Regulation of T cell Receptor Activation by Dynamic Membrane Binding of the CD3ε Cytoplasmic Tyrosine-Based Motif (original) (raw)
. Author manuscript; available in PMC: 2009 Nov 15.
Summary
Many immune system receptors signal through cytoplasmic tyrosine-based motifs (ITAMs), but how receptor ligation results in ITAM phosphorylation remains unknown. Live cell imaging studies showed a close interaction of the CD3ε cytoplasmic domain of the T cell receptor (TCR) with the plasma membrane through fluorescence resonance energy transfer (FRET) between a C-terminal fluorescent protein and a membrane fluorophore. Electrostatic interactions between basic CD3ε residues and acidic phospholipids enriched in the inner leaflet of the plasma membrane were required for binding. The nuclear magnetic resonance (NMR) structure of the lipid-bound state of this cytoplasmic domain revealed deep insertion of the two key tyrosines into the hydrophobic core of the lipid bilayer. Receptor ligation thus needs to result in unbinding of the CD3ε ITAM from the membrane to render these tyrosines accessible to Src kinases. Sequestration of key tyrosines into the lipid bilayer represents a previously unrecognized mechanism for control of receptor activation.
Introduction
T cell receptor (TCR) recognition of MHC-bound peptides is an essential step for the initiation of adaptive immune responses to invading pathogens and induces a complex signaling cascade that results in clonal expansion, cytokine production and other effector functions (Davis and Chien, 2003). The TCR is one of the most complex multi-protein cell surface receptors and is composed of the ligand-sensing TCR heterodimer and three signaling subunits - CD3γε, CD3δε and ζζ - that associate with the TCR in the membrane through a set of conserved ionizable transmembrane residues (Call et al., 2002; Davis and Chien, 2003). These signaling subunits contain cytoplasmic immunoreceptor tyrosine-based activation motifs (ITAMs) with a consensus sequence of YxxL/Ix6–12YxxL/I (Reth, 1989). The two tyrosines are phosphorylated by a Src kinase, Lck or Fyn, and a dually phosphorylated ITAM recruits the ZAP-70 protein tyrosine kinase, which in turn phosphorylates downstream components of the signaling pathway (Weiss and Littman, 1994). The phospho-tyrosine and leucine/isoleucine residues of each YxxL/I hemi-motif are bound by deep pockets of the tandem SH2 domains of ZAP-70 (Hatada et al., 1995). Binding of a dually phosphorylated ITAM peptide reorients the two SH2 domains relative to each other, destabilizing inhibitory interactions made by the linker connecting the SH2 and kinase domains. These conformational changes explain how ITAM binding induces activation of the ZAP-70 kinase domain (Deindl et al., 2007).
The central unresolved question in this field is how ligand engagement by the TCR ectodomains results in phosphorylation of the cytoplasmic domains of the three dimeric signaling subunits. This question is of general significance because many different receptors in cells of hematopoietic origin signal through cytoplasmic ITAMs, including the B cell receptor, several Fc receptors, a group of NK cell receptors and many others (Reth, 1989; Weiss and Littman, 1994). A common feature among these receptors is that the ligand binding subunit assembles with one or more dimeric signaling subunits with cytoplasmic ITAMs (Call and Wucherpfennig, 2007). ITAMs are, however, evolutionarily more ancient than the immune systems of mammals. The C. elegans engulfment receptor CED-1 and its Drosophila ortholog, Draper, are expressed by glial cells in the central nervous system and mediate phagocytosis of apoptotic neurons. Draper contains an ITAM motif (YxxIx11YxxL) that is phosphorylated by the Src kinase Src42A, resulting in recruitment of a tyrosine kinase (Shark) homologous to mammalian Syk and ZAP-70 (Ziegenfuss et al., 2008). Furthermore, ITAMs have been identified in membrane proteins of several viruses that can cause cellular transformation, including the envelope protein of mouse mammary tumor virus (MMTV), the LMP2A protein of Epstein-Barr virus (EBV) and the gp30 protein of bovine leukemia virus (Lu et al., 2006; Reth, 1989; Ross et al., 2006). MMTV uses the ITAM to activate Syk and mutation of its ITAM substantially reduces the ability of this virus to induce transformation (Ross et al., 2006).
Most models of TCR triggering postulate a ligand-induced conformational change, but the structural consequences of such a conformational change remain unknown. Furthermore, textbooks depict the cytoplasmic ITAMs as flexible chains that float in the cytoplasm because peptides representing the cytoplasmic domains are unstructured in solution. However, a previous study showed that the cytoplasmic domain of the TCR ζ chain can bind to synthetic lipid vesicles that contain acidic phospholipids (Aivazian and Stern, 2000). Acidic phospholipids were tested because the lipid distribution of the plasma membrane is asymmetric and the most abundant negatively charged lipid, phosphatidylserine, is primarily localized to the cytoplasmic face through active transport mechanisms (Devaux, 1991). That paper reported that binding to synthetic acidic lipid vesicles substantially increased the α-helical content of the ζ cytoplasmic domain based on circular dichroism (CD) measurements, which suggested a α-helical conformation of the ζ chain ITAMs (Aivazian and Stern, 2000). However, a follow-up study by the same group concluded - again based on CD measurements - that the ζ cytoplasmic domain remained unstructured when bound to acidic lipid vesicles and that lipid binding only partially inhibited ITAM phosphorylation (Sigalov et al., 2006). The physiological relevance of lipid binding by ITAMs remained uncertain because membrane binding has never been demonstrated in cells, an issue that is critical because membrane binding by the ζ cytoplasmic domain primarily involved electrostatic interactions, which can be non-specific.
In this study we have examined the functional relevance of membrane binding by ITAMs, using the CD3ε cytoplasmic domain (CD3εCD) of the TCR as an example. Binding of CD3εCD to the inner leaflet of the plasma membrane was visualized in live cells by fluorescence resonance energy transfer (FRET) between a fluorescent protein attached to the C-terminus of CD3εCD and a fluorescent membrane dye. The NMR structure of CD3εCD in its lipid-bound state showed that the two tyrosines of the ITAM are inserted into the hydrophobic core of the lipid bilayer. TCR triggering therefore has to result in dissociation of the ITAM from the membrane to render the tyrosines accessible to kinases.
Results
Identification of CD3εCD sequence elements involved in membrane binding
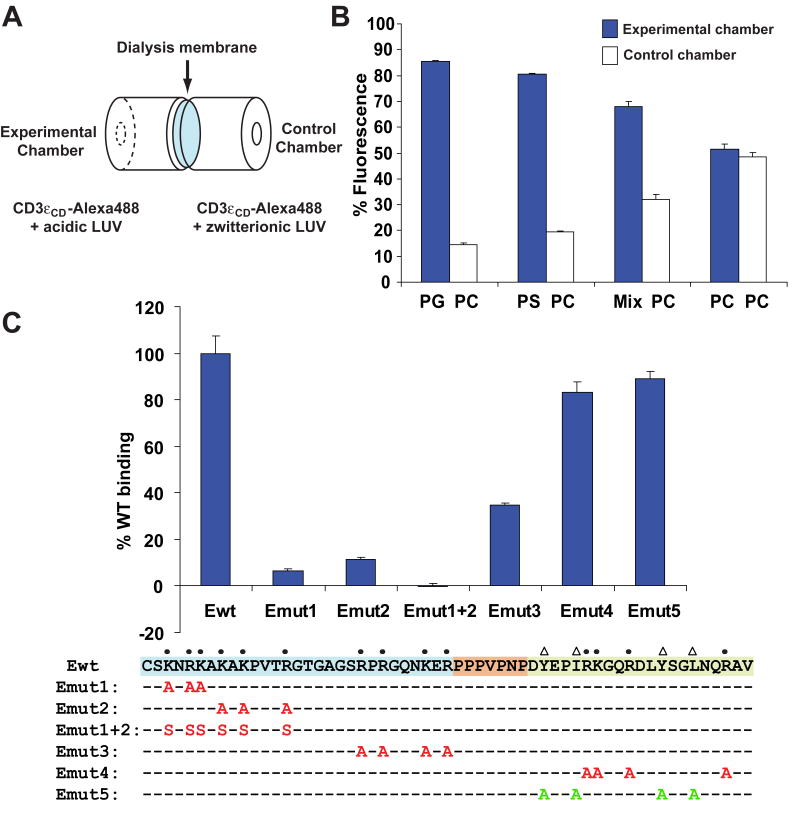
The CD3ε cytoplasmic domain can be divided into three segments: a positively charged N-terminal segment with 10 basic residues, a proline-rich stretch and the C-terminal ITAM (Fig. 1). For quantitative comparison of a panel of CD3εCD mutants, we developed an equilibrium-based micro-dialysis assay (Fig. 1A) in which movement of the large unilamellar lipid vesicles (LUVs) between the chambers was blocked by a dialysis membrane but Alexa-488 labeled CD3εCD could diffuse between the two chambers. When equilibrium had been reached the relative distribution of labeled peptide was determined in both the experimental chamber (LUVs containing acidic phospholipids) and the control chamber (LUVs composed of the zwitterionic lipid dimyristoyl-phosphatidylcholine, DMPC). The labeled CD3εCD peptide showed strong enrichment in the experimental chamber when it contained LUVs composed of acidic phospholipids, dimyristoyl-phosphatidylserine (DMPS) or dimyristoyl-phosphatidylglycerol (DMPG). Binding was also observed to LUVs formed with a mixture of lipids designed to approximate the composition of the inner leaflet of the plasma membrane (denoted as ‘Mix’ in Fig. 1B) (Fridriksson et al., 1999).
