Deletion of Cavin/PTRF causes global loss of caveolae, dyslipidemia and glucose intolerance (original) (raw)
. Author manuscript; available in PMC: 2009 Oct 1.
SUMMARY
Caveolae are specialized invaginations of the plasma membrane found in numerous cell types. They have been implicated as playing a role in a variety of physiological processes and are typically characterized by their association with the caveolin family of proteins. We show here by means of targeted gene disruption in mice, that a distinct caveolae-associated protein, Cavin/PTRF, is an essential component of caveolae. Animals lacking Cavin have no morphologically detectable caveolae in any cell type examined and have markedly diminished protein expression of all three caveolin isoforms whilst retaining normal or above normal caveolin mRNA expression. Cavin knockout mice are viable and of normal weight but have higher circulating triglyceride levels, significantly reduced adipose tissue mass, glucose intolerance and hyperinsulinemia, which characteristics constitute a lipodystrophic phenotype. Our results underscore the multi-organ role of caveolae in metabolic regulation and the obligate presence of Cavin for caveolae formation.
INTRODUCTION
Caveolae are 50–100 nm invaginations of cell surface membranes that have been extensively studied for their possible function in signal transduction, membrane and lipid trafficking. Their numbers are especially abundant in endothelial cells, adipocytes and skeletal muscle (Parton and Simons, 2007; Pilch et al., 2007). Caveolae require the expression of caveolins, a family of 22–24 kD integral membrane proteins consisting of caveolin-1 (Rothberg et al., 1992) (Cav1), which is predominantly expressed in adipocytes, fibroblasts, and endothelial cells (Parton and Simons, 2007; Pilch et al., 2007); Cav2, which is expressed in the same cells that express Cav1 (Scherer et al., 1996); and Cav3, which is specifically expressed in skeletal, cardiac, and certain vascular smooth muscle beds (Tang et al., 1996; Way and Parton, 1995). Under most experimental conditions, the ectopic expression of Cav1 or Cav3 in caveolin deficient cultured cells appears to be sufficient for the formation of morphologically defined caveolae, whereas ectopic expression of Cav2 is insufficient in this regard and requires the co-expression of Cav1, with which it forms hetero-oligomers in caveolae (Parton et al., 2006). Thus, the accepted paradigm is that caveolins are the sole or major structural component of caveolae whose expression is necessary and sufficient for their biogenesis. However, recent results from several labs have identified Cavin (also known as PTRF for Polymerase I and Transcript Release Factor (Jansa et al., 1998)) as an abundant peripheral membrane protein that is resident on the cytoplasmic face of caveolae (Aboulaich et al., 2004; Pilch et al., 2007; Vinten et al., 2001). It is detected only in this plasma membrane domain when visualized by immune-electron microscopy of gold-labeled anti-Cavin antibody (Vinten et al., 2005; Vinten et al., 2001). Moreover, the distribution of Cavin in rodents coincides with those tissues that express both Cav1 and Cav3 (Vinten et al., 2001). We and others have recently shown that over or under expression of Cavin in cultured cells leads to a parallel change in Cav1 protein (Hill et al., 2008; Liu and Pilch, 2008) and caveolae abundance (Hill et al., 2008). Thus, we hypothesized that Cavin may be of physiological importance and a requisite protein component of caveolae.
Specific biochemical and physiological roles for caveolae have been difficult to ascribe with certainty and many aspects of their putative functions are controversial. However, mice and humans deficient in their expression display numerous abnormalities that underscore their physiological importance (Le Lay and Kurzchalia, 2005). Cav3 deficiency causes limb girdle muscular dystrophy and rippling muscle disease in humans and in mice (Betz et al., 2001; McNally et al., 1998; Minetti et al., 1998; Vorgerd et al., 2001; Woodman et al., 2004), and very recently, Cav1 deficiency in humans has been shown to cause a type X lipodystrophy (Cao et al., 2008; Kim et al., 2008). Deficiencies of caveolae in mice have also been shown to play a role in additional pathologies including cardiovascular disease, diabetes, cancer and pulmonary fibrosis (Cohen et al., 2004).
Thus, to explore the importance of Cavin in vivo, we generated knockout mice lacking this protein. The resultant animals lack caveolae in all tissues examined, underscoring a previously unrecognized obligatory role for Cavin in Cav3-dependent caveolae formation in skeletal muscle. Cavin deficient mice have a unique metabolic phenotype of reduced adipose tissue content and hyperlipidemia while expending less energy than their wild type counterparts, apparently due to an inability to maintain equivalent physical activity. Consistent with the observed reduced fat mass, the RNA levels are diminished for important adipocyte genes such as the adipokines leptin and adiponectin. The observed dyslipidemia is also consistent with our in vitro data showing that caveolin expression allows enhanced fatty acid partitioning into cells (Meshulam et al., 2006). In addition to hyperlipidemia, Cavin null mice exhibit glucose intolerance, hyperinsulinemia and a reduction in proteins associated with insulin signaling in adipocytes and skeletal muscle. Taken together, impairment of these processes causes a lipodystrophic and insulin resistant phenotype not unlike that very recently described for humans deficient in Cav1 (Cao et al., 2008; Kim et al., 2008).
