawler, T. Zioncheck, and R. Derynck. 1993. Determination of type I receptor specificity by the type II receptors for TGF-β or activin. Science 262900-902. [DOI] [PubMed] [Google Scholar]
- 11.Feng, X. H., and R. Derynck. 1997. A kinase subdomain of transforming growth factor-β (TGF-β) type I receptor determines the TGF-β intracellular signaling specificity. EMBO J. 163912-3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng, X. H., and R. Derynck. 2005. Specificity and versatility in TGF-β signaling through Smads. Annu. Rev. Cell Dev. Biol. 21659-693. [DOI] [PubMed] [Google Scholar]
- 13.Germain, S., M. Howell, G. M. Esslemont, and C. S. Hill. 2000. Homeodomain and winged-helix transcription factors recruit activated Smads to distinct promoter elements via a common Smad interaction motif. Genes Dev. 14435-451. [PMC free article] [PubMed] [Google Scholar]
- 14.Goumans, M. J., F. Lebrin, and G. Valdimarsdottir. 2003. Controlling the angiogenic switch: a balance between two distinct TGF-β receptor signaling pathways. Trends Cardiovasc. Med. 13301-307. [DOI] [PubMed] [Google Scholar]
- 15.Goumans, M. J., G. Valdimarsdottir, S. Itoh, F. Lebrin, J. Larsson, C. Mummery, S. Karlsson, and P. ten Dijke. 2003. Activin receptor-like kinase (ALK)1 is an antagonistic mediator of lateral TGFβ/ALK5 signaling. Mol. Cell 12817-828. [DOI] [PubMed] [Google Scholar]
- 16.Goumans, M. J., G. Valdimarsdottir, S. Itoh, A. Rosendahl, P. Sideras, and P. ten Dijke. 2002. Balancing the activation state of the endothelium via two distinct TGF-β type I receptors. EMBO J. 211743-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groppe, J., C. S. Hinck, P. Samavarchi-Tehrani, C. Zubieta, J. P. Schuermann, A. B. Taylor, P. M. Schwarz, J. L. Wrana, and A. P. Hinck. 2008. Cooperative assembly of TGF-β superfamily signaling complexes is mediated by two disparate mechanisms and distinct modes of receptor binding. Mol. Cell 29157-168. [DOI] [PubMed] [Google Scholar]
- 18.Hill, C. S., J. Wynne, and R. Treisman. 1995. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell 811159-1170. [DOI] [PubMed] [Google Scholar]
- 19.Inman, G. J., F. J. Nicolas, J. F. Callahan, J. D. Harling, L. M. Gaster, A. D. Reith, N. J. Laping, and C. S. Hill. 2002. SB-431542 is a potent and specific inhibitor of transforming growth factor-β superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol. Pharmacol. 6265-74. [DOI] [PubMed] [Google Scholar]
- 20.Inman, G. J., F. J. Nicolas, and C. S. Hill. 2002. Nucleocytoplasmic shuttling of Smads 2, 3, and 4 permits sensing of TGF-β receptor activity. Mol. Cell 10283-294. [DOI] [PubMed] [Google Scholar]
- 21.Korchynskyi, O., and P. ten Dijke. 2002. Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J. Biol. Chem. 2774883-4891. [DOI] [PubMed] [Google Scholar]
- 22.Lebrin, F., M. J. Goumans, L. Jonker, R. L. Carvalho, G. Valdimarsdottir, M. Thorikay, C. Mummery, H. M. Arthur, and P. ten Dijke. 2004. Endoglin promotes endothelial cell proliferation and TGF-β/ALK1 signal transduction. EMBO J. 234018-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levy, L., and C. S. Hill. 2005. Smad4 dependency defines two classes of transforming growth factor β (TGF-β) target genes and distinguishes TGF-β-induced epithelial-mesenchymal transition from its antiproliferative and migratory responses. Mol. Cell. Biol. 188108-8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levy, L., M. Howell, D. Das, S. Harkin, V. Episkopou, and C. S. Hill. 2007. Arkadia activates Smad3/Smad4-dependent transcription by triggering signal-induced SnoN degradation. Mol. Cell. Biol. 276068-6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, X., J. Yue, R. S. Frey, Q. Zhu, and K. M. Mulder. 1998. Transforming growth factor β signaling through Smad1 in human breast cancer cells. Cancer Res. 584752-4757. [PubMed] [Google Scholar]
- 26.Macias-Silva, M., P. A. Hoodless, S. J. Tang, M. Buchwald, and J. L. Wrana. 1998. Specific activation of Smad1 signaling pathways by the BMP7 type I receptor, ALK2. J. Biol. Chem. 27325628-25636. [DOI] [PubMed] [Google Scholar]
- 27.Massagué, J. 2000. How cells read TGF-β signals. Nat. Rev. Mol. Cell Biol. 1169-178. [DOI] [PubMed] [Google Scholar]
- 28.Massagué, J., S. W. Blain, and R. S. Lo. 2000. TGFβ signaling in growth control, cancer, and heritable disorders. Cell 103295-309. [DOI] [PubMed] [Google Scholar]
