Contact Guidance Mediated Three-Dimensional Cell Migration is Regulated by Rho/ROCK-Dependent Matrix Reorganization (original) (raw)
Abstract
Cells generate mechanical force to organize the extracellular matrix (ECM) and drive important developmental and reparative processes. Likewise, tumor cells invading into three-dimensional (3D) matrices remodel the ECM microenvironment. Importantly, we previously reported a distinct radial reorganization of the collagen matrix surrounding tumors that facilitates local invasion. Here we describe a mechanism by which cells utilize contractility events to reorganize the ECM to provide contact guidance that facilitates 3D migration. Using novel assays to differentially organize the collagen matrix we show that alignment of collagen perpendicular to the tumor-explant boundary promotes local invasion of both human and mouse mammary epithelial cells. In contrast, organizing the collagen matrix to mimic the ECM organization associated with noninvading regions of tumors suppresses 3D migration/invasion. Moreover, we demonstrate that matrix reorganization is contractility-dependent and that the Rho/Rho kinase pathway is necessary for collagen alignment to provide contact guidance. Yet, if matrices are prealigned, inhibiting neither Rho nor Rho kinase inhibits 3D migration, which supports our conclusion that Rho-mediated matrix alignment is an early step in the invasion process, preceding and subsequently facilitating 3D migration.
INTRODUCTION
Cell migration within three-dimensional (3D) microenvironments is an important in vivo event during multiple physiologic and pathological processes, such as tissue morphogenesis and cancer metastasis. In contrast to cell migration on two-dimensional (2D) surfaces, 3D migration requires that cells progress through a dense extracellular matrix (ECM) by varying cell morphology and actively remodeling the matrix (1–6). Contact guidance in 3D is the phenomena by which the matrix provides directional cues to the cells and directs the motility response via anisotropy in the microenvironment (7–9). In fibroblasts, it has been shown that contact guidance cues from collagen alignment promote 3D cell migration along the axis of collagen alignment (7,8). Herein, we examine the role of cell-generated contact guidance in promoting 3D cell migration during local invasion of mammary epithelial cells.
The microenvironment surrounding normal mammary epithelium and noninvasive breast tumors is largely composed of a dense collagen matrix organized parallel to the epithelial-stromal interface. We previously reported that a radial (perpendicular to the tumor boundary) realignment of this matrix in vivo facilitates local invasion (10), which is consistent with findings that metastatic epithelial cells directly migrate along stromal collagen fibers (11). Furthermore, we reported that in noninvading regions of tumors the collagen matrix remains organized parallel to the tumor boundary (10). Importantly, although proteases are known to play a role in matrix remodeling and tumor progression (4,12,13), tumor cells remain capable of matrix deformation (6) and invasion when proteases are inhibited (3,5); and matrix metalloproteases (MMPs) are not required for radial alignment to facilitate local invasion (14).
In this article we engineer assays to generate the differential matrix alignment surrounding mammary carcinomas in vivo. These assays allow us to investigate the mechanism by which cell contractility is utilized to reorganize the collagen matrix, which provides contact guidance that promotes 3D cell migration. We show that cell invasion is greater into regions of the collagen matrix that have fibers aligned perpendicular to the tumor-explant boundary than those regions with parallel organization. Moreover, we demonstrate that matrix reorganization is contractility-dependent and that the Rho GTPase pathway is necessary for collagen realignment to provide contact guidance and promote 3D migration. However, neither Rho nor the Rho-effector, Rho kinase (ROCK), are required for 3D migration, which supports our conclusion that matrix reorganization is an early step in the invasion process that facilitates 3D migration through the ECM.
METHODS AND MATERIALS
Cell culture, tumor explant harvest, and reagents
MDA-MB-231 human breast carcinoma cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) Fetal Bovine Serum (FBS) and cultured at 37°C with 10% CO2. Tumor explants (TE) were harvested as previously described (10). Briefly, fixed volume tumor explants were harvested from the central region of late-stage (invasive) polyoma middle-T mouse mammary tumors using a 3mm biopsy punch, and maintained briefly in DMEM containing 5% (v/v) FBS plus penicillin/streptomycin/Amphotericin B solution (Cellgro, Hemdon, VA). The Rho inhibitor C3 exoenzyme was obtained from Cytoskeleton (Denver, CO); the pharmacological inhibitors H1152, Blebbistatin, GM6001, Aprotinin, and Leupeptin were obtained from Calbiochem (La Jolla, CA).
3D migration/invasion assay
Two tumor explants (described above) or two MDA-MD-231 cell-seeded collagen gels (CSCGs) were organized within a 3D collagen gel as depicted in Fig. 1 A. To make CSCGs, 5 × 105 cells were suspended within a 1.0 mg/mL rat-tail collagen solution (BD Biosciences, San Jose, CA), which was neutralized with an equal volume of 100 mM HEPES in 2× concentrated phosphate buffered saline. CSCGs were formed by placing a 250 _μ_L volume of the cell/collagen solution into wells of 96-well culture plates and allowing them to polymerize at 37°C for at least 2 h. CSCGs (or TEs) were briefly floated in DMEM containing 10% FBS before being placed into the larger collagen matrix. To generate the embedding collagen matrix, an acellular 2.0 mg/mL (neutralized) collagen solution was added to culture dishes and allowed to partially polymerize for 15 min at room temperature. Then using a custom grid reference that is placed under the culture dish and identifies proper explant location along the major axis of the culture well, TEs or CSCGs were floated ∼7 mm apart along the maximal well diameter and the TEs or CSCGs encased in a 2.0 mg/mL collagen gel. After two hours, DMEM containing 5% or 10% (v/v) FBS was added to the assay for TEs or CSCGs, respectively, and changed every 3–4 days over the course of the assays.
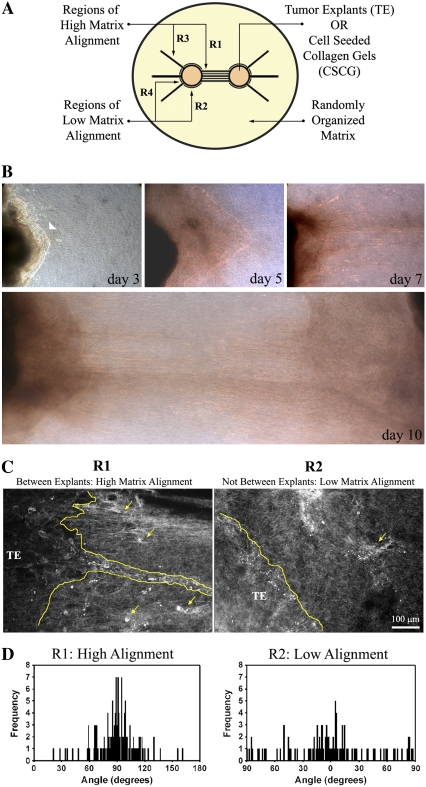
FIGURE 1.