RESULTS
Generation of Cavin null mice
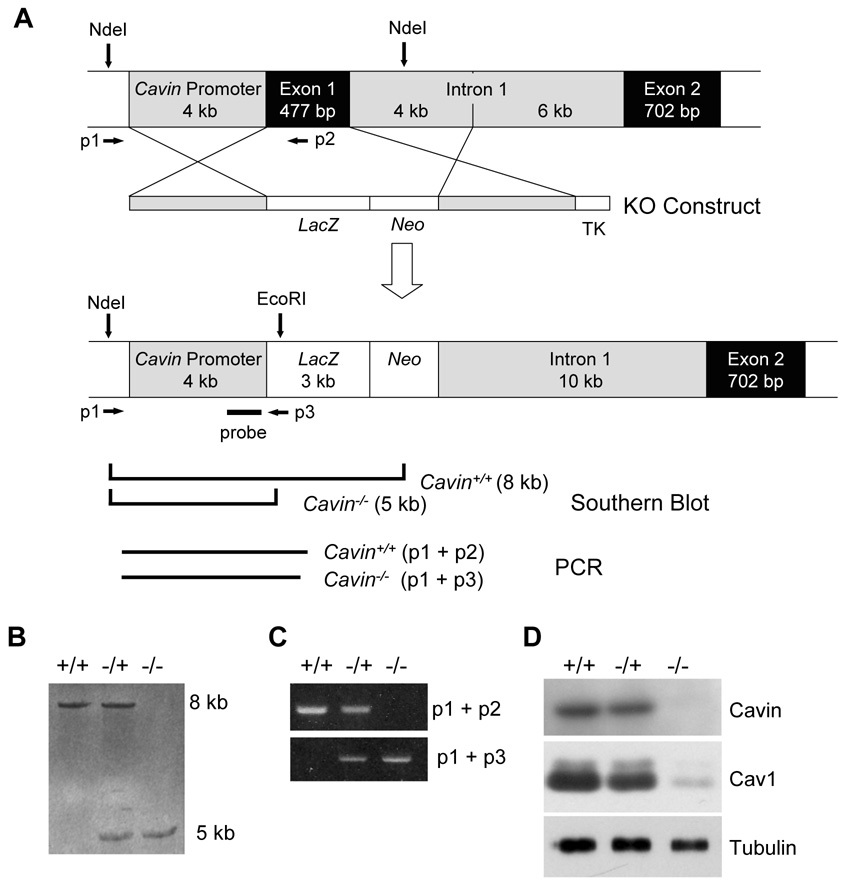
We replaced part of exon 1 of the Cavin gene (Ptrf) with a LacZ/Neo fusion cassette as shown in Figure 1A. The resultant expression of β-galactosidase was observed in muscle and endothelial cells but was absent in liver (data not shown), consistent with the known tissue distribution of Cavin protein (Vinten et al., 2001) (see also Figure 2D). Analyses of genomic DNA by Southern blot (Figure 1B), RNA by PCR (Figure 1C) and protein by Western blot (Figure 1D) confirmed the gene disruption and absence of Cavin gene products in mice homozygous for the targeted allele (Cavin−/−). Mice heterozygous for the targeted allele (_Cavin_−/+) were grossly normal and exhibit reduced amounts of Cavin and Cav1 protein (Figure 1D). Homozygous animals are viable and fertile but exhibit a number of metabolic abnormalities (see below). In all subsequent studies, gender-, strain- and age-matched homozygous knockout animals were compared to wild type controls.
Figure 1.
Generation of Cavin knockout mice. The Cavin knockout mice were generated using a previously described strategy involving lacZ insertion (Yang et al., 2006). (A) Schematic representation of the structure of the targeting vector and restriction enzyme sites of the Cavin gene locus before and after homologous recombination. Exon 1 in Cavin gene was replaced by the gene encoding prokaryotic β-galactosidase, designated here as LacZ. (B) Genotype analysis by Southern blotting of offspring from Cavin heterozygote (Cavin+/−) mice intercrosses to generate homozygote (_Cavin_−/−) mice is described in Experimental Procedures. Probing of EcoRI and NdeI digested genomic DNA revealed 8.4 Kb and 5.3 Kb fragments for wild-type and knockout genes, respectively. (C) PCR analysis was also devised as a 3-primer PCR-based screening strategy. The common forward primer (p1) was derived from the Cavin promoter region before the recombination site, and the wild-type specific reverse primer was derived from Cavin exon 1 (p2). The knockout-specific reverse primer was derived from the LacZ cassette (p3). Primer sequences are provided in Supplemental Experimental Procedures. (D) Total protein from lung and adipocytes of wild type (Cavin+/+), heterozygote (Cavin+/−) and knockout (_Cavin_−/−) mice was examined by Western blotting with anti-Cavin, anti-Cav1 and anti-tubulin antibodies. See Supplemental Experimental Procedures for details.
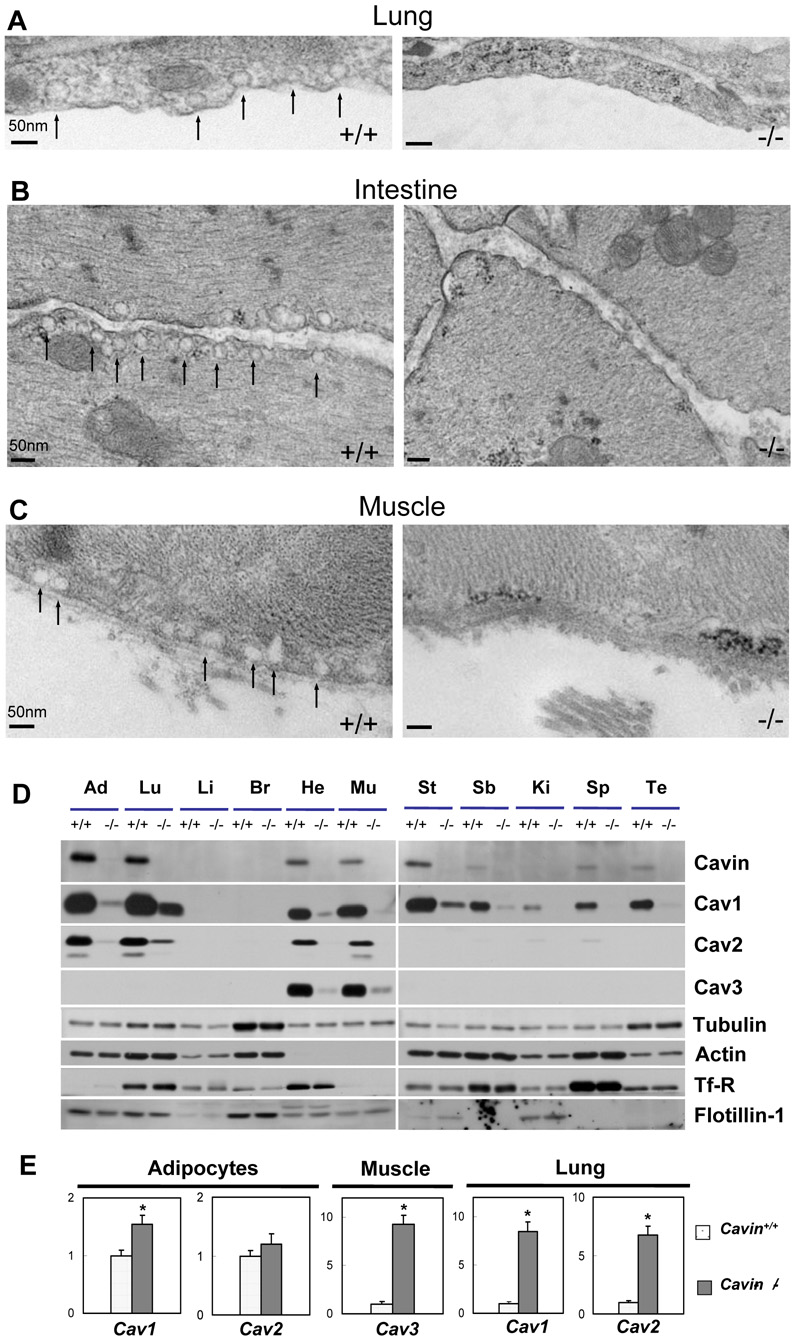
Figure 2.
