Cathepsin L Proteolytically Processes Histone H3 During Mouse Embryonic Stem Cell Differentiation (original) (raw)
. Author manuscript; available in PMC: 2009 Oct 17.
Summary
Chromatin undergoes developmentally-regulated structural and chemical changes as cells differentiate, which subsequently lead to differences in cellular function by altering patterns of gene expression. To gain insight into chromatin alterations that occur during mammalian differentiation, we turned to a mouse embryonic stem cell (ESC) model. Here we show that histone H3 is proteolytically cleaved at its N-terminus during ESC differentiation. We map the sites of H3 cleavage and identify Cathepsin L as a protease responsible for proteolytically processing the N-terminal H3 tail. In addition, our data suggest that H3 cleavage may be regulated by covalent modifications present on the histone tail itself. Our studies underscore the intriguing possibility that histone proteolysis, brought about by Cathepsin L and potentially other family members, plays a role in development and differentiation that was not previously recognized.
Introduction
Embryonic stem cells (ESCs) undergo dramatic changes in morphology, cell cycle, and gene expression as they differentiate into defined cell types (Kim et al., 2008; Murry and Keller, 2008). Since eukaryotic genomes are intimately associated with histone proteins to form chromatin, this physiologically-relevant structure must be remodeled as part of a large-scale mechanism to achieve rapid and drastic changes in gene expression (Arney and Fisher, 2004; Gan et al., 2007). For example, undifferentiated ESCs typically display increased physical plasticity and less compacted chromatin than their differentiated counterparts (Meshorer et al., 2006; Pajerowski et al., 2007). ESCs also undergo radical changes in gene expression as they differentiate, conveniently providing markers of “stemness” whose expression dramatically decreases (e.g. Oct 3/4) as differentiation progresses. Such changes suggest that cells undergo a significant reorganization of their genome during the differentiation process and that, moreover, this transition must be carefully regulated in order for the cell to differentiate properly and adopt a specific lineage.
Recent studies have shown that histone covalent modification patterns change significantly upon ESC differentiation (Giadrossi et al., 2007). For example, core histones (H2A, H2B, H3 and H4) are largely deacetylated upon differentiation and histone deacetylase activity may be required for ESC differentiation (Lee et al., 2004). Chromatin-immunoprecipitation (ChIP) experiments have also identified specific genes and/or genomic regions that change their “epigenetic signature” upon differentiation (Azuara et al., 2006; Bernstein et al., 2006a).
Despite a wealth of emerging data describing changing patterns of epigenetic signatures during ESC differentiation, very little is known about the mechanisms used to achieve such change. Several possible mechanisms for removing the more stable histone modifications, e.g. lysine methylation, include enzymatic demethylation, histone replacement, and regulated histone proteolysis (Bannister and Kouzarides, 2004). Enzymatic activities have been identified that carry out the first two mechanisms (Ahmad and Henikoff, 2002; Shi et al., 2004) and there is precedence for controlled histone H3-specific proteolysis (Allis et al., 1980; Falk et al., 1990); however, to our knowledge, specific, regulated, endogenous proteolysis has not been well documented in mammalian cells.
Here we show that ESCs employ regulated histone proteolysis in order to change their “epigenetic signature” upon differentiation. Furthermore, we have identified Cathepsin L as a developmentally-regulated histone H3 protease and demonstrated that its activity may be modulated by the modification of the histone tail itself. Taken together, our studies reveal novel nuclear functions of this family of cysteine proteases in histone and stem cell biology and suggest that controlled histone proteolysis may be part of a mechanism for introducing variation into the chromatin polymer.
Results
A Faster Migrating H3 Species Is Detected in Differentiating Mouse ESCs
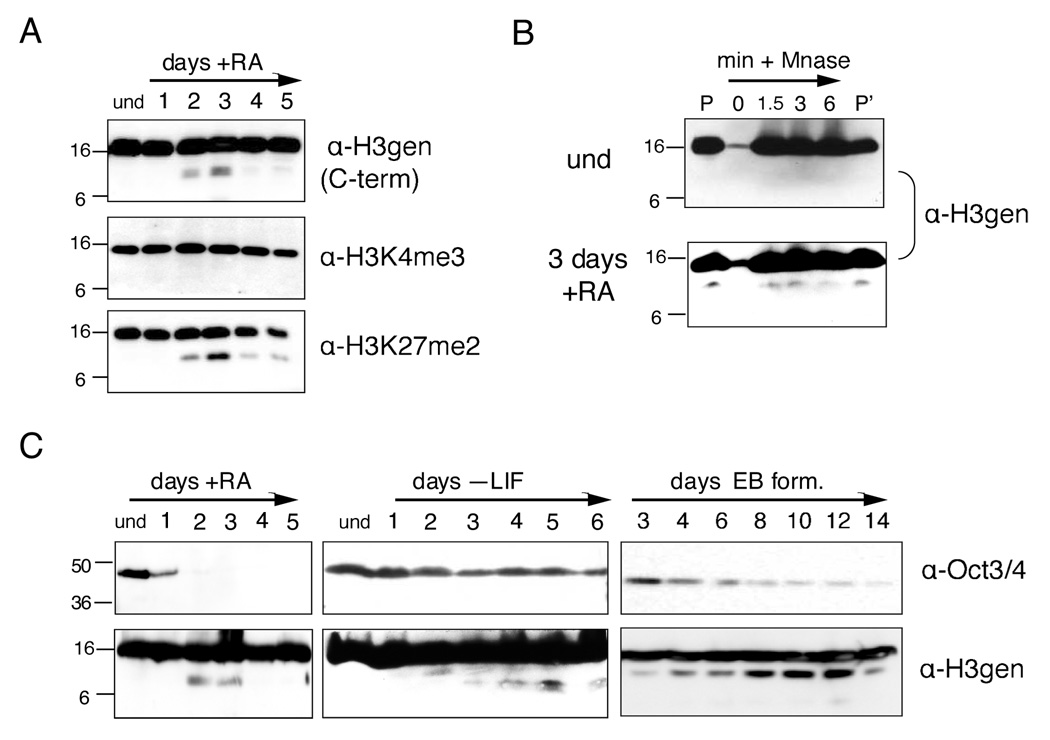
To survey changes in histone proteins and their modifications during mouse ESC differentiation, we used immunoblotting to probe whole-cell extracts (WCEs) with various histone antibodies. When probing with specific histone H3 antibodies (e.g. including the H3 general C-terminal, H3K27me2, and H3K27me1 antibodies), we observed reproducibly a faster migrating band in samples taken at time points corresponding to days two and three post-induction with retinoic acid (RA). Notably, this band(s) was observed using an H3 general antibody generated against the C-terminus of histone H3 (Figure 1A and Supplemental Figure S1), but not with an H3 general antibody generated against the first six N-terminal amino acids (Supplemental Figure S1). The faster migrating H3 species was also observed when probing immunoblots with an H3-K27me2 antibody (Figure 1A); in contrast, it was not recognized when replicate immunoblots were probed with the H3-K4me3 antibody (Figure 1A). Taken together, the results of these experiments suggested that an extreme amino-terminal fragment of H3 was missing in the faster-migrating H3 sub-band.
Figure 1. A distinct histone H3 species is detected in chromatin during ESC differentiation.
(A) Undifferentiated (und) ESCs were differentiated with RA in a monolayer, harvested for WCEs at the time points indicated, and analyzed by immunoblotting with the antibodies indicated to the right of each panel; H3gen refers to the H3 general C-terminal antibody unless otherwise indicated. Molecular weights (in kD) are indicated to the left in all subsequent gels and immunoblots.
(B) Chromatin was isolated from undifferentiated ESCs and ESCs differentiated with RA for 3 days, digested with micrococcal nuclease for the indicated times, and analyzed by immunoblotting; P=chromatin pellet input, P’=post-Mnase pellet.
(C) ESCs were differentiated using three basic methods: monolayer differentiation with RA (left), monolayer differentiation with LIF withdrawal (middle), and embryoid body (EB) formation by cell aggregation (right); WCEs were analyzed for both a marker of pluripotency (Oct 3/4) and the histone H3 sub-band (H3gen).
We then asked if the H3 sub-band was chromatin associated. Micrococcal-digested chromatin was prepared by standard methods (Wysocka et al., 2001) from both undifferentiated ESCs and ESCs undergoing differentiation with RA, and soluble mononucleosomes were probed with the H3 general antibody. As shown in Figure 1B, the faster migrating H3 band was seen in the undigested chromatin pellet as well as in the solubilized mononucleosomes derived from differentiating cells, but not the chromatin isolated from undifferentiated ESCs (Figure 1B).
