The phage abortive infection system, ToxIN, functions as a protein–RNA toxin–antitoxin pair (original) (raw)
Abstract
Various mechanisms exist that enable bacteria to resist bacteriophage infection. Resistance strategies include the abortive infection (Abi) systems, which promote cell death and limit phage replication within a bacterial population. A highly effective 2-gene Abi system from the phytopathogen Erwinia carotovora subspecies atroseptica, designated ToxIN, is described. The ToxIN Abi system also functions as a toxin–antitoxin (TA) pair, with ToxN inhibiting bacterial growth and the tandemly repeated ToxI RNA antitoxin counteracting the toxicity. TA modules are currently divided into 2 classes, protein and RNA antisense. We provide evidence that ToxIN defines an entirely new TA class that functions via a novel protein-RNA mechanism, with analogous systems present in diverse bacteria. Despite the debated role of TA systems, we demonstrate that ToxIN provides viral resistance in a range of bacterial genera against multiple phages. This is the first demonstration of a novel mechanistic class of TA systems and of an Abi system functioning in different bacterial genera, both with implications for the dynamics of phage-bacterial interactions.
Keywords: Bacteriophage, Bacteriostasis, Erwinia, plasmid, resistance
Bacteria, the most abundant organisms on the planet, constantly face challenges from their own viral parasites, bacteriophages. Outnumbered approximately 10 to 1 by the estimated ≥1030 phages on Earth (1, 2), bacteria become infected at rates of 1025 per second (3). The rapid turnover of such large quantities of organic material impacts on nutrient cycling and the global climate (4, 5). This global predator–prey relationship is an evolutionary clash that has forced bacteria to develop multiple methods of protection (6). These include surface alterations to avoid phage adsorption, prevention of phage DNA injection, restriction of incoming DNA, acquiring phage-specific immunity through clustered regularly interspaced short palindromic repeats (7) and abortive infection (Abi). Abi systems provide population protection by promoting “altruistic suicide” of an infected bacterium (8). The majority of Abis have been found on plasmids of Gram-positive lactococcal strains (9), but some have been found in Gram-negative species, including Escherichia coli, Vibrio cholerae, and Shigella dysenteriae (10–12). Abi systems often are highly toxic when activated; they have varied targets and can act on central cellular processes to inhibit phage DNA replication, transcription, and protein synthesis (9). Specific effects include premature cell lysis by AbiZ (13) and interference of a phage RuvC-like endonuclease by AbiD1 (14).
Toxic proteins play many roles within bacteria. The recent influx of genomic information has allowed frequent identification of multiple “toxin–antitoxin” (TA) loci on the chromosomes of both bacteria and archaea (15). Although originally identified as plasmid addiction systems (16), the apparent widespread nature of these TA operons has led to discussion of the biological role of such systems (17). TA systems rely on the dual activity of a toxin and an antagonistic antitoxin (18). Antitoxins are labile compared with their toxins, and when production of both components is inhibited, the antitoxin is turned over preferentially, allowing the toxin to take effect (18). The toxins of known TA systems, similar to Abi proteins, can target central cellular processes such as DNA replication and translation by inhibiting DNA gyrase (ccd and parDE loci) and causing mRNA degradation (relBE and mazEF loci), respectively (18). TA loci are thought to fall into 2 categories, protein–protein systems, such as Phd-Doc (18), and RNA–RNA systems, such as hok (host killing)/sok (suppressor of killing) (19).
Here we identify a cryptic plasmid of the Gram-negative phytopathogen Erwinia carotovora subspecies atroseptica (Eca) 1039, carrying a gene encoding a protein with sequence identity to AbiQ of Lactococcus lactis (20). This Eca homologue, designated ToxN (for toxin), protects against multiple phages through abortive infection. Controlled expression of ToxN is bacteriostatic, and toxicity of the ToxN protein is suppressed by the product of an upstream gene, toxI (ToxN inhibitor), encoding an antitoxic RNA. Therefore, together, ToxIN acts as a novel protein–RNA TA pair, the first described TA system of this type. We further show that there are widespread homologues of this new class of TA system in diverse phyla, and that the antiphage activity is maintained within multiple enteric genera.
Results
An Eca Plasmid Provides Phage Resistance.
Eca causes soft-rot and blackleg disease of potatoes (21). A 5620-bp cryptic plasmid, pECA1039, was isolated from Eca 1039 and sequenced. Bioinformatic searches identified a ColE1-type replication origin and up to 11 predicted ORFs (supporting information (SI) Table S1 and Fig. 1A). The product of the third predicted ORF, designated ToxN, has 31% identity to AbiQ, an Abi protein from L. lactis W-37 plasmid pSRQ900 (20). The toxN gene was 3′ of a gene annotated de novo (designated toxI) and predicted to be operonic with toxN.
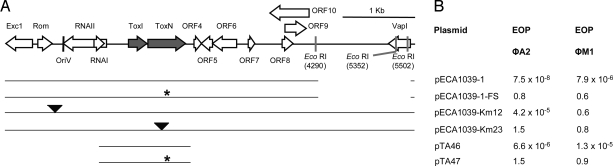
Fig. 1.
toxIN on plasmid pECA1039 encodes a phage-resistance system. (A) Linear map of pECA1039 and subsequent constructs, together with (B) EOP data of each construct versus φA2 and φM1. Here * denotes a frameshift mutation, and ▼ denotes a transposon insertion site.
