Estrogen directly activates AID transcription and function (original) (raw)
Abstract
The immunological targets of estrogen at the molecular, humoral, and cellular level have been well documented, as has estrogen's role in establishing a gender bias in autoimmunity and cancer. During a healthy immune response, activation-induced deaminase (AID) deaminates cytosines at immunoglobulin (Ig) loci, initiating somatic hypermutation (SHM) and class switch recombination (CSR). Protein levels of nuclear AID are tightly controlled, as unregulated expression can lead to alterations in the immune response. Furthermore, hyperactivation of AID outside the immune system leads to oncogenesis. Here, we demonstrate that the estrogen–estrogen receptor complex binds to the AID promoter, enhancing AID messenger RNA expression, leading to a direct increase in AID protein production and alterations in SHM and CSR at the Ig locus. Enhanced translocations of the c-myc oncogene showed that the genotoxicity of estrogen via AID production was not limited to the Ig locus. Outside of the immune system (e.g., breast and ovaries), estrogen induced AID expression by >20-fold. The estrogen response was also partially conserved within the DNA deaminase family (APOBEC3B, -3F, and -3G), and could be inhibited by tamoxifen, an estrogen antagonist. We therefore suggest that estrogen-induced autoimmunity and oncogenesis may be derived through AID-dependent DNA instability.
Humoral immune responses triggered by foreign antigens require B cell activation. The activated B cell undergoes antibody affinity maturation, which in higher vertebrates includes somatic hypermutation (SHM), gene conversion of antigen-binding V regions, and class switching (1); all of these processes require activation-induced deaminase (AID) (2–5). AID initiates these events by deaminating deoxycytosine to deoxyuracil in DNA (for review see reference [6–8]). The resulting dU:dG lesion can be recognized by several different DNA repair pathways to create the aforementioned antibody diversifications. This necessity for immune diversification and sufficiency for genome instability also highlights AID as an important pathogenic regulator. In the immune system, hyper- or hypoexpression of AID can alter autoimmune pathologies (9–11). Furthermore, SHM, by way of AID, may contribute to lymphomagenesis by mutating (proto-)oncogenes and tumor suppressor genes, or by promoting chromosomal translocations (12–14). There is strong evidence that AID is required for c-myc translocation, leading to tumorgenesis in a murine model for Burkitt's lymphoma (15, 16). Other nonphysiological AID targets include BCL6, CD95/Fas, RHO/TTF, PAX-5, and PIM1 (12, 17–19). Outside the immune system, there are indications that systemic hyperexpression of AID can induce non–B cell cancers from lung (20), lymphatic (20), and liver (21) tissues.
AID has also been implicated as a developmental epigenetic reprogramming factor, and its expression levels in oocytes is almost equivalent to that in lymph nodes (22), suggesting that AID could be regulated by pathways other than B cell activation pathways (e.g., E-box proteins [23], NF-κB [24], and Pax5 [25]), with hormones being plausible candidates.
Several clinical and epidemiological studies have indicated that females can have stronger and more rapid immune responses upon antigen encounter (26, 27). This gender bias is also reflected in the occurrence of pathogenic immune responses, as found in asthma and other autoimmune diseases (28–31). Several nonimmune pathologies are also strongly influenced by the activity of sex hormones, most notably certain types of cancer. Estrogen and its biological and synthetic derivatives are thought to be oncogenic for breast and ovarian tissue (32, 33), most often being associated with their growth-promoting and differentiating capacity.
To further elucidate how AID can be regulated, both within and outside the immune system, and to determine which signaling pathways could use DNA deaminases as DNA instability factors, we analyzed the effect of estrogen on AID's expression and on downstream pathways such as SHM and class switch recombination (CSR). We show that AID can be up-regulated by estrogen, whereas tamoxifen (Tam) can inhibit this stimulation. This effect was most pronounced, but not limited to regulation at the level of transcription. Treatment with estrogen increased AID protein expression, enhanced CSR, augmented mutation frequency in Ig and non-Ig genes, and increased the translocation frequency of c-myc. Estrogen-induced AID messenger RNA (mRNA) production was independent of other B cell stimulatory pathways and could be observed outside immune tissue. We were able to identify two potential estrogen response elements (EREs) near the AID promoter, and determined enhanced ERα binding to the promoter after estrogen treatment in vitro and in vivo. APOBEC3, the evolutionarily related DNA deaminases (34), were also responsive to estrogen treatment in different tissues and cell types. Our data indicate that the mutagenic DNA deaminases are potentially an important target for hormonal regulation.
RESULTS
Differential effect of sex hormones on AID mRNA
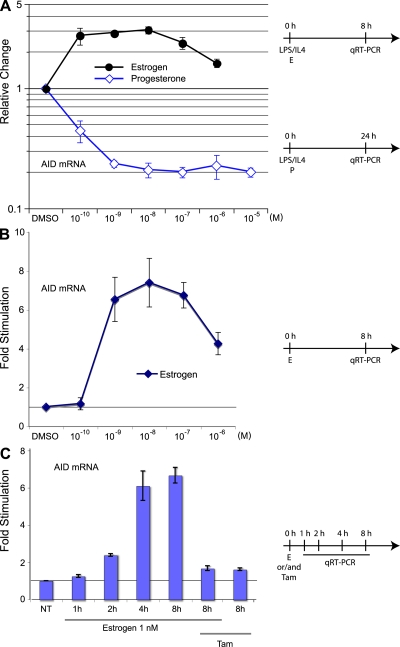
Hormones such as estrogen and progesterone exert their biological effects through binding to their intracellular receptors and, upon entering the nucleus, act as transcription factors (35). To determine the effects of hormones on AID expression, we stimulated isolated murine splenic B cells with IL-4 and LPS, which are known to induce AID (24), while adding physiological amounts of progesterone and estrogen. Progesterone addition reduced AID mRNA levels by fivefold, as revealed by quantitative real-time PCR (qRT-PCR; Fig. 1 A). This and all subsequent qRT-PCR analyses were normalized to GAPDH expression, and all enhancements or repressions were analyzed as relative changes to DMSO. To observe a stimulatory effect of estrogen, cells were only treated for a short time period (8 h) rather than the usual 24–48 h. This was done to avoid a possible maximal induction of LPS/IL-4 caused by longer treatment. In contrast to progesterone, physiological amounts of estrogen were able to enhance AID expression threefold in these cells (Fig. 1 A). This antithetical effect of estrogen and progesterone indicated that AID gene regulation is embedded within a systemic sex hormone pathway.
Figure 1.
The effects of estrogen and progesterone on AID mRNA in murine splenic B-cells. (A) AID mRNA in response to estrogen and progesterone treatment in stimulated B cells. Isolated mouse spleen B cells were stimulated with LPS and IL-4 and treated with different physiological concentrations of estrogen for 8 h or progesterone for 24 h. Unless indicated, DMSO is set to 1, and treatments are represented as relative change to DMSO. (B) AID mRNA in response to estrogen treatment in unstimulated B cells after 8 h treatment with physiological concentrations of estrogen. (C) AID mRNA induction upon different treatment. Cells were treated with 1 nM estrogen and/or 50 nM Tam (Tam) for up to 8 h. DMSO at 0 h is set to 1. All qRT-PCR data are representative of three independent experiments, and error bars indicate standard deviations from the mean. Timelines of cell treatments are indicated next to the graphs. NT, not treated. For A and B, absolute values as compared with GAPDH mRNA are shown in Fig. S10, available at http://www.jem.org/cgi/content/full/jem.20080521/DC1.
We focused our subsequent experiments on the verification that the estrogen-induced stimulation on AID was analogous to another known estrogen response gene. Because we could determine that the expression of gene regulated in breast cancer 1 (a known estrogen response gene) (36) had a similar stimulation profile in B cells (unpublished data), it seemed likely that the AID gene could also be activated in uninduced B cells with estrogen. Stimulation of isolated splenic B cells with physiological amounts of estrogen produced a sevenfold increase in AID mRNA (Fig. 1 B). This induction began to plateau at 4 h, with the earliest indication of an increase detectable at ∼2 h (Fig. 1 C).
In most systems, the synthetic hormone Tam acts as an antagonist to estrogen stimulation, presumably by binding to the estrogen receptor (ER) and altering its DNA binding capacity (37). The presence of Tam during the estrogen treatment of isolated splenic B cells inhibited the stimulatory activity, whereas Tam on its own had only a limited effect on AID mRNA expression at the concentration used (Fig. 1 C, Estrogen 1 nM/Tam and Tam, respectively). Interestingly, at very low concentrations, Tam can have a stimulatory effect on AID mRNA (Fig. S1 A, available at http://www.jem.org/cgi/content/full/jem.20080521/DC1), which may reflect Tam's agonistic activity (see Discussion).
