Cell culture-produced hepatitis C virus does not infect peripheral blood mononuclear cells (original) (raw)
. Author manuscript; available in PMC: 2009 Dec 1.
Published in final edited form as: Hepatology. 2008 Dec;48(6):1843–1850. doi: 10.1002/hep.22550
Abstract
Hepatitis C virus (HCV) replicates primarily in the liver, but HCV RNA has been observed in association with other tissues and cells including B and T lymphocytes, monocytes, and dendritic cells. We have taken advantage of a recently described, robust system that fully recapitulates HCV entry, replication and virus production in vitro to re-examine the issue of HCV infection of blood cell subsets. The HCV replicase inhibitor 2’C-methyladenosine was used to distinguish HCV RNA replication from RNA persistence. While cell culture-grown HCV replicated in Huh-7.5 hepatoma cells, no HCV replication was detected in B or T lymphocytes, monocytes, macrophages, or dendritic cells from healthy donors. No blood cell subset tested expressed significant levels of Claudin-1, a tight junction protein needed for HCV infection of Huh-7.5 cells. A B cell line expressing high levels of Claudin-1, CD81, and SR-BI remained resistant to HCV pseudoparticle infection. We bypassed the block in HCV entry by transfecting HCV RNA into blood cell subsets. Transfected RNA was not detectably translated and induced high levels of IFNα. Supernatants from HCV RNA-transfected macrophages inhibited HCV replication in Huh-7.5 cells.
Conclusion
We conclude that multiple blocks prevent blood cells from supporting HCV infection.
Introduction
The hepatitis C virus (HCV) is an important human pathogen, infecting an estimated 120-180 million individuals worldwide1. Although HCV is recognized and targeted by innate, cellular, and humoral immune mechanisms, persistent infection is common. Infected individuals may develop cirrhosis, liver failure, hepatocellular carcinoma, and extrahepatic diseases including mixed cryoglobulinemia and non-Hodgkin lymphoma.
The major site of HCV replication is the liver. However, HCV RNA has been detected in association with peripheral blood mononuclear cells (PBMCs) of infected individuals. B cells, monocytes, dendritic cells, and T cells have each been reported to have detectable levels of HCV RNA or proteins2-9. In some reports10,11, but not others12,13, HCV RNA was associated with PBMCs after successful antiviral therapy. Some reports indicated that blood cells could be infected in vitro6,14,15. Distinguishing RNA association from true HCV replication has been problematic. Multiple artifacts complicate detection and quantitation of the replicative intermediate minus strand RNA16,17. In previous studies, retroviral18 and lentiviral19 pseudoparticles bearing HCV envelope glycoproteins (HCVpp) did not infect primary B cells or B cell lines.
It has been proposed that HCV infection of immune cells could contribute to viral persistence by altering the ability of these cells to mount an immune response2; this hypothesis is not supported by the very low levels of HCV RNA, far below one copy per cell, that are measured in association with PBMCs2,3,10,20. In addition, HCV infection of lymphocytes could contribute to the pathogenesis of extrahepatic disease. If PBMCs serve as a reservoir for HCV, they might contribute to re-infection of the graft following liver transplant. Furthermore, reports that HCV RNA persists in association with PBMCs after successful antiviral therapy raise the possibility that PBMC-associated HCV could contribute to virologic relapse. Thus, it is important to clarify the mechanisms by which HCV RNA may associate with PBMCs and to determine whether these cell subsets support viral replication.
We have re-examined the issue of HCV infection of PBMC subsets using a robust in vitro system that fully recapitulates HCV entry, replication and virus production21-24. We used a sensitive and quantitative real-time RT-PCR assay to measure changes in HCV RNA after infecting with cell culture-derived HCV (HCVcc). Translation of viral RNA was also assessed with a stable secreted luciferase reporter and by immunostaining for the non-structural protein, NS5A. We reasoned that if HCV infects B cells, T cells, monocytes, macrophages (Mϕ), or DCs, we should observe a time-dependent increase in HCV RNA that is sensitive to a specific antiviral drug. Furthermore, we tested the ability of PBMC subsets to support infection by HCVpp bearing diverse HCV envelope glycoproteins. We measured expression of known HCV entry factors in PBMC subsets, and prepared a B cell line expressing multiple entry factors to evaluate its ability to support HCV entry. Furthermore, we evaluated expression of HCV genomes transfected into DCs and Mϕ in order to test the ability of HCV to replicate in cells lacking known HCV entry factors.
Materials and methods
Cell preparation, purification and culture are detailed in Supplemental Material.
HCVcc constructs
The following HCVcc viruses and RNAs were used. J6/JFH21 was used for the experiments shown in Figures 2 and 3. Jc1FLAG(p7-nsGluc2A), used for the experiments shown in Figures 5 and 6, is a J6/JFH genome with the intragenotypic break point at the C3 position25 in NS2, and a Flag epitope, followed by a Gly-Ser-Gly-Ala linker, fused to E2’s N-terminus. The Gaussia luciferase (Gluc) reporter, in tandem with the foot and mouth disease autoproteolytic peptide sequence (2A), was inserted between p7 and NS2 using a strategy similar to that previously described for J6/JFH(p7-Rluc2A)26. The native signal sequence of Gluc (Gluc aa 1-16) was deleted and the resultant reporter sequence was fused to the C-terminus of p7. The C-terminus of p7 functions as a signal sequence resulting in Gluc secretion. Jc1FLAG(p7-nsGluc2A)/GNN carries a mutation in the NS5B RNA-dependent RNA polymerase, rendering it replication-defective.