As we have no phages able to infect Eca 1039, 3 pECA1039 subclones (pECA1039–1, -2, and -3) were used to transform Eca strain 1043, and the transformants were tested for phage resistance against phages φA2 and φM1 (22). Plasmid pECA1039–1 provided protection from φA2 and φM1 (Fig. 1B), but pECA1039–2 and pECA1039–3 did not (data not shown). Furthermore, a toxN frameshift (FS) mutation in pECA1039–1 abolished the phage-resistance phenotype (Fig. 1B). To determine whether the native replicon provided phage resistance, in vitro transposon mutants of pECA1039 were generated. In Eca 1043, plasmid pECA1039-Km12 provided resistance to φA2 (Fig. 1B), but only a reproducible reduction in plaque size for φM1 (data not shown). Why pECA1039-Km12 affects the size but not number of φM1 plaques is unclear. Plasmid pECA1039-Km23 has a transposon insertion within toxN, which abolished phage resistance (Fig. 1B). A smaller subcloned region consisting of toxN and toxI provided protection from phage infection, and this response was removed by a FS mutation within toxN (pTA46 and pTA47, respectively; Fig. 1B). Therefore, the toxIN locus on pECA1039 encodes an effective phage-resistance system.
toxIN Encodes an Abi System.
The protein sequence identity between ToxN and AbiQ from L. lactis suggests that the toxIN locus might encode a phage Abi system, and this hypothesis was tested (Table S2). Adsorption of phages φA2 and φM1 to Eca 1043 was unaffected by the presence of a toxIN plasmid, compared with toxI, _toxN_-FS, and vector-only control plasmids. Moreover, φA2 and φM1 “escape” mutant phages were isolated at a low frequency and were nonresponsive to toxIN. Therefore, to evaluate whether toxIN encoded a restriction-modification system, phages that were able to overcome the resistance system were passaged twice through Eca 1043 before retitrating on Eca 1043 containing toxIN. These phages retained their insensitivity to toxIN, indicating a stable genetic resistance mechanism and not phenotypic escape from a restriction-modification system (data not shown). The survival of Eca 1043 infected with φA2 and φM1 was unaffected by the presence of toxIN. Finally, toxIN dramatically reduced the burst size and efficiency of center of infection (ECOI) formation of φA2 and φM1 on Eca 1043. All of our data are consistent with ToxIN functioning as a phage Abi system (Table S2).
ToxN Is a Toxin in E. coli.
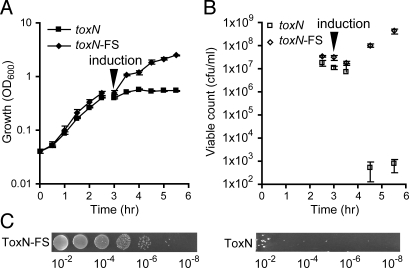
Initial attempts to clone toxN under nonnative promoters yielded only mutant toxN clones, suggesting that the gene product was toxic in E. coli. Indeed, induction of toxN expression using the P_araBAD_ promoter in E. coli resulted in growth cessation, as measured by OD600 (Fig. 2A), along with a ≈1 × 106 reduction in colony- forming units (cfu) per mL (Fig. 2 B and C). Induction of a _toxN_-FS strain had no effect, demonstrating that the ToxN protein was required for growth inhibition. Furthermore, E. coli expressing ToxN displayed no obvious morphological differences compared with the _toxN_-FS control under combined bright field and fluorescence microscopy (data not shown). Therefore, expression of the ToxN protein was growth-inhibiting to E. coli and did not result in cell lysis.
Fig. 2.
ToxN is growth-inhibiting. (A) Growth (OD600) and (B) viable counts of E. coli DH5α were measured after induction of the toxN gene (pTA49) or a _toxN_-FS control (pTA50); for details, see Materials and Methods. (C) Serial dilutions of exponentially grown cultures of E. coli DH5α with ToxN-FS (pTA50) or ToxN (pTA49) plated on LBA, Ap, and L-ara (inducing conditions).
toxIN Is Bicistronic.
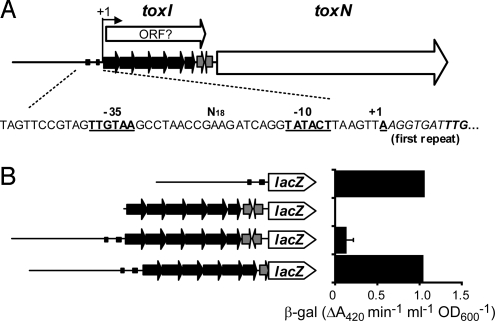
The toxI gene is composed of 5.5 almost-identical direct repeats of 36 nucleotides, followed by a predicted rho-independent transcriptional terminator (Fig. 3A). Within this region is a predicted ORF with a rare TTG start codon but no similarity to any other predicted protein. The transcriptional start site (+1) of toxI was mapped and found to be preceded by putative -10 and -35 promoter elements related to the E. coli σ70 consensus (Fig. 3A). In a low-copy lacZ promoter probe vector, the promoter had moderate expression in E. coli (Fig. 3B). Further experiments indicated no detectable toxN promoter within toxI. Transcriptional read-through into toxN from the toxI promoter past the terminator was detected, however (Fig. 3B). The presence of a read-through transcript from the mapped +1 into toxN was confirmed by RT-PCR (data not shown); thus, these 2 genes are cotranscribed, with the majority (≈90%) of transcriptions terminating at the rho-independent hairpin.
Fig. 3.
Organization of the toxIN locus. (A) Schematic representation of the toxIN locus. The transcription start (+1), toxI tandem repeats (black arrows), rho-independent terminator (gray arrows), toxN gene (white arrow), and promoter -35 and -10 elements are indicated. The hypothetical toxI ORF also is shown. (B) toxIN promoter lacZ transcriptional fusions in E. coli DH5α using plasmids (from top to bottom) pTA104, pTA105, pTA106, and pTA119.
toxI Encodes an Antitoxin.