The absence of Cavin leads to global loss of caveolae, down-regulation of protein and up-regulation of mRNA expression levels for caveolin isoforms and caveolae resident proteins. Electron micrographs of lung (A), smooth muscle (B) and skeletal muscle (C) tissues in wild type (Cavin+/+) and Cavin knockout mice (_Cavin_−/−). Analysis was performed on 8 weeks old male Cavin+/+ or _Cavin_−/− mice of identical strain. The caveolae from wild type mice showed the normal small invaginations (arrows). The caveolae structures have disappeared from lung, skeletal muscle and smooth muscle. (D) Whole cell lysates from wild type (Cavin+/+) and Cavin knockout mice (_Cavin_−/−) (10 weeks old strain-matched males) from the indicated tissues were prepared in RIPA buffer. Protein (10–50 µg) was subjected to SDS-PAGE, and immunoblotted with the indicated antibodies. Abbreviations are: Ad, Lu, Li, Br, He, Mu, St, SB, Ki, Sp, and Te, which represent adipocytes, lung, liver, brain, heart, muscle (mixed hind limb skeletal), stomach, small bowel, kidney, spleen and testis, respectively. (E) Quantitative RT-PCR (Supplemental Experimental Procedures) was performed in adipocytes, lung and muscle tissue by amplification with primers specific for Cav1, Cav2 and Cav3. Data were normalized to 18S rRNA and expressed as fold change relative to the mRNA levels in wild type samples. The statistical significance of differences between wild type and Cavin knockout mice (asterisks) was determined with Student's t test, *P < 0.01. Each result represents the average value and SE of at least three independent experiments.
Cavin deficient mice lack caveolae
Analysis of selected tissues from homozygous Cavin null mice by electron microscopy revealed that caveolae are completely absent from lung epithelium (Figure 2A), intestinal smooth muscle (Figure 2B) and skeletal muscle (Figure 2C) as well as in endothelial cells from all of these tissues. Caveolae were abundantly present in these same tissues in wild type mice (Cavin+/+) (Figure 2A–C, left hand panels). The absence of caveolae in both lung epithelium and skeletal muscle is particularly notable because different caveolin isoforms, Cav1 and Cav3 respectively, are required for caveolae formation in these tissues (Parton and Simons, 2007). Thus, Cavin is required for the formation and/or stabilization of morphologically defined caveolae in all tissues, regardless of the caveolin isoforms expressed.
Ablation of Cavin reduces expression of all caveolin isoforms
At the protein level, the amount of caveolin was markedly reduced in all tissues examined from Cavin knockout mice including in lung and adipocytes where caveolae are most abundant. It is again notable that Cav3 protein was virtually absent from heart and skeletal muscle (Figure 2D). Note that the expression of Cav1 in these tissues from wild type mice (Figure 2D) is due to its presence in vascular cells, not in myocytes. As expected, Cav2 protein expression is reduced in the Cavin null mice to essentially the same magnitude as Cav1 although there is some variation in their ratio depending on the tissue. There were no differences in total actin and tubulin levels between wild type and knockout animals (Figure 2D), similar to our results obtained by knocking down Cavin in cultured adipocytes (Liu and Pilch, 2008). There was also no change in transferrin receptor (Tf-R) expression, as would be expected for a plasma membrane marker known to be absent from caveolae (Liu and Pilch, 2008), nor was there any change in flotillin expression, a marker for non-caveolae lipid raft domains (Bickel et al., 1997). Interestingly, the expression of Cav1 & 2 mRNAs in adipocytes from Cavin null mice was slightly increased and was markedly higher in lung, as was Cav3 expression in skeletal muscle, all as compared to wild type animals (Figure 2E). Thus in selected tissues, caveolin gene expression is significantly increased under these circumstances, a possible compensatory response to the lack of caveolae.
