Control of c-myc mRNA stability by IGF2BP1-associated cytoplasmic RNPs (original) (raw)
Abstract
The RNA-binding protein IGF2BP1 (IGF-II mRNA binding protein 1) stabilizes the c-myc RNA by associating with the Coding Region instability Determinant (CRD). If and how other proteins cooperate with IGF2BP1 in promoting stabilization of the c-myc mRNA via the CRD remained elusive. Here, we identify various RNA-binding proteins that associate with IGF2BP1 in an RNA-dependent fashion. Four of these proteins (HNRNPU, SYNCRIP, YBX1, and DHX9) were essential to ensure stabilization of the c-myc mRNA via the CRD. These factors associate with IGF2BP1 in a CRD-dependent manner, co-distribute with IGF2BP1 in non-polysomal fractions comprising c-myc mRNA, and colocalize with IGF2BP1 in the cytoplasm. A selective shift of relative c-myc mRNA levels to the polysomal fraction is observed upon IGF2BP1 knockdown. These findings suggest that IGF2BP1 in complex with at least four proteins promotes CRD-mediated mRNA stabilization. Complex formation at the CRD presumably limits the transfer of c-myc mRNA to the polysomal fraction and subsequent translation-coupled decay.
Keywords: IGF2BP1, IMP1, ZBP1, CRD-BP, c-myc, mRNP, mRNA stability
INTRODUCTION
The IGF2BP-proteins (also termed IGF-II mRNA binding proteins [IMPs], zipcode binding proteins [ZBPs], or coding region instability determinant binding proteins [CRD-BPs]), comprise three members (IGF2BP1-3) that regulate the localization, translation, or turnover of their target transcripts (Yisraeli 2005). Members of the protein family show an oncofetal expression declining toward the end of embryogenesis, barely detectable expression in the postnatal phase, but rapid de novo synthesis in various neoplasias (Nielsen et al. 1999; Yaniv and Yisraeli 2002; Kobel et al. 2007).
A major function of IGF2BP1 is the control of target mRNA translation, as demonstrated for the β-actin and IGF-II mRNAs. While inhibition of IGF-II mRNA translation by IGF2BP1 was suggested to rely on binding of the protein to the 5′-UTR, hindering translation of the β-actin mRNA requires association of the protein with the Zipcode in the 3′-UTR of the transcript (Nielsen et al. 1999; Hüttelmaier et al. 2005). Inhibition of β-actin mRNA translation is abrogated upon Src-mediated tyrosine-phosphorylation of IGF2BP1 (ZBP1) by releasing the protein–RNA complex (Hüttelmaier et al. 2005). Besides a target-mRNA specific role in controlling translation, IGF2BP1 also promotes mRNA stabilization. While IGF2BP1 stabilizes the c-myc and βTrCP1 mRNAs by associating with a sequence in the open reading frame, termed the coding region determinant (CRD), stabilization of the CD44 mRNA was suggested to require binding of IGF2BP1 to the 3′-UTR of the transcript (Lemm and Ross 2002; Noubissi et al. 2006; Vikesaa et al. 2006; Kobel et al. 2007). In the case of c-myc mRNA stabilization, the association of IGF2BP1 with the CRD, which is located in the last 249 nucleotides (nt) of the c-myc coding sequence, was suggested to prevent mRNA degradation by polysome-associated endonucleases (Bergstrom et al. 2006; Sparanese and Lee 2007). These were proposed to cleave the c-myc mRNA at the CRD once ribosomes are slowed down during translation by entering a region of rare codons at the beginning of the CRD (Lemm and Ross 2002; Sparanese and Lee 2007). Hence, IGF2BP1 was suggested to promote c-myc mRNA stability via blocking endonucleolytic cleavage by associating at the CRD. However, it remained elusive if IGF2BP1 prevents c-myc mRNA degradation by hindering endonucleolytic cleavage co-translationally or by sequestering the transcript in cytoplasmic mRNPs.
IGF2BP1 containing cytoplasmic mRNPs were recently characterized by a proteomic approach revealing various RNA-binding proteins to assemble with IGF2BP1 in granule-like complexes with a diameter of ∼100–300 nm (Jonson et al. 2007). Notably, these “IGF2BP1-RNPs” were suggested to comprise mRNAs that were never translated, since they contain CBP80, factors of the exon-junction complex, but no eIF4E and eIF4G (Jonson et al. 2007). Despite this correlative evidence, it remained elusive if cytoplasmic IGF2BP1-RNPs stabilize c-myc mRNA via the CRD and which of the proteins co-purifying with IGF2BP1 promote this function.
Here, we report the identification of protein factors that associate with IGF2BP1 and promote c-myc mRNA stability via the CRD. Using a comprehensive knockdown analysis, we identify IGF2BP1, HNRNPU, YBX1, DHX9, and SYNCRIP as factors involved in promoting c-myc mRNA stabilization via the CRD. Destabilization of the c-myc mRNA upon IGF2BP1 knockdown is correlated with the loss of c-myc mRNA from non-translating cytoplasmic mRNPs. This suggests that retention of the mRNA in these mRNPs by a multiprotein complex comprising IGF2BP1 prevents translation-coupled decay of the c-myc mRNA via the CRD.
RESULTS
Identification of IGF2BP1-associated proteins
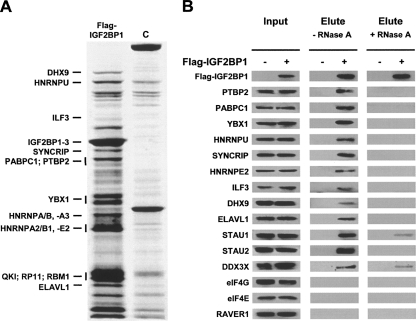
Aiming to identify proteins associating with IGF2BP1, we immunopurified Flag-tagged IGF2BP1 from cytoplasmic extracts of stably transfected U2OS cells. Co-purified proteins were fractionated by SDS-PAGE electrophoresis, and several protein bands specifically enriched from the stably expressing cells were excised and characterized by MS-MS based mass spectrometry (Fig. 1A; Supplemental Fig. S1). These analyses identified several RNA-binding proteins and ribosomal proteins (data not shown) that associate with Flag-tagged IGF2BP1, as previously reported (Jonson et al. 2007).
FIGURE 1.
IGF2BP1-associated proteins. (A) Colloidal Coomassie blue-stained SDS-polyacrylamide gel of proteins isolated from (C) wild-type or U2OS cells stably transfected with Flag-IGF2BP1 by anti-Flag agarose beads. Proteins identified by MS/MS-analyses are indicated by horizontal bars. Vertical bars indicate that analyzed gel slices comprising more than one visible protein band were excised. A complete list of identified RNA-binding proteins is summarized in Supplemental Figure S1. (B) Western blot analysis of indicated proteins in input and elute upon co-purification with Flag-tagged IGF2BP1 from stably transfected U2OS cells (Flag-IGF2BP1, +). Wild-type U2OS cells (Flag-IGF2BP1, −) served as controls. Analyses were performed in the (−) absence or (+) presence of 10 μg/mL RNase A.
