An efficient and low immunostimulatory nanoparticle formulation for systemic siRNA delivery to the tumor (original) (raw)
. Author manuscript; available in PMC: 2009 Oct 6.
Abstract
We have developed a nanoparticle formulation [liposomes-protamine-hyaluronic acid nanoparticle (LPH-NP)] for systemically delivering siRNA into the tumor. The LPH-NP was prepared in a self-assembling process. Briefly, protamine and a mixture of siRNA and hyaluronic acid were mixed to prepare a negatively charged complex. Then, cationic liposomes were added to coat the complex with lipids via charge-charge interaction to prepare the LPH-NP. The LPH-NP was further modified by DSPE-PEG or DSPE-PEG-anisamide by the post-insertion method. Anisamide is a targeting ligand for the sigma receptor over-expressed in the B16F10 melanoma cells. The particle size, zeta potential and siRNA encapsulation efficiency of the formulation were approximately 115 nm, +25 mV and 90%, respectively. Luciferase siRNA was used to evaluate the gene silencing activity in the B16F10 cells, which were stably transduced with a luciferase gene. The targeted LPH-NP (PEGylated with ligand) silenced 80% of luciferase activity in the metastatic B16F10 tumor in the lung after a single i.v. injection (0.15 mg siRNA/kg). The targeted LPH-NP also showed very little immunotoxicity in a wide dose range (0.15 – 1.2 mg siRNA/kg), while the previously published formulation, LPD-NP (liposome-protamine-DNA nanoparticle), had a much narrow therapeutic window (0.15–0.45 mg/kg).
Keywords: siRNA, tumor, metastasis, tumor targeted delivery, nanoparticles
1. Introduction
Our lab has previously demonstrated that the LPD-NP (liposome-protamine-DNA nanoparticle) surface-modified with a targeting ligand could deliver siRNA selectively to the sigma receptor positive cells and induce significant RNAi and antitumor effects [1–5]. Although the formulation showed a great potential of delivering siRNA, the therapeutic window was relatively narrow (0.15–0.45 mg/kg) [5]. The narrow window of the formulation might be a consequence of using the calf thymus DNA as a carrier DNA which enhances the particle condensation and stability.
In general, methods of forming nanoparticles with siRNA are usually through a self-assembling process mediated by charge-charge interaction, in which cationic carriers bind with the anionic nucleic acid. We and another group found that cationic carriers, especially the polymers, form a looser complex with siRNA than with the plasmid DNA, resulting in unstable particle formulation and reduced delivery efficacy [3, 6]. This finding may be based on the fact that the molecular weight of siRNA is too low for an efficient polymer interaction. Bolcato-Bellemin et al. [6] added short complementary A5–8/T5–8 overhangs to make the siRNA bind to each other and form a large "gene-like" structure. They showed that the siRNA with sticky overhangs had increased complex stability with polyethylenimine, improved RNase protection and enhanced gene silencing. We incorporated a high-molecular-weight carrier DNA, calf thymus DNA (50 kbp) in our LPD-NP formulation to enhance the particle condensation [3]. This carrier DNA containing formulation showed 10–30% decreased particle size and 20–80% increased delivery efficiency compared with the formulation without the carrier DNA. However, calf thymus DNA as a foreign DNA to human may cause unexpected toxicity and immune response when used clinically [7]. Alternatively, a formulation containing plasmid DNA instead of the calf thymus DNA can be prepared with similar particle characteristics. However, plasmid DNA contains a high amount of CpG motifs and the resulting formulation was highly immunogenic [3, 4]. In this study, we investigated the feasibility of using hyaluronic acid (HA) to improve the nanoparticle formation and reduce the immunotoxicity of the formulation. Since HA is a polyanionic polysaccharide, it provides multivalent charges to enhance the particle condensation but contains no immunostimulatory CpG motifs. HA is also a biogenic component, distributed widely in the extracellular matrix and found in the viscous fluid of the mammalian joints [8]. More recently, HA has been investigated as a drug delivery agent for various routes of administration including ophthalmic, nasal, pulmonary, oral, parenteral and topical [9]. Moreover, it is a polymer of relatively low toxicity and has been approved by the FDA for injections [10].
Basically, the LPD-NP and LPH-NP (liposomes-protamine-HA nanoparticle) were both prepared by mixing the formulation components in a salt free solution and the nanoparticles were formed in a self-assembling process. For the LPH-NP formulation development, particles made of different ratios of the three major components, siRNA/HA, protamine and liposomes, were prepared and evaluated by their particle size, zeta potential and in vitro delivery efficiency. The optimized formulation was named LPH-NP. The LPH-NP was then coated by PEG-lipid containing a targeting ligand, anisamide, and thus was modified for targeting sigma receptor expressing B16F10 tumor. The delivery efficiency and gene silencing activity of the LPD-NP and LPH-NP formulations were compared in vitro and in vivo in a lung metastasis model. The immunotoxicity of the two formulations was also analyzed.