We next asked if the appearance of the faster migrating H3 species was dependent on the methods employed to trigger ESC differentiation. To address this question, ESCs were differentiated using three different methods: monolayer differentiation with RA, monolayer differentiation by withdrawal of leukemic inhibitory factor (LIF), and embryoid body formation (EB formation) by cell aggregation. As shown in Figure 1C (upper panels), the time course for expression of pluripotency marker Oct3/4 differs for each of the induction methods employed, suggesting differences in the timing and progression of differentiation. Timing of the appearance of the faster migrating H3 band was also dependent on the method used to induce ESC differentiation (lower panels), suggesting that this event is also dependent on the progress of differentiation. The faster migrating H3 was observed at days two and three post RA induction during monolayer differentiation (Figure 1A and Figure 1C, left), but delayed until day five following withdrawal of LIF (Figure 1C, middle), the latter correlating with the similarly delayed decrease of Oct3/4. During EB formation, the faster migrating H3 species appeared early and then peaked between days eight and twelve (Figure 1C, right), suggesting a slower and more complex differentiation progression. We chose to use monolayer differentiation with RA for subsequent experiments in order to minimize differences in the timing and heterogeneity of differentiation.
Histone H3, Marked by Both “Active” and “Silent” Modifications, Is Proteolytically Cleaved in The N-terminal Tail During ESC Differentiation
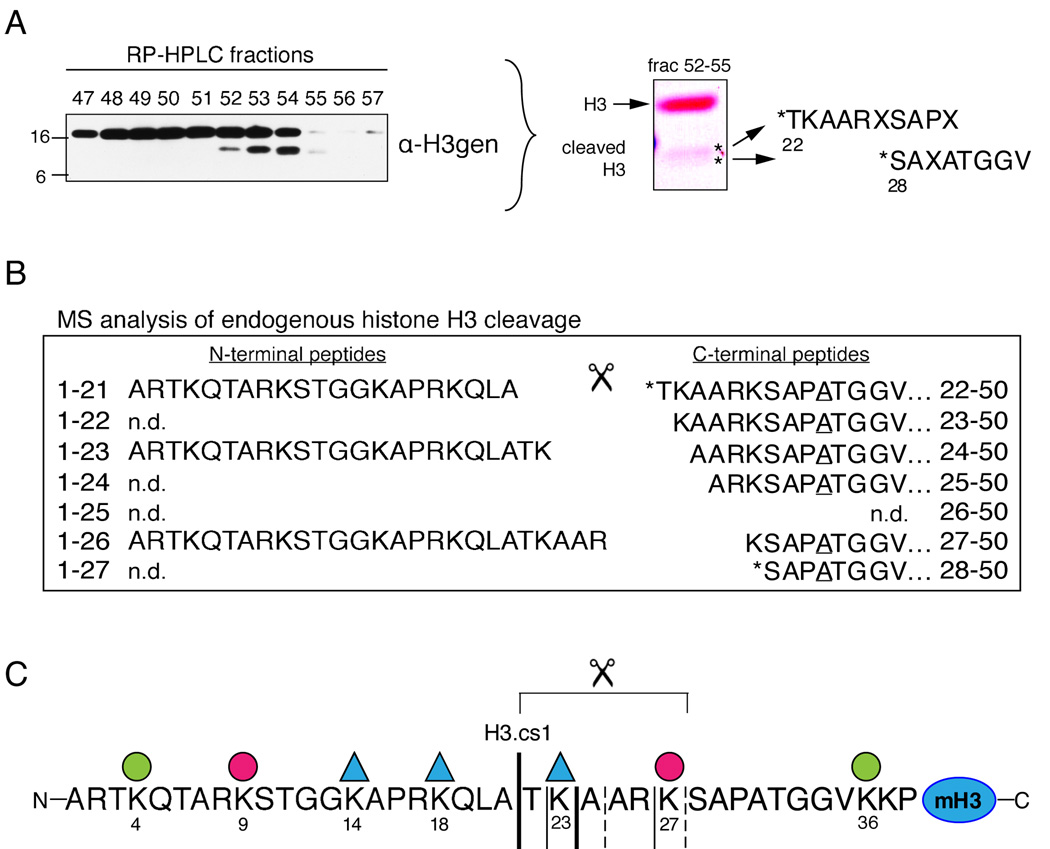
To determine the nature of the faster migrating H3 sub-species, histones were extracted from differentiating ESC nuclei three days post RA induction and separated by reverse phase high pressure liquid chromatography (RP-HPLC, C8 column, data not shown). Fractions containing the H3 sub-band were pooled and further resolved by RP-HPLC (C18 column), and the resulting fractions were screened by immunoblot as shown in Figure 2A (left). We then subjected C18-RP-HPLC fractions containing the H3 sub-band to two separate sequencing methods. First, equal amounts of fractions 52–54 were pooled, separated by SDS-PAGE, and transferred to immunoblot; the material in each of in the two sub-bands labeled by asteriks was then subjected to Edman degradation (Figure 2A, right). Although multiple amino acids were released during each cycle for both samples, the observed data strongly suggested that the top and bottom sub-bands contained sequences derived from cleavage following residues A21 and K27, respectively, in the N-terminal tail of H3.
Figure 2. Histone H3 is N-terminally cleaved during ESC differentiation.
(A) RP-HPLC fractions were screened for the H3 sub-band by immunoblotting (left). Equal amounts of fractions 52–55 were then pooled, separated by SDS-PAGE, and blotted to PVDF (middle). Each sub-band (asteriks) was excised and subjected to Edman degradation, which suggested that H3 had been N-terminally cleaved between residues A21 and T22 and between K27 and S28 (right). Residues not clearly identified in the generated sequence are denoted “X.”
(B) Sample in fraction 54 was digested with GluC to generate intact, N-terminal peptides that terminate with E50; peptides were then analyzed by MS. Six highly modified, truncated fragments of the GluC-generated 1–50 peptide were observed (right). Note that the two sequences detected by Edman degradation were also detected by MS (asterisks). All six of the truncated 1–50 peptides contain Ala at position 31 (underlined) and are thus derived from the H3 isoform H3.2. Three highly modified forms of the complementary N-terminal fragments, A1-A21, A1-K23, and A1-R26 (left) are also present in the same HPLC fraction.
(C) The sequence of the mammalian histone H3 tail and the cleavage sites mapped in (A) and (B); the bold solid line indicates the “primary” cleavage site mapped by both Edman degradation and MS (H3.cs1); additional significant cleavage sites are marked with regular solid lines; less abundant sites are marked by dashed lines. Lysines found by MS to be highly acetylated (ac) or methylated (me) are marked by a triangle or circle, respectively (see Supplemental Figure S2 for details).
Second, to define in greater detail the peptide sequences and post-translational modifications present in the faster migrating H3 species, additional sample from fraction 54 (Figure 2A) was digested with GluC to produce N-terminal H3-fragments ending in E50 and the resulting peptides were then analyzed by MS. Spectra recorded detected six, highly-modified, truncated, N-terminal, histone H3-peptides beginning at residues T22, K23, A24, A25, K27 and S28 (Figure 2B, right). Relative abundances of the six peptides suggest that the preferred cleavage sites are C-terminal to A21 and K23. These results suggest that the primary H3 cleavage site is between amino acids 21 and 22 of the amino terminus (Figure 2C, H3.cs1) and that the final cleavage site is between amino acids 27 and 28. Notably, the list of cleavage sites detected by MS contains the two sites detected by Edman degradation (asterisks).
In addition, new mass spectrometric methods using a combination of electron transfer dissociation/proton transfer charge reduction and accurate mass measurements were employed to characterize the modification patterns on the proteolyically cleaved H3 (Coon et al., 2005; Taverna et al., 2007). Interestingly, the data reveals that the cleaved H3 sub-species has a distinct covalent modification profile, suggesting that the H3 sub-band may be preferentially marked, before or after proteolytic processing, with a specific epigenetic signature (Figure 2C and Supplemental Figure S2). Specifically, marks of both “active” (e.g. H3K23Ac and H3K36me) and “silent” transcription (e.g. H3K27me) were reproducibly detected on a single GluC-digested peptide derived from the proteolytically-processed H3 fragment. Moreover, all six of the truncated peptides contained Ala at position 31, revealing that the cleaved H3 peptide is H3 isoform H3.2, not H3.3. Although non-cleaved H3.3 peptide was detected in the same RP-HPLC fraction as the cleaved H3 species, cleaved H3.3 was not detected. Since H3.2 and H3.1 elute in two separate peaks, we cannot definitively conclude that H3.2 is preferentially cleaved over H3.1, however significantly less H3 cleavage was detected in peak 2 (H3.1) than in peak 1 (H3.2+H3.3) by immunoblotting. Although it is unclear how this particular pattern of modifications and H3 isoform affect the mechanism of proteolysis, these data suggest the intriguing possibility that that regulation via post-translational modification of the substrate and/or isoform preference may regulate the proteolytic processing (see Figure 7).