- 29.Miettinen, P. J., R. Ebner, A. R. Lopez, and R. Derynck. 1994. TGF-β induced transdifferentiation of mammary epithelial cells to mesenchymal cells: involvement of type I receptors. J. Cell Biol. 1272021-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicolás, F. J., K. De Bosscher, B. Schmierer, and C. S. Hill. 2004. Analysis of Smad nucleocytoplasmic shuttling in living cells. J. Cell Sci. 1174113-4125. [DOI] [PubMed] [Google Scholar]
- 31.Oft, M., K. H. Heider, and H. Beug. 1998. TGFβ signaling is necessary for carcinoma cell invasiveness and metastasis. Curr. Biol. 81243-1252. [DOI] [PubMed] [Google Scholar]
- 32.Oft, M., J. Peli, C. Rudaz, H. Schwarz, H. Beug, and E. Reichmann. 1996. TGF-β1 and Ha-Ras collaborate in modulating the phenotypic plasticity and invasiveness of epithelial tumor cells. Genes Dev. 102462-2477. [DOI] [PubMed] [Google Scholar]
- 33.Persson, U., H. Izumi, S. Souchelnytskyi, S. Itoh, S. Grimsby, U. Engstrom, C. H. Heldin, K. Funa, and P. ten Dijke. 1998. The L45 loop in type I receptors for TGF-β family members is a critical determinant in specifying Smad isoform activation. FEBS Lett. 43483-87. [DOI] [PubMed] [Google Scholar]
- 34.Petritsch, C., H. Beug, A. Balmain, and M. Oft. 2000. TGF-β inhibits p70 S6 kinase via protein phosphatase 2A to induce G(1) arrest. Genes Dev. 143093-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pyrowolakis, G., B. Hartmann, B. Muller, K. Basler, and M. Affolter. 2004. A simple molecular complex mediates widespread BMP-induced repression during Drosophila development. Dev. Cell 7229-240. [DOI] [PubMed] [Google Scholar]
- 36.Röhnelt, R. K., G. Hoch, Y. Reiss, and B. Engelhardt. 1997. Immunosurveillance modelled in vitro: naive and memory T cells spontaneously migrate across unstimulated microvascular endothelium. Int. Immunol. 9435-450. [DOI] [PubMed] [Google Scholar]
- 37.Ross, S. J., and C. S. Hill. 2008. How the Smads regulate transcription. Int. J. Biochem. Cell Biol. 40383-408. [DOI] [PubMed] [Google Scholar]
- 38.Scharpfenecker, M., M. van Dinther, Z. Liu, R. L. van Bezooijen, Q. Zhao, L. Pukac, C. W. Lowik, and P. ten Dijke. 2007. BMP-9 signals via ALK1 and inhibits bFGF-induced endothelial cell proliferation and VEGF-stimulated angiogenesis. J. Cell Sci. 120964-972. [DOI] [PubMed] [Google Scholar]
- 39.Schmierer, B., and C. S. Hill. 2005. Kinetic analysis of Smad nucleocytoplasmic shuttling reveals a mechanism for transforming growth factor β-dependent nuclear accumulation of Smads. Mol. Cell. Biol. 259845-9858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmierer, B., and C. S. Hill. 2007. TGFβ-SMAD signal transduction: molecular specificity and functional flexibility. Nat. Rev. Mol. Cell Biol. 8970-982. [DOI] [PubMed] [Google Scholar]
- 41.Seki, T., J. Yun, and S. P. Oh. 2003. Arterial endothelium-specific activin receptor-like kinase 1 expression suggests its role in arterialization and vascular remodeling. Circ. Res. 93682-689. [DOI] [PubMed] [Google Scholar]
- 42.ten Dijke, P., H. Yamashita, H. Ichijo, P. Franzen, M. Laiho, K. Miyazono, and C. H. Heldin. 1994. Characterization of type I receptors for transforming growth factor-β and activin. Science 264101-104. [DOI] [PubMed] [Google Scholar]
- 43.ten Dijke, P., H. Yamashita, T. K. Sampath, A. H. Reddi, M. Estevez, D. L. Riddle, H. Ichijo, C. H. Heldin, and K. Miyazono. 1994. Identification of type I receptors for osteogenic protein-1 and bone morphogenetic protein-4. J. Biol. Chem. 26916985-16988. [PubMed] [Google Scholar]
- 44.Wu, X., J. Ma, J. D. Han, N. Wang, and Y. G. Chen. 2006. Distinct regulation of gene expression in human endothelial cells by TGF-β and its receptors. Microvasc. Res. 7112-19. [DOI] [PubMed] [Google Scholar]
- 45.Yao, L. C., I. L. Blitz, D. A. Peiffer, S. Phin, Y. Wang, S. Ogata, K. W. Cho, K. Arora, and R. Warrior. 2006. Schnurri transcription factors from Drosophila and vertebrates can mediate Bmp signaling through a phylogenetically conserved mechanism. Development 1334025-4034. [DOI] [PubMed] [Google Scholar]
- 46.Yu, P. B., C. C. Hong, C. Sachidanandan, J. L. Babitt, D. Y. Deng, S. A. Hoyng, H. Y. Lin, K. D. Bloch, and R. T. Peterson. 2008. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat. Chem. Biol. 433-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]