2. Materials and Methods
2.1. Materials
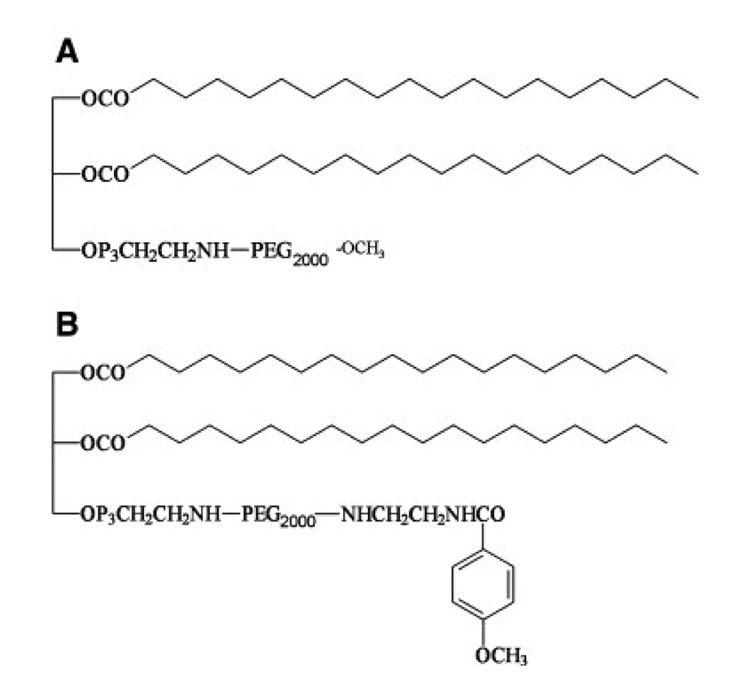
1,2-dioleoyl-3-trimethylammonium-propane chloride salt (DOTAP), cholesterol, and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy (polyethyleneglycol)-2000] ammonium salt (DSPE-PEG2000) (Fig. 1A) were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL). Protamine sulfate (fraction X from salmon), hyaluronic acid sodium salt from Streptococcus equi (HA) and calf thymus DNA (for hybridization, phenol-chloroform extracted and ethanol precipitated) were from Sigma-Aldrich (St. Louis, MO). DSPE-PEG2000-anisamide was synthesized in our lab using the methods described previously [11] and the structure is shown in Fig. 1B.
Fig. 1.
Chemical structures of DSPE-PEG2000 (A) and DSPE-PEG2000-anisamide (B).
Anti-luciferase siRNA (GL3) (target sequence 5’- CTT ACG CTG AGT ACT TCG A -3’) was purchased from Dharmacon (Lafayette, CO) in deprotected, desalted, annealed form. For in vitro intracellular siRNA delivery study and determination of siRNA encapsulation efficiency, fluorescein-labeled siRNA (3’end of the sense strand, FAM-siRNA) provided by Dharmacon was used.
B16F10 cells, murine melanoma cells, were obtained from the American Type Cell Collection and were stably transduced with GL3 firefly luciferase gene using a retroviral vector in Dr. Pilar Blancafort’s lab at the University of North Carolina at Chapel Hill (UNC). The gene silencing activity in the cells can be easily assessed by analyzing luciferase activity when siRNA against luciferase gene is used. The cells were maintained in Dulbecco’s modified eagle medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Invitrogen). B16F10 cells expressing sigma receptors [12] were used as model cells in our study.
2.2. Experimental animals
Female C57BL/6 mice of age 6–8 week (16–18 g) were purchased from Charles River Laboratories (Wilmington, MA). All work performed on animals was in accordance with and approved by the IACUC committee at UNC.
2.3. Optimization of the LPH-NP formulation
Liposome-protamine-HA nanoparticles (LPH-NP) were prepared as follows. Briefly, small unilamellar liposomes consisting of DOTAP and cholesterol (molar ratio = 1:1) were prepared by thin film hydration followed by membrane extrusion (400 nm × 10 times, 200 nm × 10 times, 100 nm × 10 times and 50 nm × 15 times). The total lipid concentration of the liposomes was fixed at 40 mM. To prepare complex of siRNA/HA and protamine, 150 µl of protamine (200 µg/mL) and 150 µl of a mixture of siRNA and HA (160–210 µg/ml, weight ratio = 1:1) were mixed in a 1.5 ml tube. The complex was allowed to stand at room temperature for 10 min before analysis of the size and zeta potential. The complex prepared by the optimal ratio of siRNA/HA and protamine was mixed with 0–50 µl of DOTAP/cholesterol liposomes (total lipid concentration = 40 mM). Again, the particle size and zeta potential of the resulting particles were analyzed. Additionally, delivery efficiency of the LPH-NP of different lipid/siRNA ratios was determined as described in section 2.4. The optimal ratio of the LPH-NP formulation was determined by the results from particle size, zeta potential and in vitro delivery efficiency. The optimized formulation was termed as the naked LPH-NP. Non-targeted LPH-NP and targeted LPH-NP were prepared by incubating the naked LPH-NP suspension (330 µl) with 36.6 µl micellar solution of DSPE-PEG2000 or DSPE-PEG2000-anisamide (10 mg/ml) at 50°C for 10 min, respectively, and then allowed to stand at room temperature for 10 min. The resulting LPH-NP formulations were used within 20 min for the following experiments.
Liposome-protamine-DNA nanoparticles (LPD-NP) were prepared as previously described [4, 5]. Briefly, naked LPD-NP were obtained by quickly mixing 150 µl suspension A (8.3 mM liposomes (DOTAP/cholesterol molar ratio = 1: 1) and 200 µg/ml protamine) with 150 µl solution B (160 µg/ml siRNA and 160 µg/ml calf thymus DNA) followed by incubation at room temperature for 10 min. Non-targeted LPD-NP and targeted LPD-NP were prepared by incubating the naked LPD-NP suspension (300 µl) with 37.8 µl micellar solution of DSPE-PEG2000 or DSPE-PEG2000-anisamide (10 mg/ml) at 50°C for 10 min, respectively, and then allowed to stand at room temperature for 10 min. The resulting LPD-NP formulations were used within 20 min for the following experiments.