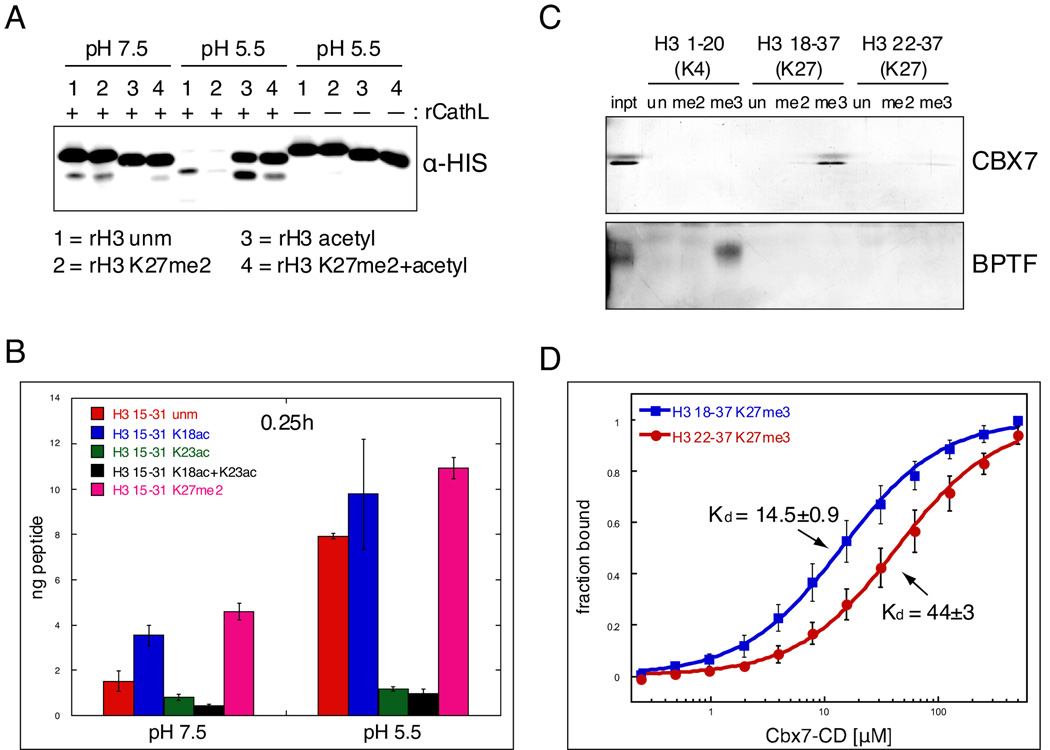
Figure 7. Covalent histone modifications modulate Cathepsin L activity and its downstream effects.
(A) rCathepsin L was incubated with each of four rH3 substrates: 1= rH3 unmodified, 2= rH3 alkylated to K27me2, 3= rH3 pan-acetylated with acetic anhydride, 4= rH3+K27me2 pan-acetylated with acetic anhydride.
(B) H3 cleavage reactions were performed as in (A) using synthesized peptides that represent the H3 tail from amino acids 15 to 31. Reactions were incubated with ~250pmol peptide and quenched with 0.1% TFA before being plated in duplicate for ELISA with the H3cs.1 antibody. Signal was normalized to that of mock reactions for each peptide. Results represent the mean of three independent experiments ± SD.
(C) Peptide pull-down assays were performed with the chromodomain of mouse CBX7 and the PHD finger of human BPTF.
(D) Fluorescence anisotropy Cbx7-CD protein binding to non-cleaved peptide (18–37) vs. cleaved peptide (22–37); binding decreases 3 fold with cleaved peptide, p<0.01. Kds are in µM±SEM. Data points represent the mean±SD.
Also detected in fraction 54 (Figure 2A) were highly modified forms for three of the most abundant, complementary, N-terminal fragments generated by proteolytic cleavage of H3 (Figure 2B, left). The detection of these intact, complementary cleavage products indicates that the cleavage sites mapped above are the result of endopeptidase activity alone. Post-translational modifications detected on these three N-terminal peptides again include marks of both “active” (e.g. H3K14Ac) and “silent” transcription (e.g. H3K9me) as seen on the C-terminal fragments (Supplemental Figure S2). These findings support the conclusion that a small fraction of total histone H3.2 undergoes highly specific endoproteolytic cleavage during ESC differentiation that may be regulated by unique patterns of covalent modifications.
The Lysosomal Cysteine Protease Cathepsin L is Present in Fractions That Are Enriched with H3 Protease Activity
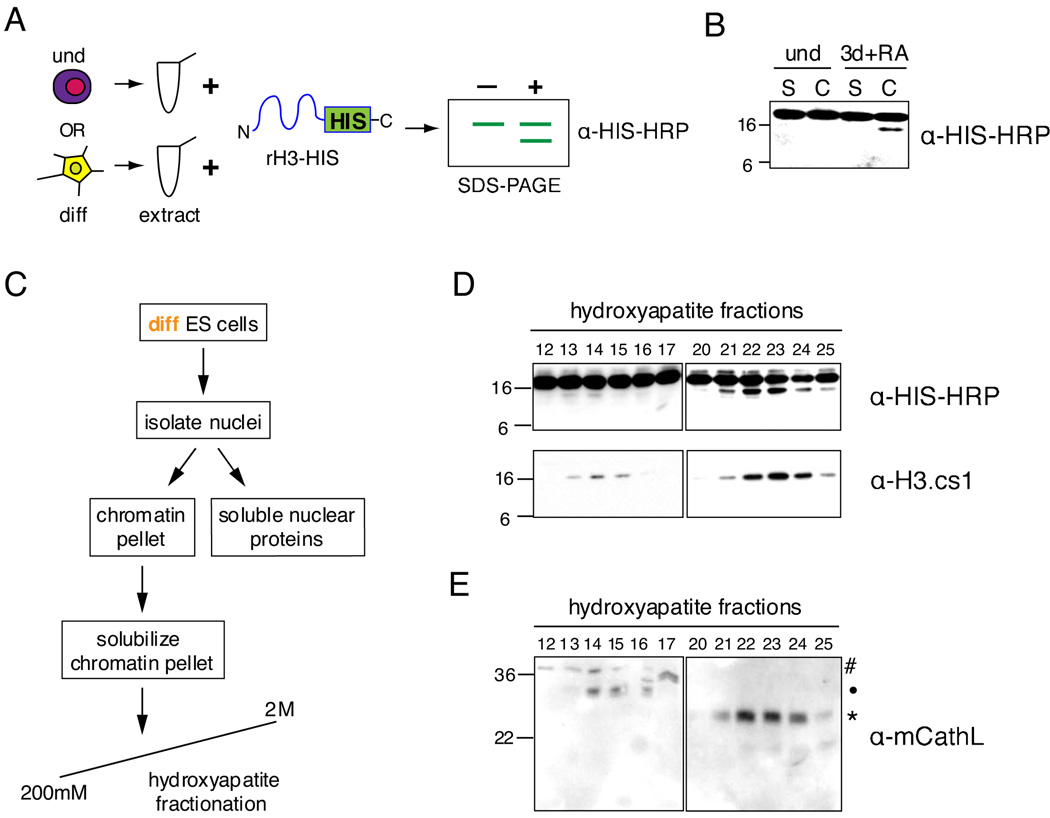
To identify and characterize the putative H3 protease(s), we first established an in vitro H3 cleavage assay. Undifferentiated or differentiating ESCs were harvested and their proteins extracted; these lysates were then incubated with C-terminally 6xHIS tagged, full-length recombinant histone H3 (rH3-HIS), and the reaction products were analyzed by immunoblotting with HIS-HRP antibody (Figure 3A). Although this assay has the potential to detect any N-terminal H3 cleavage, chromatin isolated from differentiating ESCs at the time point when in vivo cleavage was observed (~3 days +RA) consistently possessed strong in vitro H3 cleavage activity that reproduced the size difference of the endogenously cleaved H3 (Figure 3B).
Figure 3. The cysteine protease Cathepsin L is detected in fractions enriched for histone H3 cleavage activity.
(A) Schematic of in vitro H3 cleavage assay (see text for details).
(B) A representative example of the H3 cleavage assay comparing soluble cytosolic + nuclear protein extract (S) and solubilized chromatin extract (C) from undifferentiated to those prepared from 3 days + RA differentiating ESCs.
(C) Schematic of extract fractionation for protease enrichment.
(D) H3 cleavage assay of hydroxyapatite fractions generated by scheme shown in (C); assay reactions were analyzed by immunoblotting with both HIS-HRP and H3.cs1 antibodies.
(E) Hydroxyapatite fractions assayed in (D) were analyzed for the presence of Cathepsin L by immunoblotting with Cathepsin L antibody; # designates proprotein (~37kD), • indicates intermediate processed form (~30kD), and * indicates mature processed form (~25kD).
To better ensure that the cleaved H3 product observed in our in vitro cleavage assay was physiologically relevant, we generated an antibody that recognized the primary site of H3 cleavage, H3.cs1 (Figure 2C; see Experimental Procedures and Supplemental Figure S4 for details on antibody generation and characterization). Our results indicated that this antibody (“H3.cs1”) was specific for the primary site of in vivo generated H3 cleavage product (A21/T22) and was not sensitive to the acetylation status of H3K23 (see characterization by ELISA and immunoblotting in Supplemental Figure S4). Importantly, the H3.cs1 antibody fails to react with full length H3 and its signal is highly enriched at the same time point at which we detect an H3 sub-band using the H3-general or H3 K27me2 antibodies during our standard RA-induced ESC differentiation time course (Supplemental Figure S4B).
