Multiple genes of apparent algal origin suggest ciliates may once have been photosynthetic (original) (raw)
. Author manuscript; available in PMC: 2009 Jul 8.
Published in final edited form as: Curr Biol. 2008 Jul 8;18(13):956–962. doi: 10.1016/j.cub.2008.05.042
Summary
Plantae (sensu Cavalier-Smith 1981) [1] plastids evolved via primary endosymbiosis whereby a heterotrophic protist enslaved a photosynthetic cyanobacterium. This 'primary' plastid spread into other eukaryotes via secondary endosymbiosis. An important but contentious theory in algal evolution is the chromalveolate hypothesis that posits chromists (cryptophytes, haptophytes, and stramenopiles) and alveolates (ciliates, apicomplexans, and dinoflagellates) share a common ancestor that contained a red algal derived 'secondary' plastid [2]. Under this view, the existence of several later-diverging plastid-lacking chromalveolates such as ciliates and oomycetes would be explained by plastid loss in these lineages. To test the idea of a photosynthetic ancestry for ciliates we used the 27,446 predicted proteins from the macronuclear genome of Tetrahymena thermophila to query prokaryotic and eukaryotic genomes. We identified 16 proteins of possible algal origin in the ciliates Tetrahymena and Paramecium tetraurelia. Fourteen of these are present in other chromalveolates. Here we compare and contrast the likely scenarios for algal gene origin in ciliates either via multiple rounds of horizontal gene transfer (HGT) from algal prey or symbionts, or through endosymbiotic gene transfer (EGT) during a putative photosynthetic phase in their evolution.
Results and Discussion
Rationale for Study
Multiple sources of data reveal the evolutionary history of photosynthesis in eukaryotes has followed a circuitous path via serial plastid captures; i.e., endosymbioses [2, 3]. The story begins with the primary plastids of Plantae (i.e., glaucophyte, green, and red algae) that resulted from the ancient (putatively single) engulfment and enslavement of a cyanobacterial endosymbiont [4–6]. Thereafter this plastid was transferred at least three times through eukaryote-eukaryote (secondary) endosymbioses [3]. It has been suggested that the chromist algae (the chlorophyll _c_-containing haptophytes, cryptophytes, photosynthetic stramenopiles such as chrysophytes, diatoms and phaeophytes), the non-photosynthetic stramenopiles (e.g., oomycetes and bicosoecids), and the alveolates (dinoflagellates, apicomplexans, and ciliates) evolved from a single common ancestor that contained a secondary plastid of red-algal origin. These taxa are postulated as the supergroup Chromalveolata [2]. Ciliates are an independent branch within alveolates [7, 8] and in contrast to most dinoflagellates and apicomplexans (that form sister groups of each other) there is currently no evidence of a plastid or a plastid-derived compartment in this group. The question remains unanswered whether ciliates once harbored a secondary algal endosymbiont, putatively like other alveolates and were therefore also once photosynthetic. This issue was recently clarified for apicomplexans with the finding of the marine protist Chromera velia that is the closest known photosynthetic relative of these parasitic taxa [9].
The chromalveolate hypothesis [2] is primarily supported by phylogenetic analysis of plastid-encoded [10] and plastid-targeted proteins [11], which most often show a red algal origin of this organelle. However, some analyses also provide evidence for green algal genes of plastid function that are encoded in the nucleus of chromalveolates [12, 13]. Due to their sporadic distribution, it is unclear whether these genes have arisen through independent HGTs or from EGT. Recent multi-gene analyses of nuclear genes support the monophyly of cryptophytes and haptophytes [14, 15] and the surprising inclusion of the supergroup Rhizaria within chromalveolates [14, 16]. Most single- and multi-gene analyses support alveolate monophyly and their sister group relationship to stramenopiles [17–19] (see Figure 1). Therefore, although the overall chromalveolate phylogenetic framework remains to be elucidated, the evidence is reasonably strong that many of its constituent members had a photosynthetic ancestry. This hypothesis is supported by analysis of nuclear genome data from the parasitic, plastid-lacking oomycetes (stramenopiles) in which 30 candidate genes of putative cyanobacterial and algal (i.e., endosymbiotic) origin were found in two Phytophthora species [20]. Similarly, phylogenomic analyses of apicomplexan complete genomes turned up dozens of nuclear genes of putative endosymbiotic origin in Cryptosporidium parvum (219 genes), Plasmodium falciparum (207 genes), Theileria parva (180 genes), and Toxoplasma gondii (87 genes), including 21 genes shared between the four species [21]. Surveys of the non-photosynthetic alveolates Perkinsus marinus (oyster parasite) [22] and Oxyrrhis marina (early diverging dinoflagellate) [23] also uncovered genes of putative secondary endosymbiotic origin in the nuclear genomes. Although these numbers are difficult to compare with each other due to widely different bioinformatic approaches that were used, the data clearly demonstrate that even in the absence of a plastid (e.g., C. parvum), secondary endosymbionts leave detectable ‘footprints’ in the nuclear genomes of chromalveolates. Given this observation, we asked the question, do ciliates also contain genes of algal origin that may bespeak a photosynthetic past for this lineage? Existing studies of the macronuclear genome of the ciliates Tetrahymena thermophila and Paramecium tetraurelia have provided negative results in this respect [24, 25]. Here we re-investigate this issue using a phylogenomic approach followed by detailed searches of public databases and phylogenetic analysis of target genes on a gene-by-gene basis. Our goal was twofold, first to identify ciliate genes of putative algal origin, and second to investigate how these genes originated, either via EGT due to a photosynthetic past or recurrent HGT from different algal sources.