Figure 1. Definition of CD3εCD residues that contribute to lipid binding.
A. Microdialysis system to measure equilibrium binding of Alexa-488 labeled CD3εCD to experimental and control lipid vesicles.
B. Binding of Alexa-488 labeled CD3εCD to lipid vesicles containing acidic phospholipids. LUVs were composed of either 100% of the acidic phospholipids DMPG (PG) or DMPS (PS), 100% of the zwitterionic lipid DMPC (PC) or a lipid mixture (Mix) designed to approximate the composition of the inner leaflet of the plasma membrane (40% POPC, 20% POPS, 20% POPE, 10% PI, 10% POPG). For each condition, binding was compared to a control chamber containing LUVs composed of 100% DMPC. CD3εCD-Alexa488 was used at a concentration of 4nM and LUVs at 5 mM. Triplicate measurements were made and results (mean ± SD) plotted as the percentage of Alexa-488 fluorescence in experimental and control chambers.
C. Basic residues contribute to membrane binding by CD3εCD. Clusters of basic residues (marked by dots) were mutated to alanine or serine and the four conserved hydrophobic residues of the ITAM (marked by triangles) to alanine. Alexa-488 labeled mutants were tested for lipid binding using the microdialysis assay with 100% DMPS in the experimental chamber and 100% DMPC in the control chamber (each at 1 mM). The basic residue rich segment (cyan), proline rich segment (orange) and the ITAM (green) of CD3εCD are highlighted. Triplicate measurements were made and results (mean ± SD) plotted as the percentage of WT CD3εCD lipid binding
The 14 basic residues, of which ten are located in the N-terminal segment and four in the ITAM, were divided into four clusters (Emut1-4). Mutation of the basic residues located closest to the N-terminus of CD3εCD (Emut1, Emut2) resulted in the most substantial reduction in lipid binding. Basic residues in the center of the CD3εCD (Emut3) also contributed substantially to membrane binding, whereas mutation of the basic residues within the ITAM (Emut4) had little impact (Fig. 1C). Furthermore, substitution of the four functionally important hydrophobic residues of the ITAM (Y38, I41, Y49, L52) by alanine (Emut5) did not substantially reduce lipid binding.
Membrane binding by the CD3ε cytoplasmic domain in T cells
Given the important contribution of electrostatic interactions it was critical to examine membrane binding in cells. Fluorescence resonance energy transfer (FRET) requires close spatial proximity between donor and acceptor fluorophores, typically a distance of <100 Å (Kenworthy, 2001). The CD3ε cytoplasmic domain is 57 residues in length, and in a fully extended conformation its C-terminus would be at a distance of ~200 Å from the membrane. Significant FRET between a fluorescent membrane dye and a fluorescent protein attached to the C-terminus of the CD3ε cytoplasmic domain should thus only be detected if the cytoplasmic domain indeed interacts with the lipid bilayer. We chose the monomeric teal fluorescent protein (mTFP1, abbreviated as TFP) as the FRET donor, one of the brightest and most photo-stable known fluorescent proteins (quantum yield of 0.85) (Ai et al., 2006). TFP has excitation and emission maxima at 462 nm and 492 nm, respectively, making it an ideal donor for yellow or orange acceptors (Ai et al., 2006). As the FRET acceptor, we selected octadecyl rhodamine B (R18), a plasma membrane dye with excitation and emission maxima at 540 nm and 565 nm (Fig. S1). Other studies have shown that fluorescent membrane dyes can be used to examine conformational changes of membrane proteins by FRET (Chigaev et al., 2003; Jiao et al., 2005).
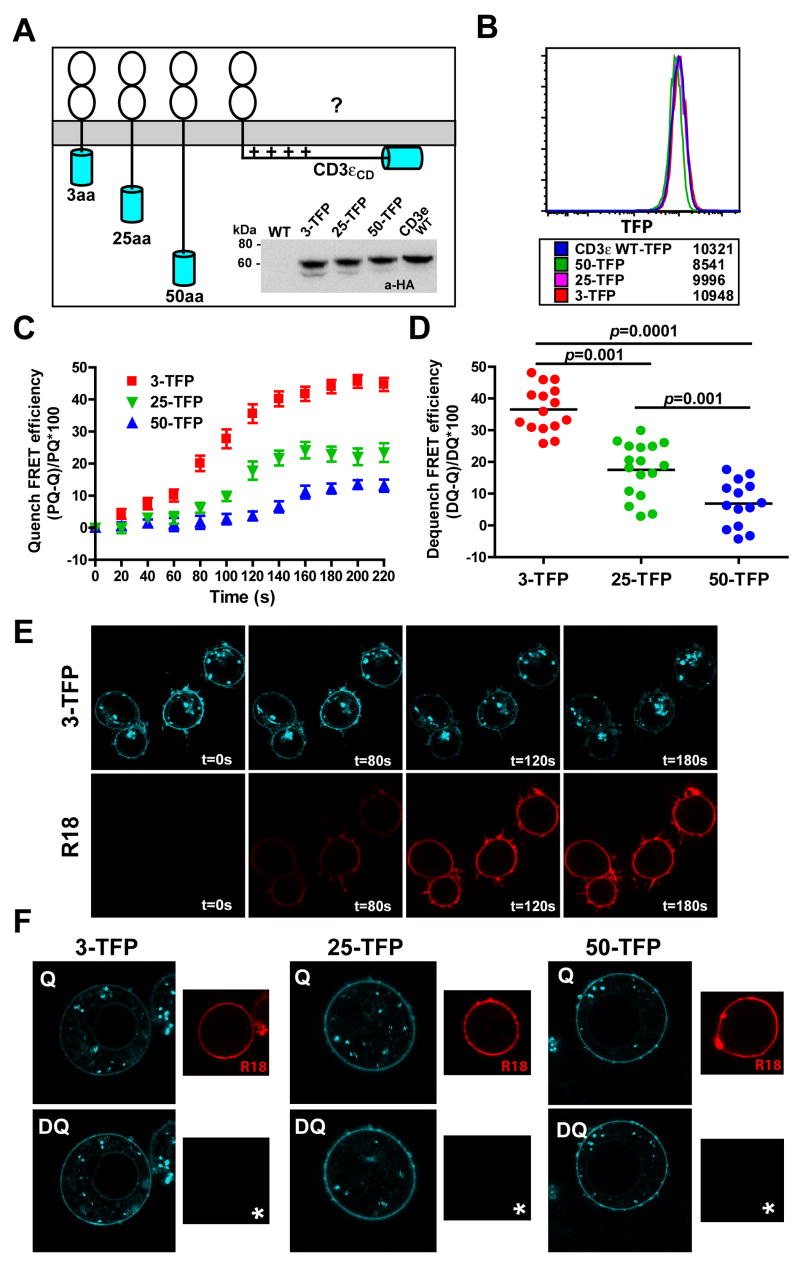
We tested this strategy using constructs in which flexible linkers of increasing length (3, 25 and 50 residues) were placed between the transmembrane (TM) and TFP domains (Fig. 2A). The construct with the 3-amino acid (aa) linker served as a positive control and the 25 and 50-aa constructs were used to determine the FRET efficiency that might result when either the C-terminal ITAM or the entire cytoplasmic domain dissociate from the membrane, respectively. A residual FRET signal is expected even with a 50-aa linker because the TFP group can freely move within the space defined by the maximal length of the flexible linker. These cytoplasmic domains were attached to the TM domain of an irrelevant monomeric membrane protein, the inhibitory NK cell receptor KIR2DL3, with two extracellular immunoglobulin domains and no known interactions with the TCR. Jurkat cell transfectants that expressed these fluorescent proteins were cloned by single cell sorting, and clones with similar expression levels were selected for analysis (Fig. 2B).
