Bacterial species exhibit diversity in their mechanisms and capacity for protein disulfide bond formation (original) (raw)
Abstract
Protein disulfide bond formation contributes to the folding and activity of many exported proteins in bacteria. However, information about disulfide bond formation is limited to only a few bacterial species. We used a multifaceted bioinformatic approach to assess the capacity for disulfide bond formation across this biologically diverse group of organisms. We combined data from a cysteine counting method, in which a significant bias for even numbers of cysteine in proteins is taken as an indicator of disulfide bond formation, with data on the presence of homologs of known disulfide bond formation enzymes. These combined data enabled us to make predictions about disulfide bond formation in the cell envelope across bacterial species. Our bioinformatic and experimental results suggest that many bacteria may not generally oxidatively fold proteins, and implicate the bacterial homolog of the enzyme vitamin K epoxide reductase, a protein required for blood clotting in humans, as part of a disulfide bond formation pathway present in several major bacterial phyla.
Keywords: cysteine, genomics, protein folding, vitamin K epoxide reductase
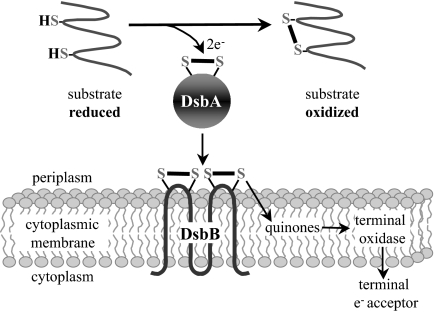
Disulfide bonds, formed by the oxidation of pairs of cysteines, assist the folding and stability of many exported proteins. In Escherichia coli, the periplasmic protein DsbA and the membrane-bound protein DsbB promote the introduction of disulfide bonds into proteins (Fig. 1) (1). DsbA, with the active site motif, Cys-X-X-Cys, embedded in a thioredoxin fold, introduces disulfide bonds into proteins that are translocated into the periplasm. The active site cysteines of DsbA must be reoxidized for the enzyme to regain activity, a step catalyzed by DsbB. DsbB then shuttles electrons received from DsbA to the electron transport chain via membrane-bound quinones.
Fig. 1.
Disulfide bond formation pathway of E. coli. (arrows indicate flow of electrons)
For most organisms, disulfide-bonded proteins are restricted to noncytoplasmic compartments. However, Mallick et al. (2) found that cytoplasmic proteins from some hyperthermophilic archaea contain disulfide bonds. Furthermore, they showed that the presence of disulfide-bonded proteins in the cytoplasm correlates with a bias for even numbers of cysteines in the archaeal proteome. One explanation for an enrichment of even numbers of cysteines in proteins with disulfide bonds is that odd numbers of cysteines in a protein could allow the formation of inappropriate disulfide bonds, resulting in a misfolded protein (3). In fact, organisms from bacteria to eukaryotes express disulfide bond isomerases that ensure the correct array of disulfide bonds in a protein after such “mistakes” are made (1, 4). To avoid the problem of mismatched cysteines, there may be evolutionary pressure to select for an even number of cysteines in proteins with disulfide bonds.
We reasoned that a bioinformatic analysis to determine whether proteins in the cell envelope of different bacteria have significant biases for even numbers of cysteines could indicate whether this compartment contains disulfide-bonded proteins. Given the considerable biological diversity within the domain Bacteria, and given that oxidative protein folding has been studied extensively only in a small fraction of bacterial species, a more extensive analysis of this group of organisms may reveal novel aspects of disulfide bond formation.
Here, we show that a significant bias for even numbers of cysteine does correlate with the location of disulfide bond formation in E. coli, the cell envelope. We then analyzed the cysteine content of predicted cell envelope proteins from each of 375 other bacterial genomes, to assess whether each of these organisms may have disulfide-bonded proteins. We also used homology searches in each genome to identify members of the DsbA and DsbB protein families. The merging of these data enabled us to generate predictions as to whether oxidative folding is likely to occur in the cell envelope of each of the bacteria examined, and, if so, whether the organism uses the Dsb pathway.
Our results lead us to propose that oxidative folding of cell envelope proteins may not be a well conserved feature of bacterial cell biology. To support this hypothesis, we present experimental data from Bacteroides fragilis NCTC9343. In addition, we found many bacteria that were predicted by our analysis to carry out disulfide bond formation, but lack a homolog of DsbB. This observation has led us to the identification of a candidate for a novel disulfide bond formation enzyme, the bacterial homolog of the eukaryotic enzyme vitamin K epoxide reductase (VKOR). We present experimental evidence for a DsbB-like activity of the Mycobacterium tuberculosis H37Rv homolog of VKOR.
Results
Cysteine Composition of Cell Envelope Proteins in E. coli.
We examined the E. coli proteome to determine whether differences in patterns of cysteine distribution correlate with the compartment in which disulfide bond formation takes place, the cell envelope. We divided the proteome into 5 classes, based on subcellular location that was predicted by bioinformatic approaches for analyzing the ORFs in the genome (see Methods). These protein classes are cytoplasmic (class 1), membrane spanning segments of transmembrane proteins (TM-membrane spanning) (class 2), cytoplasmic loops and domains of transmembrane proteins (TM-cytoplasmic) (class 3), periplasmic loops and domains of transmembrane proteins (TM-periplasmic) (class 4), and exported proteins, which we define as the mature parts of signal-sequence directed exported proteins, including most periplasmic, outer membrane-bound, and extracellular proteins (class 5).