We then used both the HIS-HRP antibody and the H3.cs1 antibody in the above H3 cleavage assay to follow the biochemical enrichment of H3 protease activity in nuclear extract derived from differentiating ESCs. We fractionated nuclear extract (Figure 3C) and detected two peaks of putative H3 cleavage activity by both the HIS-HRP antibody and the H3.cs1 antibody (Figure 3D). We then subjected two activity-containing fractions (#22 and 23, Figure 3D) and an adjacent non-activity-containing fraction (#20) to MS analysis. Four peptides were detected for the cysteine protease Cathepsin L in both activity-containing fractions 22 and 23, yet none of these peptides were detected in the neighboring non-activity containing fraction 20 (see Supplemental Figure S5 for details). To validate this identification further, we used a commercially available antibody to screen for the presence of Cathepsin L in the hydroxyapatite fractions (mCathL, Figure 3E). Importantly, reactivity with mCathL correlates well with the detection of H3 cleavage activity in the fractions aligned above (compare Figure 3D to 3E).
Cathepsin L is known to exist in three forms: a proenzyme running at ~37kD, a single chain intermediate at ~30kD, and a double chain mature form at ~25kD and ~5kD (Ishidoh et al., 1998). The pro form must be cleaved to become active, whether by self-cleavage or by another enzyme (Turk et al., 2000); in contrast, both the “intermediate” and “mature” forms have been shown to be active (Mason et al., 1989). The hydroxyapatite fractions of the stronger peak of activity (~21–25) correlate with the detection of the mature form of Cathepsin L by immunoblot (Figure 3E, marked by asterisk), while the weaker activity fractions (~13–15) correlate closely with the detection of the intermediate form of Cathepsin L (Figure 3E, marked by dot). This correlation not only supports a causal relationship between the presence of Cathepsin L and the H3 cleavage activity, but also helps to explain the difference in elution and activity between the two peaks of H3 cleavage activity generated by the hydroxyapatite chromatography.
Fractions Enriched in H3 Cleavage Activity Exhibit Cathepsin L-like Activity
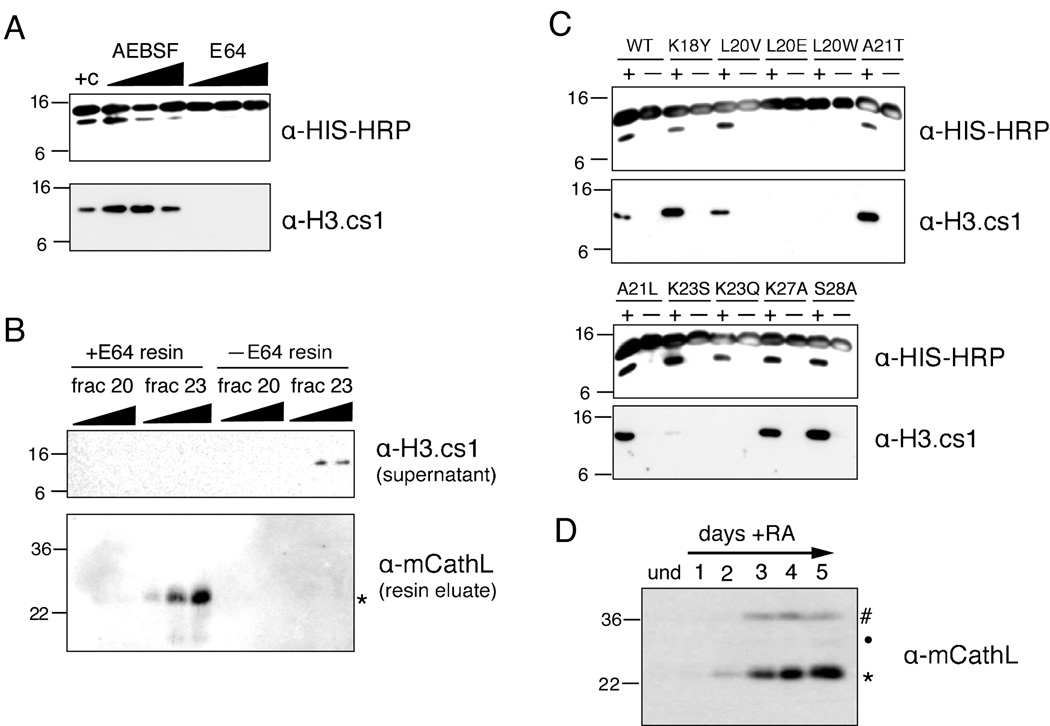
To validate further the identification of Cathepsin L as a histone H3 protease, we tested our enriched fractions with different classes of protease inhibitors in our H3 cleavage assay. As shown in Figure 4A, adding increasing amounts of serine protease inhibitor AEBSF produced a modest reduction in cleavage activity at a high concentration (20mM), but not at the lower concentrations tested (2mM and 10mM). However, when using the cysteine protease inhibitor E64, all cleavage was abolished at even the lowest concentration tested (10µM).
Figure 4. Cathepsin L cleaves histone H3 in vitro and associates with chromatin in vivo.
(A) Hydroxyapatite fraction #23 (Figure 3D) was assayed +/− protease inhibitors in the H3 cleavage assay; cysteine protease inhibitor E64 is a potent inhibitor of the H3 protease activity in fraction #23.
(B) Immobilized E64 was incubated with both an active hydroxyapatite fraction (23) and one without enzymatic activity (20); control resin was incubated with each fraction in parallel. Resins were then analyzed for bound proteins by immunoblotting (bottom panel) and the supernatant was tested for H3 protease activity (top panel).
(C) Hydroxyapatite fraction #23 (Figure 3D) was assayed with various rH3-HIS point mutants in the H3 cleavage assay.
(D) Chromatin from undifferentiated ESCs and ESCs differentiated +RA for the number of days indicated was digested with micrococcal nuclease; the solubilized mononucleosomes were analyzed by immunoblotting with Cathepsin L antibody.
Noting the strong inhibition of H3 cleavage activity by E64, an irreversible inhibitor that binds covalently to its substrate and has been shown to inhibit Cathepsin L (see Supplemental Figure S6 and Barrett et al., 1982), we then took advantage of a commercially available E64-bound resin to test whether it could precipitate Cathepsin L from our active hydroxyapatite fractions and, subsequently, remove the soluble H3 cleavage activity. Both E64 bound and control resins were added to fractions 20 (in which little to no H3 cleavage activity detected) and 23 (in which strong H3 cleavage activity was detected), incubated to allow binding and potential inhibition, and then pelleted by centrifugation. The cleared supernatant was subsequently tested for H3 cleavage activity. As expected, E64 resin successfully removed H3 cleavage activity from fraction 23, while control resin did not (Figure 4B, top). Importantly, as shown using the cleavage-site specific H3.cs1 antibody, the activity removed by the E64 resin included the specific A21/T22 cleavage site activity.
We then eluted any bound proteins from the E64 and control resins and probed the eluates for the presence of Cathepsin L. As hypothesized, mCathL was detected on the +E64 resin that had been incubated with fraction 23, but not on control resin nor on resin incubated with fraction 20 (Figure 4B, bottom). As a control, we also analyzed the above eluates by immunoblotting with an antibody to another cathepsin family member, Cathepsin B, but did not detect any immunoreactive species (data not shown). Taken together, the loss of activity from solution paired with the presence of Cathepsin L protein bound to the corresponding +E64 resin strongly suggests a causal relationship between this cysteine protease and the primary H3 protease activity characterized above (H3.cs1).
Cathepsin L is also known to preferentially cleave proteins that contain hydrophobic residues in their P2 position (two residues N-terminal from the cleaved bond, as originally defined (Schechter and Berger, 1967)), specifically leucine and phenylalanine (Rawlings and Barrett; Rawlings et al., 2008). Importantly, and in keeping with this characteristic, the sequence surrounding the primary H3 cleavage site (A21/T22) mapped from ES cells by both MS and Edman degradation (Figure 2C, solid line) includes a leucine at the P2 position (L20). To verify the significance of this leucine in the H3 sequence and support the identification of Cathepsin L in fraction 23, we mutated it (L20V, L20E & L20W) as well as neighboring residues and tested the mutated recombinant proteins in our H3 cleavage assay (Figure 4C). Although the more conservative L20V mutant showed little change in H3 cleavage, L20E and L20W mutants completely abolished H3 cleavage as assayed by both the HIS and H3.cs1 antibodies (top panels). Moreover, the loss of H3 cleavage upon mutation of L20, compared to little or no loss of signal upon mutation of neighboring residues (K18 or A21), suggests that L20 is particularly important to the enzymatic activity in fraction 23. Together, this mutational analysis also supports the identification of Cathepsin L as the enzyme responsible for the histone H3 cleavage characterized in the above in vitro assays.
Interestingly, mutating K23 to either S or Q also had a significant effect on the H3 cleavage activity (Figure 4C, bottom panels). Although no difference was seen in H3 cleavage by assaying with HIS, these mutations greatly diminished the cleavage site recognized by the H3.cs1 antibody, suggesting a possible role for this residue in regulating the precision of H3 proteolysis by Cathepsin L. It is interesting to note that none of the N-terminal peptides identified by MS were acetylated at K23 while C-terminal peptides acetylated at K23 were consistently detected (Supplemental Figure S2). Together, these data suggest the intriguing possibility that the acetylation of K23 may serve to inhibit cleavage at certain sites.