Figure 2. FRET approach to examine CD3εCD membrane binding in cells.
A. Approach for measuring FRET efficiency as a function of the distance of TFP (FRET donor, labeled cyan) from a membrane labeled with R18 (FRET acceptor). Inset: Western blot analysis with HA tag mAb showing expression of all constructs in transfected Jurkat clones.
B. FACS analysis of TFP fluorescence for clones expressing the different constructs. Numbers represent the mean fluorescence intensity for each clone.
C & E. Measurement of FRET efficiency with the donor quenching approach. Cells were adhered to glass coverslips, loaded into a flow chamber and Ringer solution (37°C) was injected over the cells (Prequench: PQ). R18 was then injected and images taken at 20 second intervals for both channels. FRET efficiency was calculated based on the reduction of TFP fluorescence (Quenched state: Q) due to energy transfer to the R18 acceptor (10 cells/construct, error bars indicate SEM).
D & F. R18 labeled cells (Quenched state: Q) were imaged in both channels and the acceptor was then photobleached (*) to release the quenched fluorescence (Dequench: DQ). D. FRET efficiencies were measured and plotted (14–17 cells shown here from one experiment).
FRET results in a reduction in the fluorescence intensity of the donor (TFP) due to energy transfer to the acceptor (R18), a process referred to as donor quenching (Kenworthy, 2001). Cells were captured onto glass coverslips and imaged using a flow chamber to enable real time FRET measurements during R18 injection. R18 labeling of the plasma membrane resulted in substantial quenching of TFP fluorescence for the 3-aa linker positive control construct, indicative of a high FRET efficiency (>40%, Fig. 2C & E, Supplemental movie M1). Minimal photobleaching of the donor (approximately 5%) was observed during these experiments due to repeated image acquisition during R18 labeling. This degree of photobleaching was observed for all constructs, and FRET values were therefore not corrected for this factor. Constructs with 25 and 50 residue linkers exhibited lower FRET efficiencies (23% and 12%, respectively) that correlated inversely with the distance of the TFP domain from the plasma membrane (Movies M2–3). This reduction in donor fluorescence was only observed at the plasma membrane where TFP and R18 were co-localized, but not in intracellular compartments that only had a TFP signal (Fig. 2E).
A second approach for FRET measurements is to photobleach the acceptor and measure the increase in donor fluorescence when its energy is no longer transferred to the acceptor (donor dequenching). Images of the donor fluorophore (TFP) were acquired prior to and after photobleaching of the acceptor (Fig. 2D & F). Again, lengthening of the linker resulted in a stepwise reduction in FRET efficiencies. FRET efficiencies were moderately underestimated with this approach due to some bleaching of the donor (~5%) during acceptor photobleaching.
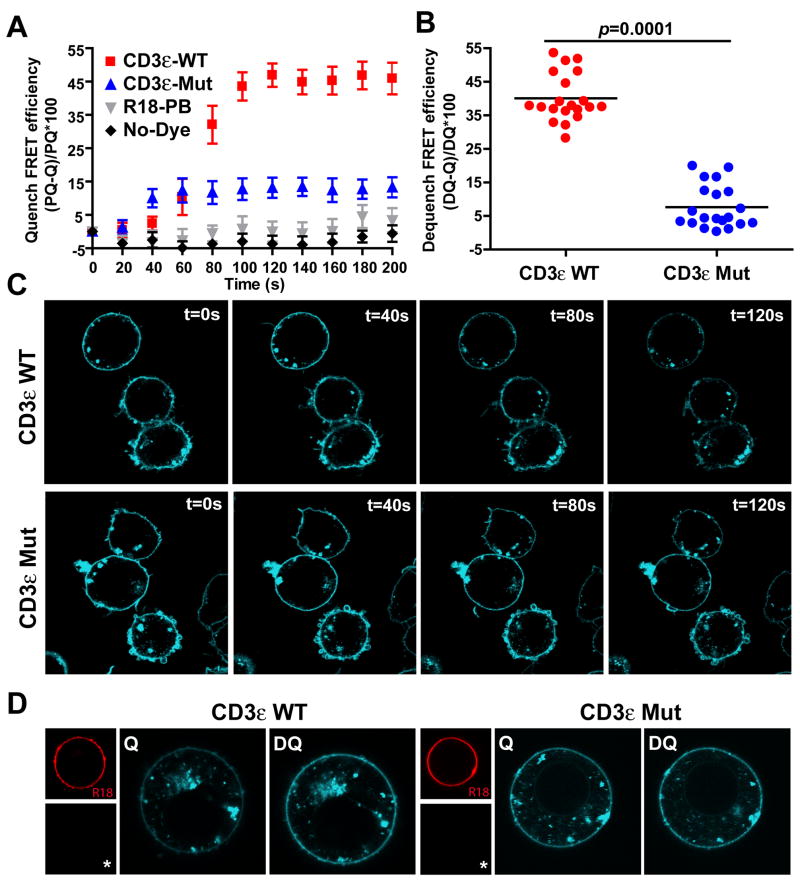
We then placed the wild-type CD3εCD into this reporter construct (HA-KIR-CD3εCD-TFP) and measured a high FRET efficiency similar in magnitude to the 3-aa linker control using both quenching and dequenching approaches (Fig. 3A–D, Movie M4). To demonstrate specificity of binding, we analyzed a mutant in which six basic residues in the N-terminal segment of the CD3ε cytoplasmic domain were mutated to serine (Emut1+2, Fig. 1). Indeed, low FRET values similar to the 50-aa linker were obtained for this mutant (Fig. 3, Movie M5). Also, the kinetics of R18 labeling and increases in FRET were closely correlated (Fig. S2). Furthermore, the incorporation of photobleached R18 dye did not change the TFP fluorescence intensity demonstrating that the change in TFP fluorescence caused by fluorescent R18 was due to FRET (Fig. 3A, S2). These results firmly establish a close interaction of the CD3ε cytoplasmic domain with the plasma membrane in T cells.
Figure 3. The CD3ε cytoplasmic domain is bound to the inner leaflet of the plasma membrane.
A & C. The FRET efficiency was determined for transfectants expressing the CD3ε WT or a CD3ε mutant cytoplasmic domain (Emut1+2) (10 cells/construct, error bars indicate SEM) using the quenching approach, as in Figure 2C & E. R18-PB: R18 photobleached.
B & D. The donor dequenching method was used to measure the FRET efficiency for the same transfectants (20 cells shown here from one experiment), as in Figure 2D & F.
Lipid binding induces partial folding of CD3εCD
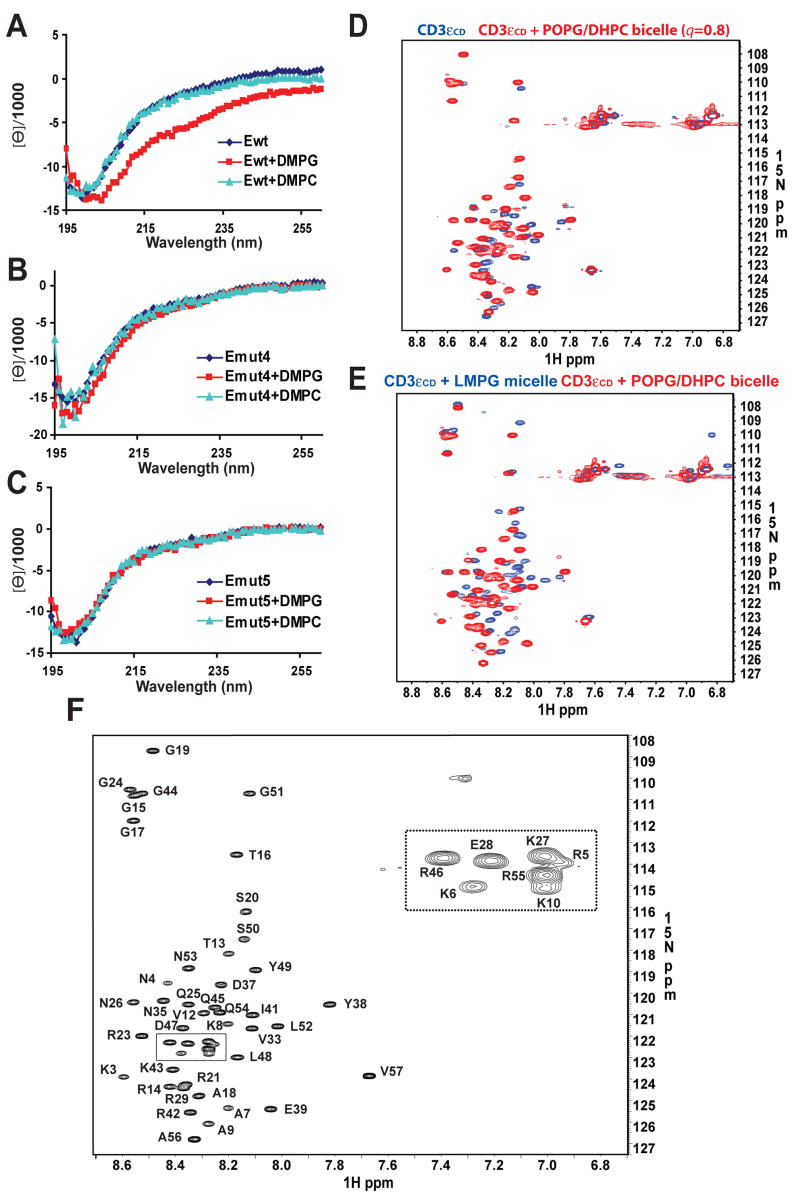
Based on far UV Circular Dichroism (CD), CD3εCD behaves as an unstructured protein in solution. However, the presence of acidic DMPG LUVs induced a shift in the CD spectrum consistent with partial folding (Fig. 4A). Such a change was not observed with control DMPC LUVs, indicating a dependence on the interaction with the lipid. Interestingly, Emut4 and Emut5 lost their secondary structure even though these mutations within the ITAM did not substantially reduce lipid binding. These results indicate that both basic and hydrophobic residues of the ITAM contribute to the local structure of this segment (Fig. 4B, 4C). The other mutants were not included in these experiments because of their poor binding to LUVs.