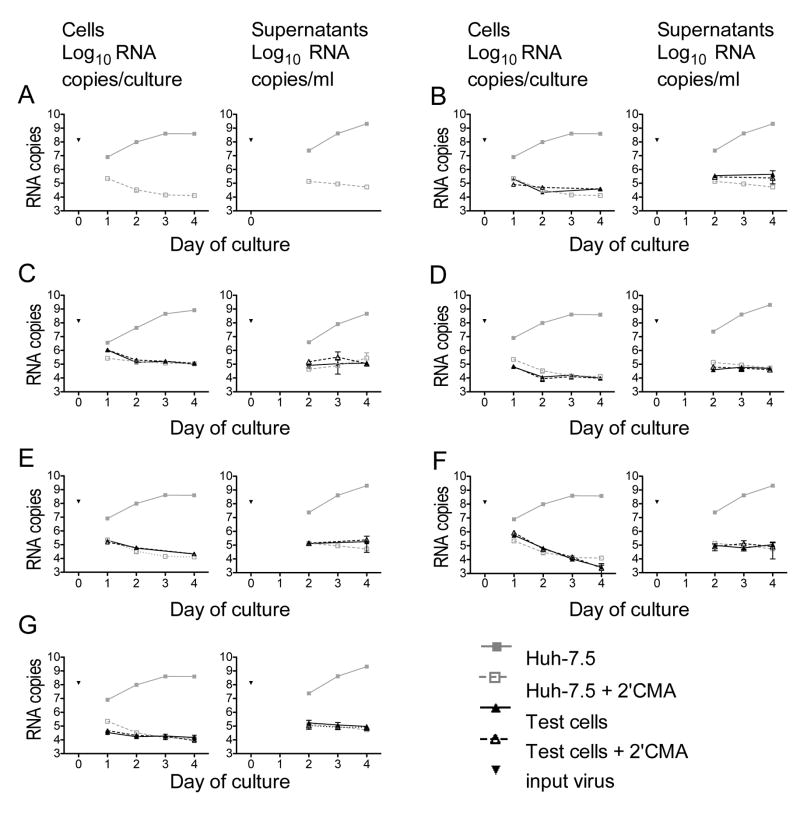
Figure 2. HCVcc infection of Huh-7.5 cells and PBMC subsets in the absence and presence of the NS5B inhibitor, 2’CMA.
HCVcc (5×105 TCID50 units) was added to 106 cells; cultures were washed after 24 hr. RNA was prepared from cells and culture supernatants at the indicated times after infection. The amount of input HCV RNA is shown for each experiment. HCV RNA levels in parallel cultures of Huh-7.5 cells are shown in grey for each plot. Each point represents the mean ± SD of three replicates; experiment shown is representative of 10 similar experiments. (A) Huh-7.5 cells. (B) B cells. (C) Monocyte-derived DCs. (D) Mϕ. (E) pbDCs. (F) Monocytes. (G) T and NK cells.
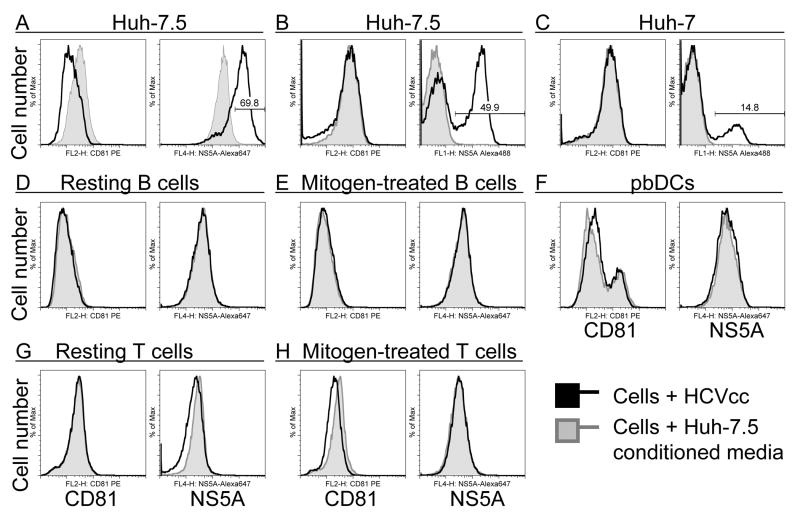
Figure 3. HCVcc infection of Huh-7.5 and PBMC subsets as assessed by NS5A staining.
Cells were cultured with HCVcc as in Figure 2. After 72 hours, fixed and permeabilized cells were immunostained for CD81 and NS5A. 20,000 cells/sample were analyzed by flow cytometry. (A), Huh-7.5 cells stained with Alexa 647-conjugated anti-NS5A. (B), Huh-7.5 and (C), Huh-7 cells stained with Alexa 488-conjugated anti-NS5A. (D), Resting and (E), mitogen-stimulated B cells. (F), pbDCs including myeloid (CD81high) and plasmacytoid (CD81low) subsets. (G), Resting and (H) mitogen-stimulated T/NK cells. Data represent five similar experiments.
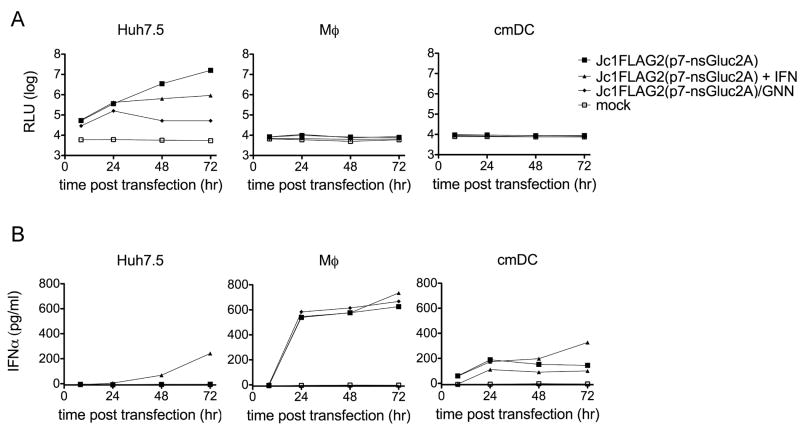
Figure 5. HCV RNA transfection into Mϕ and cmDCs.
RNA encoding HCV with a secreted Gluc reporter was introduced into cells by transfection. Supernatants were collected at the indicated times after transfection. 80 pg/ml (500 U/ml) human IFNα was added to the indicated cultures at 24 hr and again at 48 hr after transfection. (A) Gluc activity in supernatants from cells transfected with functional HCV RNA Jc1FLAG2(p7-nsGluc2a), the polymerase-defective HCV genome Jc1FLAG2(p7-nsGluc2a)/GNN, or mock transfected (no RNA). (B) IFNα levels in supernatants from the transfected cells shown in (A). Each point represents the mean ± SD of three measurements; data are representative of four similar experiments.