Association of identified proteins with IGF2BP1 was further evaluated by Western blotting. In addition to proteins identified by MS-analyses, STAU1, STAU2, and DDX3X, previously determined as components of transported neuronal mRNPs, were included in these studies (Kanai et al. 2004; Elvira et al. 2006). The translation initiation factors eIF4E and eIF4G as well as the PTBP2-associated splicing regulator RAVER1 served as negative controls, since these proteins were not detected here or in the recently presented proteomic analysis of IGF2BP1-containing mRNPs (Hüttelmaier et al. 2001; Jonson et al. 2007). Except eIF4E, eIF4G, and RAVER1, all analyzed proteins co-purified with IGF2BP1 from cytoplasmic extracts of stably transfected U2OS cells (Fig. 1B, −RNaseA). However, in contrast to previous studies suggesting that IGF2BP1-containing mRNPs are distinct from STAUFEN-containing RNA-granules, co-purification of IGF2BP1 with both STAUFEN isoforms was observed in U2OS cells (Jonson et al. 2007). The majority of proteins associated with IGF2BP1 in an RNA-dependent fashion, since co-purification was abolished or severely reduced in the presence of RNaseA (Fig. 1B, +RNaseA). These findings suggest that the association of proteins with IGF2BP1 is indirect (via RNA) or that RNA binding is required to induce conformations favoring complex assembly (Fig. 1B, +RNaseA).
Control of c-myc mRNA turnover by IGF2BP1-associated protein factors
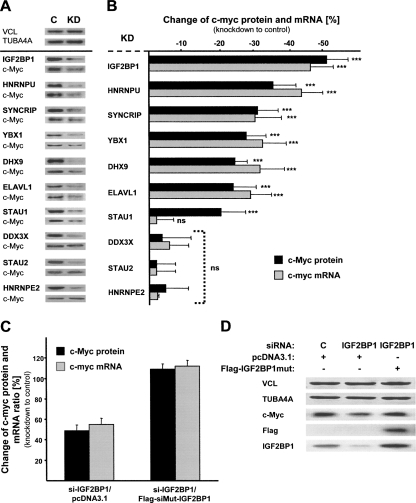
To identify if IGF2BP1-associated proteins serve a function in regulating c-myc mRNA turnover, RNAi based “loss-of-function” studies were performed in U2OS cells. IGF2BP1 knockdown caused a significant reduction of c-myc protein and mRNA levels at steady state, as previously demonstrated in ovarian carcinoma-derived cells (Kobel et al. 2007). Significantly decreased c-myc protein and mRNA levels were also observed upon knockdown of HNRNPU, DHX9, SYNCRIP, YBX1, and ELAVL1, whereas no such decrease was determined using STAU2-, DDX3X-, and HNRNPE2-directed siRNAs (Fig. 2A,B; Supplemental Fig. S2). Notably, reduced c-Myc protein levels were determined upon the knockdown of STAU1, but c-myc mRNA amounts remained unaffected. This exception could indicate a role of STAU1 in promoting translation of distinct mRNAs via their 5′-UTR, as previously suggested (Dugre-Brisson et al. 2005).
FIGURE 2.
Control of c-myc mRNA levels by IGF2BP1 and associated proteins. (A) Western blot analyses of indicated proteins in U2OS cell extracts upon knockdown (KD) of factors indicated in bold or transfection of control siRNAs (C). (B, black bars) Altered c-Myc protein levels upon indicated knockdown (KD) were determined by Western blotting (A, shows representative analyses) and cross-normalization to VCL and TUBA4A. For linearity of c-Myc, TUBA4A, and VCL detection by semiquantitative Western blotting, see Supplemental Figure S2. Altered c-myc mRNA levels were analyzed by qRT-PCR using the ΔΔCt method and cross-normalization to PPIA, RPLP0, and 28S RNA serving as internal controls. Error bars indicate the SD determined from at least five independent experiments. (***) p ≤ 0.0005; (ns) not significant (p ≥ 0.05). (C,D) Altered (black bars) c-myc protein and (gray bars) mRNA levels in U2OS cells transfected for 72 h with indicated siRNAs and for 24 h with indicated plasmids were determined as in _A_and B. The change of c-myc mRNA and protein ratios is shown relative to controls transfected with control siRNAs (72 h) and empty pcDNA3.1 plasmid (24 h). (D) Representative Western blotting for indicated proteins. Error bars indicate the SD determined from two independent experiments.
To further validate IGF2BP1 as a regulator of c-myc mRNA turnover in cells, decreased c-myc mRNA levels upon IGF2BP1 knockdown were recovered by expressing an siRNA-insensitive IGF2BP1 mutant protein (Fig. 2C,D). As expected, c-myc protein and mRNA levels remained largely unaffected in cells transfected with IGF2BP1-directed siRNAs for 72 h and the siRNA-insensitive IGF2BP1 mutant (Flag-siMUT-IGF2BP1) for 24 h. Moreover, detailed analysis of IGF2BP1 knockdown revealed that the siRNAs used barely affected IGF2BP2 and IGF2BP3 (Supplemental Fig. S3A). Notably, β-actin mRNA stability remained unaffected by IGF2BP1 knockdown; however, β-Actin protein levels were elevated upon knockdown (Supplemental Fig. S3B,C). Together these analyses identified that IGF2BP1 stabilized the c-myc mRNA and inhibited β-actin mRNA translation in U2OS cells as previously demonstrated in ovarian carcinoma-derived cells and neurons, respectively (Hüttelmaier et al. 2005; Kobel et al. 2007).
Aiming to validate HNRNPU, SYNCRIP, YBX1, DHX9, and ELAVL1 as factors involved in controlling c-myc mRNA stability, alternative siRNAs were used to monitor effects on c-myc mRNA and protein levels at steady state (Supplemental Fig. S4A–C). These analyses confirmed reduced c-myc mRNA and protein amounts upon knockdown and thus indicated that the observed decrease in protein and mRNA levels is unlikely to result from off-target effects.
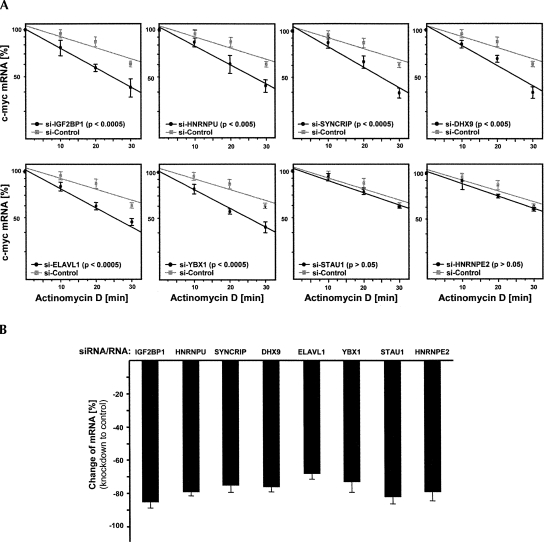
Since the decrease of mRNA amounts at steady state could result from altered transcription or mRNA stability, RNA decay analyses were performed. To allow for an accurate and rapid investigation of IGF2BP1-associated proteins involved in c-myc mRNA stabilization, mRNA degradation was analyzed by quantitative RT-PCR in Actinomycin D (ActD)-treated U2OS cells (Fig. 3A). Cells were transfected with indicated siRNAs 72 h before the decay analyses. Total RNA was isolated at indicated time points after ActD application, and knockdown of each analyzed factor was validated by quantitative RT-PCR for each experiment (Fig. 3B). Quantitative RT-PCR analyses were based on the ΔΔCt method using PPIA, RPLP0, and 28S RNA as internal controls, since these transcripts remained stable during the 30–45-min decay analyses (Livak and Schmittgen 2001; Kobel et al. 2007). The relative amount of c-myc mRNA in each cell population and at each time point was determined in three independent experiments by cross-normalization using the internal controls and setting the input amount of c-myc mRNA to 100%.