The distribution of particle size of the samples was measured using a submicron particle sizer (NICOMP particle sizing systems, AutodilutePAT Model 370, Santa Barbra, CA) in the NICOMP mode. The polydispersity index was also checked to evaluate distribution characteristics. The zeta potential of the samples diluted in 1 mM KCl was determined by the Zeta Plus zeta potential analyzer (Brookhaven Instruments Corporation, Holtsville, NY).
Transmission electron microscope (TEM) images of the resulting LPH-NP and LPD-NP were acquired using a Phillips CM12 (FEI, Hillsboro, OR). Briefly, freshly prepared nanoparticle samples (5 µl) were dropped onto a 300-mesh carbon-coated copper grid (Ted Pella, Inc., Redding, CA) and allowed for a short incubation (5 min) at room temperature. Grids were then stained with 1% uranyl acetate (40 µl) and wicked dry. All images were acquired at an accelerating voltage of 100 kV. Gatan DigitalMicrograph software was used to analyze the images.
The siRNA encapsulation efficiency in the LPH-NP and LPD-NP was determined by passing the FAM-siRNA containing formulations through a Sepharose CL4B size exclusion column (Pharmacia Biotech, Uppsala, Sweden). Unencapsulated FAM-siRNA was separated from encapsulated one and the incorporation efficiency was determined as previously described [1].
2.4. In vitro intracellular siRNA delivery study
B16F10 cells (0.5 × 105 cells/0.25 ml/well) were seeded in 48-well plates (Corning Inc., Corning, NY) 20 h before experiments. Cells were treated with different formulations containing 500 nM FAM-siRNA in the culture medium at 37°C for 4 h. Cells were washed three times with PBS followed by incubation with 200 µl lysis buffer (0.3% Triton X-100 in PBS) at 37°C for 0.5 h. Fluorescence intensity of 100 µl cell lysate was determined by a plate reader (λex: 485 nm, λem: 535 nm) (PLATE CHAMELEON Multilabel Detection Platform, Bioscan Inc., Washington, DC). For free ligand competition study, cells were co-incubated with 50 µM haloperidol with different formulations.
2.5. In vitro luciferase gene silencing study
B16F10 cells (1 × 105 cells/0.5 ml/well) were seeded in 24-well plates (Corning Inc., Corning, NY) 20 h before experiments. Cells were treated with different formulations containing 250 nM siRNA in the culture medium at 37°C for 24 h. Cells were washed three times with PBS followed by incubation with 100 µl lysis buffer (0.05% Triton X-100 and 2 mM EDTA in 0.1 M Tris-HCl) at 37°C for 0.5 h. Ten µl lysate was mixed with 100 µl substrate (Luciferase Assay System, Promega Co., Madison, WI) and the luminescence was measured by a plate reader. The protein concentrations of the samples were determined by using a protein assay kit (Micro BCA™ protein assay kit, Pierce). Luciferase activity of a sample was normalized with the protein content and expressed as percent luminescence intensity compared to the untreated control.
2.6. In vivo luciferase gene silencing study
C57BL/6 mice were i.v. injected with 2 × 105 B16F10 cells via the tail vein. Seventeen days later, mice were given i.v. injections of anti-luciferase siRNA at the dose of 0.15 mg/kg formulated in different formulations. Control siRNA (target sequence: 5’-AATTCTCCGAACGTGTCACGT-3’) [13] formulated in targeted LPH-NP or targeted LPD-NP was also prepared to verify if the silencing effect was sequence dependent. For dose response study, tumor bearing mice were i.v. injected with siRNA in targeted LPH-NP at dose of 0.0375, 0.075, 0.15, 0.45 and 1.5 mg/kg. After 24 h, mice were euthanized and the lungs were collected. The tumor nodules in the lung were isolated and homogenized in 300 µl lysis buffer (0.05% Triton X-100 and 2 mM EDTA in 0.1 M Tris-HCl) followed by centrifugation at 10,000 rpm for 10 min. Ten µl supernatant was mixed with 100 µl luciferase substrate and the luciferase activity was measured by a plate reader. The protein concentrations of the samples were determined by using a protein assay kit described above. Luciferase activity of a sample was normalized with the protein content and expressed as percent luminescence intensity compared to the untreated control.
2.7. Cytokine induction assay
C57BL/6 mice were i.v. injected with anti-luciferase siRNA formulated in the targeted LPH-NP and the targeted LPD-NP at various doses. Targeted LPD-NP formulated with plasmid DNA (pNGVL-Luc prepared by Bayou Biolabs) [14] instead of calf thymus DNA was prepared and used as a positive control. Two h after the injections, blood samples were collected from the tail artery and allowed to stand at room temperature for 0.5 h for coagulation. Serum was obtained by centrifuging the clotted blood at 16,000 rpm for 20–40 min. Cytokine levels were determined by using ELISA kits for IL6 and IL12 (BD Biosciences, San Diego, CA).
2.8. Statistical analysis
Data are presented as the mean ± SD. The statistical significance was determined by using the analysis of variance (ANOVA). P values of <0.05 were considered significant.