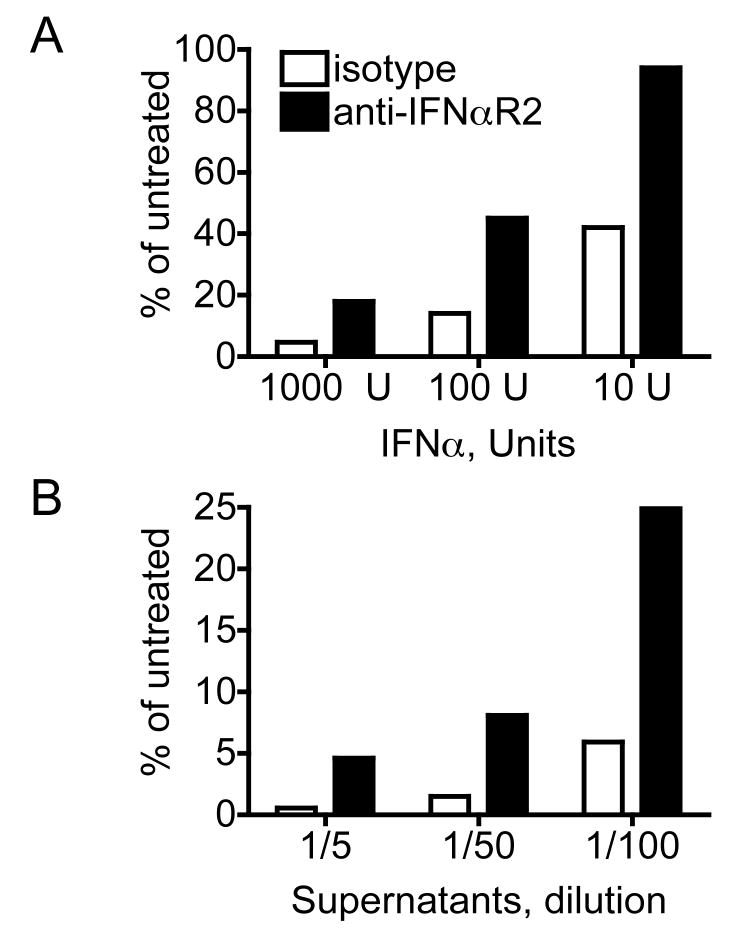
Figure 6. IFNα produced by HCV RNA-transfected Mϕ blocks HCV replication.
Huh-7.5 cells were pre-treated for 18 hr with either (A) recombinant human IFNα2a or (B) diluted supernatants from Mϕ transfected 24 hr earlier with HCV RNA Jc1FLAG2(p7-nsGluc2a), plus 2 μg/ml monoclonal anti-IFNαR2 or isotype control as indicated. Huh-7.5 cells were washed three times and infected with Jc1FLAG2(p7-nsGluc2a) virus. Gluc was measured in cell culture supernatants 72 hr after infection. Percentage of HCV replication was calculated using the formula: 100×(mean RLUtreated)/(mean RLUAntibody only). Data are representative of three similar experiments.
Cell culture-grown HCV (HCVcc)
HCVcc was obtained as previously described21. HCVcc was collected from supernatants 48-72 hr after transfection. Infectious units (TCID50) were quantified by limiting dilution titration on naïve Huh-7.5 cells21.
Pseudoparticles
Production and characterization of pseudoparticles are described in Supplemental Material.
Preparation of CLDN1+Ramos cells is described in Supplemental Material.
Infection and transfection of cells
HCVcc was added to PBMC subsets at a ratio of 0.5 TCID50 units per test cell. After 24 hours at 37°C, cells were washed three times with warmed media. Cultures were continued for the indicated times. To bypass entry, we delivered 0.2 μg HCV RNA (Jc1FLAG2(p7-nsGluc2a) or Jc1FLAG2(p7-nsGluc2a)/GNN) by transfection with the Mirus TransIT mRNA transfection kit (Mirus Bio Corporation, Madison, WI).
HCV inhibitors are described in Supplemental Material.
Results
Isolation of blood cell subsets
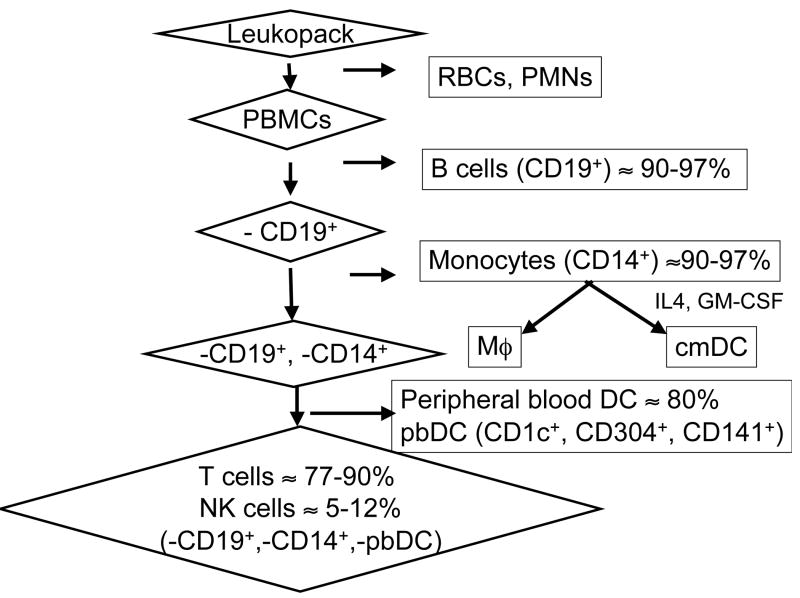
HCV RNA has been reported to associate with various blood cell subsets, including B cells, monocytes/Mϕ, and DCs. To test the ability of cell culture-produced HCV (HCVcc) to replicate in these subsets, we performed immunomagnetic separation of subsets from healthy donor leukocyte preparations as summarized in Figure 1. cmDCs and Mϕ were prepared from purified monocytes as detailed in Supplemental Material.