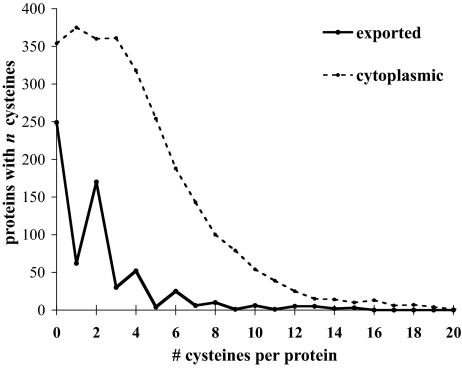
We analyzed all proteins from compartments 1–5 both for the bias for even numbers of cysteines and for overall cysteine content. First, we found that the percentage of cysteines in exported proteins (class 5) is considerably smaller than the percentage of cysteines in the entire proteome; 39% of exported proteins have cysteine as compared with 87% of cytoplasmic proteins. This strong bias against cysteine in exported proteins has been noted before in the analysis of a much smaller subset of proteins (5). Second, a substantial majority of cysteine-containing exported proteins (71%) have even numbers of cysteines, reflected in the saw tooth shape of the plot in Fig. 2.
Fig. 2.
Exported proteins show a unique bias for even numbers of cysteines. Cysteine distribution in E. coli K12 proteins - cytoplasmic and exported (classes 1 and 5).
To determine the significance of this latter finding, we tested these data against the predictions of a null hypothesis in which cysteines are distributed randomly among ORFs for exported proteins. This test takes into account the cysteine composition of the compartment, as well as the length of each protein within the compartment, and then distributes the cysteines at random within each protein (according to a Poisson distribution, see Methods). For each class of proteins, we compared the actual fraction of proteins with even numbers of cysteines to the fraction predicted by the random model (Table 1). The fraction of proteins predicted by the random model to have an even number of cysteines is different for each class because the amino acid composition of each class is different and the distribution of total number of residues per protein varies in different classes.
Table 1.
Cysteine distribution in E. coli proteins
| Predicted subcellular location | Percent of cys-containing proteins with even numbers of cysteine | Percent of cys-containing proteins with even numbers of cysteine predicted by random model (mean) | Standard deviations from random mean | Bias for even cysteines (observed values/random model values) |
|---|---|---|---|---|
| Cytoplasmic (class 1) | 45.7 | 46.4 ± 1.0 | −0.7 | 0.98 |
| TM–cytoplasmic (class 2) | 31 | 30.2 ± 2.1 | +0.37 | 1.03 |
| TM–membrane spanning (class 3) | 41.9 | 40.6 ± 1.8 | +0.66 | 1.01 |
| TM–periplasmic (class 4) | 42.9 | 26.5 ± 2.3 | +7.05 | 1.62 |
| Exported (class 5) | 71.4 | 40.2 ± 2.2 | +14.04 | 1.77 |
Whereas proteins or portions of proteins that are predicted to be localized to or pass through the periplasm have a highly significant bias for even numbers of cysteines (classes 4 and 5) (Table 1), membrane-spanning segments, portions of membrane proteins predicted to be exposed to the cytoplasmic compartment, as well as soluble cytoplasmic proteins do not (classes 1,2, and 3) (Table 1). Our analysis shows for E. coli that cysteine distributed according to the random model would result in 40.2% (mean value) of exported cysteine-containing proteins having an even number of cysteines, in contrast to the 71% actually observed, the latter number being 14 standard deviations above the mean of the random model value. The bias for (ratio of observed to expected) even numbers of cysteines in both the mature exported (class 5) and TM-periplasmic (class 4) classes is similar (Table 1). Their z scores are different because the standard deviation of the TM-periplasmic class is larger, a consequence of the smaller number of residues per protein in the TM-periplasmic class. For this reason we used the exported class of proteins, class 5, for all of our subsequent analyses.
We compared the data for other amino acids in exported proteins to see whether these features are unique to cysteine [supporting information (SI) Fig. S1]. As with cysteine, for each of the other amino acids we calculated the fraction of exported proteins with even numbers of that amino acid, as well as the predicted fraction of proteins containing an even number of cysteines according to the random model of amino acid distribution, as described above. The number of standard deviations of the actual data from the mean of the random model calculation gave us a z score. The z score is plotted against the AApref, a measure of the preference for or against incorporating the amino acid in an exported protein. AApref is the ratio of the frequency of the amino acid in the class to the frequency in the proteins encoded in the entire genome. Of all of the amino acids, cysteine shows the strongest bias against incorporation into exported proteins. Additionally, for all other amino acids, the fraction of exported proteins with an even number of the amino acid is approximately what would be expected from the random model.
Based on previous results of Mallick et al. (2), we suspect that these values for cysteine in exported proteins that differentiate it from other amino acids reflect the active formation of disulfide bonds in the E. coli periplasm. Therefore, a preference for even numbers of cysteines in exported proteins may be an indicator of disulfide bonds in the cell envelope proteins of a given organism.
Computational Analysis of Disulfide Bond Formation in Other Sequenced Bacterial Genomes.
