XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease (original) (raw)
. Author manuscript; available in PMC: 2009 Sep 5.
Summary
Inflammatory bowel disease (IBD) has been attributed to aberrant mucosal immunity to the intestinal microbiota. The transcription factor XBP1, a key component of the endoplasmic reticulum (ER) stress response, is required for development and maintenance of secretory cells and linked to JNK activation. We report that XBP1 deletion in intestinal epithelial cells (IEC) results in spontaneous enteritis and increased susceptibility to induced colitis secondary to both Paneth cell deficiency and overactive responses of the intestinal epithelial cell (IEC) to the IBD-inducers, TNFα and flagellin. An association of XBP1 variants with human IBD was identified and replicated (rs35873774, P-value 1.6×10−5) with novel, private hypomorphic variants identified as susceptibility factors. Hence, intestinal inflammation can originate solely from XBP1 abnormalities in IEC thus linking cell-specific ER stress to the induction of organ-specific inflammation. We report the first mouse model of spontaneous intestinal inflammation arising from alterations in a genetic risk factor for human IBD.
Introduction
In eukaryotes, signals emanating from the ER induce a transcriptional program that enables cells to survive ER stress. This highly coordinated response, the Unfolded Protein Response (UPR), facilitates the folding, processing, export and degradation of proteins emanating from the ER during stressed conditions (Ron and Walter, 2007). Three distinct UPR signalling pathways exist in mammalian cells that include ER transmembrane inositol-requiring enzyme-1α and β (IRE1α and β), protein kinase-like ER kinase (PERK), and activating transcription factor 6 (ATF6) (Wu and Kaufman, 2006). The most evolutionarily conserved of these is the kinase/endoribonuclease IRE1 whose activation by ER stress results in the excision of a 26 bp fragment from the mRNA encoding the transcription factor X-box-binding protein-1 (XBP1) by an unconventional splicing event that generates XBP1s, a potent inducer of a subset of UPR target genes (Calfon et al., 2002). XBP1s is required for ER expansion (Shaffer et al., 2004), the development of highly secretory cells such as plasma cells, pancreatic and salivary gland epithelial cells, adaptation of tumor cells to hypoxic conditions and glucose deprivation (Reimold et al., 2001; Lee et al., 2005). XBP1s directs transcription of a core group of genes involved in constitutive maintenance of ER function in all cell types, and a remarkably diverse set of tissue- and condition-specific targets (Acosta-Alvear et al., 2007; Lee et al., 2003b; Shaffer et al., 2004). IECs additionally express IRE1β whose deletion results in increased ER stress, and exacerbated dextran sodium sulfate (DSS)-induced colitis (Bertolotti et al., 2001).
We hypothesized that a stressful environmental milieu in cells with high secretory activity might induce inflammation. If so, inducing ER stress in vivo by cell-specific XBP1 deletion might lead to organ-specific inflammation providing a mechanistic explanation for the initiation of proinflammatory diseases. We focused on intestinal epithelium which contains four highly secretory epithelial cell lineages that are exposed to high concentrations of exogenous antigens; absorptive epithelium, goblet, Paneth and enteroendocrine cells that are derived from a common, constantly renewing intestinal epithelial stem cell (Barker et al., 2007). We show that induction of ER stress in intestinal epithelium through tissue (and cell type)-specific disruption of XBP1 results in spontaneous enteritis due to inability of XBP1-deficient IECs to properly generate antimicrobial activity and respond appropriately to inflammatory signals in the local milieu. Several single nucleotide polymorphisms (SNPs) within the XBP1 gene locus on chromosome 22q12.1 confer risk for both types of IBD, Crohn’s disease (CD) and ulcerative colitis (UC), establishing the ER stresss pathway as a common genetic contributor to IBD in the human population.
Results
XBP1 deletion in IEC leads to ER stress and spontaneous enteritis
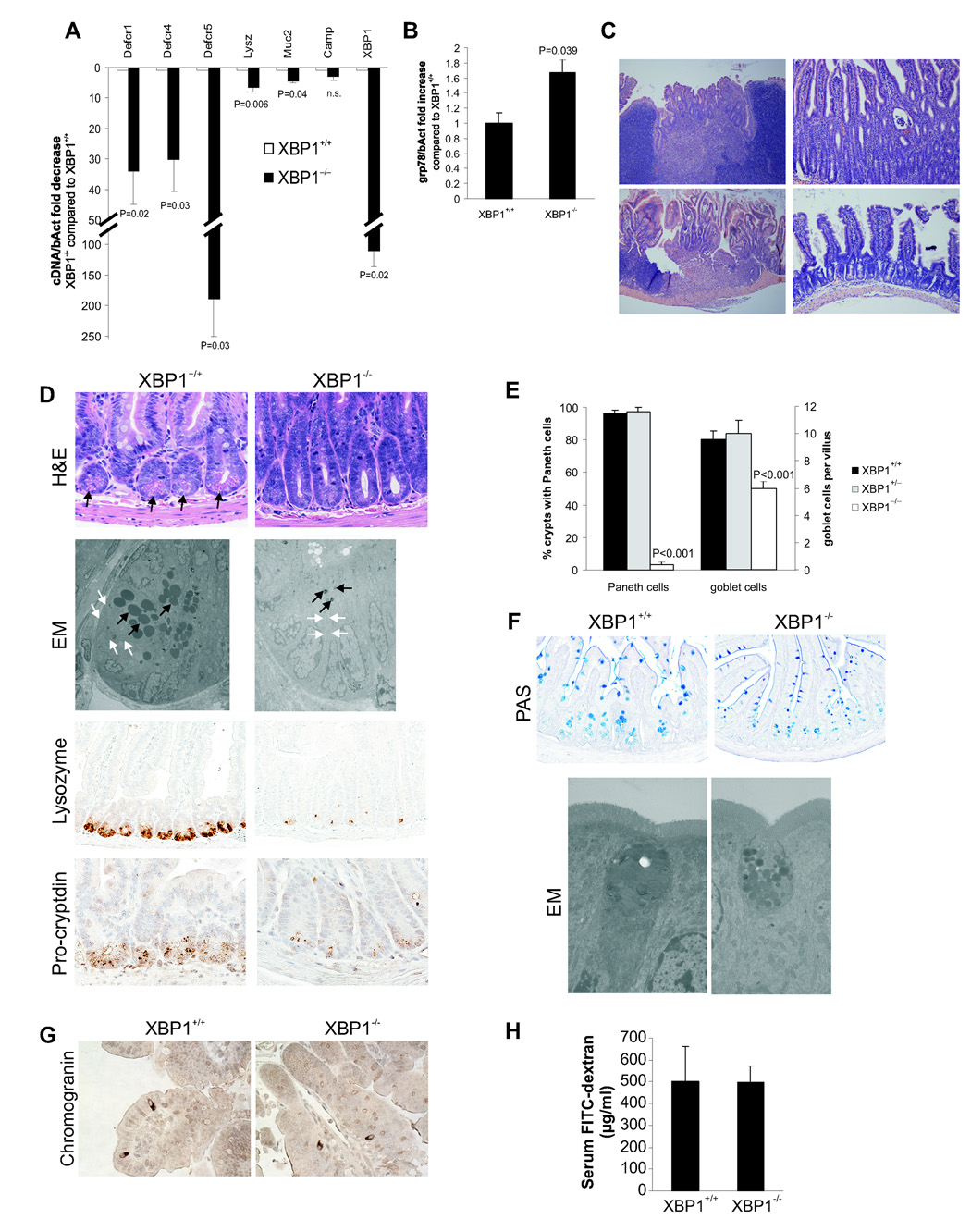
XBP1flox/flox mice were generated by targeting loxP sites to introns flanking exon 2, and bred onto Villin (V)-Cre transgenic mice (Supplementary Fig. 1abc), that directs Cre recombinase activity specifically to small and large intestinal epithelium (Madison et al., 2002). XBP1flox/floxVCre (XBP1−/−) offspring were born at a Mendelian ratio and developed normally. XBP1 exon 2 was efficiently and functionally deleted specifically within the intestinal epithelium (99% in small intestine, 87±4% in colon) (Fig. 1A and Supplementary Fig. 1DE). Elevated basal grp78 levels in XBP1−/− small intestinal epithelia indicated increased ER stress (Fig. 1B) that was confirmed by microarray analysis showing both increased grp78 (Haspa5) and Chop (Ddit3) (_P_=0.02) (Supplementary Table 1 and Supplementary Fig. 2A). Spontaneous small intestinal mucosal inflammation, in association with increased ER stress occurred in 19/31 (61%) adult XBP1−/− but not in (0/20) XBP1+/+ mice (X2 _P_=9.87×10−6; Fig. 1C). Notably, 5/16 (31%) heterozygous XBP1flox/wtVCre “XBP1+/−” mice displayed mild spontaneous small intestinal inflammation (X2 _P_=0.007; Supplementary Fig. 2B). The inflammatory changes were patchy and ranged in severity from lamina propria polymorphonuclear infiltrates, to crypt abscesses and frank ulcerations without granulomas (Fig. 1C and Supplementary Fig. 2B).
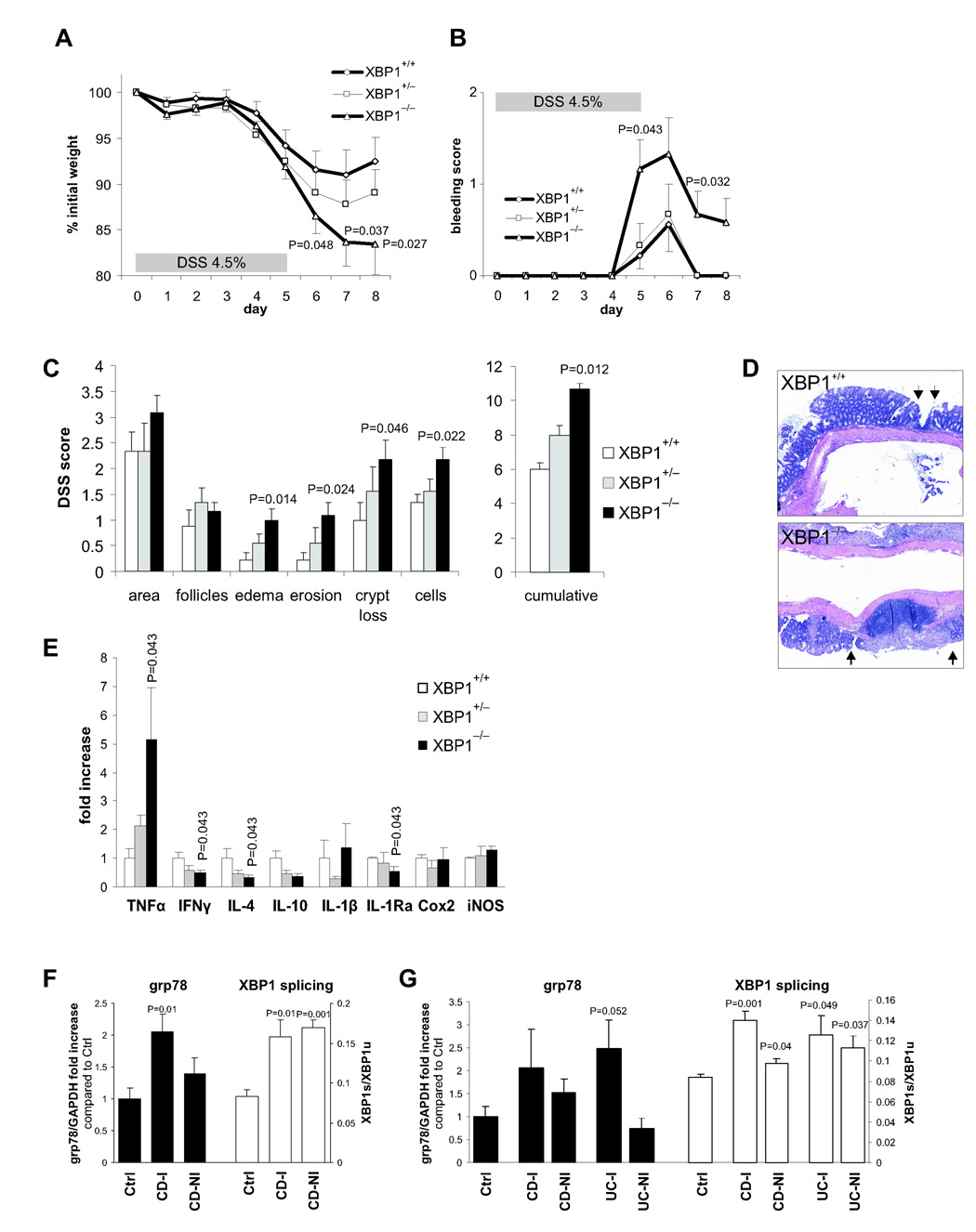
Figure 1. Spontaneous enteritis and Paneth cell loss in XBP1−/− mice.