FIGURE 3.
Control of c-myc mRNA decay. (A) Decay of c-myc mRNA was monitored in U2OS cells treated with Actinomycin D and transfected with indicated siRNAs. RNA levels were determined by quantitative RT-PCR from three independent experiments as in Figure 2B. (B) Knockdown efficiency for each target was monitored by quantitative RT-PCR for input controls (see A). The decrease of mRNA levels was determined by quantitative RT-PCR for each time point as in Figure 2B. (A) Error bars indicate SD, and data were fitted by linear regression. (A) _P_-values were determined by Student's _t_-testing.
Confirming a role in stabilizing the c-myc mRNA, the knockdown of IGF2BP1 led to accelerated decay of the c-myc mRNA (Fig. 3A). While in cells transfected with control siRNAs, the c-myc mRNA decayed with a half-life of ∼41 min, its half-life was reduced to 24 min upon IGF2BP1 knockdown. In agreement with previous analyses in distinct cells, these findings indicated that c-myc mRNA degradation was accelerated approximately twofold upon IGF2BP1 knockdown (Kobel et al. 2007). Accelerated decay of the c-myc mRNA was also observed upon the knockdown of HNRNPU, YBX1, SYNCRIP, ELAVL1, and DHX9 (Fig. 3A). In contrast, no significant change in the rate of c-myc mRNA degradation was observed in response to STAU1 or HNRNPE2 knockdown. These findings are in agreement with the analyses of c-myc protein and mRNA levels at steady state (Fig. 2B). Notably, the knockdown of each regulatory factor alone led to an approximately twofold accelerated decay of the c-myc mRNA, suggesting that each factor is essential and promotes stabilization through the same complex or mechanism. Summarized, these analyses confirm IGF2BP1, HNRNPU, YBX1, SYNCRIP, DHX9, and ELAVL1 as factors involved in controlling c-myc mRNA turnover. In contrast, STAU1 presumably promotes c-myc translation, whereas HNRNPE2, although associated with IGF2BP1, apparently serves no role in controlling c-myc mRNA translation or turnover.
Control of c-myc mRNA turnover via the CRD
Regulation of c-myc mRNA turnover was proposed to depend on at least two cis determinants, one of which is the AU-rich element (ARE) in the 3′-UTR of the mRNA (Brewer and Ross 1988, 1989; Swartwout and Kinniburgh 1989). Regulation via the ARE relies on at least two classes of _trans_-acting proteins. While AUF-1 (HNRNPD) is thought to promote destabilization of the transcript, members of the ELAV-like protein family stabilize the mRNA via the ARE (Chagnovich and Cohn 1996; DeMaria and Brewer 1996; Manohar et al. 2002). In contrast, IGF2BP1 was suggested to protect the c-myc mRNA from endonucleolytic cleavage by associating with the CRD (Lemm and Ross 2002; Kobel et al. 2007; Sparanese and Lee 2007).
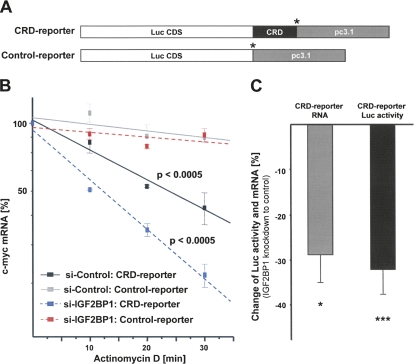
With the aim to analyze if factors identified here regulate c-myc mRNA turnover via the CRD, luciferase reporter assays were performed. These were previously shown to recapitulate IGF2BP1-facilitated stabilization of the c-myc mRNA in HEK293 cells (Kobel et al. 2007). The human c-myc CRD was fused in frame to the Firefly-luciferase coding sequence, and a transcript lacking the CRD served as a control (Fig. 4A). The suitability of the reporter system for recapitulating CRD-mediated mRNA destabilization was validated by mRNA decay analyses (Fig. 4B). For these, HEK293 cells were transfected with IGF2BP1-directed or control siRNAs for 48 h before the transfection of reporter plasmids. At 72 h post-transfection of siRNAs (24 h post-transfection of plasmids), cells were exposed to ActD, and degradation of the reporter mRNAs was monitored by quantitative RT-PCR based on isolated and DNAse-treated total RNA. Degradation of the CRD-reporter transcript was significantly elevated compared to the control-reporter transcript, indicating that inserting the c-myc CRD led to a significant destabilization of the reporter transcript. Similar to the endogenous c-myc mRNA decay of the CRD-reporter mRNA was elevated about twofold upon IGF2BP1 knockdown (Fig. 4B). This confirmed that CRD-mediated degradation can be recapitulated by the CRD reporter, as previously demonstrated (Kobel et al. 2007). On the protein level, the accelerated decay of the reporter mRNA resulted in reduced luciferase activity at steady state, although the observed reduction of luciferase activity (∼30%–40% decrease upon IGF2BP1 knockdown) was slightly reduced compared to the observed decrease of c-Myc protein (∼50% reduction) upon IGF2BP1 knockdown (Fig. 4C). Summarized, these analyses confirmed that the luciferase activity at steady state can be used as a “fast” read-out to determine CRD-mediated destabilization of the CRD-reporter transcript, although effects on luciferase activity were reduced compared to the decrease of endogenous c-Myc protein levels (also see Supplemental Fig. S7A,B).
FIGURE 4.
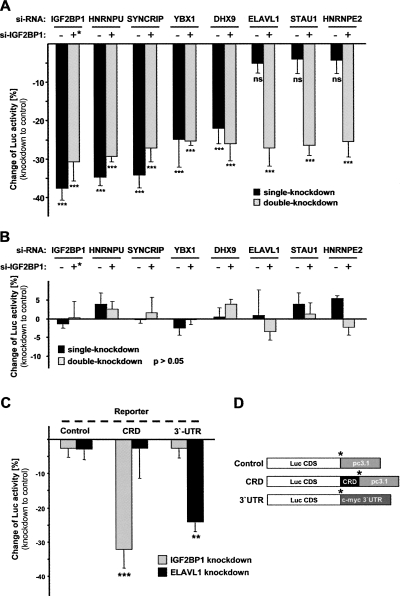
Control of CRD-mediated c-myc mRNA degradation by IGF2BP1. (A) Schematic of reporter transcripts (5′–3′), as follows: (Luc CDS) Firefly luciferase CDS; human c-myc CRD in frame with Luc CDS; and/or pcDNA3.1 plasmid-encoded 3′-UTR. (*) Stop codons. (B) Degradation of the indicated reporter transcripts was analyzed in HEK293 cells transfected with IGF2BP1 or control siRNAs by quantitative RT-PCR, essentially as described in Figure 3A. The change of RNA levels was determined in three independent experiments, and statistical significance was analyzed by Student's _t_-testing. (C) The change of steady-state CRD-RNA levels and correlating luciferase activity were determined between HEK293 cells transfected with IGF2BP1 or control siRNAs. Error bars indicate SD. (*) p < 0.05; (***) p < 0.0005.