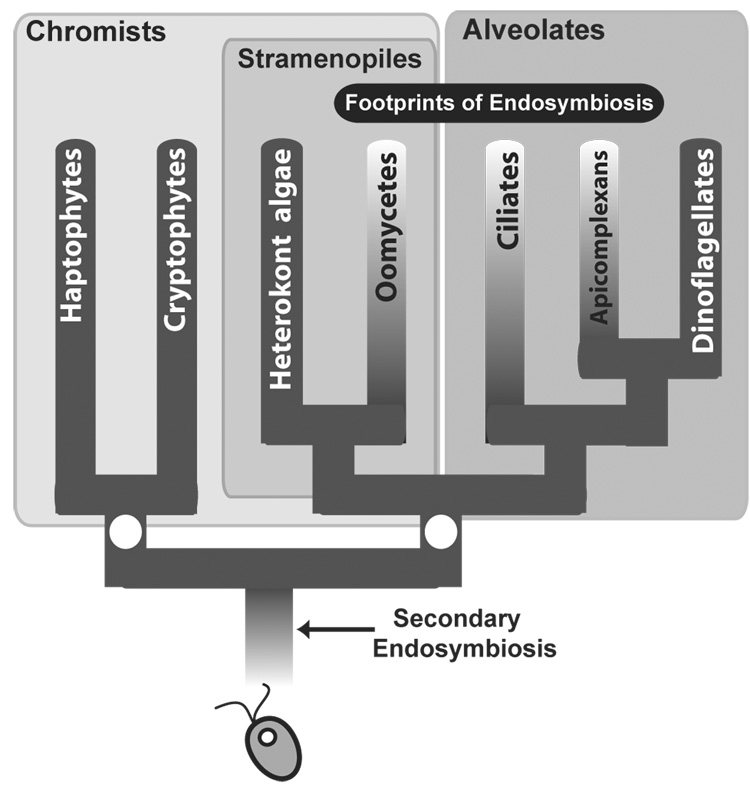
Figure 1. The chromalveolate hypothesis and secondary endosymbiotic gene transfer.
The current phylogenetic framework for this supergroup based on multi-gene analyses is shown with the filled circles marking well-supported deep nodes. The chromists do not form a monophyletic group in these trees. Genome analyses demonstrate the footprint of a plastid-containing ancestry in non-photosynthetic groups via the existence of remnant, putative endosymbiont genes in their nucleus (e.g., oomycetes [20], apicomplexans [21, 36], and ciliates [this paper]). The chromalveolates clearly had a red algal secondary endosymbiont but evidence also exists for green algal derived genes in these taxa [12].
Phylogenomic Analysis
We identified 16 trees that contain branches with moderate to high (≥70% bootstrap probability with at least one maximum likelihood approach; see Experimental Procedures section) uniting ciliates (i.e., Tetrahymena and Paramecium) with other chromalveolates and Plantae (primary algae and land plants) (Table 1). These algal genes have functions that go beyond plastid metabolism and here are interpreted as markers of algal gene transfer rather than strictly as evidence of a former plastid in ciliates. Six (Fig. 2A and Supplemental Data Figs. S1, S2, S4, S6, S7) of the 16 trees contain a Euglena gracilis homolog that branches within Plantae (Fig. 2A and Figs. S1, S2, S6, S7) and chromalveolates (Fig. S4). These euglenid genes are most likely derived via EGT from the green algal-derived secondary endosymbiont in this lineage; i.e., in 4/5 cases the Euglena homolog is nested within the green clade. In addition, in the proton-translocating pyrophosphatase tree (Fig. S14), a homolog (derived from a partial EST) from the heterolobosean Stachyamoeba lipophora branches within chromalveolates, suggesting a recent HGT into this species from a chromalveolate source. Importantly, most of the 16 ciliate proteins have homolog in apicomplexans, oomycetes (non-photosynthetic) (12/16; Table 1 and Supplemental Data Figs. S2, S5, S7–S11, S12–S16), and/or diatoms, haptophytes, dinoflagellates, and cryptophytes (photosynthetic) (11/16; Table 1 and Figs. S1, S2, S4, S5, S7, S9–S11, S13, S14, S16). Both the tree topology and gene distribution data imply therefore an ancient shared ancestry of these sequences in chromalveolates. In some trees (4/16; Figs. S5, S11, S12, S15), the branch containing ciliates, chromalveolates, and Plantae is related to other eukaryotic homologs (e.g., opisthokonts and excavates). The HNH endonuclease (Fig. S3) is unique to Plantae and ciliates, and the PA domain containing protein (Fig. S10) is exclusively found in Plantae and chromalveolates (including ciliates). We suggest the origin of these latter two genes in chromalveolates is through ancient EGT or HGT. In either case, the direction of transfer is likely to be from Plantae to chromalveolates because of the absence of phagotrophy in Plantae, combined with the well-known predatory (ciliates, dinoflagellates, bicosoecids) and saprophytic (oomycetes) life styles in chromalveolates. Interestingly, in two trees (Figs. S1 and S2) chromalveolate (including ciliates) and Plantae proteins are associated with cyanobacterial homologs (see below).
Table 1.