A. Small intestinal mucosal scrapings (n = 8 per group) from _Xbp1_-deleted (“XBP1−/−”) and _Xbp1_-sufficient (“XBP1+/+”) mouse intestinal epithelium were analyzed for cryptdin-1 (Defcr1), cryptdin-4 (Defcr4), cryptdin-5 (Defcr5), lysozyme (Lysz), mucin-2 (Muc2), cathelicidin (Camp1), and XBP1 (primers binding in the floxed region) mRNA expression. Data are expressed as fold decrease in XBP1−/− compared to XBP1+/+ specimens, normalized to β-actin (Student’s t test). B. Fold increase in grp78 mRNA expression in XBP1−/− compared to XBP1+/+ epithelium, normalized to β-actin (n = 3 per group, Student’s t test). C. Spontaneous enteritis in XBP1−/− mice (upper panels and lower left panel), and normal histology of XBP1+/+ mice (lower right panel). Upper left, cryptitis with villous shortening, crypt regeneration and architectural distortion; upper right, neutrophilic crypt abscesses; lower left, duodenitis with surface ulceration and granulation tissue. D. Paneth cells with typical eosinophilic granules on H&E stained sections at the base of crypts in XBP1+/+, but not XBP1−/− epithelium. Electron microscopy (EM) with only rudimentary electron-dense granules and a contracted ER in XBP1−/− basal crypt epithelial cells, normal configuration in XBP+/+ mice. Immunohistochemistry (IHC) for the granule proteins lysozyme and pro-cryptdin in XBP1+/+ and XBP1−/− epithelia. E. Enumeration of Paneth cells and goblet cells in small intestines (n = 5 per group, Student’s t test). F. Goblet cell staining by PAS in XBP1+/+ and XBP1−/− epithelia. EM exhibited smaller cytoplasmic mucin droplets and a contracted ER in XBP1−/− goblet cells. No structural abnormalities were found in neighboring absorptive epithelia in XBP1−/− mice. G. The marker for enteroendocrine cells, chromogranin, was detected by IHC in small intestines of XBP1+/+ and XBP1−/− mice. H. XBP1+/+ and XBP1−/− mice were orally administered FITC-dextran, and FITC-dextran serum levels assayed 4h later.
Absent Paneth cells and reduced goblet cells in XBP1−/− epithelium
XBP1−/− intestine was completely devoid of Paneth cells (Fig. 1DE), compared to XBP1+/+ and XBP1+/− mice (Fig. 1E and Supplementary Fig. 2B). Paneth cell granules store lysozyme and pro-forms of cryptdins, which were barely detectable in XBP1−/− crypts (Fig. 1D) and electron microscopy (EM) confirmed few rudimentary electron-dense granules of minute size, and a compressed ER in XBP1−/− Paneth cells (Fig. 1D). mRNA expression of cryptdins-1, -4, and -5 and lysozyme, but not cathelicidin, were substantially reduced (Fig. 1A). We also noted reduced numbers and size of goblet cells within the small intestine but not colon with reduced secretory granules by EM and reduced mRNA for the goblet cell protein Muc2 in XBP1−/− small intestinal epithelia (Fig. 1AF and Supplementary Fig. 2C). Enteroendocrine cells were unaffected (Fig. 1G and Supplementary Fig. 2D) and the epithelial barrier function of absorptive epithelia was normal (Fig. 1H). Thus, XBP1−/− mice exhibited a major defect in Paneth cells and a minor defect in goblet cells in the small intestine with an unperturbed epithelial barrier.
XBP1 deletion results in apoptosis of differentiated Paneth cells and exhibits signs of a regenerative response
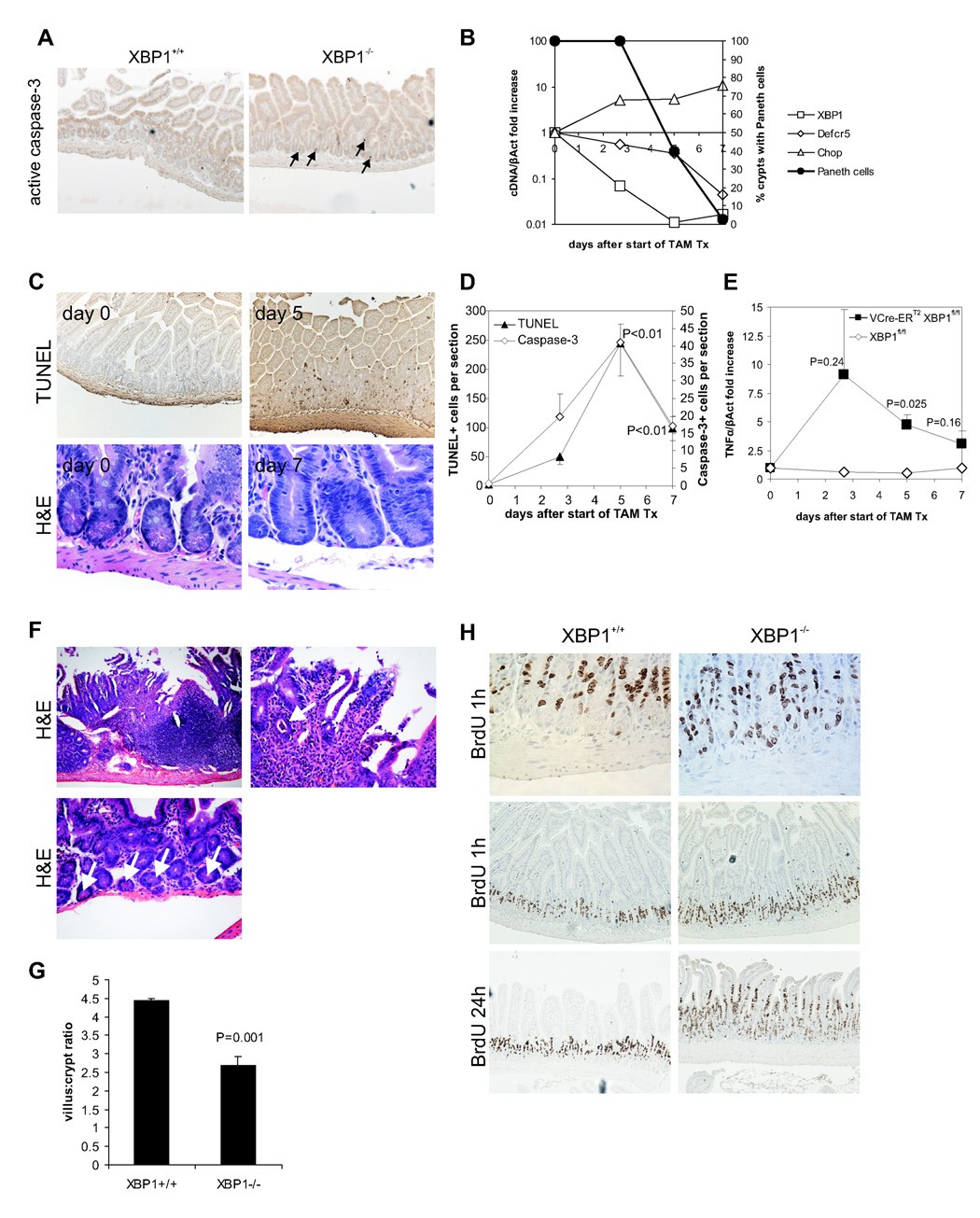
Quantitative PCR (β-catenin, Tcf4, Math1, Hes1; Supplementary Fig. 3A), microarray analysis (Supplementary Table 1) and β-catenin distribution (Supplementary Fig. 3B) of XBP1−/− and XBP1+/+ intestinal epithelial mRNA did not reveal significant alterations in factors involved in intestinal cell fate decisions (Barker et al., 2007). We hypothesized that the highly secretory Paneth cell might undergo programmed cell death from failure to manage ER stress as observed in pancreatic acinar cells (Lee et al., 2005). Indeed, a few pyknotic, apoptotic cells were detected in XBP1−/− crypts (anti-active caspase-3+ and TUNEL+; Fig. 2A and Supplementary Fig. 4A). To circumvent the problems associated with detecting a low-frequency event (apoptosis) in a slowly replenishing cell population, we generated XBP1floxneo/floxneoVillinCre-ERT2 mice (Supplementary Fig. 1A). Along with efficient deletion of XBP1 after initiation of tamoxifen treatment (Fig. 2B), Paneth cell numbers were reduced by 98% on day 7, paralleled by a similar decrease in cryptdin-5 mRNA transcripts. Apoptotic epithelial nuclei (Fig. 2C and Supplementary Fig. 4B) were observed after 2.7 days, peaked at day 5, and declined on day 7 (Fig. 2D). Apoptotic cells were present at the base of crypts (Paneth cells), and in villous epithelium (goblet cells) (Fig. 2C). We observed a gradual increase of TNFα and Chop (Ddit3) mRNA (Fig. 2BE), similar to XBP1−/− mice (Supplementary Table 1 and Supplementary Fig. 2A). We noted small intestinal inflammation in individual XBP1floxneo/floxneoVillinCre-ERT2 mice at all time-points analyzed (2.7, 5, and 7 days); focal enteritis was present in 4 of 9 mice at day 5 (44%) ranging from lamina propria polymorphonuclear infiltrates, to crypt abscesses and frank ulcerations (Fig. 2F, upper two panels), despite only minor reductions in Paneth cells (Fig. 2F, lower panel). Cumulatively, at all time points examined, we observed enteritis in 7/18 (39%) XBP1floxneo/floxneoVillinCre-ERT2 and 0/7 controls after induction with tamoxifen. The small intestinal epithelium exhibited villus shortening with a reduction of the villus:crypt ratio (Fig. 2G), indicative of a regenerative response in XBP1−/− mice. A 1h pulse of bromodeoxyuridine (BrdU) labelled the proliferative pool of intestinal stem cells, and was similar in XBP1+/+ and XBP1−/− mice (Fig. 2H). However, 24h after BrdU injection, labeled cells were detected higher up in the crypt-villus axis in XBP1−/− mice, indicating an increased migration rate (Fig. 2H). Thus XBP1 affects IEC homeostasis both through controlling cell renewal and cell death.