To analyze if factors promoting c-myc mRNA stability on the endogenous level act via the CRD, both reporter constructs were co-transfected for 24 h with Renilla luciferase 48 h post-transfection of siRNAs. Firefly activities were normalized by Renilla activities, and the change of normalized Firefly activity was determined between cells transfected with indicated and control siRNAs (Fig. 5A, black bars, CRD reporter; Fig. 5B, black bars, control reporter). A significant reduction of CRD- but not the control-reporter activity was observed upon IGF2BP1 knockdown (Fig. 5, cf. A and B). These findings indicated that a decrease of normalized luciferase activity was due to reduced stability of the reporter transcript in these analyses, as demonstrated before (Fig. 4B; Kobel et al. 2007). Significantly reduced activity of the CRD but not the control reporter was also observed upon the knockdown of HNRNPU, SYNCRIP, YBX1, and DHX9. The knockdown of ELAVL1, STAU1, or HNRNPE2 had no impact on either of the two reporter transcripts, but ELAVL1 knockdown induced accelerated decay of endogenous c-myc mRNA. This is in agreement with previous findings, suggesting that ELAVL1 promotes c-myc mRNA stability via the 3′-UTR. To validate 3′-mediated stabilization by ELAVL1, changes of luciferase activity for the CRD reporter and a reporter containing the c-myc 3′-UTR were analyzed in response to IGF2BP1 and ELAVL1 knockdown (Fig. 5C). Knockdown of IGF2BP1 led to reduced luciferase activity of the CRD but not the 3′-UTR reporter, whereas the opposite was observed upon ELAVL1 knockdown. Together these findings indicate that HNRNPU, SYNCRIP, YBX1, and DHX9 prevent c-myc mRNA degradation via the CRD, whereas ELAVL1 stabilizes the c-myc mRNA via the 3′-UTR. To evaluate if factors controlling c-myc mRNA stability via the CRD act in an additive manner, double-knockdown analyses were performed. Notably, destabilization of the CRD reporter was not elevated upon the combined knockdown of factors promoting c-myc or CRD-reporter mRNA stability (Fig. 5A, gray bars; alternative presentation in Supplemental Fig. S5A). However, while knockdown of STAU1 or HNRNPE2 alone had no effect on the CRD reporter, the combined knockdown with IGF2BP1 decreased luciferase activity to levels obtained by IGF2BP1 knockdown alone. Together these findings suggest that:
FIGURE 5.
Control of CRD-mediated c-myc mRNA degradation by IGF2BP1-associated proteins. (A,B) Changes of (A) normalized CRD-reporter or (B) control-reporter luciferase activity were determined upon single or double knockdown of indicated factors compared to controls (+* for IGF2BP1 indicates half the amount of transfected siRNA compared to single knockdown). (C) Changes of luciferase activity expressed from reporter transcripts depicted in D were analyzed upon IGF2BP1 or ELAVL1 knockdown as described in A and B. (D) Schematic of reporter transcripts (5′–3′), as follows: (Luc CDS) Firefly luciferase CDS; human c-myc CRD in frame with Luc CDS; and/or pcDNA3.1 plasmid encoded 3′-UTR; (3′-UTR) c-myc 3′-UTR. (*) Stop codons. Error bars indicate the SD. (**) p < 0.005; (***) _p_ < 0.0005; (ns) not significant (_p_ > 0.05). Note that no significant changes were determined in B(p > 0.05).
- IGF2BP1, HNRNPU, SYNCRIP, YBX1, and DHX9 are essential for CRD-mediated stabilization of the c-myc mRNA;
- ELAVL1 promotes c-myc mRNA stability via the 3′-UTR;
- Accelerated decay of the endogenous c-myc mRNA upon knockdown of regulatory factors is not due to altered transcription or nuclear mRNA processing, since exogenous reporter transcripts were affected in a CRD-dependent manner; and
- Stabilization via the CRD is likely to occur via one regulatory complex, since double knockdown of regulatory factors did not yield additive effects.
Stabilization of the c-myc mRNA by IGF2BP1-associated cytoplasmic mRNPs
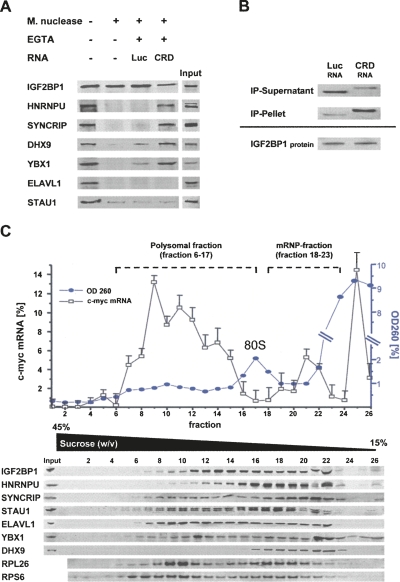
To validate if proteins essential to stabilize the c-myc mRNA via the CRD associate with IGF2BP1 in a CRD-dependent fashion, complex formation was analyzed in Micrococcal nuclease-treated cytoplasmic extracts of HEK293 cells (Fig. 6A). Upon nuclease treatment, the association of HNRNPU, SYNCRIP, DHX9, YBX1, and ELAVL1 with IGF2BP1 was lost, supporting the finding that association of analyzed proteins with IGF2BP1 is RNA-dependent (see Fig. 1B). However, upon blocking nuclease activity by EGTA and the addition of in vitro synthesized CRD-RNA (CRD), all proteins except ELAVL1 and STAU1 were again co-purified with IGF2BP1. In contrast, no or severely reduced association was observed upon the addition of Firefly-luciferase RNA (Luc). The analysis of CRD- and luciferase-RNA levels in the pellet and supernatant after IGF2BP1 immunopurification confirmed that both RNAs remained stable during purification, but the CRD-RNA was selectively enriched by immunopurification of IGF2BP1 (Fig. 6B). These findings indicate that HNRNPU, SYNCRIP, DHX9, and YBX1 but not ELAVL1 and STAU1 associate with IGF2BP1 in a CRD-dependent manner, although it remained elusive if the identified proteins associate at the CRD at distinct binding sites or indirectly via IGF2BP1. However, it appears unlikely that the analyzed proteins with the exception of IGF2BP1 bind unspecifically to any RNA, since the luciferase RNA was not enriched upon immunopurification of any factor (Supplemental Fig. S6A). Hence, together these analyses suggest that:
FIGURE 6.