Dyslipidemia and glucose intolerance in Cavin deficient mice
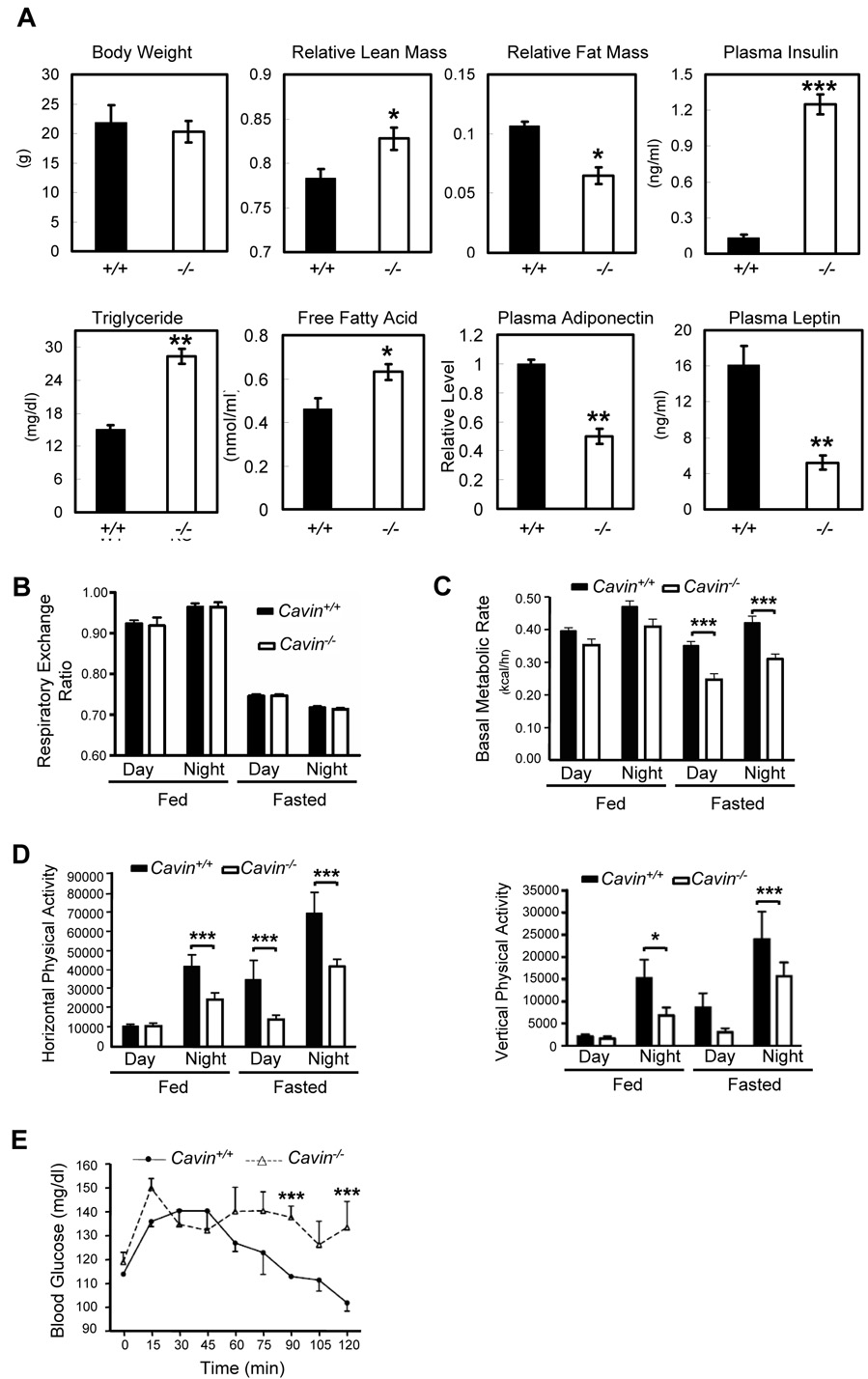
Given the abnormalities observed in caveolin deficient mice and humans, we next measured a number of metabolic parameters associated with carbohydrate and lipid metabolism (Figure 3A). Cavin null mice are viable and fertile and exhibit essentially the same weight, but they display a leaner body mass than wild type animals. Moreover, the knockout mice have significantly elevated serum triglycerides and free fatty acids as well as dramatically increased levels of insulin. They also exhibit reduced leptin and adiponectin expression and together, these parameters are characteristic of certain congenital lipodystrophies (Agarwal and Garg, 2006) and insulin resistance. Consistent with this interpretation, the animals have an impaired whole body glucose tolerance (Figure 3E) as compared to gender- and age-matched wild type animals, and interestingly, display a lower resting metabolic rate (Figure 3C) and decreased habitual physical activity (Figure 3D) with equivalent utilization of fuel sources (Figure 3B). Thus the lean phenotype of the Cavin null mice and reduced metabolic capacity despite hyperlipidemia most likely results from the reduced capacity for physical activity that these animals display.
Figure 3.
Analysis of metabolic parameters in cavin null and wild type mice. (A) Body weight, body composition, plasma insulin, triglyceride, free fatty acid, adiponectin, leptin levels were measured in Cavin+/+ and _Cavin_−/− mice as described in Experimental Procedures. Mice were individually housed in metabolic chambers under fed and fasted conditions and oxygen consumption and carbon dioxide production were measured for determination of (B) respiratory exchange ratio (RER), (C) basal metabolic rate and (D) infrared beam breaks were used to quantify ambulatory activity. (E) Glucose tolerance was assessed in Cavin+/+ and _Cavin_−/− mice and values are expressed relative to basal concentrations. Results are presented as means ± standard error. The statistical significance of differences was determined by Student's _t_-test. The effects of group and time were determined by two-way analysis of variance. When group effects were significant (p < 0.05), Tukey-Kramer sub-analyses were performed to identify statistically significant differences between Cavin+/+ and _Cavin_−/− mice. (4-male and 4-female per group, *P < 0.05, **P < 0.01, ***P < 0.001).
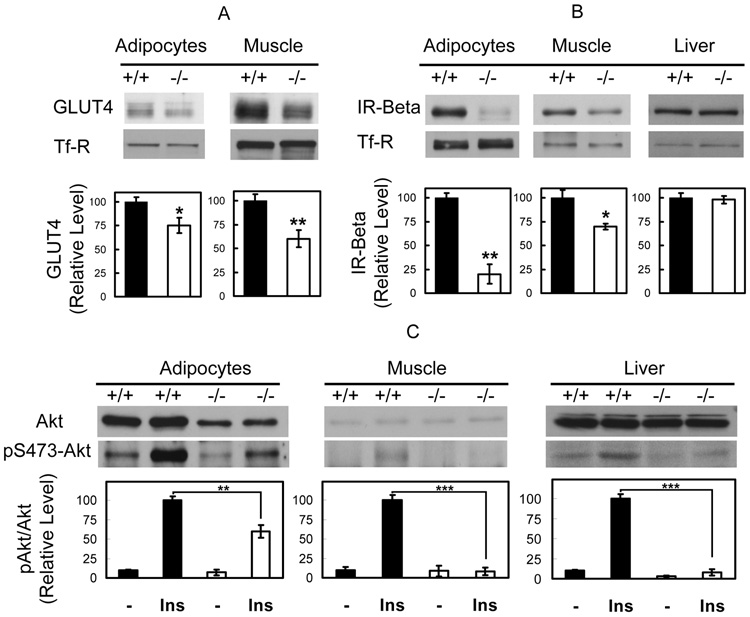
Insulin action is impaired in Cavin null mice
Glucose intolerance is associated with insulin resistance and an inability of normal circulating insulin levels to clear blood glucose, primarily into skeletal muscle (Petersen and Shulman, 2002). In addition, since insulin stimulated glucose transport is required for triglyceride formation in adipocytes, reduced insulin action in fat cells also contributes to insulin resistance (Abel et al., 2001). Accordingly, we measured the level and activity of several proteins involved in insulin action in these tissues, namely Glut4 (Figure 4A), the insulin receptor beta subunit (Figure 4B) and Akt2 (Figure 4C), the latter being the ultimate kinase target of insulin signaling to glucose transport (Ng et al., 2008). Glut 4 levels were slightly diminished in adipocytes (75%) and muscle (60%) compared to wild type animals. As expected, transferrin receptors were unchanged in null versus wild type mice. Insulin receptor levels were markedly reduced in fat, less so in muscle and not at all in liver. Insulin-dependent Akt phosphorylation (S473) in fat, muscle and liver was significantly reduced in the Cavin null as compared to wild type animals (Figure 4C), consistent with the overall insulin resistant phenotype. Glut4 is the transporter isoform that is responsible for insulin-dependent glucose uptake in fat and muscle (Pilch, 2008), and therefore a reduction in insulin signaling together with diminished Glut4 levels can easily account for the observed dyslipidemia, glucose intolerance and insulin resistance seen for Cavin null mice.