Figure 2. XBP1 deletion results in apoptotic Paneth cell loss, inflammation, a distorted villus:crypt ratio, and IEC hyperproliferation.
A. Apoptotic nuclei were identified in XBP1+/+ (XBP1flox/floxVCre) and XBP1−/− (XBP1flox/flox) sections with anti-active (cleaved) caspase-3. Arrows point to apoptotic cells. B. XBP1floxneo/floxneoVCre-ERT2 mice were administered 5 daily doses of 1 mg tamoxifen to induce deletion of the XBP1floxneo gene in the intestinal epithelium. XBP1, cryptdin-5 (Defcr5), and Chop mRNA (all expressed normalized to β-actin; left y axis) expression in epithelium during and after tamoxifen treatment. Percentage of crypts with Paneth cells on H&E staining is shown (right y axis). Representative experiment of 3 performed. C. TUNEL and H&E staining on small intestinal sections of tamoxifen-treated XBP1floxneo/floxneoVCre-ERT2 mice collected at the indicated days. D. TUNEL+ and caspase-3+ cells were enumerated by light microscopy (3 mice per time-point with ileal and jejunal sections each; P values indicate comparisons to time-point 0; Student’s t test). E. TNFα mRNA was quantified by qPCR in small intestinal epithelial scrapings from ileum harvested at the indicated time-points after start of tamoxifen administration from VCre-ERT2 XBP1floxneo/floxneo (n = 4 per time-point) or XBP1floxneo/floxneo (n = 1 per time-point) mice. P values indicate comparisons to time-point 0; Student’s t test. F. Enteritis in the small intestine in VCre-ERT2 XBP1floxneo/floxneo mice on day 5 after TAM administration. Upper left panel, 100×; upper right panel, same section, 400×, arrow points to a crypt abscess; lower panel 100×, crypts with Paneth cells (arrows). G. Jejunal sections of XBP1flox/flox (XBP1+/+; n = 7) and XBP1flox/floxVCre (XBP1−/−; n = 8) mice were assessed for their villus:crypt ratio on H&E stainings (ratios of ≥ 4:1 are considered normal for jejunum). H. XBP1flox/flox (XBP1+/+) and XBP1flox/floxVCre (XBP1−/−) mice were administered bromodeoxyuridine (BrdU) i.p., and small intestinal sections harvested after 1 h and 24 h (n = 3 per genotype per time-point). The 1 h time-point labels the pool of proliferating IEC in the crypts (mostly transit amplifying IEC), whereas the 24 h time-point assesses the migration along the crypt-villus axis indicating the turn-over of the IEC compartment.
XBP1 deletion impairs mucosal defence to oral Listeria monocytogenes infection
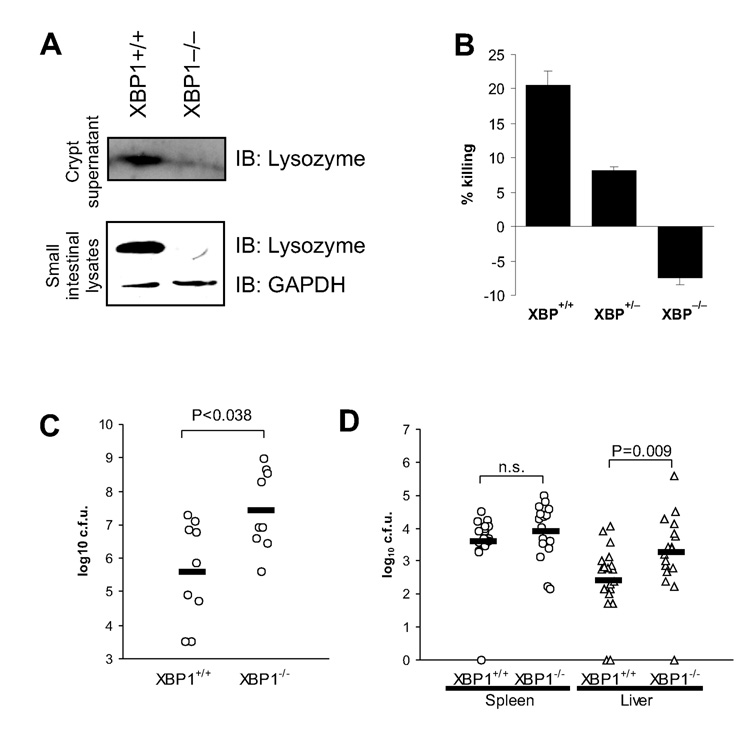
XBP1−/− small intestinal lysates and supernatants lacked detectable lysozyme in response to carbamyl choline (Fig. 3A) or LPS (not shown), and LPS-elicited XBP1−/− crypt supernatants lacked bactericidal killing activity (Fig. 3B). Oral infection with Listeria monocytogenes, a gram positive intracellular pathogen that is affected by Paneth cell defects (Kobayashi et al., 2005) revealed that 10 hours after infection, 100-fold higher numbers of colony forming units (c.f.u.) of L. monocytogenes were recoverable from faeces of XBP1−/− compared to XBP1+/+ mice (Fig. 3C). Translocation into liver and spleen after 72h revealed a 10-fold higher number of L. monocytogenes recovered from XBP1−/− livers, but similar numbers from spleen (Fig. 3D). These data suggest that XBP1 in Paneth cells is required to decrease the luminal burden of L. monocytogenes.
Figure 3. XBP1 deficiency in epithelium results in impaired antimicrobial function.
A. Lower panel. Small intestinal tissue from XBP1+/+ and XBP1−/− mice was homogenized, resolved on SDS-PAGE and detected by anti-lysozyme IgG and GAPDH to ensure equal loading. Upper panel. Small intestinal crypts isolated from XBP1+/+ and XBP1−/− animals were stimulated with 10µM carbamyl choline (CCh). Supernatants were precipitated, resolved on SDS-PAGE and detected by anti-lysozyme IgG. Blots are representative of 2 independent experiments. B. Small intestinal crypts were stimulated with LPS for 30min, and supernatants assayed for bactericidal activity. Data are expressed as % killing compared to unstimulated crypts, and are representative of 2 independent experiments. C. Intestinal epithelial cell-specific XBP1−/− mice (n = 9) and XBP1+/+ littermates (n = 9; 5–10 weeks of age) were perorally infected with 3.6×108 L. monocytogenes. Faeces was aseptically collected 10h after infection and colony forming units (c.f.u.) of L. monocytogenes determined. Data are presented as c.f.u. per mg dry weight of faeces. D. Oral infection with L. monocytogenes was performed as in (C), liver and spleen aseptically harvested 72h after infection, and c.f.u. of L. monocytogenes determined (XBP1+/+ n = 20; XBP1−/− n = 17). Data are expressed as c.f.u. per organ. Two-tailed Mann-Whitney test was performed for (C) and (D).
XBP1 deficiency results in enhanced responses of IECs to typical mucosal inflammatory signals
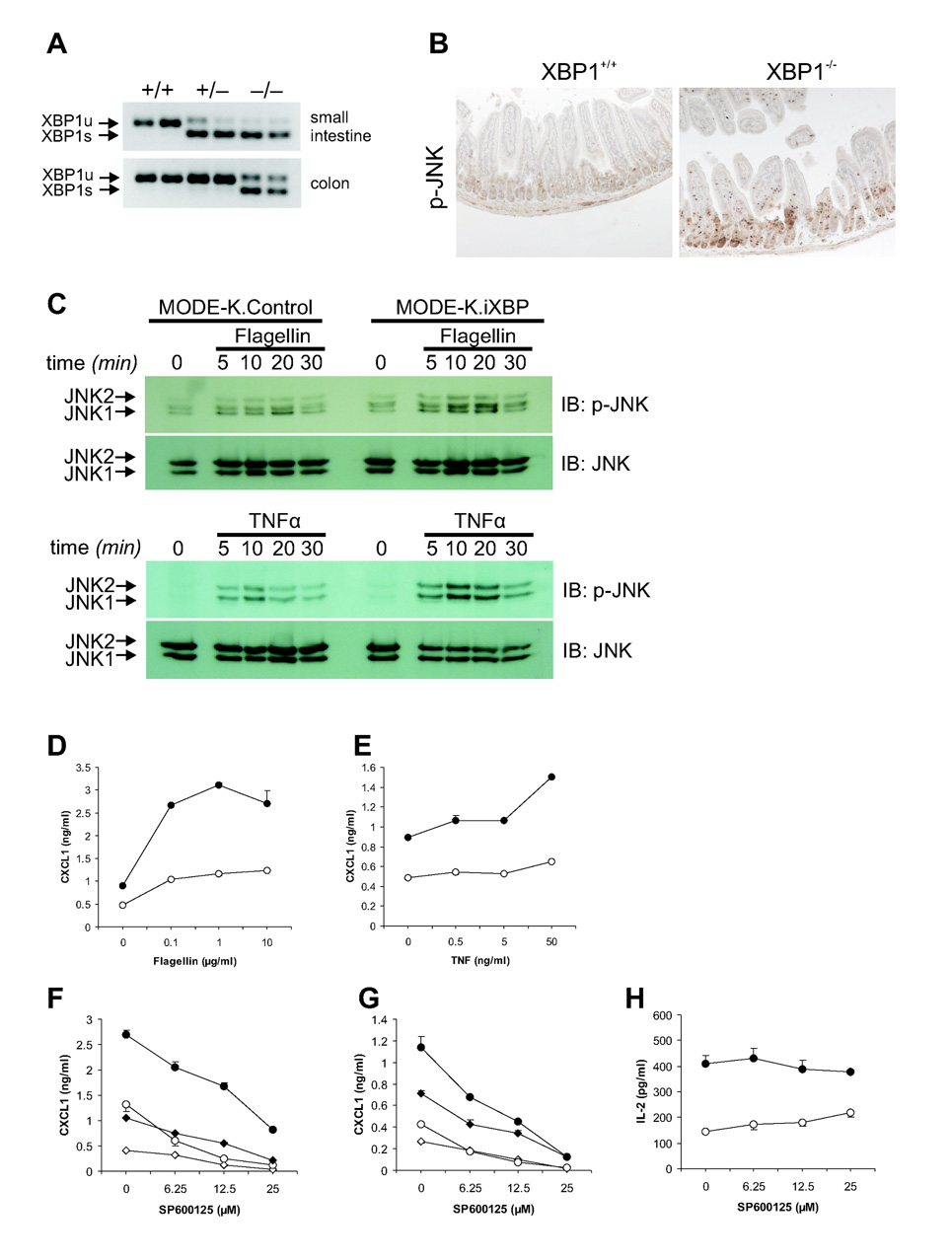
XBP1 mRNA splicing is a marker of IRE1 activation and ER stress (Calfon et al., 2002; Lin et al., 2007). Virtually complete splicing of mutant XBP1 mRNAs in XBP1−/− small and large intestine and partial splicing in XBP1+/− small intestine was observed in contrast to barely detectable splicing in XBP1+/+ mice (Fig. 4A) indicating IRE1 hyperactivation. JNK phosphorylation was increased in XBP1−/− small intestinal epithelia compared to controls consistent with the described TRAF2-dependent function of IRE1 to activate JNK (Urano et al., 2000) (Fig. 4B). To test whether XBP1 mediated intestinal inflammation arose from increased JNK activity in a microbiota and cytokine free system, we silenced XBP1 expression in the mouse IEC line MODE-K with a siRNA retrovirus (iXBP) and used flagellin and TNFα, as proinflammatory stimulants (Lodes et al., 2004). TNFα and flagellin increased JNK phosphorylaton and CXCL1 production from MODE-K.iXBP (50–90% reduction of XBP1) compared to MODE-K.Ctrl cells (Fig. 4CDE) that was dose-dependently and specifically (Supplementary Fig. 5AB) blocked by the JNK inhibitor, SP600125 (Fig. 4FG) but did not affect CD1d-restricted MODE-K antigen presenting function (van de Wal et al., 2003) (Fig. 4H). We conclude that impaired XBP1 expression directly heightens proinflammatory JNK/SAPK signaling in IECs in response to environmental stimuli and may contribute to Paneth, goblet cell and MODE-K.iXBP apoptosis (Fig. 2ACD, Supplementary Fig. 6AB).