CRD-mediated complex formation. (A) Western blot analysis of indicated proteins co-purified with IGF2BP1 from HEK293 cell extracts upon Micrococcal nuclease treatment. The nuclease was inactivated by EGTA before the addition of indicated in vitro transcribed RNAs as indicated; (Luc) Firefly-luciferase coding sequence; (CRD) human c-myc CRD. (B) RT-PCR analysis of Luc- and CRD-RNA in supernatant and pellet after immunopurification of IGF2BP1 from nuclease pre-treated HEK293 cell extracts as described in A. (C) Relative c-myc mRNA levels (total RNA in gradient was set to 100%) were determined for individual fractions of polyribosome gradients by qRT-PCR. (Blue) OD260 readings are depicted for each fraction. (Lower panel) Western blot analyses of indicated proteins (RP: ribosomal protein) for each fraction. The monosomal fraction is indicated as 80S. Fractions defined as polysomal (6–17) or mRNP (18–23) are indicated. Error bars indicate the SD from two independent gradient analyses.
- IGF2BP1, HNRNPU, SYNCRIP, DHX9, and YBX1 associate in a multi-protein complex via the CRD.
- This complex formation is essential to prevent CRD-dependent degradation of the c-myc mRNA.
- The regulatory function of the CRD-associated complex is lost upon knockdown of each component, suggesting that each associated protein is essential (see Fig. 5A, double knockdown).
To validate these conclusions in cells, the distribution of IGF2BP1-associated proteins and c-myc mRNA was analyzed by polysome-gradient centrifugation (Fig. 6C). All of the proteins analyzed codistributed with IGF2BP1-containing fractions. Notably, a significant overlap with the non-polysome-associated fraction of cellular c-myc mRNA was observed (Fig. 6C). Since this fraction migrated at a lower density as the monosomal (80S) fraction, it was classified as the “mRNP fraction.”
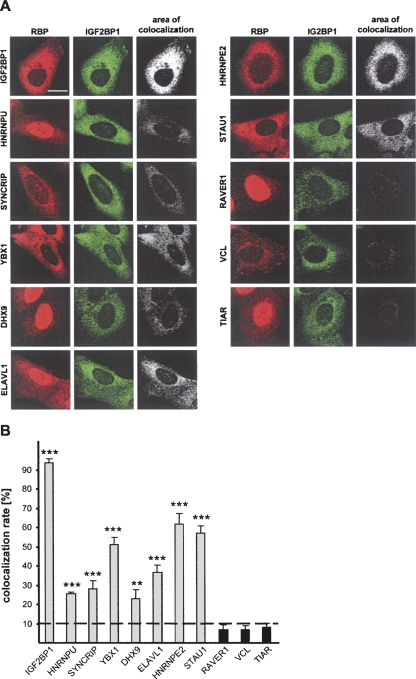
Co-purification of all analyzed proteins with IGF2BP1 from cytoplasmic extracts in a CRD-dependent manner suggested that all regulatory factors colocalize in the cytoplasm. Colocalization of the identified proteins with IGF2BP1 was analyzed by counterstaining using indirect immunofluorescence (Fig. 7A). The colocalization rate was quantified using the colocalization algorithm provided with the Leica-LSM SP5 software. As expected, double staining for IGF2BP1 using a polyclonal anti-ZBP1 (red, Cy3) and a monoclonal anti-IGF2BP1 (green, Alexa488) antibody revealed a colocalization of >90%. In contrast, colocalization of IGF2BP1 with RAVER1, VCL, or TIAR, which were not identified as binding partners of IGF2BP1, was reduced beyond 10%. Supporting the association of identified regulatory factors with IGF2BP1 in cytoplasmic mRNPs, mainly cytoplasmic colocalization at efficiency ranging from ∼25% to 65% was identified for all analyzed proteins. Notably, colocalization with proteins not involved in controlling c-myc mRNA stability via the CRD but identified to associate with IGF2BP1 (STAU1, HNRNPE2, ELAVL1) was observed as well. This suggests that alternative cytoplasmic mRNPs exist that are not essential for promoting c-myc mRNA stabilization. Hence, although these analyses do not allow us to conclude whether the analyzed proteins directly associated with IGF2BP1 and/or the c-myc mRNA, the studies provided further evidence for regulatory cytoplasmic mRNPs that contain IGF2BP1, HNRNPU, SYNCRIP, YBX1, DHX9, ELAVL1, HNRNPE2, and STAU1.
FIGURE 7.
Cytoplasmic colocalization of IGF2BP1 with RNA-binding proteins controlling c-myc mRNA turnover. (A) Double immunostaining for indicated RNA-binding proteins (RBP) or (red) VCL and (green) IGF2BP1. (Left, _upper_panel) The subcellular distribution of IGF2BP1 was traced by a (red) polyclonal and a (green) monoclonal antibody. Images were acquired with a Leica LSM-SP5, and the area of colocalization was determined using the colocalization tool provided with the Leica-SP5 software. Bar, 10 μm. (B) The area of colocalization for each counterstain was determined for 50 cells in two independent experiments. The maximum colocalization is given for IGF2BP1 (double staining by a polyclonal and a monoclonal antibody), whereas a background threshold determined by background colocalization of IGF2BP1 with TIAR, RAVER1, and VCL is indicated as a dotted line. Error bars indicate standard deviation for 100 cells analyzed in two experiments in total. (**) p < 0.005; (***) _p_ < 0.0005; no* (_p_ > 0.05).
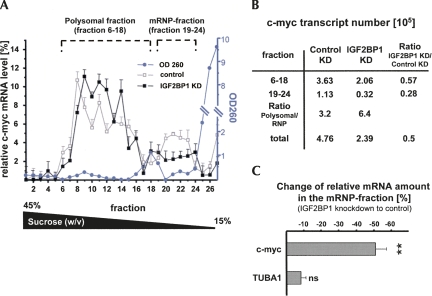
With the aim to monitor how IGF2BP1 knockdown affects the ratio of c-myc mRNA in the polysomal and mRNP fraction, U2OS cells transfected with control or IGF2BP1-directed siRNAs were analyzed by gradient centrifugation in the presence of cycloheximide (Fig. 8A–C). Gradient inputs were normalized by OD260 and total protein concentrations, both determined prior to gradient loading (data not shown). Consistent with the observation that IGF2BP1 knockdown caused a reduction of c-myc mRNA and protein levels by ∼50%, the total amount of c-myc mRNA in the knockdown gradient was reduced by 50% (total c-myc copy numbers in Fig. 8B; see Supplemental Fig. S7A). However, determining the relative amount of mRNA in each fraction (sum of copy numbers over all fractions equals 100%) revealed that the relative level of c-myc mRNA in the mRNP fraction was reduced, whereas it was increased in the polysomal fraction (Fig. 8A). Notably, the OD260 profile and the distribution of ribosomal proteins remained unaffected upon IGF2BP1 knockdown (data not shown). To prove that the observed shift of c-myc mRNA ratios was transcript specific, the changes of relative c-myc and TUBA1B mRNA levels were quantified for the mRNP fraction upon IGF2BP1 knockdown (Fig. 8C). These analyses revealed a specific and significant decrease of relative c-myc mRNA levels in the mRNP fraction by ∼50%, whereas the ratio remained largely unaffected for the TUBA1B mRNA. This indicates that IGF2BP1 and presumably the regulatory proteins identified here determine the relative ratio of c-myc mRNA levels in the polysomal versus the mRNP fraction.