Ciliate genes of putative endosymbiotic origin identified in our study.
| Fig | Accession | Annotation | BS | pCH | npCH | Bact | Pt T |
|---|---|---|---|---|---|---|---|
| S1 | XP_001023477 | Delta 12 fatty acid desaturase | 93/93 | • | |||
| S2 | XP_001019413 | Folate/biopterin transporter family protein | 79/90 | • | • | • • | |
| S3 | XP_001023127 | HNH endonuclease family protein | O | • • | |||
| S4 | XP_001008692 | PPI-Phosphofructokinase family protein | 91/78 | • | • | ||
| S5 | XP_001024446 | Protein phosphatase 2A regulatory B subunit | 97/84 | • | • | • | |
| S6 | XP_001019769 | Aminotransferase class IV family protein | 70/- | • | • • | ||
| S7 | XP_001007903 | Hypothetical protein. (MinD-like ATPase) | 100/100 | • | • | • | • • |
| S8 | XP_001033476 | Kinase pfkB family protein (ribokinase) | 100/100 | • | • | • | |
| S9 | XP_001021395 | RNA methyltransferase. TrmH family protein | 50/79 | • | • | • | |
| S10 | XP_001027786 | PA domain containing protein | O | • | • | ||
| S11 | XP_001031541 | Aldehyde dehydrogenase (NAD) family protein | 100/100 | • | • | ||
| S12 | XP_001022369 | Hypothetical protein | 99/100 | • | |||
| S13 | XP_001030231 | Glucose-6-phosphate isomerase family protein | 100/89 | • | • | • | |
| S14 | XP_001031634* | Inorganic H+ PPi family protein (vacuolar-type) | 98/- | • | • | • | |
| S15 | XP_001024882 | ATPase, AAA family protein | 100/100 | • | |||
| S16 | XP_001031763 | Hypothetical protein (glycosyl-transferase) | 100/100 | • | • | • |
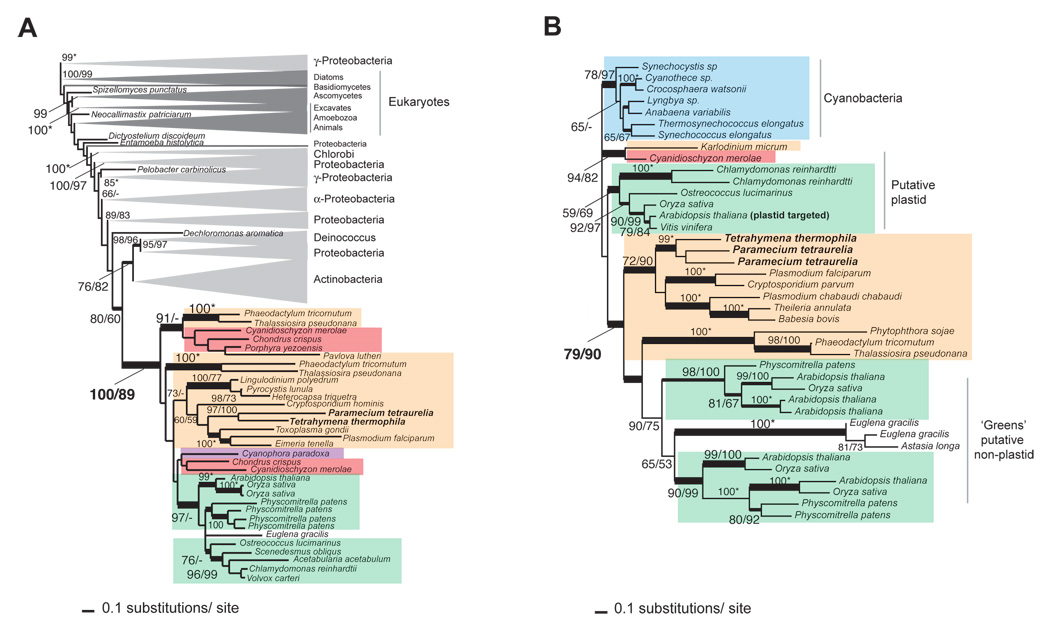
Figure 2. Maximum likelihood (RAxML) trees of algal-derived ciliate proteins.
A) The protein of the glucose-6-phosphate isomerase family (see Figure S13 for full tree) from Plantae (green boxes for green algae and land plants, red for red algae, and purple for glaucophytes) and chromalveolates (orange boxes) is closely related to bacterial (light grey triangles) homologs. Other non-Plantae or non-chromalveolate eukaryote clades are indicated (dark gray triangles). B) This is the tree of the folate-biopterin transporter (FBT) and provides evidence of a cyanobacterial gene origin (blue box) in Plantae (see above) and then its transfer into chromalveolates via EGT. In both trees the RAxML bootstrap values are shown on the left of the slash mark and PHYML bootstrap values on the right. Only bootstrap values >50% are shown. The asterisks indicate that these nodes have the same bootstrap support from both RaxML and PHYML analyses. The thick lines indicate branches with a Bayesian posterior probability >0.95. Branch lengths are proportional to the number of substitutions per site (see the scale bars).