Figure 4. XBP1 deficiency results in increased inflammatory tone of the epithelium.
A. Small intestinal and colonic epithelial mRNA scrapings from XBP1+/+, XBP1+/−, and XBP1−/− mice were analyzed for XBP1 mRNA splicing status. B. Small intestinal formalin-fixed sections were stained with rabbit anti-phospho-JNK antibody, and revealed a patchy staining pattern in XBP1−/−, but not XBP1+/+ sections. Control rabbit mAb was negative (not shown). Representative of n = 5 per group. C. MODE-K.iXBP and MODE-K.Control were stimulated for the indicated time periods with flagellin (1 µg/ml) and TNFα (50 ng/ml) and analyzed for P-JNK and total JNK by Western. D. MODE-K.iXBP (filled circles) and MODE-K.Ctrl (open circles) cells were stimulated for 4h with flagellin, and supernatants assayed by ELISA for CXCL1. E. Experiment as in (D), with TNFα. F. MODE-K.iXBP (circles) and MODE-K.Ctrl (diamonds) cells were stimulated with either 10µg/ml flagellin (filled symbols) or media alone (open symbols) for 4h, with the JNK inhibitor SP600125 and supernatants assayed for CXCL1. G. As in (F), MODE-K cells were stimulated with 50 ng/ml TNF-α (filled symbols) or media alone (open symbols). H. MODE-K.iXBP (filled circles) and MODE-K.Ctrl (open circles) cells loaded with the glycolipid antigen α-galactosylceramide (αGC), fixed, and co-cultured with the CD1d-restricted NKT cell hybridoma DN32.D3 and antigen presentation measured as IL-2 release from DN32.D3.
XBP1 deficiency leads to increased susceptibility to experimental colitis
The XBP1−/− colon, unlike the small intestine, did not exhibit spontaneous colitis but colonic IECs displayed evidence of increased ER stress (Fig. 4A). We therefore examined the in vivo effects of DSS, a toxin for mucosal epithelial cells that disrupts barrier function (Strober et al., 2002). XBP1−/− mice given 4.5% DSS in the drinking water exhibited more severe wasting and rectal bleeding than XBP1+/+ littermates (Fig. 5AB). Histologically, XBP1−/− colons displayed increased areas of mucosal erosions, edema, and cellular infiltration along with increased crypt loss compared to XBP1+/+ littermates (Fig. 5CD). XBP1+/− mice exhibited an intermediate phenotype (Fig. 5ABC). Antibiotic treatment abrogated the differences in severity of DSS colitis between XBP1+/+ and XBP1−/− mice (Supplementary Fig. 7AB) highlighting the importance of the commensal flora in the colitis observed (Fig. 5A–D). Levels of TNFα, a central mediator of inflammation in DSS colitis (Kojouharoff et al., 1997), were elevated in DSS treated XBP1−/− vs XBP1+/+ colonic tissues with intermediate TNFα expression in XBP1+/− mice (Fig. 5E).
Figure 5. XBP1 deficiency increases susceptibility to DSS colitis.
A. 4.5% DSS was administered in drinking water for 5 days and then replaced by regular drinking water in XBP1+/+ (n = 9), XBP1+/− (n = 9) and XBP1−/− (n = 12) littermates (age 6–12 weeks). Wasting is presented as % of initial weight. One-tailed Student’s t test was performed. B. Presence of rectal bleeding during DSS colitis was assessed daily and scored as in Methods. Mean ± s.e.m.; XBP1+/+ (n = 9), XBP1+/− (n = 9) XBP1−/− (n = 12). Two-tailed Mann-Whitney test was peformed. C. Individual signs of inflammation of colonic tissue harvested on day 8 of DSS colitis were scored blindly. Two-tailed Mann-Whitney test was performed. D. Typical colonic histology on day 8 of DSS colitis. Arrows, borders of ulcers. E. mRNA expression (normalized to βactin) of inflammatory mediators was quantified by qPCR in colonic specimens on day 8 of DSS colitis. n = 4 per group. Mean ± s.e.m analyzed by two-tailed Mann-Whitney test. F. Human ileum in Crohn’s disease exhibits signs of ER stress. Inflamed (“CD-I”, n = 3) and non-inflamed (“CD-NI”, n = 3) ileal biopsies from CD patients and healthy control (“Ctrl”, n = 4) subjects were analyzed for grp78 mRNA expression (levels in Controls were arbitrarily set at 1, and CD-I and CD-NI levels expressed as ratio to Controls; left y axis). XBP1 mRNA splicing is expressed as ratio of XBP1s/XBPu (right y axis). G. Human colon mucosa in Crohn’s disease (“CD”) and ulcerative colitis (“UC”) exhibits signs of ER stress. Colonic biopsies from inflamed (“−I”) and non-inflamed (“−NI”) CD and UC patients (n = 3 each) and healthy control subjects (“Ctrl”, n = 4) were analyzed for grp78 mRNA expression and XBP1 splicing as described in (F).
Human ileal and colonic mucosa in CD and UC exhibit signs of ER stress
We analyzed UPR activation in the intestine of healthy individuals, CD and UC in ileal and colonic biopsies. Grp78 expression was increased in inflamed ileal CD mucosa and XBP1s levels were increased in both inflamed and non-inflamed ileal CD biopsies (Fig. 5F). Similarly, XBP1s levels in inflamed and non-inflamed colonic CD and UC mucosa were increased compared to those from healthy individuals, and there was increased grp78 in inflamed UC and a trend toward increased grp78 expression in inflamed CD specimens (Fig. 5G). These data indicate the presence of ER stress and increased IRE1 activity in the ileum and colon of CD and UC patients.
SNPs within the XBP1 gene region are associated with IBD
Three previously reported genome-wide linkage studies independently suggested linkage of the 22q12 region with IBD (Hampe et al., 1999; Barmada et al., 2004; Vermeire et al., 2004), with signals as close as 0.3 Mb from the XBP1 gene. We examined a German patient cohort of 1103 controls, 550 CD, and 539 UC patients (panel 1), genotyping for twenty tagging SNPs (average SNP distance 5.25 kb; Supplementary Fig. 8A–E), selected from HapMap data of individuals of European ancestry using de Bakkers algorithm as implemented in Haploview (Barrett et al., 2005). Three SNPs were significantly associated with IBD: rs5997391, rs5762795, and rs35873774 (Table 1). The latter SNP, located in intron 4 of XBP1, remained significantly associated after correcting for multiple testing using Bonferroni correction (P-valuecorr. = 0.0011).
Table 1. Association results of twenty SNPs genotyped across the XBP1 gene region.
Single-point association results for the three independent IBD, i.e. CD+UC) panels are shown. Inter-marker distances are shown in kilobases (kb) and positions refer to NCBI’s build 35. Minor allele frequencies (MAF) are listed for unaffected controls (U), Crohn disease (CD), and ulcerative colitis (UC) patients. _P_-values are shown for the standard χ2-test with one degree of freedom and _P_-values <0.05 are highlighted by bold type. Odds ratios (OR) including 95% confidence intervals (95% CI) are shown for carriership of the rarer allele A1 and _P_-values <0.05 are highlighted by bold type. For detailed genotype counts see Supplementary Table 2.