Development of the 3D migration/invasion assay. (A) Diagram illustrating the 3D migration/invasion assay used in this study. In the assay, two TEs or CSCGs are placed at a fixed distance and encased in a randomly organized collagen matrix. Note that Region 1 (R1) consists of the volume in which the cells align the matrix and describes the width, height, and distance between the two TEs or CSCGs, which is defined as the primary axis. Regions 2 (R2) are “above” and “below” the explants and are regions in which the matrix orientation distributes around 0°, due to growth induced explant volume expansion pushing the matrix, and as such these regions do not substantially realign the matrix perpendicular to the explant boundary and are designated “low alignment regions.” Regions 3 (R3) are aligned regions due to force balance within the matrix that act as a control relative to the low alignment region (R4) to separate out possible chemoattraction effects of one explant on the other. (B) Phase contrast microscopy of the dominant axis of 3D invasion (Region R1) for PyVT tumor explants over 10 days. Note the early collective migration (highlighted with an arrowhead in the upper left panel of B) that progresses and leads to lines of collective invasion and single cell invasion. Results shown are representative of 10 assays. (C) MPSLM of the TE-collagen gel boundary at day 14, showing collective (outlined) and single cell (examples marked with arrows) migration into cell-aligned collagen matrix (R1), consistent with the in vivo condition (10). Note that in low alignment regions (R2), the few cells that move into the collagen gel lack guidance from the aligned matrix and do not continue to invade through the matrix. (D) Quantification of cell-mediated matrix alignment in regions R1 and R2 using SHG imaging of the collagen matrix at the explant-collagen gel boundary (n = 3–4 assays). Collagen alignment was quantified by measuring the fiber angle relative to the tumor boundary and shown as a population distribution of the measured angles. The fiber angle was significantly different between regions R1 and R2 (p < 0.00001).
Generation of aligned collagen matrices
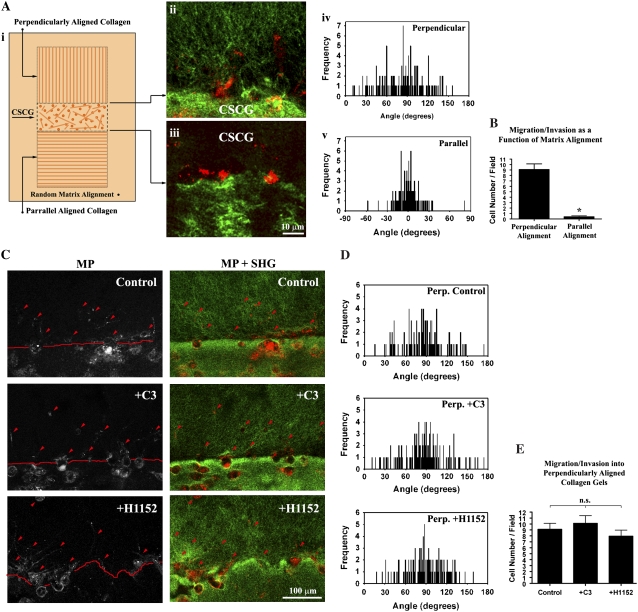
Collagen was magnetically aligned following the procedures outlined in Guo and Kaufman (15). Briefly, 1.5 _μ_m diameter streptavidin coated iron oxide beads (Bangs Labs: BM551) were added to 2.0 mg/mL collagen solution at a concentration of 0.1 mg/mL. Collagen solution was pipetted into a rectangular mold and collagen aligned by placing a cylindrical magnet (∼2G; McMaster: 5862K32) under the mold during collagen gel polymerization (i.e., collagen fibrillogenesis). Collagen alignment was confirmed by second harmonic imaging of the collagen matrix. To study cell migration in 3D, as a function of matrix alignment, the assay depicted in Fig. 5 A was utilized. Aligned collagen was arranged either perpendicular or parallel to a rectangle CSCG and encased within a 2.0 mg/mL collagen matrix. The migration assay was performed in the presence of DMEM containing 10% FBS.
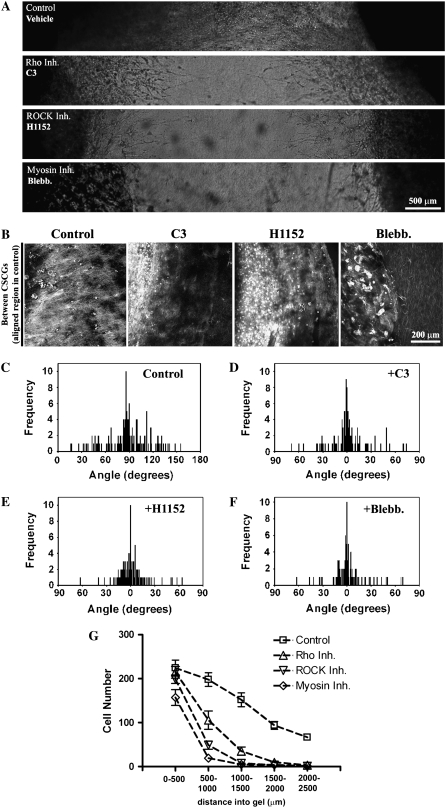
FIGURE 5.
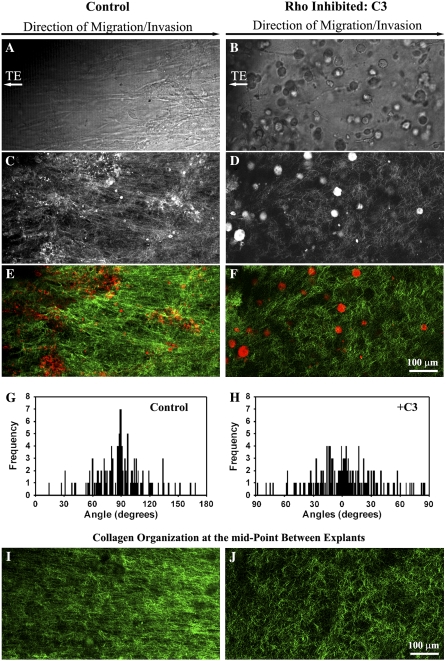
Rho and ROCK are required for contractility-mediated matrix reorganization to produce migration promoting contact guidance (A) Phase contrast microscopy images showing 3D invasion into region R1 after Rho (10 _μ_g/mL), ROCK (2.5 _μ_M), and myosin-driven contractility (10 _μ_M) inhibition. (B) MPLSM images of the CSCG-collagen gel interface under control, Rho inhibited (C3; 10 _μ_g/mL), ROCK inhibited (H1152; 2.5 _μ_M), and myosin contractility inhibited (Blebbistatin; 10 _μ_M) conditions, showing decreased matrix reorganization in region R1 after 14 days. (C_–_F) Quantification of matrix alignment in region R1 from second harmonic imaging of the collagen matrix at the CSCG-collagen gel boundary at day 14 (n = 3–4 assays) when cells were treated with vehicle (C), Rho inhibitor C3 (10 _μ_g/mL; D), ROCK inhibitor (2.5 _μ_M; E), or the myosin inhibitor blebbistatin (10 _μ_M; F). Note that all three inhibitors cause a loss in collagen fiber alignment, as evidenced by a distribution of fibers around 0° in the presence of inhibitors. (G) Quantification of the number of cells (at day 14) invading a defined distance into region R1 after matrix realignment was abolished after treatment with C3 (10 _μ_g/mL), H1152 (2.5 _μ_M), or blebbistatin (10 _μ_M).