Figure 4. Membrane binding induces partial folding of CD3.
εCD
A–C. Far UV CD spectra of CD3εCD and mutants (20μM) in solution or in the presence of DMPG or DMPC LUVs (5mM). Lipid binding of wild-type CD3εCD to DMPG LUVs induced partial folding. No secondary structure was observed for mutants Emut4 and Emut5, even when bound to DMPG LUVs.
D. Superimposed 15N-1H HSQC spectra of CD3εCD in phosphate buffer (blue) and in the presence of POPG/DHPC bicelles (_q_=0.8, red). Changes in chemical shift and/or signal intensity were evident for all backbone amide resonances upon addition of bicelles, and 18 additional resonances became observable.
E. Superimposed 15N-1H HSQC spectra of CD3εCD in the presence of 100 mM LMPG micelles or 100 mM POPG/DHPC bicelles (_q_=0.8). The spectra differ substantially, even though micelles and bicelles had the same headgroup (phosphoglycerol).
F. The high-resolution 15N-1H TROSY-HSQC spectrum of POPG/DHPC bicelle-bound CD3εCD is shown with backbone assignments. All of the CD3εCD residues were assigned unambiguously, except the first two N-terminal residues and the proline residues.
The structure of the lipid-bound state of CD3εCD was then determined by multidimensional nuclear magnetic resonance (NMR) spectroscopy. The critical aspect for determination of a physiologically relevant structure of the cytoplasmic domain was to provide a membrane surface suitable for solution NMR techniques. Detergent micelles do not adequately mimic the planar surface of the inner leaflet of the plasma membrane because they have a strong curvature. This is particularly true for polypeptides that reside on the surface of the membrane (Chou et al., 2002). Bicelles are disc-shaped structures composed of one or more long-chain phospholipids (such as 1-palmitoyl-2-oleoyl-phosphatidylglycerol, POPG) that assemble into a central bilayer and a short-chain lipid (such as dihexanoyl-phosphatidylcholine, DHPC) that forms the rim. The core region of bicelles thus mimics a section of natural membrane much better than micelles (Prosser et al., 2006).
The two-dimensional 1H-15N heteronuclear single-quantum coherence (HSQC) spectrum of CD3εCD in the absence of phospholipids showed moderately dispersed amide resonances, but inhomogeneous signal intensities indicated little or no structure (Fig. S3A). For example, 4 out of 6 expected amide resonances from glycines were not observable due to fast exchange with solvent. In the presence of bicelles the spectrum showed greater peak homogeneity and all expected amide resonances were observed. This indicated that the protein was more structured and that the amide protons were protected by hydrogen bonds and the lipid bilayer environment (Fig. 4D). Interestingly, arginine sidechain NH, which exchange too quickly with solvent at pH 6.7 to be detectable in the HSQC experiment, appeared upon binding to bicelles. A control experiment using bicelles composed of the zwitterionic lipid POPC and DHPC yielded a spectrum virtually identical to free CD3εCD (Fig. S3C), consistent with the biochemical data. As expected, the spectrum obtained in the presence of micelles formed by the detergent lysomyristoyl-phosphatidylglycerol (LMPG) was substantially different, confirming that micelles do not provide the same environment as bicelles even when both have the same acidic headgroup (PG) (Fig. 4E, S3B).
Smaller bicelles tumble faster and yield more favorable relaxation properties for solution NMR studies, but they may not be large enough to accommodate a membrane-surface binding protein such as CD3εCD with an elongated conformation. Larger bicelles resemble true membranes more closely but broaden the NMR resonances due to slower reorientation in solution. For structure determination, it was thus important to identify the optimal size of POPG/DHPC bicelles. Bicelle size is determined by the ratio of long-chain to short-chain lipids (the q value). We tested bicelles with q values of 0.5, 0.8 and 1.0. Chemical shift changes were observed between _q_=0.5 and 0.8, but not between _q_=0.8 and _q_=1.0, indicating that the bilayer region at _q_=0.8 was large enough to accommodate CD3εCD. A _q_=0.8 bicelle is estimated to have a diameter of ~72 Å according to the corrected Vold-Prosser model (Glover et al., 2001).
The key hydrophobic residues of the ITAM are deeply embedded in the membrane
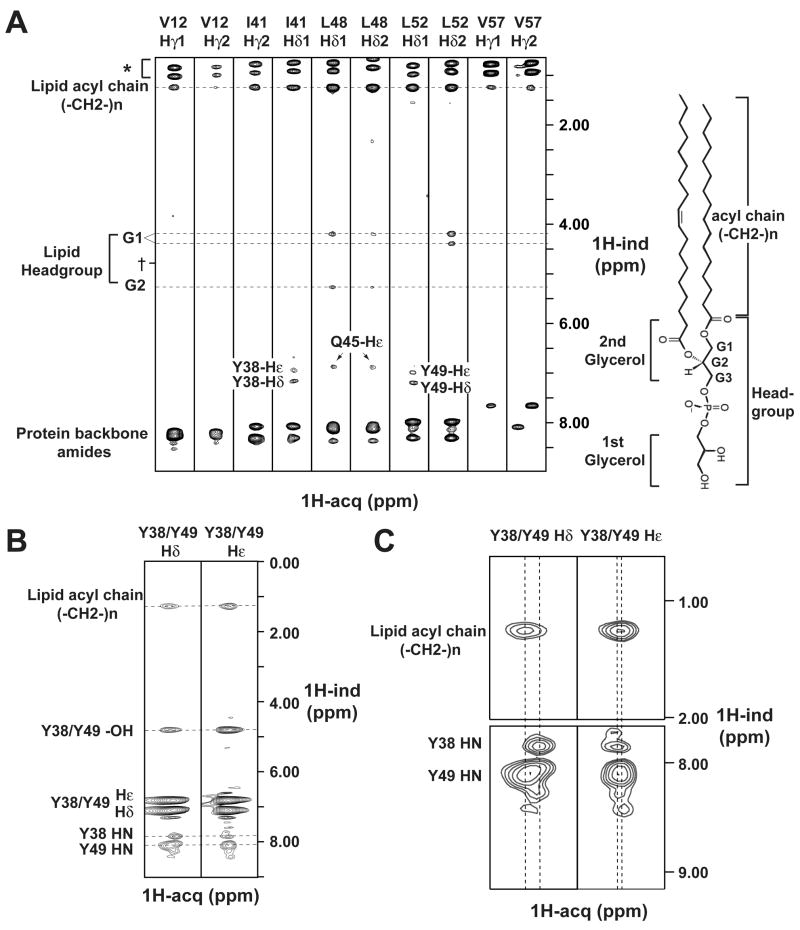
All CD3εCD residues were assigned with standard sequential assignment techniques except for the two N-terminal residues, which are in fast exchange with water, and the proline residues lacking backbone amide protons (Fig. 4F). To investigate the interaction of CD3εCD with the lipid, we measured nuclear overhauser effects (NOEs) between the lipid (POPG) and the peptide backbone as well as hydrophobic side chains. Bicelles were generated with deuterated DHPC and protonated POPG so that NOEs arose only from the interaction of the peptide with the POPG bilayer portion. Protons belonging to different chemical groups of POPG (the first headgroup glycerol, the second headgroup glycerol and the acyl chains, Fig. 5A) have distinct chemical shifts and could thus be used to position CD3εCD with respect to the lipid bilayer.