Cathepsin L Is Associated with Chromatin In Vivo
Although there is previous evidence that Cathepsin L and its activity exists in the cell nucleus of MEFs (Goulet et al., 2004), we set out to document this nuclear localization in ESCs using a biochemical approach: we isolated chromatin and then solubilized it by micrococcal nuclease digestion as described above (Figure 1C). As shown by probling with Cathepsin L antibody, Cathepsin L was indeed associated with chromatin fragments released by nuclease digestion (Figure 4D). Importantly, chromatin association only begins to appear upon differentiation and is not apparent in undifferentiated ES cells. We note as well that the mature form of Cathepsin L (marked by asterisk) was enriched over the full length (hash) or intermediate (dot) forms.
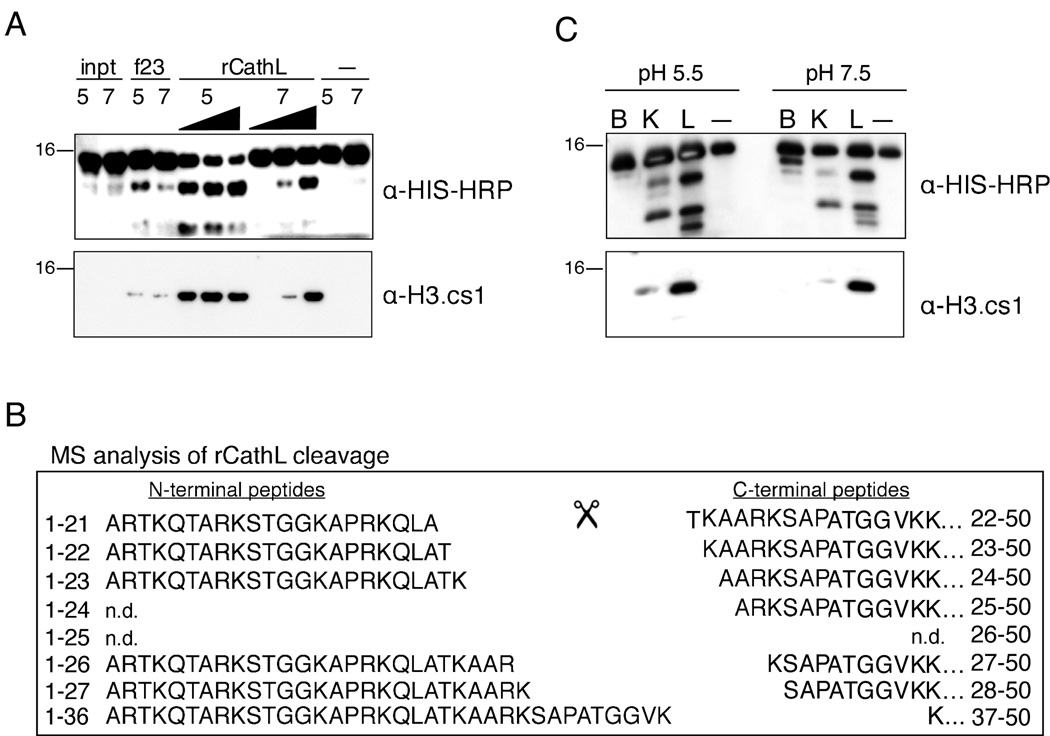
rCathepsin L Reproduces the In Vivo Histone H3 Cleavage Pattern
To further validate the identification of Cathepsin L as an H3 protease, we tested recombinant mouse Cathepsin L enzyme in our H3 cleavage assay (Figure 5). Since Cathepsin L displays optimal activity at the more acidic pH of the lysosome (i.e. ~pH 5, (Barrett et al., 2003)), and since we suspected that Cathepsin L is acting within the nucleus at a higher pH (~pH 7–8), we tested the recombinant enzyme in both pH environments. Importantly, recombinant mouse Cathepsin L was able to cleave recombinant histone H3 at both pH 5.5 and pH 7.4. Moreover, the recombinant enzyme cleaved H3 and created the specific epitope recognized by the cleavage site-specific H3.cs1 antibody (Figure 5A). To further characterize the sites of proteolytic cleavage, we analyzed the reaction products of rCathepsin L and rH3-HIS by MS. Importantly, MS analysis revealed that the six sites of cleavage mapped in the endogenous H3 samples (Figure 2) were also produced by rCathepsin L in vitro (Figure 5B). Summed ion currents for all charge states of the unmodified sequences were employed to estimate the relative abundances of the six peptides beginning with the indicated residues as follows: T22 > K23, A24, and S28 > A25 and K27. The most abundant site of cleavage in vitro was between A21 and T22, the same site that was shown to be most abundant in the MS analysis of endogenous histone H3. In addition, the relative abundances of the in vivo H3 cleavage sites and those produced by Cathepsin L in vitro are highly correlative. One exception is cleavage between residues K27 and S28, which is more abundant in the in vitro assay compared to that mapped from in vivo samples (the latter shows a preference for cleavage between residues R26/K27 over K27/S28). We speculate that the presence of histone modifications in vivo play a role in regulating the preferred cleavage site of Cathepsin L, particularly at sites surrounding H3K27, which was shown to be preferentially methylated on cleaved H3 by both immunoblot and MS analysis (Figure 1A and Supplemental Figure S2). Also observed in both reactions are five of the six complementary N-terminal peptides generated by rCathepsin L cleavage of H3 (left), and their abundances correlated with that of their C-terminal counterparts. Taken together, the above data support the conclusion that Cathepsin L is capable of generating all of the histone H3.2 fragments observed at day three following induction of ESC differentiation by retinoic acid.
Figure 5. rCathepsin L cleaves histone H3 in vitro.
(A) Recombinant Cathepsin L cleaves rH3 in vitro at both pH 5.5 and pH 7.4 and generates a fragment that is recognized by both α-HIS-HRP (top panel) and α-H3.cs1 (middle panel) antibodies.
(B) Recombinant mouse Cathepsin L was incubated with recombinant H3-HIS at both pH 5.5 and pH 7.4; after 2 hours, the reaction products were subjected to analysis by mass spectrometry. Both N-terminal (left) and C-terminal (right) fragments of the rH3 cleavage were detected; note the similarity to the pattern of in vivo cleavage shown in Figure 2B.
(C) Recombinant Cathepsins B, K, and L were pre-activated and incubated with recombinant H3-HIS at both pH 5.5 and pH 7.4 for 15 minutes; the reactions were separated by SDS-PAGE and analyzed by immunoblotting.
We then wanted to test other cathepsin family members in the H3 cleavage assay. We chose to test rCathepsin B, one of the most abundant lysosomal proteases, and rCathepsin K, which is reported to have a significant preference for leucine in the P2 position of its substrates much like Cathepsin L (Cathepsin S also shares this preference but was not found to have significant expression in our ESC model). Following pre-activation the recombinant enzymes, Cathepsins B, K and L were incubated with rH3-HIS at both pH 5.5. and pH 7.5, and the reactions were then analyzed by immunoblotting with both HIS and H3.cs1 antibodies. As shown in Figure 5C, pre-activated Cathepsin L cleaves rH3-HIS robustly and produces a pattern that is similar to that observed in vivo. In contrast, Cathepsin B cleaves rH3 with a distinct pattern from Cathepsins K or L and does not significantly produce the H3.cs1 epitope. Cathepsin K produces a similar pattern to Cathepsin L under these assay conditions, although they appear to differ in their preferences for specific sites (e.g. the cleavage site recognized by the H3cs.1 antibody is less abundant in the Cathepsin K reactions).
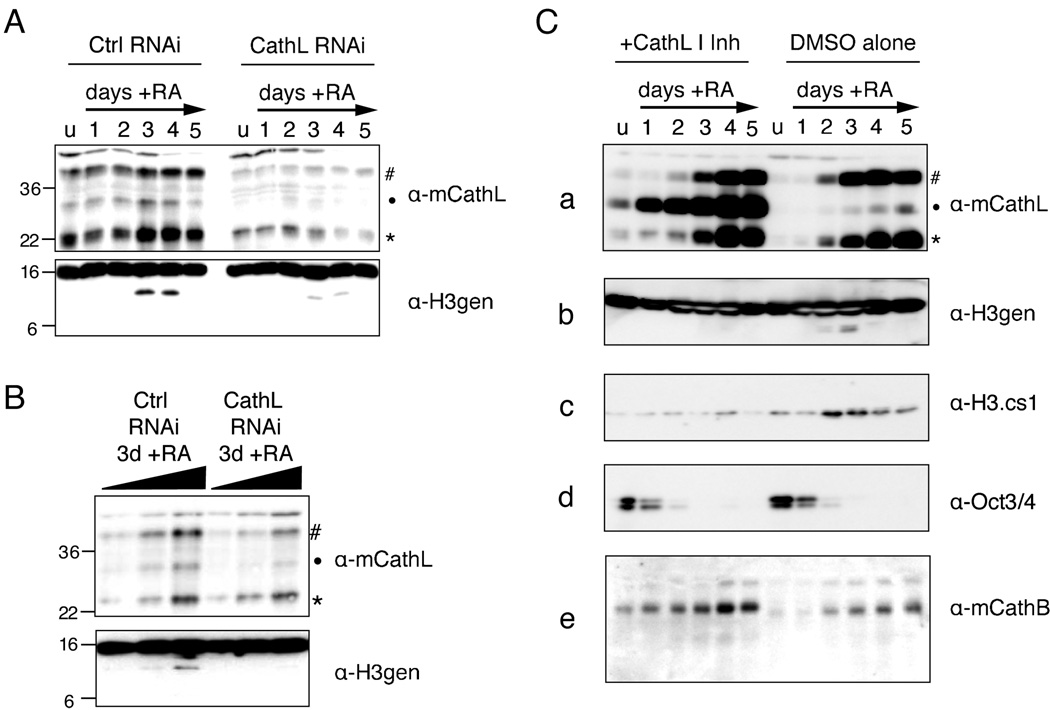
Both RNAi-mediated and Chemical Inhibition of Cathepsin L Inhibits H3 Cleavage In Vivo
We then sought to show that knockdown of Cathepsin L in vivo led to a decrease in H3 cleavage. Stable cells lines constitutively expressing short RNAi hairpins to a control gene and the Cathepsin L gene (Ctsl) were differentiated with RA, as usual, and cells were harvested at various time points. In parallel, samples taken at day 3 post-induction with RA were also evaluated for knockdown efficiency and H3 cleavage by titration of sample (Figure 6B). As shown in Figure 6A and B, knockdown of Cathepsin L (shown by immunoblotting with Cathepsin L antibody, upper panels) led to reproducible decrease in H3 cleavage as detected by the H3 C-terminal antibody (lower panels).