| Panel 1(1103 controls, 550 CD, 539 UC) | Panel 2(2042 controls, 1303 CD, 551 UC) | Panel 3(2177 controls, 909 CD, 537 UC) | Panel 1 + 2 +3(5322 controls, 2762 CD, 1627 UC) | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| # | dbSNP ID | Position | Distance [kb] | A1 | MAF U | MAF CD | MAF UC | P-Value CD | P-Value UC | P-Value IBD | MAF U | MAF CD | MAF UC | P-Value CD | P-Value UC | P-Value IBD | MAF U | MAF CD | MAF UC | P-Value CD | P-Value UC | P-Value IBD | P-Value IBD | OR (95% CL) IBD |
| 1 | rs714191 | 27,469,965 | - | C | 0.18 | 0.19 | 0.18 | 0.45 | 0.79 | 0.53 | 0.18 | 0.18 | 0.18 | 0.39 | 0.84 | 0.45 | 0.18 | 0.20 | 0.19 | 0.078 | 0.48 | 0.097 | 0.086 | 1.07 (0.98–1.16) |
| 2 | rs5997391* | 27,470,093 | 0.13 | T | 0.07 | 0.06 | 0.05 | 0.15 | 0.012 | 0.014 | 0.06 | 0.05 | 0.05 | 0.057 | 0.036 | 0.014 | 0.07 | 0.06 | 0.06 | 0.15 | 0.12 | 0.058 | 0.000072†‡ | 0.79 (0.69–0.89) |
| 3 | rs5752792* | 27,478,313 | 8.22 | C | 0.16 | 0.17 | 0.18 | 0.50 | 0.085 | 0.15 | 0.17 | 0.18 | 0.19 | 0.45 | 0.099 | 0.19 | 0.18 | 0.19 | 0.18 | 0.21 | 0.58 | 0.22 | 0.031 | 1.08 (0.99–1.18) |
| 4 | rs6005863* | 27,481,002 | 2.69 | G | 0.41 | 0.41 | 0.39 | 0.84 | 0.25 | 0.41 | 0.41 | 0.40 | 0.37 | 0.43 | 0.032 | 0.12 | 0.42 | 0.44 | 0.39 | 0.07 | 0.11 | 0.62 | 0.22 | 0.98 (0.90–1.07) |
| 5 | rs5762788 | 27,499,324 | 18.32 | A | 0.33 | 0.30 | 0.31 | 0.10 | 0.44 | 0.12 | 0.32 | 0.29 | 0.28 | 0.0032 | 0.024 | 0.00081†‡ | 0.32 | 0.33 | 0.28 | 0.70 | 0.012 | 0.30 | 0.00042†‡ | 0.85 (0.78–0.92) |
| 6 | rs6005879 | 27,502,868 | 3.54 | A | 0.05 | 0.04 | 0.05 | 0.17 | 0.94 | 0.39 | 0.05 | 0.04 | 0.05 | 0.016 | 0.52 | 0.032 | 0.05 | 0.05 | 0.05 | 0.45 | 0.55 | 0.39 | 0.22 | 0.92 (0.80–1.06) |
| 7 | rs5752797 | 27,504,552 | 1.68 | T | 0.17 | 0.18 | 0.17 | 0.85 | 0.83 | 0.98 | 0.18 | 0.16 | 0.16 | 0.11 | 0.18 | 0.065 | 0.18 | 0.21 | 0.17 | 0.00089†‡ | 0.70 | 0.025 | 0.86 | 1.00 (0.91–1.09) |
| 8 | rs5997403 | 27,505,938 | 1.39 | T | 0.05 | 0.04 | 0.05 | 0.25 | 0.99 | 0.48 | 0.04 | 0.04 | 0.04 | 0.091 | 0.76 | 0.15 | 0.04 | 0.04 | 0.05 | 0.79 | 0.53 | 0.60 | 0.34 | 0.94 (0.81–1.09) |
| 9 | rs5762795* | 27,507,054 | 1.12 | A | 0.33 | 0.30 | 0.30 | 0.10 | 0.084 | 0.039 | 0.32 | 0.29 | 0.28 | 0.0082 | 0.016 | 0.0016†‡ | 0.32 | 0.33 | 0.28 | 0.73 | 0.00469† | 0.22 | 0.00012†‡ | 0.84 (0.78–0.92) |
| 10 | rs2267131* | 27,515,025 | 7.97 | C | 0.12 | 0.14 | 0.13 | 0.39 | 0.84 | 0.51 | 0.14 | 0.12 | 0.12 | 0.12 | 0.096 | 0.051 | 0.13 | 0.16 | 0.11 | 0.00353† | 0.20 | 0.13 | 0.92 | 0.96 (0.87–1.05) |
| 11 | rs2097461 | 27,516,433 | 1.41 | C | 0.33 | 0.30 | 0.30 | 0.14 | 0.16 | 0.077 | 0.32 | 0.29 | 0.27 | 0.012 | 0.0044 | 0.0013†‡ | 0.32 | 0.33 | 0.28 | 0.67 | 0.00573† | 0.25 | 0.00023†‡ | 0.85 (0.78–0.92) |
| 12 | rs35873774* | 27,516,485 | 0.05 | C | 0.08 | 0.05 | 0.06 | 0.0028 | 0.028 | 0.0011†‡ | 0.07 | 0.07 | 0.06 | 0.82 | 0.30 | 0.53 | 0.08 | 0.05 | 0.05 | 0.00342† | 0.00147†‡ | 0.000092†‡ | 0.000016†‡ | 0.74 (0.66–0.84) |
| 13 | rs2269578 | 27,521,397 | 4.91 | C | 0.12 | 0.13 | 0.13 | 0.71 | 0.52 | 0.54 | 0.13 | 0.12 | 0.11 | 0.10 | 0.089 | 0.043 | 0.13 | 0.15 | 0.12 | 0.017 | 0.27 | 0.24 | 0.76 | 0.94 (0.85–1.04) |
| 14 | rs3788409 | 27,522,705 | 1.31 | G | 0.33 | 0.30 | 0.31 | 0.12 | 0.39 | 0.11 | 0.32 | 0.30 | 0.28 | 0.025 | 0.029 | 0.0065 | 0.33 | 0.33 | 0.28 | 0.79 | 0.00493† | 0.19 | 0.00097†‡ | 0.85 (0.78–0.92) |
| 15 | rs6005893 | 27,524,344 | 1.64 | T | 0.05 | 0.04 | 0.04 | 0.23 | 0.17 | 0.11 | 0.05 | 0.03 | 0.04 | 0.043 | 0.18 | 0.028 | 0.04 | 0.05 | 0.05 | 0.27 | 0.18 | 0.13 | 0.19 | 0.91 (0.79–1.06) |
| 16 | rs35679096 | 27,524,947 | 0.60 | A | 0.03 | 0.03 | 0.03 | 0.45 | 0.32 | 0.28 | 0.04 | 0.03 | 0.03 | 0.43 | 0.18 | 0.24 | 0.03 | 0.04 | 0.03 | 0.39 | 0.97 | 0.55 | 0.38 | 0.93 (0.78–1.10) |
| 17 | rs133440 | 27,535,932 | 10.99 | C | 0.04 | 0.04 | 0.03 | 0.44 | 0.19 | 0.21 | 0.05 | 0.05 | 0.03 | 0.96 | 0.099 | 0.54 | 0.05 | 0.04 | 0.06 | 0.40 | 0.060 | 0.68 | 0.47 | 0.97 (0.84–1.12) |
| 18 | rs5762839* | 27,556,001 | 20.07 | T | 0.17 | 0.16 | 0.17 | 0.72 | 0.82 | 0.90 | 0.17 | 0.15 | 0.14 | 0.0039 | 0.032 | 0.0011†‡ | 0.17 | 0.19 | 0.15 | 0.092 | 0.100 | 0.70 | 0.055 | 0.88 (0.80–0.96) |
| 19 | rs5762852 | 27,567,027 | 11.03 | C | 0.17 | 0.18 | 0.17 | 0.59 | 0.65 | 0.54 | 0.17 | 0.16 | 0.15 | 0.14 | 0.11 | 0.064 | 0.17 | 0.19 | 0.15 | 0.13 | 0.090 | 0.81 | 0.46 | 0.94 (0.86–1.03) |
| 20 | rs5762853 | 27,569,785 | 2.76 | G | 0.24 | 0.24 | 0.24 | 0.94 | 0.73 | 0.80 | 0.24 | 0.22 | 0.22 | 0.029 | 0.11 | 0.017 | 0.24 | 0.27 | 0.23 | 0.050 | 0.48 | 0.28 | 0.25 | 0.94 (0.86–1.02) |
We replicated this index finding by genotyping two additional independent patient panels (panel 2: 1854 patients; 2042 controls, panel 3: 1446 patients; 2177 controls) and reproduced the significant association with two of these three index SNPs in each panel (rs5997391 and rs5762795 in panel 2; rs5762795 and rs35873774 in panel 3; Table 1 and Supplementary Table 2). Significant associations of 11 and 8 additional SNPs in panels 2 and 3, respectively, with CD, UC, and/or IBD were obtained. Several association signals in each panel were robust to correction for multiple testing and remained significant after 10,000 permutations. The combined analysis of panels 1, 2 and 3 in a total of 5322 controls, 2762 CD, and 1627 UC patients identified a total of 6 SNPs associated with IBD that were robust after correction for multiple testing and significant after 10,000 permutations; the minor alleles of these six SNPs conferred protection from IBD (Table 1 and Supplementary Fig. 8A). The strongest associated variant was rs35873774 (P-value = 1.6×10−5). The odds ratio (OR) for carriership of the rarer C allele of rs35873774 in the combined IBD panel was 0.74 (95% confidence interval [CI] 0.66–0.84; Table 1).
Among the 20 SNPs tested, markers 2–5, 7, 9–14, and 18–20 (Table 1) are located in a 99 kb large block. Supplementary Table 3 summarizes the haplotype analysis results of the 7-marker haplotype tagging SNPs of this block (2–4, 9–10, 12, 18). Three of the 8 haplotypes were significantly associated with IBD after 10,000 permutations. Haplotypes #5 and #7 were protective, whereas #4 was a risk haplotype. Multiple logistic regression analysis of the entire IBD panel including gender as a covariate, revealed a best model fit with SNPs rs5997391 and rs35873774 (intron 4/5 of XBP1). Logistic regression analysis did not reveal epistatic effects.
Deep sequencing reveals multiple rare variants including two hypomorphic variants that might confer risk
LD around the XBP1 gene, flanked by two recombination hotspots (Supplementary Fig. 8B), is generally weak (Supplementary Fig. 8E). The complex haplotype structure of the locus (Supplementary Table 3) suggested that multiple rare, private SNPs might contribute to its IBD association. We re-sequenced all exons, splice sites, and promoter regions in 282 unaffected controls, 282 CD, and 282 UC patients, and exons and splice sites only in an additional 282 UC patients (Supplementary Table 4 and Supplementary Fig. 9). Apart from verifying 15 already annotated variants, 51 new polymorphisms were identified, among them 39 rare SNPs detected once in either the CD, UC, and/or control cohort. The discovery frequency for rare SNPs was 5, 16, and 18 for 282 controls, CD, and UC patients. Sequencing of the coding region in another 282 UC patients yielded another 3 novel SNPs. Five novel non-synonymous SNPs (nsSNPs; XBP1snp8, XBP1snp17, XBP1snp22, XBP1snp29, XBP1snp30) were discovered in the sequencing cohort of 1128 patients but not controls. Taqman genotyping revealed the actual frequencies of these 5 novel nsSNPs in panels 1+2. Notably, heterozygous individuals were only observed among the case groups for 4 of the 5 rare nsSNPs, while the fifth nsSNP (XBP1snp22) occurred at equal frequencies in all groups (Supplementary Table 4 and Supplementary Fig. 9). The novel nsSNPs were too rare to warrant formal statistical analysis.