Figure 5. Aromatic and aliphatic ITAM sidechains of bicelle-bound CD3εCD reside in the hydrophobic lipid bilayer interior.
A. Selected strips showing methyl NOEs from a 13C-separated, double-13C-filtered NOESY spectrum of 15N,-13C-labeled CD3εCD bound to POPG/DHPC bicelles (_q_=0.8). Methyl-bearing sidechains in the CD3εCD N-terminal (V12) and ITAM (I41, L48, L52) regions as well as the C-terminus (V57) show strong NOEs to methylene protons from the lipid acyl chains and weak NOEs to lipid headgroup protons, but none to water, indicating a strong propensity to be located in the lipid bilayer hydrophobic core. Bicelles were prepared using protonated POPG and deuterated DHPC. Therefore, all NOEs to acyl methylene protons are to bilayer lipids rather than to bilayer-stabilizing short-chain lipids. * incompletely suppressed protein methyl signals. † water proton chemical shift.
B. Strips from an aromatic 13C-separated, double-13C-filtered NOESY spectrum of the same sample as in (A) demonstrating NOEs from the sidechain δ and ε aromatic protons of Y38 and Y49 to methylene protons from the acyl chains in the interior of the lipid bilayer. The observation of NOEs to the tyrosine hydroxyl protons, which are normally in fast exchange with water, indicates that the hydroxyls are now protected by the lipid bilayer.
C. Enlarged strips from B. Although the δ and ε aromatic protons of Y38 and Y49 are overlapped, the two tyrosines could be distinguished by their backbone amide proton resonances.
We first measured NOEs between the peptide backbone amides and POPG protons in a 3D 15N-separated NOESY experiment (200 ms mixing time). CD3εCD and DHPC were deuterated to eliminate any amide-to-sidechain and amide-to-short lipid signals (Fig. S4). A number of hydrophobic residues only showed NOEs (distance of < 5 Å) to lipid acyl chains and the POPG headgroup, but not to water, indicating that these backbone atoms are close to the membrane interior and the lipid headgroups. These include key residues of the ITAM, such as the first tyrosine and isoleucine residues of the ITAM (Y38 and I41). The second tyrosine of the ITAM may be less deeply embedded in the hydrophobic core of the lipid bilayer: Y49 HN showed a strong NOE to lipid methylenes, but also weaker NOEs to the lipid headgroup and water, suggesting that this backbone atom may be positioned at the interface between the hydrated headgroup region and the hydrophobic interior.
We next conducted 3D 13C-separated and double-13C-filtered NOESY experiments (170 ms mixing time) to exclusively observe NOEs from the methyl and aromatic protons of the protein sidechains to the protons of POPG (Fig. 5A, B). Almost all methyl protons from Val, Ile and Leu showed strong NOEs to lipid acyl chains but not to water. Leu48 and Leu52 also showed NOEs to the POPG headgroup but the signals were much weaker than the NOEs to the lipid acyl chains. The aromatic protons of the two tyrosines showed substantial NOEs to lipid acyl chains. Although the aromatic protons Hδ and Hε from the two tyrosines were overlapped in the aromatic NOESY spectrum, they were distinguishable based on their backbone amide proton chemical shifts (Fig. 5C). It is intriguing that the Ile41 and Leu52 methyl protons also had NOEs to the Tyr38 and Tyr49 aromatic protons, respectively, indicating that the two hydrophobic residues of each ITAM hemi-motif (YxxI and YxxL) are within close proximity to each other. Therefore, the four functionally critical hydrophobic residues of the ITAM are deeply inserted in the hydrophobic interior of the lipid bilayer.
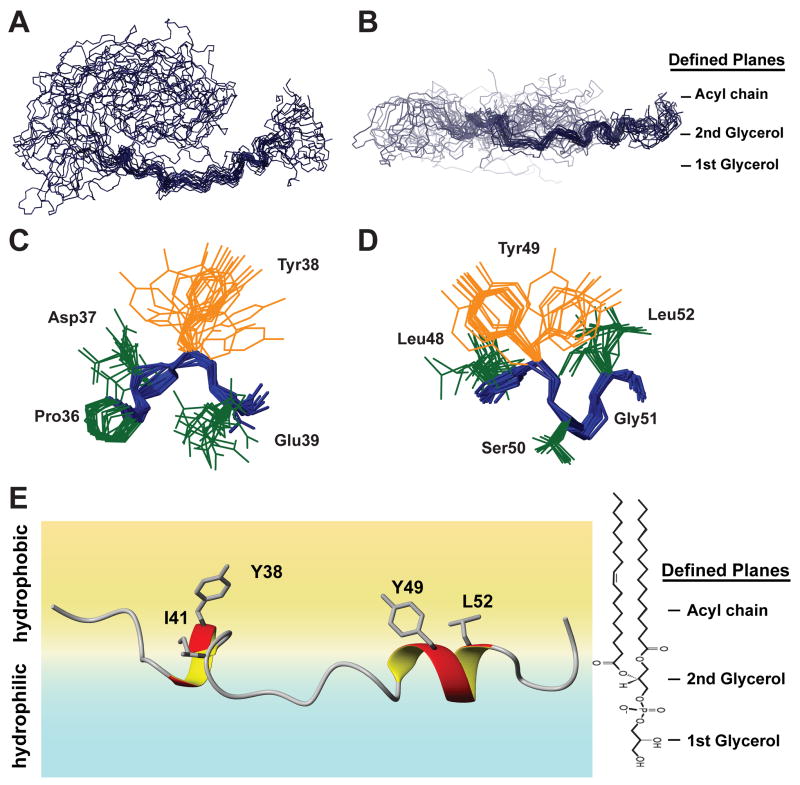
NMR structure of the membrane-bound state of CD3εCD
Due to the dynamic nature of CD3εCD in the membrane, a structure ensemble calculated in the absence of a lipid bilayer is of questionable relevance. Therefore, the membrane-bound CD3εCD structure was calculated and positioned in a lipid bilayer model by combining two major sets of restraints: protein NOE restraints measured by 15N and 13C-separated NOESY experiments and protein-lipid restraints measured by the protein-lipid NOESY experiments. The conventional NMR structure calculation protocol does not support localization of a molecule relative to a lipid bilayer. We therefore introduced an atom-to-plane distance potential into the Xplor-NIH refinement program (Version 2.19, http://nmr.cit.nih.gov/xplor-nih/). By modeling the lipid bilayer as three parallel planes corresponding to the first POPG headgroup glycerol, the second POPG headgroup glycerol and the hydrophobic acyl chains, we used the observed CD3ε-lipid NOEs to position CD3εCD with respect to the lipid bilayer (see Experimental Procedures below for details). An ensemble of 200 structures was calculated from 239 protein NOE-derived restraints and 142 plane restraints. Several dihedral angles predicted by TALOS were also used for the structure refinement (Supplemental Table 1). Overall, the CD3εCD is deeply embedded into the polar headgroup and hydrophobic acyl chain regions of the lipid, with many peptide backbone atoms being located close to the interface between the polar headgroup region and the non-polar core of the lipid. This general position of the peptide enables interaction of the basic side chains with the charged headgroups of the lipid while simultaneously permitting deep insertion of hydrophobic residues into the hydrophobic interior of the bilayer. The ITAM segment (Asn35 to Asn53) whose hydrophobic residues (YxxI and YxxL) are inserted into the lipid bilayer is structured with helical turns surrounding the two tyrosines (Fig. 6A, 6E). The aromatic rings of the two tyrosines are both inserted into the hydrophobic core of the lipid bilayer, which induces helical turn structures in the Pro36-Glu39 and Leu48-Leu52 stretches (Fig. 6C, 6D, 6E). Mutation of the four hydrophobic residues of the ITAM resulted in a loss of secondary structure based on CD measurements, indicating that lipid insertion of these side chains induces folding of the ITAM. In contrast, the long N-terminal segment shows substantial lateral flexibility (Fig. S5), even though it is restrained with respect to the bilayer plane and positioned at the interface between the POPG headgroup region and the acyl chains (Fig. 6B). The insertion of the key hydrophobic residues of the ITAM into the lipid bilayer thus renders the two tyrosines inaccessible to Src kinases in the lipid-bound state.