Figure 6. Both RNAi and chemical inhibition of Cathepsin L reduce histone H3 cleavage in vivo.
(A) Control and Ctsl RNAi cells lines were differentiated with RA as usual and harvested at the indicated time points; WCEs were then separated by SDS-PAGE and analyzed for both Cathepsin L expression (upper panel) and histone H3 cleavage (lower panel) by immunoblotting.
(B) A serial two-fold dilution of samples from day 3 post-induction with RA were resolved by SDS-PAGE gel and analyzed by immunoblotting as in (A).
(C) The addition of Cathepsin L Inhibitor I to the cell media of differentiating ESCs (left side) inhibits the processing of Cathepsin L itself (a) as well as that of histone H3 (b, c) as compared to DMSO alone treated control cells (right side). Loss of pluripotency marker Oct 3/4 was not affected (d) nor was the self-processing of another cathepsin family member, Cathepsin B (e).
To further assess whether Cathepsin L causes histone H3 cleavage in vivo, we chose to use a commercially available, cell-permeable Cathepsin L specific inhibitor, Cathepsin L Inhibitor I. Inhibition of Cathepsin L using a chemical inhibitor allowed us to assess the effect of Cathepsin L inhibition within a single, wild-type cell line rather than comparing individual cell lines created by drug selection. Undifferentiated ESCs were treated with or without inhibitor for 24 hours prior to plating for differentiation. Cells were then differentiated with RA while either Cathepsin L Inhibitor I (10µM) or DMSO alone was maintained in the media. As shown in Figure 6C, cells treated with inhibitor showed significant failure to fully process Cathepsin L from its intermediate form (marked by dot) into its mature form (asterisk), demonstrating that its activity had been inhibited (Figure 6C, panel a, left). In contrast, no significant accumulation of the intermediate form was seen in the DMSO alone treated cells (right). Notably, histone H3 cleavage also decreased significantly in the cells treated with the inhibitor, but not in cells treated with DMSO alone, as shown by probing immunoblots with both H3-general and H3.cs1 antibodies (Figure 6C, panels b and c). Interestingly, all cleavage detected by the H3-general antibody was abolished in the cells treated with inhibitor, suggesting that Cathepsin L is responsible for all sites of cleavage (supported by MS data in Figure 5).
Importantly, neither Oct-3/4 nor Cathepsin B levels (Figure 6C, panels d and e) appeared to change upon addition of the inhibitor. Since Oct-3/4 is a marker of pluripotency that is normally lost rapidly upon differentiation with RA (Figure 1C), the fact that this pattern is unchanged in the presence of the Cathepsin L Inhibitor I indicates that the inhibitor does not alter the pluripotency or differentiation capacity of ESCs prior to the time point at which H3 cleavage is observed. This conclusion is supported by Q-PCR data for the expression of the pluripotency marker Nanog (Supplemental Figure S8), which also remains unaffected by Cathepsin L inhibition. The consistency of Cathepsin B levels between those cells treated with Cathepsin L Inhibitor I and control cells suggests that the inhibitor is indeed specific for Cathepsin L. Markers of cell lineage were also analyzed by Q-PCR, and suggest modest changes in neural/ectodermal expression patterns and levels of endodermal marker expression between inhibitor treated and control cells (Supplemental Figure S8).
Histone Tail Modifications Can Modulate Cathepsin L Activity and Its Downstream Effects
We then asked whether known covalent modifications on the histone tail affect the cleavage activity of Cathepsin L. As depicted in Figure 2C, several amino acids near the sites of Cathepsin L cleavage are known to be modified by either acetylation (triangles) or methylation (circles). To test the potential effects of these modifications on the cleavage activity of Cathepsin L, we first turned to the H3 cleavage assay described in Figure 3. Four different recombinant H3 substrates were prepared as follows: unmodified rH3, rH3 dimethylated specifically at K27me2, rH3 “pan-acetylated” by treatment with acetic anhydride and rH3 with both K27me2 and pan-acetylation. These substrates were shown to have the specific modifications of interest by immunoblot (Supplemental Figure S7) and were verified by MS (≥ 90%, data not shown). As shown in Figure 7A, this assay demonstrated that acetylation of lysine residues greatly reduced cleavage of H3 by rCathepsin L at both pH 7.5 and 5.5 (compare substrate 1 to 3). In contrast, K27me2 increases H3 cleavage (compare substrate 1 to 2; greater depletion of full-length rH3+K27me2 suggesting increased cleavage activity at pH 5.5).
In order to test the effect of these modifications more quantitatively and specifically, we synthesized a set of five peptides with identical backbone sequence flanking the H3 cleavage site (H3 15–31) modified as follows: unmodified, K18 acetyl, K23 acetyl, K18ac+K23ac, and K27me2. We then employed an ELISA based assay using the H3cs.1 antibody to quantitatively measure the H3cs.1 cleavage activity of rCathepsin L on these peptides. As shown in Figure 7B, clear differences in rCathepsin L activity were observed at both pH 7.5 and pH 5.5. As suggested by the rH3 cleavage assay described above, K27me2 (magenta) strongly increases the ability of Cathepsin L to cleave at H3cs.1 as compared to the matched unmodified H3 15–31 peptide (red). Interestingly, acetylation at K18 (blue) also increases this activity, suggesting that the acetylation of another lysine(s) must be responsible for the abrogation of cleavage by acetylation demonstrated above. Our data suggest that acetylation at K23ac is at least partly responsible for this effect, as the K23ac peptide shows very little cleavage activity at H3cs.1, both alone (green) and in combination with K18ac (black).
We then asked what effect of the cleavage of the H3 tail might have on the binding of effector proteins. Since K27 methylation is a well-documented binding site for the chromodomain-containing protein Polycomb (Bernstein et al., 2006b; Fischle et al., 2003), we sought to test the effect of H3 cleavage on Pc binding to H3K27 methylation. Using peptides that mimicked either the “non-cleaved” H3 tail (H3 18–37) or the “cleaved” H3 tail (H3 22–37), we assayed the effects of H3 cleavage on the ability for the chromodomain of mouse Pc (CBX7-CD) to bind to K27 methylation (Figure 7C); the PHD-finger of the known H3K4 methyl binding protein BPTF was used as a positive control and to demonstrate specificity. As shown in Figure 7C and quantified in 7D, H3 cleavage significantly diminished the ability of CBX7 to bind to H3K27 methylation, suggesting that proteolytic modification introduced by Cathepsin L could lead to significant downstream effects.
Discussion
A rapidly growing literature demonstrates that cells undergo dramatic developmentally-regulated changes in gene expression and cellular morphology upon transition from undifferentiated stem cells to specific lineages (Giadrossi et al., 2007; Kim et al., 2008; Murry and Keller, 2008). Despite remarkable progress made in documenting epigenetic signatures such as “bivalent domains” i.e. an H3 tail bearing both H3K4me3 and H3K27me3 marks (Bernstein et al., 2006a), little is known as to what mechanisms may function to bring about the above changes at a chromatin level. Our data suggest that mouse ESCs employ a novel, regulated histone H3 proteolysis mechanism that may serve to alter epigenetic signatures upon differentiation. Other means of actively removing histone methyl marks have been well documented, including enzymatic demethylation (Anand and Marmorstein, 2007; Shi et al., 2004) and selective histone variant replacement (Ahmad and Henikoff, 2002). However, considerably less evidence documenting endogenous histone proteolysis has been reported (Allis et al., 1980; Falk et al., 1990). The identification and characterization of developmentally-regulated H3 cleavage by Cathepsin L during ESC differentiation is an important step in understanding limited nuclear histone proteolysis as a potential mode of transcriptional regulation.