Figure 1. Enrichment of PBMC subsets.
PBMCs obtained by density gradient centrifugation were subjected to sequential rounds of positive selection for B cell, monocyte, and DC markers. Monocytes were further differentiated in vitro to yield populations of Mϕ and cmDC. T and NK cells made up the bulk of the remaining cells. Percentages indicate the frequency of the indicated cells in the final enriched population.
No evidence for robust HCVcc replication in primary blood cells
We tested the ability of cell culture-produced HCV, HCVcc21, to infect and replicate in isolated PBMC subsets (Figure 2). Huh-7.5 cells and blood cells were cultured for 24 hours with HCVcc at a ratio of 0.5 TCID50 unit/cell (~1.3×108 copies of HCV RNA/106 cells). After three washes, cells were cultured for three more days. Cells were harvested daily for RNA quantitation. Supernatants were harvested daily beginning at day 2 of culture; supernatant viral RNA was not measured at day 1 because the media at this point contained input virus. As a negative control, we measured HCVcc replication in the presence of 2’CMA, an inhibitor of the NS5B RNA-dependent RNA polymerase, at 80 times the IC50 for this drug21. The amount of HCV RNA associated with Huh-7.5 cells was approximately 0.1% of input HCV RNA after 24 hours of infection plus extensive washing (Figure 2A). HCV replication was detected as a time-dependent increase in HCV RNA in Huh-7.5 cell cultures. Cell-associated HCV RNA increased over the next three days in Huh-7.5 cells and supernatants. When 2’CMA was present, cell-associated HCV RNA did not increase during culture.
In primary B cell cultures, the amount of HCV RNA associated with cells was comparable to that seen in 2’CMA-treated Huh-7.5 cell cultures, in which no replication was evident (Figure 2B). However, HCV RNA did not increase over time in the cells or supernatants. 2’CMA did not affect the level of HCV RNA. In the experiment shown, limitations in B cell numbers prevented us from collecting RNA at day 3 of culture. Mitogen addition did not change these results (data not shown).
We tested the ability of HCVcc to replicate in cmDCs (Figure 2C), Mϕ (Figure 2D), pbDCs (Figure 2E), monocytes (Figure 2F), and T/NK cells (Figure 2G). As for B cells, HCV RNA was detected in cultures of these cell types after 24 hours incubation with HCVcc and extensive washing. The level of HCV RNA associated with the cultures was <1% of the input; this declined or remained stable after washing away input viral RNA. In all PBMC subsets, the level of HCV RNA associated with cells and tissue culture supernatants was unaffected by 2’CMA. Although no increase in HCV RNA was detected in PBMC subsets or in 2’CMA-treated Huh-7.5 cells, viral RNA persisted for many days in culture. Longer cultures of up to 14 days also showed no increase in HCV RNA (data not shown).
The HCV nonstructural protein NS5A was readily detected in Huh-7.5 cells (Figure 3A-B) and less permissive Huh-7 cells (Figure 3C) 72 hours after HCVcc infection. In contrast, no NS5A was detected in resting or activated B cells (Figure 3D-E), pbDCs (Figure 3F), or resting or activated T/NK cells (Figure 3G-H) 72 hours after HCVcc addition. Together, these results demonstrate that HCVcc did not replicate in the PBMC subsets tested. Mitogen stimulation did not render lymphocytes permissive for HCVcc infection.
No infection of PBMC subsets with HCVpp
To determine whether PBMC subsets could support HCV entry, we performed HCVpp infection assays (Table 1). Pseudoparticles bearing HCV E1E2 glycoproteins from HCV genotypes 1a, 1b, 2a, 3a, and 4a readily infected Huh-7.5 cells, as expected. Incorporation of envelope glycoproteins in HCVpp varies substantially among HCV isolates18; therefore, the maximum infectivity of different HCVpp variants bearing different glycoproteins varied despite use of equal pseudoparticle amounts. Primary fetal hepatoblasts also supported HCVpp entry. 293T cells expressing CLDN1 supported HCVpp entry at a low level, as previously reported27. However, HCVpp did not infect any PBMC subset. Although others have reported that mitogen-treated lymphocytes can harbor replicating HCV10,28,29, we found that mitogen stimulation did not make lymphocytes permissive for HCVpp entry. Together, these results indicate that these PBMC populations are not susceptible to infection mediated by HCV glycoproteins representing a variety of genotypes. VSV-Gpp efficiently infected all subsets except for B cells and unstimulated T cells. The lack of HCVpp entry into B and T cells is consistent with previous published reports18,19.
TABLE 1.