3. Results and discussion
The LPH-NP nanoparticles were formed solely by charge-charge interaction among the three major components. First, siRNA/HA mixture was condensed by protamine into a negatively charged complex. Given a fixed amount of siRNA/HA, the complex should only contain a minimal required amount of protamine that can provide sufficient complexation but keep the complex negatively charged. Second, the complex interacted with the cationic liposome to form the LPH-NP. Again, the LPH-NP should only contain slightly excess amount of lipids that allow full coating of the complex with the cationic lipids. The ratio of the three components determines the charge, size and delivery efficiency of the formulations.
3.1. Development of the LPH-NP formulation
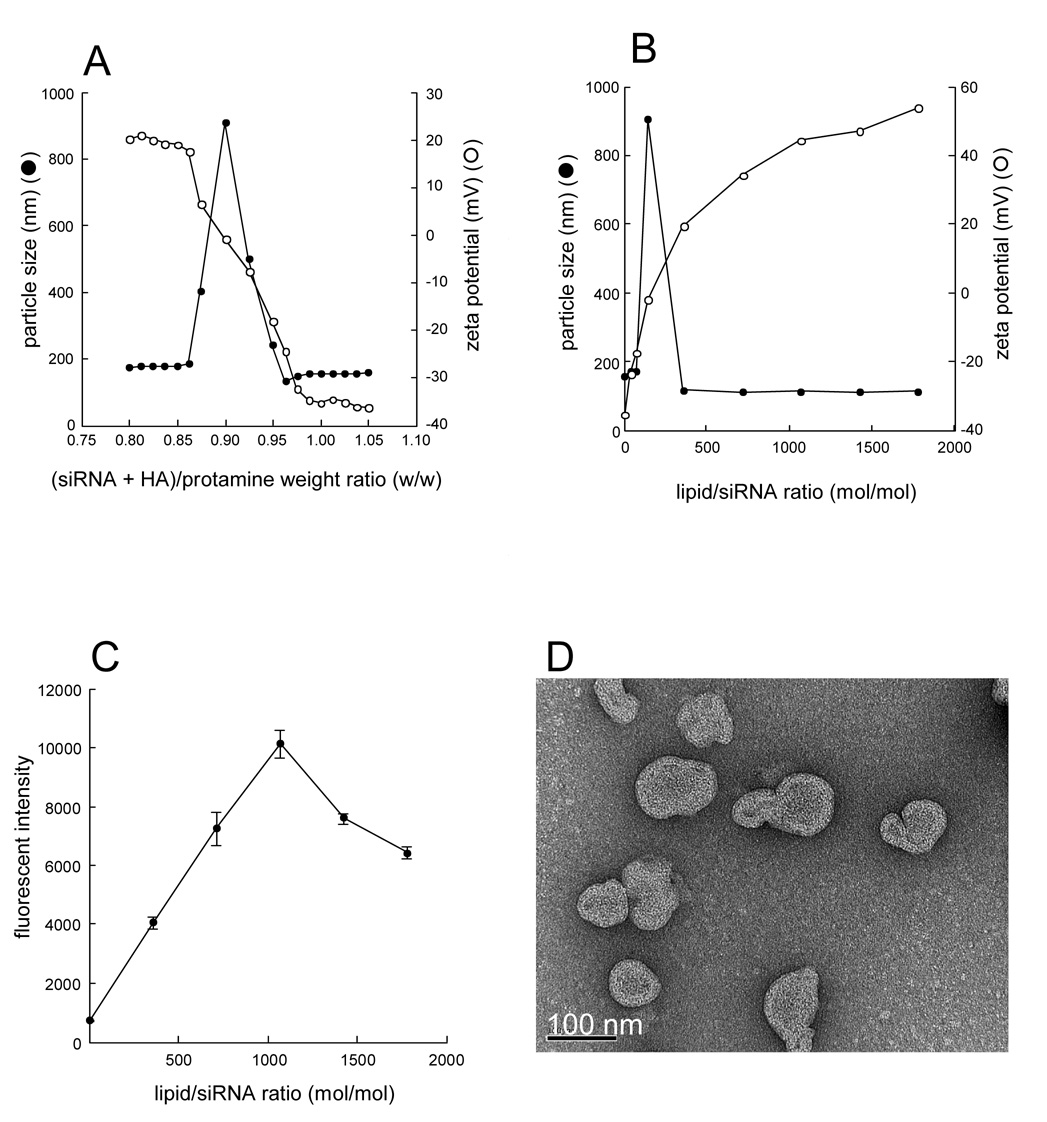
First, we prepared the complex of siRNA/HA and protamine in different ratios and measured their particle size and zeta potential. As shown in Fig. 2A, particle size and zeta potential changed according to the ratio of siRNA/HA to protamine. Large aggregates were found at the ratio around 0.9 (siRNA/HA: protamine, weight ratio). At this ratio, a neutral complex was formed (zeta potential ~0 mV), which tended to aggregate. Increase of protamine amount in the complex increased the zeta potential from −35 mV to 20 mV with a sharp change at the ratio between 0.865–0.9625. We chose 1.0 as the optimal ratio, as the complex stayed negatively charged with a relatively small size (~150 nm).
Fig. 2. Formulation optimization of the LPH-NP.