Another intriguing observation is that in 8 trees (Figs. S4, S6–S9, S13, S14, S16) the branch including ciliates, other chromalveolates, and Plantae is closely related to homologs from non-cyanobacterial prokaryotes. An example is glucose-6-phosphate isomerase type I-B [26] (Fig. 2A or Fig. S13 for a detailed tree) that was previously used to identify a well-supported branch uniting chromalveoltes and Plantae [26]. Grauvogel et al. [26] interpreted this result as support for the monophyly of these supergroups [26]. In contrast, we postulate here that the type I-B clade indicates gene transfer between Plantae and chromalvelates rather than support for ‘host’ monophyly. Under our preferred view, bacterial genes originated in Plantae via a single ancient HGT and then were transferred to the chromalveolate nucleus via secondary endosymbiotic EGT. In support of this view, molecular phylogenetic analyses [14–16] until now fail to provide convincing evidence for a common ancestry of these supergroups. If chromalveolates and Plantae were monophyletic, our phylogenomic approach should have identified a large number of trees (inferred from conserved proteins) with well-supported branches uniting these supergroups, rather than the 16 proteins we found. An alternate more complex scenario for these bacterial-derived genes involves multiple independent HGTs in chromalveolates from different Plantae. Finally, the bacterial-derived genes may have been present in the ancestral eukaryote (e.g., derived from the proto-mitochondrion) and over time were lost from all other supergroups except Plantae and chromalveolates, thereby generating their monophyly due solely to a shared gene presence. Although we cannot convincingly prove (or disprove) any of these competing scenarios, we suggest that the most likely explanation for Plantae-chromalveolate gene monophyly observed here is secondary endosymbiotic EGT via the substantiated connection between these two supergroups. Three (Figs. S6, S7, S8) of these eight bacterial-derived proteins are putatively plastid-targeted in Arabidopsis (see Experimental Procedures and Cellular Localization sections), reflecting a possible ancestral association with plastid endosymbiosis (i.e., organelle function) in other lineages. Finally, it should be noted that the number of identified trees is by definition a minimal estimate using our data set due to the loss of phylogenetic signal (i.e., trees) in anciently diverged sequences (e.g., [27]).
Evaluating the Strength of the Ciliate-Plantae Phylogenetic Relationship
Eight trees (Figs. S1, S7, S8, S11, S12, S13, S15, S16) contain branches that unite Plantae and chromalveolate (including ciliates) sequences with strong support (i.e., ≥ 89% bootstrap probability, BP, with both RAxML and PhyML, and Bayesian posterior probability, PP = 1.0; see red circles in supplemental figures). In addition, in four other trees (Figs. S2, S4, S5, S14) the branch uniting chromalveolates (including ciliates) and Plantae is highly supported (≥ 90 BP, PP = 1.0) using at least one maximum likelihood approach. The interrelationships within these clades are however unresolved. As described above, in 7/12 trees the Plantae-chromalveolate branch includes other non-Plantae, non-chromalveolate eukaryotes (i.e., Euglena in Fig. S1 and Stachyamoeba in Fig. S14), or prokaryotes (e.g., Leptospira in Fig. S15), that we attribute to independent HGT or EGT events.
To assess ciliate-chromalveolate-Plantae monophyly, we used the approximately unbiased (AU-) test to generate likelihoods for trees that repositioned ciliates with non-Plantae and forced the monophyly of ciliates with other chromalveolates (see Experimental Procedures). For this analysis, nine RAxML trees were selected in which chromalveolates were non-monophyletic and/or their relationship within or as sister to the Plantae was not robustly supported (i.e., Supplemental Data Figs. S1, S2, S4, S5, S7, S8, S9, S12, S13). The results of this analysis (using a significance value of p < 0.01) show that in 8/9 cases (i.e., excluding protein phosphatase 2A regulatory B subunit; Fig. S5, Table S4) disruption of the Plantae-chromalveolate clade by placing members outside of this group produced tree topologies that were significantly worse than the best maximum likelihood (i.e. RAxML) tree. In 8/9 trees, forcing chromalveolate monophyly was not significantly rejected (Supplemental Table S1–S8). Only for glucose-6-phosphate isomerase (G6PF, Figure S13 and Table S9) did the AU-test reject this topological rearrangement. These results provide two key insights: 1) it would be unwise to over-interpret the internal branching patterns within the chromalveolate-Plantae clades using these single-protein trees, however, 2) the monophyly of these supergroups (which we interpret as EGT or HGT from Plantae to chromalveolates) is not rejected. It is worth considering therefore that what on the surface appears to be examples of multiple HGTs from Plantae may simply reflect the inability to capture ancient phylogenetic signal from single proteins to substantiate EGT (for discussion, see [28]). This is particularly true for ciliates [29] and parasitic taxa such as apicomplexans [30] and oomycetes, many of which have undergone rapid and heterogeneous rates of protein evolution. These single-protein trees may however prove significantly more conclusive in the future with the addition of a broader taxonomic diversity of Plantae. For example, the red algae are represented in our analysis by two thermoacidophiles with highly reduced genomes (Cyanidioschyzon merolae [16.5 Mb; 5,331 genes] and Galdieria sulphuraria [ca. 15 Mb]; Cyanidiales). Lack of a red algal homolog in some trees (e.g., Fig. S10, S16) could be explained by the loss of homologs only in Cyanidiales; i.e., addition of data from mesophilic reds would change our interpretation.