Infectivity of pseudoparticles bearing diverse HCV glycoproteins for Huh7.5, 293T, and PBMC subsets
| Infection of cells (% GFP positive) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Viral Glycoprotein (genotype) → | NE | VSV-G | H77 (1a) | Con1 (1b) | OH8 (1b) | JFH1 (2a) | J6 (2a) | S52 (3a) | ED43 (4a) |
| Target Cells ↓ | |||||||||
| Huh-7.5 | 1.4 | 61.7 | 29.3 | 36.3 | 59.7 | 36.5 | 9.88 | 4.4 | 12.3 |
| 293T | 0.16 | 50 | 0.14 | 0.14 | 0.25 | 0.22 | 0.16 | 0.14 | 0.15 |
| 293T-CLDN1 | 0.16 | 48.6 | 3.19 | 2.14 | 5.57 | 0.8 | 0.34 | 0.25 | 1.18 |
| Ramos | 0.01 | 12 | 0.01 | 0.01 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 |
| Ramos-CLDN1 | 0.01 | 13.1 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 |
| B cells | 0.01 | 0.00 | 0.01 | 0.00 | 0.00 | 0.01 | 0.00 | 0.00 | 0.00 |
| B cells + PWM | 0.00 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| T cells | 0.01 | 0.01 | 0.00 | 0.01 | 0.00 | 0.00 | 0.00 | 0.01 | 0.00 |
| T cells + PHA | 0.00 | 3.83 | 0.00 | 0.00 | 0.01 | 0.00 | 0.01 | 0.00 | 0.00 |
| Monocytes | 0.01 | 2.68 | 0.01 | 0.01 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 |
| Mϕ | 0.37 | 1.94 | 0.32 | ND | ND | ND | ND | ND | ND |
| cmDCs | 0.00 | 3.18 | 0.00 | ND | ND | ND | ND | ND | ND |
| pbDC | 0.01 | 5.64 | 0.00 | 0.00 | 0.01 | 0.00 | 0.01 | 0.00 | 0.01 |
| Fetal hepatocytes | 0.27 | 39.2 | 2.63 | ND | ND | ND | ND | ND | ND |
Expression of HCV entry factors in primary blood cells
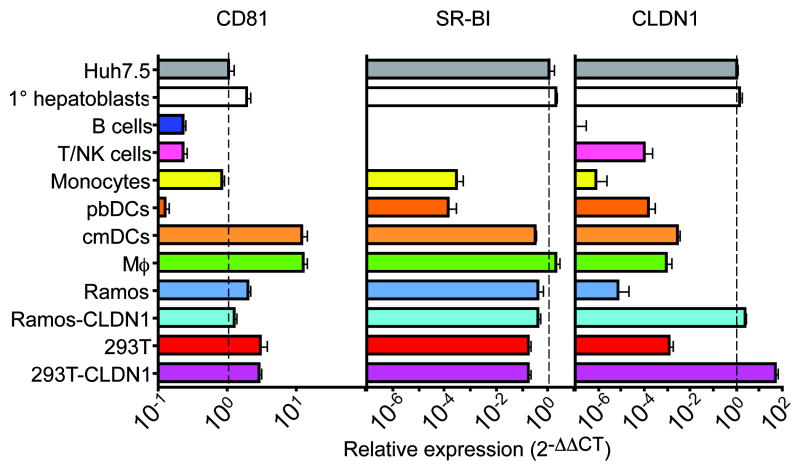
Several factors have been determined to play a role in HCV entry into susceptible cells. These include CD8118,30,31, the scavenger receptor SR-BI32-35, and the tight-junction protein Claudin-1 (CLDN1)27,36. Importantly, all three entry factors are required for HCVcc or HCVpp infection of Huh-7.5 cells27. We performed quantitative real-time RT-PCR to determine whether PBMCs expressed these entry factors. Results are summarized in Figure 4. High levels of CD81 RNA were detected in all PBMC subsets. However, SR-BI RNA was undetectable in B or T/NK cells and was low in monocytes and pbDC. CLDN1 was not expressed at high levels in any PBMC subset tested. Of note, as little as a five-fold reduction in CLDN1 expression abrogates HCVcc infection in Huh-7.5 cells27. Our results indicate that CLDN1 expression in B cells is at least six orders of magnitude lower than in Huh-7.5 cells. CLDN1 levels in other PBMC subsets were also below CLDN1 levels in non-permissive 293T cells, and therefore too low to support HCV entry. Relative levels of CD81, SR-BI, and CLDN1 protein were also assessed by immunoblotting and flow cytometry; results were consistent with our RNA measurements (Figure 3 and data not shown). Thus, primary B cells, T cells, monocytes, pbDCs, Mϕ and cmDCs all lack at least one known HCV entry factor. Primary fetal hepatocytes expressed CLDN-1, SR-BI, and CD81 at levels comparable to Huh-7.5. CLDN6 and CLDN9 RNA37,38 were detected at low levels in all cells tested (Figure S1A-B). A splice variant of SR-BI (termed SR-BII in reference39) was detected in B and T cells (Figure S1C).
Figure 4. Expression of HCV entry factors.
RNA levels were measured by quantitative real-time RT-PCR using SYBR Green. The housekeeping gene RPS11 was used for normalization. The ratio of entry factor RNA to RPS11 RNA was calculated for each cell type, and expression relative to Huh-7.5 is shown. Data represent the mean ± SD of three measurements, and are representative of three similar experiments (three blood donors). On each graph, the dotted line indicates the RNA level in Huh-7.5 cells for comparison. (A) CD81 RNA levels. (B) SR-BI RNA levels. (C) CLDN1 RNA levels.
CLDN1 expression does not make B cells permissive for HCV
In the kidney epithelial cell line 293T, CLDN1 overexpression can confer susceptibility to HCVpp infection27. PBMCs do not express CLDN1 (Figure 4). Although Mϕ and the B cell line Ramos expressed at least as much CD81 and SR-BI as do permissive Huh-7.5 cells, neither cell type supported HCVpp infection (Table 2). We tested whether ectopic expression of CLDN1 could render these cells permissive to HCVpp infection. We prepared stable lines of Ramos B cells expressing CLDN1 by retroviral transduction. Transduced Ramos cells (>95% CLDN1+) and transduced Mϕ did not support HCVpp entry even when expressing high levels of CD81, SR-BI, and CLDN1 (Figure 4, Table S1, and data not shown). While cell surface expression of CLDN1 was confirmed by flow cytometry (Figure S2), it is possible that CLDN1 would not function normally in blood cells since they do not form tight junctions. CLDN1+ Ramos cells also were not permissive for HCVcc infection (data not shown). VSV-Gpp infection was readily detected in Ramos cells and Mϕ regardless of CLDN1 expression. These results suggest that Ramos cells and Mϕ lack additional, as yet undefined, entry factors for HCVpp.