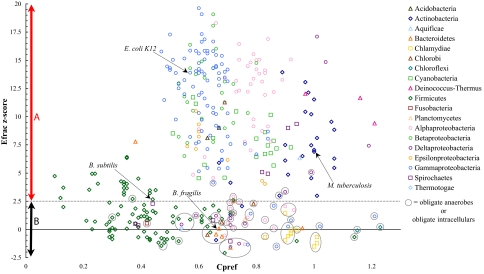
We extended this analysis of cysteine patterns in exported proteins to 375 fully sequenced bacterial genomes. As with E. coli, we predicted the subcellular localization of all proteins for each genome and then calculated the fraction of cysteine-containing exported proteins with even numbers of cysteines, as well as the fraction if cysteine were distributed according to our random model in that set of proteins. The number of standard deviations of the actual data from the mean obtained with the random model gave us a z score for each genome which is then plotted versus the bias against incorporating cysteine into exported proteins (Cpref) for that genome (Fig. 3).
Fig. 3.
Cysteine counting in exported proteins of all bacterial genomes. The z score for the fraction of exported proteins with even numbers of cysteine (Efrac), is plotted against the Cpref (frequency of cysteine in exported proteins/frequency of cysteine in all proteins) for each genome (a Cpref of <1.0 indicates a bias against incorporation of cysteine into exported proteins). Region A: Disulfide bond formation is predicted to occur in exported proteins in these organisms. These genomes (_z_ > 2.57) have significantly more exported proteins with even numbers of cysteine than is predicted by the random model. (i.e., there is a probability of 0.01% that the genome would have a z score of >2.57 if the cysteines were distributed at random.) Region B: These organisms are predicted to have no, or limited, disulfide bond formation in exported proteins. These genomes (2.57 > z > −2.57) do not have a significant fraction of exported proteins with even numbers of cysteine.
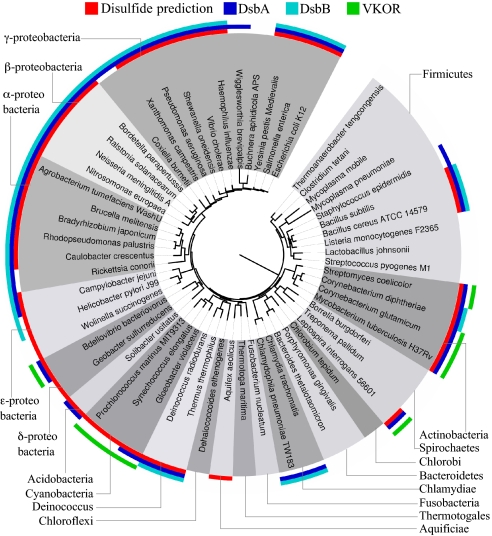
We classified bacteria according to whether their exported proteins showed a significant bias for even numbers of cysteine: We hypothesized that (i) bacteria that oxidatively fold cell envelope proteins should have significantly more even numbers of cysteines in exported proteins than is predicted by our random model (z >2.57) (Fig. 3, Region A), and (ii) bacteria that do not generally make disulfide bonds would lack significant bias for even number of cysteines in exported proteins (2.57 > z > −2.57) (Fig. 3, Region B). These data were then combined with the results of homology searches for members of the DsbA and DsbB family of proteins to allow further deductions about the biology of disulfide bond formation in each bacterium (a representative dataset is presented as Fig. 4; for complete dataset, see Table S1).
Fig. 4.
Combined results of disulfide predictions based on cysteine counting and homology searches. Genomes with significant numbers of exported proteins with even numbers of cysteines (z > 2.57) are indicated in red, and the distribution of DsbA (dark blue) and DsbB (light blue) homologs are shown in a representative subset of all organisms analyzed. The genomes containing a homolog of VKOR are indicated in green.
Bacteria Predicted to Catalyze Disulfide Bond Formation in Cell Envelope Proteins.
We find that most bacteria belonging to the same phylum as E. coli, the Proteobacteria, have DsbA and DsbB homologs, as has been observed previously (6, 7), as well as similarly high fractions of exported proteins with even numbers of cysteines and low cysteine content. These results suggest similar redox biology of cysteine-containing proteins in the cell envelope of most of these organisms. Exceptions include the obligate intracellular proteobacteria, the obligate anaerobic proteobacteria, and the delta-proteobacteria. Surprisingly, there was only one other group of bacteria (members of the phylum Deinococcus-Thermus) showing high fractions of exported proteins with even numbers of cysteines and homologs of both DsbA and DsbB.
One striking pattern that emerges from this analysis is that no homolog of DsbB is found in a large number of bacteria that we predicted should catalyze disulfide bond formation, many of which do have a DsbA homolog. This group includes most Actinobacteria, Cyanobacteria (including chloroplasts), aerobic delta-proteobacteria, Spirochaetes in the genus Leptospira, and the Bacteroidete Salinibacter ruber. The bioinformatic prediction of disulfide bond formation for some of these organisms is consistent with in vitro and in vivo studies that directly identified disulfide bond-containing exported proteins (8–10). Thus, these organisms would need an alternative to DsbB for the reoxidation of DsbA.