Figure 6. NMR Structure of the bicelle-bound CD3εCD peptide.
A & B. Top and side view of the bundles of the 20 lowest-violation structures of POPG/DHPC bicelle-bound CD3εCD peptide aligned on all heavy atoms. The bundles are positioned into three planes representing the membrane bilayer. While the C-terminal segment contains obvious helical turns localized around the conserved ITAM sequences, the N-terminal region has no stable structure. The entire structure approximates a horseshoe shape that is highly dynamic within the plane of the bicelle. The side view in panel B illustrates that the peptide backbone resides within a narrow plane of depth in the lipid bilayer.
C & D. Preferred sidechain orientations of Y38, L48, Y49 and L52 of the ITAM pointing into the plane of the lipid bilayer.
E. NOEs observed from backbone/sidechain protons to solvent/lipid protons were used as structural restraints in an additional refinement step to relate the peptide structure to a multi-limit plane representing the lipid bilayer region of the POPG/DHPC bicelle. The POPG structure graphic to the right indicates the position of the lipid molecule in the lipid bilayer and the type of NOE restraints utilized to localize backbone and sidechain elements to a particular region. While the peptide backbone resides primarily at the interface between the hydrated PG headgroup region and the hydrophobic bilayer interior, the aromatic and aliphatic ITAM sidechains protrude into the acyl lipid interior.
Functional relevance of lipid binding by the CD3ε ITAM
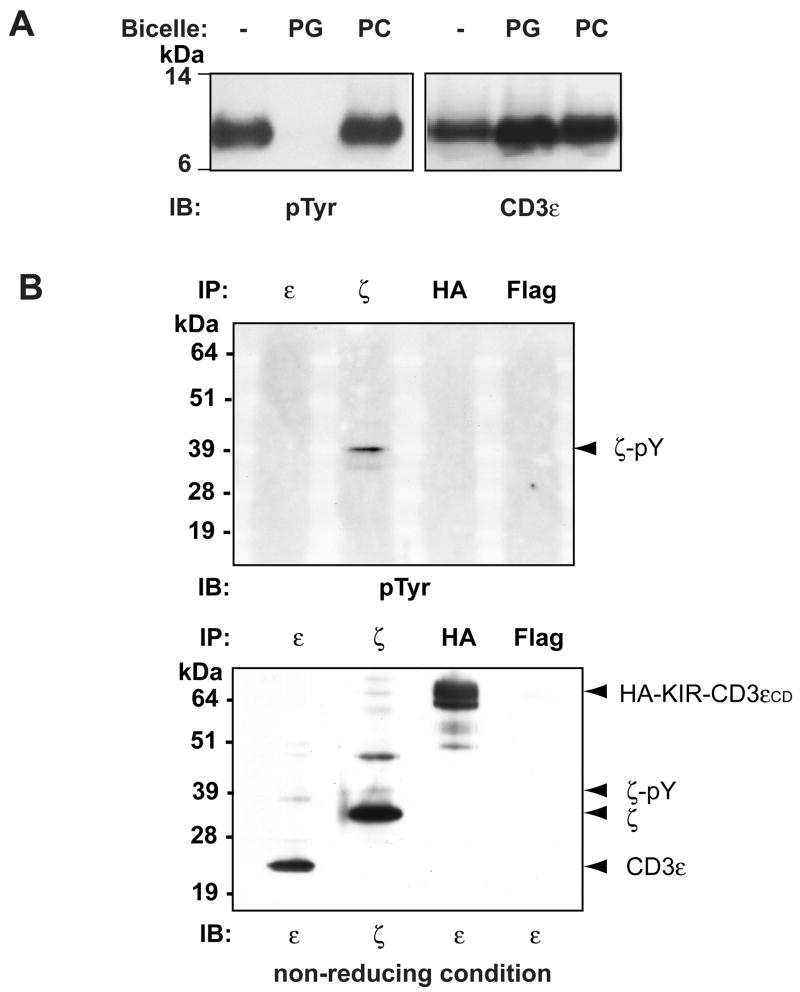
The structural data indicate that the tyrosines should not be phosphorylated when the ITAM is lipid-bound. An in vitro kinase assay showed that addition of POPG/DHPC bicelles, but not POPC/DHPC control bicelles, prevented phosphorylation of CD3εCD by Lck (Fig. 7A). Also, no basal phosphorylation of endogenous CD3ε or the HA-KIR-CD3εCD-TFP reporter protein could be detected, but basal phosphorylation of the TCR ζ chain was observed, consistent with previous studies (Kersh et al., 1998). Based on the size difference between phosphorylated and non-phosphorylated ζ chains, it appears that only a small fraction is basally phosphorylated (Fig. 7B).
Figure 7. Membrane binding renders the tyrosines of CD3ε inaccessible to Lck.
A. Lipid binding prevents tyrosine phosphorylation of CD3εCD. The tyrosine residues of CD3εCD were efficiently phosphorylated by Lck in an in vitro kinase assay. Addition of POPG/DHPC bicelles but not control POPC/DHPC bicelles prevented tyrosine phosphorylation. Western blots were probed with a phospho-tyrosine antibody (biotinylated P-Tyr-100 and streptavidin-horseradish peroxidase, SA-HRP) and a CD3εCD antibody (CD3ε m20 and anti-goat IgG-HRP).
B. Neither endogenous CD3ε nor the HA-KIR-CD3εCD-TFP reporter protein are basally phosphorylated. HA-KIR-CD3εCD-TFP transfected Jurkat cells were solubilized using 1% NP40 and proteins were immunoprecipitated with CD3ζ(6B10.2), CD3ε (UCHT1), HA (3F10) or FLAG (M2, isotype control) antibodies. Phosphorylated and total proteins were detected with a phospho-tyrosine antibody (biotinylated P-Tyr-100 and SA-HRP, upper panel), a CD3εCD antibody (CD3ε m20 and anti-goat IgG-HRP, which detects CD3ε and the reporter protein) or an antibody to the ζ chain (anti-CD3ζ 6B10.2-HRP, lower panel). SDS-PAGE was performed under non-reducing conditions.
Discussion
The NMR structure of the CD3ε cytoplasmic domain in its lipid-bound state showed that the signature tyrosine and leucine/isoleucine residues of the ITAM are inserted into the hydrophobic interior of the lipid bilayer where they are inaccessible to Src kinases. Dissociation of the CD3ε ITAM from the membrane is thus required to render the tyrosines accessible to kinases. The FRET experiments unequivocally demonstrated that the ITAM is membrane-bound in live, non-activated T cells. Such a demonstration of membrane binding in live cells was critical for establishing the physiological significance of lipid binding because electrostatic interactions between acidic lipid headgroups and basic residues of CD3ε play a prominent role in membrane association. These findings may be relevant for many other receptors that signal through cytoplasmic ITAMs.
An overall structural model of the ITAM-lipid interaction was developed using NOEs between the CD3ε cytoplasmic domain and the lipid headgroup/acyl chain regions as structural restraints. Rather than binding to the surface of the membrane through interaction of basic residues with the headgroups of acidic phospholipids, the peptide backbone is embedded in the hydrophilic and hydrophobic layers of the membrane. This positioning enables deep insertion of the hydrophobic ITAM residues into the acyl chain region of the membrane. Studies on other membrane-interacting peptides, such as the MARCKS peptide and Ras family members (Heo et al., 2006; Yeung et al., 2006; Zhang et al., 2003), have shown that both hydrophobic and basic residues contribute to membrane binding, which is consistent with the lipid-CD3ε ITAM interactions described here. The MARCKS peptide has five phenylalanine residues and NMR measurements showed NOEs between the phenylalanine rings and the acyl chain protons of the lipid (Zhang et al., 2003). The membrane insertion model of the CD3ε ITAM may thus be relevant to other membrane-interacting signaling domains.
Three functional segments of the CD3ε cytoplasmic domain can be distinguished. The N-terminal half of the cytoplasmic domain contains a large number of basic residues that are critical for membrane binding, in particular the clusters of basic residues located in closest proximity to the TM domain. A polyproline stretch that has been shown to bind Nck is located between this N-terminal domain and the ITAM (Gil et al., 2002). Neither hydrophobic nor basic residues of the ITAM segment were found to make substantial contributions to membrane binding, but CD measurements indicated that they are nevertheless essential for the secondary structure of the ITAM in the lipid-bound state. It thus appears that the ITAM is less strongly bound to the plasma membrane than the N-terminal domain, suggesting that it is poised to dissociate from the membrane upon receptor ligation. The structural features of the ITAM support this conclusion: the ITAM is not folded into a complete α-helix which presumably would represent a more stable structure, but rather only contains helical turns centered on the critical membrane-embedded tyrosines. The Stern lab had first suggested that the ITAMs of the ζ chain fold into α-helices upon lipid binding, and later revised this model by proposing that the cytoplasmic domain is completely unstructured even in the lipid-bound state (Aivazian and Stern, 2000; Sigalov et al., 2006). This study focused on a different cytoplasmic domain of the TCR-CD3 complex, but indicates that aspects of both models are correct and likely to be of functional significance: the long N-terminal segment is highly flexible, while the close interaction of the hydrophobic residues of the ITAM with the lipid induces partial folding of the ITAM centered on the critical residues of the motif.