HCV RNA is not translated and does not replicate when transfected into Mϕ or DCs
It is possible that the lack of HCV replication observed in PBMC subsets resulted from the inability of HCVcc to enter these cells. An alternative virus entry pathway could exist in some PBMC subsets. To bypass entry, we transfected HCV Jc1FLAG2(p7-nsGluc2a) RNA into readily-transfected Mϕ and cmDCs. Jc1FLAG2(p7-nsGluc2a) allows sensitive detection of HCV RNA translation by measurement of the secreted Gluc reporter. We tested these cells for the ability to support HCV RNA translation and replication (Figure 5A). As a positive control, the same RNA was transfected into Huh-7.5 cells. As a negative control, we transfected cells with Jc1FLAG2(p7-nsGluc2a)/GNN, which does not replicate due to a lethal mutation in NS5B. In Huh-7.5 cells transfected with either Jc1FLAG2(p7-nsGluc2a) or Jc1FLAG2(p7-nsGluc2a)/GNN, translation of input RNA was readily detected as an increase in secreted Gluc activity by 8 hours after transfection. In contrast, Mϕ and DCs transfected with Jc1FLAG2(p7-nsGluc2a) or Jc1FLAG2(p7-nsGluc2a)/GNN RNA showed no Gluc activity over background at any time after transfection. Control experiments using a polio replicon encoding GFP confirmed that Mϕ and cmDCs were efficiently transfected (data not shown). High levels of input HCV RNA were detected in all transfected cells by quantitative real time RT-PCR; HCV RNA declined during culture in transfected Mϕ and cmDCs (data not shown).
HCV RNA-transfected cmDC and Mϕ produce IFNα
Since viral RNA is a potent stimulus for innate antiviral mechanisms, we hypothesized that HCV RNA-transfected cmDC and Mϕ might produce IFNs after transfection, and that this could block translation and replication. Supernatants from HCV RNA-transfected Huh-7.5, Mϕ, and cmDC were tested by ELISA for IFNα (Figure 5B). While IFNα was not detected in the supernatants of transfected Huh-7.5 cells, cmDC and Mϕ produced between 100-800 pg/ml IFNα following HCV RNA transfection. This level of IFNα is sufficient to block HCV replication in Huh-7.5 cells (Figure 5B). Moreover, supernatants from Jc1FLAG2(p7-nsGluc2a)-transfected Mϕ inhibited HCV replication in Huh-7.5 cells in a dose-dependent manner (Figure 6). The inhibition was partially abrogated by a blocking antibody to IFNα receptor. This implies that recognition of the transfected RNA induced Mϕ to produce IFNα at levels sufficient to block HCV replication in a permissive cell line. We also tested IFNα levels in supernatants from Mϕ, B cells, T/NK cells, monocytes, and cmDCs treated with HCVcc. While these cells did not support HCV replication, they also did not produce IFNα in response to added HCVcc (data not shown). Thus, the inability of these cells to support HCVcc infection and replication is not due to IFNα production.
Discussion
We have used a system that recapitulates HCV entry, replication, and production of infectious virus to examine the ability of HCV to productively infect immune cells from peripheral blood. Here we show that B cells, T/NK cells, monocytes, Mϕ and DCs do not support productive infection with cell culture-produced HCV of genotype 2a. Several lines of evidence support this conclusion. Levels of HCV RNA did not increase in cultures of PBMC subsets at any time following culture with HCVcc. In addition, the low level of HCV RNA detected in PBMC cultures was not affected by 2’CMA, an inhibitor of HCV’s RNA-dependent RNA polymerase. Although NS5A was readily observed in HCVcc-infected Huh-7.5 and Huh-7 cells, we did not detect NS5A in any PBMC subset cultured with HCVcc. Moreover, retroviral pseudoparticles bearing diverse HCV glycoproteins did not infect any PBMC subset tested. All PBMC subsets tested lacked expression of at least one known HCV entry factor. We did not detect translation of HCV RNA introduced into Mϕ and DC by transfection. Instead, the transfected cells produced substantial amounts of IFNα.
In Huh-7.5 cells, HCVcc and HCVpp infection are dependent on at least three proteins: CD81, SR-BI, and CLDN127. Co-expression of these three molecules renders the epithelial cell line 293T permissive for HCVpp entry27. No PBMC subset tested expressed all of these entry factors (Figure 4). This is not surprising since CLDN1 is a tight junction protein with no known functions in lymphoid cells. Ectopic expression of CLDN1 in a B cell line that already expresses high levels of CD81 and SR-BI (Figure 4) did not render the cells permissive to HCVpp entry (Table 2). The Ramos cell line may lack other, as yet unidentified, HCV entry factors. Alternatively, it is possible that Ramos and other cell types express a factor that inhibits HCV entry. In contrast to primary PBMC subsets, a preparation of primary fetal hepatoblasts was permissive for HCVpp entry (Table 1) and low-level HCVcc replication (data not shown). Our studies did not evaluate the potential contribution of other putative HCV binding or entry factors40,41.
The possibility remains that lymphotropic strains or quasispecies5,7,9 of HCV exist in infected individuals. Approximately 0.1% of the input RNA remained associated with PBMC cultures after input virus was washed away (Figure 2). Detection of HCV RNA associated with blood cells is not by itself proof that the virus replicates in these cells4. Viral RNA may be bound or taken up by cells without undergoing a complete infectious cycle. CD81, an entry factor for HCV, is expressed by many cell types in the blood and in other tissues. Therefore, CD81 could mediate PBMC capture of HCV from the environment. Anti-HCV antibodies may permit HCV-specific B cells to capture HCV RNA from the circulation. In this regard, it was reported that HCV RNA was found specifically in PBMC populations containing B cells capable of producing HCV-specific antibodies8. Furthermore, antibody binding to HCV may permit HCV capture by Fc receptors, complement receptors, or rheumatoid factor. Consistent with this notion, it has been reported that HCV RNA was specifically enriched in B cells bearing rheumatoid factor-like immunoglobulin in HCV patients42. Since we did not use PBMCs or serum from HCV patients in this study, we conclude that HCV RNA can associate with PBMCs independently of HCV-specific or autoreactive antibodies. However, despite the association of HCV RNA with these cells, they do not demonstrate efficient HCV envelope-mediated entry or HCV RNA replication. Our results indicate that multiple factors prevent PBMCs from supporting robust HCV infection.
Supplementary Material
Figure 1 A B
Figure 1 C
Figure 2
Supp Table 1
Acknowledgments
We acknowledge Jodie Tassello and Michelle Hunter for preparation of viral pseudoparticle stocks. We thank Margaret MacDonald for antibodies, Jens Bukh for E1E2 clones used in some HCVpp, and David Olsen and Steven Carroll for 2’CMA.
Financial Support: Supported by the NIH (AI60561), the Gates Foundation, the Greenberg Medical Research Institute, and the Irma T. Hirschl/Monique Weill Caulier Trust.