Transmitted light microscopy
Assays were imaged with a 10× objective on a TE300 Nikon inverted microscope (Nikon, Melville, NY) controlled by Slidebook software (Intelligent Imaging Innovations, Denver, CO). Composite images were created by imaging adjacent regions of the assay and reconstructing the image with Adobe Photoshop (Adobe Systems, San Jose, CA). Additionally, transmitted light images were acquired concurrently with multiphoton images using the system described below.
Multiphoton laser-scanning microscopy (MPLSM)
Live cell and collagen imaging was performed with MPLSM to generate multiphoton excitation (MPE) and second harmonic generation (SHG) as described previously (10). Briefly, the custom system utilizes a mode-locked Ti:sapphire laser (Millennium/Tsunami, Spectra-Physics, Mountain View, CA) excitation source producing around 100 fs pulse widths, which was tuned to 890 nm to generate both MPE autofluorescence signals from cells or green fluorescent protein (GFP) excitation and SHG signals from collagen. The beam was focused onto the sample with a 20× Plan Apo Multi-immersion lens (Nikon, NA = 0.75, WD 0.17 for water). The presence of collagen was confirmed using fluorescence lifetime imaging microscopy (FLIM) on the same system, since the SHG from collagen has no lifetime. Furthermore, because of the fundamentally different physical properties of MPE and SHG, signals could be discriminated by filtering the emission signal. We used a 464 nm (cut-on) long pass filter and a 480–550 nm (bandpass) filter to isolate the MPE emission from autofluorescence or GFP, respectively, from the conserved 445 nm SHG emission. A 445 nm (narrow bandpass) filter was therefore used to isolate the SHG emission (all filters: TFI Technologies, Greenfield, MA). The number of cells invading into the collagen gel was collected within three focal plains (center ± 30 _μ_m) over the entire length between the TEs or CSCGs, or up to 250 _μ_m into prealigned collagen matrices. Quantification of the collagen fiber angle relative to the explant or CSCG boundary was performed with ImageJ (National Institutes of Health) as previously described (10).
Statistical analysis
For two-group comparisons _t_-test was used. For multi-group comparisons, one-way analysis of variance (ANOVA) with a post-hoc Tukey test was used.
RESULTS AND DISCUSSION
An important unanswered question is how cells realign the collagen matrix to promote 3D migration and local invasion. Therefore, to recapitulate the behaviors that we observed in vivo and to study the molecular mechanisms by which they are regulated, we engineered a novel assay of 3D migration/invasion to study the differential effects of matrix alignment and the invasion of cells relative to such alignment. In this system, either two TEs (3 mm biopsy) of fixed diameter or two cell-seeded collagen gels (CSCGs; 5 × 105 cells/CSCG) are arranged in a defined manner within a collagen matrix (Fig. 1 A). Initial studies with tumor explant culture systems utilized a single explant (10); however, preliminary studies led us to the use of two explants as a means to enhance the matrix alignment between the explants in a reproducible geometry due to opposing forces from cell contraction. Whereas one explant can provide collagen alignment (particularly with the use of preferential gel or culture dish geometries) and may resemble alignment in vivo with cell contraction being resisted by stromal or connective tissue components, two explants dramatically improve the degree of alignment and are therefore very useful for in vitro experiments.
Overall, this system is a scaled-down model of the tumor environment, within which there is a fraction of the tumor cell population and an initial collagen matrix of 2 mg/mL, which is consistent with the concentration commonly used with 3D collagen gel cultures containing either epithelial or fibroblast cells (6,16–18) but is lower than average concentrations determined in tumor xenografts that may be ≥ 10 mg/mL (19). However, because cells in the explant reorganize the collagen matrix, the local collagen concentration is likely much greater than 2 mg/mL. To determine if this is the case, collagen levels within the gels and around the cells were determined optically by analysis of the mean SHG signal, which scales linearly with collagen concentration and is a good approximation of collagen concentration (20). Comparison of the mean SHG signal intensity between randomly organized 2 mg/mL collagen matrix and regions where cells have contracted and organized the matrix demonstrate that the SHG signal is 4.3–5.3 times stronger in contracted regions near the cell populations (Table 1), which suggests local collagen densities approaching 8.5–10.5 mg/mL.
TABLE 1.
Average SHG signal in collagen matrices (mean ± SD)
| Matrix region | SHG intensity (a.u.) |
|---|---|
| Randomly organized (no cell contraction) | 1.0 ± 0.4 |
| Contracted and aligned by CSCGs (Region R1) | 4.3 ± 1.4 |
| Contracted and aligned by TEs (Region R1) | 5.3 ± 1.6 |
As is demonstrated in Figs. 1 and 2, the transformed cells exert contractile forces on the matrix between the two explants and align the collagen matrix with fibers distributed ∼90° relative to the explant boundary (Fig. 1 A; Regions R1). In contrast, the regions on “top” and “bottom” of the explant or plug did not display a high degree of radial alignment (Regions R2). In fact, these regions display a trend toward parallel organization (distributed around 0° relative to the explant boundary and referred to here as “low alignment regions” as they are not realigned to the radial orientation) likely because of growth expansion of the TE/CSCGs pushing the collagen, which resembles the in vivo morphology we previously reported for noninvasive growing tumors (10). Likewise because of force balance resistance to contraction, certain regions not between the TEs or CSCGs also aligned radially (Regions R3) whereas adjacent regions (Regions R4) did not. As such, comparison of regions R3 and R4 provide a valuable control to rule out the possibility that migration between the two explants is being driven solely by chemoattraction from one explant to another (Supplementary Material, Fig. S1 and Fig. S2).