Functional studies support the conclusions based on the NMR structure. Phosphorylation of CD3εCD in an in vitro kinase assay was inhibited by addition of POPG/DHPC bicelles, consistent with the NMR data that the tyrosine aromatic rings are deeply inserted into the lipid interior. A basal level of ζ chain phosphorylation has been observed in non-stimulated T cells and a previous study showed that it only involves one of two tyrosines of a given ITAM (Kersh et al., 1998). We were able to confirm basal ζ chain phosphorylation, but did not detect basal phosphorylation of CD3ε or the reporter protein with the CD3ε cytoplasmic domain. Our experiments also indicate that only a small fraction of ζ chains is phosphorylated in non-stimulated cells, consistent with previous studies (Kersh et al., 1998). The CD3ε cytoplasmic domain binds with higher affinity to lipid vesicles than the ζ cytoplasmic domain (Sigalov et al., 2006), which may explain the difference in basal phosphorylation between CD3ε and ζ chains. Particular tyrosines of ζ may thus become transiently accessible to kinases in a small subset of TCRs. A recent study has suggested that the proline-rich motif of CD3ε is accessible to a mAb in both resting and activated thymocytes (Mingueneau et al., 2008). Due to the intracellular location of the antibody target, the cells had to be fixed and permeabilized by detergent treatment prior to antibody labeling. It is likely that CD3εCD-lipid interactions were disrupted by detergent treatment, accounting for this result.
There are two general possibilities to explain the dissociation of the CD3ε cytoplasmic domain from the membrane when TCRs are triggered by pMHC: a mechanical force exerted on the TCR or a change in its microenvironment. A number of different conformational change/mechanical force models have been proposed but there is no agreement whether a pushing, pulling and/or twisting force is involved (Kuhns et al., 2006; Ma et al., 2008; Sun et al., 2001). Due to the highly flexible nature of the N-terminal polybasic domain of CD3ε a mechanical force cannot be efficiently transmitted to the C-terminal ITAM, and such a mechanical force would therefore have to be strong and/or sustained. Alternatively, receptor triggering could induce changes in the microenvironment around ligand-engaged TCRs, for example by causing transient alterations in the partitioning behavior of the TCR in the lipid bilayer. Given the importance of the charge-charge interactions between the CD3ε cytoplasmic domain and acidic phospholipids, transient changes in the local lipid environment could allow release of the CD3ε ITAM from the membrane.
In summary, this study demonstrates that the ITAM of an activating receptor in the immune system is lipid bound in nonactivated cells. The sequestration of the key tyrosines into the hydrophobic core of the lipid bilayer visualized by the NMR structure represents a novel structural mechanism for regulation of receptor activation. Control over the phosphorylation state of this and other ITAMs is essential to regulate the induction of immune effector functions that need to be engaged with great spatial and temporal resolution so that they are effective in protecting the host from invading pathogens without causing harm.
Experimental Procedures
Binding of CD3εCD to synthetic lipid vesicles
CD3εCD was expressed as a GST fusion protein in E. coli, separated from GST by enterokinase cleavage, purified by reverse-phase HPLC and labeled at an N-terminal cysteine residue with an Alexa-488 maleimide derivative (Invitrogen, Carlsbad, CA). LUVs composed of different phospholipids were generated as previously described (Wang et al., 2001) and binding of labeled CD3εCD to LUVs was measured by microdialysis. At equilibrium (24 hours), the Alexa-488 fluorescence was measured in chambers containing LUVs composed of the zwitterionic lipid DMPC (control chamber) versus acidic phospholipids (experimental chamber).
NMR structure of CD3εCD bound to bicelles
All NMR experiments were conducted on Bruker spectrometers at 27°C. Details of chemical resonance assignment and measurement of structural restraints are given in Supplementary Data.
Structures were calculated using the program Xplor-NIH Version 2.19 (Schwieters et al., 2003) using high temperature simulated annealing (from 1000 to 200 K) of an extended CD3εCD polypeptide to satisfy protein NOEs and dihedral angles and protein-POPG NOEs. The protein NOE restraints were enforced by flat-well harmonic potential, with the force constant ramped from 5 to 30 kcal mol−1 Å−2. In positioning the dynamic polypeptide in the lipid bilayer, three parallel planes given by y = −22 Å (the first POPG headgroup glycerol), −17 Å (the second POPG headgroup glycerol), and −11 Å (the hydrophobic acyl chain) were used to represent a leaflet of the POPG bilayer. Distance restraints between protein protons and lipid planes were enforced with flat-well harmonic potentials (±Δ), with the force constant ramped from 2 to 30 kcal mol−1 Å−2. The value of Δ, or the size of the flat well in Å, was assigned based on the NOE intensity and the dynamic properties of phospholipid bilayer obtained from X-ray and neutron diffraction data (Wiener and White, 1992). In the current structure refinement, Δ = f σ(x), where f is a scaling factor qualitatively assigned based on the relative NOE intensities, and σ(x) is the width of the probability distribution of lipid component x (σ(Plane1) = 8, σ(Plane2) = 3, σ(Plane3) = 8). In addition to the distance restraints, a number of backbone dihedral restraints derived from chemical shifts (predicted by TALOS as “Good”) were enforced by flat-well (±30°) harmonic potentials, with force constant fixed at 30 kcal mol−1 rad−2. An ensemble of 200 structures was calculated from 239 protein NOE-derived restraints, 7 protein backbone dihedral restraints, and 142 protein-lipid restraints. We selected 20 structures with the lowest number of violations to describe the conformational sampling of CD3εCD in the POPG lipid bilayer (Fig. 6).
Accession Numbers: The structure described in this paper has been deposited and assigned the RCSB ID code rcsb100662 and the PDB ID code 2k4f.
FRET measurements
Constructs were transfected into human Jurkat cells by electroporation and stable clones were isolated by single cell sorting. FRET between the C-terminal TFP domain and the membrane dye octadecyl rhodamine B (R18, Invitrogen, Carlsbad, CA) was measured by fluorescence microscopy (Zeiss LSM 510) using a flow chamber set-up to measure TFP quenching in real time. TFP fluorescence intensity in segments of the plasma membrane was measured before and during R18 injection and the quenching FRET efficiency was calculated as E = (PQ−Q/PQ) × 100, where PQ and Q are Prequenced and Quenched donor fluorescence. FRET measurements were also made using a donor dequenching approach in which TFP fluorescence was measured before and after photobleaching of the R18 acceptor. The dequenching FRET efficiency was calculated as E = (DQ−Q/DQ) × 100, where DQ and Q are Dequenched and Quenched donor fluorescence.
Supplementary Material
Sup. Movie1
Sup. Movie2
Sup. Movie3
Sup. Movie4
Sup. Movie5
supplement
Acknowledgments
This work was supported by a grant from the NIH (AI054520 to KW), the Pew Scholars Program in Biomedical Sciences (to JJC), a grant (to CC) and a fellowship (to CX) from the Arthritis Foundation, a Benacerraf Fellowship and a fellowship from the Canadian Institute for Health Research (to EG), a Helen Hay Whitney Foundation fellowship (to MC), as well as the Intramural Research Program of CIT, NIH (to CS).