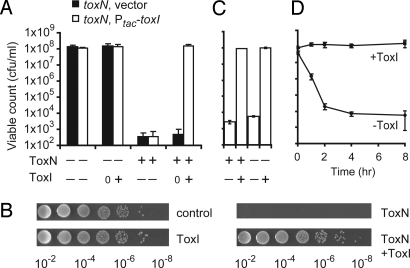
The toxIN genetic organization and toxic nature of toxN suggested that this locus might encode a TA system with toxI providing the antitoxin function. When cloned under an inducible promoter (P_tac_), induction of toxI expression provided antitoxin activity, demonstrating that toxI transcription was necessary for the repression of ToxN toxicity (Fig. 4 A and B). The toxicity of ToxN in Eca 1043 also was prevented by toxI expression, and co-overexpression of ToxI and ToxN from separate plasmids was found to confer phage resistance (data not shown). Next, to examine whether the antitoxin, ToxI, was less stable than ToxN, both components were expressed, and then either component or both components were switched off. As expected, when ToxN was either continuously expressed or turned off in the presence of ToxI, no decrease in viable count was observed (Fig. 4C); however, when both components were switched off, the viable count decreased by >1 × 104 (Fig. 4C), suggesting that ToxI is less stable than ToxN. In summary, these results demonstrate that the toxIN operon encodes a TA system.
Fig. 4.
The toxIN locus encodes a bacteriostatic TA system. (A) Protection of E. coli DH5α from ToxN inhibition by transcription of toxI. Protection assays were conducted as described in Materials and Methods, and the strains shown are E. coli DH5α, pTA49, pTA100 (toxN, vector) and E. coli DH5α, pTA49, pTA76 (toxN, P__tac-toxI_). The symbols “+” and “−” refer to induction or repression of toxN (L-ara and glu) and toxI (+/− IPTG). Empty vector induction is indicated by 0. (B) Serial dilutions of E. coli DH5α, pTA49, pTA76 on LBA plates with Ap, Sp and, glu (control), glu and IPTG (ToxN), L-ara (ToxN), and L-ara and IPTG (ToxN+ToxI). (C) ToxI is less stable than ToxN. E. coli DH5α, pTA49, pTA76 was grown expressing both ToxI and ToxN, as described in Materials and Methods, and plated under conditions resulting in continued expression or repression of either ToxI or ToxN or both (+ and −). (D) ToxN is bacteriostatic. ToxN was induced in E. coli DH5α, pTA49, pTA76 for different times, and viable counts were determined on LBA Ap, Sp, and glu plates without (ToxN) or with (ToxN+ToxI) IPTG. Time (hr) refers to hours after ToxN induction.
ToxIN Is a Reversible Bacteriostatic TA System.
The lack of ToxN-induced bacterial lysis prompted an investigation into whether or not ToxN is bacteriostatic. Delayed overexpression of toxI enabled full recovery of E. coli cells that had been expressing ToxN for at least 8 h (Fig. 4D). Furthermore, cells that had transiently expressed ToxN did not grow on plates within 1 day, due to the presence of the toxin. But after 2 days of incubation, colonies arose, presumably due to turnover of ToxN (data not shown). This finding indicates that ToxN functions through a reversible growth-inhibiting (bacteriostatic) mechanism.
ToxIN Is a Novel Protein–RNA TA System.
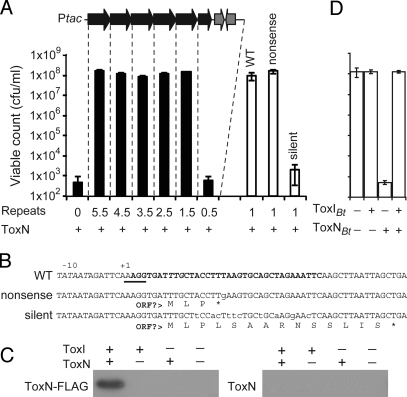
TA systems are broadly classified into protein–protein and RNA antisense groups. Whether or not toxI encoded a protein was examined. First, toxI was tagged with C-terminal hexahistidine sequences. These constructs were functional in protection assays, but no ToxI protein was detectable by Western blot analysis (data not shown). Next, plasmids were constructed that enabled the expression of RNA species with 5.5, 4.5, 3.5, 2.5, 1.5, and 0.5 repeats, followed by the native rho-independent transcriptional terminator. All plasmids with at least 1.5 “repeats” could protect E. coli from ToxN (Fig. 5A). In addition, expression of only a single 36-nucleotide (nt) repeat could inhibit ToxN toxicity (Fig. 5 A and B). It was possible that the single 36-nt RNA was encoding a small peptide that inhibits ToxN. To test this, a single nt nonsense mutation was generated in this 36-nt sequence that would terminate translation at the putative fourth codon. This plasmid could still protect E. coli from ToxN, whereas a plasmid with multiple silent point mutations in the ToxI RNA, which in theory could still code for the same hypothetical peptide, was nonfunctional (Fig. 5 A and B). Therefore, toxI encodes an RNA antitoxin, with the minimum functional unit currently defined as a single 36-nt repeat.
Fig. 5.
ToxIN is an RNA-protein TA system. (A) Protection of E. coli DH5α from ToxN inhibition (pTA49) by expression of toxI deletions composed of 5.5 (pTA76), 4.5 (pTA78), 3.5 (pTA79), 2.5 (pTA80), 1.5 (pTA81), 0.5 (pTA93), a WT single 36-nt repeat (pTA103), a nonsense 36-nt repeat mutant (pTA122), or a silent 36-nt repeat mutant (pTA107). (B) Sequences of the 36-nt inserts in pTA103 (WT), pTA122 (nonsense), and pTA107 (silent) and the putative short peptides that they may encode. A putative ribosome binding site is underscored, the possible start codon is in italic type, and the single repeat is in bold type. (C) Expression of the ToxI RNA results in the stable production of ToxN. E. coli DH5α, pTA76, pTRB1 and E. coli DH5α, pTA76, pTA49 were grown, and samples were probed with a polyclonal anti-FLAG antibody as described in Materials and Methods. (D) Protection of E. coli DH5α from ToxN_Bt_ inhibition by transcription of toxI Bt. Protection assays were performed as described in Materials and Methods; the strain shown here is E. coli DH5α, pTA117, pTA115 (toxN Bt, P__tac-toxIBt_). The symbols “+” and “−” refer to induction or repression of toxN (L-ara and glu) and toxI (+/− IPTG).
RNA antitoxins function through an antisense base-pairing mechanism that results in inhibition of toxin translation. To enable analysis of ToxN protein levels, we extended the toxN ORF to encode a C-terminal FLAG epitope. We then performed toxicity and phage-resistance assays, which confirmed that this C-terminal FLAG epitope does not affect ToxN toxicity or reduce phage resistance conferred by ToxN (data not shown). Surprisingly, coexpression of ToxI and ToxN resulted in detectable levels of ToxN protein when assessed by SDS/PAGE (data not shown) and Western blot analysis (Fig. 5C). The identity of the ToxN band separated by SDS/PAGE was verified by MS analysis (70% coverage). Overexpression of toxN alone did not lead to detectable levels of the FLAG-tagged ToxN protein (Fig. 5C). Therefore, toxI encodes an RNA (ToxI) that inhibits the toxic effects of the ToxN protein. To the best of our knowledge, this is the first reported example of a protein–RNA TA system.
ToxIN Defines a New TA Family.
The novel mechanism of ToxIN prompted us to examine the phylogenetic distribution of this TA module, which resulted in the identification of at least 19 ToxN homologues, including ToxN itself (Table S3). Genes predicted to encode ToxN-like proteins were found in Gram-positive and -negative bacteria across a range of bacterial phyla. ToxN has no sequence similarity to characterized protein domains, and thus it is the defining member of this new toxin class. Of the 18 _toxN_-like genes with 5′ sequences available, 16 had putative RNA antitoxin species with near-consensus promoters and predicted rho-independent terminators, and 13 of these had distinctive repeats (Table S3). The antitoxin sequences are not conserved in sequence or predicted structure (data not shown), although some closely related _toxN_-like genes are associated with near-identical repeats (e.g., Haemophilus influenzae plasmids) (Table S3). The TA gene organization is conserved in all detected members of this proposed novel class of protein–RNA TA systems.
To test whether the toxN_-like genes and their predicted antitoxin RNAs also functioned as protein-RNA TA systems, we examined toxIN from Bacillus thuringiensis serovar kurstaki strain HD73, plasmid pAW63 (toxIN Bt). Expression of ToxN_Bt in E. coli caused a dramatic reduction in viable count that could be inhibited by expression of the antitoxin RNA, ToxI_Bt_ (Fig. 5D), which has 3.1 near-identical repetitive sequences of 34 bp (Table S3).
toxIN Acts as an Abi System in Multiple Enteric Bacteria.
The distribution of toxIN_-like loci motivated us to test the host range of ToxIN activity. First, we demonstrated that ToxIN could provide resistance in Eca 1043 against a suite of phages. Of 25 Eca phages (including φA2 and φM1), 13 were sensitive [here defined as an efficiency of plaquing (EOP) ≤ 1 × 10−2] to ToxIN (pTA46), with the full-suite EOP ranging from 1 to ≤ 1 × 10−10. Next, we found that in E. coli DH5α, none of the 4 coliphages tested (Mu, λ_vir, P1_vir_, and T4) was affected by toxIN (pTA46), but 5 of 10 newly isolated coliphages (φTB16–25) were sensitive to ToxIN, with EOPs ≤ 1 × 10−8. Finally, in Serratia marcescens Db11, of 11 phages tested, 3 were sensitive to toxIN (pECA1039Km12 and Km15; reduced plaque size, EOP = 0.5 to < 1 × 10−7). This clearly indicates that toxIN encodes a TA/Abi system active against a range of phages within multiple enteric genera.
Discussion
We have identified and characterized the Eca plasmid-encoded toxIN locus, which directs abortive infection of multiple phages in various enteric bacteria. Our data demonstrate that toxIN functions as an Abi system (Fig. 1 and Table S2). Furthermore, we have shown that toxIN defines a new family of novel TA systems proposed to act through an RNA–protein interaction, an entirely new mechanistic class of TA systems separate from the well-characterized proteic (15, 18) and RNA antisense groups (19).
The toxic effects of ToxN were inhibited by expression of a gene transcribed immediately 5′ of toxN, designated toxI. This genetic organization is common to many TA modules. Together, the ToxI and ToxN products constitute a TA system with an antitoxin product, ToxI, apparently of lower stability than ToxN (Fig. 4). The toxIN genes are bicistronic, with most expression terminating after toxI, leaving ≈10% of read-through into toxN. Termination might be important for obtaining an appropriate ToxI:ToxN stoichiometry. Surprisingly, the growth-inhibiting effect of ToxN was bacteriostatic (Fig. 4), and ToxI could resuscitate E. coli in the absence of new ToxN synthesis, suggesting that ToxI may be able to compete efficiently with the cellular target(s) of ToxN. Our results are similar to those observed for RelE and MazF toxins expressed for up to 4 or 5 h, respectively (23); however, another study demonstrated a “point of no return” after overexpression of MazF, whereby MazE could rescue most bacterial growth only within 6 h (24).
The toxI gene is composed of 5.5 directly repeated sequences and contains a small putative ORF. But evidence supports the assignment of an RNA species as the active antitoxin component: (i) transcription of only a single repeat RNA of 36 nt retained full activity (Fig. 5); (ii) a nonsense point mutant variant that terminated the putative ORF remained functional, whereas a silent mutant variant that had unaltered peptide coding was inactive (Fig. 5); (iii) the predicted transcribed regions 5′ of most other toxN genes contain no predicted ORFs (data not shown); (iv) ToxI_Bt_ from Bacillus thuringiensis was an active antitoxin against ToxN_Bt_ but had no putative coding region (Fig. 5); and, finally, (v) overexpression of hexahistidine-tagged ToxI protected against ToxN with no detectable ToxI protein production.
The RNA nature of ToxI suggests that the toxIN system is an RNA antisense TA module; however, when ToxI RNA was overexpressed, the ToxN protein was detectable (Fig. 5). Clearly, the toxIN system cannot be described by RNA antisense or proteic TA systems and thus requires a new classification. We propose a new class of TA modules requiring an RNA antitoxin predicted to interfere with the biochemical activity of the protein toxin. Our bioinformatic analyses identified numerous ToxIN-type loci in diverse bacterial phyla and supported an organizational and functional link between the predicted RNA antitoxins and the ToxN-like genes. For example, 2 toxN pseudogenes identified in Histophilus somni strain 129PT displayed antitoxin degeneration (data not shown) and another putative ToxIN system, that from B. thuringiensis plasmid pAW63, functioned as a TA module in E. coli (Fig. 5).
Most _toxIN_-like genes are plasmid-encoded, and their phylogenetic distribution implies a role of horizontal gene transfer in their dissemination. Indeed, the B. thuringiensis plasmid pAW63 is conjugal (25). We were interested in mimicking the acquisition of the toxIN system by different bacterial genera and the effects on phage resistance. Our study provides the first evidence that an Abi (or TA) system can impart protection in different genera against multiple phages. This has implications for the dissemination and action of Abi/TA systems, and the technical utility of such a broad-spectrum system (with respect to both host and phages) has not escaped our attention.
Despite extensive recent research, debate continues about the biological roles of TA systems (reviewed in ref. 17). One suggested function of TA systems is to reduce phage infection; in fact, the hok/sok loci from E. coli plasmid R1 excludes phage T4 (26), and the chromosomal TA system, mazEF, protects against phage P1 (27). The toxIN genes abort the infections of different phages and function in a number of different genera; therefore, chromosomal or plasmid-encoded representatives across 3 TA classes [proteic (mazEF), RNA antisense (hok/sok), and protein–RNA (toxIN)] can function to limit phage infection. This study demonstrates that the effect of TA modules as antiphage elements may be an evolutionarily important, widespread phenomenon. This is particularly important in light of the huge numbers of phages in the environment (2) and the strong selective pressure on bacteria to develop phage-resistance mechanisms (28). But TA elements can have biological roles in the absence of phages (e.g., plasmid stabilization) (17), and phages may provide further selective pressure for the maintenance of these genes in some circumstances. Interestingly, toxIN also can provide plasmid stabilization (P.F., unpublished data).
To the best of our knowledge, this is the first case of an Abi system that functions as a TA module, blurring the boundary between these systems. Some features of other Abi systems show similarities to TA modules. Some Abi systems require 2 protein components [e.g., AbiE (29), AbiG (30), AbiL (31), and AbiT (32)], and others may have RNA antitoxins presumably overlooked by standard gene sequence analysis [e.g., _abiQ_ (20)]. In addition, numerous Abi proteins are toxic when expressed in the absence of phages [e.g., AbiD1 (33), AbiB (9), and AbiK (34)]. Analogously, restriction-modification systems, thought to function primarily in phage defense, also can function as TA modules for plasmid stabilization (35).
The antiphage activity of the ToxN homologue AbiQ is characterized by a late-acting step that prevents the processing of accumulated phage DNA (20). ToxN also prevents mature phage particle formation after normal phage DNA accumulation (Table S2 and T.B., unpublished data). Based on our current understanding, we propose an extended model for ToxIN antiphage activity. Before phage infection, transcription of the toxIN locus results in an excess of the unstable ToxI RNA relative to the ToxN protein. The ToxI RNA is predicted to interact directly with ToxN and inhibit toxicity. On phage infection, alterations in host transcription or translation or the degradation of bacterial DNA could destabilize the ToxI:ToxN ratio, freeing ToxN to interact with its target(s) to inhibit growth. Alternatively, a specific phage product could interact directly with ToxIN and trigger this system; for example, a phage antitermination mechanism may act to increase ToxN levels. We favor the first hypothesis because of the action of toxIN against multiple phages in different hosts, the observed sequence diversity in the phage gene pool (3), and the observation of no change in ToxN levels after phage infection (T.B., unpublished data). Due to its putative ToxI RNA binding, ToxN might function in the absence of ToxI to target cellular RNA [e.g., MazF from _E. coli_ (36)]. It remains unclear how this can lead to the prevention of phage DNA processing. Research into toxIN will provide novel information about the mechanistic features of this new class of TA systems and insight into phage–host interactions.
Materials and Methods
Bacterial Strains, Plasmids, and Culture Conditions.
Eca 1043 (21) and 1039 (37) were grown at 25°C, E. coli DH5α (Gibco/BRL) was grown at 37°C, and S. marcescens Db11 (38) was grown at 30°C in Luria broth (LB) at 300 rpm or on LB agar (LBA) containing 1.5% (w v−1) agar, and growth (OD600) was measured as described previously (39). When required, LB was supplemented with the following antibiotics: kanamycin 50 μg mL−1, spectinomycin (Sp) 50 μg mL−1, ampicillin (Ap) 100 μg mL−1, and tetracycline 35 μg mL−1. When required, D-glucose (glu) at 0.2% (w v−1), L-arabinose (L-ara) at 0.1% (w v−1), and isopropyl-β-D-thiogalactopyranoside (IPTG) at 1 mM were used, unless stated otherwise. All experiments were performed at least in triplicate (unless stated otherwise) and plotted as mean ± SD.
DNA Manipulations and Sequence Analysis.
Molecular biology techniques and sequencing were performed as described previously (39). Named primers are listed in Table S4. All plasmids were verified by DNA sequencing. Sequence data were analyzed using GCG (Genetics Computer Group, University of Wisconsin), and ToxN homologues were identified using BLAST and PSI-BLAST. Direct repeats were identified using Tandem Repeats Finder (40), and transcriptional terminators were detected using STEMLOOP in GCG. ORFs were predicted by Genemark.hmm (41) and ORF Finder (http://www.ncbi.nlm.nih.gov/projects/gorf/).
Subcloning and Sequencing of pECA1039.
Plasmid pECA1039 was extracted from Eca 1039 and cloned into EcoRI-digested pUC19. Three pECA1039 EcoRI subclones in pUC19 (pECA1039–1, -2, and -3) were sequenced. pECA1039 was completed by sequencing across the EcoRI junctions. pECA1039–1 was digested with BsmI, the 3′ overhang was removed with T4 polymerase, and ligated. The resulting plasmid (pECA1039–1-FS) has a 2-bp deletion, causing a premature stop codon after L114 in the ToxN protein. The toxIN genes and toxI, _toxN_-FS controls were cloned by PCR into pBR322 EcoRI and HindIII sites with primers MJ7 and KD02 using pECA1039–1 and pECA1039–1-FS, respectively, as template DNA, producing pTA46 (toxIN) and pTA47 (toxI, _toxN_-FS).
In Vitro Mutagenesis of pECA1039.
In vitro transposon mutagenesis was performed on plasmid pECA1039 with EZ::TN™ <NotI/KAN-3>, following the manufacturer's instructions (Epicentre). Insertion sites of the EZ::TN™ <NotI/KAN-3> transposons were mapped by sequencing using primer PF134.
Phage Techniques.
Phage resistance (i.e., EOP) was calculated after overnight incubation of phages in a 0.35% agar lawn of bacterial host using (phage titer on test host/phage titer on control host). Adsorption assays were performed as follows. A 10-mL bacterial culture was adjusted to OD600 = 1, inoculated with phages at a multiplicity of infection of 0.01, and incubated at 25°C at 300 rpm. Samples were obtained at 0 min and 30 min (φA2) or at 40 min (φM1), and each sample was added to 900 μL of phage buffer. After centrifugation for 10 min at 16,200 × g, 10 μL of the supernatant was taken for titer determination. Percentage adsorption was calculated as the percent change in supernatant titer from 0 to 30/40 min. Cell survival, ECOI, and burst size assays were performed as described previously (20).
ToxN Toxicity Assays.
The toxN and toxN_-FS genes were cloned by PCR into pBAD30 (42) EcoRI and HindIII sites using primers PF137 and KD02, with pECA1039–1 and pECA1039–1-FS, respectively, as template DNA. Transformants of all pBAD30 clones were selected on LBA, Ap, and glu to repress expression of the P_araBAD promoter. The resulting constructs enabled controlled expression of native, untagged ToxN (pTA49) and ToxN-FS (pTA50).
Cultures of E. coli DH5α, pTA49 and E. coli DH5α, pTA50 were grown overnight with Ap and glu. These cultures were then incubated in 25 mL of LB, Ap, and glu in 250-mL flasks at 37°C and 300 rpm from a starting OD600 ≈ 0.04 until the cultures were in the exponential phase (≈1 × 108 cfu mL−1). Then the bacteria were resuspended in LB, Ap, and L-ara and incubated as described earlier. At specified times, the OD600 was measured and samples were removed, washed with PBS, and plated for viable counts at 37°C on LBA, Ap, and glu.
ToxIN Bacteriostatic, Protection, and Stability Assays.
Bacteriostatic assays were performed using E. coli DH5α, pTA49, pTA76 exactly as for the aforementioned toxicity assays, except that cfu were determined at different times at 37°C on (i) LBA, Ap, Sp, and glu and (ii) LBA, Ap, Sp, glu, and IPTG. ToxIN protection assays were performed as described for the toxicity assays with the following modifications: A range of different ToxI expression plasmids was used with appropriate antibiotic selection (see Results for details), the L-ara induction step was omitted, and the cells were enumerated on LBA, Ap, and Sp plates supplemented with (i) glu, (ii) glu and IPTG, (iii) L-ara, and (iv) L-ara and IPTG. ToxIN stability assays using E. coli DH5α, pTA49, pTA76 were carried out similarly to the ToxIN protection assays but with cultures grown in LB, Ap, Sp, L-ara, and IPTG (instead of in LB, Ap and glu) before plating for viable counts.
Mapping the Transcriptional Start of toxIN.
RNA was extracted from Eca 1043, pTA46, and 5′ RACE of toxIN was performed using the Roche 5′/3′ second-generation RACE kit. cDNA was synthesized using random hexamers and SuperScript II RT (Invitrogen) and the specific primers used were PF146 and PF147. The transcriptional start site of toxIN was confirmed by sequencing five 5′ RACE clones. As a further confirmation, RT-PCR was used to validate the location of the 5′ end of the transcript (data not shown).
toxIN Promoter lacZ Fusion Experiments.
The toxIN promoter was amplified using primers PF186 and PF187. To test for a separate toxN promoter, the toxI region was amplified using primers PF188 and PF189. To examine transcriptional read-through into the toxin gene, the toxIN promoter and toxI were amplified by PCR using primers PF186 and either PF189 or PF202. The resulting PCR products were cloned into the EcoRI and HindIII sites of pRW50 (39), giving plasmids pTA104, pTA105, pTA106, and pTA119. Promoter expression was determined as described previously (39).
Construction of ToxI Expression Vectors.
For ToxI expression vectors compatible with pTA49, a Sm/Sp-resistant derivative of pQE-80L (QIAGEN) was created as follows. First, the Sm/Sp-resistance cassette from miniTn_5_Sm/Sp in strain LIS (39) was cloned by PCR into pQE-80L BspHI sites, using primers PF172 and PF173, resulting in plasmid pTA100. Then a series of toxI IPTG-inducible expression vectors with varying numbers of DNA sequence repeats were created, as described below. The toxI gene was cloned by PCR into pTA100 EcoRI and HindIII sites using primers PF164 and MJ12. Plasmids were isolated with 5.5 (pTA76), 4.5 (pTA78), 3.5 (pTA79), 2.5 (pTA80), and 1.5 (pTA81) repeats. A 0.5-repeat construct (pTA93) was constructed in the same manner but with PF183 instead of PF164. To generate a plasmid with a single 36-nt repeat unit transcribed precisely from the +1 in the IPTG-inducible promoter in pTA100, PCR was performed with PF185 and PF184 using pQE-80L as the template. The product was digested with XhoI and HindIII and ligated into pTA100 cut with the same enzymes, giving plasmid pTA103. Plasmids that expressed a single mutant repeat containing either silent mutations relative to the predicted peptide coding sequence (pTA107) or a nonsense mutation (pTA122) were created in the same manner as pTA103 but with primers PF190 or PF260, respectively, instead of PF184.
Western Blot Analysis for ToxN.
A C-terminally FLAG-tagged toxN gene was cloned by PCR into pBAD30 EcoRI and HindIII sites using primers PF137 and MJ13, resulting in pTRB1. For protein samples, cultures of E. coli DH5α, pTA76, pTRB1 and E. coli DH5α, pTA76, pTA49 were grown as described for the protection assays until reaching an OD600 ≈ 0.5–0.8. Then cultures of each strain were resuspended into 25 mL of LB, Ap, and Sp supplemented with (i) glu, (ii) glu and IPTG, (iii) L-ara, or (iv) L-ara and IPTG and grown at 15°C and 300 rpm for 18 h. Western blot analysis was performed against samples from the resulting cultures (normalized to OD600), using a primary rabbit polyclonal anti-FLAG antibody (Sigma–Aldrich) and a goat anti-rabbit HRP conjugated secondary antibody (Sigma–Aldrich) as directed by the manufacturer. Mass spectrometry of ToxN was performed at PNAC, University of Cambridge.
Construction of Bacillus thuringiensis toxIN Plasmids.
The B. thuringiensis toxIBt and toxNBt genes were cloned by PCR from plasmid pAW63 into EcoRI and HindIII sites of pTA100 and pBAD30, respectively, using primer pairs PF194–PF196 and PF197–PF195. The resulting plasmids were pTA115 (toxIBt) and pTA117 (toxNBt).
Supplementary Material
Supporting Information
Acknowledgments.
We thank members of the Salmond and Welch groups, Dr N. Crickmore, Dr L. Gumy, Dr M. Johnson, C. Kirkpatrick, and Dr I. Toth for discussions, strains, phages, and microscopy assistance. This work was supported by the Biotechnology and Biological Sciences Research Council and performed under a Department for Environment, Food and Rural Affairs plant health license. T.B. also was supported by a Collaborative Award in Science and Engineering studentship from UCB-Celltech Ltd.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data Deposition: The sequence reported in this paper has been deposited in the GenBank database (Plasmid pECA1039, accession no. FJ176937).
References
- 1.Chibani-Chennoufi S, Bruttin A, Dillmann ML, Brussow H. Phage–host interaction: An ecological perspective. J Bacteriol. 2004;186:3677–3686. doi: 10.1128/JB.186.12.3677-3686.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wommack KE, Colwell RR. Virioplankton: Viruses in aquatic ecosystems. Microbiol Mol Biol Rev. 2000;64:69–114. doi: 10.1128/mmbr.64.1.69-114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lima-Mendez G, Toussaint A, Leplae R. Analysis of the phage sequence space: The benefit of structured information. Virology. 2007;365:241–249. doi: 10.1016/j.virol.2007.03.047. [DOI] [PubMed] [Google Scholar]
- 4.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: The unseen majority. Proc Natl Acad Sci U S A. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuhrman JA. Marine viruses and their biogeochemical and ecological effects. Nature. 1999;399:541–548. doi: 10.1038/21119. [DOI] [PubMed] [Google Scholar]
- 6.Petty NK, Evans TJ, Fineran PC, Salmond GP. Biotechnological exploitation of bacteriophage research. Trends Biotechnol. 2007;25:7–15. doi: 10.1016/j.tibtech.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 7.Sorek R, Kunin V, Hugenholtz P. CRISPR: A widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. 2008;6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- 8.Forde A, Fitzgerald GF. Bacteriophage defence systems in lactic acid bacteria. Antonie Van Leeuwenhoek. 1999;76:89–113. [PubMed] [Google Scholar]
- 9.Chopin MC, Chopin A, Bidnenko E. Phage abortive infection in lactococci: Variations on a theme. Curr Opin Microbiol. 2005;8:473–479. doi: 10.1016/j.mib.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 10.Snyder L. Phage-exclusion enzymes: A bonanza of biochemical and cell biology reagents? Mol Microbiol. 1995;15:415–420. doi: 10.1111/j.1365-2958.1995.tb02255.x. [DOI] [PubMed] [Google Scholar]
- 11.Chowdhury R, Biswas SK, Das J. Abortive replication of choleraphage phi 149 in Vibrio cholerae biotype el tor. J Virol. 1989;63:392–397. doi: 10.1128/jvi.63.1.392-397.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith HS, Pizer LI, Pylkas L, Lederberg S. Abortive infection of Shigella dysenteriae P2 by T2 bacteriophage. J Virol. 1969;4:162–168. doi: 10.1128/jvi.4.2.162-168.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durmaz E, Klaenhammer TR. Abortive phage-resistance mechanism AbiZ speeds the lysis clock to cause premature lysis of phage-infected Lactococcus lactis. J Bacteriol. 2007;189:1417–1425. doi: 10.1128/JB.00904-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bidnenko E, Ehrlich D, Chopin MC. Phage operon involved in sensitivity to the Lactococcus lactis abortive infection mechanism AbiD1. J Bacteriol. 1995;177:3824–3829. doi: 10.1128/jb.177.13.3824-3829.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buts L, Lah J, Dao-Thi MH, Wyns L, Loris R. Toxin-antitoxin modules as bacterial metabolic stress managers. Trends Biochem Sci. 2005;30:672–679. doi: 10.1016/j.tibs.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 16.Yarmolinsky MB. Programmed cell death in bacterial populations. Science. 1995;267:836–837. doi: 10.1126/science.7846528. [DOI] [PubMed] [Google Scholar]
- 17.Magnuson RD. Hypothetical functions of toxin-antitoxin systems. J Bacteriol. 2007;189:6089–6092. doi: 10.1128/JB.00958-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerdes K, Christensen SK, Lobner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat Rev Microbiol. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 19.Gerdes K, Wagner EG. RNA antitoxins. Curr Opin Microbiol. 2007;10:117–124. doi: 10.1016/j.mib.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 20.Emond E, et al. AbiQ, an abortive infection mechanism from Lactococcus lactis. Appl Environ Microbiol. 1998;64:4748–4756. doi: 10.1128/aem.64.12.4748-4756.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bell K, et al. Genome sequence of the enterobacterial phytopathogen Erwinia carotovora subsp. atroseptica and characterization of virulence factors. Proc Natl Acad Sci U S A. 2004;101:11105–11110. doi: 10.1073/pnas.0402424101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toth IK, et al. Evaluation of phenotypic and molecular typing techniques for determining diversity in Erwinia carotovora subspp. atroseptica. J Appl Microbiol. 1999;87:770–781. doi: 10.1046/j.1365-2672.1999.00929.x. [DOI] [PubMed] [Google Scholar]
- 23.Pedersen K, Christensen SK, Gerdes K. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol Microbiol. 2002;45:501–510. doi: 10.1046/j.1365-2958.2002.03027.x. [DOI] [PubMed] [Google Scholar]
- 24.Amitai S, Yassin Y, Engelberg-Kulka H. MazF-mediated cell death in Escherichia coli: a point of no return. J Bacteriol. 2004;186:8295–8300. doi: 10.1128/JB.186.24.8295-8300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilcks A, Jayaswal N, Lereclus D, Andrup L. Characterization of plasmid pAW63, a second self-transmissible plasmid in Bacillus thuringiensis subsp. kurstaki HD73. Microbiology. 1998;144(Pt 5):1263–1270. doi: 10.1099/00221287-144-5-1263. [DOI] [PubMed] [Google Scholar]
- 26.Pecota DC, Wood TK. Exclusion of T4 phage by the hok/sok killer locus from plasmid R1. J Bacteriol. 1996;178:2044–2050. doi: 10.1128/jb.178.7.2044-2050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hazan R, Engelberg-Kulka H. Escherichia coli mazEF-mediated cell death as a defense mechanism that inhibits the spread of phage P1. Mol Genet Genom. 2004;272:227–234. doi: 10.1007/s00438-004-1048-y. [DOI] [PubMed] [Google Scholar]
- 28.Hoskisson PA, Smith MC. Hypervariation and phase variation in the bacteriophage “resistome”. Curr Opin Microbiol. 2007;10:396–400. doi: 10.1016/j.mib.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 29.Garvey P, Fitzgerald GF, Hill C. Cloning and DNA sequence analysis of two abortive infection phage-resistance determinants from the lactococcal plasmid pNP40. Appl Environ Microbiol. 1995;61:4321–4328. doi: 10.1128/aem.61.12.4321-4328.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Connor L, Coffey A, Daly C, Fitzgerald GF. AbiG, a genotypically novel abortive infection mechanism encoded by plasmid pCI750 of Lactococcus lactis subsp. cremoris UC653. Appl Environ Microbiol. 1996;62:3075–3082. doi: 10.1128/aem.62.9.3075-3082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng YM, Liu CQ, Dunn NW. Genetic organization and functional analysis of a novel phage abortive infection system, AbiL, from Lactococcus lactis. J Biotechnol. 1999;67:135–149. doi: 10.1016/s0168-1656(98)00175-8. [DOI] [PubMed] [Google Scholar]
- 32.Bouchard JD, Dion E, Bissonnette F, Moineau S. Characterization of the two-component abortive phage infection mechanism AbiT from Lactococcus lactis. J Bacteriol. 2002;184:6325–6332. doi: 10.1128/JB.184.22.6325-6332.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anba J, Bidnenko E, Hillier A, Ehrlich D, Chopin MC. Characterization of the lactococcal abiD1 gene coding for phage abortive infection. J Bacteriol. 1995;177:3818–3823. doi: 10.1128/jb.177.13.3818-3823.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emond E, et al. Phenotypic and genetic characterization of the bacteriophage abortive infection mechanism AbiK from Lactococcus lactis. Appl Environ Microbiol. 1997;63:1274–1283. doi: 10.1128/aem.63.4.1274-1283.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kulakauskas S, Lubys A, Ehrlich SD. DNA restriction-modification systems mediate plasmid maintenance. J Bacteriol. 1995;177:3451–3454. doi: 10.1128/jb.177.12.3451-3454.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Engelberg-Kulka H, Hazan R, Amitai S. mazEF: A chromosomal toxin-antitoxin module that triggers programmed cell death in bacteria. J Cell Sci. 2005;118:4327–4332. doi: 10.1242/jcs.02619. [DOI] [PubMed] [Google Scholar]
- 37.Bell KS, et al. Sample sequencing of a selected region of the genome of Erwinia carotovora subsp. atroseptica reveals candidate phytopathogenicity genes and allows comparison with Escherichia coli. Microbiology. 2002;148:1367–1378. doi: 10.1099/00221287-148-5-1367. [DOI] [PubMed] [Google Scholar]
- 38.Flyg C, Kenne K, Boman HG. Insect pathogenic properties of Serratia marcescens: Phage-resistant mutants with a decreased resistance to Cecropia immunity and a decreased virulence to Drosophila. J Gen Microbiol. 1980;120:173–181. doi: 10.1099/00221287-120-1-173. [DOI] [PubMed] [Google Scholar]
- 39.Fineran PC, Everson L, Slater H, Salmond GP. A GntR family transcriptional regulator (PigT) controls gluconate-mediated repression and defines a new, independent pathway for regulation of the tripyrrole antibiotic, prodigiosin, in Serratia. Microbiology. 2005;151:3833–3845. doi: 10.1099/mic.0.28251-0. [DOI] [PubMed] [Google Scholar]
- 40.Benson G. Tandem Repeats Finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lukashin AV, Borodovsky M. GeneMark.hmm: New solutions for gene finding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information