Hormonal regulation of AID mRNA is predominantly via transcription
Although we were able to observe an increase in AID mRNA at 2 h (Fig. 1), we needed to determine if this regulation was direct or indirect. We pretreated splenic B cells with the translational inhibitor cycloheximide (CHX), followed by stimulation with estrogen, and observed an increase in mRNA production equivalent to treatment without inhibitor (Fig. S1 B). This suggested that the effect of estrogen was directly mediated on AID's mRNA synthesis. Because qRT-PCR of cDNA is a readout of steady-state mRNA, we also tested if the increase by estrogen was caused by transcription or mRNA metabolism. Treatment of cells with transcription inhibitors (actinomycin D [ACT] and α-amanitin [AMA]) abrogated the effect of estrogen, indicating that this alteration in AID's mRNA was not caused by message stability (Fig. S1 B). As estrogen can affect pre-mRNA to mRNA processing (38), we designed qRT-PCR primers to span the complete transcription unit of AID (Fig. S2 A, available at http://www.jem.org/cgi/content/full/jem.20080521/DC1). When we compared the relative change in expression of the various pre-mRNA's exons and introns to that of the mature mRNA, we found only a minor effect caused by estrogen treatment (the 5′-most PCR unit, <100 bp away from the start of AID gene, was up-regulated to almost the same extent as the mature mRNA [Fig. S2 B]). The intron between exon 3 and 4 (7,538–7,689 bp) showed a marginal increase in response to estrogen over that of the mature RNA, indicating a potential region for estrogen-induced splicing of AID RNA. Because the relative change did not significantly alter the overall effect, we did not pursue this analysis further, although recent data suggests that alternative splicing may influence AID expression (39). The experiments substantiate the notion that estrogen's main mode of action on AID is through transcriptional regulation and not mRNA metabolism.
Identification of hormone response elements in the AID promoter regions
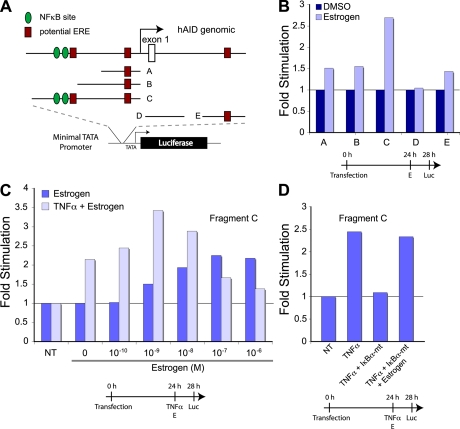
The rapid effect of the hormones on AID message via transcription suggested that the AID gene is a direct target for hormonal regulation. Using bioinformatic analysis (Fig. 2 A), we were able to identify putative EREs in the context of other response elements, such as NF-κB. We dissected the 1.5 kb upstream and the 2 kb downstream of the ATG regions for hormone-responsive elements in a heterologous transcription assay. The potential response regions were placed into a luciferase reporter construct and transfected into human SiHa cells, followed by treatment with the indicated hormone (or cotransfected with expression plasmids), and then analyzed for luciferase activity. As we were primarily interested in the effect of hormones on expression, we used relative change as a readout rather than absolute values, which provided a more direct evaluation of the hormone treatment but potentially obscured the individual effect of the various DNA elements. When compared with DMSO treatment, estrogen responsiveness was most significant with Fragment C, indicating that this contained the predominant estrogen-responsive DNA element. Comparable to the mRNA production of AID in B cells, Fragment C also responded in a dose-dependent manner to estrogen (Fig. 2 C). Aside from the putative ERE, Fragment C also harbored the two published NF-κB binding sites (24). As indicated by the qRT-PCR analysis in Fig. 1 A, estrogen and the LPS/IL-4–induced NF-κB stress-response pathway could act synergistically on AID mRNA production. To more directly stimulate the NF-κB pathway in SiHa cells, we used the cell-autonomous activator TNF-α (Fig. 2 C). Interestingly, aside from the synergy (e.g., 10−9 M), the two maxima of the dose response were offset (TNF-α treated, 10−9 M; untreated, 10−7 M), indicating a higher complexity of the two interacting pathways. To demonstrate independence of the two pathways, we analyzed the response of Fragment C to treatment with TNF-α and estrogen upon cotransfection of the dominant-negative mutant of IκBα (IκBα S32A/S36A dominant mutant [IκBα-mt]), which is known to inhibit the release of NF-κB from the cytoplasm into the nucleus after stimulation. As shown in Fig. 2 D, the TNF-α activation was inhibited in the presence of IκBα-mt, yet estrogen was able to independently activate the transcription. This indicated that in the AID promoter, the NF-κB site and its proximal ERE could act independently as well as synergistically.
Figure 2.
Human AID promoter analysis for hormone response elements. (A) Schematic representation of potential EREs (square) and NF-κB sites (circle) and their respective locations in the human promoter. The indicated promoter regions (marked A–E) were inserted into a luciferase reporter construct with a minimal promoter. The vectors were transfected into SiHa cells, incubated for 24 h, treated for 4 h with hormones or TNF-α, and analyzed for luciferase activity. (B) Relative luciferase activity after estrogen treatment. Cells were transfected with constructs containing AID promoter fragments and treated with estrogen for 4 h. (C) Effect of TNF-α and estrogen on the human AID promoter. Expression construct with an AID promoter region containing NF-κB sites and putative ERE (Fragment C) were transfected into cells, followed by TNF-α and/or estrogen treatment for 4 h. (D) Estrogen can act independently from NF-κB. Cells were cotransfected with Fragment C and an IκBα-mt expression vector. After 24 h, cells were treated with TNF-α and/or 100 nM estrogen for 4 h. Timelines of cell treatments are indicated below the graphs. NT, not treated.
ERE binding in B cell extracts
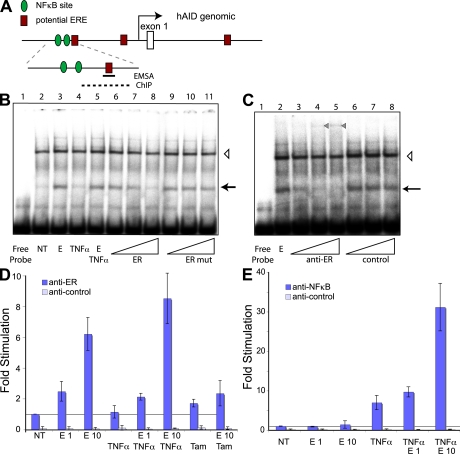
The transient transfection assay indicated that a predicted ERE was subject to estrogen regulation. Thus, we analyzed ER binding to the AID promoter and focused on the more widely expressed receptor subtype ERα. To analyze the binding of ER to parts of the AID promoter in vitro, we prepared nuclear extracts from treated and untreated cells and performed electromobility shift assays (EMSA). For the biochemical analysis of the ER binding to the ERE, we focused on NF-κB proximal ERE of Fragment C. A 34-bp fragment containing the 5′-most proposed ER binding site was incubated with untreated and treated extract (Fig. 3 B, lanes 2 and 3). The estrogen treatment clearly induced a protein that could bind the fragment (arrow), which was not induced when treating the B cells with TNF-α before extract preparation (lane 4). Cotreatment with estrogen and TNF-α had the same effect as estrogen alone (Fig. 3 B, lane 5), indicating that the two pathways act through different nuclear proteins. Competition experiments with the ER binding site (Fig. 3 B, lanes 6–8) or a mutation of the proposed ER site (Fig. 3 B, lanes 9–11) showed a specific competition, indicating ER-like binding kinetics. Using antibodies to the DNA-binding domain of ERα strongly inhibited the formation of the estrogen-induced band (Fig. 3 C, lanes 2 vs. 3–5), but the shift was unaffected by a control antibody (Fig. 3 C, lanes 2 vs. 6–8, arrow). The anti-ERα antibody did induce the appearance of a high molecular weight complex (Fig. 3 C, triangle in lanes 4 and 5), which could be caused by either the supershift of a dimerized ERα or heterodimer ERα/ERβ; however, this needs to be analyzed further.
Figure 3.
Identification of ER binding to human AID promoter by EMSA and ChIP. (A) Schematic representation of human AID promoter region (as in Fig. 2 A). The position of the oligonucleotide used for EMSA and the region amplified by qRT-PCR for ChIP are marked as a black line and a dashed line, respectively. (B) Estrogen (denoted as E) -induced oligonucleotide shift (marked with an arrow) in Ramos nuclear extracts. Cells were treated for 72 h in hormone-depleted serum, followed by 4-h treatment with 10 nM estrogen (lanes 3 and 5–11) and/or TNF-α (lanes 4 and 5), and nuclear extract preparation. Different concentrations of unlabeled competitors ER (lanes 6–8) and mutated ER mut (lanes 9–11) were added to the binding reaction. Open triangle, nonspecific DNA binding band. (C) EMSA with anti-ERα antibodies. Increasing concentrations of anti-ERα antibody and a nonspecific antibody were added to the binding reaction (see Materials and methods). The estrogen-induced band is marked with an arrow, and a super-shifted band appearing upon anti-ERα antibody addition is marked with a closed triangle. Open triangle, nonspecific DNA-binding band. (D) ERα binds to upstream region of human AID promoter. Cells were treated as in B. Data are representative of three independent experiments and error bars indicate standard deviations from the mean. ChIP was performed using anti-ERα or control antibodies, and the bound DNA was subjected to qRT-PCR. Estrogen and Tam treatments are marked with E1 (estrogen 1 nM), E10 (estrogen 10 nM), and Tam, respectively. (E) Estrogen can cooperate with TNF-α in recruiting NF-κB to AID promoter. ChIP is as in D, using anti–NF-κB or control antibodies. NT, not treated.