Figure 4.
GLUT4 expression is reduced and Insulin signaling is impaired in Cavin null mice. After insulin injection (i.p. 0.75 U/kg), animals were sacrificed and whole cell lysates of wild type (Cavin+/+) and Cavin knockout mice (_Cavin_−/−) (16 weeks old strain-matched males) from adipocyte, muscle and liver tissues were prepared in RIPA buffer. Representative western blots and quantitative bar graphs are shown for the detection of (A) GLUT4, (B) IR-beta protein levels and (C) total Akt and Akt S473 phosphorylation levels. The statistical values are displayed as means and SE of 3 independent experiments. (*P < 0.05, **P < 0.01, ***P < 0.001).
DISCUSSION
Since the discovery that caveolin (now Cav1) is a protein component of caveolae (Rothberg et al., 1992), the scientific community has been intensely interested in these structures, and this interest is underscored by the more recent revelations of human pathology due to “caveolinopathies” (Betz et al., 2001; Cao et al., 2008; Kim et al., 2008; Le Lay and Kurzchalia, 2005; McNally et al., 1998; Minetti et al., 1998; Vorgerd et al., 2001; Woodman et al., 2004). Despite this robust research activity, there has been little consensus as to how caveolae function and previous hypotheses, such as their role as a general scaffold for transmembrane signal transduction pathways (Shaul and Anderson, 1998), have been called into question by the relatively normal signaling seen in Cav1 knockout mice. One aspect of caveolae biology, however, that is supported by a variety of recent studies from numerous groups is a role in adipocytes for lipid storage and release (Pilch et al., 2007), and our current study further supports this hypothesis. Thus, Cav1 has been found in association with lipid droplets (Liu et al., 2002; Liu et al., 2004; Parton and Simons, 2007; van Meer, 2001) and has been postulated to traffic to these structures from the cell surface (Le Lay et al., 2006). Ectopic Cav1 expression in HEK293 cells enhances their ability to move exogenously added free fatty acids to the cytosol (Meshulam et al., 2006) and also enhances the ability of these cells to store triglyceride (Meshulam et al, unpublished). We have suggested that caveolins function in this regard by their ability to form detergent resistant membranes domains that “buffer” high fatty acid concentrations (Meshulam et al., 2006). This can protect cells against the potential cytotoxic effects of high levels of fatty acids, which are themselves mild detergents. The lipodystrophy we see in Cavin deficient mice, which is very similar to that recently been reported for Cav1 deficient humans (Cao et al., 2008; Kim et al., 2008), is entirely consistent with this hypothesis.
Our data show that the absence of Cavin eliminates caveolae thus impairing the ability of adipocyte to store triglycerides, which in turn leads to the observed increase in circulating lipids, glucose intolerance and insulin resistance. Because the animals have a reduced respiratory capacity and are less active, the concentration of circulating lipids remains high. Elevated levels of fatty acids correlate with insulin resistance in numerous animal models as well as in humans (Boden, 1997; Kraegen et al., 2001), and this is also the situation in the Cav1 deficient lipodystrophic patients (Cao et al., 2008; Kim et al., 2008). Thus, while there are likely to be multiple molecular mechanisms of insulin resistance (Hoehn et al., 2008; Muoio and Newgard, 2008), reduced insulin signaling and GLUT4 levels in muscle, such we observe in the Cavin null mice, are one such mechanism.
How do Cavin null animals compare with Cav1 and Cav3 null mice regarding metabolic parameters? Cav1 null mice of an age comparable to those in the present study (8–12 weeks) have a normal respiratory capacity, normal basal insulin and glucose levels (Razani et al., 2002). At this age, they do have hypertriglyceridemia with normal free fatty acid levels, but they are not glucose intolerant. Cav1 null mice go on to show adipocyte abnormalities that are revealed at >6 months of age, at which time, they are significantly leaner and are more resistant to diet induced obesity than their wild type littermates (Razani et al., 2002). Thus, the Cav1 deficient phenotype is significantly milder than that we see for Cavin deficiency, where metabolic defects are observed at a much younger age. Cav3 null animals, on the other hand, are fatter than wild type mice, even at 8–9 weeks, and they do exhibit impaired glucose tolerance and insulin resistance without an increase in circulating lipids (Capozza et al., 2005). They also have a reduced insulin signaling capacity similar to what we show here. Thus the Cav3 deficient animals also differ significantly from the Cavin nulls and although double Cav1/3 knockout animals have also been described (Park et al., 2002) and might be expected to resemble Cavin knockout mice, their metabolic parameters have not been determined.
In summary, the data presented here document that Cavin/PTRF expression is an essential requirement in mammals for the formation of caveolae in vivo in all tissues. Our results would therefore suggest the need for a revised model to explain caveolae structure and biogenesis because Cavin, and probably other cytoplasmic and/or membrane proteins in addition to Cav1 or Cav3, is absolutely necessary for the formation and/or stability of caveolae. Our Cavin deficient animals represent a model of metabolic, adipocyte and muscle dysfunction for exploring the role of caveolae in a variety of physiological processes in vivo and in vitro. They further suggest the possibility that the Cavin gene locus may be a site of heretofore ungenotyped lipodystrophies present in the human population.