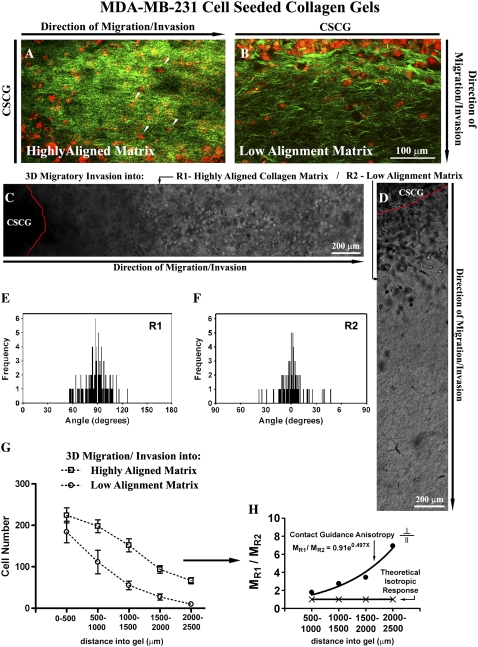
FIGURE 2.
Contact guidance-mediated 3D migration/invasion of human breast carcinoma cells. (A) MPLSM image of MDA-MB-231 invasive breast carcinoma cells invading from gels CSCGs into the aligned (Region R1) collagen matrix (examples highlighted with arrows) at day 14. MPE of cells are pseudo-colored red and SHG signals from collagen are pseudo-colored green. Note that the primary axis is from left to right with the CSCG to the left of the image. Signals were separated into MPE and SHG by filtering the emission signal as described in the Methods section. (B) MPLSM image of MDA-MB-231 invasive breast carcinoma cells invading from CSCGs into the matrix that is not realigned (Region R2) at day 14. MPE of cells are pseudo-colored red and SHG signals from collagen are pseudo-colored green. Note that the axis of invasion for region R2 is from top to bottom of the image with the CSCG to the top of the image. (C and D) Phase contrast microscopy of cells invading into the aligned region R1 (C) and the region of low-alignment R2 (D).(E and F) Quantification of matrix alignment in regions R1 (E) and R2 (F) using second harmonic imaging of the collagen matrix at the CSCG-collagen gel boundary (n = 3–4 assays) at day 14, as in Fig. 1. (G) Quantification of the number of cells invading a defined distance as a function of matrix alignment at day 14. Note that the distance spanning 0–500 _μ_m includes outward growth of the CSCG whereas cells found beyond 500 _μ_m are solely due to 3D migration. Note that migration into aligned regions is significantly (p < 0.009) greater than migration into low-alignment regions for all distances ≥ 500 _μ_m. (H) Differential 3D invasion due to contact guidance anisotropy, with the ratio of cells migration into region R1 (MR1) to region R2 (MR2) showing a positive and nonlinear response that differs from the isotropic case in which the ratio is one. The direction and shape of the anisotropic ratio suggests contact guidance cues from the aligned matrix promote 3D migration.
Metastatic tumor cells invade through aligned collagen fibers
Using late-stage tumors from the polyoma-middle-T (PyVT) transgenic model of human breast cancer, which is reliably invasive and metastatic with a 100% incidence of pulmonary metastasis (21–23), we examined 3D migration/invasion in vitro. When tumor explants from late-stage PyVT tumors were seeded into the 3D migration assay shown in Fig. 1 A, collective sheet-like volumes of cells invaded predominantly into Region 1 at early time points (days 1–5; see arrow in Fig. 1 B). This was followed (days 5–10) by continuation of the sheet-like collective migration as well as tracks of collectively invading cells (as seen in Fig. 1, B and C) and individual cell migration/invasion, which is consistent with previous reports demonstrating both individual and collective migration associated with carcinoma cell invasion (4,10,24).
To further investigate how organization of the matrix influenced 3D migration, we used live imaging of invading cells and the collagen matrix. To address the inherent challenges associated with imaging both cells and the ECM in live 3D microenvironments, live-cell and 3D matrix imaging was performed using MPLSM (25), which produces MPE of engineered or endogenous fluorophores (26,27) and SHG from noncentrosymmetric structures such as collagen (28,29). This approach allows visualization of invading cells within the matrix and quantification of matrix alignment within the planes of invasion. Using an MPLSM excitation wavelength of 890 nm, which excites endogenous fluorescence from flavin adenine dinucleotide (FAD) (30) while simultaneously producing a strong collagen SHG signal to image the matrix, we observed that the dominant region of invasion was into highly aligned collagen (Regions R1: Fig. 1, C and D and R3: data not shown) relative to regions of low alignment (R2 and R4, respectively). Quantification of fibrillar collagen organization from SHG images adjacent to the explant in Region R1 revealed a collagen alignment distributed around ∼90° perpendicular to the tumor-stromal boundary, whereas low alignment regions (R2 in Fig. 1 A) were significantly different (p < 0.00001) and displayed a wide distribution around 0°, or parallel to the tumor boundary (Fig. 1 D). Hence, 3D cell invasion was enhanced in regions where the cells had perpendicularly aligned the collagen matrix, whereas few cells invaded into low alignment regions (Fig. 1 C). These observations are consistent with our previous report that when a single tumor explant is seeded into a 3D collagen matrix, groups of epithelial cells directly interact with collagen fibers and begin to organize the matrix to support invasion into the collagen gel at early (24 h) time points (10). Importantly, the invasion of metastatic cells followed interaction with, and reorganization of, the collagen matrix.
Collagen matrix alignment promotes 3D migration of human breast carcinoma cells
Using the highly migratory and invasive human breast carcinoma MDA-MB-231 cell line as a model of metastasis, we seeded cells into collagen plugs to form the CSCGs depicted in Fig. 1 A. Consistent with our findings from tumor explants, the human breast carcinoma cells underwent 3D migration/invasion predominantly into aligned regions of the matrix (Fig. 2, A_–_F). After 14 days, cells had invaded over 2500 _μ_m into the aligned matrix (Fig. 2 G), resulting in an average migration speed of ∼7.44 _μ_m/h, which is consistent with data demonstrating 3D migration speeds in the range of ∼4–12 _μ_m/h in the DU-145 prostate cancer cell line (2). Importantly, significantly fewer cells had invaded into regions of low realignment (p < 0.009 for all distances ≥ 500 _μ_m; Fig. 2, D and G; for similar results in the chemoattraction control regions R3 vs. R4 see Fig. S2). This finding suggests that radially aligned collagen fibers provide contact guidance via differential alignment, or anisotropy, of the collagen matrix (7,8). With further analysis we found that due to structural anisotropy in the collagen matrix, the degree of migration into the aligned region (MR1) was significantly greater than migration for cells that invaded into the regions of low alignment (MR2; Fig. 2 H). If the matrix structure was isotropic (i.e., had the same organization in all directions) then the migration ratio would be unity (Fig. 2 H), which further supports our conclusion that the degree of tumor cell invasion is a function of matrix organization. Hence, we propose that the matrix anisotropy provides contact guidance that is sensed by the cells and utilized to regulate 3D migration.
