Highly parallel identification of essential genes in cancer cells (original) (raw)
Abstract
More complete knowledge of the molecular mechanisms underlying cancer will improve prevention, diagnosis and treatment. Efforts such as The Cancer Genome Atlas are systematically characterizing the structural basis of cancer, by identifying the genomic mutations associated with each cancer type. A powerful complementary approach is to systematically characterize the functional basis of cancer, by identifying the genes essential for growth and related phenotypes in different cancer cells. Such information would be particularly valuable for identifying potential drug targets. Here, we report the development of an efficient, robust approach to perform genome-scale pooled shRNA screens for both positive and negative selection and its application to systematically identify cell essential genes in 12 cancer cell lines. By integrating these functional data with comprehensive genetic analyses of primary human tumors, we identified known and putative oncogenes such as EGFR, KRAS, MYC, BCR-ABL, MYB, CRKL, and CDK4 that are essential for cancer cell proliferation and also altered in human cancers. We further used this approach to identify genes involved in the response of cancer cells to tumoricidal agents and found 4 genes required for the response of CML cells to imatinib treatment: PTPN1, NF1, SMARCB1, and SMARCE1, and 5 regulators of the response to FAS activation, FAS, FADD, CASP8, ARID1A and CBX1. Broad application of this highly parallel genetic screening strategy will not only facilitate the rapid identification of genes that drive the malignant state and its response to therapeutics but will also enable the discovery of genes that participate in any biological process.
Keywords: oncogene, pooled library, RNAi, screen, shRNA
Although human cancers harbor hundreds of genetic alterations, only a subset of these alterations is likely to impact tumor initiation or maintenance. Furthermore, genes that are not altered at the genomic level may play essential roles in tumor development. Thus, to identify genes with important roles in cancer, systematic functional assessment of genes for their contribution to specific cancer phenotypes is complementary to structural characterization of the cancer genome. Integrating both structural and functional approaches will provide insight into therapeutic targets for treating cancer.
The recent development of RNAi libraries targeting the human and mouse genomes has enabled systematic genetic studies in mammalian cells by using arrayed and pooled screens (1–8). However, scaling up the application of this methodology to identify all essential genes across a diverse range of human cancers requires an integrated experimental and computational approach that is efficient, robust, and economical. Here, we describe the development and application of genome-scale high-throughput methods using our lentiviral RNAi library to systematically assess cancer gene function and to integrate structural and functional approaches in the study of cancer.
Results and Discussion
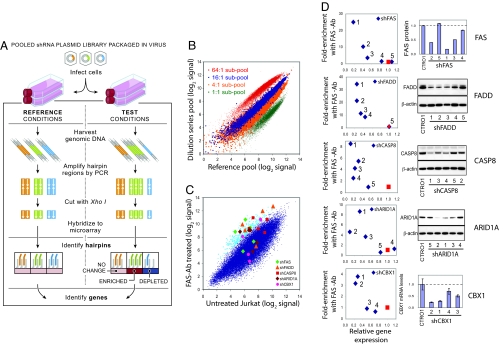
To apply RNA interference at genome scale, we developed a highly parallel “pooled screening” strategy that employs the previously described library created by The RNAi Consortium (TRC) (9, 10). The TRC library contains ≈170,000 lentivirally encoded short hairpin RNAs (shRNAs), with 5 or more independent shRNAs targeting each of 17,200 human genes, as well as an equivalent collection targeting each of 16,000 mouse genes. The pooled screening approach involves infecting cultured cells with a pool of shRNAs, allowing the cells to proliferate for a period, isolating the shRNA sequences from the resulting cells by PCR amplification, and measuring the relative abundance of the shRNAs (by cleaving the hairpins with a restriction enzyme and hybridizing them to a microarray complementary to the half-hairpin sequences) [Fig. 1A, supporting information (SI) Fig. S1 and SI Methods]. In the experiments below, we used a sublibrary containing 45,000 shRNAs corresponding to ≈9,500 human genes (45k shRNA pool). We demonstrated that 4-fold changes in relative shRNA abundance are easily resolved (Fig. 1B) using this approach.
Fig. 1.