A), Effect of (siRNA+HA)/protamine weight ratio on particle size and zeta potential of siRNA/HA/protamine complex. Protamine (200 µg/mL, 150 µl) and a mixture of siRNA and HA (160–210 µg/ml, weight ratio = 1:1, 150 µl) were mixed in a 1.5 ml tube and kept at room temperature for 10 min. Then particle size and zeta potential were measured. B), Effect of lipid/siRNA molar ratio on particle size and zeta potential of LPH-NP. Complex of siRNA/HA and ((siRNA+HA)/protamine weight ratio = 1.0, 300 µl) and DOTAP/cholesterol liposomes (total lipid concentration = 40 mM, 0–50 µl) were mixed and kept at room temperature for 10 min. Then particle size and zeta potential were measured. C), Effect of lipid/siRNA molar ratio of LPH-NP on the in vitro intracellular siRNA delivery in B16F10 cells. LPH-NP containing 500 nM FAM-siRNA was added to B16F10 cells (0.5 × 105 cells/well) and incubated for 4 h. Then fluorescent intensity in cells was measured. Each value represents the mean ± S.D. (n=4). D), TEM images of optimal LPD-NP (lipid/siRNA molar ratio = 1067). Bar indicates 100 nm.
Second, we mixed siRNA/HA/protamine complex with different amounts of the cationic lipid to prepare the naked LPH-NP and analyzed their particle size and zeta potential. As mentioned earlier, slightly excess amount of the cationic lipid is required to obtain fully coated LPH-NP. At the ratio of 142 (lipid: siRNA, molar ratio), large aggregates with a neutral charge were detected (Fig. 2B). Increase of the lipid/siRNA ratio increased zeta potential. To further investigate what ratio resulted in the optimal LPH-NP formulation, we encapsulated FAM-siRNA in the LPH-NP formulations prepared with different lipid/siRNA ratios and determined their in vitro delivery efficiency. As shown in Fig. 2C, at the ratio of 1.067, the formulation had the highest cellular delivery efficiency. Further increase of the cationic lipid decreased the delivery efficiency. This may be due to the competitive binding with the cells from the excess cationic liposomes. At the optimal ratio, the particle size was around 120 nm and the zeta potential was about 45 mV. TEM examination confirmed that the size of the optimized LPH-NP was around 100 nm (Fig. 2D).
3.2. Characteristics of PEGlyated LPH-NPs and LPD-NPs
PEGlyated LPH-NPs, with or without a targeting ligand, anisamide, were prepared to maintain pharmaceutical stability in a physiological condition and selective delivery into the target cells expressing the sigma receptor [2]. Characteristics of the PEGylated formulations are summarized in Table 1. The particle size was all around 120 nm with a narrow size distribution (polydispersity index < 0.2). PEGlyation significantly reduced the zeta potential of the naked LPH-NP (from 45 mV to about 25 mV) because of the steric hindrance provided by the PEG. The zeta potential of the targeted LPH-NP was slightly higher than that of the non-targeted LPH-NP due to the positively charged anisamide ligand. The siRNA encapsulation efficiency of PEGlyated LPH-NPs was more than 90%. The properties of LPD-NP and LPH-NP were similar to each other. Additionally, no difference was found in the TEM examination between the LPH-NP and LPD-NP formulations (data not shown).
Table 1.
Characteristics of PEGlyated LPH-NPs and LPD-NPs
| formulations | particle size a) (nm) | zeta potential a) (mV) | encapsulation efficiency b) (%) |
|---|---|---|---|
| non-targeted LPH-NP | 114.4 ± 16.2 | 22.4 ± 2.01 | 92.2 ± 3.2 |
| targeted LPH-NP | 117.9 ± 15.7 | 27.5 ± 1.96 | 92.3 ± 1.2 |
| non-targeted LPD-NP | 117.5 ± 17.1 | 21.1 ± 1.56 | 92.6 ± 3.2 |
| targeted LPD-NP | 114.3 ± 16.7 | 25.4 ± 1.82 | 91.8 ± 0.6 |
3.3. In vitro intracellular siRNA delivery of different formulations
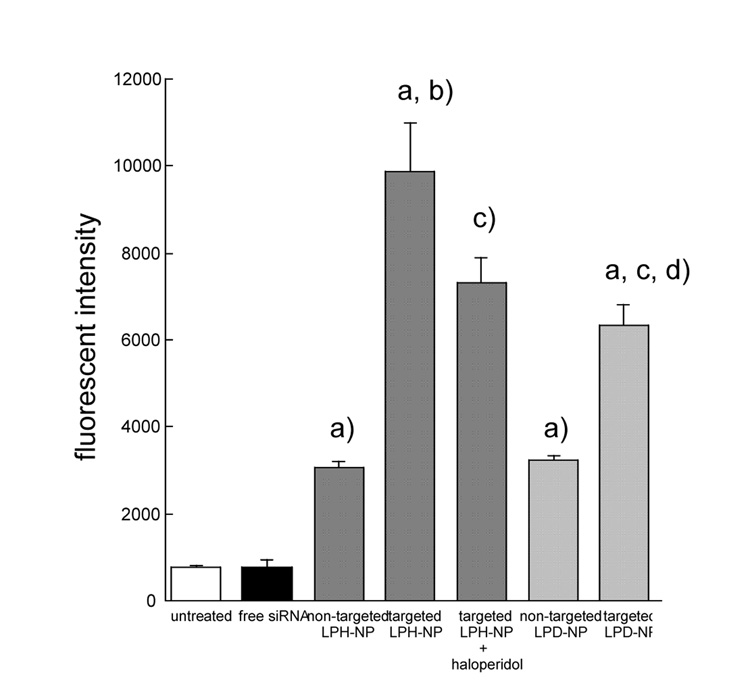
In vitro intracellular siRNA delivery study was performed in B16F10 cells (Fig. 3). The fluorescence intensity in cells treated with free siRNA showed background fluorescence. This indicates that free siRNA could hardly penetrate through the cell membrane due to its highly hydrophilic nucleic acid backbone. The siRNA delivery efficiency of the targeted LPH-NP (PEGylated and with ligand) was significantly higher than that of the non-targeted LPH-NP (PEGylated but without ligand), and inhibited by co-incubation with haloperidol, a known ligand for sigma receptor. This indicates that targeted LPH-NP could deliver siRNA to the B16F10 cells through sigma receptor mediated endocytosis, similar to the targeted LPD-NP [2]. Interestingly, the delivery efficiency of targeted LPH-NP was significantly higher than that of the targeted LPD-NP. The reason for the observation is not clear at the present time. This could be due to different release rate of siRNA from the LPD-NP as compared to the LPH-NP.