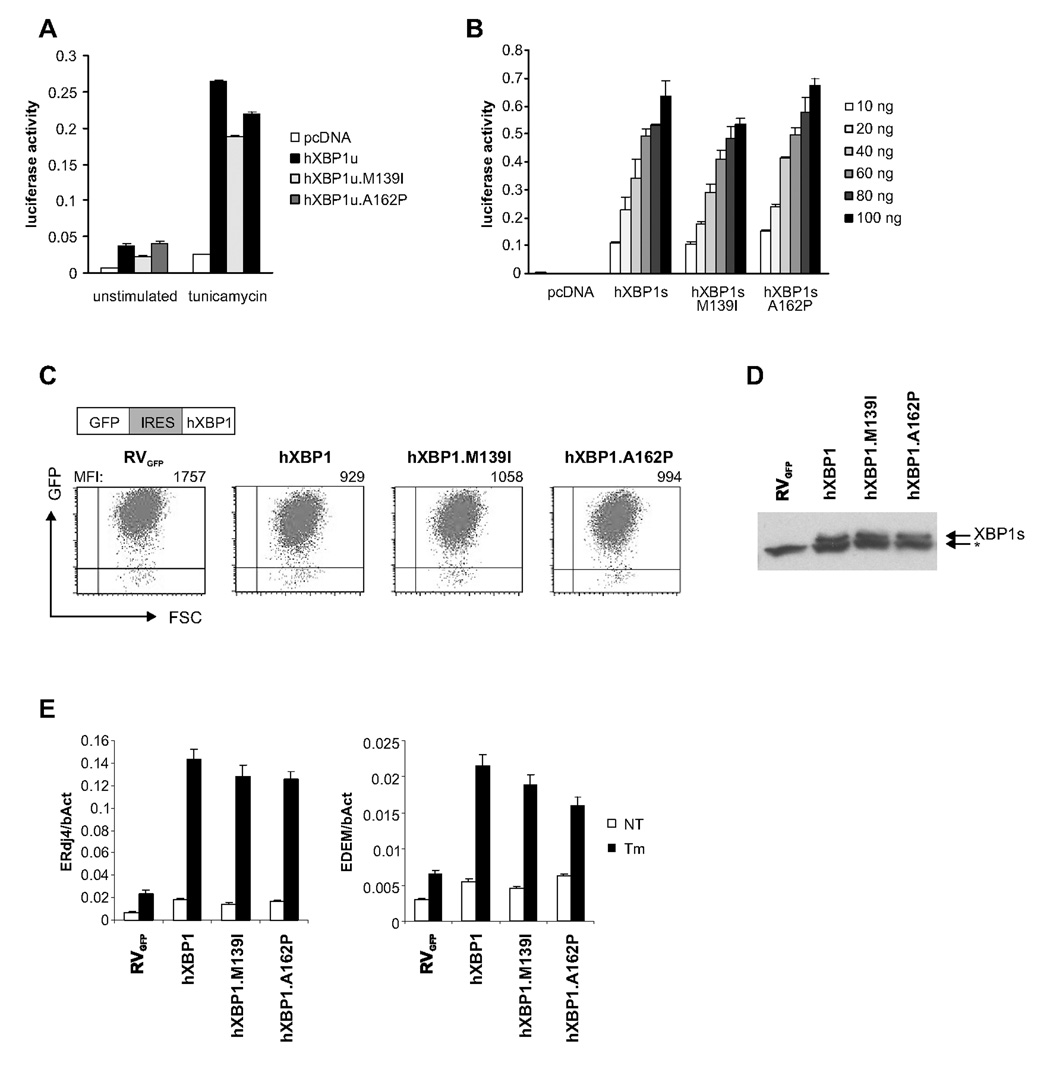
The nsSNPs, XBP1snp8 (M139I) and XBP1snp17 (A162P), present in IBD patients but not controls (Supplementary Table 4) lead to amino acid changes in the XBP1 hinge region between the bZiP and transactivation domains. XBP1snp17 in exon 4 is 10 bp upstream of the XBP1 mRNA splice site recognized by IRE1. We engineered the respective mutations into unspliced (hXBP1u) and spliced (hXBP1s) versions and transiently cotransfected MODE-K cells with wildtype or mutant XBP1 plasmids and an UPRE-luciferase reporter construct (Lee et al., 2003b). hXBP1u.M139I and hXBP1s.M139I had diminished UPRE transactivating function compared to wildtype plasmids in untreated and tunicamycin (Tm) treated MODE-K (Fig. 6AB). hXBP1u.A162P displayed impaired UPRE transactivation only in Tm-treated MODE-K cells (Fig. 6A), while hXBP1s.A162P transactivation was unaltered (Fig. 6B). To test ability to induce XBP1s target genes, we reconstituted XBP1−/− mouse embryonic fibroblasts (MEFs) (Iwakoshi et al., 2003) with either wildtype or mutant hXBP1u-GFP retroviral constructs, obtaining similar GFP fluorescence and comparable protein levels (Fig. 6CD). hXBP1.M139I induced less ERdj4 (DNAJB9) and EDEM (EDEM1) mRNA than wildtype both at baseline and upon Tm treatment, while hXBP1u.A162P was hypomorphic only under conditions of ER stress (Fig 6E) as above (Fig 6B). hXBP1s.P15L (XBP1snp22), the only rare nsSNP present at similar frequencies in IBD patients and controls, was not hypomorphic in these assays (Supplementary Fig. 10AB).
Figure 6. Rare XBP1 variants are hypomorphic.
A. MODE-K cells were transfected with UPRE-luciferase and unspliced hXBP1u expression plasmids encoding the rare, IBD-associated minor alleles XBP1snp8 (M139I) and XBP1snp17 (A162P) and treated with 1 µg/ml Tm. Values represent luciferase activities normalized to cotransfected Renilla reporter. B. Experiments as in (a), with indicated amounts of spliced hXBP1s cDNA variants C. Transduction efficiency measured by FACS of XBP1−/− MEF cells reconstituted with human XBP1 wildtype or SNP variants bi-cistronic retroviral vectors (RVGFP) (MFI, mean fluorescence intensity). D. XBP1s protein levels were determined by western blot of Tm-treated cells (*, non-specific band). E. ERdj4 and EDEM mRNA levels (normalized to β-actin mRNA expression) in untreated (NT) or Tm (1 µg/ml)-treated cells for 6 hrs.
Discussion
We present the first spontaneous mouse model of intestinal inflammation that arises from a gene defect in an actual genetic risk factor for human IBD. We suggest that XBP1 unifies key elements of IBD pathogenesis within the IEC compartment, pointing toward a primary defect in IEC function in IBD pathogenesis. Our results introduce the ER stress response as a likely integral component of organ-specific inflammation. XBP1 controls organ-specific inflammation through two major mechanisms that are probably codependent. First, Paneth cell function was strikingly impaired in XBP1−/− mice as evidenced by diminished antimicrobial peptide secretion and a compromised response to pathogenic bacteria. Second, XBP1 deficiency itself induced ER stress that led to a heightened pro-inflammatory response of the epithelium to known IBD inducers flagellin and TNFα (Supplementary Fig. 11).
XBP1 and environmental factors
Consistent with our results, increased grp78 expression has recently been reported in IBD patients (Shkoda et al., 2007; Heazlewood et al., 2008). While Shkoda et al suggested that ER stress occurs secondary to an inflammatory insult to IECs, our data instead point to specific impairment of the ER stress response as a cause, rather than a consequence, of intestinal inflammation. This might be obvious in the context of the genetic association of XBP1 variants with IBD reported here, but we speculate that environmental factors may also impair XBP1 function (and hence the ER stress response). Monozygotic twin studies have highlighted the importance of as yet unknown environmental and/or epigenetic factors in the development of IBD (Halfvarson et al., 2003). One might speculate that microbial- or food-derived XBP1 inhibitors could interfere with the pathways described herein, particularly in a genetically susceptible host, thus contributing to the development of intestinal inflammation. Along those lines, a recent report found that a 21-membered macrocyclic lactam termed ‘trierixin’ isolated from Streptomyces sp. potently inhibits endogenous XBP1 splicing in an epithelial cell line (Tashiro et al., 2007).
Paneth cell deficiency, IEC inflammatory tone and enteritis
Although Paneth and absorptive epithelial cells have been linked to intestinal inflammation (Kobayashi et al., 2005; Zaph et al., 2007; Nenci et al., 2007; Wehkamp et al., 2005), neither Paneth cell depletion (Garabedian et al., 1997), inability to convert pro-cryptdins to cryptdins (Wilson et al., 1999) nor Nod2 deletion (Kobayashi et al., 2005) cause spontaneous or induced intestinal inflammation. A recent study reported development of spontaneous UC that is dependent on a specific ‘colitogenic’ microbial milieu arising in a genetically altered host, that is vertically and horizontally transmissible to genetically intact mice (Garrett et al., 2007). However, such ‘colitogenic’ microbiota does not apparently arise in Paneth cell- or cryptdin-deficient mice (Garabedian et al., 1997; Wilson et al., 1999; Kobayashi et al., 2005). We conclude that bacterial “dysbiosis” alone is insufficient to cause intestinal inflammation if unaccompanied by a pro-inflammatory state including that primarily of the epithelium.
XBP1 deficiency in IECs resulted in IRE1α hyperactivation through an unidentified mechanism and increased JNK phosphorylation in the epithelial compartment in vivo. An increased susceptibility to DSS colitis was reported in IRE1β−/− mice (Bertolotti et al., 2001). Although IRE1β-deficiency did not lead to spontaneous enteritis, colitis or Paneth cell depletion, baseline levels of grp78 were elevated consistent with an active UPR in the absence of IRE1β. IECs are currently emerging as key mediators of inflammatory and immune mechanisms in mucosal tissues. IEC deletion of IKKβ (Zaph et al., 2007) or NEMO (Nenci et al., 2007), both upstream of NFκB, resulted in mucosal immune dysfunction and spontaneous colitis, respectively, the latter as a consequence of IEC barrier dysfunction. We find that even minor deficiencies in XBP1 expression within IECs lead to spontaneous enteritis, while leaving the intestinal barrier largely intact.
Genetic association of XBP1 polymorphisms with IBD
IBD is a complex polygenetic disease as evidenced by the recent discovery and replication of several genetic risk factors that include NOD2, the 5q31 haplotype (SLC22A4, SLC22A5), the 5p13.1 locus (PTGER4), DLG5, the IL23 receptor, ATG16L1, IRGM and IL12B on 5q33, NKX2-3, PTPN2, the 17q23.2 and the 17q11.1 loci, and NELL1 (Mathew, 2008). Since the functionally relevant variants for most of these loci and their role in IBD pathogenesis remain to be identified, a coherent model from gene to intestinal inflammation has yet to be developed although some of these risk alleles point towards abnormalities of innate immune responses (e.g. NOD2) and autophagy (e.g. ATG16L1, IRGM), adaptive immune functions (e.g. IL23R) and the intestinal epithelial barrier (e.g. DLG5) in human IBD. Our studies reveal abnormalities of the ER stress response as another pathway for the development of intestinal inflammation and IBD.
We suggest that the linkage results obtained on chromosome 22 from three independent microsatellite-based genome scans (Hampe et al., 1999; Barmada et al., 2004; Vermeire et al., 2004) could reflect the associations of rare and common variants of the XBP1 gene region reported here. A currently emerging concept is that rare sequence variants with strong phenotypic effects might contribute substantially to variation in complex traits, and the aggregated risk contribution may result in common traits (Cohen et al., 2004; Gorlov et al., 2008) a view strongly supported by analyzing frequencies of synonymous and non-synonymous SNPs in an extensive data set. The authors found that the distribution of SNPs predicted to be ‘possibly’ and ‘probably’ damaging was shifted toward rare SNPs compared with the MAF distribution of benign and synonymous SNPs that are not likely to be functional (Gorlov et al., 2008). We found rare SNPs 3 times more frequently in the CD and UC sequencing cohorts than the control cohort and validated five rare non-synonymous coding variants, 4 of them present only in IBD patients.