We noticed that in the genomes of S. ruber, Leptospira interrogans and the delta-proteobacterium, Bdellovibrio bacteriovorus, the DsbA homolog is fused to an integral membrane protein—a homolog of the eukaryotic VKOR. VKOR was only recently identified in mammals as the enzyme that maintains the cellular pool of vitamin K, a quinone, in the reduced state, and is the target of the widely used anticoagulant warfarin, also known as coumadin (11, 12). In mammals, reduced vitamin K (as opposed to the oxidized form, vitamin K 2,3 epoxide) is required as a cofactor for the gamma-carboxylation of glutamic acid residues in the blood clotting protein prothrombin. VKOR has four highly conserved cysteine residues, two of which are in a Cys-X-X-Cys motif and are essential for catalytic activity in vitro (13, 14). Recent work suggests that eukaryotic VKOR activity is stimulated by the presence of protein disulfide isomerase (PDI) (15), which is the thioredoxin-like protein that catalyzes disulfide bond formation in eukaryotes (4). Strikingly, this passage of electrons from PDI to VKOR to a quinone is exactly analogous to the passage of electrons from DsbA to DsbB to quinones in E. coli during disulfide bond formation. Our genomic analysis indicated that most of the bacteria that are missing a DsbB homolog, but which we predict make disulfide bonds, have a VKOR homolog (Fig. 4 and Table S1), and that the VKOR gene in bacteria is mostly limited to these genomes. We found a number of examples where a VKOR homolog was fused to either a DsbA homolog or a thioredoxin homolog in bacteria that were missing DsbB. Such fusions have been noted previously (14).
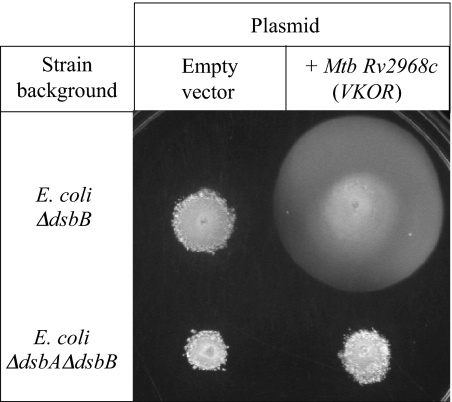
We hypothesized that the bacterial VKOR homolog serves the role of DsbB in these bacteria. As a preliminary test of this hypothesis, we cloned one of the bacterial homologs of VKOR, from M. tuberculosis H37Rv, and tested for its ability to complement a strain of E. coli K12 deleted for dsbB. We used a motility assay to assess complementation for disulfide bond formation because one of the flagellar structural proteins, FlgI, requires a disulfide bond for stability. The M. tuberculosis VKOR homolog complements the motility defect of the Δ_dsbB_ strain, albeit not to wild type levels (Fig. 5). This complementation depends on the presence of dsbA, indicating that VKOR is restoring disulfide bond formation to FlgI, not by acting as a general oxidant, but rather through the intermediary of DsbA, as does DsbB. Thus, the bacterial VKOR may be a key disulfide bond formation enzyme in bacteria lacking DsbB, which includes a large and diverse collection of bacterial species.
Fig. 5.
A bacterial VKOR homolog restores disulfide bond formation to E. coli deleted for dsbB. Disulfide bond formation was assayed by using motility plates, because motility requires active disulfide bond formation. Expression of the M. tuberculosis VKOR homolog restores motility to an E. coli Δ_dsbB_ strain, but not a Δ_dsbA_Δ_dsbB_ strain.
Bacteria That Are Predicted to Have Limited Disulfide Bond Formation.
Our data suggests that several groups of bacteria have no or very few proteins with disulfide bonds based on their low fractions of exported proteins with even number of cysteines (Fig. 3, Region B). These bacteria comprise a phylogenetically diverse set of organisms, with species from the phyla Proteobacteria, Actinobacteria, Bacteroidetes, Firmicutes, and Spirochaetes as well as all sequenced species from the phyla Chlorobi, Fusobacterium, Thermotogae, and Chloroflexi. Many of these bacteria also lack homologs of DsbA and DsbB, consistent with the prediction that they do not oxidatively fold exported proteins. Thus, a potentially novel type of cell envelope biology may be present in this group of organisms, because the bacterial cell envelope is generally thought of as an oxidizing environment.
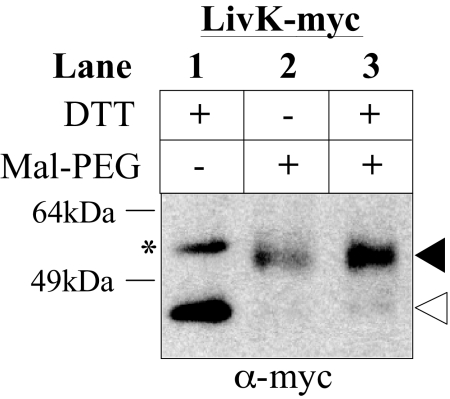
We performed a preliminary test of the hypothesis that these organisms do not have an oxidizing cell envelope using a bacterium, B. fragilis, predicted by our analysis to fall into this class. To assess disulfide bond formation in this organism, we cloned and expressed in B. fragilis an E. coli K12 protein known to have a disulfide bond, LivK (16). We used cysteine alkylation experiments to determine whether the protein acquired a disulfide bond when expressed in B. fragilis. If the cysteines of LivK are free (not disulfide-bonded), they will react with the alkylating agent, resulting in an increase in molecular weight. When expressed in B. fragilis, LivK is exported (data not shown), and the cysteines of the protein can be alkylated, indicating the absence of disulfide bonds in the protein (Fig. 6). We also expressed E. coli alkaline phosphatase in B. fragilis from the same vector, but failed to detect the protein, suggesting that the protein may have been degraded due to a lack of disulfide bonds or was not well expressed. Thus, unlike E. coli DsbA, which forms disulfide bonds in a wide range of substrates from both eukaryotes and bacteria without any apparent specificity toward different substrates, B. fragilis may either have a very limited ability to make disulfide bonds or lack such a system altogether.