Fig. 3.
In vitro intracellular siRNA delivery of different formulations in B16F10 cells. Formulations containing FAM-siRNA (500 nM) were added to B16F10 cells (0.5 × 105 cells/well) and then incubated at 37°C for 4 h. Fluorescent intensity in cells was measured. For free ligand competition study, cells were co-incubated with 50 µM haloperidol with formulations. Each value represents the mean ± S.D. (n=4). a) p<0.05, significantly different compared with the free siRNA. b) p<0.05, significantly different compared with the non-targeted LPH-NP. c) p<0.05, significantly different compared with the targeted LPH-NP. d) p<0.05, significantly different compared with the non-targeted LPD-NP.
3.4. In vitro luciferase gene silencing of different formulations
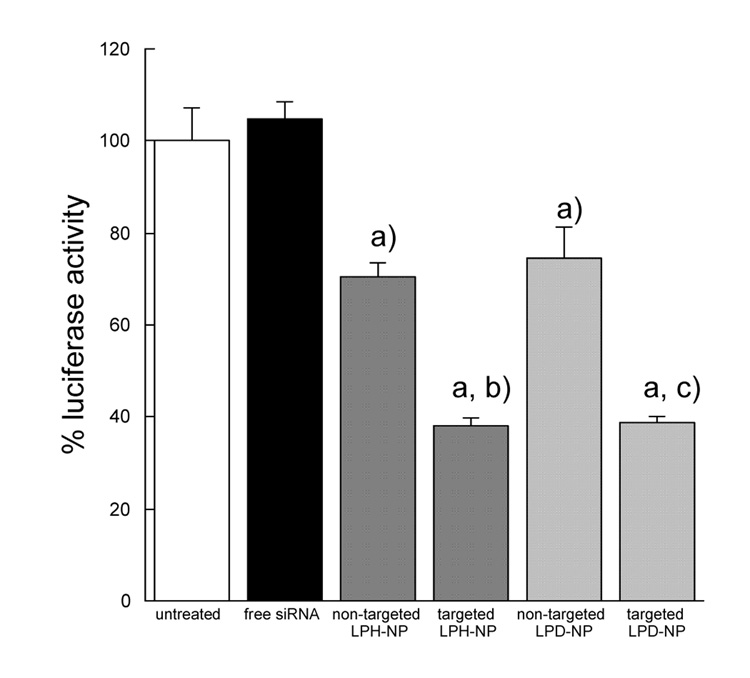
In vitro gene silencing study was performed in B16F10 cells, which were stably transduced with the firefly luciferase gene by using a retroviral vector (Fig. 4). Free siRNA showed no gene silencing effect because the permeability of siRNA itself through the cell membrane was very low (Fig. 4). The data were consistent with that of the in vitro intracellular siRNA delivery study (Fig. 4). Gene silencing effect of the targeted LPH-NP was similar to that of the targeted LPD-NP although the siRNA delivery efficiency of the targeted LPH-NP was higher than that of the targeted LPD-NP (Fig. 3 and 4). This could be due to the saturation of gene silencing effect inside the cells. The silencing activity of the targeted LPH-NP leveled off, when siRNA concentration was more than 250 nM (data not shown).
Fig. 4.
In vitro luciferase gene silencing effects of different formulations in B16F10 cells. Each formulation containing anti-luciferase siRNA (250 nM) were added to B16F10 cells (1 × 105 cells/well) and then incubated at 37°C for 24 h. Luciferase activity in cells was measured. Each value represents the mean ± S.D. (n=3). a) p<0.05, significantly different compared with the untreated control. b) p<0.05, significantly different compared with the non-targeted LPH-NP. c) p<0.05, significantly different compared with the non-targeted LPD-NP.
