Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection (original) (raw)
. Author manuscript; available in PMC: 2009 Dec 26.
SUMMARY
Although many transcription factors are known to control important aspects of neural development, the genome-wide programs that are directly regulated by these factors are not known. We have characterized the genetic program that is activated by MEF2, a key regulator of activity-dependent synapse development. These MEF2 target genes have diverse functions at synapses, revealing a broad role for MEF2 in synapse development. Several of the MEF2 targets are mutated in human neurological disorders including epilepsy and autism-spectrum disorders, suggesting that these disorders may be caused by disruption of an activity-dependent gene program that controls synapse development. Our analyses also reveal that neuronal activity promotes alternative polyadenylation site usage at many of the MEF2 target genes, leading to the production of truncated mRNAs that may have different functions than their full-length counterparts. Taken together, these analyses suggest that the ubiquitously expressed transcription factor MEF2 regulates an intricate transcriptional program in neurons that controls synapse development.
INTRODUCTION
In the central nervous system (CNS), environmental stimuli regulate the development and function of neural circuits that underlie behavior and cognition. Sensory experience, which leads to an increase in neurotransmitter release onto individual neurons in the CNS, promotes both the maturation of synapses and the elimination of excess synapses within various neural circuits during postnatal development (Hua and Smith, 2004), and drives experience-dependent changes in synaptic connectivity that underlie learning and memory (Malinow and Malenka, 2002). One way increased neurotransmitter release triggers changes in circuit connectivity is through new gene transcription. Increased synaptic activity leads to calcium influx into the postsynaptic cell, which activates calcium-dependent signaling pathways that in turn regulate transcription factors within the nucleus (Flavell and Greenberg, 2008). Several transcription factors that mediate neuronal activity-dependent transcription in neurons, including CREST and NeuroD, control early steps of neural circuit development such as dendritic outgrowth (Aizawa et al., 2004; Gaudilliere et al., 2004). Other activity-regulated transcription factors, including CREB, SRF, NeuroD2, and MEF2 family members, regulate later aspects of circuit development by controlling synaptic development and function (Barco et al., 2002; Etkin et al., 2006; Flavell et al., 2006; Ince-Dunn et al., 2006; Ramanan et al., 2005; Shalizi et al., 2006).
Despite evidence that individual activity-regulated transcription factors control specific aspects of neural circuit development, the molecular mechanisms by which these factors coordinate complex processes such as dendritic outgrowth and synaptic development remain unclear. Previous studies have for the most part identified the target genes of activity-dependent transcription factors one at a time. Thus, the diversity and complexity of the activity-regulated gene networks remain to be investigated. For example, except perhaps for CREB which has been suggested to control hundreds of target genes in neuronal cell lines (Impey et al., 2004), it is not known if a given activity-regulated transcription factor regulates the expression of several or perhaps hundreds of target genes in order to coordinate a specific aspect of neural circuit development.
MEF2 family transcription factors are critical for the development and function of many types of cells, including those found in the musculoskeletal, cardiac, vascular, immune and nervous systems (Potthoff and Olson, 2007). In all of these contexts, MEF2 transcriptional activity is tightly regulated by extracellular stimuli. In neurons, MEF2 can be activated by neurotrophin stimulation as well as calcium influx resulting from increased neurotransmitter release at synapses. The neuronal activity-dependent activation of MEF2 induces a program of gene expression that restricts the number of excitatory synapses formed onto hippocampal neurons, cerebellar granule neurons and medium spiny neurons of the nucleus accumbens both in vitro and in vivo (Barbosa et al., 2008; Flavell et al., 2006; Pulipparacharuvil et al., 2008; Shalizi et al., 2006). Furthermore, the disruption of MEF2 expression in vivo in the hippocampus or the nucleus accumbens results in deficits in behavioral plasticity that are correlated with an increase in excitatory synapse number (Barbosa et al., 2008; Pulipparacharuvil et al., 2008). Consistent with a widespread role for MEF2 in synapse development, a similar function for MEF2 has also been identified in species as distant from mammals as the nematode C. elegans, where mef-2 negatively regulates excitatory synaptic function at the cholinergic neuromuscular synapse (Simon et al., 2008). Despite the importance of MEF2 as a mediator of activity-dependent synaptic development in a wide range of species, the mechanisms by which MEF2 orchestrates synaptic maturation are not known.
To examine how MEF2 coordinates synapse development in response to neuronal activity, we have applied genome-wide strategies to identify the full complement of target genes that are controlled by MEF2 in neurons during the process of activity-dependent synapse development. This approach of understanding the function of a transcription factor through the identification of its target genes has the potential to reveal mechanistic insights into transcription factor function that might be missed by traditional loss-of-function experiments (through RNAi or gene targeting). Using the loss-of function approach, it is often difficult to discern the direct effects of transcription factor removal (resulting from altered expression of its direct target genes) from the homeostatic effects that arise as an indirect consequence of altering the expression of an entire transcriptional program for several days.