Our finding that Cathepsin L cleaves histone H3 in mouse ES cells was unexpected as this enzyme was originally described as a lysosomal protease (Barrett et al., 2003). However, Cathepsin L has been shown to localize to nuclei where it plays a role in the proteolytic processing of transcription factor CDP/Cux (Goulet et al., 2004); biochemical studies have also shown that pro-Cathepsin L is localized to the nucleus in _ras_-transformed mouse fibroblasts (Hiwasa and Sakiyama, 1996). In addition, evidence of endogenous, nuclear serpin inhibitors with the ability to inhibit Cathepsin L activity, e.g. MENT (Bulynko et al., 2006) and Cystatin B (Riccio et al., 2001), further supports the notion that Cathepsin L, and potentially other cysteine proteases, play important but poorly understood roles in regulated nuclear proteolysis. As far as we are aware, transcription factor CDP/Cux is the only nuclear substrate of Cathepsin L identified to date, and histones have not yet been identified as substrates of this class of proteases in mammalian cells. Interestingly, however, recent studies in sea urchin have suggested that a Cathepsin L-like cysteine protease may be responsible for the degradation of sperm histones during a key chromatin remodeling event after fertlization (Morin et al., 2008). Our data support other studies that demonstrate the nuclear localization of Cathepsin L and provide the first indication that cellular histones, H3 in particular, are key substrates of this family of proteases in mammalian cells.
Our finding that Cathepsin L is an H3 protease is interesting when considering the phenotype common to the Cathepsin L knockout mouse (Nakagawa et al., 1998) and the Cathepsin L mutant mouse, furless (Roth et al., 2000). These mice exhibit periodic hair loss due to the improper cycling and morphogenesis of their hair follicles, suggesting a defect in stem cell renewal and/or differentiation. Cathepsin L knockout mice are viable and fertile, however, indicating that its functions are nonessential and/or redundant. Interestingly, Cathepsin L/Cathepsin B double knockout mice exhibit severe brain atrophy and die two to four weeks after birth (Felbor et al., 2002). The severity and selectivity of this phenotype suggests that these two enzymes overlap in their specific functions. Although we do not observe significant inhibition of Cathepsin B upon in vivo chemical inhibition of Cathepsin L, nor do we observe the same pattern of H3 cleavage with recombinant Cathepsin B compared to Cathepsin L in vitro, we cannot exclude the possibility that redundancy in H3 cleavage function may exist between these or other related enzymes at other stages or lineages of differentiation.
Limited proteolysis of nuclear proteins is an important means of regulating transcription and other cell processes (Goulet and Nepveu, 2004; Vogel and Kristie, 2006). Here we show that limited proteolysis of histone H3 by Cathepsin L occurs during the differentiation of ESCs and may be regulated both positively and negatively by covalent modifications on the H3 tail itself. These findings require a revaluation as to the function of this family of enzymes in transcriptional and epigenetic regulation; many important questions remain. First, how is the protease cleavage activity regulated? Along this line, we show that covalent modifications (such as acetylation and/or methylation of nearby lysines) serve to regulate, positively and negatively, the H3 proteolytic processing event. We note that proteolytic processing of H3 in the ciliate model occurs selectively in a hypoacetylated, transcriptionally silent (micronuclear) genome, while processing of H3 fails to occur in hyperacetylated, transcriptionally active macronuclei (Allis et al., 1980). These data raise the question as to whether other chromatin-modifying enzyme complexes, such as HATs, HDACs, or ubiquitinating enzymes, play a role in regulating histone proteolysis.
Second, by what mechanism is the cleaved H3 replaced, and is DNA replication and chromatin assembly required? Our findings that the histone variant H3.2 is preferably cleaved as compared to H3.3 suggests that S-phase/replication-coupled replacement may be involved (Loyola and Almouzni, 2007). Along this line, we note that proteolytic processing of transcription factor CDP/Cux by Cathepsin L occurs during the G1/S-phase transition and is coupled to cell cycle progression (Goulet et al., 2004). Whether proteolytic processing of H3 is cell cycle dependent remains unclear, although it is interesting to note that our studies were done in ESCs, which cycle rapidly and spend a high percentage of time in S-phase as measured by flow cytometry (data not shown).
Third, and more broadly, does this proteolytic mechanism apply to other histone substrates or to other cathepsin family members or cathepsin-like proteases, and is the general mechanism conserved in other organisms or operating in other stages of development or differentiation? Although Cathepsin L is the only member of this family shown to localize to the nucleus, the conservation in genomic structure and sequence between this protease and its related family members suggest that other cathepsins may function similarly.
Finally, with other mechanisms of histone demethylation well documented (i.e. enzymatic demethylation, variant replacement, etc.), does histone proteolysis serve other purposes that are not appreciated, such as the generation of new N-termini? Alternatively, histone cleavage may remove critical recognition elements and thereby block the binding of downstream effectors (see Figure 7D). Such questions will be the aims of future studies.
Experimental Procedures
Cell culture and differentiation
ES cells (LF2 line, gift from Austin Smith) were cultured and differentiated with RA as previously described (Bernstein et al., 2006b; Smith, 1991). For more detail, see Supplemental Experimental Procedures.
Cellular extract preparation
Whole cell extracts were prepared by resuspending cell pellets in SDS-Laemmli sample buffer and boiling immediately. Chromatin extracts were prepared by sonicating chromatin pellets in buffer (10mM HEPES, 10mM KCl, 1.5mM MgCl2, 0.34M sucrose, 10% glycerol, 5mM β-mercaptoethanol) after isolation by various methods (Dignam et al., 1983; Mendez and Stillman, 2000). Digestion of chromatin fractions into mononucleosomes was accomplished by treatment with microccocal nuclease as described (Wysocka et al., 2001). For more detail, see Supplemental Experimental Procedures.
Histone purification and mapping of H3 cleavage sites
Histones were acid extracted from nuclei and purified by RP-HPLC as described (Shechter et al., 2007). H3 sub-bands were excised and subjected to Edman degradation as described previously (Strahl et al., 1999). Fraction 54 was digested and analysed by MS-MS as described in Supplemental Experimental Procedures.
In vitro H3 cleavage assay
Extracts were incubated in buffer (10mM HEPES, 10mM KCl, 1.5mM MgCl2, 0.34M sucrose, 10% glycerol, 5mM β-mercaptoethanol, all final) with 0.1µg/µl rH3-HIS (purified from E. coli), incubated at 37°C for 1–3 hours, and analyzed by immunoblotting with HIS-HRP and/or H3.cs1 antibody. For MS analysis, reactions were quenched by addition of 0.1% (final) TFA. rCathepsin L (R&D Systems 1515-CY-010), rCathepsin B (R&D Systems BAF965), and rCathepsin K (Calbiochem 342001) were purchased from commercial sources.
Enzyme enrichment and identification
3 days +RA differentiating ESC chromatin pellets were solubilized by sonication in buffer A (80mM NaPO−4, 200mM NaCl), injected onto a 2 or 5mL hydroxyapatite column (BioRad), and fractionated with a 200mM to 2M NaCl gradient. Each fraction was then assayed as described above and subjected to MS analysis (see Supplemental Experimental Procedures).
Antibodies
The following antibodies were purchased from commercial vendors: Cathepsin L (R&D Systems AF1515), Cathepsin B (R&D Systems AF965), Penta-HIS HRP Conjugate Kit (Qiagen 34460), Oct3/4 (BD Transduction Laboratories 611202), H3gen (Abcam 1791), H3K4me3 (Abcam 8580), H3K27me2 (Millipore 07–452). See Supplemental Experimental Procedures for details on the generation of the H3cs.1 and N-terminal H3gen antibodies.
Plasmid construction and recombinant protein purification
See Supplemental Experimental Procedures.
RNAi knockdown
Short hairpins were purchased from Open Biosystems (RHS3979–9628371 and RMM3981–9597987) and transfected into 293T cells to produce Lentiviral particles. Heterogeneous ESC lines were created by infection and selection with puromycin.
Binding Assays
Peptide pull-down assays were performed as described (Wysocka, 2006) using biotinylated peptides conjugated to streptavidin agarose beads (Pierce 20349). Elutions were loaded onto 10 or 15% SDS-PAGE gels and silver stained. Fluorescence anisotropy was performed as described (Bernstein et al., 2006b) using FAM-labeled peptides. For details on quantification, see Supplemental Experimental Procedures.