3.5. In vivo luciferase gene silencing of different formulations
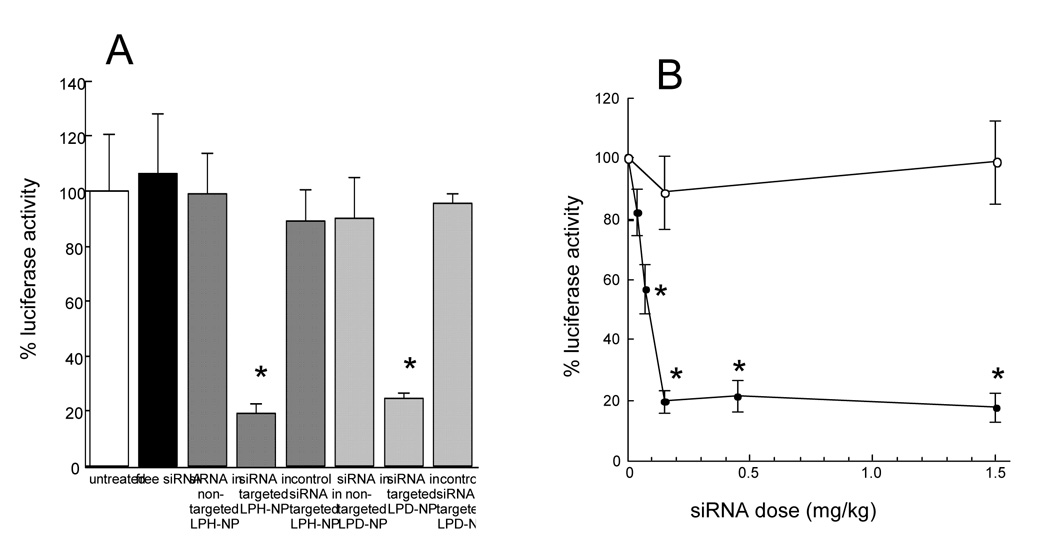
In vivo gene silencing study was performed in the B16F10 lung metastasis model (Fig. 5). Anti-luciferase siRNA formulated in the targeted LPH-NP silenced 80% luciferase activity (Fig. 5A). This effect was similar to that of anti-luciferase siRNA formulated in the targeted LPD-NP (Fig. 5A). The other control treatments, including free siRNA, siRNA in the non-targeted LPD-NP or the non-targeted LPH-NP and control siRNA in the targeted LPD-NP or LPH-NP, showed no RNAi effect. The data shown in Fig. 4 and Fig. 6 suggest that the enhanced gene silencing activity of the targeted LPH-NP was mainly due to the significantly improved tumor uptake and use of the correct siRNA sequence in the formulations containing siRNA. The ED50 of targeted LPH-NP was 0.075 mg/kg (Fig. 5B), which was the same as the targeted LPD-NP formulation [4]. Optimal dose for maximum gene silencing effect (80%) was 0.15 mg/kg, and further increase of dose did not result in any higher activity (Fig. 5B). Both the in vitro and in vivo gene silencing data showed that LPD-NP and LPH-NP formulations were equivalent for gene silencing activity.
Fig. 5.
In vivo luciferase gene silencing effects of different formulations at the dose of 0.15 mg/kg (A) and that of the targeted LPH-NP at various doses (B) in the pulmonary metastatic tumors. C57BL/6 mice were i.v. injected with 2 × 105 B16F10 cells via the tail vein. Seventeen days later, mice were given i.v. injections of different siRNA formulations. After 24 h, luciferase activity in lung tumor was measured. In panel B: (closed circle), luciferase siRNA in targeted LPH-NP; (open circle), control siRNA in targeted LPH-NP. Each value represents the mean ± S.D. (n=3–4). *p<0.05, significantly different compared with the untreated control.
Fig. 6.
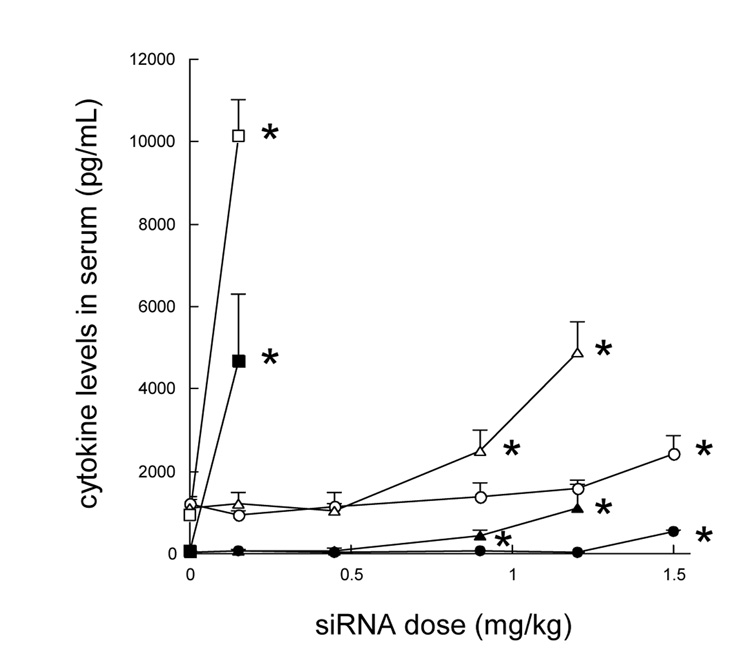
The serum cytokine levels of the mice 2 h after the i.v. injections of siRNA in different formulations. C57BL/6 mice were i.v. injected with different siRNA formulations. After 2 h, blood was collected from the tail artery and serum IL6 (closed symbols) and IL12 (open symbols) levels were analyzed. Formulations: (circles), siRNA and HA in targeted LPH-NP; (triangles), siRNA and calf thymus DNA in targeted LPD-NP; (squares), siRNA and plasmid DNA in the targeted LPD-NP. Each value represents the mean ± S.D. (n=4). *p<0.05, significantly different compared with the untreated control (siRNA dose = 0 mg/kg).