In this study, we have used genome-wide approaches to identify 182 activity-dependent MEF2 target genes that regulate a variety of different aspects of synaptic function including excitatory synapse weakening, excitatory synapse maturation, inhibitory synapse development and presynaptic vesicle release. Mutations in several of the MEF2 target genes in the human population result in an increased susceptibility to neurological disorders such as epilepsy and autism-spectrum disorders, suggesting that these disorders may be caused at least in part by disruption of activity-dependent gene networks that control synapse development. One striking feature of the gene network controlled by MEF2 is that in response to neuronal activity there is a switch in polyadenylation site usage at many MEF2 target genes that results in the activity-dependent production of truncated mRNAs, many of which are predicted to have functions that are distinct from their pre-existing, full-length counterparts. Taken together, these experiments reveal new mechanisms by which the activity-regulated transcription factor MEF2 coordinates synaptic maturation during brain development.
RESULTS AND DISCUSSION
mRNA expression profiling reveals activity-regulated MEF2 target genes in hippocampal neurons
As a first step towards defining the activity-regulated MEF2-dependent transcriptional program that controls synapse development, we conducted an mRNA profiling experiment to identify activity-regulated genes in hippocampal neurons undergoing synapse development. Hippocampal neurons harvested from embryonic day 18 (E18) rats were grown in dissociated cultures for 10 days in vitro (DIV). Hippocampal neurons at this stage in their development are undergoing extensive changes in both synapse number and synaptic strength, and we have previously shown that the manipulation of MEF2 activity in these cells alters the course of synapse development (Flavell et al., 2006). To identify activity-regulated genes that might control synapse development, we exposed neurons to elevated levels of extracellular potassium chloride (KCl), which leads to membrane depolarization and calcium influx via L-type voltage-gated calcium channels. Using Affymetrix RAE230.2 microarrays that include ~30,000 probe sets, we identified 643 probe sets that are upregulated after one or six hours of membrane depolarization (Fig. 1A). Importantly, a large number of the genes we identified are also activated by physiological levels of neurotransmitter release in vivo, including c-fos, bdnf, gadd45b, nur77, egr1, arc and many others (Majdan and Shatz, 2006).
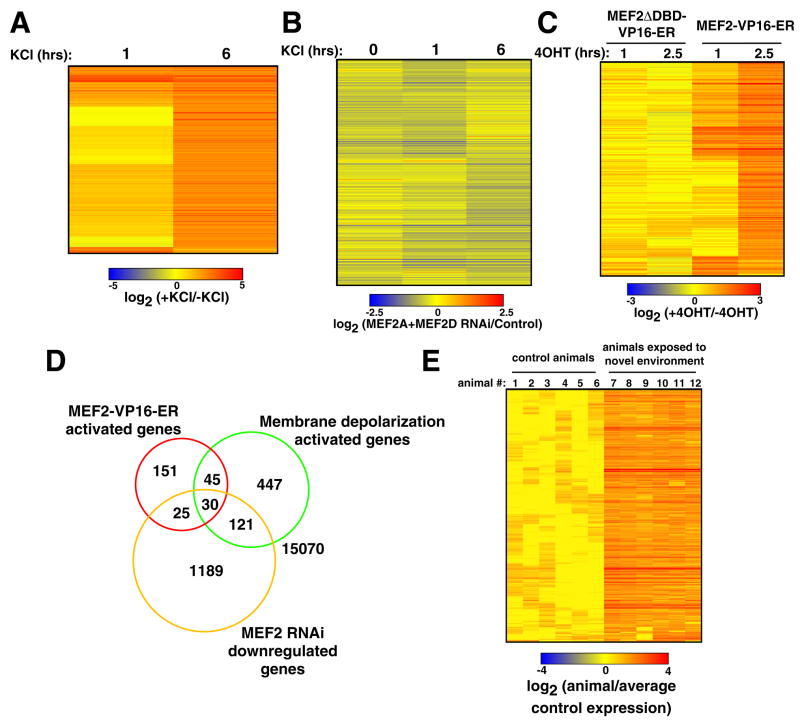
Figure 1. mRNA profiling analysis reveals activity-regulated MEF2 target genes.
(A) 643 probe sets whose expression is upregulated in E18+10 days in vitro (DIV) hippocampal neurons upon KCl-mediated membrane depolarization were identified by microarray analysis. Probe sets with similar expression profiles were clustered together using a Pearson correlation-based method and the expression levels of each of these probe sets (each line represents a single probe set) are displayed as a log2ratio of their expression values after one or six hours after KCl depolarization divided by their expression value in unstimulated (0 hour) cells. (B) 1365 probe sets whose expression is downregulated in E18+10 DIV hippocampal neurons in the presence of MEF2 RNAi were identified by microarray analysis. These probe sets were clustered as in (A) and expression levels are displayed as a log2ratio of their expression values in the presence of MEF2 RNAi divided by their expression values under control conditions for each of the three time points examined. (C) 251 probe sets whose expression is upregulated by 4OHT application to MEF2-VP16-ER-expressing hippocampal neurons at E18+10 DIV were identified by microarray analysis. Probe sets were clustered as in (A) and their levels of expression are displayed as the log2ratio of the expression value one or 2.5 hours after 4OHT application divided by the expression value prior to 4OHT application. (D) Venn Diagram showing the overlap of the probe sets identified in the three gene sets shown in (A–C). The levels of overlap between each of these three gene sets are statistically significant (p<0.0001 for all three comparisons, Fisher Exact Test). (E) 304 probe sets whose expression is upregulated in forebrains of rats exposed to a novel environment for three hours were identified by microarray analysis. These probe sets were clustered as in (A) and their levels of expression are displayed as the log2ratio of their expression values in each individual animal divided by the mean expression value of all control animals.