Potential Cyanobacteria-Derived Ciliate Genes
The phylogenetic tree of the putative folate/biopterin transporter (pFBT; Fig. 2B, Fig. S2) is intriguing because it points to a possible cyanobacterial gene origin in ciliates. This putative vitamin transporter is present in plants as both plastid-targeted and non-plastid isoforms. In our tree (Fig. 2B), pFBT from chromalveolates groups with the non-plastid targeted Plantae proteins (including the euglenids Astasia longa and Euglena gracilis) as sister to the plastid-targeted (i.e., in Arabidopsis) and cyanobacterial homologs. This topology suggests that cytosolic pFBT evolved in Plantae via duplication of the cyanobacterial gene, followed by co-option of one gene for cytosolic functions. Cytosolic pFBT has not yet been detected in red algae. These results may indicate a possible plastid (cyanobacterial) ancestry of chromalveolate pFBT. Distantly related pFBT homologs exist in trypanosomatids (ca. 25% similarity using BLASTP over a 200 amino acid region) but these sequences give rise to unreliable, partial protein alignments and were excluded from the final analysis. Another interesting result is the delta-12 fatty acid desaturase (FAD2) tree (Fig. S1). The ciliate homolog is included in a highly supported (93% BP) clade that includes cyanobacteria, diatoms, Isochrysis, Ostreococcus spp., Cyanidioschyzon, and Euglena. The ciliate and two diatom proteins are robustly (>95% BP) separated from the remaining sequences in this clade. The cyanobacterial-derived protein has a putative non-plastid function in Ostreococcus spp. and Cyanidioschyzon. A possible explanation for this result is that the Plantae ancestor recruited cyanobacterial FAD2 for lipid metabolism and later the gene was transferred to the chromalveolate ancestor. There is however another FAD2 clade that is clearly of non-cyanobacterial origin with homologs shared with the green lineage, fungi, and other protists. This group of enzymes has a cytosolic function. The phylogenetic affiliation of Plantae and cyanobacterial proteins for pFBT fits well with an ancient origin through EGT [27, 31, 32], whereas the FAD2 tree topology suggests gene gains through HGT.
Cellular Localization
Cellular targeting predictions using the Arabidopsis homologs (see Experimental Procedures) revealed that six (i.e., putative folate/biopterin transporter, putative HNH endonuclease, subunit B of the protein phosphatase 2A, aminotransferase class IV, MinD-like hypothetical protein, kinase of the pfkB family) of the 16 proteins are likely to be plastid targeted in plants (see Table 1). The putative functions of these proteins are diverse, including membrane transport, modulation of protein activity, carbohydrate metabolism, and amino acid biosynthesis-degradation. Closer inspection of the non-Plantae sequences uncovered that the folate/biopterin transporter proteins (Fig. S2) from Phaeodactylum, Phytophthora and some apicomplexans, but not ciliates, have amino (N-) terminal extensions (ranging in size from 19 – 46 aa) when compared to the cyanobacterial homologs. These N-terminal extensions do not contain potential cleavable sites according to SignalP (www.cbs.dtu.dk/services/SignalP). Similarly, MinD-like proteins (Fig. S7) from ciliates, Phaeodactylum, and Phytophthora have N-terminal extensions (20 – 80 aa) in comparison to the prokaryotic homologs. However, none of these proteins appear to be organelle (i.e., apicoplast or mitochondrial) targeted. The remaining chromalveolate homologs of the Arabidopsis plastid-targeted proteins apparently do not contain protein extensions. These results may be explained by the possible re-targeting of former plastid proteins to different cell locations (e.g., cytosol) to express novel functions. The other 10 Arabidopsis homologs are unlikely to be plastid-localized (Table 1) and are apparently involved in a broad range of functions including carbohydrate metabolism (PPI-phosphofructokinase, Fig. S4; glucose-6-phosphate isomerase type I-B, Fig. S13), lipid biosynthesis (delta 12 fatty acid desaturase, Fig. S1), RNA processing (RNA methyltransferase, Fig. S9), oxidoreductase activity (NAD-dependent aldehyde dehydrogenase, Fig. S11), and bionergetic metabolism (inorganic H+ pyrophosphatase, Fig. S14). Glucose-6-phosphate isomerase type I-B is an interesting case because the Arabidopsis homolog is a cytosolic protein, whereas the Chlamydomonas and red algal homologs have evolved secondarily into plastid-targeted isoforms [26].
EGT vs. HGT for Algal Gene Origin in Ciliates
Although it is currently impossible to prove conclusively which fraction of the 16 genes of algal origin in ciliates originated via HGT vs. EGT, the branching pattern for many protein trees suggests an ancient origin in alveolates with several genes being shared with other chromalveolates. This result is explicable under the prevailing views of chromalveolate evolution [2, 8, 10] although ancient HGTs would also lead to a taxonomically broad distribution of algal genes in chromalveolates. Under the competing model of random gene introductions into ciliates over their long history, we might expect to find fewer examples of the monophyly of alveolates and chromalveolates with Plantae (as seen here) and more evidence for sporadic, recent HGTs from algal sources in these taxa. In this regard, the ciliate proteins shared only with Plantae (HNH endonuclease; Fig. S3), and with Plantae and Euglena among eukaryotes (aminotransferase class IV; Fig. S6), are likely candidates for origin through independent HGTs from Plantae sources.
Under the EGT scenario it is not surprising that virtually all of the algal-derived plastid targeted proteins have been lost from ciliates. An analogous example is the ‘loss’ (i.e., deletion or high divergence) of the vast majority of genes encoding mitochondrial proteins in the nuclear genome of parasitic protists like Entamoeba histolytica [33] and Giardia lamblia [34]. These species have secondarily lost most of the mitochondrial functions and retain a remnant organelle (mitosomes) with limited metabolic roles (e.g., Fe-S cluster biosynthesis). Thus, it is not surprising that once an organelle is lost or degenerates, the nuclear genes associated with its canonical function (e.g., oxidative phosphorylation in mitochondria and photosynthesis in plastids) and maintenance are also lost, leaving behind only vestiges of the ancestral condition. In our study this vestige includes a set of genes (11/14, discounting two potentially HGT-derived candidates) that are shared with another previously photosynthetic lineage, the oomycetes [20]. Therefore when the putative plastid was lost in the ciliate ancestor, most of the genes (likely several hundred) associated with the function of this organelle would also be expected to be jettisoned, with the exception of those recruited for non-plastid functions. This is essentially what we find with remnant algal genes involved in general processes like amino acid, nucleic acid, and lipid metabolism (Table 1).