3.6. Immunotoxicity
Systemic immunotoxicity study was performed in C57BL/6 mice (Fig. 6). The immunotoxicity of the formulations was evaluated by their induction of the proinflammatory cytokines (IL6 and IL12) in the serum. The targeted LPH-NP did not induce significant production of IL6 and IL12 over a wide dose range of 0.15–1.2 mg/kg. On the other hand, the targeted LPD-NP containing calf thymus DNA significantly increased IL6 and IL12 levels at doses higher than 0.45 mg/kg. The data suggest that the therapeutic window of the targeted LPH-NP was greatly improved as compared to the targeted LPD-NP. Thus, the targeted LPH-NP formulation shows a greater potential for clinical use as compared to the targeted LPD-NP.
4. Conclusion
We have developed the LPH-NP formulation for systemically delivering siRNA into a metastatic tumor model. The targeted LPH-NP showed similar characteristics and gene silencing activity compared to the targeted LPD-NP, while significantly improved the therapeutic window by at least 2.7-fold. Containing no foreign DNA in the LPH-NP formulation also promises its potential use in human.
Acknowledgements
The authors would like to thank Dr. Joyeeta Sen for synthesizing the DSPE-PEG2000-anisamide and the Electron Microscopy Center at the Dental Research Center at UNC for the assistance on the TEM imaging. B16F10 cells were transduced with the luciferase gene in Dr. Pilar Blancafort’s lab. This project was supported by institutional funds at UNC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Li SD, Huang L. Surface-modified LPD nanoparticles for tumor targeting. Ann. N.Y. Acad. Sci. 2006;1082:1–8. doi: 10.1196/annals.1348.001. [DOI] [PubMed] [Google Scholar]
- 2.Li SD, Huang L. Targeted delivery of antisense oligodeoxynucleotide and small interference RNA into lung cancer cells. Mol.Pharm. 2006;3:579–588. doi: 10.1021/mp060039w. [DOI] [PubMed] [Google Scholar]
- 3.Li SD, Chen YC, Hackett MJ, Huang L. Tumor-targeted delivery of siRNA by self-assembled nanoparticles. Mol. Ther. 2008;16:163–169. doi: 10.1038/sj.mt.6300323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li SD, Chono S, Huang L. Efficient gene silencing in metastatic tumor by siRNA formulated in surface-modified nanoparticles. J. Control. Rel. 2008;126:77–84. doi: 10.1016/j.jconrel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li SD, Chono S, Huang L. Efficient oncogene silencing and metastasis inhibition via systemic delivery of siRNA. Mol. Ther. doi: 10.1038/mt.2008.51. (online available 2008 Mar 18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolcato-Bellemin AL, Bonnet ME, Creusat G, Erbacher P, Behr JP. Sticky overhangs enhance siRNA-mediated gene silencing. Proc. Natl. Acad. Sci. U.S.A. 2007;104:16050–16055. doi: 10.1073/pnas.0707831104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwartz DA, Quinn TJ, Thorne PS, Sayeed S, Yi AK, Krieg AM. CpG motifs in bacterial DNA cause inflammation in the lower respiratory tract. J. Clin. Invest. 1997;100:68–73. doi: 10.1172/JCI119523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito T, Iida-Tanaka N, Niidome T, Kawano T, Kubo K, Yoshikawa K, Sato T, Yang Z, Koyama Y. Hyaluronic acid and its derivative as a multi-functional gene expression enhancer: Protection from non-specific interactions, adhesion to targeted cells, and transcriptional activation. J. Control. Rel. 2006;112:382–388. doi: 10.1016/j.jconrel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 9.Kafedjiiski K, Jetti RKR, Foger F, Hoyer H, Werle M, Hoffer M, Bernkop-Schnurch A. Synthesis and in vitro evaluation of thiolated hyaluronic acid for mucoadhesive drug delivery. Int. J. Pharm. 2007;343:48–58. doi: 10.1016/j.ijpharm.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 10.U.S. Food and Drug Administration. New Device Approval: Restylane™ Injectable Gel - P020023. 2003 http://www.fda.gov/cdrh/MDA/DOCS/p020023.html.
- 11.Banerjee R, Tyagi P, Li S, Huang L. Anisamide-targeted stealth liposomes: a potent carrier for targeting doxorubicin to human prostate cancer cells. Int. J. Cancer. 2004;112:693–700. doi: 10.1002/ijc.20452. [DOI] [PubMed] [Google Scholar]
- 12.Pham TQ, Berghofer P, Liu X, Greguric I, Dikic B, Ballantyne P, Mattner F, Nguyen V, Loc'h C, Katsifis A. Preparation and biologic evaluation of a novel radioiodinated benzylpiperazine, 123I-MEL037, for malignant melanoma. J. Nucl. Med. 2007;48:1348–1356. doi: 10.2967/jnumed.107.041673. [DOI] [PubMed] [Google Scholar]
- 13.Zhang X, Chen ZG, Choe MS, Lin Y, Sun SY, Wieand HS, Shin HJ, Chen A, Khuri FR, Shin DM. Tumor growth inhibition by simultaneously blocking epidermal growth factor receptor and cyclooxygenase-2 in a xenograft model. Clin. Cancer Res. 2005;11:6261–6269. doi: 10.1158/1078-0432.CCR-04-2102. [DOI] [PubMed] [Google Scholar]
- 14.Zhang JS, Liu F, Conwell CC, Tan Y, Huang L. Mechanistic studies of sequential injection of cationic liposome and plasmid DNA. Mol. Ther. 2006;13:429–437. doi: 10.1016/j.ymthe.2005.08.021. [DOI] [PubMed] [Google Scholar]