To determine which of these activity-regulated genes might be targets of MEF2 that control synapse development, MEF2 expression in neurons was reduced by introducing into the neurons MEF2A- and MEF2D-specific shRNAs using lentiviral vectors. By comparing the microarray results between the control (scrambled shRNA expression) and MEF2 RNAi conditions both before and after membrane depolarization, we were able to identify a large set of genes whose expression is altered as a result of MEF2 knockdown (1365 probe sets identified; Fig. 1B).
When the genes whose expression is downregulated in the presence of MEF2 RNAi (at any time point) are pooled together into a single gene set, there is a highly significant level of overlap between this gene set and the activity-regulated gene set (151 overlapping probe sets, p<0.0001, Fisher Exact Test; Fig. 1D). The genes that are present in both gene sets are most often decreased in their expression in the presence of MEF2 RNAi either one or six hours after membrane depolarization (147/151, or 97%), but only rarely in unstimulated cells (25/151, or 17%). Importantly, the majority (70%) of the activity-regulated genes identified in our initial analysis are induced normally even in the presence of MEF2 RNAi. This indicates that the knockdown of MEF2 does not prevent activity-dependent gene expression generally, but instead alters the expression of a subset of activity-regulated genes.
It is possible that in the MEF2 loss-of-function experiments we might have failed to identify particular activity-regulated MEF2 target genes because another transcription factor compensates for the loss of MEF2 at the promoters of these genes. To circumvent this potential problem, we performed a MEF2 gain-of-function experiment in hippocampal neurons at the time that synapses are forming and maturing. For these experiments we employed an inducible form of MEF2 in which the ligand binding domain of the estrogen receptor (ER) is fused to the C-terminus of the constitutively active MEF2-VP16 fusion protein to generate MEF2-VP16-ER. We have previously shown that in the absence of the ER ligand 4-OH-Tamoxifen (4OHT), MEF2-VP16-ER remains sequestered in the cytoplasm of hippocampal neurons and is inactive (Flavell et al., 2006). However, upon addition of 4OHT to the culture media MEF2-VP16-ER rapidly translocates to the nucleus and activates MEF2-dependent transcription. Hippocampal neurons were transduced with a lentivirus that drives the expression of MEF2-VP16-ER and mRNA profiling experiments were performed in the absence or presence of 4OHT using the same microarray platform described above. To control for the non-specific effects that 4OHT application or MEF2-VP16-ER expression might have on gene expression, we generated a lentivirus that expresses a DNA binding-deficient control version of MEF2-VP16-ER (MEF2ΔDBD-VP16-ER) and performed a parallel series of experiments using neurons transduced with this virus. Probe sets that were upregulated in response to 4OHT application in both MEF2-VP16-ER- and MEF2ΔDBD-VP16-ER-expressing neurons were excluded from further analysis.
We identified 251 probe sets that display increased expression in neurons expressing MEF2-VP16-ER within one or 2.5 hours of 4OHT application, but not in neurons expressing MEF2ΔDBD-VP16-ER (Fig. 1C). We asked whether these probe sets overlap with the activity-regulated gene set or the set of genes whose expression is downregulated in the presence of MEF2 RNAi. In both cases there was a highly significant degree of overlap (75 and 55 probe sets, respectively, p<0.0001 for both comparisons, Fisher Exact Test; Fig. 1D), whereas the group of probe sets that were downregulated as a consequence of 4OHT application to MEF2-VP16-ER-expressing neurons did not significantly overlap with either of the other gene sets. Moreover, the MEF2-VP16-ER-induced gene set did not overlap with genes whose expression is increased in the presence of MEF2 RNAi or genes whose expression decreased upon KCl treatment. These data demonstrate that the gain-of-function approach to MEF2 target gene identification largely corroborates the loss-of-function approach.
Because the genes identified by the KCl depolarization, MEF2 RNAi and MEF2-VP16-ER experiments overlap in a manner that is statistically significant and converge on a common set of genes (Fig. 1D), it is likely that the genes that are present in more than one of these gene sets include the bona fide activity-regulated MEF2 target genes in hippocampal neurons. In our subsequent studies we only considered genes identified in at least two of these three gene sets. This approach allows for the possibilities that any single one of the approaches to MEF2 target gene discovery might not have detected all of the activity-regulated MEF2 target genes and that some of the genes present in one particular gene set might not be bona fide activity-regulated MEF2 target genes. We found that 221 probe sets were present in at least two of these three gene sets (Table S1). These probe sets map to 182 non-redundant genes that can be confidently mapped in the rat genome and were therefore the subject of further analysis. We randomly selected 15 of these genes for validation by an independent method, reverse transcription-quantitative PCR (RT-qPCR), and found that over 95% of the expression changes that were initially identified by the microarray analysis were confirmed by RT-qPCR (Fig. S1–S3). These data indicate that the results from these microarray experiments have a low false discovery rate.