Interestingly, on 2D surfaces, substrate stiffness is known to regulate cell migration, a behavior known as durotaxis (31). Although studies have explored the mechanisms associated with durotaxis on 2D substrates (32–34), the mechanisms of durotaxis in 3D microenvironments are even less understood; but computational modeling predicts that the speed of cell migration will exhibit a matrix stiffness-dependent biphasic response (35). Importantly, in collagen matrices in vivo, matrix alignment significantly increases the strength and stiffness of the matrix relative to a matrix that is not organized along the principal axis or a disordered matrix (36,37). We interpret this to mean that in our assay the collagen matrix in the aligned region has greater stiffness due to better structural order. As such, cells probing the matrix and contracting along the primary axis may be encountering a stiffer environment, which implies that part of the physical mechanism of the contact guidance cues is, by definition, a 3D durotaxis response. However, the complex balance of a biphasic response to matrix stiffness reported on 2D substrates (34) and predicted in 3D (35) requires further study in 3D collagen matrices, and the likely influence of a haptotactic component should not be understated. Differing organization of collagen likely results in differential presentation or changes in the local density of adhesive ligands, which are known to influence cell motility on 2D substrates and within 3D matrices (32,34,35). Hence, it seems reasonable to speculate that contact guidance cues are influencing cell behavior by a coupled durotactic-haptactic response.
Matrix reorganization and 3D invasion is protease independent in vitro
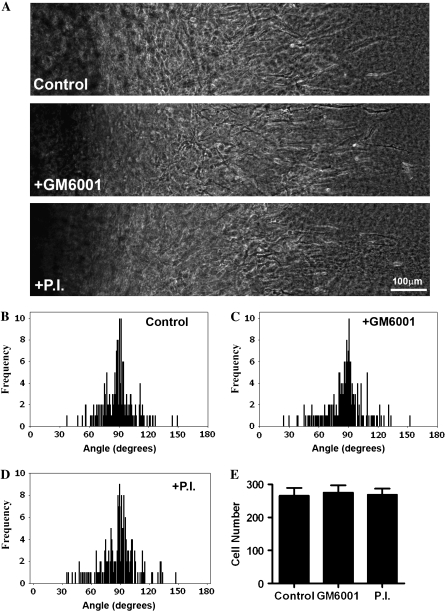
Proteases are recognized to play a role in tumor progression (4,12,13). However, in vitro, tumor cells remain capable of matrix deformation (6) and invasion (3,5) when proteases are inhibited. Furthermore, recent work using a mouse model heterozygous for collagen that is resistant to MMP degradation demonstrated no inhibition of collagen realignment during local invasion (14). Therefore, to investigate whether or not extracellular proteases are required for collagen matrix realignment and 3D migration in vitro, we treated cultures with GM6001, a broad spectrum MMP inhibitor, or GM6001 + Aprotinin + Leupeptin, which additionally block a broad range of serine and cysteine proteases. Inhibition of protease activity did not inhibit invasion of MDA-MB-231 cells into collagen gels (Fig. 3 A). Furthermore, collagen matrix realignment was not inhibited by protease inhibition (Fig. 3, B_–_D), and quantitative analysis of 3D cell migration showed no difference in the number of cells migrating into the aligned collagen matrix (Fig. 3 E). Hence, in vitro, MDA-MB-231 cells remain capable of matrix reorganization and 3D migration in the presence of the protease inhibitors used in this study. However, the in vivo condition may be more complex with the presence of a developmentally established and highly organized ECM that must be “broken-down” to facilitate substantial reorganization. In collagen gels the fiber lengths are considerably shorter and the collagen less organized than the collagen structure surrounding a normal gland before tumor growth and progression (see mammary collagen anatomy in Provenzano et al. (10).). As such, tumor growth may require ECM degradation to accommodate early tumor progression. In fact we suspect that this loosening of the collagen matrix may help set in motion the collagen reorganization associated with local invasion. Future in vivo studies examining the temporal aspects of protease activation and reorganization over the time course of tumor progression will provide further insight into these open questions.
FIGURE 3.
3D Invasion and matrix reorganization are protease independent in vitro. (A) Transmitted light microscopy of MDA-MB-231 cells invading into region R1 from CSCGs (located to the left of the images) exposed to vehicle (top), 10 _μ_M GM6001 (middle), or the protease inhibitor (P.I.) cocktail containing 10 _μ_M GM6001 plus 2 _μ_g/mL aprotinin and 100 _μ_M leupeptin (bottom) at day 10, representative of n ≥ 5. (B–D) Quantification of matrix alignment in region R1 from second harmonic imaging of the collagen matrix at the CSCG-collagen gel boundary at day 10 (n ≥ 5 assays). Cells were treated with vehicle (B), GM6001 (C), or the P.I. cocktail (D). (E) 3D migration/invasion of cells treated with vehicle alone (control), GM6001, or the P.I. cocktail was not significantly different for the number of cells that had invaded up to 1 mm into the collagen gel after 10 days.
Rho/ROCK dependent matrix reorganization regulates contact guidance driven 3D invasion in tumor cells
The Rho pathway effector, ROCK, is required for breast epithelial cell contractility to locally deform the collagen matrix in a protease-independent manner (6), and Rho and ROCK regulate whole gel contraction by breast carcinoma cells (18). We therefore hypothesized that the force needed to deform and reorganize the collagen matrix was generated by cell contractility events governed by the Rho pathway. Addition of the Rho inhibitor, exoenzyme C3 transferase (C3), substantially reduced the degree of matrix reorganization (Fig. 4). Associated with this decreased matrix alignment (i.e., loss of anisotropy; Fig. 4, G and H), tumor cells were not aligned along the primary axis of invasion (Fig. 4 A), and invasion into the matrix between the explants (Region R1 in Fig. 1 A) was decreased (Fig. 4, B and C). Matrix realignment at the explant-matrix interface was significantly reduced (p < 0.0001) in region R1 after Rho inhibition such that the matrix alignment frequency distribution resembled Region 2 in control explants (compare Fig. 1 D Region 2 with Fig. 4 H). Thus, Rho inhibition caused a loss of anisotropy that extended through the entire volume between explants (Fig. 4, I and J).
FIGURE 4.