EXPERIMENTAL PROCEDURES
Generation and genotyping of Cavin knockout mice
The Cavin knockout mice were generated using a previously described strategy (Yang et al., 2006). Briefly for the construction of the targeting vector, mouse BAC clone bMQ-107C23 (derived from AB2.2 ES cells that are derived from the 129S7 strain) containing the Cavin/Ptrf gene, which encodes the Cavin protein, was obtained from The Wellcome Trust Sanger Institute (Cambridge, UK). The targeting vector was constructed from the BAC clone using a PCR strategy. Two 4-kb fragments from immediately 5’ and 3’ to the exon-1 region in Cavin gene (NCBI GenBank accession number AF458959) were generated by PCR and sub-cloned into upstream and downstream of neomycin cassette (Neo) in the pPNT targeting vector respectively. Then a 3-kb fragment encoding prokaryotic β-gal (LacZ) from pcDNA3.1/Hygro/LacZ vector (Invitrogen) was sub-cloned 5' of the Neo cassette to generate the final targeting vector.
The linearized targeting construct was electroporated into TC1 ES cells, and incorporation of the vector by homologous recombination was selected using G418 and FIAU. ES clones (200) were assessed for correct targeting using PCR and Southern blot analysis. Genomic DNA was digested with EcoRI or NdeI and hybridized with a 1.3 kb XbaI-XhoI probe spanning promoter region.
Cavin+/− clones produced an 8.0 kb wild type and a 5.3 kb knockout band. Six correctly targeted clones were identified. Two were microinjected into C57BL/6 blastocysts, and one gave rise to a female chimera with significant ES cell contribution (as determined by Agouti coat color). By mating with C57BL/6 males and genotyping the offspring by Southern blotting and PCR analysis, germ line transmission was confirmed (Figure 1). Male and female heterozygous F1 animals were interbred to obtain Cavin knockout (_Cavin_−/−) animals. Genotypic verification by PCR is described in detail in Supplemental Experimental Procedures.
Gender- and strain-matched (about 85% C57BL/6, as per PCR-based gene marker analysis MAX-BAX (Charles River Laboratories)) animals were analyzed at about 6–8 weeks of age. Experiments were conducted under the direct supervision of trained veterinarians of the Boston University Laboratory Animal Science Center, and animal protocols were approved by the Institutional Animal Care and Use Committee.
Body Composition Analysis
Quantitative magnetic resonance was performed on 8-week old wild type (n = 6) and Cavin knockout (n = 6) mice to directly measure total body fat and lean mass (EchoMRI-900, Echo Medical Systems). Relative lean mass and fat mass values were determined by dividing absolute values by body weight.
Metabolic Chamber Studies
8-week old wild type (n = 8) and Cavin knockout (n = 8) mice were individually housed in metabolic chambers and basal metabolic rate and habitual physical activity were measured using a comprehensive laboratory animal monitoring system (Columbus Instruments). Basal metabolic rate (kcal/hr) was derived from measures of oxygen consumption (VO2) and carbon dioxide production (VCO2) by open circuit indirect calorimetry. Horizontal, or ambulatory, physical activity was quantified by the number of infrared beams (emitted by photocells) that were disrupted in the horizontal plane of the chambers. Metabolic and activity data were first captured for 24 hours under fed conditions and then food was removed from the chambers and the animals were monitored for another 24 hours under fasted conditions.
Analysis of Plasma Parameters
Mouse blood was drawn from the tail vein and decanted directly into heparinized capillary tubes (Fisher Scientific). Triglyceride, and free fatty acid levels were measured with standard enzymatic colorimetric assays (Pointe Scientific, Inc. and BioVision). Insulin and leptin levels were determined by ELISA with kits (Alpco Diagnostics, and GenWay, respectively). Adiponectin levels were determined by quantitative immunoblotting with an anti-adiponectin antibody (Affinity BioReagents), with the value for the wild type mice set to 1.0.
Glucose tolerance test (GTT)
Wild type (n = 8) and Cavin knockout (n = 8) mice were fasted overnight (12 hours), weighed and administered with 1 g/kg body weight of 20% D-glucose by intraperitoneal cavity injection. Tail vein blood was collected before and at various times (Figure 3E) after injection for measurement of glucose concentration (Accu-Chek, Roche).
Supplemental Experimental Procedures
The details of Western Blot analysis, Quantitative RT-PCR and Transmission Electronic Microscope Examination are described in Supplemental Experimental Procedures.
Statistical Analyses
We expressed data as arithmetic means ± SE and performed statistical analysis as indicated in the figure legends. Significant differences are indicated in the figures.