Supplementary Material
01
Acknowledgements
We thank J. Wysocka, T. Swigut, A. Ruthenburg, D. Shechter, L, Banaszynski, and B. Ueberheide for their advice on biochemical and ESC methods, and J. Wysocka and D. Shechter for their critical reading of the manuscript. Peptides were synthesized at the proteomics facilities at RU and Tufts. E.M.D. is an Anderson Cancer Center fellow (The Rockefeller University). This work was funded by institutional support from The Rockefeller University and grants from the Tri-Institutional Stem Cell Initiative (funded by the Starr Foundation) and National Institutes of Health Grants GM49351 (to J.R.P), GM37537 (to D.F.H), and GM53512 (to C.D.A).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- Allis CD, Bowen JK, Abraham GN, Glover CV, Gorovsky MA. Proteolytic processing of histone H3 in chromatin: a physiologically regulated event in Tetrahymena micronuclei. Cell. 1980;20:55–64. doi: 10.1016/0092-8674(80)90234-2. [DOI] [PubMed] [Google Scholar]
- Anand R, Marmorstein R. Structure and mechanism of lysine-specific demethylase enzymes. J Biol Chem. 2007;282:35425–35429. doi: 10.1074/jbc.R700027200. [DOI] [PubMed] [Google Scholar]
- Arney KL, Fisher AG. Epigenetic aspects of differentiation. J Cell Sci. 2004;117:4355–4363. doi: 10.1242/jcs.01390. [DOI] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. Histone methylation: recognizing the methyl mark. Methods Enzymol. 2004;376:269–288. doi: 10.1016/S0076-6879(03)76018-2. [DOI] [PubMed] [Google Scholar]
- Barrett AJ, Kembhavi AA, Brown MA, Kirschke H, Knight CG, Tamai M, Hanada K. L-trans-Epoxysuccinyl-leucylamido(4-guanidino)butane (E-64) and its analogues as inhibitors of cysteine proteinases including cathepsins B, H and L. Biochem J. 1982;201:189–198. doi: 10.1042/bj2010189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett AJ, Rawlings ND, Woessner JF, editors. Handbook of Proteolytic Enzymes. 2 addition edn. Academic Press; 2003. [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006a;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol. 2006b;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulynko YA, Hsing LC, Mason RW, Tremethick DJ, Grigoryev SA. Cathepsin L stabilizes the histone modification landscape on the Y chromosome and pericentromeric heterochromatin. Mol Cell Biol. 2006;26:4172–4184. doi: 10.1128/MCB.00135-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coon JJ, Ueberheide B, Syka JE, Dryhurst DD, Ausio J, Shabanowitz J, Hunt DF. Protein identification using sequential ion/ion reactions and tandem mass spectrometry. Proc Natl Acad Sci U S A. 2005;102:9463–9468. doi: 10.1073/pnas.0503189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk MM, Grigera PR, Bergmann IE, Zibert A, Multhaup G, Beck E. Foot-and-mouth disease virus protease 3C induces specific proteolytic cleavage of host cell histone H3. J Virol. 1990;64:748–756. doi: 10.1128/jvi.64.2.748-756.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felbor U, Kessler B, Mothes W, Goebel HH, Ploegh HL, Bronson RT, Olsen BR. Neuronal loss and brain atrophy in mice lacking cathepsins B and L. Proc Natl Acad Sci U S A. 2002;99:7883–7888. doi: 10.1073/pnas.112632299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Q, Yoshida T, McDonald OG, Owens GK. Concise review: epigenetic mechanisms contribute to pluripotency and cell lineage determination of embryonic stem cells. Stem Cells. 2007;25:2–9. doi: 10.1634/stemcells.2006-0383. [DOI] [PubMed] [Google Scholar]
- Giadrossi S, Dvorkina M, Fisher AG. Chromatin organization and differentiation in embryonic stem cell models. Curr Opin Genet Dev. 2007;17:132–138. doi: 10.1016/j.gde.2007.02.012. [DOI] [PubMed] [Google Scholar]
- Goulet B, Baruch A, Moon NS, Poirier M, Sansregret LL, Erickson A, Bogyo M, Nepveu A. A cathepsin L isoform that is devoid of a signal peptide localizes to the nucleus in S phase and processes the CDP/Cux transcription factor. Mol Cell. 2004;14:207–219. doi: 10.1016/s1097-2765(04)00209-6. [DOI] [PubMed] [Google Scholar]
- Goulet B, Nepveu A. Complete and limited proteolysis in cell cycle progression. Cell Cycle. 2004;3:986–989. [PubMed] [Google Scholar]
- Hara K, Kominami E, Katunuma N. Effect of proteinase inhibitors on intracellular processing of cathepsin B, H and L in rat macrophages. FEBS Lett. 1988;231:229–231. doi: 10.1016/0014-5793(88)80737-3. [DOI] [PubMed] [Google Scholar]
- Hiwasa T, Sakiyama S. Nuclear localization of procathepsin L/MEP in ras-transformed mouse fibroblasts. Cancer Lett. 1996;99:87–91. doi: 10.1016/0304-3835(95)04041-2. [DOI] [PubMed] [Google Scholar]
- Hsieh JJ, Cheng EH, Korsmeyer SJ. Taspase1: a threonine aspartase required for cleavage of MLL and proper HOX gene expression. Cell. 2003;115:293–303. doi: 10.1016/s0092-8674(03)00816-x. [DOI] [PubMed] [Google Scholar]
- Ishidoh K, Saido TC, Kawashima S, Hirose M, Watanabe S, Sato N, Kominami E. Multiple processing of procathepsin L to cathepsin L in vivo. Biochem Biophys Res Commun. 1998;252:202–207. doi: 10.1006/bbrc.1998.9613. [DOI] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Hart SR, Skalnik DG. Histone deacetylase activity is required for embryonic stem cell differentiation. Genesis. 2004;38:32–38. doi: 10.1002/gene.10250. [DOI] [PubMed] [Google Scholar]
- Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyola A, Almouzni G. Marking histone H3 variants: how, when and why? Trends Biochem Sci. 2007;32:425–433. doi: 10.1016/j.tibs.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Mason RW, Wilcox D, Wikstrom P, Shaw EN. The identification of active forms of cysteine proteinases in Kirsten-virus-transformed mouse fibroblasts by use of a specific radiolabelled inhibitor. Biochem J. 1989;257:125–129. doi: 10.1042/bj2570125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin V, Sanchez A, Quinones K, Huidobro JG, Iribarren C, Bustos P, Puchi M, Geneviere AM, Imschenetzky M. Cathepsin L inhibitor I blocks mitotic chromosomes decondensation during cleavage cell cycles of sea urchin embryos. J Cell Physiol. 2008 doi: 10.1002/jcp.21459. [DOI] [PubMed] [Google Scholar]
- Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Roth W, Wong P, Nelson A, Farr A, Deussing J, Villadangos JA, Ploegh H, Peters C, Rudensky AY. Cathepsin L: critical role in Ii degradation and CD4 T cell selection in the thymus. Science. 1998;280:450–453. doi: 10.1126/science.280.5362.450. [DOI] [PubMed] [Google Scholar]
- Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher DE. Physical plasticity of the nucleus in stem cell differentiation. Proc Natl Acad Sci U S A. 2007;104:15619–15624. doi: 10.1073/pnas.0702576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ. MEROPS: the peptidase database. Cambridge CB10 1SA, UK: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Morton FR, Kok CY, Kong J, Barrett AJ. MEROPS: the peptidase database. Nucleic Acids Res. 2008;36:D320–D325. doi: 10.1093/nar/gkm954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccio M, Di Giaimo R, Pianetti S, Palmieri PP, Melli M, Santi S. Nuclear localization of cystatin B, the cathepsin inhibitor implicated in myoclonus epilepsy (EPM1) Exp Cell Res. 2001;262:84–94. doi: 10.1006/excr.2000.5085. [DOI] [PubMed] [Google Scholar]
- Roth W, Deussing J, Botchkarev VA, Pauly-Evers M, Saftig P, Hafner A, Schmidt P, Schmahl W, Scherer J, Anton-Lamprecht I, et al. Cathepsin L deficiency as molecular defect of furless: hyperproliferation of keratinocytes and pertubation of hair follicle cycling. FASEB J. 2000;14:2075–2086. doi: 10.1096/fj.99-0970com. [DOI] [PubMed] [Google Scholar]
- Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- Shechter D, Dormann HL, Allis CD, Hake SB. Extraction, purification and analysis of histones. Nat Protoc. 2007;2:1445–1457. doi: 10.1038/nprot.2007.202. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Smith A. Journal of Tissue Culture Methods. 1991;13:89–94. [Google Scholar]
- Strahl BD, Ohba R, Cook RG, Allis CD. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc Natl Acad Sci U S A. 1999;96:14967–14972. doi: 10.1073/pnas.96.26.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna SD, Ueberheide BM, Liu Y, Tackett AJ, Diaz RL, Shabanowitz J, Chait BT, Hunt DF, Allis CD. Long-distance combinatorial linkage between methylation and acetylation on histone H3 N termini. Proc Natl Acad Sci U S A. 2007;104:2086–2091. doi: 10.1073/pnas.0610993104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk B, Turk D, Turk V. Lysosomal cysteine proteases: more than scavengers. Biochim Biophys Acta. 2000;1477:98–111. doi: 10.1016/s0167-4838(99)00263-0. [DOI] [PubMed] [Google Scholar]
- Vogel JL, Kristie TM. Site-specific proteolysis of the transcriptional coactivator HCF-1 can regulate its interaction with protein cofactors. Proc Natl Acad Sci U S A. 2006;103:6817–6822. doi: 10.1073/pnas.0602109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J. Identifying novel proteins recognizing histone modifications using peptide pull-down assay. Methods. 2006;40:339–343. doi: 10.1016/j.ymeth.2006.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Reilly PT, Herr W. Loss of HCF-1-chromatin association precedes temperature-induced growth arrest of tsBN67 cells. Mol Cell Biol. 2001;21:3820–3829. doi: 10.1128/MCB.21.11.3820-3829.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01