In summary, we stress that our study does not address the evolution of photosynthesis in the entire chromalveolate group but rather uses this hypothesis to guide our work with alveolates. There is much still left to be learned about plastid gain and loss in this lineage and many of these insights will come not from analyses of algae but nuclear genes in currently non-photosynthetic (e.g., Perkinsus [22, 35], Oxyrrhis [23], katablepharids, telonemids [14]) and plastid-lacking (Cryptosporidium parvum [36]) taxa to unearth information about their past ‘lives’. In summary, our analyses show that Tetrahymena and Paramecium contain algal-derived genes whose presence do not prove but fit well with the modus operandi for photosynthetic algae that have secondarily lost the canonical plastid or its ancestral functions (e.g., oomycetes and apicomplexans). Proof for an algal past for ciliates would come from the finding of an as-yet undescribed photosynthetic ancestor for this lineage. The recent description of the plastid-bearing Chromera velia [9] as a relative of apicomplexan parasites suggests that this development is formally possible.
Experimental Procedures
Phylogenomics
To identify genes of putative algal origin in ciliates, we screened the 27,466 predicted proteins from the Tetrahymena thermophila complete macronuclear genome [25] using reciprocal BLAST (WU-BLAST with e-value < 0.001) against a 13-species Plantae data set assembled from completed genomes and EST libraries (274,434 sequences). Our data set included 6 green algae and land plants (Arabidopsis thaliana, Chlamydomonas reinhardtii, Oryza sativa, Physcomitrella patens, Ostreococcus spp., and Volvox carteri), 5 red algae (Chondrus crispus, Cyanidioschyzon merolae, Gracilaria gracilis, Porphyra yezoensis, and Galdieria sulphuraria), and 2 glaucophytes (Cyanophora paradoxa and Glaucocystis nostochinearum). This search identified 3,997 candidate proteins. We excluded proteins at this step that had significant BLAST e-values but only partial (i.e., domain) conservation over the entire sequence alignment.
We used PhyloGenie [37] to run a phylogenomic analysis of the 3,997 Tetrahymena candidates against a local database comprised of >500 genomes (2 ciliates, 13 Plantae, 14 chromalveolates, 14 cyanobacteria, 4 animals, 6 fungi, 500 bacteria, 3 Amoebozoa, and 5 excavates; the complete taxon list available upon request from DB) for a total of 2,558,167 protein sequences. The PhyloGenie BLAST e-value cut-off was set at < 1e−6 and distance trees were generated using neighbor-joining (NJ) with Poisson distance correction and 100 bootstrap replicates. We used our tree-topology-search tool PhyloSort [38] to identify all NJ trees that showed monophyly of ciliates and Plantae (with or without chromalveolates included within the clade). Considering a minimum of 50% BP, we found 246 trees (representing 184 unique gene families) matching the topological constraint. After a manual review of the 246 alignments and trees of the matching Tetrahymena genes, we selected a set of 133 genes for a second round of phylogenetic analysis using PHYML and the JTT model of amino acid substitution, gamma distribution with 4 substitution rate categories, and 100 bootstrap replicates.
Manual inspection of the 133 PHYML trees revealed 25 topologies that showed unambiguous clades that included ciliates, chromalveolates, and Plantae. We reevaluated these 25 candidate trees by including homologous proteins not included in our local genome database. This was done through BLAST searches against the GenBank, JGI, and TBestBD databases to address as broad a set of target taxa as possible. To ensure accuracy, translated sequences from EST databases (e.g., TBestBD) were included in our final alignments only if they contained >50% of the total number of characters. The protein data sets were re-aligned using ClustalX [39] and manually refined. The alignments are available in the supplemental tables. The final ML trees were estimated with RAxML (VI-HPC, v2.2.1) [40] using the WAG substitution model, gamma distribution (‘PROTGAMMA’ implementation), with 4 discrete rate categories, and starting from a random tree. The branch support was evaluated with 100 bootstrap replicates using both RAxML (WAG substitution model and the ‘PROTCAT’ implementation) and PhyML (WAG + Γ substitution model, and parameters estimated during the tree search). Posterior probabilities of tree nodes were calculated with MrBayes 3.1 [41] running a MC3 for 1 million generations using 1 cold and 3 heated chains starting with a random tree. The pool of trees was sampled every 100th generation. Final posterior probabilities were estimated after discarding the trees of the first 2.5×105 generations.
Approximately Unbiased Test
We generated alternative hypothesis to assess the monophyly of chromalveolates and their relationship to Plantae. Prior to generating the alternative trees, we removed the long-branched Euglena gracilis sequences (genes of known secondary EGT origin) and other partial sequences generated from ESTs from the data sets. Nine ML trees were used as starting points to identify likely alternative topologies. First, we generated a monophyletic chromalveolate branch, then the ‘new’ clade was removed and added to other likely branches (see table S1) in the respective backbone tree. The site-by-site likelihoods were estimated for each alternative tree with TreePuzzle [42] using the WAG + Γ (four rate categories) substitution model and the –wsl option. The AU test was done with CONSEL V0.1i [43] to identify the set of plausible tree topologies for each protein data set
Protein Targeting Predictions
To gain insights into the identity and function of the algal proteins, we assessed their putative cellular locations. Given that the available computational tools have been ‘trained’ extensively with land plant sequences, we used the Arabidopsis thaliana (if present in the tree) proteins that group with ciliates and chromalveolates to predict the cellular location of the plant homolog using Predotar V1.03 (http://urgi.versailles.inra.fr/predotar/predotar.html) and TargetP 1.1 Server (www.cbs.dtu.dk/services/TargetP).