Supplementary Material
01
ACKNOWLEDGMENTS
We thank Greg Martin from the Boston University School of Medicine Transgenic Core, Michelle Ganno from the Metabolic Phenotypic Core, and Maria Makitalo for assistance in producing targeted ES cells. Mouse BAC clones were kindly provided by The Wellcome Trust Sanger Institute (Cambridge, UK). This work was supported by National Institutes of Health grants DK38452 and DK42596 to DB, AG031154 to NKL, NH13262 to KR, HD42779 to KHA, and DK30425 and DK56935 to PFP. DB is also supported by the Microscopy Core Facility of the MGH Program in Membrane Biology and by the Boston Area Diabetes and Endocrinology Research Center grant, DK 57521.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abel ED, Peroni O, Kim JK, Kim YB, Boss O, Hadro E, Minnemann T, Shulman GI, Kahn BB. Adipose-selective targeting of the GLUT4 gene impairs insulin action in muscle and liver. Nature. 2001;409:729–733. doi: 10.1038/35055575. [DOI] [PubMed] [Google Scholar]
- Aboulaich N, Vainonen JP, Stralfors P, Vener AV. Vectorial proteomics reveal targeting, phosphorylation and specific fragmentation of polymerase I and transcript release factor (PTRF) at the surface of caveolae in human adipocytes. Biochem J. 2004;383:237–248. doi: 10.1042/BJ20040647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal AK, Garg A. Genetic disorders of adipose tissue development, differentiation, and death. Annu Rev Genomics Hum Genet. 2006;7:175–199. doi: 10.1146/annurev.genom.7.080505.115715. [DOI] [PubMed] [Google Scholar]
- Betz RC, Schoser BG, Kasper D, Ricker K, Ramirez A, Stein V, Torbergsen T, Lee YA, Nothen MM, Wienker TF, et al. Mutations in CAV3 cause mechanical hyperirritability of skeletal muscle in rippling muscle disease. Nat Genet. 2001;28:218–219. doi: 10.1038/90050. [DOI] [PubMed] [Google Scholar]
- Bickel PE, Scherer PE, Schnitzer JE, Oh P, Lisanti MP, Lodish HF. Flotillin and epidermal surface antigen define a new family of caveolae-associated integral membrane proteins. J Biol Chem. 1997;272:13973–13802. doi: 10.1074/jbc.272.21.13793. [DOI] [PubMed] [Google Scholar]
- Boden G. Role of fatty acids in the pathogenesis of insulin resistance and NIDDM. Diabetes. 1997;46:3–10. [PubMed] [Google Scholar]
- Breton S, Lisanti MP, Tyszkowski R, McLaughlin M, Brown D. Basolateral distribution of caveolin-1 in the kidney. Absence from H+- atpase-coated endocytic vesicles in intercalated cells. J Histochem Cytochem. 1998;46:205–214. doi: 10.1177/002215549804600209. [DOI] [PubMed] [Google Scholar]
- Cao H, Alston L, Ruschman J, Hegele RA. Heterozygous CAV1 frameshift mutations (MIM 601047) in patients with atypical partial lipodystrophy and hypertriglyceridemia. Lipids Health Dis. 2008;7:3. doi: 10.1186/1476-511X-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capozza F, Combs TP, Cohen AW, Cho YR, Park SY, Schubert W, Williams TM, Brasaemle DL, Jelicks LA, Scherer PE, et al. Caveolin-3 knockout mice show increased adiposity and whole body insulin resistance, with ligand-induced insulin receptor instability in skeletal muscle. Am J Physiol Cell Physiol. 2005;288:C1317–C1331. doi: 10.1152/ajpcell.00489.2004. [DOI] [PubMed] [Google Scholar]
- Cohen AW, Hnasko R, Schubert W, Lisanti MP. Role of caveolae and caveolins in health and disease. Physiol Rev. 2004;84:1341–1379. doi: 10.1152/physrev.00046.2003. [DOI] [PubMed] [Google Scholar]
- Hill MM, Bastiani M, Luetterforst R, Kirkham M, Kirkham A, Nixon SJ, Walser P, Abankwa D, Oorschot VM, Martin S, et al. PTRF-Cavin, a conserved cytoplasmic protein required for caveola formation and function. Cell. 2008;132:113–124. doi: 10.1016/j.cell.2007.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehn KL, Hohnen-Behrens C, Cederberg A, Wu LE, Turner N, Yuasa T, Ebina Y, James DE. IRS1-Independent Defects Define Major Nodes of Insulin Resistance. Cell Metab. 2008;7:421–433. doi: 10.1016/j.cmet.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James DE, Brown R, Navarro J, Pilch PF. Insulin-regulatable tissues express a unique insulin-sensitive glucose transport protein. Nature. 1988;333:183–185. doi: 10.1038/333183a0. [DOI] [PubMed] [Google Scholar]
- Jansa P, Mason SW, Hoffmann-Rohrer U, Grummt I. Cloning and functional characterization of PTRF, a novel protein which induces dissociation of paused ternary transcription complexes. Embo J. 1998;17:2855–2864. doi: 10.1093/emboj/17.10.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CA, Delepine M, Boutet E, El Mourabit H, Le Lay S, Meier M, Nemani M, Bridel E, Leite CC, Bertola DR, et al. Association of a homozygous nonsense caveolin-1 mutation with Berardinelli-Seip congenital lipodystrophy. J Clin Endocrinol Metab. 2008;93:1129–1134. doi: 10.1210/jc.2007-1328. [DOI] [PubMed] [Google Scholar]
- Kraegen EW, Cooney GJ, Ye J, Thompson AL. Triglycerides, fatty acids and insulin resistance--hyperinsulinemia. Exp Clin Endocrinol Diabetes. 2001;109:S516–S526. doi: 10.1055/s-2001-15114. [DOI] [PubMed] [Google Scholar]
- Le Lay S, Hajduch E, Lindsay MR, Le Liepvre X, Thiele C, Ferre P, Parton RG, Kurzchalia T, Simons K, Dugail I. Cholesterol-induced caveolin targeting to lipid droplets in adipocytes: a role for caveolar endocytosis. Traffic. 2006;7:549–561. doi: 10.1111/j.1600-0854.2006.00406.x. [DOI] [PubMed] [Google Scholar]
- Le Lay S, Kurzchalia TV. Getting rid of caveolins: phenotypes of caveolin-deficient animals. Biochim Biophys Acta. 2005;1746:322–333. doi: 10.1016/j.bbamcr.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Liu L, Pilch PF. A critical role of cavin (polymerase I and transcript release factor) in caveolae formation and organization. J Biol Chem. 2008;283:4314–4322. doi: 10.1074/jbc.M707890200. [DOI] [PubMed] [Google Scholar]
- Liu P, Rudick M, Anderson RG. Multiple functions of caveolin-1. J Biol Chem. 2002;277:41295–41298. doi: 10.1074/jbc.R200020200. [DOI] [PubMed] [Google Scholar]