Functional studies revealed that two of these IBD-restricted non-synonymous SNPs behaved as hypomorphs as evidenced by decreased transactivation of the UPR and induction of XBP1s target genes, either in all conditions tested (XBP1snp8), or in response to exogenous induction of ER stress (XBP1snp17). This pattern of decreased transactivation upon transfection of mutant XBP1 cDNAs was observed in IEC lines with endogenous (wildtype) XBP1, as well as XBP1−/− MEFs reconstituted with mutant or wildtype XBP1. Hence, these rare, IBD-associated variants are indeed hypomorphic as would be predicted for risk conferring variants from the mechanisms established through our mouse model. While the functional impact of non-synonymous SNPs can be estimated by in vitro studies as presented herein, the biological significance and contribution to disease risk of the other associated as well as rare SNPs located outside the coding region is hard to predict; nonetheless, there are excellent examples that those variants could have important functional consequences (Birney et al., 2007; Libioulle et al., 2007). The phenomenon that multiple rare variants contribute to the overall risk at a particular locus most likely represents a common situation in many complex polygenic diseases (i.e. every patient has a “private” risk SNP). This is also exemplified by NOD2, which not only harbors few common alleles strongly associated with CD, but also multiple rare alleles that – taken together – account for a substantial proportion of disease risk attributed to that locus. It cannot be excluded though, taking the results of the haplotype analysis into account, that common variants contribute to disease risk at the XBP1 locus in addition to the excess of private variants in patients. We assume that most given disease-associated genes will have a wide spectrum of allelic variants, both common and rare/private.
Experimental Procedures
Mice
The generation of XBP1flox/floxVCre and VCreERT2 transgenic mice is detailed in Supplementary Data. All mouse protocols were approved by the Harvard Standing Committee on Animals.
Reagents
The source of antibodies, proteins and inhibitors are as follows: rabbit phospho-JNK, total-JNK, active (cleaved) caspase-3 (Cell Signaling Technology), anti-lysozyme (DakoCytomation), anti-procryptdin (Ayabe et al., 2002) generously provided by A. Ouellette (UC Irvine), flagellin (Invivogen), TNFα (Peprotech). The JNK-1,-2,-3 inhibitor SP600125 (Sigma), p38 inhibitor SB203580, MEK inhibitors PD98059 and U0126 (Calbiochem) were dissolved in DMSO as recommended. Carbamyl choline and lipopolysaccharide (LPS; from Escherichia coli 0111:B4) (Sigma), were used at final concentrations of 10µM and 1µg/ml, respectively.
Immunohistochemistry, TUNEL and Electron microscopy
Tissues were handled by standard methods as detailed in Supplementary Methods. Apoptotic cells were detected on paraffin embedded small intestine using TUNEL-POD kit (Roche Applied Sciences). Small intestinal tissue from sex-matched XBP1+/+ and XBP1−/− littermates was fixed as previously described (see Supplemental Methods) and observed with a JEOL 1200EX TEM at 60 kV operating voltage.
Oral L. monocytogenes infection
Sex and age matched groups of XBP1+/+ and XBP1−/− littermates were infected under BL2 conditions using gastric gavage at 3.6×108 L. monocytogenes strain 10403s per mouse. CFU assays (faeces c.f.u./mg dry weight; liver and spleen c.f.u./organ) were performed as in (Kobayashi et al., 2005) and Supplementary methods.
Dextran sodium sulphate colitis
Sex and age-matched littermates (8 to 12 wks) received 4.5% DSS (ICN Biomedicals Inc.) in drinking water for 5 days then regular water thereafter, or neomycin sulfate and metronidazole (1.5 g/L) (Sigma). Antibiotic treated mice received 7% DSS. Weight was recorded daily and rectal bleeding assessed as (0, absent; 1, traces of blood at anus or the base of the tail; 2, clearly visible rectal blood). Histological and mRNA expression studies on RNeasy kit isolated colon RNA (Qiagen) used mice sacrificed on day 8 after DSS treatment. Histological scoring of colons was as in (Garrett et al., 2007).
Crypt isolation, stimulation, and bactericidal activity assays
Small intestinal crypts were isolated, stimulated with 10µM CCh or 1µg/ml LPS and lysozyme levels and bactericidal activity against 1 × 103 c.f.u. Salmonella typhimurium cs015 following published protocols (Ayabe et al., 2000) and Supplementary methods.
Bromodeoxyuridine (BrdU) incorporation
XBP1+/+ and XBP1−/− littermates were injected with 1mg BrdU (Becton Dickinson) in 500µl PBS, and small intestinal tissue harvested after 1h or 24h and paraffin embedded tissue sectioned and stained with anti-BrdU antibody (Becton Dickinson).
Epithelial RNA isolation and quantification
XBP1+/+ and XBP1−/− intestines were opened longitudinally, rinsed with cold PBS, everted on a plain surface, RNAlater added and epithelium immediately scraped off using RNAse-free glass slides. Total RNA isolated using RNAeasy columns (Qiagen) was reverse transcribed and quantified by SYBR green PCR (Biorad). For microarray analysis, RNAs isolated from 3 specimens per genotype were pooled, and microarray carried out at the Biopolymers Core Facility (Harvard Medical School) with mouse genome 430 2.0 array (Affymetrix, Santa Clara, CA). Data analysis was performed with Agilent GeneSpring GX and Affymetrix GCOS software under default parameter setting. Quantitative PCR was performed as in (Lee et al., 2003b). See Supplementary Table 5 for PCR primers.
XBP1 splicing assay
XBP1 splicing was measured by specific primers flanking the splicing site yielding PCR product sizes of 164 and 138bp for human XBP1u and XBP1s, 171bp and 145bp, for mouse XBP1. Products were resolved on 2% agarose gels, and band intensity determined densitometrically (Optiquant Software, Perkin Elmer).
XBP1 silencing in MODE-K cells
The SV40 large T antigen-immortalized small intestinal epithelial cell line MODE-K (gift of D. Kaiserlian, Institute Pasteur) was transduced as described (Iwakoshi et al., 2003) with an XBP1-specific RNAi vector and a control vector identical to (Lee et al., 2003a) except that SFGΔU3hygro was used, and knockdown confirmed by qPCR. MODE-K.iXBP and MODE-K.Ctrl were seeded for CXCL1 experiments as described (Song et al., 1999) at 1×105 cells/well in 96 well plates, adhered for 2–4 hours, supernatant removed and stimulated with flagellin and TNFα for 4h or preincubated for 30min with JNK, p38, and MEK inhibitors, supernatants removed and cells stimulated in fresh media with flagellin and TNFα CD1d-restricted antigen presentation by MODE-K cells (van de Wal et al., 2003) is in Supplementary methods. JNK phosphorylation was assessed in MODE-K cells seeded at 1×106 per well 6 well plates, allowed to form confluent mono-layers over 48–72h, stimulated with flagellin and TNFα for the indicated time periods, washed in ice-cold PBS and lysed in 500µl RIPA buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with protease (Complete®, Roche Applied Science) and Ser/Thr and Tyr phosphatase (Upstate) inhibitors.
Western blot
Protein content of lysates was determined by BCA assay, and equal amounts of lysates containing Laemmli buffer were boiled at 95°C for 5min, resolved on 10% SDS-PAGE (for MODE-K cell lysates) or 12% SDS-PAGE (for TCA precipitates of purified crypts), transferred to Protran membranes (Whatman), blocked with 5% milk in TBS-T, incubated with primary antibody in 3–5% BSA in TBS-T at 4°C overnight, washed, and incubated with a 1:2,000 dilution of HRP-conjugated anti-rabbit secondary antibody in 3–5% milk in TBS-T for 45min at room temperature. Bands were visualized using SuperSignal chemoluminescent substrate (Pierce).
Human biopsy samples
Ileal and colonic biopsies were obtained from randomly selected patients with clinically, endoscopically and histologically confirmed diagnosis of CD and UC, as well as healthy control patients without any signs of intestinal inflammation. The diagnosis of CD and UC was confirmed by established criteria of clinical, radiological and endoscopic analysis, and from histology reports. Informed consent was obtained and procedures performed according to the approval by the local ethics committee of the Innsbruck Medical University. Biopsies were collected in RNAlater (Ambion), RNA isolated using RNAeasy columns (Qiagen), reverse transcribed, and used for quantitative PCR and XBP1 splicing assays.
Patient recruitment
German patients and controls in panels 1 and 2 almost completely overlap with the panels termed A and B in two recently published studies (Franke et al., 2007; Hampe et al., 2007). Panel 3 is unpublished. All patients were recruited at the Charité University Hospital (Berlin, Germany) and the Department of General Internal Medicine of the Christian-Albrechts-University (Kiel, Germany), with support from the German Crohn and Colitis Foundation and BMBF competence network “IBD”. Clinical, radiological and endoscopic (i.e. type and distribution of lesions) studies unequivocally confirmed the diagnosis of CD or UC, with confirmative or compatible histological findings. In the case of uncertainty, patients were excluded from the study. German healthy control individuals were obtained from the popgen biobank (Krawczak et al., 2006). Informed written consent was obtained from all study participants. All collection protocols were approved by the Charité University Hospital and the Department of General Internal Medicine of the Christian-Albrechts-University ethics committees.
Genotyping and Sequencing
Genomic DNA was prepared and amplified as in Supplementary Methods, genotyping performed using the SNPlex™ Genotyping System (Applied Biosystems, Foster City, CA) on an automated platform, genotypes generated by automatic calling using the Genemapper 4.0 software (Applied Biosystems) and all cluster plots reviewed manually. Prior to statistical analyses, quality checks (PHWE>0.01, MAFcontrols>1%, callrate≥90%) were applied to the SNPs under study. Single-marker association and haplotype analyses, permutation tests, calculation of pairwise LD, and SNP selection were performed using Haploview 4.0 (Barrett et al., 2005). Haplotype blocks were automatically defined as in (Gabriel et al., 2002). Only haplotypes with population frequencies >1.0 % were included in the final association analysis.
Single-marker disease associations and possible marker-marker interactions were assessed for statistical significance by means of logistic regression analysis (forward selection), as implemented in the procedure LOGISTIC of the SAS software package (SAS Institute, Cary, NC). Prior to analyses, individuals with missing data were removed and genotypes coded numerically.
Genomic DNA sequencing was performed using Applied Biosystems’ BigDye™ chemistry according to the supplier’s recommendations (for primer sequences, see Supplementary Table 6) and analyzed as described in Supplementary methods. Authenticity of the five novel discovered rare nsSNPs and rs5762809 was checked by TaqMan genotyping (Applied Biosystems) on an automated platform. For primer and probe sequences see Supplementary Table 6 and Supplementary Table 7; for genotype counts see Supplementary Table 2 and Supplementary Table 4.
UPRE reporter assays
Expression plasmids hXBP1u and hXBP1s were engineered to incorporate the XBP1snp17 (A162P_)_, XBP1snp8 (M139I) and XBP1snp22 (P15L) minor variants using the GeneTailor site directed mutagenesis system (Invitrogen). See Supplementary Data for primers used. Transient transfection of MODE-K cells followed by luciferase assays was performed as in (Lee et al., 2003b). Reconstitution of XBP1−/− MEF cells was with bicistronic retroviral vectors expressing GFP and human XBP1 constructed by inserting PCR amplified cDNAs for human wildtype and XBP1snp17 and XBP1snp8 variants into RVGFP vector between BglII and SalI sites, as described previously (Iwakoshi et al., 2003) and transduced as described (Lee et al., 2003b).