Supplementary Material
02
Acknowledgements
This work was supported by grants to DB from the National Science Foundation (EF-043117, EF-0625440) and the National Institutes of Health (R01ES013679). We are grateful to anonymous reviewers for their constructive criticisms.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cavalier-Smith T. Eukaryote kingdoms: seven or nine? Biosystems. 1981;14:461–481. doi: 10.1016/0303-2647(81)90050-2. [DOI] [PubMed] [Google Scholar]
- 2.Cavalier-Smith T. Principles of protein and lipid targeting in secondary symbiogenesis: Euglenoid, dinoflagellate, and sporozoan plastid origins and the eukaryote family tree. J Eukaryot Microbiol. 1999;46:347–366. doi: 10.1111/j.1550-7408.1999.tb04614.x. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharya D, Yoon HS, Hackett JD. Photosynthetic eukaryotes unite: endosymbiosis connects the dots. Bioessays. 2004;26:50–60. doi: 10.1002/bies.10376. [DOI] [PubMed] [Google Scholar]
- 4.Delwiche CF. Tracing the Thread of Plastid Diversity through the Tapestry of Life. Am Nat. 1999;154:S164–S177. doi: 10.1086/303291. [DOI] [PubMed] [Google Scholar]
- 5.McFadden GI. Primary and secondary endosymbiosis and the origin of plastids. J Phycol. 2001;37:951–959. [Google Scholar]
- 6.Palmer JD. The symbiotic birth and spread of plastids: how many times and whodunit? J Phycol. 2003;39:4–12. [Google Scholar]
- 7.Fast NM, Xue L, Bingham S, Keeling PJ. Re-examining alveolate evolution using multiple protein molecular phylogenies. J Eukaryot Microbiol. 2002;49:30–37. doi: 10.1111/j.1550-7408.2002.tb00336.x. [DOI] [PubMed] [Google Scholar]
- 8.Harper JT, Waanders E, Keeling PJ. On the monophyly of chromalveolates using a six-protein phylogeny of eukaryotes. Int J Syst Evol Microbiol. 2005;55:487–496. doi: 10.1099/ijs.0.63216-0. [DOI] [PubMed] [Google Scholar]
- 9.Moore RB, Obornik M, Janouskovec J, Chrudimsky T, Vancova M, Green DH, Wright SW, Davies NW, Bolch CJ, Heimann K, et al. A photosynthetic alveolate closely related to apicomplexan parasites. Nature. 2008;451:959–963. doi: 10.1038/nature06635. [DOI] [PubMed] [Google Scholar]
- 10.Yoon HS, Hackett JD, Pinto G, Bhattacharya D. The single, ancient origin of chromist plastids. Proc Natl Acad Sci U S A. 2002;99:15507–15512. doi: 10.1073/pnas.242379899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harper JT, Keeling PJ. Nucleus-encoded, plastid-targeted glyceraldehyde-3-phosphate dehydrogenase (GAPDH) indicates a single origin for chromalveolate plastids. Mol Biol Evol. 2003;20:1730–1735. doi: 10.1093/molbev/msg195. [DOI] [PubMed] [Google Scholar]
- 12.Li S, Nosenko T, Hackett JD, Bhattacharya D. Phylogenomic analysis identifies red algal genes of endosymbiotic origin in the chromalveolates. Mol Biol Evol. 2006;23:663–674. doi: 10.1093/molbev/msj075. [DOI] [PubMed] [Google Scholar]
- 13.Petersen J, Teich R, Brinkmann H, Cerff R. A "green" phosphoribulokinase in complex algae with red plastids: evidence for a single secondary endosymbiosis leading to haptophytes, cryptophytes, heterokonts, and dinoflagellates. J Mol Evol. 2006;62:143–157. doi: 10.1007/s00239-004-0305-3. [DOI] [PubMed] [Google Scholar]
- 14.Hackett JD, Yoon HS, Li S, Reyes-Prieto A, Rummele SE, Bhattacharya D. Phylogenomic analysis supports the monophyly of cryptophytes and haptophytes and the association of rhizaria with chromalveolates. Mol Biol Evol. 2007;24:1702–1713. doi: 10.1093/molbev/msm089. [DOI] [PubMed] [Google Scholar]
- 15.Patron NJ, Inagaki Y, Keeling PJ. Multiple gene phylogenies support the monophyly of cryptomonad and haptophyte host lineages. Curr Biol. 2007;17:887–891. doi: 10.1016/j.cub.2007.03.069. [DOI] [PubMed] [Google Scholar]
- 16.Burki F, Shalchian-Tabrizi K, Minge M, Skjaeveland A, Nikolaev SI, Jakobsen KS, Pawlowski J. Phylogenomics reshuffles the eukaryotic supergroups. PLoS ONE. 2007;2:e790. doi: 10.1371/journal.pone.0000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science. 2000;290:972–977. doi: 10.1126/science.290.5493.972. [DOI] [PubMed] [Google Scholar]
- 18.Gajadhar AA, Marquardt WC, Hall R, Gunderson J, Ariztia-Carmona EV, Sogin ML. Ribosomal RNA sequences of Sarcocystis muris, Theileria annulata and Crypthecodinium cohnii reveal evolutionary relationships among apicomplexans, dinoflagellates, and ciliates. Mol Biochem Parasitol. 1991;45:147–154. doi: 10.1016/0166-6851(91)90036-6. [DOI] [PubMed] [Google Scholar]