In vivo validation of mRNA profiling results
Because these initial experiments were performed using cultured neurons, we examined whether the set of candidate MEF2 target genes identified in these experiments are also induced upon physiological stimulation of neurons in the intact brain. To address this issue, we conducted a microarray analysis to identify genes whose expression is increased in response to sensory experience. To enhance levels of sensory experience, rats that had been housed in a standard laboratory cage (control) were placed for three hours in a novel cage containing objects of different shapes and colors that induce exploratory behavior, a procedure that has previously been shown to lead to new gene transcription in neurons in the forebrain (novel environment; (Nithianantharajah and Hannan, 2006; Ramanan et al., 2005)). We identified 304 probe sets that display increased expression in the forebrains of rats exposed to a novel environment (n = 6 animals per condition; Fig. 1E). These microarray results were also validated by RT-qPCR (Fig. S4). The candidate MEF2 target genes characterized in cultured hippocampal neurons represent a large fraction (16%, or 48/304) of the genes that are upregulated in vivo in response to new sensory experience. Moreover, 22% (48/221) of the candidate MEF2 target genes that are upregulated as a result of membrane depolarization of cultured neurons are also induced by new sensory experiences in vivo, a level that is far greater than would be expected by chance alone (p<0.0001, Fisher Exact test). Consistent with these results, a large number of the candidate MEF2 target genes were also identified in a screen for genes whose expression is increased in neurons in primary visual cortex in response to visual stimulation (including c-fos, bdnf, gadd45b, nur77, egr1, arc and many others; (Majdan and Shatz, 2006)).
For individual genes, the fold increase in gene expression in response to KCl treatment in vitro was highly correlated with the fold increase in gene expression in response to exposure to a novel environment (R2=0.36; p<0.0001), although the level of induction was consistently less robust in the novel environment experiment. This suggests that these two types of stimulation activate gene expression through similar mechanisms. However, the in vivo experiment appears to be less sensitive at detecting activity-regulated changes in gene expression, likely due to the heterogeneity of the cell population. Nevertheless, because the gene expression changes detected by these two approaches are highly correlated and overlap significantly, we conclude that a large fraction of the candidate MEF2 target genes identified in cultured neurons are induced in the brains of mice exposed to novel sensory experiences.
ChIP-chip to identify MEF2 binding sites in the genome: direct MEF2 target genes
The mRNA profiling experiments described above do not address whether the genes identified are direct or indirect targets of MEF2. To determine which of these genes are directly regulated by MEF2 and to provide additional confirmation that our microarray experiments had indeed identified MEF2 target genes, we performed chromatin immunoprecipitation followed by tiling microarray analysis (ChIP-chip) to identify MEF2D binding sites within the regulatory regions of these 182 genes. We developed ChIP conditions under which a MEF2D-specific antibody immunoprecipitated DNA fragments of the nur77 proximal promoter, a genomic region known to be bound by MEF2 (Fig. S5; (Youn et al., 2000)). Preabsorption of the anti-MEF2D antibody with the peptide antigen used to generate the antibody blocked its ability to immunoprecipitate MEF2D proteins as well as nur77 promoter DNA fragments (Fig. S5). Because these polyclonal anti-MEF2D antibodies might non-specifically immunoprecipitate some DNA fragments and therefore lead to the mis-identification of MEF2D binding sites, we used the MEF2 peptide-absorbed anti-MEF2D antibody as a negative control in all of the MEF2D ChIP-chip experiments described below.
We performed MEF2D ChIP-chip using a custom tiling microarray containing the genomic loci of the 182 genes identified in our mRNA profiling experiments (including 10 kb 5′ and 3′ to the genes). In these experiments, we also mapped the transcriptional start sites (TSSs) within the putative MEF2 target genes by performing ChIP-chip with an antibody that specifically recognizes RNA polymerase II when bound to sites of transcription initiation (Fig. 2C–D; (Mikkelsen et al., 2007)). To identify regions of MEF2D and RNA polymerase II occupancy, we developed a “peak-calling” algorithm that identified reproducible regions of MEF2D or RNA polymerase II binding that were not detected in negative control ChIP experiments conducted in parallel. Binding of MEF2D to a subset of the genomic regions identified through ChIP-chip was verified by subsequent ChIP-qPCR experiments (92%, or 12 of the 13 regions tested; Fig. S6). These data indicate that the ChIP-chip approach can be used to identify regions of MEF2D occupancy in the rat genome.