- Liu P, Ying Y, Zhao Y, Mundy DI, Zhu M, Anderson RG. Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J Biol Chem. 2004;279:3787–3792. doi: 10.1074/jbc.M311945200. [DOI] [PubMed] [Google Scholar]
- McNally EM, de Sa Moreira E, Duggan DJ, Bonnemann CG, Lisanti MP, Lidov HG, Vainzof M, Passos-Bueno MR, Hoffman EP, Zatz M, Kunkel LM. Caveolin-3 in muscular dystrophy. Hum Mol Genet. 1998;7:871–877. doi: 10.1093/hmg/7.5.871. [DOI] [PubMed] [Google Scholar]
- Meshulam T, Simard JR, Wharton J, Hamilton JA, Pilch PF. Role of caveolin-1 and cholesterol in transmembrane fatty acid movement. Biochemistry. 2006;45:2882–2893. doi: 10.1021/bi051999b. [DOI] [PubMed] [Google Scholar]
- Minetti C, Sotgia F, Bruno C, Scartezzini P, Broda P, Bado M, Masetti E, Mazzocco M, Egeo A, Donati MA, et al. Mutations in the caveolin-3 gene cause autosomal dominant limb-girdle muscular dystrophy. Nat Genet. 1998;18:365–368. doi: 10.1038/ng0498-365. [DOI] [PubMed] [Google Scholar]
- Muoio DM, Newgard CB. Mechanisms of disease: molecular and metabolic mechanisms of insulin resistance and beta-cell failure in type 2 diabetes. Nat Rev Mol Cell Biol. 2008;9:193–205. doi: 10.1038/nrm2327. [DOI] [PubMed] [Google Scholar]
- Ng Y, Ramm G, Lopez JA, James DE. Rapid Activation of Akt2 Is Sufficient to Stimulate GLUT4 Translocation in 3T3-L1 Adipocytes. Cell Metab. 2008;7:348–356. doi: 10.1016/j.cmet.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Park DS, Woodman SE, Schubert W, Cohen AW, Frank PG, Chandra M, Shirani J, Razani B, Tang B, Jelicks LA, et al. Caveolin-1/3 double-knockout mice are viable, but lack both muscle and non-muscle caveolae, and develop a severe cardiomyopathic phenotype. Am J Pathol. 2002;160:2207–2217. doi: 10.1016/S0002-9440(10)61168-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton RG, Hanzal-Bayer M, Hancock JF. Biogenesis of caveolae: a structural model for caveolin-induced domain formation. J Cell Sci. 2006;119:787–796. doi: 10.1242/jcs.02853. [DOI] [PubMed] [Google Scholar]
- Parton RG, Simons K. The multiple faces of caveolae. Nat Rev Mol Cell Biol. 2007;8:185–194. doi: 10.1038/nrm2122. [DOI] [PubMed] [Google Scholar]
- Petersen KF, Shulman GI. Pathogenesis of skeletal muscle insulin resistance in type 2 diabetes mellitus. Am J Cardiol. 2002;90:11G–18G. doi: 10.1016/s0002-9149(02)02554-7. [DOI] [PubMed] [Google Scholar]
- Pilch PF. The mass action hypothesis: formation of Glut4 storage vesicles, a tissue-specific, regulated exocytic compartment. Acta Physiol (Oxf) 2008;192:89–101. doi: 10.1111/j.1748-1716.2007.01788.x. [DOI] [PubMed] [Google Scholar]
- Pilch PF, Souto RP, Liu L, Jedrychowski MP, Berg EA, Costello CE, Gygi SP. Cellular spelunking: exploring adipocyte caveolae. J Lipid Res. 2007;48:2103–2111. doi: 10.1194/jlr.R700009-JLR200. [DOI] [PubMed] [Google Scholar]
- Razani B, Combs TP, Wang XB, Frank PG, Park DS, Russell RG, Li M, Tang B, Jelicks LA, Scherer PE, Lisanti MP. Caveolin-1-deficient mice are lean, resistant to diet-induced obesity, and show hypertriglyceridemia with adipocyte abnormalities. J Biol Chem. 2002;277:8635–8647. doi: 10.1074/jbc.M110970200. [DOI] [PubMed] [Google Scholar]
- Rothberg KG, Heuser JE, Donzell WC, Ying YS, Glenney JR, Anderson RG. Caveolin, a protein component of caveolae membrane coats. Cell. 1992;68:673–682. doi: 10.1016/0092-8674(92)90143-z. [DOI] [PubMed] [Google Scholar]
- Scherer PE, Okamoto T, Chun M, Nishimoto I, Lodish HF, Lisanti MP. Identification, sequence, and expression of caveolin-2 defines a caveolin gene family. Proc Natl Acad Sci U S A. 1996;93:131–135. doi: 10.1073/pnas.93.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaul PW, Anderson RG. Role of plasmalemmal caveolae in signal transduction. Am J Physiol. 1998;275:L843–L851. doi: 10.1152/ajplung.1998.275.5.L843. [DOI] [PubMed] [Google Scholar]
- Tang Z, Scherer PE, Okamoto T, Song K, Chu C, Kohtz DS, Nishimoto I, Lodish HF, Lisanti MP. Molecular cloning of caveolin-3, a novel member of the caveolin gene family expressed predominantly in muscle. J Biol Chem. 1996;271:2255–2261. doi: 10.1074/jbc.271.4.2255. [DOI] [PubMed] [Google Scholar]
- van Meer G. Caveolin, cholesterol, and lipid droplets? J Cell Biol. 2001;152:F29–F34. doi: 10.1083/jcb.152.5.f29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinten J, Johnsen AH, Roepstorff P, Harpoth J, Tranum-Jensen J. Identification of a major protein on the cytosolic face of caveolae. Biochim Biophys Acta. 2005;1717:34–40. doi: 10.1016/j.bbamem.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Vinten J, Voldstedlund M, Clausen H, Christiansen K, Carlsen J, Tranum-Jensen J. A 60-kDa protein abundant in adipocyte caveolae. Cell Tissue Res. 2001;305:99–106. doi: 10.1007/s004410100389. [DOI] [PubMed] [Google Scholar]
- Vorgerd M, Ricker K, Ziemssen F, Kress W, Goebel HH, Nix WA, Kubisch C, Schoser BG, Mortier W. A sporadic case of rippling muscle disease caused by a de novo caveolin-3 mutation. Neurology. 2001;57:2273–2277. doi: 10.1212/wnl.57.12.2273. [DOI] [PubMed] [Google Scholar]
- Way M, Parton RG. M-caveolin, a muscle-specific caveolin-related protein. FEBS Lett. 1995;376:108–112. doi: 10.1016/0014-5793(95)01256-7. [DOI] [PubMed] [Google Scholar]
- Woodman SE, Sotgia F, Galbiati F, Minetti C, Lisanti MP. Caveolinopathies: mutations in caveolin-3 cause four distinct autosomal dominant muscle diseases. Neurology. 2004;62:538–543. doi: 10.1212/wnl.62.4.538. [DOI] [PubMed] [Google Scholar]
- Yang D, Zhang Y, Nguyen HG, Koupenova M, Chauhan AK, Makitalo M, Jones MR, St Hilaire C, Seldin DC, Toselli P, et al. The A2B adenosine receptor protects against inflammation and excessive vascular adhesion. J Clin Invest. 2006;116:1913–1923. doi: 10.1172/JCI27933. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01