Pooled RNAi screening strategy and performance using pools of 45,000 shRNA-expressing viruses. (A) Schematic of pooled shRNA library screens. (B) Performance evaluation of half-hairpin barcodes (hhbs) using pools containing known relative proportions of DNA. Two 45,000-shRNA pools were created by combining 4 subsets of the shRNA library plasmids (labeled in green, orange, blue, and red, each consisting of ≈11,000 different plasmids) in a 1:1:1:1 ratio of concentration for the “Reference pool” and in a 1:4:16:64 ratio for the “Dilution series pool.” To measure relative shRNA abundance in each pool, hhbs were hybridized to a custom Affymetrix barcode array. The observed separation of the 4 subsets of shRNAs according to their known relative proportions in the 2 pools illustrates the ability of hhbs to deconvolute the pooled shRNA library. (C) Primary screen results for genes required for FAS-induced apoptosis in Jurkat T cells. Cells were infected with the 45k pool viral library and cultured in the presence or absence of activating FAS antibody CH11 (FAS-Ab) for 3 weeks. Hybridization signals for hhbs amplified from the FAS-Ab treated group (average of 5 replicates) are plotted against those from the untreated group (average of 10 replicates). Array data for the 400 shRNAs (0.9% of pool) exhibiting highest enrichment in FAS-Ab treated group relative to untreated group are depicted in light blue. Array data for the shRNAs targeting the 5 hit genes are shown by distinct symbols. (D) Plot of target gene knockdown versus enrichment of shRNAs in FAS-treated samples for hit genes. FAS resistance was measured by relative proliferation rate of cells infected by individual candidate shRNA viruses (targeting FAS, FADD, CASP8, ARID1A, or CBX1) versus cells infected with a mixture of control shRNA viruses. Target gene suppression was measured by FACS (FAS), immunoblotting (FADD, CASP8, and ARID1A), or quantitative PCR (CBX1).
As an initial test of the system, we performed 2 positive-selection screens. The first screen was designed to identify genes whose inhibition renders T cells resistant to apoptosis induced by the activation of FAS, which functions in immune cell homeostasis (11). We infected Jurkat T cells with the 45k shRNA pool, so that the typical cell received 0.3 shRNAs [multiplicity of infection (MOI) = 0.3] and each shRNA was introduced to ≈200 independent cells. After selection to eliminate uninfected cells, the remaining cells were treated for 21 days with an activating anti-FAS antibody (12) at a dose sufficient to deplete the number of uninfected cells by a factor of ≈105. To identify shRNAs that confer resistance, we measured the overrepresentation of shRNAs in the surviving treated cells relative to untreated cells. To filter out shRNAs acting through off-target effects, we defined genes to be “hits” if at least 2 independent shRNAs against the gene were ranked in the top 0.9% of overrepresented shRNAs (Fig. 1C). There were 11 hits, of which 9 were confirmed by testing the shRNAs individually. We were able to reliably measure gene expression levels for 7 of these genes and found that 5 showed strong correlation between the level of resistance to FAS-induced apoptosis and the level of gene knockdown—confirming that the shRNA effect is “on-target” (Fig. 1D). The 5 genes include 3 with well-established roles in FAS-induced apoptosis (11, 13–15) (FAS, FADD, and CASP8) and 2 previously undescribed genes—ARID1A, a SWI/SNF chromatin remodeling complex component (16, 17), and CBX1, a chromatin silencing protein (18). For all 5 genes, the effective shRNAs also inhibited both FAS-induced CASPASE 8 cleavage and FAS-induced mitochondrial leakage (Fig. S2), indicating that, like the 3 known genes, the 2 previously undescribed genes act upstream of CASPASE 8 activation. The lack of downstream apoptosis genes among the hits could be due to false negative results (missed active genes) or a true finding that stems from individual downstream genes not being absolutely required for apoptosis in these cells because of functional redundancy or the activation of compensatory processes.
The second positive selection screen sought to identify genes whose inhibition renders H82 small-cell lung cancer cells resistant to etoposide, a small molecule that alters the activities of topoisomerase IIA (TOPOIIA) (19, 20) and is used to treat small-cell lung and other cancers (21). By using a high dose of etoposide sufficient to eliminate unmodified H82 cells, 1 confirmed suppressor gene emerged: TOPOIIA itself (Fig. S3). Consistent with this observation, reduced TOPOIIA expression has been shown to confer etoposide resistance in SCLC lines (22). Together, these 2 positive selection screens demonstrate the utility of our approach in studying genes involved in cell viability.
We then turned to the more difficult challenge of identifying the genes that are essential for the proliferation of specific cancer cell lines, which involves the infection of cell lines with a pool of shRNAs and the identification of underrepresented shRNAs among surviving cells. These negative-selection screens require more precise quantification of shRNA abundance than positive-selection screens that seek to identify shRNAs that are dramatically overrepresented.
We performed negative-selection RNAi screens with the 45k shRNA pool in 12 cancer cell lines representing diverse cancer types, including small-cell lung cancer (H82, H187), non-small-cell lung cancer (A549, H1650, H1975, HCC827), glioblastoma (LN229, U251), CML (K562), and lymphocytic leukemia (Jurkat, SUPT1, REH). For each of the cell lines, we performed at least 10 independent infections and compared the abundance of each shRNA at ≈28 days after infection to the initial abundance in the DNA plasmid pool from which the lentiviral vectors were produced. For 2 of the cell lines (K562 (Fig. S4) and U251), we confirmed that the abundance at 3–4 days after infection was highly similar to the abundance in the plasmid pool, demonstrating that representation is preserved after viral packaging, viral transduction, and initial infection (Fig. 2A). In contrast, the representation is strikingly different at later time points (2–4 weeks), reflecting unequal survival of cells with different hairpins (Fig. S4 and Fig. 2A).
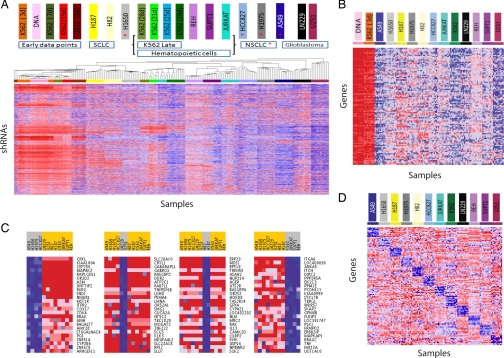
Fig. 2.
Screens for essential genes in 12 cancer cell lines. The 45K pool viral library was used to infect 12 cancer cell lines in multiple replicates. Heat maps depict relative abundance of shRNAs, individually or combined by their gene target (red, high; blue, low). (A) Unsupervised hierarchical clustering of the hhb array data for 175 samples from screens of 12 cell lines (10 replicates per cell line for 4-week time points; 5 or 10 replicates for earlier time points, as noted), and the initial 45k shRNA DNA plasmid pool (10 replicates). The 10,117 shRNAs with the highest coefficient of variation in signal across all 175 samples (CV >0.30) were included in the clustering analysis. (B) Commonly essential genes. The average of “leading edge” shRNA signals for each of the top-100 commonly essential genes (requiring a minimum of 8 of 12 cell lines to contribute to the essentiality enrichment score) exhibits extensive depletion after 4 weeks. (C) Top cell lineage-specific essential genes for cell lines derived from: (i) 4 non-small-cell lung cancers, (ii) 2 glioblastomas, (iii) 2 small-cell lung cancers, and (iv) 4 leukemias. (D) Identification of cell line-specific essential genes based on relative shRNA depletion in 1 cell line versus the other 11 cell lines. Average signals for leading edge shRNAs for the top-10 specific essential genes for each cell line are displayed. ABL1 and BCR are 1st and 5th best-scoring genes, respectively, in K562 cells.
In total, we generated a database of 5.4 million measurements of the relative abundance of the 45,000 shRNAs across the 12 cell lines and 10 replicates. Both unsupervised clustering and consensus clustering of these data clustered the replicates together, supporting the robustness of the results, and furthermore grouped the cell lines according to their developmental lineage (Fig. 2A and Fig. S5). To define genes as hits based on shRNA depletion data, we developed a statistic called an RNAi gene enrichment ranking (RIGER) score. Briefly, we examine the position of the 5 shRNAs targeting the gene in the full ranked list of the 45,000 shRNAs, assess whether the set is biased toward the top of the list based on a KS statistic, and calculate an enrichment score and gene ranking based on a permutation test (see Materials and Methods). The inclusion of all shRNAs targeting each gene increases the power of the screen, compensating for variation in gene suppression and off-target effects. We applied RIGER to each of the ≈9,500 genes, to identify the cancer-cell essential genes (Dataset S1).
The 12 cancer cell lines showed substantial correlation in their gene requirements for proliferation. For example, 530 genes ranked in the top 5% for essentiality in 5 or more cell lines, whereas only 2 genes would be expected if the cell lines were uncorrelated (Dataset S2). We identified “commonly essential” genes using a second application of RIGER to find genes enriched for essentiality among the 12 cell lines; we found 268 commonly essential genes with an FDR <25% (Fig. 2B and Dataset S3). Using gene-set enrichment analysis (GSEA) (23), we observed that the commonly essential genes showed a strong enrichment for certain molecular pathways including ribosomal proteins, mRNA processing and splicing, translation factors, and proteasome degradation (Dataset S4). For selected genes in these highly enriched pathways, we validated target specificity by comparing proliferation to target gene suppression for multiple shRNAs (Fig. S6).
In addition to these commonly essential genes, we identified “cell lineage-specific” essential genes (Fig. 2C and Dataset S5), which we defined as genes that exhibited a stronger phenotype in cell lines derived from a particular cancer type than in other cancer types. A total of 63 genes exhibited specific essentiality for the 4 non-small-cell lung cancer (NSCLC) cell lines that was significantly stronger than observed for randomly selected subsets of 4 cell lines (P < 0.05). Similarly, 32 genes showed significant differential essentiality for the 4 lymphocytic and myelogenous leukemia lines. This type of analysis thus enables a systematic approach to identify an important class of genes that are differentially required for proliferation in a cancer-specific manner.
We also identified “cell line-specific” genes, which showed specific differential requirement in 1 cell line versus the other 11 (Fig. 2D, Fig. S7, and Dataset S6). Such genes can generate initial hypotheses about cancer-specific gene dependencies, but confirmation in additional cell lines would be required to define the cancer-specificity of these gene requirements. For example, chronic myelogenous leukemia is represented by only a single cell line (K562) among the 12. In K562 cells, we found that the 5 top-scoring genes included ABL1 and BCR (ranked 1st and 5th, respectively, of ≈9,500 genes); these 2 genes are involved in the BCR-ABL translocation harbored by this cell line. We retested individually the 13 shRNAs against these genes, confirming that the inhibition is cell line-specific and that the level of inhibition is strongly correlated with the level of gene inhibition (Fig. S8). Thus, ABL1 is readily identified as a selectively highly required gene in the K562 cell line, and in this positive-control case, we know that follow-up experiments would confirm this trait to be shared among CML cell lines, demonstrating the utility of this approach to identify bona fide oncogenes.
A particularly powerful way to characterize cancer cells may be to combine information about both structure (genomic mutation) and function (gene essentiality) to reveal oncogenes. Several recent studies have illustrated the ability to identify key cancer genes in this manner (6, 24, 25). Indeed, when we searched for known oncogenes among the highest scoring genes in each cell line, we found several common oncogenes. For example, KRAS, MYC, and MYB were in the top 1% of essential genes in at least 1 cancer cell line (KRAS in LN229, A549, Jurkat, and H1650, and MYC and MYB in K562; Figs. S9 and S10). KRAS was found to be required in a KRAS mutant cell line, A549 (9th ranking gene, Dataset S1).
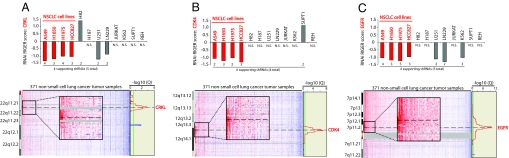
To extend this approach to genes resident in regions of copy number gain in human cancers, we intersected (i) the list of genes in regions of genomic amplification identified in a recent study of 371 NSCLC tumors (26) and (ii) the list of cell lineage-specific essential genes with strong preferential essentiality in the 4 NSCLC cell lines (Dataset S5). The top-scoring gene, CRKL (P = 0.010; Fig. 3A), a member of an adapter protein family that activates the RAS signaling pathway (27); falls in one of the most significantly amplified regions in NSCLC 22q11.21, for which no oncogenes were previously known (26). We confirmed the essentiality of CRKL in A549 and H1975 NSCLC cells through: (i) a competitive cell survival experiment and (ii) experiments demonstrating that the level of CRKL knockdown was correlated with the level of growth inhibition (Fig. S11). The second-best-scoring gene was CDK4 (P = 0.014; Fig. 3B), which modulates the p16INK4a-cyclin D1-CDK4-RB growth regulatory pathway. This pathway is altered in the majority of NSCLCs, and high levels of CDK4 are associated with tumor progression (26, 28, 29). The third-best-scoring gene was EGFR (P = 0.03; Fig. 3C), a gene frequently amplified or mutated in NSCLC that has been successfully targeted by small-molecule inhibitors (26, 30–32). Although we screened only 4 NSCLC cell lines, the intersection of structural and functional data readily identifies 2 known oncogenes (EGFR and CDK4) and implicates an additional likely oncogene (CRKL) in human NSCLC. These observations suggest that the combination of large-scale structural and function data will accelerate the comprehensive identification of genes essential for the malignant state.
Fig. 3.
Identification of known and putative oncogenes by integrating functional and structural genomics. RNAi RIGER scores for CRKL (A), CDK4 (B), and EGFR (C) in each of the 12 cell lines relative to control and copy number changes in NSCLC tumors (26) at the loci encoding these genes. The number of shRNAs ranked in the leading edge of the RIGER analysis is noted. Two or more shRNAs for each gene were required to be in the RIGER leading edge to obtain a RIGER score for that gene; otherwise the RIGER result is labeled N.S. (no score). Significance of the observed copy number changes based on frequency and magnitude was calculated by using the GISTIC algorithm (41). False-discovery rates (red line, −LOG10 Q values for amplification; blue line, −LOG10 Q values for deletion; green line is 0.25 cutoff for significance) are depicted vertically along each chromosomal position.
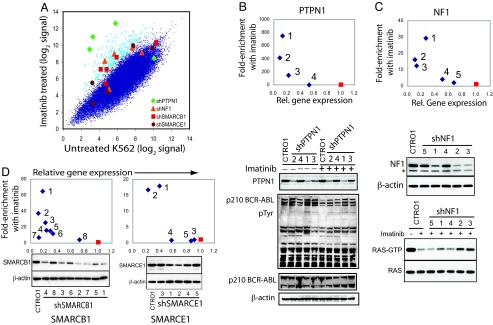
A further application of pooled shRNA screening is to perform suppressor and enhancer screens to identify genes that interact with known genes, pathways, and drugs. To test this approach, we screened for genes that modulate the response of CML cells to imatinib, a clinically approved inhibitor of BCR-ABL (33, 34). Such screens have the potential not only to identify genes that interact with BCR-ABL but also, importantly, highlight genes that may influence the development of imatinib resistance. We performed a positive-selection screen in which we infected K562 cells with the 45k pool, exposed these cells to a lethal dose of imatinib, and identified genes whose inhibition conferred survival (Fig. 4A). By using the same criteria as for the FAS-Ab modifier screen, 10 genes were identified as hits, 2 of which failed to be confirmed in tests of individual shRNAs. Target knockdown measurements were obtained for 7 of these genes, of which the shRNAs for 4: PTPN1, NF1, SMARCB1, and SMARCE1 showed strong correlation between the level of resistance and the level of gene knockdown (Fig. 4 B–D). One of these genes, PTPN1, has previously been reported to be a negative regulator of BCR-ABL signaling, because the expression of a dominantly interfering mutant of PTPN1 rendered BCR-ABL-dependent cells resistant to imatinib (35–37). We found that shRNA-mediated inhibition of PTPN1 leads to increased tyrosine phosphorylation of BCR-ABL in the presence or absence of imatinib (Fig. 4B Lower). Further confirming this finding, we also performed a separate screen to identify genes that permit cells to survive RNAi-mediated suppression of BCR-ABL and identified PTPN1 as the top-scoring hit (Fig. S12). Among the other genes, NF1 is a Ras GTPase that suppresses tumor formation by inhibiting ras activation (38, 39), and it is a tumor suppressor for both type 1 neurofibromatosis and juvenile myelomonocytic leukemia, a childhood leukemia with characteristics similar to CML (40). We found that shRNA-mediated inhibition of NF1 partially restored levels of active RAS in imatinib-treated cells (Fig. 4C Bottom). Both SMARCB1 and SMARCE1 encode subunits of the SWI1/SNF5 matrix-associated actin-binding chromatin-remodeling complex (17), and SMARCB1 has been implicated as a tumor-suppressor gene in infantile malignant rhabdoid tumors and epithelioid sarcomas. These observations suggest a previously unrecognized role for this chromatin remodeling complex in imatinib-sensitivity of CML cells. Moreover, this screen suggests that this approach can be used to systematically identify genes and pathways that interact with a specific gene, pathway, or small-molecule perturbation.
Fig. 4.
Screen for modifiers of the response to imatinib in K562 cells. K562 cells were infected with the 45k pool shRNA viral library and treated in the presence or absence of imatinib for 21 days (10 replicate infections for each group). (A) Averaged microarray hybridization signals for each shRNA in the imatinib-treated cell samples are plotted versus average hybridization signals for the untreated samples. The 400 shRNAs yielding the greatest resistance to imatinib are indicated in light blue. The shRNAs targeting 4 hit genes are labeled. (B–D) Knockdown validation of shRNAs conferring resistance to imatinib. The enrichment of shRNA-infected cells in response to imatinib was tested by coculturing GFP-labeled shRNA-infected cells with control cells for 3 weeks, followed by FACS analysis. Target gene knockdown by the shRNAs was determined by immunoblotting. (B) Cells infected with shPTPN1 were untreated or treated with imatinib, followed by immunoblotting for PTPN1, phosphotyrosine, ABL1 and β-actin. (C) Cells infected with shNF1 were treated with imatinib, followed by immunoprecipitation of GTP-bound RAS and immunoblotting for RAS. (D) Knockdown validation of shRNAs targeting SMARCB1 and SMARCE1.
Conclusions
Extending the application of the experimental and analytical strategies described here to a much larger set of cancer cell lines will permit systematic discovery of genes involved in cancer cell proliferation and survival. The inclusion of 5 independent shRNAs targeting each human gene in this shRNA library provides power to discriminate specific from off-target effects in the primary screen and different levels of on-target knockdown, whereas the RIGER algorithm provides the means to rank genes based on these multiple shRNAs. Increasing the number of shRNAs available per gene and measuring the knockdown performance of each shRNA will further improve both the sensitivity of this approach to detect hit genes and the ability to discriminate against off-target effects. Although we have assessed only a single phenotype (proliferation) in a limited number of cell lines, this method may be applied to other phenotypes and cell types including more “normal” cultured cells. We anticipate that systematic efforts to apply these approaches to study other cancer phenotypes will eventually lead to a more complete view of the Achilles' heels of different types of cancers. Our initial efforts suggest that such studies can be performed at a relatively modest cost, although they will require larger, validated shRNA libraries than we are currently generating, and an extensive collection of cell lines.
When combined with the increasingly complete structural analyses of cancer genomes by The Cancer Genome Atlas and other such efforts, the experimental and analytical strategies for pooled shRNA screens described herein provide a feasible strategy to systematically identify the key genes involved in cancer initiation, maintenance, and progression and likely targets for therapeutic intervention. Moreover, although we have used cancer cell proliferation to develop and validate these methodologies, the broad application of these approaches in other experimental contexts promises to provide insights into a wide range of biological phenotypes in mammalian cells.
Materials and Methods
A genome-scale pooled shRNA library of 45,000 shRNAs in viral vectors (45k pool) was produced from the sequence-validated arrayed TRC shRNA library and used for all of the screens reported here. The shRNA representation of the library was measured by using the half of the shRNA sequence as a molecular barcode (a “half-hairpin barcode”, hhb), which was obtained by restriction enzyme digestion of PCR-amplified shRNA sequences from library-infected cells. The hhb representation was assessed by hybridizing the hhbs to a high-density Affymetrix custom microarray. The shRNA hhb hybridization data were preprocessed with modified Dchip software, and analyzed by using the RIGER algorithm. These computational analysis tools, dCHIP for RNAi and RIGER, are available online at http://www.broad.mit.edu/rnai_analysis. Detailed methods for all experiments are provided in SI Methods. For primers used in SYBR assays and TaqMan probes, see Table S1. For a key to the shRNA labels used in the figures, see Dataset S7. For analyses used to assess essential genes, see SI Methods and Scheme S1.
Supplementary Material
Supporting Information
Acknowledgments.
We thank Preeti Gupta and Mike Mittman for assistance with Affymetrix microarray design; Jinyan Du, Casey Gates, Leigh Brody, Nathan Berkowitz, Neelum Khattak, Daniel Lam, Brian Wong, Jordi Barretina, and Cindy Nguyen for technical assistance and materials; and Pablo Tamayo, Preeti Gupta, Mike Mittman, Serena Silver, Jennifer Grenier, and Glenn Cowley for helpful discussions. This work is a project of The RNAi Consortium (TRC), financially supported by past and present consortium members: Academia Sinica, Bristol–Myers Squibb, Broad Institute, Eli Lilly, Novartis, Ontario Institute for Cancer Research, and Sigma–Aldrich. H.W.C. was supported by the Croucher Foundation.
Footnotes
The authors declare no conflict of interest.
References
- 1.Schlabach MR, et al. Cancer proliferation gene discovery through functional genomics. Science. 2008;319:620–624. doi: 10.1126/science.1149200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silva JM, et al. Profiling essential genes in human mammary cells by multiplex RNAi screening. Science. 2008;319:617–620. doi: 10.1126/science.1149185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brummelkamp TR, et al. An shRNA barcode screen provides insight into cancer cell vulnerability to MDM2 inhibitors. Nat Chem Biol. 2006;2:202–206. doi: 10.1038/nchembio774. [DOI] [PubMed] [Google Scholar]
- 4.Berns K, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- 5.Berns K, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 6.Westbrook TF, et al. A genetic screen for candidate tumor suppressors identifies REST. Cell. 2005;121:837–848. doi: 10.1016/j.cell.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 7.Kolfschoten IG, et al. A genetic screen identifies PITX1 as a suppressor of RAS activity and tumorigenicity. Cell. 2005;121:849–858. doi: 10.1016/j.cell.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 8.Ngo VN, et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441:106–110. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
- 9.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 10.Root DE, Hacohen N, Hahn WC, Lander ES, Sabatini DM. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat Methods. 2006;3:715–719. doi: 10.1038/nmeth924. [DOI] [PubMed] [Google Scholar]
- 11.Itoh N, et al. The polypeptide encoded by the cDNA for human cell surface antigen Fas can mediate apoptosis. Cell. 1991;66:233–243. doi: 10.1016/0092-8674(91)90614-5. [DOI] [PubMed] [Google Scholar]
- 12.Yonehara S, Ishii A, Yonehara M. A cell-killing monoclonal antibody (anti-Fas) to a cell surface antigen co-downregulated with the receptor of tumor necrosis factor. J Exp Med. 1989;169:1747–1756. doi: 10.1084/jem.169.5.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boldin MP, Goncharov TM, Goltsev YV, Wallach D. Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1- and TNF receptor-induced cell death. Cell. 1996;85:803–815. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- 14.Chinnaiyan AM, O'Rourke K, Tewari M, Dixit VM. FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell. 1995;81:505–512. doi: 10.1016/0092-8674(95)90071-3. [DOI] [PubMed] [Google Scholar]
- 15.Muzio M, et al. FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death-inducing signaling complex. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 16.Dallas PB, et al. p300/CREB binding protein-related protein p270 is a component of mammalian SWI/SNF complexes. Mol Cell Biol. 1998;18:3596–3603. doi: 10.1128/mcb.18.6.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang W, et al. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 18.Furuta K, et al. Heterochromatin protein HP1Hsbeta (p25beta) and its localization with centromeres in mitosis. Chromosoma. 1997;106:11–19. doi: 10.1007/s004120050219. [DOI] [PubMed] [Google Scholar]
- 19.Nitiss JL, Liu YX, Hsiung Y. A temperature sensitive topoisomerase II allele confers temperature dependent drug resistance on amsacrine and etoposide: A genetic system for determining the targets of topoisomerase II inhibitors. Cancer Res. 1993;53:89–93. [PubMed] [Google Scholar]
- 20.Osheroff N. Effect of antineoplastic agents on the DNA cleavage/religation reaction of eukaryotic topoisomerase II: Inhibition of DNA religation by etoposide. Biochemistry. 1989;28:6157–6160. doi: 10.1021/bi00441a005. [DOI] [PubMed] [Google Scholar]
- 21.Hande KR. Etoposide: Four decades of development of a topoisomerase II inhibitor. Eur J Cancer. 1998;34:1514–1521. doi: 10.1016/s0959-8049(98)00228-7. [DOI] [PubMed] [Google Scholar]
- 22.Andoh T, et al. Reduced expression of DNA topoisomerase II confers resistance to etoposide (VP-16) in small cell lung cancer cell lines established from a refractory tumor of a patient and by in vitro selection. Oncol Res. 1996;8:229–238. [PubMed] [Google Scholar]
- 23.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boehm JS, et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129:1065–1079. doi: 10.1016/j.cell.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 25.Firestein R, et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455:547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weir BA, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Senechal K, Halpern J, Sawyers CL. The CRKL adaptor protein transforms fibroblasts and functions in transformation by the BCR-ABL oncogene. J Biol Chem. 1996;271:23255–23261. doi: 10.1074/jbc.271.38.23255. [DOI] [PubMed] [Google Scholar]
- 28.Wikman H, et al. CDK4 is a probable target gene in a novel amplicon at 12q13.3-q14.1 in lung cancer. Genes Chromosomes Cancer. 2005;42:193–199. doi: 10.1002/gcc.20122. [DOI] [PubMed] [Google Scholar]
- 29.Zhao X, et al. Homozygous deletions and chromosome amplifications in human lung carcinomas revealed by single nucleotide polymorphism array analysis. Cancer Res. 2005;65:5561–5570. doi: 10.1158/0008-5472.CAN-04-4603. [DOI] [PubMed] [Google Scholar]
- 30.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 31.Paez JG, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 32.Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–1167. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- 33.Deininger M, Buchdunger E, Druker BJ. The development of imatinib as a therapeutic agent for chronic myeloid leukemia. Blood. 2005;105:2640–2653. doi: 10.1182/blood-2004-08-3097. [DOI] [PubMed] [Google Scholar]
- 34.Wong S, Witte ON. The BCR-ABL story: Bench to bedside and back. Annu Rev Immunol. 2004;22:247–306. doi: 10.1146/annurev.immunol.22.012703.104753. [DOI] [PubMed] [Google Scholar]
- 35.Koyama N, et al. Inhibition of phosphotyrosine phosphatase 1B causes resistance in BCR-ABL-positive leukemia cells to the ABL kinase inhibitor STI571. Clin Cancer Res. 2006;12:2025–2031. doi: 10.1158/1078-0432.CCR-04-2392. [DOI] [PubMed] [Google Scholar]
- 36.LaMontagne KR, Jr, Flint AJ, Franza BR, Jr, Pandergast AM, Tonks NK. Protein tyrosine phosphatase 1B antagonizes signalling by oncoprotein tyrosine kinase p210 bcr-abl in vivo. Mol Cell Biol. 1998;18:2965–2975. doi: 10.1128/mcb.18.5.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.LaMontagne KR, Jr, Hannon G, Tonks NK. Protein tyrosine phosphatase PTP1B suppresses p210 bcr-abl-induced transformation of rat-1 fibroblasts and promotes differentiation of K562 cells. Proc Natl Acad Sci USA. 1998;95:14094–14099. doi: 10.1073/pnas.95.24.14094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu GF, et al. The neurofibromatosis type 1 gene encodes a protein related to GAP. Cell. 1990;62:599–608. doi: 10.1016/0092-8674(90)90024-9. [DOI] [PubMed] [Google Scholar]
- 39.Basu TN, et al. Aberrant regulation of ras proteins in malignant tumour cells from type 1 neurofibromatosis patients. Nature. 1992;356:713–715. doi: 10.1038/356713a0. [DOI] [PubMed] [Google Scholar]
- 40.Side LE, et al. Mutations of the NF1 gene in children with juvenile myelomonocytic leukemia without clinical evidence of neurofibromatosis, type 1. Blood. 1998;92:267–272. [PubMed] [Google Scholar]
- 41.Beroukhim R, et al. Assessing the significance of chromosomal aberrations in cancer: Methodology and application to glioma. Proc Natl Acad Sci USA. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information