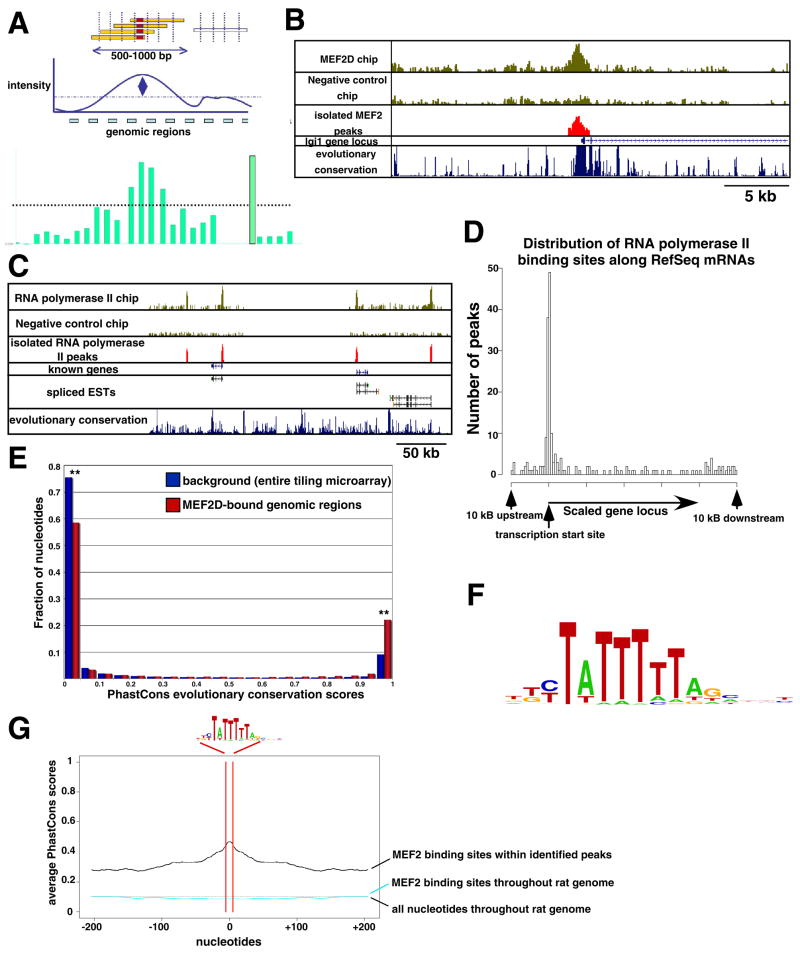
Figure 2. MEF2D ChIP-chip identifies MEF2 binding sites surrounding the candidate target genes.
(A) Method for the identification of MEF2 binding sites by ChIP-chip. Sonicated DNA fragments enriched by MEF2D immunoprecipitation (yellow) will center on the site of MEF2D binding (red). The intensity of the signals from several consecutive probe sets on the tiling microarray that correspond to this genomic region will be identified as a peak of MEF2D binding. (B) Example of a genomic region of MEF2D occupancy identified at the lgi1 gene locus. Raw data from one representative experiment are shown in the two upper panels. The genomic region identified by our peak calling algorithm is shown in red, and the annotated RefSeq lgi1 mRNA (exons shown as blue boxes, introns shown as blue lines with arrows that indicate direction of transcription) as well as level of evolutionary conservation within this genomic region are shown below. (C) An example of genomic regions of RNA Polymerase II occupancy is shown as in (B). In addition, rat expressed sequence tags (ESTs) are shown to indicate where un-annotated mRNAs are likely to exist in this region of the rat genome. (D) Distribution of the regions of RNA Polymerase II occupancy relative to annotated RefSeq mRNAs. For this graph, the genomic loci of all genes were scaled to 40 kilobases (kb) and regions of RNA polymerase II occupancy are plotted according to their positions at or near the genes. (E) Histogram of the PhastCons evolutionary conservation scores within the entire custom tiling microarray (blue) or within the MEF2D-bound regions of the custom tiling microarray (red). These distributions are bimodal, as the majority of the nucleotides in the rat genome are either non-conserved (close to zero) or highly conserved (close to one). Asterisks are shown to indicate that these two distributions are significantly different (p<0.001, Chi-Square test). (F) Logo of the MEF2 response element (MRE) that was identified as significantly enriched within the MEF2-bound genomic regions. (G) Mean PhastCons scores of genomic regions surrounding each occurrence of the MRE in (F) are shown. The nucleotides corresponding to the MRE are between the red lines, whereas the 400 nucleotides adjacent to each MRE surround the red lines. Mean PhastCons scores are shown for regions surrounding MREs within the MEF2D-bound regions of the tiling microarry (black) or within random regions of the rat genome (cyan). The mean PhastCons scores of all the nucleotides in the rat genome are also displayed (dotted black line). The PhastCons scores of the MREs within the MEF2D-bound regions are significantly higher than those within random regions of the rat genome (p<0.01, Chi-Square test).