Rho is Required for matrix reorganization that facilitates 3D migration. (A and B) Transmitted light microscopy of cells invading into region R1 from PyVT tumor explants (located to the left of the images) exposed to 10 _μ_g/mL of the Rho inhibitor C3 exoenzyme (B) or vehicle alone (A) at 14 days. (C–F) Combined (C and D) and MPE/SHG signal separated (E and F) MPLSM images near the TE-collagen interface at region R1 showing decreased cell-induced matrix reorganization when explants were treated with C3 at 10 _μ_g/mL (D and F) when compared to control conditions (C and E). (G and H) Quantification of matrix alignment in region R1 using second harmonic imaging of the collagen matrix at the CSCG-collagen gel boundary at day 14 (n = 3–4 assays) when cells were treated with the Rho inhibitor C3 at 10 _μ_g/mL (H) or vehicle (G). (I) SHG image of collagen alignment at the midpoint between explants after 14 days under control conditions. (J) SHG image of collagen alignment at the midpoint between explants after 14 days when cells are treated with 10 _μ_g/mL concentration of C3 exoenzyme.
To gain further insight into Rho pathway regulation of matrix reorganization and to further confirm that the process of reorganization requires contractility-based force generation, we treated human MDA-MB-231 cells with C3, the ROCK inhibitor H1152 (a more potent and selective inhibitor than Y27632), and blebbistatin, a nonmuscle myosin ATPase inhibitor. Inhibition of Rho with C3 significantly reduced 3D migration into the collagen matrix (Fig. 5, A, B, and G), and simultaneously decreased matrix organization (Fig. 5 D) when compared to control values (Fig. 5 C), which is consistent with our observations in tumor explants (Fig. 4).
Cellular contractility is regulated by myosin-light chain (MLC), which when phosphorylated increases myosin ATPase activity to promote actin–myosin interactions and force generation. ROCK, an effector of Rho, promotes cellular contractility by increasing phosphorylation of MLC and inhibiting MLC phosphatase activity (38–40). Inhibition of ROCK resulted in a similar inhibition of both invasion (Fig. 5, A, B, and G) and alignment (i.e., loss of contact guidance anisotropy; Fig. 5 E), which suggests it is the effects of Rho on this pathway that mediate these processes.
To further establish a role for contractility in matrix reorganization, myosin-based contractility was inhibited with blebbistatin. As observed above, inhibition of actin-myosin based contractility inhibited matrix reorganization (Fig. 5 F) and 3D migration (Fig. 5, A, B, and G). These results suggest that Rho/ROCK-mediated contractility is important to establish matrix alignment and raise the possibility that the contribution of Rho/ROCK in this 3D migration/invasion assay is due primarily to the role of cellular contractility on creating an anisotropic matrix that mediates 3D migration via contact guidance.
Rho/ROCK inhibition does not repress 3D migration into prealigned collagen gels
To separate the effects of Rho-mediated contractility on matrix reorganization and 3D migration and determine if the primary role of the Rho/ROCK pathway herein is to generate matrix anisotropy, we next set out to determine whether the Rho/ROCK pathway was necessary for migration into a prealigned 3D collagen matrix. To accomplish this, we magnetically prealigned the collagen matrix to mimic the perpendicular matrix orientation associated with invasion and the parallel matrix organization associated with low migration/invasion (Fig. 6 A i). Consistent with the results above, a prealigned matrix promoted 3D migration (Fig. 6 A). In contrast, invasion into the parallel matrix, which mimics noninvading regions in tumors, was significantly less (p < 0.0001; Fig. 6 A ii_–_v). Importantly, addition of the Rho inhibitor, C3, or the ROCK inhibitor, H1152, did not significantly alter 3D invasion into the prealigned matrix (Figs. 6 C_–_E), suggesting that the main role of Rho/ROCK mediated cellular contractility in the 3D invasion assay is to create the initial alignment of the 3D matrix. These results are consistent with recent reports that ROCK inhibition actually promotes migration in MCF-7 cells expressing active Rho (41) and that Rho inhibition induces migration of mesenchymal stromal cells (42). Moreover, neither Rho nor ROCK inhibition alters 2D random migration in colon carcinoma cells, whereas elevated Rho activity impairs random migration (43). Thus, neither Rho nor ROCK were required for MDA-MB-231 3D cell migration through the collagen matrix but were required for matrix alignment that creates the contact guidance to promote 3D migration/invasion.
FIGURE 6.
3D Migration in preligned collagen gels is Rho/ROCK Independent. (A) Diagram illustrating the second 3D migration/invasion assay used in this study (i). In the assay, collagen was magnetically aligned as described in the Methods section, and then aligned collagen matrix was connected to a CSCG so that the collagen alignment was either perpendicular (ii and iv) or parallel (iii and v) to the CSCG boundary. (B) 3D migration/invasion of MDA-MB-231 cells as a function of matrix alignment, confirming that contact guidance from perpendicularly aligned collagen significantly (*p < 0.0001; n = 6) promotes 3D migration. Data represent the number of cells per field up to 250 _μ_m into the aligned collagen gel after 72 h. (C) MPE of MDA-MB-231 cells (left) and combined (right) MPE (cells: red) + SHG (collagen: green) showing control and Rho (C3: 10 _μ_g/mL) and ROCK (H1152: 2.5 _μ_M) inhibited cells that have migrated into the aligned matrix after 72 h (representative of 6 assays). Note that due to the 3D nature of the environment not all cells are completely within the optical section. (D) Quantification of matrix alignment in prealigned (perpendicular) matrices from second harmonic imaging of the collagen matrix at the CSCG-collagen gel boundary (n = 3 assays), showing no significant difference in the continued alignment of the prealigned matrix after Rho inhibition Rho (C3: _μ_g/mL) or ROCK inhibition (H1152: 2.5 _μ_M). (E) 3D migration/invasion of control (vehicle alone), Rho inhibited (C3: 10 _μ_g/mL) and ROCK inhibited (H1152: 2.5 _μ_M) cells into aligned collagen matrix after 72 h. No significant difference was present between the three groups, indicating that neither Rho nor ROCK inhibition inhibits 3D migration of MDA-MB-231 breast carcinoma cells into prealigned collagen matrices. Data represent the number of cells per field up to 250 _μ_m into the aligned collagen gel.
Interestingly, a recent report by Gaggioli et al. (44) shows that fibroblasts can promote invasion of squamous cell carcinoma (SCC) cells that retain epithelial markers (and not mesenchymal markers as the MDA-MB-231 cells do) by leading SCC down invasion tracks. Of further interest is the finding that inhibition of the Rho/ROCK pathway did not inhibit fibroblast invasion, yet force-mediated matrix remodeling by fibroblasts is dependent on Rho-mediated MLC-activity and necessary for track formation to facilitate invasion of SCC cells (44). Within the context of this study, these findings raise the interesting possibility that fibroblasts may play a role in generating matrix alignment that provides contact guidance. However, unlike previous reports on invasive carcinoma cells (6), and data herein, invasion of the modestly invasive SCC12 cell line was not dependent on Rho function (44), thus highlighting the complexity of local invasion and its dependence on the diverse characteristics of invasive epithelial cells.