FIGURE 8.
Stabilization of the c-myc mRNA by cytoplasmic mRNPs. (A) Relative c-myc mRNA levels (copy numbers of all fractions sum up to 100%) in response to (black square) IGF2BP1 or (empty square) control knockdown were determined for individual fractions of polyribosome gradients by qRT-PCR, as described in Figure 6C. Polysomal and mRNP-fractions are indicated. (Blue) The OD260 for one gradient (knockdown gradient) for each fraction. (B) c-myc transcript numbers for the polysomal and mRNP-fraction in _A_were determined by qRT-PCR. (C) The relative change of c-myc and TUBA1B mRNA levels in the mRNP fraction (indicated in A) was determined by qRT-PCR from three independent gradients. Error bars indicate SD from at least three independent analyses. (**) p < 0.005; (ns) _p_ > 0.05.
DISCUSSION
In conclusion, our studies indicate that IGF2BP1 is essential to promote c-myc mRNA stabilization via the CRD in complex with at least four additional proteins: HNRNPU, SYNCRIP, YBX1, and DHX9. Each of these proteins associates with IGF2BP1 in a CRD-dependent manner and is essential to prevent CRD-mediated degradation of the c-myc mRNA. Hence, although various RNA-binding proteins associate with IGF2BP1 in an RNA-dependent manner, only subsets of these proteins appear essential for regulating the fate of specific target transcripts of IGF2BP1. Interestingly, regulatory proteins of these transcript-specific subsets do not necessarily act through the same cis determinant, since, for example, IGF2BP1, HNRNPU, SYNCRIP, DHX9, and YBX1 stabilize the c-myc mRNA via the CRD, versus ELAVL1 promoting stability via the 3′-UTR and potentially STAU1, which presumably promotes c-myc mRNA translation through the 5′-UTR. Despite these distinct correlations of function, factor, and cis determinant, IGF2BP1-associated proteins apparently associate in cytoplasmic mRNPs, but the “molecular architecture” of each mRNP as well as the number of distinct mRNPs remain elusive. Nonetheless, it appears likely that each component of the c-myc CRD-associated regulatory complex is essential, since stabilization of the c-myc mRNA is abrogated upon the knockdown of each factor, and combined knockdown does not yield additive destabilization of c-myc CRD-containing transcripts. Moreover, the identified regulatory factors associated with IGF2BP1 in a CRD-dependent manner in vitro. However, whether proteins identified to control c-myc mRNA turnover via the CRD facilitate their role via distinct RNA-binding motifs in the CRD or require IGF2BP1 as a scaffolding factor remains to be elucidated.
Previous reports suggested that IGF2BP1 prevents c-myc mRNA degradation by hindering endonucleolytic cleavage at the CRD when ribosomes are stalled by entering a region of rare codons at the beginning of the CRD (Lemm and Ross 2002; Sparanese and Lee 2007). This suggested a translation-coupled decay of the c-myc mRNA via the CRD, an assumption supported by the finding that CRD-directed endonucleolytic activity is associated with polyribosomes (Bergstrom et al. 2006; Tafech et al. 2007). In agreement, we demonstrate that the increase of CRD-mediated RNA decay upon IGF2BP1 knockdown correlates with altered c-myc mRNA ratios between the polysomal and non-polysomal fraction. This suggests that the IGF2BP1-mediated association of cellular c-myc mRNA with the mRNP fraction appears essential to prevent accelerated translation-coupled decay of the c-myc mRNA. Hence, association of the c-myc mRNA with the mRNP fraction via the CRD and IGF2BP1 appears to be the rate-limiting step in controlling CRD-mediated decay of the c-myc mRNA. Nonetheless, IGF2BP1 and some of the associated protein factors involved in controlling c-myc mRNA turnover via the CRD are also found in “light” polysomal fractions. Hence, one could hypothesize that CRD-associated regulatory complexes stay at least partially associated with the c-myc mRNA until the first ribosomes enter the rare codon region at the beginning of the CRD, as previously proposed for IGF2BP1 (Lemm and Ross 2002). By staying associated with the CRD into the first round of translation, the regulatory complex could thus shield the CRD from endonucleolytic attack until elongation proceeds upon loading of ribosomes with rare codon complementary tRNAs. Summarized, one would therefore expect that stability control of the c-myc mRNA via the CRD relies on two sequential events:
- The c-myc mRNA is associated with IGF2BP1-containing cytoplasmic mRNPs in which the transcript is shielded from endonucleolytic attack at the CRD.
- During the first round of translation, a regulatory protein complex associated with the CRD could prevent endonucleolytic attack at the CRD.
Hence, to efficiently prevent accelerated decay of the c-myc mRNA, its amount in the actively translating fraction should be limited proportionally to the availability of loaded tRNAs fitting the rare-codon requirements at the beginning of the CRD. As discussed, association of the c-myc mRNA with cytoplasmic mRNPs via IGF2BP1 and associated proteins could provide the regulatory mechanism to balance this supply/demand ratio. Support for this assumption is also provided by the finding that the de novo synthesis of IGF2BP1 in malignant neoplasias is correlated with elevated c-myc mRNA levels in tumor-derived cells (Kobel et al. 2007). Hence, IGF2BP1-facilitated association of the c-myc mRNA with cytoplasmic mRNPs is likely to provide a mechanism for sustaining the elevated c-Myc expression observed in various tumors.
MATERIALS AND METHODS
Cell culture and transfection
Human osteosarcoma-derived U2OS and human embryonic kidney HEK293 cells were grown in DMEM medium supplemented with 10% fetal bovine serum (FBS). Stable cell lines were generated by transfection of expression vector pcDNA3.1/Zeo (Invitrogen) containing the Flag-IGF2BP1 construct into U2OS cells. Stable clones were selected by Zeocin at 400 μg/mL. U2OS cells stably expressing Flag-IGF2BP1 were cultured at 50 μg/mL Zeocin. U2OS and HEK293 were transiently transfected with DNA or siRNA at ∼80% confluence with Lipofectamine 2000, essentially as previously described (Stohr et al. 2006; Kobel et al. 2007). For DNA transfection, a 2:1 Lipofectamine (μL)/DNA (μg) ratio was used. For the transfection of siRNAs, a 1:50 Lipofectamine (μL)/RNA (pmol) ratio was used. For sequential transfection, cells were transfected with siRNAs for 48 h before transfection with reporter plasmids for 24 h.
Plasmids and siRNAs
The human ZBP1 (IGF2BP1) was cloned by RT-PCR from HepG2 cells and subcloned in pcDNA3.1/Zeo (Invitrogen) equipped with a Flag-M2 epitope tag. The luciferase reporter plasmids and the RNAi-insensitive IGF2BP1 mutant were as previously described (Hüttelmaier et al. 2005; Kobel et al. 2007). siRNAs were designed using the Dharmacon siDESIGN Center and diluted to a final concentration of 50 pmol (Supplemental Table S1). The efficiency of each siRNA was validated based on Western blotting and quantitative RT-PCR (Figs. 2, 3B; Supplemental Figs. S3, S4).