Of the 182 genes examined, 78 (43%) have MEF2D binding sites within the gene (including gene exons and introns) or within 10 kB 5′ or 3′ of the gene (Table S2). This represents a notable increase over the background level of MEF2 binding detected on a separate chromosomal tiling microarray that was carried out as a control (8.8%) and MEF2 binding to arbitrarily selected control genes that were included on our custom tiling microarray (21%). These experiments demonstrate that the genes identified in our expression analyses display an enrichment of nearby regions of MEF2D occupancy and indicate that a large number of the genes identified by the gene expression analyses are likely to be direct targets of MEF2.
Bioinformatic analyses of the genomic regions of MEF2D occupancy
To determine whether the genomic regions of MEF2D occupancy that we identified serve as functionally important MEF2 binding sites, we performed several analyses. First, we reasoned that if these genomic regions play critical roles in gene regulation, the nucleotide sequences within these regions should be highly conserved through vertebrate evolution. Indeed, compared to other sequences included on the tiling microarray, the MEF2D-bound regions are signficantly enriched for evolutionarily conserved nucleotides (p<0.001, Chi-Square test; Fig. 2E). Second, we asked whether a specific nucleotide sequence motif was more commonly found within the MEF2D-bound genomic regions than elsewhere in the rat genome. An unbiased analysis of these genomic regions identified a 10-nucleotide motif that is significantly enriched within these regions and can be found in 80% of the regions of MEF2D occupancy. This sequence ((C/T)TA(T)4TA(G/A)) is nearly identical to the previously characterized MEF2 binding site, (C/T)TA(T/A)4TA(G/A), also termed the MEF2 response element (MRE; Fig. 2F; (Potthoff and Olson, 2007)). Compared to identical sequences found elsewhere in the rat genome (this sequence is predicted to occur randomly every ~350 base pairs), the MREs found within regions of MEF2D binding are significantly more likely to be conserved through vertebrate evolution (p<0.01, Chi-Square test; Fig. 2G), suggesting that they are likely to be functionally important. Taken together with the mRNA profiling experiments, these analyses strongly suggest that MEF2, by binding to evolutionarily conserved MREs near MEF2 target genes, directly regulates the expression of a large set of activity-regulated genes in neurons at the time that synapses are developing and maturing.
The sites of MEF2D binding were often (28% of the sites) located close to the 5′ ends of Reference Sequence (RefSeq)-annotated mRNAs transcribed from the MEF2-regulated genes, suggesting that these MEF2D binding sites form part of the proximal promoters of these genes (within 2.5 kb; Fig. 3A–B). In addition to these binding sites, we also identified several MEF2D binding sites (8% of the sites) that were located near TSSs identified by RNA polymerase II ChIP-chip but were not close to the 5′ ends of any RefSeq-annotated mRNAs (Fig. 3C). It is likely that these MEF2D binding sites also form part of the proximal promoters of these genes, but that the mRNAs produced by these genes have not yet been fully annotated.
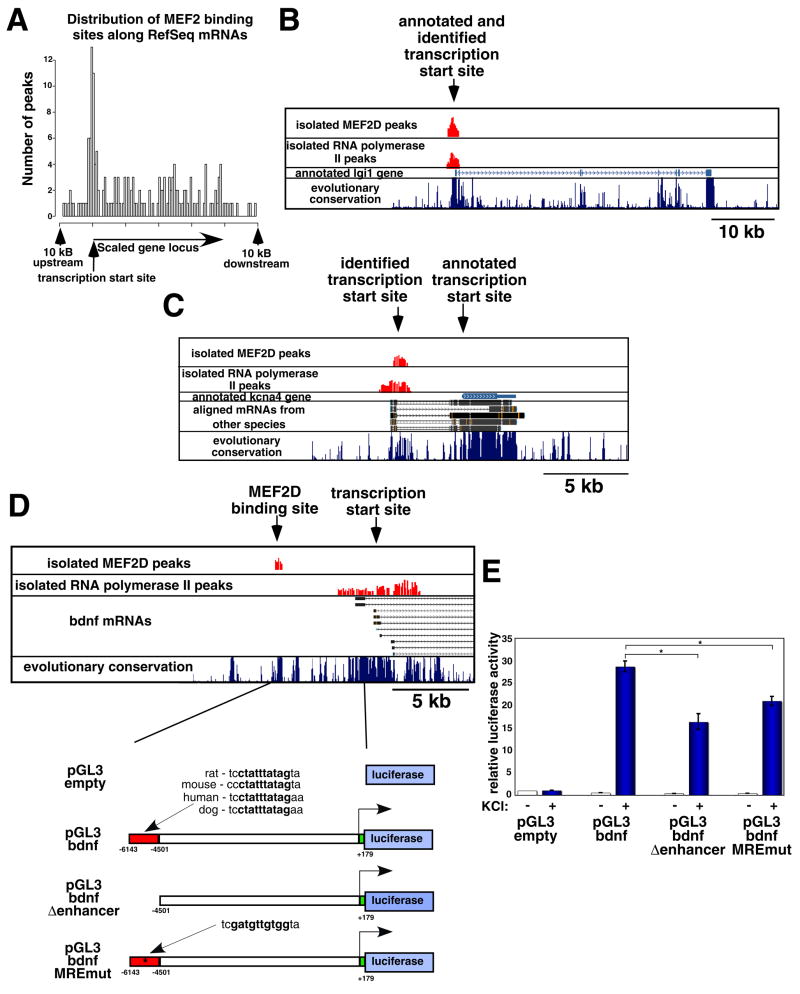
Figure 3. Location of the genomic regions of MEF2D occupancy at MEF2-regulated genes.