Because estrogen and TNF-α co-stimulation did not alter ER binding to the ERE, we wanted to determine if the reciprocal of NF-κB binding to the NF-κB site, after the combined treatment, was also unaffected. To that end, we probed the published NF-κB site (24) with our extracts. Using cold competitors (Fig. S3 B, lane 4 vs. 6–11, available at http://www.jem.org/cgi/content/full/jem.20080521/DC1), as well as anti–NF-κB antibodies (Fig. S3 C), we could demonstrate the specificity of the NF-κB binding site. As with the ER binding site, NF-κB binding was not altered by cotreatment with estrogen and TNF-α (Fig. S3 B, lanes 4 vs. 5). This indicated that the respective treatments did not alter the general DNA-binding properties of ER or NF-κB proteins. Because the distance, on the AID promoter, between the ER and NF-κB sites was larger than our EMSA probes (i.e., neither probe contained both binding sites), we could not study the effect of cooperative binding.
ER binding to AID promoter in vivo
Although the in vitro binding of the ER to the AID promoter indicated a direct binding, we also wanted to probe this interaction in vivo. To this end, we used the constitutive AID-expressing and mutating Burkitt lymphoma Ramos cells and performed chromatin immunoprecipitation (ChIP) assay, followed by qRT-PCR. Treated cells were fixed, lysed, DNA sheared, and ERα or NF-κB immunoprecipitated, and the DNA was released for PCR. As shown in Fig. 3 D, the anti-ER antibody specifically immunoprecipitated the AID promoter upon estrogen stimulation in a dose-dependent manner. A control antibody was unable to precipitate this region. Analogous to the EMSA assay, we did not detect a significant increase in ER binding to AID promoter when we co-stimulated with TNF-α, just as TNF-α treatment alone did not enhance ER binding. Again, cotreatment of cells with Tam and estrogen before ChIP did reduce the binding of ERα to the AID promoter, whereas Tam on its own did not significantly alter ERα binding (Fig. 3 D, E 10/Tam and Tam, respectively), providing further evidence for a direct binding of ERα to the AID promoter. Treatment of the cells with TNF-α increased the binding of NF-κB to the AID promoter by more than sevenfold (Fig. 3 E). Interestingly, co-stimulation of the cells with TNF-α and estrogen had a synergistic effect on the NF-κB binding; the binding increased to >30-fold above unstimulated treatment (4.5-fold above TNF-α alone).
Estrogen up-regulates AID protein production
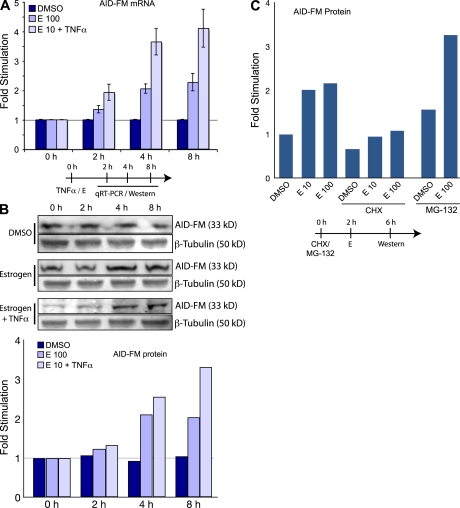
For us to determine if the effect of estrogen on AID would also extend to the protein level, we developed a quantitative approach for measuring AID protein. We generated a DT40 cell line (a cell line derived from a chicken B cell lymphoma that constitutively expresses AID and undergoes Ig diversification) to express a double tag (3xFLAG-2xTEV-3xc-Myc) fused to the C terminal exon of endogenous AID (AID FLAG-Myc–tagged AID protein [AID-FM]; Fig. S4, available at http://www.jem.org/cgi/content/full/jem.20080521/DC1). The modified DT40 express a WT AID protein and an AID-FM fusion protein, both transcribed from the endogenous AID locus. Comparing the qRT-PCR induction kinetics with that of the AID-FM expression, we determined that the steady-state levels of AID protein correlated to those of the mRNA (Fig. 4, A and B). Using the translation inhibitor CHX (Fig. 4 C), we showed that estrogen was not able to increase AID-FM expression in the absence of translation. In the presence of the proteasome inhibitor MG-132 (Fig. 4 C), estrogen increased AID-FM production above that of the MG-132 alone, indicating that estrogen was not acting on the proteasome to increase AID-FM activity. The aforementioned co-stimulatory effects of TNF-α were also observed at the level of protein production (Fig. 4, E 10/TNF-α).
Figure 4.
The effects of estrogen on AID protein in DT40. (A) Estrogen induces AID mRNA expression. AID-FM–tagged DT40 cells were treated with DMSO, 100 nM estrogen, and 10 nM estrogen with TNF-α, lysed, and analyzed for AID mRNA expression with pRT-PCR at various time-points. (B) Estrogen induces AID-FM fusion protein expression. Treatment as in A, but lysates were analyzed by quantitative Western blot. For each sample, FLAG and Tubulin expression was quantitated. The graph is derived from correlating the FLAG expression to Tubulin expression, and then determining the ratio of estrogen-induced FLAG expression to untreated DMSO samples. (C) Estrogen does not affect AID-FM fusion protein stability. Cells were incubated with CHX or MG-132 for 2 h, followed by estrogen treatment for 4 h. Protein levels were determined by quantitative Western blot. For all experiments, cells were grown in hormone depleted media for 48 h. Results are normalized to control treatments as indicated on each graph. Timelines of cell treatments are indicated below the graphs.
Hormonal regulation of CSR and SHM
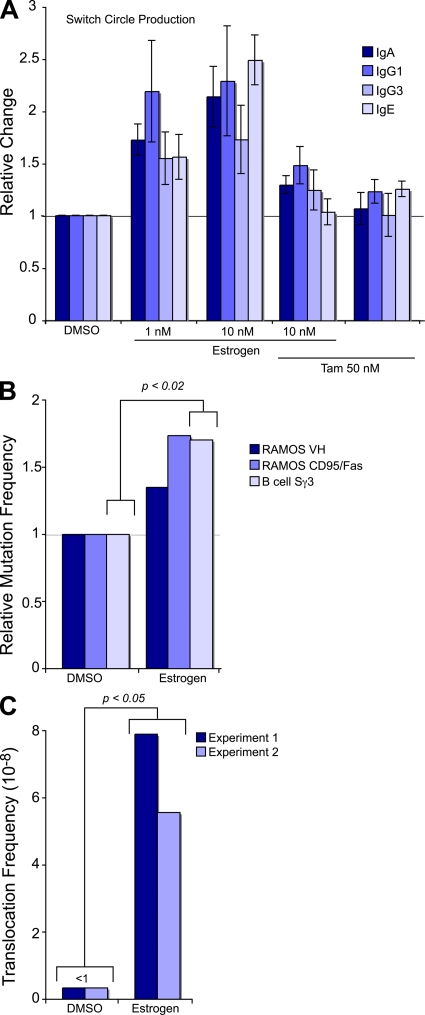
On the molecular level, CSR requires AID, yet regulation can be achieved on multiple levels, some of which may be hormonally influenced. To analyze CSR, isolated mouse splenic B cells were treated with different combinations of cytokines and hormones. B cell stimulation with LPS, LPS and IL-4, LPS and IFN-γ, or LPS and TGF-β results in switching to IgG3, IgG1, IgG2a, or IgG2b/IgA, respectively. To ensure we could correlate AID activity precisely, and to avoid possible proliferative and antiapoptotic effects of estrogen on stimulated B cells, we monitored the early molecular events of class switching. One of the molecular intermediates during class switching is the generation of a looped-out circular DNA called a switch circle (Fig. S5, available at http://www.jem.org/cgi/content/full/jem.20080521/DC1). The circular DNA contains a recombined transcription unit that produces switch circle transcripts; it is generated from the promoter of the downstream switch region and the Igμ switch region. Using a previously described qRT-PCR approach (40), we were able to show enhanced switching to IgG1, IgG3, IgA, or IgE after hormone treatment of splenic B cells (Fig. 5 A). Presumably through the increased production of AID, estrogen was able to enhance switch circle formation for all subclasses tested. As with the AID production, this link was perturbed with the addition of Tam during estrogen treatment (Fig. 5 A, Estrogen 10 nM/Tam and Tam).
Figure 5.