Fig. 6.
The E. coli protein LivK does not become disulfide bonded when expressed in B. fragilis. Determination of the redox state of the E. coli protein LivK-myc expressed in B. fragilis. Samples are TCA precipitated then treated as follows, Lane 1: DTT was added to fully reduce the sample to provide a control for unlabeled protein (position of band is indicated with an open arrow), Lane 2: Mal-PEG alkylation. If the cysteines in the protein are not disulfide bonded, they will react with the 2kD alkylating agent, resulting in an increase in molecular weight. The filled arrow indicates the position of the expected shift as a result of alkylation of both cysteines, Lane 3: Control for full alkylation of the protein, samples were first reduced with DTT, then alkylated with Mal-PEG. The asterisk indicates a cross-reacting band, which also shifts upon alkylation (data not shown).
Although the bacteria that we predict lack protein disulfide bonds are phylogenetically diverse, a common trait of many of them is their classification as obligate anaerobes or obligately intracellular organisms (Fig. 3, circled data points). In some cases, the closest relatives of these bacteria are aerobic or free-living bacteria that are predicted to have an oxidizing envelope. The indication that these groups of functionally, but not necessarily phylogenetically, related bacteria may lack disulfide bond formation suggests that they may share some common environmental and/or genetic influences. For instance, the generally reducing environments (e.g., anaerobic sediments or host cell cytoplasm) that these organisms inhabit may be unfavorable for disulfide bond formation. In addition, a number of the obligate anaerobes are obligate fermenting organisms, including members of the genera Clostridia and Lactobacillales, within the phylum Firmicutes. These bacteria are generally thought to lack an electron transport chain (17). Because disulfide bond formation in E. coli is linked to the electron transport chain, an obligately fermentative metabolism may be incompatible with the ability to form disulfide bonds.
Dsb Homologs in Genomes Predicted to Lack Disulfide Bond Formation.
The combination of cysteine counting and analysis of genomes for DsbA/DsbB homologs did not always allow clear predictions of the general redox state of cysteine-containing proteins in the cell envelope. For example, Bacillus subtilis has slightly more even numbers of cysteines in exported proteins than is predicted by the random model (z = 2.67). Yet, systems (Bdb proteins) for disulfide bond formation in exported proteins have been described in this organism that are related by sequence homology to DsbA/DsbB. However, only two native substrates of these disulfide bond formation pathways are known (18, 19).
We also predict that the members of the phylum Chlamydiales do not generally make disulfide bonds, yet these bacteria all have DsbA and DsbB homologs, and have at least one disulfide-bonded protein, the major outer membrane protein (20). However, a recent paper reported in vitro evidence that an exported thioredoxin-like protein from Chlamydia pnuemoniae, DsbH, has reducing activity, and suggested that this organism may actively reduce proteins in the cell envelope (21).
The delta-proteobacterium Geobacter metallireducens also has a low number of exported proteins with even numbers of cysteines and yet has DsbA/DsbB homologs encoded in its genome. The dsbA and dsbB of this bacterium are linked in a putative operon, rather than located at different chromosomal sites as they are in E. coli K12 and most other organisms. This putative operon is occasionally found as an additional copy of dsbA/B in some close relatives of E. coli K12, including pathogenic E. coli (CFT073, UTI89, APEC 01, 536), and some Salmonella species. All organisms with this conserved dsbAB putative operon also include a tightly linked gene astA, encoding the secreted enzyme arylsulfotransferase, an enzyme that in Enterobacter amnigenus has a disulfide bond essential for its activity (22). It may be that this DsbA/B is present in organisms such as G. metallireducens specifically to act on arylsulfotransferase or a small subset of proteins.
Because, for each of these genomes, we find a conflict between the cysteine counting data and the presence of DsbA and DsbB homologs, the balance between reduced and oxidized proteins in the cell envelope of these bacteria may be a complex issue. Surprisingly, examination of the genomic context of the DsbA and DsbB homologs in all of these genomes shows that they are located in a putative operon, which in some cases is found on a prophage or plasmids. This observation may be of interest from an evolutionary perspective, because it suggests that DsbA and DsbB homologs found outside of Proteobacteria may have moved into these genomes via horizontal gene transfer.
Discussion
We have used a multifaceted bioinformatic approach to generate predictions regarding the capacity of different bacteria to catalyze disulfide bond formation. Our work suggests that there is considerable diversity in mechanism and capacity for disulfide bond formation across bacterial species.