Immunopurification
U2OS cells were washed with TapI buffer (10 mM HEPES at pH 7.6, 10 mM KCl, 2 mM MgCl2) before lyses in ice-cold TapII buffer (10 mM HEPES at pH 7.6, 3 mM MgCL2, 10 mM KCl, 5% glycerol, 0.5% NP-40) supplemented with Protease Inhibitor Cocktail (Sigma Aldrich), 50 U/mL RNasin (Promega), or 10 μg/mL RNaseA where indicated. Lysates (volume ratio 1:5 with Tap II) were dounced 20 times and incubated for 10 min on ice. Nuclei and cell debris were pelleted upon centrifugation at 3000_g_ for 10 min at 4°C; pellets were resuspended in TAPII buffer (volume ratio 1:5) and centrifuged again at 3000_g_ for 10 min at 4°C. Supernatants of both extractions were combined, adjusted to 150 mM KCl, and cleared at 13,000_g_ for 10 min at 4°C. Immunopurification of Flag-IGF2BP1 was performed by using 50 μL of antiFlagM2 agarose beads (100 μL of a 1:1 slurry on 5 mL of cytoplasmic extract at ∼4–5 mg/mL in Tap-II adjusted to 150 mM KCl) and extensive washing with TapII buffer adjusted to 150 mM KCl. Proteins were eluted with 0.2 M glycine (pH 2.5) for 20 min on ice. For Western blot analyses, immunopurification was performed by using ∼25 μL of 1:1 slurry of anti-FlagM2 agarose on 1 mL of cytoplasmic lysate.
Mass spectrometry
Protein bands were excised from Coomassie-stained SDS-gels, washed, and digested with trypsin (Promega) in 30 μL of 10 mM ammonium bicarbonate (pH 8.0) overnight at 37°C. Tryptic peptides were extracted, and liquid chromatography–tandem mass spectrometry (LC/MS/MS) analyses were performed using a Micromass CapLC column 75-mm, 3 15-cm, 3-mm dC18 Atlantis NanoEase (Waters) coupled to a quadrupole time-of-flight mass spectrometer (Q-TOF2; Micromass). Peptide fragment ion spectra were searched against the nonredundant protein database for exact matches using the MASCOT program (http://www.matrixscience.com).
Semiquantitative Western blotting
Protein extracts were isolated from cells transfected with siRNAs for 72 h. Cells were lysed in ice-cold RIPA buffer. Protein concentrations of each lysate were determined using a DC protein assay (Bio-Rad). For semiquantitative analyses, 20 and 30 μg of total protein of each sample was analyzed per lane. Western blotting was performed using the indicated primary antibodies and IRDye 800CW-labeled mouse and/or IRDye 680-labeled rabbit antibodies, respectively (Supplemental Table S3). Signal intensities were determined by an Odyssey infrared Imaging System (LI-COR Biosciences). The change of relative c-Myc protein levels at each concentration of total protein loaded was quantified by internal cross-normalization to TUBA4A and VCL protein levels from the same blot. For each experiment, two amounts of total protein (20 and 30 μg of total protein) were analyzed per sample. Each knockdown analysis was performed five times to allow for statistical analyses by Student's _t_-test.
Gradient analysis
U2OS cells were incubated with cycloheximide at 100 μg/mL for 15 min before lysis and clearing in TapII buffer supplemented with cycloheximide and RNasin. Total protein concentration, using DC protein assay (Bio-Rad), and OD260 of the supernatants were determined to allow loading of gradients with equal amounts of protein and RNA. Linear 15%–45% (w/v) sucrose gradients in TapII buffer (lacking NP-40 and supplemented with cycloheximide and RNasin) were centrifuged in a Beckman SW40 rotor at 30,000 rpm for 2 h. Gradients were fractionated, and OD260 was recorded for each 400-μL fraction before the isolation of total protein by TCA precipitation and RNA isolation by Trizol, respectively. RNA levels in each fraction were determined by quantitative RT-PCR. For determining the total number of transcripts, standard curves were recorded in each experiment, and where indicated, the relative amount of RNA in each fraction was calculated based on the total number of transcripts determined in the gradient.
Quantitative RT-PCR and mRNA decay analyses
Cells transfected with indicated siRNAs for 72 h were directly harvested (mRNA steady-state level) or treated with 5 μM Actinomycin D (mRNA decay) and harvested at indicated time points. Total RNA was isolated by Trizol and DNAse-treated for the analysis of reporter mRNA levels. Random primed reverse transcription of RNA (1.5 μg) was performed by MMLV-RT (Promega). Quantitative PCRs were performed using a 2xTaq-Master Mix (Promega) supplemented with SYBRGreen in an MX3000p cycler (Stratagene). PCR primers were designed based on the Primer3 software and validated by analyzing the melting peaks (Supplemental Table S2). Changes in c-myc or reporter mRNA levels were determined by the ΔΔCt method using PPIA, RPLP0, and 28S RNAs for internal cross-normalization. All data were analyzed from at least three independent experiments, and statistical significance was validated by Student's _t_-testing.
Luciferase reporter assay
HEK293 cells were transfected with siRNAs for 48 h before the transfection of Firefly and Renilla plasmids (1:1 ratio) for 24 h. Luciferase activities were determined by DualGlo (Promega) as previously described (Stohr et al. 2006). Firefly-luciferase activities were normalized by _Renilla_-luciferase activities (normalized activity). To determine the change of luciferase activity for each reporter, the normalized activity of the control sample, transfected with control siRNAs, was set to 100% and compared to samples transfected with siRNAs directed against indicated factors. Note that the sequential transfection of siRNAs and reporter plasmids results in reduced effects due to inefficient double-transfection efficiency.
In vitro complex formation
HEK293 cells were lysed in ice-cold TapII buffer supplemented with CaCl2 (2 mM), and protein extracts were stored at −80°C at 4 mg/mL total protein. Where indicated, 100 μL of extracts were treated with Micrococcal nuclease for 20 min at 37°C before the nuclease was deactivated with EGTA (5 mM) and in vitro transcribed RNA (5 μg) was added. Endogenous IGF2BP1 or other factors were immunopurified upon 1:10 dilution of the extract with Tap II buffer adjusted to 150 mM KCl and the addition of 10 μg/mL yeast t-RNA. Co-purified proteins were analyzed by Western blotting, and co-purified RNAs were analyzed upon Trizol-based extraction of the pellet and supernatant fraction by semiquantitative RT-PCR.
In vitro transcription
The luciferase open reading frame and the CRD (the last 249 nt of the human c-myc coding sequence) template RNAs were in vitro transcribed using the MAXIscript Kit (Ambion).
Immunofluorescence
U2OS cells were grown on coverslips, fixed, and processed for indirect immunostaining as previously described using the primary and secondary antibodies indicated in Supplemental Table S3 (Stohr et al. 2006). All image acquisition was performed on a TCS SP5 LSM-microscope (Leica) using a 60× objective (Plan Apo 60×; Leica) and standardized settings for sequential imaging. The colocalization rate was quantified using the colocalization tool provided by the LAS-AF software (Leica) under the following conditions and calculations: threshold 50%, background 50%, colocalization rate [%] = Area colocalization/Area foreground, Area foreground = Area image − Area background. For each counterstaining, 50 cells of two independent experiments were analyzed. Counterstaining for anti-IGF2BP1 by a polyclonal and a monoclonal antibody was used as a positive control for “maximal” colocalization.