- 19.Yoon HS, Grant J, Tekle YI, Wu M, Chaon BC, Cole JC, Logsdon JM, Jr, Patterson DJ, Bhattacharya D, Katz LA. Broadly sampled multigene trees of eukaryotes. BMC Evol Biol. 2008;8:14. doi: 10.1186/1471-2148-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tyler BM, Tripathy S, Zhang X, Dehal P, Jiang RH, Aerts A, Arredondo FD, Baxter L, Bensasson D, Beynon JL, et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313:1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- 21.Huang J, Mullapudi N, Sicheritz-Ponten T, Kissinger JC. A first glimpse into the pattern and scale of gene transfer in the Apicomplexa. Int J Parasitol. 2004;34:265–274. doi: 10.1016/j.ijpara.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 22.Matsuzaki M, Kuroiwa H, Kuroiwa T, Kita K, Nozaki H. A Cryptic Algal Group Unveiled: A Plastid Biosynthesis Pathway in the Oyster Parasite Perkinsus marinus. Mol Biol Evol. 2008 doi: 10.1093/molbev/msn064. [DOI] [PubMed] [Google Scholar]
- 23.Slamovits CH, Keeling PJ. Plastid-Derived Genes in the Non-Photosynthetic Alveolate Oxyrrhis marina. Mol Biol Evol. 2008 doi: 10.1093/molbev/msn075. [DOI] [PubMed] [Google Scholar]
- 24.Aury JM, Jaillon O, Duret L, Noel B, Jubin C, Porcel BM, Segurens B, Daubin V, Anthouard V, Aiach N, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- 25.Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, Badger JH, Ren Q, Amedeo P, Jones KM, et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006;4:e286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grauvogel C, Brinkmann H, Petersen J. Evolution of the glucose-6-phosphate isomerase: the plasticity of primary metabolism in photosynthetic eukaryotes. Mol Biol Evol. 2007;24:1611–1621. doi: 10.1093/molbev/msm075. [DOI] [PubMed] [Google Scholar]
- 27.Martin W, Rujan T, Richly E, Hansen A, Cornelsen S, Lins T, Leister D, Stoebe B, Hasegawa M, Penny D. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc Natl Acad Sci U S A. 2002;99:12246–12251. doi: 10.1073/pnas.182432999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deusch O, Landan G, Roettger M, Gruenheit N, Kowallik KV, Allen JF, Martin W, Dagan T. Genes of cyanobacterial origin in plant nuclear genomes point to a heterocyst-forming plastid ancestor. Mol Biol Evol. 2008 doi: 10.1093/molbev/msn022. [DOI] [PubMed] [Google Scholar]
- 29.Zufall RA, McGrath CL, Muse SV, Katz LA. Genome architecture drives protein evolution in ciliates. Mol Biol Evol. 2006;23:1681–1687. doi: 10.1093/molbev/msl032. [DOI] [PubMed] [Google Scholar]
- 30.Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, Zhu G, Lancto CA, Deng M, Liu C, Widmer G, Tzipori S, et al. Complete Genome Sequence of the Apicomplexan, Cryptosporidium parvum. Science. 2004;304:441–445. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]
- 31.Martin W, Herrmann RG. Gene transfer from organelles to the nucleus: how much, what happens, and Why? Plant Physiol. 1998;118:9–17. doi: 10.1104/pp.118.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reyes-Prieto A, Hackett JD, Soares MB, Bonaldo MF, Bhattacharya D. Cyanobacterial contribution to algal nuclear genomes is primarily limited to plastid functions. Curr Biol. 2006;16:2320–2325. doi: 10.1016/j.cub.2006.09.063. [DOI] [PubMed] [Google Scholar]
- 33.Loftus B, Anderson I, Davies R, Alsmark UC, Samuelson J, Amedeo P, Roncaglia P, Berriman M, Hirt RP, Mann BJ, et al. The genome of the protist parasite Entamoeba histolytica. Nature. 2005;433:865–868. doi: 10.1038/nature03291. [DOI] [PubMed] [Google Scholar]
- 34.Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, Olsen GJ, Best AA, Cande WZ, Chen F, Cipriano MJ, et al. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science. 2007;317:1921–1926. doi: 10.1126/science.1143837. [DOI] [PubMed] [Google Scholar]
- 35.Teles-Grilo ML, Tato-Costa J, Duarte SM, Maia A, Casal G, Azevedo C. Is there a plastid in Perkinsus atlanticus (Phylum Perkinsozoa)? Eur J Protistol. 2007;43:163–167. doi: 10.1016/j.ejop.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 36.Huang J, Mullapudi N, Lancto CA, Scott M, Abrahamsen MS, Kissinger JC. Phylogenomic evidence supports past endosymbiosis, intracellular and horizontal gene transfer in Cryptosporidium parvum. Genome Biol. 2004;5:R88. doi: 10.1186/gb-2004-5-11-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frickey T, Lupas AN. PhyloGenie: automated phylome generation and analysis. Nucleic Acids Res. 2004;32:5231–5238. doi: 10.1093/nar/gkh867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moustafa A, Bhattacharya D. PhyloSort: a user-friendly phylogenetic sorting tool and its application to estimating the cyanobacterial contribution to the nuclear genome of Chlamydomonas. BMC Evol Biol. 2008;8:6. doi: 10.1186/1471-2148-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 41.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREEPUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- 43.Shimodaira H, Hasegawa M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
02