This analysis led us to the identification of a disulfide bond formation protein, the bacterial homolog of eukaryotic VKOR, which appears to be an alternative to DsbB in several major phyla of bacteria. We show that the bacterial VKOR can restore disulfide bond formation to E. coli deleted for dsbB in a DsbA-dependent manner. DsbB and VKOR do not show obvious homology at the primary sequence level, so it is surprising that these proteins seem to perform similar reactions, that is, the passage of electrons from thioredoxin-like proteins to membrane bound quinones via redox-active cysteines in the protein. In bacteria, the important biological outcome of these reactions may be disulfide bond formation, whereas in mammals it is the production of reduced vitamin K for gamma-carboxylation of proteins such as prothrombin. Because VKOR and DsbB seem to have evolved independently to perform similar reactions, an interesting topic for future studies will be to determine whether further similarities between DsbB and VKOR exist, such as at the structural level or in reaction mechanisms. Because the mammalian VKOR is of considerable medical interest due to its role in blood clotting in humans, our current findings and future studies of the bacterial homolog could provide useful insights into the biological properties and cellular roles of this protein family. During the preparation of this manuscript, we became aware of a publication reaching a similar conclusion about the role of the VKOR homolog in disulfide bond formation in the cyanobacterium Synechocystis 6803 (23).
Our predictions also led us to test the hypothesis that some bacteria have a cell envelope in which disulfide bond formation does not occur. The results of our experiments with one of these organisms, B. fragilis, are consistent with this proposal. The absence of homologs of the DsbA/DsbB system may be a satisfactory explanation for a lack of disulfide bond formation in this organism. As further evidence, we find that B. fragilis, as well other bacteria that we predict lack protein disulfide bonds, encode homologs of E. coli alkaline phosphatase that lack cysteines, in particular those responsible for stabilizing the E. coli enzyme (data not shown) (24).
The presence of free cysteines in the exported proteins of organisms that do not make disulfide bonds may result in these proteins being susceptible to oxidative damage, such as sulfenic acid formation or inappropriate disulfide bond formation. This would be particularly problematic for the aerotolerant anaerobes (e.g., Bacteroides spp., Lactobacillus spp.) and the aerobic bacteria (e.g., Flavobacterium johnsoniae and B. subtilis) that may lack disulfide bonds, yet survive in the presence of oxygen. Such bacteria may have mechanisms in the cell envelope to prevent or repair oxidative damage to cysteine-containing proteins. Previous studies showed that B. fragilis has at least one pathway, the Batl pathway, that contributes to aerotolerance via the reduction of disulfide bonds in the cell envelope (25). In addition, we find that B. fragilis and F. johnsoniae have several exported thioredoxins and thioredoxin-like proteins of unknown function that could play a role in repairing oxidative damage to cell envelope proteins. Some bacteria may have evolved other mechanisms to prevent unwanted cysteine oxidation in exported proteins. For example, we find that the Firmicutes tend to include very little cysteine at all in exported proteins (Fig. 3, Cpref values on x axis).
Because one of the major roles of disulfide bonds is believed to be the stabilization of exported proteins in the face of a fluctuating extracellular environment, it will be interesting to see whether novel mechanisms of protein stabilization have evolved in bacteria that lack disulfide bond formation. Recently, the crystal structure of a pilin protein from one of the Lactobacillales, Streptococcus pyogenes, revealed an isopeptide bond proposed to be an alternative to disulfide bonds (26).
Although not yet tested, our analysis also led to the prediction that those bacteria such as G. metallireducens containing dsbA/dsbB homologs in an operon may use the disulfide bond-forming enzymes for only a small number of specific substrates. In contrast, most of the Proteobacteria contain dsbA and dsbB genes at different positions on their chromosomes and are predicted by our analysis to contain disulfide bonds in a high proportion of cell envelope proteins.
Last, our results show that in some cases the capacity for disulfide bond formation correlates with the environmental niche of the bacterium. The obligate anaerobes and obligate intracellular bacteria, which often live in reducing environments, generally are predicted by our analysis not to make disulfide bonds. This potential connection between the redox biology of cysteine-containing proteins in the bacterial cell envelope and the habitat of the bacterium may signal an interesting example of the evolution of bacterial genomes and protein folding with respect to particular environments.
Methods
We downloaded all 375 GenBank complete bacterial genomes available on November 5, 2006. Flavobacterium johnsonii UW101 was added on May 8, 2007. We analyzed the protein sequences of these genomes with Phobius, http://phobius.sbc.su.se (27), and a Prosite profile for lipoproteins, release 20.0, www.expasy.ch/prosite (28). Further details, as well as the methods used for assessing the significance of the even fraction (Efrac) are described in the SI Methods.
Hidden Markov models for DsbA, DsbB and VKOR were obtained from Pfam 22.0, http://pfam.sanger.ac.uk (29). Searches were run with HMMER 2.3.2 obtained from http://hmmer.janelia.org, and results above the significance cutoff from the ls_C file were used. DsbA homologs with a cytoplasmic localization, based on Phobius predictions, were excluded. Additional homology searches were performed to obtain a more complete list of homologs, as described in the SI Methods. Information about the biology of the organisms was obtained from National Center for Biotechnology Information, www.ncbi.nlm.nih.gov/genomes/lproks.cgi, and the Genomes Online Database, www.genomesonline.org. The phylogenetic tree in Fig. 4 was generated by using the Interactive Tree of Life (iTOL) web server, http://itol.embl.de (30).