The monoclonal anti-IGF2BP1 antibody used was raised by the BSBS antibody facility (TU Braunschweig) against recombinant IGF2BP1, purified as previously described (Hüttelmaier et al. 2005).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank S. Behrens, A.D. Ostareck-Lederer, and M. Kiebler for providing antibodies. This work was supported by DFG funding to S.H.
Footnotes
REFERENCES
- Bergstrom K., Urquhart J.C., Tafech A., Doyle E., Lee C.H. Purification and characterization of a novel mammalian endoribonuclease. J. Cell. Biochem. 2006;98:519–537. doi: 10.1002/jcb.20726. [DOI] [PubMed] [Google Scholar]
- Brewer G., Ross J. Poly(A) shortening and degradation of the 3′ A+U-rich sequences of human c-myc mRNA in a cell-free system. Mol. Cell. Biol. 1988;8:1697–1708. doi: 10.1128/mcb.8.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer G., Ross J. Regulation of c-myc mRNA stability in vitro by a labile destabilizer with an essential nucleic acid component. Mol. Cell. Biol. 1989;9:1996–2006. doi: 10.1128/mcb.9.5.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagnovich D., Cohn S.L. Binding of a 40-kDa protein to the N-myc 3′-untranslated region correlates with enhanced N-myc expression in human neuroblastoma. J. Biol. Chem. 1996;271:33580–33586. doi: 10.1074/jbc.271.52.33580. [DOI] [PubMed] [Google Scholar]
- DeMaria C.T., Brewer G. AUF1 binding affinity to A+U-rich elements correlates with rapid mRNA degradation. J. Biol. Chem. 1996;271:12179–12184. doi: 10.1074/jbc.271.21.12179. [DOI] [PubMed] [Google Scholar]
- Dugre-Brisson S., Elvira G., Boulay K., Chatel-Chaix L., Mouland A.J., DesGroseillers L. Interaction of Staufen1 with the 5′ end of mRNA facilitates translation of these RNAs. Nucleic Acids Res. 2005;33:4797–4812. doi: 10.1093/nar/gki794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvira G., Wasiak S., Blandford V., Tong X.K., Serrano A., Fan X., del Rayo Sanchez-Carbente M., Servant F., Bell A.W., Boismenu D., et al. Characterization of an RNA granule from developing brain. Mol. Cell. Proteomics. 2006;5:635–651. doi: 10.1074/mcp.M500255-MCP200. [DOI] [PubMed] [Google Scholar]
- Hüttelmaier S., Illenberger S., Grosheva I., Rudiger M., Singer R.H., Jockusch B.M. Raver1, a dual compartment protein, is a ligand for PTB/hnRNPI and microfilament attachment proteins. J. Cell Biol. 2001;155:775–786. doi: 10.1083/jcb.200105044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttelmaier S., Zenklusen D., Lederer M., Dictenberg J., Lorenz M., Meng X., Bassell G.J., Condeelis J., Singer R.H. Spatial regulation of β-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438:512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- Jonson L., Vikesaa J., Krogh A., Nielsen L.K., Hansen T.V., Borup R., Johnsen A.H., Christiansen J., Nielsen F.C. Molecular composition of IMP1 RNP granules. Mol. Cell. Proteomics. 2007;6:798–811. doi: 10.1074/mcp.M600346-MCP200. [DOI] [PubMed] [Google Scholar]
- Kanai Y., Dohmae N., Hirokawa N. Kinesin transports RNA: Isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Kobel M., Weidensdorfer D., Reinke C., Lederer M., Schmitt W.D., Zeng K., Thomssen C., Hauptmann S., Hüttelmaier S. Expression of the RNA-binding protein IMP1 correlates with poor prognosis in ovarian carcinoma. Oncogene. 2007;26:7584–7589. doi: 10.1038/sj.onc.1210563. [DOI] [PubMed] [Google Scholar]
- Lemm I., Ross J. Regulation of c-myc mRNA decay by translational pausing in a coding region instability determinant. Mol. Cell. Biol. 2002;22:3959–3969. doi: 10.1128/MCB.22.12.3959-3969.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔC(T) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Manohar C.F., Short M.L., Nguyen A., Nguyen N.N., Chagnovich D., Yang Q., Cohn S.L. HuD, a neuronal-specific RNA-binding protein, increases the in vivo stability of MYCN RNA. J. Biol. Chem. 2002;277:1967–1973. doi: 10.1074/jbc.M106966200. [DOI] [PubMed] [Google Scholar]
- Nielsen J., Christiansen J., Lykke-Andersen J., Johnsen A.H., Wewer U.M., Nielsen F.C. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol. Cell. Biol. 1999;19:1262–1270. doi: 10.1128/mcb.19.2.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noubissi F.K., Elcheva I., Bhatia N., Shakoori A., Ougolkov A., Liu J., Minamoto T., Ross J., Fuchs S.Y., Spiegelman V.S. CRD-BP mediates stabilization of βTrCP1 and c-myc mRNA in response to β-catenin signaling. Nature. 2006;441:898–901. doi: 10.1038/nature04839. [DOI] [PubMed] [Google Scholar]
- Sparanese D., Lee C.H. CRD-BP shields c-myc and MDR-1 RNA from endonucleolytic attack by a mammalian endoribonuclease. Nucleic Acids Res. 2007;35:1209–1221. doi: 10.1093/nar/gkl1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohr N., Lederer M., Reinke C., Meyer S., Hatzfeld M., Singer R.H., Hüttelmaier S. ZBP1 regulates mRNA stability during cellular stress. J. Cell Biol. 2006;175:527–534. doi: 10.1083/jcb.200608071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartwout S.G., Kinniburgh A.J. c-myc RNA degradation in growing and differentiating cells: Possible alternate pathways. Mol. Cell. Biol. 1989;9:288–295. doi: 10.1128/mcb.9.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafech A., Bennett W.R., Mills F., Lee C.H. Identification of c-myc coding region determinant RNA sequences and structures cleaved by an RNase1-like endoribonuclease. Biochim. Biophys. Acta. 2007;1769:49–60. doi: 10.1016/j.bbaexp.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Vikesaa J., Hansen T.V., Jonson L., Borup R., Wewer U.M., Christiansen J., Nielsen F.C. RNA-binding IMPs promote cell adhesion and invadopodia formation. EMBO J. 2006;25:1456–1468. doi: 10.1038/sj.emboj.7601039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaniv K., Yisraeli J.K. The involvement of a conserved family of RNA binding proteins in embryonic development and carcinogenesis. Gene. 2002;287:49–54. doi: 10.1016/s0378-1119(01)00866-6. [DOI] [PubMed] [Google Scholar]
- Yisraeli J.K. VICKZ proteins: A multi-talented family of regulatory RNA-binding proteins. Biol. Cell. 2005;97:87–96. doi: 10.1042/BC20040151. [DOI] [PubMed] [Google Scholar]