Hormonal effects on Ig class switching, hypermutation, and translocation. (A) Estrogen induces isotype switching. Isolated mouse splenic B cells were stimulated for 48 h with LPS + IL-4 for switching to IgG1 and IgE, LPS + TGF-β for switching to IgA, and LPS for switching to IgG3. Indicated amounts of estrogen and/or Tam were added to the cells together with cytokines. Relative efficiency of class switching was determined by detecting circle transcripts with qRT-PCR, and data are normalized to the control treatment with DMSO from three independent experiments (error bars indicate standard deviations). (B) Estrogen increases the mutation frequency in VH and CD95/Fas loci of Ramos, and in Sγ3 of splenic mouse cells. Ramos cells were grown in the presence of 100 nM estrogen for ∼20 doublings, followed by sequencing of 341 bp from human VH or 750 bp from human CD95/Fas locus. Splenic mouse cells were treated for 6 d with LPS and 10 nM estrogen, and switch gamma3 loci amplified and sequenced (Fig. S7). Mutation frequencies are normalized to the control treatments with DMSO. A standard unpaired two-tailed Student's t test showed a significant difference in mutation frequency in the Sγ3 loci of DMSO- and estrogen-treated spleen cells. (C) Estrogen enhances the c-myc/IgH translocations in splenic B cells from p53+/− mice. In each experiment, 2 spleens per sample were treated with or without 50 nM estrogen in the presence of LPS for 72 h. More than 7 × 107 cells were analyzed by long-range PCR (5 × 104 cells/PCR; Fig. S8 and Supplemental materials and methods). Frequency was determined as c-myc/IgH translocation events per cell number analyzed. Statistics was performed on the results of the pooled experiments (two-tailed, unpaired Student's t test: P = 0.026). Fig. S7, Fig. S8, and Supplemental materials and methods are available at http://www.jem.org/cgi/content/full/jem.20080521/DC1.
To determine the effect of estrogen-induced AID on SHM, we sequenced the VH region of in vitro–cultured Ramos cells, as well as the switch region of γ3 from ex vivo–stimulated splenic B cells. A 3-wk estrogen treatment of Ramos HS13 (41), which possesses a premature stop codon embedded within the AID target motif WRC, showed enhanced surface Ig expression and VH SHM (Fig. 5 B and Fig. S6, available at http://www.jem.org/cgi/content/full/jem.20080521/DC1). Because Ramos expresses constitutive amounts of AID, estrogen treatment did not enhance AID production substantially; thus, we used the ex vivo treatment of spleens as a means to detect mutations in the switch region (42). To this end, we treated LPS-activated splenic B cells with 10 nM estrogen for 6 d and sequenced the Sγ3 region. Because AID+/− mice show a haploinsufficiency effect (43, 44), we used F1 spleens from an AID−/− and BALB/c cross for analysis, hypothesizing that the estrogen effect would be more pronounced. As can be seen in Fig. 5 B and Fig. S7, there was a significant (P < 0.02) enhancement of mutation frequency in the switch region after estrogen treatment.
Enhanced non-Ig loci targeting of AID
It is known that AID can be mistargeted to non-Ig genes (12, 17–19). The effect of aberrant AID targeting can lead to somatic mutations or even translocations of protooncogenes or tumor suppressors, and subsequently to oncogenesis (16). To determine if the activity of hormones via AID can also lead to an alteration in non-Ig loci, we chose to look at the proapoptotic tumor suppressor CD95/Fas. CD95/Fas has been shown to be somatically hypermutated in human B cells, albeit 100–1,000-fold less frequently than the Ig genes (18). We analyzed the effects of estrogen on CD95/Fas mutations in Ramos HS13. As with the physiological B cell maturation events of SHM and CSR, estrogen was able to increase the potentially pathogenic mutation frequency in CD95/Fas (Fig. 5 C and Fig. S7 B). Because of the direct effect on AID function (mutation), this data provides evidence for a novel way in which estrogen can exert a direct genotoxic effect on oncogenes or tumor suppressors.
Estrogen enhances c-myc IgH translocations
The off-target effect of AID was also detected in recent work on chromosome translocations of the c-myc oncogene into the IgH locus. More importantly, the levels of AID protein directly influenced chromosome translocation frequency. This was determined from both the analysis of AID haploinsufficiency (44) and microRNA regulation of AID protein expression (45, 46). In both cases, reduced levels of AID had a direct bearing on the number of observed c-myc/IgH translocations. Using a previously described assay (16, 47), we were able to determine that estrogen treatment of isolated splenic B cells enhances c-myc/IgH translocations in p53+/− animals (Fig. 5 C and Fig. S8, available at http://www.jem.org/cgi/content/full/jem.20080521/DC1). Although, the overall frequency of translocations was low, we have not been able to observe any translocations in LPS/DMSO-treated spleen cells, yet we observed five (two in one experiment and three in another) translocations in the LPS/estrogen treatment. This highlights the importance of regulating AID protein amounts within a cell, and the potential pathogenic consequences of unregulated AID expression.
AID induction is not limited to B cells
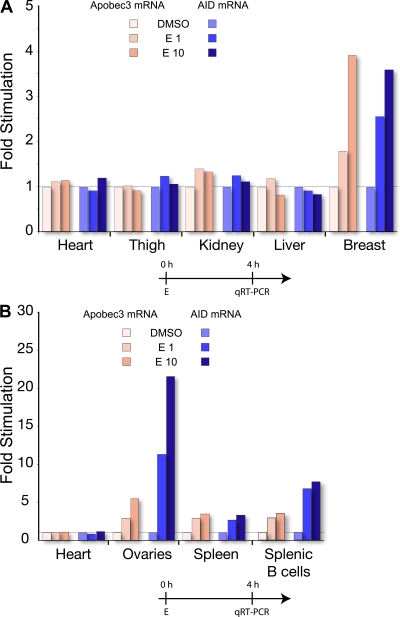
In the past, we demonstrated that AID mRNA expression is not limited to activated B cells, but can also be detected in oocytes (22). We therefore set out to determine if the increase in AID mRNA production by estrogen was also detectable in dissected tissues and various tumor cell lines. Analysis of dissected organs and tissue from mice showed AID responsiveness to hormones outside the immune system in breast and ovarian tissue (Fig. 6). As we were primarily interested in the hormone sensitivity of the transcripts, we determined the relative fold stimulation compared with DMSO, which did not reflect the absolute AID mRNA in each tissue. Interestingly, in ovaries, AID mRNA was induced almost 25-fold, which is higher than in any other organ or tissue. Because isolated oocytes produce AID mRNA (22), we suggest that the increase was predominantly caused by oocytes or other ovarian-derived tissue, rather than infiltrating lymphocytes.
Figure 6.
Estrogen induces AID and Apobec3 transcription in mouse tissue. The red and blue colors indicate the results for mApobec3 and mAID, respectively. Tissues were treated with DMSO, 1 nM estrogen (E1), or 10 nM estrogen (E10). Gene expression is normalized to the control treatments with DMSO. The tissue expression profiles represent pooled data for the respective tissues from two experiments. Timelines of cell treatments are indicated below the graphs. Absolute values as compared with GAPDH mRNA are shown in Fig. S10, available at http://www.jem.org/cgi/content/full/jem.20080521/DC1.
Gross tissue dissection can provide a good indication of possible cell types that can induce AID mRNA upon estrogen treatment, but tissue complexity could also obscure potential targets. Although we did not detect a substantial mRNA increase in hepatocyte or cervix cell lines (Fig. S9, available at http://www.jem.org/cgi/content/full/jem.20080521/DC1), we were able to show a significant increase (ranging from 2.5- to 22-fold) of AID in cell lines, including T cells, placenta, ovary, breast, and prostate. The induction observed in breast cells mimics that of the tissues isolated in Fig. 6 A, and confirms a previous report that AID has been detected in the breast cell line MCF-7 (48). Importantly, AID mRNA was not only present at basal levels but was also significantly up-regulated.
Estrogen activates APOBEC3B, 3F, and 3G mRNA transcription
AID is the ancestral member of the DNA deaminase family, and the APOBEC3 members are considered to have arisen from AID by gene duplication events (34). Within the DNA deaminase family, APOBEC3 members function predominantly in the cytoplasm and inactivate foreign DNA such as retroviruses and retrotransposable elements (49). We hypothesized that hormonal responsiveness may have been conserved among members of the APOBEC3 family. To this end, we designed qRT-PCR primers to detect APOBEC family member mRNAs in mouse and human cells. APOBEC2, a related member of the DNA deaminases without any apparent catalytic activity (34), was not affected by estrogen treatment; however, we were able to show that estrogen enhanced transcription of mouse Apobec3 from ovaries, spleen, and splenic B cells (Fig. 6). Expression of human APOBEC3B, 3F, and 3G family members was also enhanced upon estrogen treatment from several different cell lines (including those of T cell, ovarian, placental, and cervical origin; Fig. S9).
DISCUSSION
The physiological panpleiotropic effects of hormones are well documented in many aspects of development, although their effector mechanisms at the molecular level, as well as their pathogenic effects upon cancer and immunity are not well understood (i.e., the gender bias for autoimmunity [29, 31, 50] is as striking as the gender bias for some cancers [51, 52]). The correlation between the expression of hormones or their cognitive receptors and development of pathologies, has led to several different hypotheses; most of which involve hyperactivation of cell proliferation, unregulated differentiation, alteration in DNA repair, or repression of apoptosis (51). Here, we propose another means by which estrogen can be a genotoxin, by directly inducing DNA deaminases such as AID. Our initial experiment using progesterone and estrogen identified AID as being part of a hormonally regulated system. Estrogen exerted its activity through the estrogen–ER complex, directly binding to the AID promoter, whereas our preliminary data suggests that progesterone acts through an estrogen-independent pathway (unpublished data). The consequences of estrogen activation on AID mRNA could also be detected as an increase in AID protein and enhanced downstream physiological effects, such as SHM and CSR. Interestingly, AID activation (mRNA) by estrogen was more pronounced than the effect on SHM. This could indicate that although AID production is necessary for SHM, other factors (e.g., lesion processing, AID targeting, etc) can play a significant role in overall SHM efficiency (53). Similar to SHM, CSR was enhanced upon estrogen treatment, but was lagging behind AID mRNA production. The most dramatic effect from estrogen-induced overexpression of AID was seen at the level of c-myc/IgH translocations. This is analogous to the recent observations that the effect of AID mRNA down-regulation (either by microRNA targeting or haploinsufficiency) was most pronounced at the level of translocations (44–46).
Autoimmunity encompasses a broadly defined area of clinical pathologies that stem from abnormalities in numerous systemic, cellular, and molecular mechanisms, a subset of which are B cell–related pathologies (50). In systemic lupus erythematosus, abnormalities in B cell development and the production of autoreactive antibodies play an important pathological role. Overexpression of AID in autoimmune-prone mice induced a more severe systemic lupus erythematosus–like phenotype (10), whereas breeding AID-deficient mice with autoimmune-prone MRL/lpr mice significantly reduced the onset and extent of disease (11), indicating that alterations in AID can change the severity of B cell autoimmunity. Correlating our data with the known effects of estrogen on autoimmunity (28, 30, 50, 54), we propose that the effect of sex-hormones on autoimmunity could partially be through AID transcription and subsequent increase in genome instability. In addition to the direct binding of the estrogen–ER complex to the AID promoter, estrogen may also hyperstimulate AID production through the NF-κB pathway, as we were able to demonstrate that cotreatment of TNF-α and estrogen enhanced NF-κB binding to the AID promoter (Fig. 3 D). Whether this effect is caused by protein–protein interaction or estrogen-induced chromatin modification has yet to be determined, but the synergistic or even cooperative interaction of two important autoimmune modulator pathways on AID expression may have substantially pathogenic effects. There are several hypotheses on how unregulated AID can affect autoimmunity in addition to overstimulation of SHM and CSR, e.g., debilitating mutations in the signaling pathways, of tumor suppressors, or of proapoptotic genes, or alterations that activate oncogenes or antiapoptotic genes (for review see reference [55]).
Mutation and subsequent loss of growth control is usually associated with oncogenesis, and this similarity with autoimmunity has previously been noted (55) (e.g., mutations in CD95/Fas have been associated with autoimmunity, as well as B cell lymphomas). Therefore, our data on estrogen-induced mutations in CD95/Fas (Fig. 5 and Fig. S7), derived from increased AID production, may provide a novel molecular mechanism that is important for both pathologies. AID's targeting outside the Ig locus, its apparently crucial involvement with germinal center–derived B cell malignancies (43, 56), and its ability to cause various malignancies when overexpressed in transgenic mice (20, 21, 57) are strong indicators for AID's oncogenic potential.
Estrogen is one of the most important and thoroughly studied mitogenic agents in cancer, but it does not possess any direct DNA mutability, and induces transformation by proliferation. In vitro, high concentrations of estrogen derivatives (usually metabolites) can form DNA adducts or produce reactive oxygen-damaging DNA (for review see reference [58–60]), but their role as physiological genotoxins is limited. Furthermore, because the estrogen derivative Tam (61) can inhibit estrogen's oncogenesis in breast cancer, it is unlikely that estrogen (or its derivatives) form DNA adducts under those conditions. It is interesting to note that the antagonistic activity of Tam has recently been used to inhibit some of the pathologies of estrogen-induced autoimmunity (54). On the other hand, because of the pharmacological action of Tam (binding and altering the ER DNA binding capacity), under certain circumstances Tam acts as an estrogen agonist (62). This activity leads to an increased risk in secondary (e.g., ovarian) cancer after Tam treatment for breast cancer (63). Thus, our findings that low concentrations of Tam induced AID mRNA (Fig. S1) also suggest that Tam acts as an agonist and indicates that the proposed usage of synthetic estrogen derivatives as a means to inhibit AID has to be carefully evaluated. Future work on identifying a potential role of AID in mouse models of hormonally induced cancers may provide further evidence on how AID can act as an environmentally stimulated oncogene.
As our data indicate, AID's response to estrogen seems to have been evolutionarily conserved among the APOBEC3 family members, different tissues, and cell lines (Fig. 6 and Fig. S8); mouse Apobec3 and human APOBEC3B, -3F, and -3G (and -H to a lesser extent) mRNA were induced by estrogen treatment. In the past, we have hypothesized that the predominant function of AID and its evolved DNA deaminase family members was to inactivate foreign DNA in the cell (8, 34, 64, 65). It is therefore plausible that the observed estrogen response of AID, as well as its expression in oocytes (22), had served a purpose other than targeting AID to Ig genes in B cells. Data indicating that AID has retained some ability to inhibit retroviral elements have substantiated this hypothesis (66).
Our work has highlighted that there is a novel pathway by which the nonmutagenic hormone estrogen can mediate genome instability via the activation of AID and other DNA deaminases, in turn possibly altering the predispositions, induction, or severity for cancer, autoimmunity, and viral infectivity.
MATERIALS AND METHODS
Cells and tissue.
Unless indicated, mouse tissue samples and splenic B cells were derived from 8–12-wk-old female unplugged BALB/c mice, and prepared by standard protocol (see Supplemental materials and methods, available at available at http://www.jem.org/cgi/content/full/jem.20080521/DC1). Human cell lines Jurkat, T47D, Ramos HS13 (Ramos), JAR, and mouse B cells were cultivated in RPMI-1640+GlutaMax medium (Invitrogen); the cell lines MCF7, JAMA2, HeLa, and HepG2 were cultivated in E4 medium; and PC3 was cultivated in HAMS F12 medium. Chicken DT40 cells were maintained in RPMI-1640+GlutaMax with 10% FCS, 1% chicken serum, 50 μM β-mercaptoethanol, 100 U/ml penicillin, and 100 μg/ml streptomycin at 39°C with 10% CO2. For indicated experiments, cells were treated for 72 h in hormone-depleted serum (Opti-MeM Reduced Serum Medium; Invitrogen) supplemented with Charcoal Stripped Fetal Bovine Serum (Invitrogen); and nonessential amino acids (final concentration; Sigma-Aldrich), as follows: l-Alanine (8.9 μg/ml), l-Asparagine (15.0 μg/ml), l-Aspartic acid (13.3 μg/ml), l-Glutamic acid (14.7 μg/ml), Glycine (7.5 μg/ml), Proline (11.5 μg/ml), and l-Serine (10.5 μg/ml).
Reagents.
B cell stimulation was performed using 25 μg/ml LPS (Sigma-Aldrich), 50 ng/ml mouse IL-4 (R&D Systems), 20 ng/ml human TNF-α (R&D Systems), and 2 ng/ml human TGF-β1 (R&D Systems). Estrogen (17-β-estradiol; Sigma-Aldrich) and progesterone (Sigma-Aldrich) were dissolved in DMSO at a concentration of 100 mM; this solution was then diluted in DMSO to give 1,000× stock solutions, and final dilutions were made in media (final DMSO concentration was never >0.1%). The final concentrations of estrogen are indicated in the text and figure legends. The concentration of Tam (Sigma-Aldrich) was 50 nM unless otherwise stated. The final concentration for CHX (Sigma-Aldrich) was 10 μg/ml, 20 nM for ACT (Calbiochem), 4 μg/ml for AMA (Calbiochem), and 10 μM for MG-132 (Sigma-Aldrich).
RT-PCR and qRT-PCR.
qRT-PCR of mRNAs was based on a previous study (67), with modifications (see Supplemental materials and methods). The primers for qRT-PCR (Table S1, available at http://www.jem.org/cgi/content/full/jem.20080521/DC1) were designed using PrimerExpress software. qRT-PCR analysis was performed using the QuantiTect SYBR Green PCR kit (QIAGEN) according to the manufacturer's instructions, Abi 7000 Sequence Detection System (Applied Biosystems), and Abi software. Tissue or cell line hormone responsiveness was determined by qRT-PCR of gene regulated by breast cancer 1, a known estrogen-responsive gene (36).
Promoter analysis.
Human AID promoter fragments were analyzed using pE1BLuc plasmid backbone (provided by G. Akusjärvi, Uppsala University, Uppsala, Sweden) containing a luciferase ORF and a minimal promoter region. Primers (Tables S2 and S3, available at http://www.jem.org/cgi/content/full/jem.20080521/DC1) approximately every 500 bp from the human AID transcription start site were used for amplifying AID promoter regions. The obtained PCR products were cloned into pE1BLuc vector, and 3 μg DNA were transfected into SiHa cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocols. 24 h after transfection, cells were treated with the indicated concentrations of hormones or TNF-α for 4 h and analyzed thereafter for luciferase signal using Dual Luciferase Reporter Assay System (Promega) and Glomax luminometer (Promega). The expression vector containing a dominant-negative mutant for IκBα (S32A S36A; a gift from Felix Randow, Medical Research Council-Laboratory of Molecular Biology, Cambridge, England, UK) was cotransfected with pE1BLuc constructs where indicated. 1 μg CMV promoter-driven Renilla luciferase expression vector (Promega) was included in all transfections and used as an internal control for transfection efficiency.
EMSA.
Ramos cells were treated for 72 h in hormone-depleted serum before the 4-h hormone treatment. EMSA was performed by standard protocol (68) with minor modifications (for more detail see Supplemental materials and methods). 0.25 pmol of complementary oligonucleotides containing either NF-κB or putative ER binding elements were annealed, labeled, and added to 7 μg of Ramos (treated or untreated) nuclear extract. Reactions were incubated for 20 min at room temperature in the absence or presence of 1-, 3-, and 10-fold mass excess of unlabeled oligonucleotides or antibodies. Samples were electrophoresed at 4°C on 4.5% polyacrylamide gels for 120 min, followed by autoradiography on imaging plates (FujiFilm) and analysis with a FLA-5000 Scanner (FujiFilm).
ChIP.
Ramos cells were treated for 72 h in hormone-depleted serum before the 4-h hormone treatment. ChIP procedure was done as previously published, with minor modifications (69) (for further details see Supplemental materials and methods). Approximately 107 cells were fixed with 1% formaldehyde in the culture media for 10 min at 37°C, and then quenched with 0.125 M glycine for 5 min at RT. Cells were pelleted and washed twice with cold PBS. The cell pellet was resuspended in 300 μl SDS lysis buffer and incubated on ice for 10 min. Cell lysates were sonicated for 6 min with a 30-s on/off sonication cycle in 1.5 ml eppendorf tubes. Samples were centrifuged for 5 min at 13,000 rpm at 4°C, and supernatants were diluted to 1.5 ml with ChIP buffer. An aliquot was kept as input measurement. Samples were precleared with 60 μl Protein G beads (ProtG; Roche) and 1 μg salmon sperm DNA (Invitrogen) for 45 min at 4°C. Anti–NF-κB p65 antibody, rabbit anti-ERα HC-20 antibody (Santa Cruz Biotechnology, Inc.), or goat anti–mouse λ control antibody (see Supplemental materials and methods) was added to the supernatant at a concentration of 2 μg/assay and incubated for 12 h at 4°C. 60 μl of salmon sperm DNA (1 μg DNA/20 μl ProtG) was added to the samples and incubated for 1 h while rotating. The ProtG–antibody–protein complex was pelleted and washed with the following buffers: low-salt wash buffer, high-salt wash buffer, LiCl wash buffer, and standard TE buffer (twice). The sample was eluted with 250 μl of elution buffer and incubated for 15 min at room temperature with agitation. The beads were centrifuged and the elution was repeated once. Bound immunocomplexes were then reverse cross-linked with 200 mM NaCl by incubating it at 65°C for 12 h. Proteinase K was added to the sample and incubated for 1 h at 45°C. DNA was then extracted with phenol/chloroform and precipitated, resuspended in 50 μl of water and subjected to qRT-PCR, using CHIP1 and CHIP2 primers for amplifying the −1,189 to −1,039 region of the AID promoter.
sIgM fluctuation analysis.
The surface IgM (sIgM) expression was investigated in Ramos HS13 cells (41), which contain a stop codon in the λ locus, with reversion mutations resulting in sIgM production. IgM stained (anti–human IgM-FITC [Sigma, UK]) and sorted single sIgM-negative cells were grown for ∼20 doublings, with fresh hormone containing media added every 48 h. The cells were then stained (anti–human IgM-FITC) for surface expression of IgM and analyzed by flow cytometry.
Mutation analysis.
VH and C regions were cloned and sequenced from hormonally treated (20 doublings) Ramos HS13 cells, as previously described (41). The human CD95/Fas locus was PCR amplified and sequenced from isolated genomic DNA, as previously described (18). Mouse Sγ3 switch regions were analyzed from 2 AID+/− spleens and treated with either DMSO alone or 10 nM estrogen for 6 d. AID+/− were derived from breeding AID−/− (gift from T. Honjo, Kyoto University, Kyoto, Japan) with BALB/c mice. Cloning and sequencing were as previously described (42). Sequencing was performed at the LRI sequencing facility. Further details are described in the Supplemental materials and methods.
c-myc/IgH translocation.
c-myc/IgH translocations were detected by PCR as previously described (47, 70). In brief, DNA was isolated from p53+/− B cells after 72 h of LPS stimulation in the presence or absence of 50 nM estrogen. Two rounds of PCR (for primers see Supplemental materials and methods) were performed using Expand Long Template PCR system (Roche) with primers MycIg1A and primers MycIg1B in the first round, and MycIg2A and MycIg2B in the second round. PCR products were separated on agarose gels, transferred to nylon membranes, and probed with γ-[P32]-ATP–labeled oligonucleotides IgH probe and c-myc probe. P values were calculated using two-tailed unpaired Student's t test.
Class switching analysis.
Ig class switching was investigated by detecting switch-circle transcripts in stimulated mouse spleen B cells (40) by qRT-PCR (71). Isolated splenic B-cells were stimulated for up to 72 h with LPS + IL-4 for inducing switching to IgG1 and IgE, LPS + TGF-β for switching to IgA, and LPS + IFN-γ for switching to IgG3. Hormones were added to the cells together with LPS and cytokines in fresh media. Primers (Tables S4 and S5, available at http://www.jem.org/cgi/content/full/jem.20080521/DC1) that were used for detecting IgG1 (71), IgG3, IgA (40), and IgE (72), have been previously described.
Mouse tissue analysis.
Mouse tissue was dissected (from two animals) and passed through cell strainers. Cells were incubated for 6 h in hormone-depleted media before estrogen treatment for 4 h. Total RNA was extracted, cDNA was synthesized, and gene expression was analyzed by qRT-PCR. All experiments were approved by the Cancer Research UK Animal Ethics Committee and the UK Home Office.
Online supplemental material.
The Supplemental materials and methods includes details for cells and reagents, qRT-PCR, promoter analysis, EMSA, ChIP, mutation analysis, and c-myc/IgH translocations. It also describes in detail the generation of endogenously tagged AID. Fig. S1 describes the effect of Tam treatment and translation inhibitors on AID expression. Fig. S2 shows the effect of estrogen on AID splicing. Fig. S3 demonstrates the effect of estrogen on NF-kB promoter binding. Fig. S4 provides details on how we generated the DT40 AID knockin allele. Fig. S5 is a schematic of class switching and switch circle formation. Fig. S6 shows the effect of estrogen on Ramos Ig diversification. Fig. S7 provides details on the mutation analysis of IgH and CD95/Fas after estrogen treatment. Fig. S8 provides details on the c-myc/IgH translocation analysis. Fig. S9 shows the effect of estrogen on the expression of DNA deaminase family members in various cell lines. Fig. S10 provides absolute values of qRT-PCR analysis from selected experiments. We also included primer sequences used in this study in five tables (Tables S1-S5). Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20080521/DC1.
Supplementary Material
[Supplemental Material Index]
Acknowledgments
We would like to thank Göran Akusjärvi for kindly providing the pE1BLuc plasmid, Felix Randow for providing the IκBα mutant plasmid and valuable suggestions, Julian Sale for Ramos HS13, Tasuku Honjo for AID−/− mice, Cancer Research UK Cell Services, London Research Institute Sequencing facility, Maria Simon for advice, and Heather Coker for invaluable discussions and critical reading of the manuscript.
I.V. Sernandez was supported by Comunidad Autonoma de Madrid; A.R. Ramiro was supported by Ramon y Cajal program from the Ministerio de Ciencia e Innovacion; S. Pauklin was supported in part by SA Archimedes-Estonian Foundation of European Union Education and Research. Cancer Research UK funded the work.
The authors have no conflicting financial interests.
Abbreviations used: ACT, actinomycin D; AID, activation-induced deaminase; AID-FM, AID FLAG-Myc–tagged AID protein; AMA, α-amanitin; ChIP, chromatin immunoprecipitation; CHX, cycloheximide; CSR, class switch recombination; EMSA, electromobility shift assay; ER, estrogen receptor; ERE, estrogen response element; IκBα-mt, IκBα S32A/S36A dominant mutant; mRNA, messenger RNA; qRT-PCR, quantitative real-time PCR; SHM, somatic hypermutation; sIgM, surface IgM; Tam, tamoxifen.
G. Bachmann's present address is Section of Structural Biology, Institute of Cancer Research, London, SW3 6JB, UK.
References
- 1.Neuberger, M.S., R.S. Harris, J. Di Noia, and S.K. Petersen-Mahrt. 2003. Immunity through DNA deamination. Trends Biochem. Sci. 28:305–312. [DOI] [PubMed] [Google Scholar]
- 2.Arakawa, H., J. Hauschild, and J.M. Buerstedde. 2002. Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science. 295:1301–1306. [DOI] [PubMed] [Google Scholar]
- 3.Harris, R.S., J.E. Sale, S.K. Petersen-Mahrt, and M.S. Neuberger. 2002. AID is essential for immunoglobulin V gene conversion in a cultured B cell line. Curr. Biol. 12:435–438. [DOI] [PubMed] [Google Scholar]
- 4.Muramatsu, M., K. Kinoshita, S. Fagarasan, S. Yamada, Y. Shinkai, and T. Honjo. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563. [DOI] [PubMed] [Google Scholar]
- 5.Muramatsu, M., V.S. Sankaranand, S. Anant, M. Sugai, K. Kinoshita, N.O. Davidson, and T. Honjo. 1999. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 274:18470–18476. [DOI] [PubMed] [Google Scholar]
- 6.Chaudhuri, J., and F.W. Alt. 2004. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat. Rev. Immunol. 4:541–552. [DOI] [PubMed] [Google Scholar]
- 7.Di Noia, J.M., and M.S. Neuberger. 2007. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 76:1–22. [DOI] [PubMed] [Google Scholar]
- 8.Petersen-Mahrt, S. 2005. DNA deamination in immunity. Immunol. Rev. 203:80–97. [DOI] [PubMed] [Google Scholar]
- 9.Bombardieri, M., F. Barone, F. Humby, S. Kelly, M. McGurk, P. Morgan, S. Challacombe, S. De Vita, G. Valesini, J. Spencer, and C. Pitzalis. 2007. Activation-induced cytidine deaminase expression in follicular dendritic cell networks and interfollicular large B cells supports functionality of ectopic lymphoid neogenesis in autoimmune sialoadenitis and MALT lymphoma in Sjogren's syndrome. J. Immunol. 179:4929–4938. [DOI] [PubMed] [Google Scholar]
- 10.Hsu, H.C., Y. Wu, P. Yang, Q. Wu, G. Job, J. Chen, J. Wang, M.A. Accavitti-Loper, W.E. Grizzle, R.H. Carter, and J.D. Mountz. 2007. Overexpression of activation-induced cytidine deaminase in B cells is associated with production of highly pathogenic autoantibodies. J. Immunol. 178:5357–5365. [DOI] [PubMed] [Google Scholar]
- 11.Jiang, C., J. Foley, N. Clayton, G. Kissling, M. Jokinen, R. Herbert, and M. Diaz. 2007. Abrogation of lupus nephritis in activation-induced deaminase-deficient MRL/lpr mice. J. Immunol. 178:7422–7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pasqualucci, L., P. Neumeister, T. Goossens, G. Nanjangud, R.S. Chaganti, R. Kuppers, and R. Dalla-Favera. 2001. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 412:341–346. [DOI] [PubMed] [Google Scholar]
- 13.Rabbitts, T.H., A. Forster, P. Hamlyn, and R. Baer. 1984. Effect of somatic mutation within translocated c-myc genes in Burkitt's lymphoma. Nature. 309:592–597. [DOI] [PubMed] [Google Scholar]
- 14.Rabbitts, T.H., P.H. Hamlyn, and R. Baer. 1983. Altered nucleotide sequences of a translocated c-myc gene in Burkitt lymphoma. Nature. 306:760–765. [DOI] [PubMed] [Google Scholar]
- 15.Pasqualucci, L., R. Guglielmino, J. Houldsworth, J. Mohr, S. Aoufouchi, R. Polakiewicz, R.S. Chaganti, and R. Dalla-Favera. 2004. Expression of the AID protein in normal and neoplastic B cells. Blood. 104:3318–3325. [DOI] [PubMed] [Google Scholar]
- 16.Ramiro, A.R., M. Jankovic, T. Eisenreich, S. Difilippantonio, S. Chen-Kiang, M. Muramatsu, T. Honjo, A. Nussenzweig, and M.C. Nussenzweig. 2004. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 118:431–438. [DOI] [PubMed] [Google Scholar]
- 17.Kuppers, R., and R. Dalla-Favera. 2001. Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene. 20:5580–5594. [DOI] [PubMed] [Google Scholar]
- 18.Muschen, M., D. Re, B. Jungnickel, V. Diehl, K. Rajewsky, and R. Kuppers. 2000. Somatic mutation of the CD95 gene in human B cells as a side-effect of the germinal center reaction. J. Exp. Med. 192:1833–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, M., J.L. Duke, D.J. Richter, C.G. Vinuesa, C.C. Goodnow, S.H. Kleinstein, and D.G. Schatz. 2008. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 451:841–845. [DOI] [PubMed] [Google Scholar]
- 20.Okazaki, I.M., H. Hiai, N. Kakazu, S. Yamada, M. Muramatsu, K. Kinoshita, and T. Honjo. 2003. Constitutive expression of AID leads to tumorigenesis. J. Exp. Med. 197:1173–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Endo, Y., H. Marusawa, K. Kinoshita, T. Morisawa, T. Sakurai, I.M. Okazaki, K. Watashi, K. Shimotohno, T. Honjo, and T. Chiba. 2007. Expression of activation-induced cytidine deaminase in human hepatocytes via NF-kappaB signaling. Oncogene. 26:5587–5595. [DOI] [PubMed] [Google Scholar]
- 22.Morgan, H.D., W. Dean, H.A. Coker, W. Reik, and S.K. Petersen-Mahrt. 2004. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J. Biol. Chem. 279:52353–52360. [DOI] [PubMed] [Google Scholar]
- 23.Sayegh, C.E., M.W. Quong, Y. Agata, and C. Murre. 2003. E-proteins directly regulate expression of activation-induced deaminase in mature B cells. Nat. Immunol. 4:586–593. [DOI] [PubMed] [Google Scholar]
- 24.Dedeoglu, F., B. Horwitz, J. Chaudhuri, F.W. Alt, and R.S. Geha. 2004. Induction of activation-induced cytidine deaminase gene expression by IL-4 and CD40 ligation is dependent on STAT6 and NFkappaB. Int. Immunol. 16:395–404. [DOI] [PubMed] [Google Scholar]
- 25.Gonda, H., M. Sugai, Y. Nambu, T. Katakai, Y. Agata, K.J. Mori, Y. Yokota, and A. Shimizu. 2003. The balance between Pax5 and Id2 activities is the key to AID gene expression. J. Exp. Med. 198:1427–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Butterworth, M., B. McClellan, and M. Allansmith. 1967. Influence of sex in immunoglobulin levels. Nature. 214:1224–1225. [DOI] [PubMed] [Google Scholar]
- 27.Eidinger, D., and T.J. Garrett. 1972. Studies of the regulatory effects of the sex hormones on antibody formation and stem cell differentiation. J. Exp. Med. 136:1098–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, J., and R.W. McMurray. 2007. Effects of estrogen receptor subtype-selective agonists on autoimmune disease in lupus-prone NZB/NZW F1 mouse model. Clin. Immunol. 123:219–226. [DOI] [PubMed] [Google Scholar]
- 29.Nalbandian, G., and S. Kovats. 2005. Understanding sex biases in immunity: effects of estrogen on the differentiation and function of antigen-presenting cells. Immunol. Res. 31:91–106. [DOI] [PubMed] [Google Scholar]
- 30.Peeva, E., and M. Zouali. 2005. Spotlight on the role of hormonal factors in the emergence of autoreactive B-lymphocytes. Immunol. Lett. 101:123–143. [DOI] [PubMed] [Google Scholar]
- 31.Whitacre, C.C. 2001. Sex differences in autoimmune disease. Nat. Immunol. 2:777–780. [DOI] [PubMed] [Google Scholar]
- 32.Hulka, B.S., and A.T. Stark. 1995. Breast cancer: cause and prevention. Lancet. 346:883–887. [DOI] [PubMed] [Google Scholar]
- 33.Shang, Y. 2006. Molecular mechanisms of oestrogen and SERMs in endometrial carcinogenesis. Nat. Rev. Cancer. 6:360–368. [DOI] [PubMed] [Google Scholar]
- 34.Conticello, S.G., C.J. Thomas, S.K. Petersen-Mahrt, and M.S. Neuberger. 2005. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol. Biol. Evol. 22:367–377. [DOI] [PubMed] [Google Scholar]
- 35.Tsai, M.J., and B.W. O'Malley. 1994. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu. Rev. Biochem. 63:451–486. [DOI] [PubMed] [Google Scholar]
- 36.Ghosh, M.G., D.A. Thompson, and R.J. Weigel. 2000. PDZK1 and GREB1 are estrogen-regulated genes expressed in hormone-responsive breast cancer. Cancer Res. 60:6367–6375. [PubMed] [Google Scholar]
- 37.Shiau, A.K., D. Barstad, P.M. Loria, L. Cheng, P.J. Kushner, D.A. Agard, and G.L. Greene. 1998. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 95:927–937. [DOI] [PubMed] [Google Scholar]
- 38.Auboeuf, D., A. Honig, S.M. Berget, and B.W. O'Malley. 2002. Coordinate regulation of transcription and splicing by steroid receptor coregulators. Science. 298:416–419. [DOI] [PubMed] [Google Scholar]
- 39.Wu, X., J.R. Darce, S.K. Chang, G.S. Nowakowski, and D.F. Jelinek. 2008. Alternative splicing regulates activation-induced cytidine deaminase (AID): Implications for suppression of AID mutagenic activity in normal and malignant B-cells. Blood. 112:4675–4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kinoshita, K., M. Harigai, S. Fagarasan, M. Muramatsu, and T. Honjo. 2001. A hallmark of active class switch recombination: transcripts directed by I promoters on looped-out circular DNAs. Proc. Natl. Acad. Sci. USA. 98:12620–12623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sale, J.E., and M.S. Neuberger. 1998. TdT-accessible breaks are scattered over the immunoglobulin V domain in a constitutively hypermutating B cell line. Immunity. 9:859–869. [DOI] [PubMed] [Google Scholar]
- 42.Xue, K., C. Rada, and M.S. Neuberger. 2006. The in vivo pattern of AID targeting to immunoglobulin switch regions deduced from mutation spectra in msh2−/− ung−/− mice. J. Exp. Med. 203:2085–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kotani, A., N. Kakazu, T. Tsuruyama, I.M. Okazaki, M. Muramatsu, K. Kinoshita, H. Nagaoka, D. Yabe, and T. Honjo. 2007. Activation-induced cytidine deaminase (AID) promotes B cell lymphomagenesis in Emu-cmyc transgenic mice. Proc. Natl. Acad. Sci. USA. 104:1616–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takizawa, M., H. Tolarova, Z. Li, W. Dubois, S. Lim, E. Callen, S. Franco, M. Mosaico, L. Feigenbaum, F.W. Alt, et al. 2008. AID expression levels determine the extent of cMyc oncogenic translocations and the incidence of B cell tumor development. J. Exp. Med. 205:1949–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dorsett, Y., K.M. McBride, M. Jankovic, A. Gazumyan, T.H. Thai, D.F. Robbiani, M. Di Virgilio, B.R. San-Martin, G. Heidkamp, T.A. Schwickert, et al. 2008. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 28:630–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teng, G., P. Hakimpour, P. Landgraf, A. Rice, T. Tuschl, R. Casellas, and F.N. Papavasiliou. 2008. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 28:621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramiro, A.R., M. Jankovic, E. Callen, S. Difilippantonio, H.T. Chen, K.M. McBride, T.R. Eisenreich, J. Chen, R.A. Dickins, S.W. Lowe, et al. 2006. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 440:105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Babbage, G., C.H. Ottensmeier, J. Blaydes, F.K. Stevenson, and S.S. Sahota. 2006. Immunoglobulin heavy chain locus events and expression of activation-induced cytidine deaminase in epithelial breast cancer cell lines. Cancer Res. 66:3996–4000. [DOI] [PubMed] [Google Scholar]
- 49.Muckenfuss, H., M. Hamdorf, U. Held, M. Perkovic, J. Lower, K. Cichutek, E. Flory, G.G. Schumann, and C. Munk. 2006. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J. Biol. Chem. 281:22161–22172. [DOI] [PubMed] [Google Scholar]
- 50.Cohen-Solal, J.F., V. Jeganathan, C.M. Grimaldi, E. Peeva, and B. Diamond. 2006. Sex hormones and SLE: influencing the fate of autoreactive B cells. Curr. Top. Microbiol. Immunol. 305:67–88. [DOI] [PubMed] [Google Scholar]
- 51.Roy, D., and J.G. Liehr. 1999. Estrogen, DNA damage and mutations. Mutat. Res. 424:107–115. [DOI] [PubMed] [Google Scholar]
- 52.Yager, J.D., and N.E. Davidson. 2006. Estrogen carcinogenesis in breast cancer. N. Engl. J. Med. 354:270–282. [DOI] [PubMed] [Google Scholar]
- 53.Muto, T., I.M. Okazaki, S. Yamada, Y. Tanaka, K. Kinoshita, M. Muramatsu, H. Nagaoka, and T. Honjo. 2006. Negative regulation of activation-induced cytidine deaminase in B cells. Proc. Natl. Acad. Sci. USA. 103:2752–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peeva, E., J. Venkatesh, and B. Diamond. 2005. Tamoxifen blocks estrogen-induced B cell maturation but not survival. J. Immunol. 175:1415–1423. [DOI] [PubMed] [Google Scholar]
- 55.Goodnow, C.C. 2007. Multistep pathogenesis of autoimmune disease. Cell. 130:25–35. [DOI] [PubMed] [Google Scholar]
- 56.Pasqualucci, L., G. Bhagat, M. Jankovic, M. Compagno, P. Smith, M. Muramatsu, T. Honjo, H.C. Morse III, M.C. Nussenzweig, and R. Dalla-Favera. 2008. AID is required for germinal center-derived lymphomagenesis. Nat. Genet. 40:108–112. [DOI] [PubMed] [Google Scholar]
- 57.Matsumoto, Y., H. Marusawa, K. Kinoshita, Y. Endo, T. Kou, T. Morisawa, T. Azuma, I.M. Okazaki, T. Honjo, and T. Chiba. 2007. Helicobacter pylori infection triggers aberrant expression of activation-induced cytidine deaminase in gastric epithelium. Nat. Med. 13:470–476. [DOI] [PubMed] [Google Scholar]
- 58.Liehr, J.G. 1990. Genotoxic effects of estrogens. Mutat. Res. 238:269–276. [DOI] [PubMed] [Google Scholar]
- 59.Bolton, J.L., L. Yu, and G.R. Thatcher. 2004. Quinoids formed from estrogens and antiestrogens. Methods Enzymol. 378:110–123. [DOI] [PubMed] [Google Scholar]
- 60.Cavalieri, E.L., and E.G. Rogan. 2004. A unifying mechanism in the initiation of cancer and other diseases by catechol quinones. Ann. N. Y. Acad. Sci. 1028:247–257. [DOI] [PubMed] [Google Scholar]
- 61.Phillips, D.H. 2001. Understanding the genotoxicity of tamoxifen? Carcinogenesis. 22:839–849. [DOI] [PubMed] [Google Scholar]
- 62.Hodges, L.C., J.D. Cook, E.K. Lobenhofer, L. Li, L. Bennett, P.R. Bushel, C.M. Aldaz, C.A. Afshari, and C.L. Walker. 2003. Tamoxifen functions as a molecular agonist inducing cell cycle-associated genes in breast cancer cells. Mol. Cancer Res. 1:300–311. [PubMed] [Google Scholar]
- 63.Spicer, D.V., M.C. Pike, and B.E. Henderson. 1991. Ovarian cancer and long-term tamoxifen in premenopausal women. Lancet. 337:1414. [DOI] [PubMed] [Google Scholar]
- 64.Harris, R.S., K.N. Bishop, A.M. Sheehy, H.M. Craig, S.K. Petersen-Mahrt, I.N. Watt, M.S. Neuberger, and M.H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell. 113:803–809. [DOI] [PubMed] [Google Scholar]
- 65.Petersen-Mahrt, S.K., R.S. Harris, and M.S. Neuberger. 2002. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 418:99–103. [DOI] [PubMed] [Google Scholar]
- 66.Gourzi, P., T. Leonova, and F.N. Papavasiliou. 2006. A role for activation-induced cytidine deaminase in the host response against a transforming retrovirus. Immunity. 24:779–786. [DOI] [PubMed] [Google Scholar]
- 67.Vandenbroucke, I.I., J. Vandesompele, A.D. Paepe, and L. Messiaen. 2001. Quantification of splice variants using real-time PCR. Nucleic Acids Res. 29:E68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ausubel, F.M., R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl. 1987. Current Protocols in Molecular Biology. John Wiley & Sons, Inc., Boston.
- 69.Hatzis, P., and I. Talianidis. 2002. Dynamics of enhancer-promoter communication during differentiation-induced gene activation. Mol. Cell. 10:1467–1477. [DOI] [PubMed] [Google Scholar]
- 70.Kovalchuk, A.L., J.R. Muller, and S. Janz. 1997. Deletional remodeling of c-myc-deregulating chromosomal translocations. Oncogene. 15:2369–2377. [DOI] [PubMed] [Google Scholar]
- 71.Reina-San-Martin, B., S. Difilippantonio, L. Hanitsch, R.F. Masilamani, A. Nussenzweig, and M.C. Nussenzweig. 2003. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. J. Exp. Med. 197:1767–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lumsden, J.M., T. McCarty, L.K. Petiniot, R. Shen, C. Barlow, T.A. Wynn, H.C. Morse III, P.J. Gearhart, A. Wynshaw-Boris, E.E. Max, and R.J. Hodes. 2004. Immunoglobulin class switch recombination is impaired in Atm-deficient mice. J. Exp. Med. 200:1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Material Index]