The M. tuberculosis H37Rv VKOR homolog, Rv2968c, was PCR amplified with the following primers, AGCCATGGTTGCAGCGCGACCTGCCGAGCGATCC and CTGCAGTCTAGATCAGATCAGCGTCGACCAAT, and cloned in pDSW206 (31). Motility tests were performed on M63 minimal medium (32), with 0.3% agar, 0.2% glucose, 1 mM isopropyl thiogalactoside, IPTG, and 0.2 mg/ml ampicillin, and incubated for 3 days at 30°C.
The in vivo redox state of E. coli LivK-myc when expressed in B. fragilis, was determined with B. fragilis grown anaerobically at 37°C in Basal medium (33), without adding cysteine. LivK-myc was expressed from the plasmid pFD340 (34). Samples were acid trapped, and then either treated with DTT (DTT) or alkylated with MalPEG-2000MW (Sunbright ME-020MA, NOF Corporation), as described previously (35). Samples were separated with SDS/PAGE, then detected by using antibodies against the Myc epitope (Santa Cruz Biotechnology).
Supplementary Material
Supporting Information
Acknowledgments.
We thank Laurie Comstock and Mark Christopher for reagents and assistance with B. fragilis experiments, Eric Rubin for reagents to clone the VKOR homolog from M. tuberculosis, and Mark Gonzalez and Hiroshi Kadokura for critical reading of this manuscript. This work was supported by National Institute of General Medical Sciences Grant GM41883 (to J.B.). J.B. is an American Cancer Society Professor.
Note added in proof.
A recent paper (36) presents experimental evidence which is consistent with our proposal that the dsbAB homologs present in an operon in some genomes (e.g., pathogenic E. coli and G. metallireducens) show specificity in the oxidation of the protein arylsulfate sulfotransferase.
Footnotes
The authors declare no conflict of interest.
References
- 1.Nakamoto H, Bardwell JC. Catalysis of disulfide bond formation and isomerization in the Escherichia coli periplasm. Biochim Biophys Acta. 2004;1694:111–119. doi: 10.1016/j.bbamcr.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 2.Mallick P, Boutz DR, Eisenberg D, Yeates TO. Genomic evidence that the intracellular proteins of archaeal microbes contain disulfide bonds. Proc Natl Acad Sci USA. 2002;99:9679–9684. doi: 10.1073/pnas.142310499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sone M, Kishigami S, Yoshihisa T, Ito K. Roles of disulfide bonds in bacterial alkaline phosphatase. J Biol Chem. 1997;272:6174–6178. doi: 10.1074/jbc.272.10.6174. [DOI] [PubMed] [Google Scholar]
- 4.Wilkinson B, Gilbert HF. Protein disulfide isomerase. Biochim Biophys Acta. 2004;1699:35–44. doi: 10.1016/j.bbapap.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 5.Pollock MR, Richmond MH. Low cyst (e) ine content of bacterial extracellular proteins: Its possible physiological significance. Nature. 1962;194:446–449. doi: 10.1038/194446a0. [DOI] [PubMed] [Google Scholar]
- 6.Kadokura H, Katzen F, Beckwith J. Protein disulfide bond formation in prokaryotes. Annu Rev Biochem. 2003;72:111–135. doi: 10.1146/annurev.biochem.72.121801.161459. [DOI] [PubMed] [Google Scholar]
- 7.Sevier CS, et al. The prokaryotic enzyme DsbB may share key structural features with eukaryotic disulfide bond forming oxidoreductases. Protein Sci. 2005;14:1630–1642. doi: 10.1110/ps.051355705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goulding CW, et al. Gram-positive DsbE proteins function differently from Gram-negative DsbE homologs. A structure to function analysis of DsbE from Mycobacterium tuberculosis. J Biol Chem. 2004;279:3516–3524. doi: 10.1074/jbc.M311833200. [DOI] [PubMed] [Google Scholar]
- 9.Wiker HG, et al. Immunochemical characterization of the MPB70/80 and MPB83 proteins of Mycobacterium bovis. Infect Immun. 1998;66:1445–1452. doi: 10.1128/iai.66.4.1445-1452.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matoba Y, Katsube Y, Sugiyama M. The crystal structure of prokaryotic phospholipase A2. J Biol Chem. 2002;277:20059–20069. doi: 10.1074/jbc.M200263200. [DOI] [PubMed] [Google Scholar]
- 11.Li T, et al. Identification of the gene for vitamin K epoxide reductase. Nature. 2004;427:541–544. doi: 10.1038/nature02254. [DOI] [PubMed] [Google Scholar]
- 12.Rost S, et al. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature. 2004;427:537–541. doi: 10.1038/nature02214. [DOI] [PubMed] [Google Scholar]
- 13.Rost S, et al. Site-directed mutagenesis of coumarin-type anticoagulant-sensitive VKORC1: Evidence that highly conserved amino acids define structural requirements for enzymatic activity and inhibition by warfarin. Thromb Haemost. 2005;94:780–786. doi: 10.1160/TH05-02-0082. [DOI] [PubMed] [Google Scholar]