Abbreviations
HCV
hepatitis C virus
PBMCs
peripheral blood mononuclear cells
DCs
dendritic cells
HCVcc
cell culture-produced HCV
Mϕ
macrophages
pbDCs
peripheral blood dendritic cells
cmDCs
conventional monocyte-derived dendritic cells
NK
natural killer
Gluc
Gaussia luciferase
VSV-Gpp
pseudoparticles bearing the G glycoproteins from vesicular stomatitis virus
NE
no viral envelope glycoproteins
2’CMA
2’ C-methyl adenosine
CLDN1
Claudin-1
GFP
green fluorescent protein
References
- 1.Dustin LB, Rice CM. Flying under the radar: The immunobiology of hepatitis C. Annu Rev Immunol. 2007;25:71–99. doi: 10.1146/annurev.immunol.25.022106.141602. [DOI] [PubMed] [Google Scholar]
- 2.Rodrigue-Gervais IG, Jouan L, Beaule G, Sauve D, Bruneau J, Willems B, Sekaly RP, et al. Poly(I:C) and lipopolysaccharide innate sensing functions of circulating human myeloid dendritic cells are affected in vivo in hepatitis C virus-infected patients. J Virol. 2007;81:5537–5546. doi: 10.1128/JVI.01741-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zehender G, Meroni L, De Maddalena C, Varchetta S, Monti G, Galli M. Detection of hepatitis C virus RNA in CD19 peripheral blood mononuclear cells of chronically infected patients. J Infect Dis. 1997;176:1209–1214. doi: 10.1086/514114. [DOI] [PubMed] [Google Scholar]
- 4.Boisvert J, He XS, Cheung R, Keeffe EB, Wright T, Greenberg HB. Quantitative analysis of hepatitis C virus in peripheral blood and liver: replication detected only in liver. J Infect Dis. 2001;184:827–835. doi: 10.1086/323391. [DOI] [PubMed] [Google Scholar]
- 5.Caussin-Schwemling C, Schmitt C, Stoll-Keller F. Study of the infection of human blood derived monocyte/macrophages with hepatitis C virus in vitro. J Med Virol. 2001;65:14–22. doi: 10.1002/jmv.1095. [DOI] [PubMed] [Google Scholar]
- 6.Navas MC, Fuchs A, Schvoerer E, Bohbot A, Aubertin AM, Stoll-Keller F. Dendritic cell susceptibility to hepatitis C virus genotype 1 infection. J Med Virol. 2002;67:152–161. doi: 10.1002/jmv.2204. [DOI] [PubMed] [Google Scholar]
- 7.Goutagny N, Fatmi A, De Ledinghen V, Penin F, Couzigou P, Inchauspe G, Bain C. Evidence of viral replication in circulating dendritic cells during hepatitis C virus infection. J Infect Dis. 2003;187:1951–1958. doi: 10.1086/375350. [DOI] [PubMed] [Google Scholar]
- 8.Fournillier A, Freida D, Defrance T, Merle P, Trepo C, Inchauspe G. Analysis of B-lymphocyte differentiation in patients infected with hepatitis C virus. J Med Virol. 2004;72:566–574. doi: 10.1002/jmv.20039. [DOI] [PubMed] [Google Scholar]
- 9.Laporte J, Bain C, Maurel P, Inchauspe G, Agut H, Cahour A. Differential distribution and internal translation efficiency of hepatitis C virus quasispecies present in dendritic and liver cells. Blood. 2003;101:52–57. doi: 10.1182/blood-2002-03-0818. [DOI] [PubMed] [Google Scholar]
- 10.Radkowski M, Gallegos-Orozco JF, Jablonska J, Colby TV, Walewska-Zielecka B, Kubicka J, Wilkinson J, et al. Persistence of hepatitis C virus in patients successfully treated for chronic hepatitis C. Hepatology. 2005;41:106–114. doi: 10.1002/hep.20518. [DOI] [PubMed] [Google Scholar]
- 11.Pham TN, MacParland SA, Mulrooney PM, Cooksley H, Naoumov NV, Michalak TI. Hepatitis C virus persistence after spontaneous or treatment-induced resolution of hepatitis C. J Virol. 2004;78:5867–5874. doi: 10.1128/JVI.78.11.5867-5874.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wiegand J, Jackel E, Cornberg M, Hinrichsen H, Dietrich M, Kroeger J, Fritsch WP, et al. Long-term follow-up after successful interferon therapy of acute hepatitis C. Hepatology. 2004;40:98–107. doi: 10.1002/hep.20291. [DOI] [PubMed] [Google Scholar]
- 13.Bernardin F, Tobler L, Walsh I, Williams JD, Busch M, Delwart E. Clearance of hepatitis C virus RNA from the peripheral blood mononuclear cells of blood donors who spontaneously or therapeutically control their plasma viremia. Hepatology. 2008;47:1446–1452. doi: 10.1002/hep.22184. [DOI] [PubMed] [Google Scholar]
- 14.Sung VM, Shimodaira S, Doughty AL, Picchio GR, Can H, Yen TS, Lindsay KL, et al. Establishment of B-cell lymphoma cell lines persistently infected with hepatitis C virus in vivo and in vitro: the apoptotic effects of virus infection. J Virol. 2003;77:2134–2146. doi: 10.1128/JVI.77.3.2134-2146.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kondo Y, Sung VM, Machida K, Liu M, Lai MM. Hepatitis C virus infects T cells and affects interferon-gamma signaling in T cell lines. Virology. 2007;361:161–173. doi: 10.1016/j.virol.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 16.Lerat H, Berby F, Trabaud MA, Vidalin O, Major M, Trepo C, Inchauspe G. Specific detection of hepatitis C virus minus strand RNA in hematopoietic cells. J Clin Invest. 1996;97:845–851. doi: 10.1172/JCI118485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lanford RE, Chavez D, Chisari FV, Sureau C. Lack of detection of negative-strand hepatitis C virus RNA in peripheral blood mononuclear cells and other extrahepatic tissues by the highly strand-specific rTth reverse transcriptase PCR. J Virol. 1995;69:8079–8083. doi: 10.1128/jvi.69.12.8079-8083.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKeating JA, Zhang LQ, Logvinoff C, Flint M, Zhang J, Yu J, Butera D, et al. Diverse hepatitis C virus glycoproteins mediate viral infection in a CD81 dependent manner. J Virol. 2004;78:8496–8505. doi: 10.1128/JVI.78.16.8496-8505.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartosch B, Dubuisson J, Cosset F-L. Infectious hepatitis C virus pseudoparticles containing functional E1-E2 envelope protein complexes. J Exp Med. 2003;197:633–642. doi: 10.1084/jem.20021756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mellor J, Haydon G, Blair C, Livingstone W, Simmonds P. Low level or absent in vivo replication of hepatitis C virus and hepatitis G virus/GB virus C in peripheral blood mononuclear cells. J Gen Virol. 1998;79(Pt 4):705–714. doi: 10.1099/0022-1317-79-4-705. [DOI] [PubMed] [Google Scholar]