Supplementary Material
01
Acknowledgments
We thank Drs. S. Robine and N. Davidson for VCre-ERT2 mice; Dr. S. Ito for help with EM; Drs. J. Rioux and M. Starnbach for helpful discussions; P. Gupta, B. Enrich, and D. Lindenbergh-Kortleve for technical support, and Dr. S. Snapper for critical reading of the manuscript. This work was supported by grants from the Crohn’s and Colitis Foundation of America (RSB and AK), NIH grants DK44319 (RSB), P30 DK034854 (RSB; Harvard Digestive Diseases Center), AI32412, P01 AI56296 (LHG); Ellison Medical Foundation (LHG), CDA 1P50CA100707 (AHL), the Austrian Science Fund (AK and HT), and the Max Kade Foundation (AK). The authors wish to thank the patients, families and physicians for their cooperation. The cooperativeness of the German Crohn and Colitis patient association (Deutsche Morbus Crohn und Colitis Vereinigung e.V.) and of the German “Kompetenznetz Darmerkrankungen” are gratefully acknowledged. Genotyping in this study was supported by the German Ministry of Education and Research (BMBF) through the National Genome Research Network (NGFN) and the popgen biobank.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acosta-Alvear D, Zhou Y, Blais A, Tsikitis M, Lents NH, Arias C, Lennon CJ, Kluger Y, Dynlacht BD. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol. Cell. 2007;27:53–66. doi: 10.1016/j.molcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Ayabe T, Satchell DP, Pesendorfer P, Tanabe H, Wilson CL, Hagen SJ, Ouellette AJ. Activation of Paneth cell alpha-defensins in mouse small intestine. J. Biol. Chem. 2002;277:5219–5228. doi: 10.1074/jbc.M109410200. [DOI] [PubMed] [Google Scholar]
- Ayabe T, Satchell DP, Wilson CL, Parks WC, Selsted ME, Ouellette AJ. Secretion of microbicidal alpha-defensins by intestinal Paneth cells in response to bacteria. Nat. Immunol. 2000;1:113–118. doi: 10.1038/77783. [DOI] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den BM, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Barmada MM, Brant SR, Nicolae DL, Achkar JP, Panhuysen CI, Bayless TM, Cho JH, Duerr RH. A genome scan in 260 inflammatory bowel disease-affected relative pairs. Inflamm. Bowel. Dis. 2004;10:513–520. doi: 10.1097/00054725-200409000-00004. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bertolotti A, Wang X, Novoa I, Jungreis R, Schlessinger K, Cho JH, West AB, Ron D. Increased sensitivity to dextran sodium sulfate colitis in IRE1beta-deficient mice. J. Clin. Invest. 2001;107:585–593. doi: 10.1172/JCI11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- Franke A, Hampe J, Rosenstiel P, Becker C, Wagner F, Hasler R, Little RD, Huse K, Ruether A, Balschun T, et al. Systematic association mapping identifies NELL1 as a novel IBD disease gene. PLoS. ONE. 2007;2:e691. doi: 10.1371/journal.pone.0000691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Garabedian EM, Roberts LJ, McNevin MS, Gordon JI. Examining the role of Paneth cells in the small intestine by lineage ablation in transgenic mice. J. Biol. Chem. 1997;272:23729–23740. doi: 10.1074/jbc.272.38.23729. [DOI] [PubMed] [Google Scholar]
- Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, Glickman JN, Glimcher LH. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlov IP, Gorlova OY, Sunyaev SR, Spitz MR, Amos CI. Shifting paradigm of association studies: value of rare single-nucleotide polymorphisms. Am. J Hum. Genet. 2008;82:100–112. doi: 10.1016/j.ajhg.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfvarson J, Bodin L, Tysk C, Lindberg E, Jarnerot G. Inflammatory bowel disease in a Swedish twin cohort: a long-term follow-up of concordance and clinical characteristics. Gastroenterology. 2003;124:1767–1773. doi: 10.1016/s0016-5085(03)00385-8. [DOI] [PubMed] [Google Scholar]
- Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- Hampe J, Schreiber S, Shaw SH, Lau KF, Bridger S, MacPherson AJ, Cardon LR, Sakul H, Harris TJ, Buckler A, et al. A genomewide analysis provides evidence for novel linkages in inflammatory bowel disease in a large European cohort. Am. J. Hum. Genet. 1999;64:808–816. doi: 10.1086/302294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood CK, Cook MC, Eri R, Price GR, Tauro SB, Taupin D, Thornton DJ, Png CW, Crockford TL, Cornall RJ, et al. Aberrant Mucin Assembly in Mice Causes Endoplasmic Reticulum Stress and Spontaneous Inflammation Resembling Ulcerative Colitis. PLoS. Med. 2008;5:e54. doi: 10.1371/journal.pmed.0050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakoshi NN, Lee AH, Vallabhajosyula P, Otipoby KL, Rajewsky K, Glimcher LH. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat. Immunol. 2003;4:321–329. doi: 10.1038/ni907. [DOI] [PubMed] [Google Scholar]
- Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nunez G, Flavell RA. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- Kojouharoff G, Hans W, Obermeier F, Mannel DN, Andus T, Scholmerich J, Gross V, Falk W. Neutralization of tumour necrosis factor (TNF) but not of IL-1 reduces inflammation in chronic dextran sulphate sodium-induced colitis in mice. Clin. Exp. Immunol. 1997;107:353–358. doi: 10.1111/j.1365-2249.1997.291-ce1184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczak M, Nikolaus S, von Eberstein H, Croucher PJ, El Mokhtari NE, Schreiber S. PopGen: population-based recruitment of patients and controls for the analysis of complex genotype-phenotype relationships. Community Genet. 2006;9:55–61. doi: 10.1159/000090694. [DOI] [PubMed] [Google Scholar]
- Lee AH, Chu GC, Iwakoshi NN, Glimcher LH. XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. EMBO J. 2005;24:4368–4380. doi: 10.1038/sj.emboj.7600903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Anderson KC, Glimcher LH. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc. Natl. Acad. Sci. U. S. A. 2003a;100:9946–9951. doi: 10.1073/pnas.1334037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell Biol. 2003b;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libioulle C, Louis E, Hansoul S, Sandor C, Farnir F, Franchimont D, Vermeire S, Dewit O, de Vos M, Dixon A, et al. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS. Genet. 2007;3:e58. doi: 10.1371/journal.pgen.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, Walter P. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodes MJ, Cong Y, Elson CO, Mohamath R, Landers CJ, Targan SR, Fort M, Hershberg RM. Bacterial flagellin is a dominant antigen in Crohn disease. J. Clin. Invest. 2004;113:1296–1306. doi: 10.1172/JCI20295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison BB, Dunbar L, Qiao XT, Braunstein K, Braunstein E, Gumucio DL. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J. Biol. Chem. 2002;277:33275–33283. doi: 10.1074/jbc.M204935200. [DOI] [PubMed] [Google Scholar]
- Mathew CG. New links to the pathogenesis of Crohn disease provided by genome-wide association scans. Nat. Rev. Genet. 2008;9:9–14. doi: 10.1038/nrg2203. [DOI] [PubMed] [Google Scholar]
- Nenci A, Becker C, Wullaert A, Gareus R, van Loo G, Danese S, Huth M, Nikolaev A, Neufert C, Madison B, et al. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature. 2007;446:557–561. doi: 10.1038/nature05698. [DOI] [PubMed] [Google Scholar]
- Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, Friend D, Grusby MJ, Alt F, Glimcher LH. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Shapiro-Shelef M, Iwakoshi NN, Lee AH, Qian SB, Zhao H, Yu X, Yang L, Tan BK, Rosenwald A, et al. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21:81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Shkoda A, Ruiz PA, Daniel H, Kim SC, Rogler G, Sartor RB, Haller D. Interleukin-10 blocked endoplasmic reticulum stress in intestinal epithelial cells: impact on chronic inflammation. Gastroenterology. 2007;132:190–207. doi: 10.1053/j.gastro.2006.10.030. [DOI] [PubMed] [Google Scholar]
- Song F, Ito K, Denning TL, Kuninger D, Papaconstantinou J, Gourley W, Klimpel G, Balish E, Hokanson J, Ernst PB. Expression of the neutrophil chemokine KC in the colon of mice with enterocolitis and by intestinal epithelial cell lines: effects of flora and proinflammatory cytokines. J. Immunol. 1999;162:2275–2280. [PubMed] [Google Scholar]
- Strober W, Fuss IJ, Blumberg RS. The immunology of mucosal models of inflammation. Annu. Rev. Immunol. 2002;20:495–549. doi: 10.1146/annurev.immunol.20.100301.064816. [DOI] [PubMed] [Google Scholar]
- Tashiro E, Hironiwa N, Kitagawa M, Futamura Y, Suzuki S, Nishio M, Imoto M. Trierixin, a Novel Inhibitor of ER Stress-induced XBP1 Activation from Streptomyces sp. J. Antibiot. (Tokyo) 2007;60:547–553. doi: 10.1038/ja.2007.69. [DOI] [PubMed] [Google Scholar]
- Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- van de Wal Y, Corazza N, Allez M, Mayer LF, Iijima H, Ryan M, Cornwall S, Kaiserlian D, Hershberg R, Koezuka Y, et al. Delineation of a CD1d-restricted antigen presentation pathway associated with human and mouse intestinal epithelial cells. Gastroenterology. 2003;124:1420–1431. doi: 10.1016/s0016-5085(03)00219-1. [DOI] [PubMed] [Google Scholar]
- Vermeire S, Rutgeerts P, Van Steen K, Joossens S, Claessens G, Pierik M, Peeters M, Vlietinck R. Genome wide scan in a Flemish inflammatory bowel disease population: support for the IBD4 locus, population heterogeneity, and epistasis. Gut. 2004;53:980–986. doi: 10.1136/gut.2003.034033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehkamp J, Salzman NH, Porter E, Nuding S, Weichenthal M, Petras RE, Shen B, Schaeffeler E, Schwab M, Linzmeier R, et al. Reduced Paneth cell α-defensins in ileal Crohn's disease. Proc. Natl. Acad. Sci. U. S. A. 2005;102:18129–18134. doi: 10.1073/pnas.0505256102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CL, Ouellette AJ, Satchell DP, Ayabe T, Lopez-Boado YS, Stratman JL, Hultgren SJ, Matrisian LM, Parks WC. Regulation of intestinal alpha-defensin activation by the metalloproteinase matrilysin in innate host defense. Science. 1999;286:113–117. doi: 10.1126/science.286.5437.113. [DOI] [PubMed] [Google Scholar]
- Wu J, Kaufman RJ. From acute ER stress to physiological roles of the Unfolded Protein Response. Cell Death. Differ. 2006;13:374–384. doi: 10.1038/sj.cdd.4401840. [DOI] [PubMed] [Google Scholar]
- Zaph C, Troy AE, Taylor BC, Berman-Booty LD, Guild KJ, Du Y, Yost EA, Gruber AD, May MJ, Greten FR, et al. Epithelial-cell-intrinsic IKK-beta expression regulates intestinal immune homeostasis. Nature. 2007;446:552–556. doi: 10.1038/nature05590. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01