CONCLUSION
In conclusion, we developed a novel assay of 3D migration/invasion that mimics the matrix organization observed within invasive mammary tumors in vivo (10); namely, collagen alignment perpendicular to the tumor-stromal boundary in invading regions and a parallel alignment in noninvading regions. Using this assay, we demonstrate that contact guidance significantly influences cell migration, since an aligned matrix promotes 3D migration relative to regions of low alignment.
Herein, perpendicular collagen alignment by heterogeneous tumor cell populations or the highly invasive MDA-MB-231 human breast carcinoma cell line is generated by cellular contractility requiring the Rho/ROCK pathway. This finding indicates that, consistent with a previous report (6), invasive carcinoma cells can actively reorganize the matrix. However, Rho and ROCK are not required for cell migration if the matrix is prealigned, suggesting that matrix alignment is an early step in the invasion process, which subsequently facilitates local invasion through the stroma.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Supplementary Material
[Supplement]
Acknowledgments
We thank members of the Keely Laboratory and Laboratory for Optical and Computational Instrumentation for their helpful discussions regarding this work.
This work was supported by grants from the U.S. Department of Defense-CDMRP/BCRP: W81XWH-04-1-0428 (P.P.P.), National Institutes of Health-National Cancer Institute: R01-CA076537 and American Cancer Society: RSG-00-339CSM (P.J.K.), and National Institutes of Health-National Institute of Biomedical Imaging and BioEngineering : R01-EB000184 (K.W.E.).
References
- 1.Even-Ram, S., and K. M. Yamada. 2005. Cell migration in 3D matrix. Curr. Opin. Cell Biol. 17:524–532. [DOI] [PubMed] [Google Scholar]
- 2.Zaman, M. H., L. M. Trapani, A. L. Sieminski, D. Mackellar, H. Gong, R. D. Kamm, A. Wells, D. A. Lauffenburger, and P. Matsudaira. 2006. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc. Natl. Acad. Sci. USA. 103:10889–10894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolf, K., I. Mazo, H. Leung, K. Engelke, U. H. von Andrian, E. I. Deryugina, A. Y. Strongin, E. B. Brocker, and P. Friedl. 2003. Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J. Cell Biol. 160:267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolf, K., Y. I. Wu, Y. Liu, J. Gieger, E. Tam, C. Overall, M. S. Stack, and P. Friedl. 2007. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat. Cell Biol. 9:893–904. [DOI] [PubMed] [Google Scholar]
- 5.Sahai, E., and C. J. Marshall. 2003. Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat. Cell Biol. 5:711–719. [DOI] [PubMed] [Google Scholar]
- 6.Wyckoff, J. B., S. E. Pinner, S. Gschmeissner, J. S. Condeelis, and E. Sahai. 2006. ROCK- and myosin-dependent matrix deformation enables protease-independent tumor-cell invasion in vivo. Curr. Biol. 16:1515–1523. [DOI] [PubMed] [Google Scholar]
- 7.Dickinson, R. B., S. Guido, and R. T. Tranquillo. 1994. Biased cell migration of fibroblasts exhibiting contact guidance in oriented collagen gels. Ann. Biomed. Eng. 22:342–356. [DOI] [PubMed] [Google Scholar]
- 8.Guido, S., and R. T. Tranquillo. 1993. A methodology for the systematic and quantitative study of cell contact guidance in oriented collagen gels. Correlation of fibroblast orientation and gel birefringence. J. Cell Sci. 105:317–331. [DOI] [PubMed] [Google Scholar]
- 9.Dallon, J. C., J. A. Sherratt, and P. K. Maini. 1999. Mathematical modelling of extracellular matrix dynamics using discrete cells: fiber orientation and tissue regeneration. J. Theor. Biol. 199:449–471. [DOI] [PubMed] [Google Scholar]
- 10.Provenzano, P. P., K. W. Eliceiri, J. M. Campbell, D. R. Inman, J. G. White, and P. J. Keely. 2006. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 4:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang, W., J. B. Wyckoff, V. C. Frohlich, Y. Oleynikov, S. Huttelmaier, J. Zavadil, L. Cermak, E. P. Bottinger, R. H. Singer, J. G. White, J. E. Segall, and J. S. Condeelis. 2002. Single cell behavior in metastatic primary mammary tumors correlated with gene expression patterns revealed by molecular profiling. Cancer Res. 62:6278–6288. [PubMed] [Google Scholar]
- 12.Hofmann, U. B., A. A. Eggert, K. Blass, E. B. Brocker, and J. C. Becker. 2003. Expression of matrix metalloproteinases in the microenvironment of spontaneous and experimental melanoma metastases reflects the requirements for tumor formation. Cancer Res. 63:8221–8225. [PubMed] [Google Scholar]
- 13.Sabeh, F., I. Ota, K. Holmbeck, H. Birkedal-Hansen, P. Soloway, M. Balbin, C. Lopez-Otin, S. Shapiro, M. Inada, S. Krane, E. Allen, D. Chung, and S. J. Weiss. 2004. Tumor cell traffic through the extracellular matrix is controlled by the membrane-anchored collagenase MT1-MMP. J. Cell Biol. 167:769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Provenzano, P. P., D. R. Inman, K. W. Eliceiri, J. G. Knittel, L. Yan, C. T. Rueden, J. G. White, and P. J. Keely. 2008. Collagen density promotes mammary tumor initiation and progression. BMC Med. 6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo, C., and L. J. Kaufman. 2007. Flow and magnetic field induced collagen alignment. Biomaterials. 28:1105–1114. [DOI] [PubMed] [Google Scholar]
- 16.Miron-Mendoza, M., J. Seemann, and F. Grinnell. 2008. Collagen fibril flow and tissue translocation coupled to fibroblast migration in 3D collagen matrices. Mol. Biol. Cell. 19:2051–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grinnell, F., L. B. Rocha, C. Iucu, S. Rhee, and H. Jiang. 2006. Nested collagen matrices: a new model to study migration of human fibroblast populations in three dimensions. Exp. Cell Res. 312:86–94. [DOI] [PubMed] [Google Scholar]