References
- Ai HW, Henderson JN, Remington SJ, Campbell RE. Directed evolution of a monomeric, bright and photostable version of Clavularia cyan fluorescent protein: structural characterization and applications in fluorescence imaging. The Biochemical journal. 2006;400:531–540. doi: 10.1042/BJ20060874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aivazian D, Stern LJ. Phosphorylation of T cell receptor zeta is regulated by a lipid dependent folding transition. Nat Struct Biol. 2000;7:1023–1026. doi: 10.1038/80930. [DOI] [PubMed] [Google Scholar]
- Call ME, Pyrdol J, Wiedmann M, Wucherpfennig KW. The organizing principle in the formation of the T cell receptor-CD3 complex. Cell. 2002;111:967–979. doi: 10.1016/s0092-8674(02)01194-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Call ME, Wucherpfennig KW. Common themes in the assembly and architecture of activating immune receptors. Nat Rev Immunol. 2007;7:841–850. doi: 10.1038/nri2186. [DOI] [PubMed] [Google Scholar]
- Chigaev A, Buranda T, Dwyer DC, Prossnitz ER, Sklar LA. FRET detection of cellular alpha4-integrin conformational activation. Biophys J. 2003;85:3951–3962. doi: 10.1016/S0006-3495(03)74809-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou JJ, Kaufman JD, Stahl SJ, Wingfield PT, Bax A. Micelle-induced curvature in a water-insoluble HIV-1 Env peptide revealed by NMR dipolar coupling measurement in stretched polyacrylamide gel. J Am Chem Soc. 2002;124:2450–2451. doi: 10.1021/ja017875d. [DOI] [PubMed] [Google Scholar]
- Davis MM, Chien YH. T-Cell Antigen Receptors. In: Paul WE, editor. Fundamental Immunology. Philadelphia: Lippincott Williams & Wilkins; 2003. pp. 227–258. [Google Scholar]
- Deindl S, Kadlecek TA, Brdicka T, Cao X, Weiss A, Kuriyan J. Structural basis for the inhibition of tyrosine kinase activity of ZAP-70. Cell. 2007;129:735–746. doi: 10.1016/j.cell.2007.03.039. [DOI] [PubMed] [Google Scholar]
- Devaux PF. Static and dynamic lipid asymmetry in cell membranes. Biochemistry. 1991;30:1163–1173. doi: 10.1021/bi00219a001. [DOI] [PubMed] [Google Scholar]
- Fridriksson EK, Shipkova PA, Sheets ED, Holowka D, Baird B, McLafferty FW. Quantitative analysis of phospholipids in functionally important membrane domains from RBL-2H3 mast cells using tandem high-resolution mass spectrometry. Biochemistry. 1999;38:8056–8063. doi: 10.1021/bi9828324. [DOI] [PubMed] [Google Scholar]
- Gil D, Schamel WW, Montoya M, Sanchez-Madrid F, Alarcon B. Recruitment of Nck by CD3 epsilon reveals a ligand-induced conformational change essential for T cell receptor signaling and synapse formation. Cell. 2002;109:901–912. doi: 10.1016/s0092-8674(02)00799-7. [DOI] [PubMed] [Google Scholar]
- Glover KJ, Whiles JA, Wu G, Yu N, Deems R, Struppe JO, Stark RE, Komives EA, Vold RR. Structural evaluation of phospholipid bicelles for solution-state studies of membrane-associated biomolecules. Biophys J. 2001;81:2163–2171. doi: 10.1016/s0006-3495(01)75864-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatada MH, Lu X, Laird ER, Green J, Morgenstern JP, Lou M, Marr CS, Phillips TB, Ram MK, Theriault K, et al. Molecular basis for interaction of the protein tyrosine kinase ZAP-70 with the T-cell receptor. Nature. 1995;377:32–38. doi: 10.1038/377032a0. [DOI] [PubMed] [Google Scholar]
- Heo WD, Inoue T, Park WS, Kim ML, Park BO, Wandless TJ, Meyer T. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science. 2006;314:1458–1461. doi: 10.1126/science.1134389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X, Zhang N, Xu X, Oppenheim JJ, Jin T. Ligand-induced partitioning of human CXCR1 chemokine receptors with lipid raft microenvironments facilitates G-protein-dependent signaling. Mol Cell Biol. 2005;25:5752–5762. doi: 10.1128/MCB.25.13.5752-5762.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenworthy AK. Imaging protein-protein interactions using fluorescence resonance energy transfer microscopy. Methods. 2001;24:289–296. doi: 10.1006/meth.2001.1189. [DOI] [PubMed] [Google Scholar]
- Kersh EN, Shaw AS, Allen PM. Fidelity of T cell activation through multistep T cell receptor zeta phosphorylation. Science. 1998;281:572–575. doi: 10.1126/science.281.5376.572. [DOI] [PubMed] [Google Scholar]
- Kuhns MS, Davis MM, Garcia KC. Deconstructing the form and function of the TCR/CD3 complex. Immunity. 2006;24:133–139. doi: 10.1016/j.immuni.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Lu J, Lin WH, Chen SY, Longnecker R, Tsai SC, Chen CL, Tsai CH. Syk tyrosine kinase mediates Epstein-Barr virus latent membrane protein 2A-induced cell migration in epithelial cells. J Biol Chem. 2006;281:8806–8814. doi: 10.1074/jbc.M507305200. [DOI] [PubMed] [Google Scholar]
- Ma Z, Sharp KA, Janmey PA, Finkel TH. Surface-anchored monomeric agonist pMHCs alone trigger TCR with high sensitivity. PLoS Biol. 2008;6:e43. doi: 10.1371/journal.pbio.0060043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingueneau M, Sansoni A, Gregoire C, Roncagalli R, Aguado E, Weiss A, Malissen M, Malissen B. The proline-rich sequence of CD3epsilon controls T cell antigen receptor expression on and signaling potency in preselection CD4+CD8+ thymocytes. Nat Immunol. 2008;9:522–532. doi: 10.1038/ni.1608. [DOI] [PubMed] [Google Scholar]
- Prosser RS, Evanics F, Kitevski JL, Al-Abdul-Wahid MS. Current applications of bicelles in NMR studies of membrane-associated amphiphiles and proteins. Biochemistry. 2006;45:8453–8465. doi: 10.1021/bi060615u. [DOI] [PubMed] [Google Scholar]
- Reth M. Antigen receptor tail clue. Nature. 1989;338:383–384. [PubMed] [Google Scholar]
- Ross SR, Schmidt JW, Katz E, Cappelli L, Hultine S, Gimotty P, Monroe JG. An immunoreceptor tyrosine activation motif in the mouse mammary tumor virus envelope protein plays a role in virus-induced mammary tumors. J Virol. 2006;80:9000–9008. doi: 10.1128/JVI.00788-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:66–74. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- Sigalov AB, Aivazian DA, Uversky VN, Stern LJ. Lipid-binding activity of intrinsically unstructured cytoplasmic domains of multichain immune recognition receptor signaling subunits. Biochemistry. 2006;45:15731–15739. doi: 10.1021/bi061108f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun ZJ, Kim KS, Wagner G, Reinherz EL. Mechanisms contributing to T cell receptor signaling and assembly revealed by the solution structure of an ectodomain fragment of the CD3 epsilon gamma heterodimer. Cell. 2001;105:913–923. doi: 10.1016/s0092-8674(01)00395-6. [DOI] [PubMed] [Google Scholar]
- Wang J, Arbuzova A, Hangyas-Mihalyne G, McLaughlin S. The effector domain of myristoylated alanine-rich C kinase substrate binds strongly to phosphatidylinositol 4,5-bisphosphate. J Biol Chem. 2001;276:5012–5019. doi: 10.1074/jbc.M008355200. [DOI] [PubMed] [Google Scholar]
- Weiss A, Littman DR. Signal transduction by lymphocyte antigen receptors. Cell. 1994;76:263–274. doi: 10.1016/0092-8674(94)90334-4. [DOI] [PubMed] [Google Scholar]
- Wiener MC, White SH. Structure of a fluid dioleoylphosphatidylcholine bilayer determined by joint refinement of x-ray and neutron diffraction data. III. Complete structure. Biophys J. 1992;61:434–447. doi: 10.1016/S0006-3495(92)81849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung T, Terebiznik M, Yu L, Silvius J, Abidi WM, Philips M, Levine T, Kapus A, Grinstein S. Receptor activation alters inner surface potential during phagocytosis. Science. 2006;313:347–351. doi: 10.1126/science.1129551. [DOI] [PubMed] [Google Scholar]
- Zhang W, Crocker E, McLaughlin S, Smith SO. Binding of peptides with basic and aromatic residues to bilayer membranes: phenylalanine in the myristoylated alanine-rich C kinase substrate effector domain penetrates into the hydrophobic core of the bilayer. J Biol Chem. 2003;278:21459–21466. doi: 10.1074/jbc.M301652200. [DOI] [PubMed] [Google Scholar]
- Ziegenfuss JS, Biswas R, Avery MA, Hong K, Sheehan AE, Yeung YG, Stanley ER, Freeman MR. Draper-dependent glial phagocytic activity is mediated by Src and Syk family kinase signalling. Nature. 2008;453:935–939. doi: 10.1038/nature06901. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sup. Movie1
Sup. Movie2
Sup. Movie3
Sup. Movie4
Sup. Movie5
supplement