(A) Distribution of the regions of MEF2D occupancy along the MEF2-regulated genes, plotted as in Fig. 2D. (B) Example of a region of MEF2D occupancy that was identified within the proximal promoter of a MEF2-regulated gene (lgi1). The identified regions of MEF2D and RNA Polymerase II occupancy are shown in red, along with the annotated lgi1 gene and the level of evolutionary conservation within this genomic region. (C) Example of a region of MEF2D occupancy that was detected at a site of RNA Polymerase II occupancy upstream of the 5′ end of a RefSeq annotated mRNA (kcna4). (D) Example of a region of MEF2D occupancy detected far upstream (~6.5 kb) of the annotated transcriptional start site (TSS) of a MEF2-regulated gene (bndf). This genomic region was cloned into the luciferase reporter constructs that are depicted below (MEF2-bound region is depicted as a red box). (E) Normalized luciferase activity in lysates from E18+6 DIV hippocampal neurons that were transfected with the indicated luciferase reporter constructs and either treated with elevated levels of extracellular KCl (blue) or left untreated (white). Data are shown as means ± standard error of the mean (SEM). P<0.01, two-factor analysis of variance (ANOVA); asterisks indicate statistical significance in pairwise comparison: P<0.01, Bonferroni-Dunn post-hoc test.
The remaining 64% of the MEF2 binding sites that we identified using ChIP-chip occurred in regions of genes where there was no evidence to suggest that transcription is initiated nearby (within 2.5 kb; Fig. 3D). We found that the magnitude of the changes in mRNA expression observed in the gene expression analyses (Fig. 1) did not correlate with the distance of the MEF2D binding site from the TSS of the gene, suggesting that both proximally and distally located MEF2D binding sites are important for activity-dependent gene regulation. Consistent with this conclusion, luciferase reporter experiments examining the function of a MEF2 binding site located far upstream (~6.5 kb) of bdnf promoter I indicate that this MEF2 binding site is critical for activity-dependent bdnf promoter I transcription (Fig. 3D–E). We conclude from these data that MEF2 can regulate activity-dependent transcription by binding to evolutionarily conserved MREs in proximal promoters as well as distal enhancers at numerous MEF2 target genes.
Activity-dependent changes in polyadenylation site usage
One striking and unexpected feature of the activity-regulated MEF2 genetic program was revealed by an investigation of alternative exon usage within this transcriptional program before and after membrane depolarization of cultured neurons. Using the microarray data that were collected from hippocampal neurons before and after membrane depolarization, we performed a search for genes that have one or more exon that displays increased expression upon membrane depolarization while other exon(s) do not (referred to as activity-dependent exon usage). We identified ~70 genes that fulfill this criterion, including many of the newly identified MEF2 target genes. We were able to validate numerous cases of activity-dependent exon usage by performing RT-qPCR with primers that correspond to the distinct exons within these genes (Fig. 4A). Using this approach, we also verified that the activity-dependent changes in exon usage observed at several of these genes occur in neurons in primary visual cortex in response to light stimulation (Fig. 4B). Taken together, these data indicate that the microarray results reveal a genuine activity-dependent switch in exon usage that occurs in vitro and in vivo at numerous genes, including many MEF2 target genes.
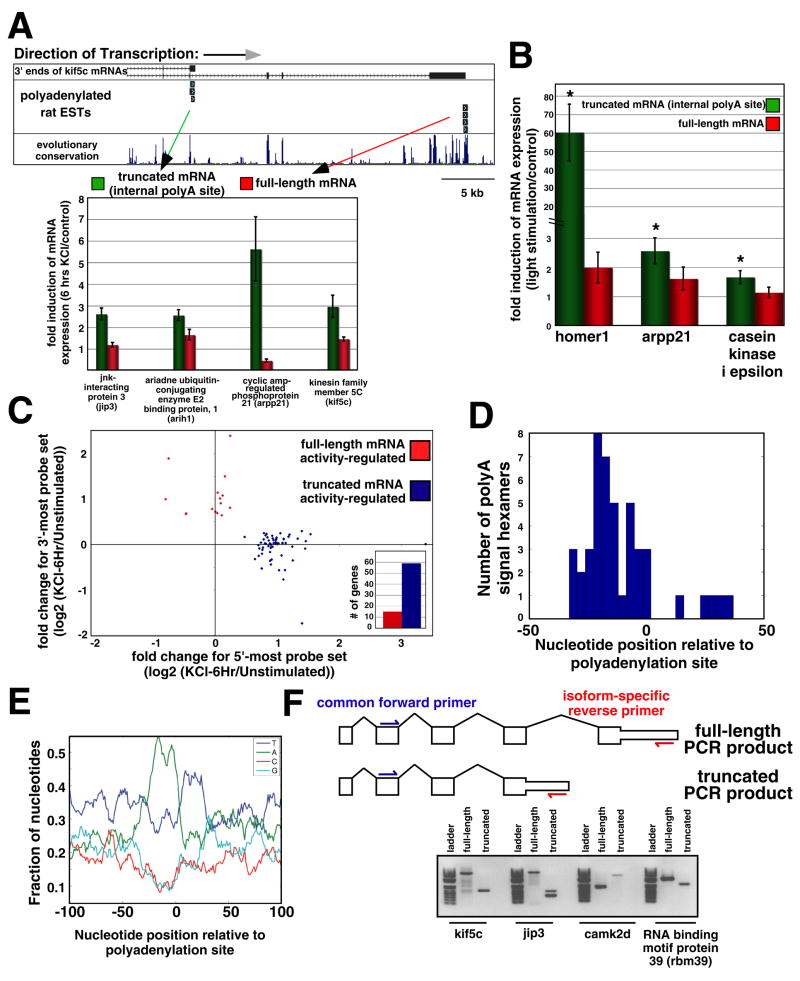
Figure 4. Neuronal activity-dependent alternative polyadenylation site usage.