- 18.Wozniak, M. A., R. Desai, P. A. Solski, C. J. Der, and P. J. Keely. 2003. ROCK-generated contractility regulates breast epithelial cell differentiation in response to the physical properties of a three-dimensional collagen matrix. J. Cell Biol. 163:583–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Netti, P. A., D. A. Berk, M. A. Swartz, A. J. Grodzinsky, and R. K. Jain. 2000. Role of extracellular matrix assembly in interstitial transport in solid tumors. Cancer Res. 60:2497–2503. [PubMed] [Google Scholar]
- 20.Brown, E., T. McKee, E. diTomaso, A. Pluen, B. Seed, Y. Boucher, and R. K. Jain. 2003. Dynamic imaging of collagen and its modulation in tumors in vivo using second-harmonic generation. Nat. Med. 9:796–800. [DOI] [PubMed] [Google Scholar]
- 21.Maglione, J. E., D. Moghanaki, L. J. T. Young, C. K. Manner, L. G. Ellies, S. O. Joseph, B. Nicholson, R. D. Cardiff, and C. L. MacLeod. 2001. Transgenic polyoma middle-T mice model premalignant mammary disease. Cancer Res. 61:8298–8305. [PubMed] [Google Scholar]
- 22.Guy, C. T., R. D. Cardiff, and W. J. Muller. 1992. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol. Cell. Biol. 12:954–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin, E. Y., J. G. Jones, P. Li, L. Zhu, K. D. Whitney, W. J. Muller, and J. W. Pollard. 2003. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am. J. Pathol. 163:2113–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hegerfeldt, Y., M. Tusch, E. B. Brocker, and P. Friedl. 2002. Collective cell movement in primary melanoma explants: plasticity of cell-cell interaction, beta1-integrin function, and migration strategies. Cancer Res. 62:2125–2130. [PubMed] [Google Scholar]
- 25.Denk, W., J. H. Strickler, and W. W. Webb. 1990. Two-photon laser scanning fluorescence microscopy. Science. 248:73–76. [DOI] [PubMed] [Google Scholar]
- 26.Zoumi, A., A. Yeh, and B. J. Tromberg. 2002. Imaging cells and extracellular matrix in vivo by using second-harmonic generation and two-photon excited fluorescence. Proc. Natl. Acad. Sci. USA. 99:11014–11019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zipfel, W. R., R. M. Williams, R. Christie, A. Y. Nikitin, B. T. Hyman, and W. W. Webb. 2003. Live tissue intrinsic emission microscopy using multiphoton-excited native fluorescence and second harmonic generation. Proc. Natl. Acad. Sci. USA. 100:7075–7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohler, W., A. C. Millard, and P. J. Campagnola. 2003. Second harmonic generation imaging of endogenous structural proteins. Methods. 29:97–109. [DOI] [PubMed] [Google Scholar]
- 29.Cox, G., E. Kable, A. Jones, I. Fraser, F. Manconi, and M. D. Gorrell. 2003. 3-dimensional imaging of collagen using second harmonic generation. J. Struct. Biol. 141:53–62. [DOI] [PubMed] [Google Scholar]
- 30.Huang, S., A. A. Heikal, and W. W. Webb. 2002. Two-photon fluorescence spectroscopy and microscopy of NAD(P)H and flavoprotein. Biophys. J. 82:2811–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lo, C. M., H. B. Wang, M. Dembo, and L. Y. Wang. 2000. Cell movement is guided by the rigidity of the substrate. Biophys. J. 79:144–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Engler, A., L. Bacakova, C. Newman, A. Hategan, M. Griffin, and D. Discher. 2004. Substrate compliance versus ligand density in cell on gel responses. Biophys. J. 86:617–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang, G., A. H. Huang, Y. Cai, M. Tanase, and M. P. Sheetz. 2006. Rigidity sensing at the leading edge through alpha-v beta-3 integrins and RPTPalpha. Biophys. J. 90:1804–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peyton, S. R., and A. J. Putnam. 2005. Extracellular matrix rigidity governs smooth muscle cell motility in a biphasic fashion. J. Cell. Physiol. 204:198–209. [DOI] [PubMed] [Google Scholar]
- 35.Zaman, M. H., R. D. Kamm, P. Matsudaira, and D. A. Lauffenburger. 2005. Computational model for cell migration in three-dimensional matrices. Biophys. J. 89:1389–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Provenzano, P. P., D. A. Martinez, R. E. Grindeland, K. W. Dwyer, J. Turner, A. C. Vailas, and R. Vanderby, Jr. 2003. Hindlimb unloading alters ligament healing. J. Appl. Physiol. 94:314–324. [DOI] [PubMed] [Google Scholar]
- 37.Lynch, H. A., W. Johannessen, J. P. Wu, A. Jawa, and D. M. Elliott. 2003. Effect of fiber orientation and strain rate on the nonlinear uniaxial tensile material properties of tendon. J. Biomech. Eng. 125:726–731. [DOI] [PubMed] [Google Scholar]
- 38.Kureishi, Y., S. Kobayashi, M. Amano, K. Kimura, H. Kanaide, T. Nakano, K. Kaibuchi, and M. Ito. 1997. Rho-associated kinase directly induces smooth muscle contraction through myosin light chain phosphorylation. J. Biol. Chem. 272:12257–12260. [DOI] [PubMed] [Google Scholar]
- 39.Amano, M., M. Ito, K. Kimura, Y. Fukata, K. Chihara, T. Nakano, Y. Matsuura, and K. Kaibuchi. 1996. Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase). J. Biol. Chem. 271:20246–20249. [DOI] [PubMed] [Google Scholar]
- 40.Kimura, K., M. Ito, M. Amano, K. Chihara, Y. Fukata, M. Nakafuku, B. Yamamori, J. Feng, T. Nakano, K. Okawa, A. Iwamatsu, and K. Kaibuchi. 1996. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase). Science. 273:245–248. [DOI] [PubMed] [Google Scholar]
- 41.Caceres, M., J. Guerrero, and J. Martinez. 2005. Overexpression of RhoA-GTP induces activation of the epidermal growth factor receptor, dephosphorylation of focal adhesion kinase and increased motility in breast cancer cells. Exp. Cell Res. 309:229–238. [DOI] [PubMed] [Google Scholar]
- 42.Jaganathan, B. G., B. Ruester, L. Dressel, S. Stein, M. Grez, E. Seifried, and R. Henschler. 2007. Rho inhibition induces migration of mesenchymal stromal cells. Stem Cells. 25:1966–1974. [DOI] [PubMed] [Google Scholar]
- 43.Vial, E., E. Sahai, and C. J. Marshall. 2003. ERK-MAPK signaling coordinately regulates activity of Rac1 and RhoA for tumor cell motility. Cancer Cell. 4:67–79. [DOI] [PubMed] [Google Scholar]
- 44.Gaggioli, C., S. Hooper, C. Hidalgo-Carcedo, R. Grosse, J. F. Marshall, K. Harrington, and E. Sahai. 2007. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat. Cell Biol. 9:1392–1400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplement]