Cohesin relocation from sites of chromosomal loading to places of convergent transcription (original) (raw)
. Author manuscript; available in PMC: 2008 Dec 27.
Published in final edited form as: Nature. 2004 Jun 30;430(6999):573–578. doi: 10.1038/nature02742
Abstract
Sister chromatids, the products of eukaryotic DNA replication, are held together after their synthesis by the chromosomal cohesin complex. This allows the spindle in mitosis to recognise pairs of replication products for segregation into opposite direction1-6. Cohesin forms large protein rings that may bind DNA strands by encircling7, but the characterisation of cohesin binding to chromosomes in vivo has remained vague. Here, we present high resolution analysis of cohesin association along budding yeast chromosomes III – VI. Cohesin localises almost exclusively between genes transcribed in converging direction. We find that not the underlying sequence, but active transcription positions cohesin at these sites. Cohesin is initially loaded onto chromosomes at separate places, marked by the Scc2/Scc4 cohesin loading complex8, from where it appears to slide to its more permanent locations. But even after sister chromatid cohesion is established changes in transcription lead to repositioning of cohesin. Thus a key architectural feature of mitotic chromosomes, the sites of cohesin binding and therefore most likely sister chromatid cohesion, display surprising flexibility. Cohesin localisation to places of convergent transcription is conserved in fission yeast, suggesting that it is a common feature of eukaryotic chromosomes.
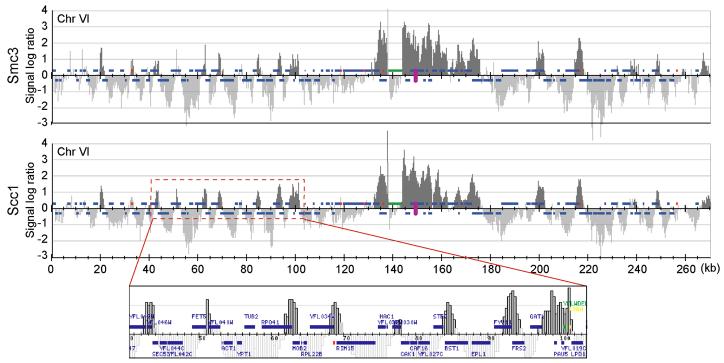
Cohesin association with yeast and human chromosomes has been studied4,9-15, but the defining characteristics of association sites, and how cohesin gets to these sites, remained unclear. We analysed cohesin binding to chromosome VI of the budding yeast Saccharomyces cerevisiae by chromatin immunoprecipitation (ChIP) followed by hybridisation to a high-density oligonucleotide array16. The pattern of association in metaphase was similar for all cohesin subunits analysed, Scc1, Scc3, Smc3, and Pds5 (Fig. 1, and Supplementary Figure S1). It was also similar before the establishment of sister chromatid cohesion, in cells arrested with the replication inhibitor hydroxyurea (Ref. 9, and Supplementary Figure S1). Cohesin bound 28 distinct sites, each spanning 1-4 kilobases (kb) in width. The intensity of association varied, with the strongest peaks found around the centromere, consistent with previous analyses9-11. The distance between neighbouring cohesin association sites ranged from 2 to 35 kb. Almost all cohesin association sites were centred in intergenic regions where genes from opposite strands converged (Fig. 1a), as previously suggested17. Using an additional high-density array, we also mapped the association of Scc1 with chromosomes III, IV, and V (Supplementary Table 1 and Figure S2). 91% (276 of 304) cohesin association sites identified lie at intergenic regions between converging genes, and of 328 convergent intergene regions 84% were bound by cohesin.
Figure 1.
Cohesin localises to convergent intergene regions along budding yeast chromosome VI. Cells containing Smc3-Flag3 or Scc1-HA6 were arrested in metaphase by nocodazole treatment and processed for ChIP against the epitope tagged subunits. Enrichment in the immunoprecipitated fraction relative to a whole genome DNA sample is shown along the length of the chromosome. Each bar represents the average of 16 oligonucleotide probes within adjacent 300 bp windows. The y-axis scale is log2. Dark grey signals represent significant binding, as detailed in the Supplementary MIAME Document. Blue bars above and below the midline are genes transcribed from left to right and opposite, respectively. The purple oval represents the centromere. Origins of replication are depicted in red, tRNA genes in yellow, and a Ty2 transposon in green.
We searched for shared motifs in the nucleotide sequences at cohesin binding sites, but failed to identify any. We therefore wondered whether the process of transcription itself was responsible for positioning cohesin towards converging intergenes. We tested this at the strong cohesin association site between STE2, a highly expressed gene on chromosome VI that is non-essential for vegetative growth, and BST1. Deletion of the STE2 promoter largely abolished cohesin association at this site (Fig. 2a). Transcription therefore appears to direct the accumulation of cohesin downstream of the gene, and not the nucleotide sequence at this site that remained unchanged.
Figure 2.
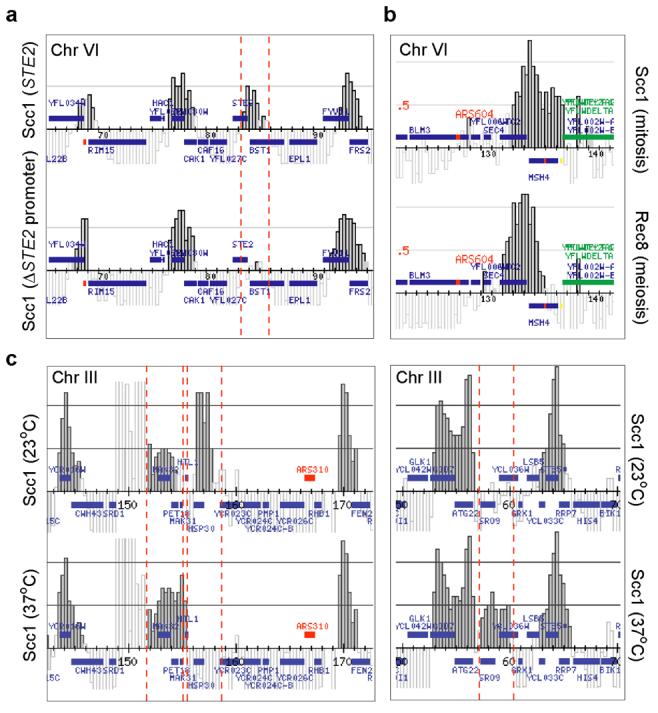
Cohesin is moved towards 3′-ends of genes by their transcription. a, Localisation of Scc1-HA6 around STE2 on chromosome VI in a nocodazole-arrested wild-type strain (top), and after deletion of the STE2 promoter (bottom). b, Localisation of Scc1-HA6 around MSH4, close to centromere VI, in mitotic cells (top), and of Rec8-Flag3 in meiosis (bottom) when MSH4 is induced. c, Localisation of Scc1-HA6 in nocodazole-arrested cells before (top) and 15 min after heat-shock (bottom). Regions around HSP30 and SRO9 on chromosome III, induced and repressed after heat-shock respectively, are shown.
Deletion of the STE2 promoter may have introduced unwanted changes to chromosomal architecture, so we addressed whether physiologic changes in transcription would have similar effects on cohesin localisation. The MSH4 gene, close to the centromere of chromosome VI, is not expressed during vegetative growth, but upregulated upon entry into meiosis18. The gene is covered by cohesin in mitosis. During meiosis, cohesin over MSH4 shifted towards the 3′-end of the gene, suggesting that MSH4 transcription causes cohesin relocalisation (Fig. 2b). This further suggests that even the abundant cohesin around centromeres is positioned by transcription. We visualised meiotic cohesin by ChIP against Rec8, a meiosis-specific Scc1 homolog. Rec8 colocalised with Scc1 that also remained detectable in meiosis (Supplementary Figure S3). Further differences compared to the mitotic cohesin pattern were all correlating with known transcriptional alterations in meiosis18. E. g. YFR022w expression is downregulated in meiosis, and the locus now was covered by cohesin extending from the neighbouring PES4/YFR024c association site (Supplementary Figure S3). Thus, decreased transcription allowed cohesin to occupy otherwise inaccessible space along the gene.
We also analysed HSP30 on chromosome III, a gene strongly induced after heat-shock19. In a metaphase arrested culture at 23°C cohesin covered much of HSP30, but after shift of the culture to 37°C for 15 min HSP30 was free of cohesin and an increased signal was observed downstream of HSP30 at the neighbouring MAK32/PET18 site (Fig. 2c, and Supplementary Figure S4). This indicates that even once cohesin is already loaded onto chromosomes, and sister chromatid cohesion is established, transcription has the ability to relocate cohesin. Another gene on chromosome III, SRO9, is downregulated in response to heat-shock19. Correspondingly, we found that cohesin covered SRO9 after, but not before, shift to 37°C (Fig. 2c). These results can be explained if cohesin rings encircle and are free to slide along chromatin. The elongating transcription machinery may push cohesin towards the 3′ end of transcriptional units, and recurring transcription may be required to maintain cohesin at these sites. Alternatively an event correlating with transcription, e. g. a certain type of chromatin modification, may be responsible for positioning cohesin.
We next addressed where cohesin is first loaded onto chromosomes after its synthesis in late G1. Cohesin interacts with, and its loading onto chromosomes depends on the Scc2/Scc4 complex8,20,21. Cytological studies suggested that cohesin does not colocalise with Scc2/Scc4 on chromosomes, which made the molecular function of this complex difficult to rationalise. We analysed the localisation of Scc2/Scc4 along chromosome VI in cells arrested in early S-phase with the replication inhibitor hydroxyurea (Fig. 3a). Scc2 and Scc4 showed an identical pattern of localisation, next to both telomeres, at the centromere, as well as at numerous places along chromosome arms. Association sites differed in width and, as expected, they were distinct from the sites occupied by cohesin. A similar pattern of Scc4 localisation was seen in G1 and metaphase arrested cells (Supplementary Figure S5). The strong Scc2/Scc4 binding site at the centromere, most prominent in S-phase cells, may be responsible for loading of abundant cohesin around this locus. Comparison of Scc2/Scc4 localisation to transcriptional activity along chromosome VI revealed that at some places Scc2/Scc4 binding coincided with strong transcription (Fig. 3b). Statistical tests suggest transcriptional activity may correlate with the binding of Scc2/Scc4 (Supplementary Note 1). This for the first time describes the chromosomal distribution of the Scc2/Scc4 complex.
Figure 3.
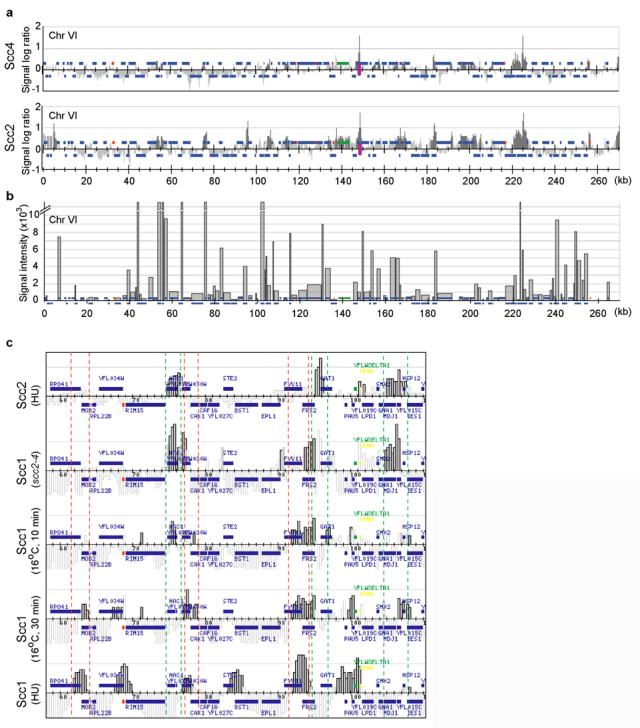
Cohesin loading at, and movement away from sites of Scc2/Scc4 binding. a, Localisation of Scc4-HA6 and Scc2-HA6 along chromosome VI in hydroxyurea-arrested cells. See the legend to chromosomal features in Fig. 1. b, Transcriptional activity along chromosome VI in wild-type logarithmically growing cells. The signal intensity reflects the abundance of transcripts, as an approximation of transcriptional activity. Broken bars exceed 10,000, see Supplementary Figure S5 for full values c, Detail of the localisation on the left arm of chromosome VI, from top to bottom, of Scc2-HA6 in hydroxyurea-arrested cells, Scc1-HA6 in scc2-4 mutant cells released from G1 into hydroxyurea block at restrictive temperature, Scc1-HA6 in wild-type cells 10 min, and 30 min following release from G1 at 16°C, and Scc1-HA6 in hydroxyurea-arrested wild-type cells. The green and red dashed lines flank sites usually seen bound by Scc2 and Scc1, respectively.
To investigate the role of the Scc2/Scc4 complex in cohesin loading, we analysed cohesin association in scc2-4 mutant cells. We were surprised to find cohesin now at the sites normally occupied by the Scc2/Scc4 complex (Fig. 3c, and Supplementary Figure S6). This suggests that an early step in the loading of cohesin onto DNA, before the step disrupted by the scc2-4 mutation, takes place at Scc2/Scc4 binding sites. Cohesin association under these conditions may be weak, which could explain why other methods failed to reveal it8. We next analysed early events of cohesin loading in wild-type cells traversing synchronously through G1. Cells were arrested with the mating pheromone α-factor and released at 16°C to slow progression through G1. The first detectable association of cohesin with chromosomes, 10 min after release, overlapped with Scc2/Scc4 binding sites (Fig. 3c, and Supplementary Figure S6). After 30 min, cohesin became detectable also at the expected converging intergenic sites. This is consistent with the idea that cohesin is loaded onto chromosomes at Scc2/Scc4 binding sites, and thereafter relocates to more permanent places.
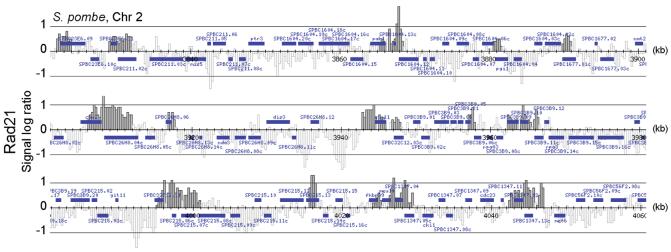
To see whether cohesin localisation by converging transcription was a general feature of eukaryotic chromosomes, we performed ChIP of cohesin in the fission yeast _Schizosaccharomyces pombe_4, followed by hybridisation to a high-density oligonucleotide array covering its chromosomes 2 and 3. Compared to budding yeast, genome organisation and chromatin biology in fission yeast resembles more closely the situation in higher eukaryotes. We found that the fission yeast Scc1 homolog, Rad21, preferentially localised to regions of convergent transcription, even though the peaks of association often extended over larger regions (Fig. 4, and Supplementary Note 2). Therefore, cohesin distribution in response to transcription appears to be a conserved mechanism.
Figure 4.
Cohesin localisation to convergent intergenic regions is conserved in fission yeast. ChIP was performed against the cohesin subunit Rad21-HA3 from logarithmically growing cells. A 240 kb long stretch from the right arm of chromosome 2 is shown.
Sister chromatid cohesion along chromosome arms is crucial for DNA repair by homologous recombination22. The prominence of cohesin at converging intergenic sites therefore raises the question whether genes are arranged in a particular order to provide for a regular pattern of cohesin association. When we analysed the succession of left or right facing genes along budding yeast chromosomes III – VI we found that it appeared random. We did not find regions on these chromosomes where the likelihood of orientation of a gene following either a left- or right-facing gene was different from equal (Supplementary Note 3). Fortuitous clusters of genes in tandem, that have been noted before23, are expected in this random distribution. The consequent chance distribution of convergence sites agrees with the observed wide range of distances between neighbouring cohesin association sites. Thus, an important structural feature of yeast chromosomes, the sites of cohesin association, and therefore most likely sister chromatid cohesion, are spaced at random. Cohesin can act as a border of transcriptional domains24. This seems now unlikely to be generally the case, as neighbouring genes in budding yeast show a high probability of co-regulation, irrespective of whether they are oriented in divergent or convergent orientation25.
We describe here that cohesin, a critical structural component of the eukaryotic chromosome, does not have a fixed pattern of localisation. The possibility that the ring shaped cohesin complex encircles chromatin offers an intriguing explanation to this finding7. After DNA is transported into the cohesin ring21,26, cohesin may be able to slide along chromatin, responding to steric requirements of transcription and maybe other forms of chromosomal metabolism. Cohesin is loaded onto chromosomes next to its loading factor Scc2/Scc4, from where it relocates to places of convergent transcription. Scc2/Scc4 has been suggested to stimulate cohesin's ATPase activity, required to open the ring for loading onto DNA. Once loaded, preventing further Scc2/Scc4 action on cohesin would be vital to secure stability of the ring21. Moving cohesin away from its loading sites, promoted by strong transcriptional activity found there, would be a safe way to achieve this. But even after sister chromatid cohesion is established cohesin is not stably trapped, and changes in transcriptional activity lead to further redistribution. This suggests cohesin provides chromosomes with an unexpectedly flexible architecture. This scenario does not exclude that places exist where cohesin engages in a more stable contact with chromatin. E. g. fission yeast cohesin's enrichment at centromeric heterochromatin is mediated by a specific interaction with the heterochromatin protein Swi6 (Ref. 13). Centromeric cohesin is crucial to promote bipolar spindle attachment in mitosis5,27, but in contrast to budding yeast, centromeres in fission yeast and higher eukaryotes cover large regions devoid of much transcriptional activity. Cohesin cannot therefore be confined there by convergent transcription, and during the evolution of larger centromeres cohesin may have acquired an alternative way to hold on to centromeric chromatin. Stable binding to heterochromatin, versus lateral mobility, may also differentiate cohesin during unloading of a subpool in prophase when chromosomes condense and transcription ceases28.
How does transcription locate cohesin towards converging intergenes? Although we cannot exclude that transcriptional termination correlates with, or induces, a specific chromatin state that attracts cohesin, it is unclear how such a feature would be restricted to convergent as opposed to other sites of transcriptional termination. We therefore imagine that the sheer size of the transcription apparatus may be too large to pass through the cohesin ring, thus pushing it while translocating along the DNA. If cohesin is sufficiently mobile, and intergenic regions sufficiently small, cohesin would be passed on along tandem genes until reaching a site of convergence. Transcription through a cohesin ring would be unwanted, as it would leave the transcript trapped. In remarkable contrast, when the replication fork travels along chromosomes during S-phase, cohesion between the replication products might be a consequence of the fork traversing through the cohesin ring. What distinguishes the replication fork from the transcription apparatus so that it does not push cohesin, but instead may slide through it, will be an interesting question to be addressed.
Methods
Yeast strains and plasmids
The strains used in this study are listed as Supplementary Table 2. S. cerevisiae strains were derivative of W303 (MATa, ade2-1 trp1-1 can1-100 leu2,3,112 his3-11,15 ura3), BY4741 (MATa, his3Δ1 leu2Δ0 met15Δ0 ura3Δ0), or SK1 (MATa/MATα, ura3/ura3 leu2/leu2 lys2/lys2 his3/his3). Endogenous genes were fused to epitope tags by gene targeting using polymerase chain reaction products (PCR). To delete the STE2 promoter, the sequence between the YFL027C and STE2 ORFs was replaced by a URA3 marker gene, flanked by 126 bp direct repeats, using a PCR product from plasmid pWJ1077 (Ref. 29, a kind gift from R. Rothstein). The URA3 gene was lost after selecting a 5-fluoroorotic acid resistant clone, leaving behind one repeat unit.
Yeast culture
S. cerevisiae cells were grown in YP medium containing 2% glucose, synchronised with α-factor (2 μM) in G1, and released into medium containing 200 mM hydroxyurea for 60 min or containing 7.5 μg/ml nocodazole for 150 min at 23°C. Cell cycle synchrony was confirmed by flow cytometry in all experiments. Heat-shock treatment was by transferring the culture to a 37°C waterbath for 15 minutes. As restrictive temperature for the scc2-4 mutant we used 37°C. Meiotic cells were collected after induction of meiosis for 4 hours at 23°C as previously described30. S. pombe cells were in logarithmic growth in minimal medium.
ChIP analysis
S. cerevisiae chromosomes III-V, chromosome VI, and S. pombe chromosomes 2-3 high-density oligonucleotide microarrays were produced by Affymetrix Custom Express Service (SC3456a520015F, P/N 520015; rikDACF, P/N 510636; and S_pombea520106F, P/N 520106, respectively). Sequence and position of oligonucleotides on the microarrays are available from Affymetrix. ChIP was carried out as previously described16. For gene expression analysis, total RNA was reverse transcribed according to the protocol provided by Affymetrix. cRNA was obtained from the cDNA by in vitro transcription, cleaned, fragmented by heating, and hybridised to YGS98 GeneChipsT oligonucleotide arrays (P/N 900256, Affymetrix). Details are available in the Supplementary MIAME Document, and at our website http://chromosomedynamics.bio.titech.ac.jp. Microarray data presented in this paper can be obtained from GEO (http://www.ncbi.nlm.nih.gov/geo) with the accession number GSE1461.
Supplementary Material
1
Acknowledgements
We are indebted to E. Schwob for triggering this collaboration. We also would like to thank A. Nakada and T. Chaplin for technical support, R. Rothstein for reagents, J. Cau, J. Sgouros, J. Svejstrup and members of our laboratories for discussions and comments on the manuscript. A. L. was supported by an EU Marie Curie Individual Fellowship and a J. Cell Sci. travelling fellowship, F. U. acknowledges supported through the EMBO Young Investigator Programme.
Footnotes
References
- 1.Michaelis C, Ciosk R, Nasmyth K. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 2.Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Losada A, Hirano M, Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomonaga T, et al. Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 2000;14:2757–2770. doi: 10.1101/gad.832000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanaka T, Fuchs J, Loidl J, Nasmyth K. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat. Cell Biol. 2000;2:492–499. doi: 10.1038/35019529. [DOI] [PubMed] [Google Scholar]
- 6.Uhlmann F. Chromosome cohesion and separation: from men and molecules. Curr. Biol. 2003;13:R104–R114. doi: 10.1016/s0960-9822(03)00039-3. [DOI] [PubMed] [Google Scholar]
- 7.Haering CH, Löwe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell. 2002;9:773–788. doi: 10.1016/s1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- 8.Ciosk R, et al. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell. 2000;5:1–20. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- 9.Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka T, Cosma MP, Wirth K, Nasmyth K. Identification of cohesin association sites at centromeres and along chromosome arms. Cell. 1999;98:847–858. doi: 10.1016/s0092-8674(00)81518-4. [DOI] [PubMed] [Google Scholar]
- 11.Megee PC, Koshland DA. functional assay for centromere-asociated sister chromatid cohesion. Science. 1999;285:254–257. doi: 10.1126/science.285.5425.254. [DOI] [PubMed] [Google Scholar]
- 12.Laloraya S, Guacci V, Koshland D. Chromosomal addresses of the cohesin component Mcd1p. J. Cell Biol. 2000;151:1047–1056. doi: 10.1083/jcb.151.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nonaka N, et al. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat. Cell Biol. 2001;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- 14.Bernard P, et al. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- 15.Hakimi M-A, et al. A chromatin remodelling complex that loads cohesin onto human chromosomes. Nature. 2002;418:994–997. doi: 10.1038/nature01024. [DOI] [PubMed] [Google Scholar]
- 16.Katou Y, et al. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature. 2003;424:1078–1083. doi: 10.1038/nature01900. [DOI] [PubMed] [Google Scholar]
- 17.Filipski J, Mucha M. Structure, function and DNA composition of Saccharomyces cerevisiae chromatin loops. Gene. 2002;300:63–68. doi: 10.1016/s0378-1119(02)00848-x. [DOI] [PubMed] [Google Scholar]
- 18.Chu S, et al. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 19.Gasch AP, et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tóth A, et al. Yeast Cohesin complex requires a conserved protein, Eco1p (Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999;13:320–333. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arumugam P, et al. ATP hydrolysis is required for cohesin's association with chromosomes. Curr. Biol. 2003;13:1941–1953. doi: 10.1016/j.cub.2003.10.036. [DOI] [PubMed] [Google Scholar]
- 22.Sjögren C, Nasmyth K. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr. Biol. 2001;11:991–995. doi: 10.1016/s0960-9822(01)00271-8. [DOI] [PubMed] [Google Scholar]
- 23.Jacq C, et al. The nucleotide sequence of Saccharomyces cerevisiae chromosome IV. Nature. 1997;387(Suppl):75–78. [PubMed] [Google Scholar]
- 24.Donze D, Adams CR, Rine J, Kamakaka RT. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 1999;13:698–708. doi: 10.1101/gad.13.6.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen BA, Mitra RD, Hughes JD, Church GM. A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nat. Genet. 2000;26:183–186. doi: 10.1038/79896. [DOI] [PubMed] [Google Scholar]
- 26.Weitzer S, Lehane C, Uhlmann F. A model for ATP hydrolysis-dependent binding of cohesin to DNA. Curr. Biol. 2003;13:1930–1940. doi: 10.1016/j.cub.2003.10.030. [DOI] [PubMed] [Google Scholar]
- 27.Toyoda Y, et al. Requirement of chromatid cohesion proteins Rad21/Scc1 and Mis4/Scc2 for normal spindle-kinetochore interaction in fission yeast. Curr. Biol. 2002;12:347–358. doi: 10.1016/s0960-9822(02)00692-9. [DOI] [PubMed] [Google Scholar]
- 28.Waizenegger IC, Hauf S, Meinke A, Peters J-M. Two distinct pathways remove mammalian cohesin complexes from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103:399–410. doi: 10.1016/s0092-8674(00)00132-x. [DOI] [PubMed] [Google Scholar]
- 29.Reid RJD, Sunjevaric I, Kedacche M, Rothstein R. Efficient PCR-based gene disruption in Saccharomyces strains using intergenic primers. Yeast. 2002;19:319–328. doi: 10.1002/yea.817. [DOI] [PubMed] [Google Scholar]
- 30.Ohta , et al. Mutations in the MRE11, RAD50, XRS2, and MRE2 genes alter chromatin configuration at meiotic DNA double-stranded break sites in premeiotic and meiotic cells. Proc Natl Acad Sci U S A. 1998;95:645–651. doi: 10.1073/pnas.95.2.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1