- 21.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 22.Lindenbach BD, Meuleman P, Ploss A, Vanwolleghem T, Syder AJ, McKeating JA, Lanford RE, et al. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proc Natl Acad Sci U S A. 2006;103:3805–3809. doi: 10.1073/pnas.0511218103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, et al. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pietschmann T, Kaul A, Koutsoudakis G, Shavinskaya A, Kallis S, Steinmann E, Abid K, et al. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc Natl Acad Sci U S A. 2006;103:7408–7413. doi: 10.1073/pnas.0504877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones CT, Murray CL, Eastman DK, Tassello J, Rice CM. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J Virol. 2007;81:8374–8383. doi: 10.1128/JVI.00690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wolk B, Hatziioannou T, et al. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801–805. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- 28.MacParland SA, Pham TN, Gujar SA, Michalak TI. De novo infection and propagation of wild-type Hepatitis C virus in human T lymphocytes in vitro. J Gen Virol. 2006;87:3577–3586. doi: 10.1099/vir.0.81868-0. [DOI] [PubMed] [Google Scholar]
- 29.Pham TN, Mulrooney-Cousins PM, Mercer SE, MacParland SA, Inglot M, Zalewska M, Simon K, et al. Antagonistic expression of hepatitis C virus and alpha interferon in lymphoid cells during persistent occult infection. J Viral Hepat. 2007;14:537–548. doi: 10.1111/j.1365-2893.2006.00834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, et al. Binding of hepatitis C virus to CD81. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 31.Koutsoudakis G, Herrmann E, Kallis S, Bartenschlager R, Pietschmann T. The level of CD81 cell surface expression is a key determinant for productive entry of hepatitis C virus into host cells. J Virol. 2007;81:588–598. doi: 10.1128/JVI.01534-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Catanese MT, Graziani R, von Hahn T, Moreau M, Huby T, Paonessa G, Santini C, et al. High avidity monoclonal antibodies against human scavenger receptor class B type I efficiently block hepatitis C virus infection in the presence of HDL. J Virol. 2007;81:8063–8071. doi: 10.1128/JVI.00193-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G, Traboni C, et al. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. Embo J. 2002;21:5017–5025. doi: 10.1093/emboj/cdf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.von Hahn T, Lindenbach BD, Boullier A, Quehenberger O, Paulson M, Rice CM, McKeating JA. Oxidized low-density lipoprotein inhibits hepatitis C virus cell entry in human hepatoma cells. Hepatology. 2006;43:932–942. doi: 10.1002/hep.21139. [DOI] [PubMed] [Google Scholar]
- 35.Zeisel MB, Koutsoudakis G, Schnober EK, Haberstroh A, Blum HE, Cosset FL, Wakita T, et al. Scavenger receptor class B type I is a key host factor for hepatitis C virus infection required for an entry step closely linked to CD81. Hepatology. 2007;46:1722–1731. doi: 10.1002/hep.21994. [DOI] [PubMed] [Google Scholar]
- 36.Timpe JM, Stamataki Z, Jennings A, Hu K, Farquhar MJ, Harris HJ, Schwarz A, et al. Hepatitis C virus cell-cell transmission in hepatoma cells in the presence of neutralizing antibodies. Hepatology. 2008;47:17–24. doi: 10.1002/hep.21959. [DOI] [PubMed] [Google Scholar]
- 37.Meertens L, Bertaux C, Cukierman L, Cormier E, Lavillette D, Cosset FL, Dragic T. The tight junction proteins claudin-1, -6, and -9 are entry cofactors for hepatitis C virus. J Virol. 2008;82:3555–3560. doi: 10.1128/JVI.01977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng A, Yuan F, Li Y, Zhu F, Hou P, Li J, Song X, et al. Claudin-6 and claudin-9 function as additional coreceptors for hepatitis C virus. J Virol. 2007;81:12465–12471. doi: 10.1128/JVI.01457-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grove J, Huby T, Stamataki Z, Vanwolleghem T, Meuleman P, Farquhar M, Schwarz A, et al. Scavenger receptor BI and BII expression levels modulate hepatitis C virus infectivity. J Virol. 2007;81:3162–3169. doi: 10.1128/JVI.02356-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barth H, Liang TJ, Baumert TF. Hepatitis C virus entry: molecular biology and clinical implications. Hepatology. 2006;44:527–535. doi: 10.1002/hep.21321. [DOI] [PubMed] [Google Scholar]
- 41.Cocquerel L, Voisset C, Dubuisson J. Hepatitis C virus entry: potential receptors and their biological functions. J Gen Virol. 2006;87:1075–1084. doi: 10.1099/vir.0.81646-0. [DOI] [PubMed] [Google Scholar]
- 42.Fornasieri A, Bernasconi P, Ribero ML, Sinico RA, Fasola M, Zhou J, Portera G, et al. Hepatitis C virus (HCV) in lymphocyte subsets and in B lymphocytes expressing rheumatoid factor cross-reacting idiotype in type II mixed cryoglobulinaemia. Clin Exp Immunol. 2000;122:400–403. doi: 10.1046/j.1365-2249.2000.01396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure 1 A B
Figure 1 C
Figure 2
Supp Table 1