- 14.Goodstadt L, Ponting CP. Vitamin K epoxide reductase: Homology, active site and catalytic mechanism. Trends Biochem Sci. 2004;29:289–292. doi: 10.1016/j.tibs.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 15.Wajih N, Hutson SM, Wallin R. Disulfide-dependent protein folding is linked to operation of the vitamin K cycle in the endoplasmic reticulum. A protein disulfide isomerase-VKORC1 redox enzyme complex appears to be responsible for vitamin K1 2,3-epoxide reduction. J Biol Chem. 2007;282:2626–2635. doi: 10.1074/jbc.M608954200. [DOI] [PubMed] [Google Scholar]
- 16.Kadokura H, et al. Snapshots of DsbA in action: Detection of proteins in the process of oxidative folding. Science. 2004;303:534–537. doi: 10.1126/science.1091724. [DOI] [PubMed] [Google Scholar]
- 17.Madigan M, Martinko J. Brock Biology of Microorganisms. 11th Ed. Upper Saddle River, NJ: Pearson Prentice Hall; 2006. [Google Scholar]
- 18.Dorenbos R, et al. Thiol-disulfide oxidoreductases are essential for the production of the lantibiotic sublancin 168. J Biol Chem. 2002;277:16682–16688. doi: 10.1074/jbc.M201158200. [DOI] [PubMed] [Google Scholar]
- 19.Meima R, et al. The bdbDC operon of Bacillus subtilis encodes thiol-disulfide oxidoreductases required for competence development. J Biol Chem. 2002;277:6994–7001. doi: 10.1074/jbc.M111380200. [DOI] [PubMed] [Google Scholar]
- 20.Yen TY, Pal S, de la Maza LM. Characterization of the disulfide bonds and free cysteine residues of the Chlamydia trachomatis mouse pneumonitis major outer membrane protein. Biochemistry. 2005;44:6250–6256. doi: 10.1021/bi047775v. [DOI] [PubMed] [Google Scholar]
- 21.Mac TT, et al. Insight into disulfide bond catalysis in Chlamydia from the structure and function of DsbH, a novel oxidoreductase. J Biol Chem. 2008;283:824–832. doi: 10.1074/jbc.M707863200. [DOI] [PubMed] [Google Scholar]
- 22.Kwon AR, Choi EC. Role of disulfide bond of arylsulfate sulfotransferase in the catalytic activity. Arch Pharm Res. 2005;28:561–565. doi: 10.1007/BF02977759. [DOI] [PubMed] [Google Scholar]
- 23.Singh AK, Bhattacharyya-Pakrasi M, Pakrasi HB. Identification of an atypical membrane protein involved in the formation of protein disulfide bonds in oxygenic photosynthetic organisms. J Biol Chem. 2008;283:15762–15770. doi: 10.1074/jbc.M800982200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Derman AI, Beckwith J. Escherichia coli alkaline phosphatase fails to acquire disulfide bonds when retained in the cytoplasm. J Bacteriol. 1991;173:7719–7722. doi: 10.1128/jb.173.23.7719-7722.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang YP, Dallas MM, Malamy MH. Characterization of the Batl (Bacteroides aerotolerance) operon in Bacteroides fragilis: Isolation of a B. fragilis mutant with reduced aerotolerance and impaired growth in in vivo model systems. Mol Microbiol. 1999;32:139–149. doi: 10.1046/j.1365-2958.1999.01337.x. [DOI] [PubMed] [Google Scholar]
- 26.Kang HJ, et al. Stabilizing isopeptide bonds revealed in gram-positive bacterial pilus structure. Science. 2007;318:1625–1628. doi: 10.1126/science.1145806. [DOI] [PubMed] [Google Scholar]
- 27.Kall L, Krogh A, Sonnhammer EL. Advantages of combined transmembrane topology and signal peptide prediction—The Phobius web server. Nucleic Acids Res. 2007;35:W429–W432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hulo N, et al. The PROSITE database. Nucleic Acids Res. 2006;34:D227–D230. doi: 10.1093/nar/gkj063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finn RD, et al. Pfam: Clans, web tools and services. Nucleic Acids Res. 2006;34:D247–51. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Letunic I, Bork P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 31.Weiss DS, et al. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J Bacteriol. 1999;181:508–520. doi: 10.1128/jb.181.2.508-520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silhavy TJ, Berman ML, Enquist LW. Experiments with Gene Fusions. Plainview, NY: Cold Spring Harbor Lab Press; 1984. pp. 102–112. [Google Scholar]
- 33.Pantosti A, Tzianabos AO, Onderdonk AB, Kasper DL. Immunochemical characterization of two surface polysaccharides of Bacteroides fragilis. Infect Immun. 1991;59:2075–2082. doi: 10.1128/iai.59.6.2075-2082.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith CJ, Rogers MB, McKee ML. Heterologous gene expression in Bacteroides fragilis. Plasmid. 1992;27:141–154. doi: 10.1016/0147-619x(92)90014-2. [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi T, Ito K. Respiratory chain strongly oxidizes the CXXC motif of DsbB in the Escherichia coli disulfide bond formation pathway. EMBO J. 1999;18:1192–1198. doi: 10.1093/emboj/18.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grimshaw JP, et al. DsbL and DsbI form a specific dithiol oxidase system for periplasmic arylsulfate sulfotransferase in uropathogenic Escherichia coli. J Mol Biol. 2008;380:667–680. doi: 10.1016/j.jmb.2008.05.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information