(A) Upper: example of a gene (kif5c) that was identified through microarray analyses to display an activity-dependent change in polyA site usage. Rat ESTs that were sequenced with polyA tails intact are shown at the 3′ ends of the two kif5c mRNAs that were independently measured on the RAE230.2 micoarray. Lower: RT-qPCR confirmation of these microarray results for four genes. Expression of each gene was measured through two distinct primer sets that correspond to different 3′ ends of mRNAs produced by the gene that were independently represented on the microarray. Expression of these genes was normalized to that of beta-3-tubulin and data are shown as means ± SEM. (B) RT-qPCR analysis conducted as in (A). Gene expression in these experiments was measured in neurons in primary visual cortex in both control animals and animals that had been acutely exposed to light stimulation. Expression of these genes was normalized to that of beta-3-tubulin and data are shown as means ± SEM. Asterisk indicates significant increase in mRNA expression (p<0.05, t-test) in animals exposed to visual stimulation versus dark-reared control animals. None of the full-length mRNAs showed a significant change by these same criteria. (C) Changes in gene expression in response to 6 hours of KCl treatment for truncated and full-length mRNAs produced by individual genes. All genes that were identified through microarray analysis as having disparate expression data between two or more probe sets mapping to the same gene are displayed. There is a strong bias for the activity-regulated probe set to correspond to the truncated mRNA (blue) rather than the full-length mRNA (red). Inset: number of genes belonging to these two categories. (D) Distribution of polyA signal hexamers surrounding the polyA regions. The exact locations of 19 polyA sites that in depth analyses indicated were high confidence sites of activity-dependent polyadenylation were used in this analysis. The polyA signal hexamers that were acceptable in our analysis were those identified by Tian et al. (2005). (E) Nucleotide density at the genomic regions surrounding the polyA sites that display activity-dependent use. Same polyA sites that were analyzed in (D) were used here. (F) Above: cartoon depicting a generic gene that has two distinct mRNA isoforms with different 3′ ends. For four such genes, RT-PCR was performed with two primer sets. For each gene, the primer sets shared the same forward primer in a 5′ exon within the gene, but had different isoform-specific reverse primers, as is shown. Using a cDNA template prepared from hippocampal neuron RNA, all RT-PCR reactions yielded products of the correct predicted length (below). Note that two distinct splice forms (corresponding to two bands) of the short jip3 transcript were amplified; also note that the 3′UTR of the truncated camk2d isoform is especially long, so that this mRNA is larger than the full-length isoform even though it lacks several 3′ protein-coding exons.
Activity-dependent changes in exon usage could be due to activity-dependent changes in the sites of transcriptional initiation, changes in splicing, or alterations in transcriptional termination and polyA tail addition. Further analysis of the genes that display an activity-dependent switch in exon usage revealed that the activity-regulated exons within these genes are most commonly found 5′ to non-activity-regulated exons within the same genes (78% of the time; Fig. 4C). We also found that the mRNAs that are produced from these genes in response to membrane depolarization have polyA tails immediately 3′ to the sequences that correspond to the activity-regulated exons within these mRNAs (Fig. 4A). In addition, hexamers corresponding to the known signal for polyA tail addition (AATAAA and closely related sequences; (Tian et al., 2005)) could be found in their characteristic position 5 to 35 base pairs 5′ to the sites of polyA tail addition in these mRNAs (Fig. 4D–E). These results indicate that the activity-regulated exons within the mRNAs that are expressed in response to membrane depolarization are the terminal (3′-most) exons within these mRNAs. Thus, prior to stimulation, the mRNAs produced from these genes are typically expressed at lower levels and are longer (including additional exon(s) or a longer 3′ UTR) than the mRNAs that are produced upon membrane depolarization. Taken together, these findings suggest that for a group of activity-regulated genes, including many MEF2 target genes, there is a switch in polyA site usage that favors the production of shorter mRNAs that utilize internal sites of polyA tail addition.
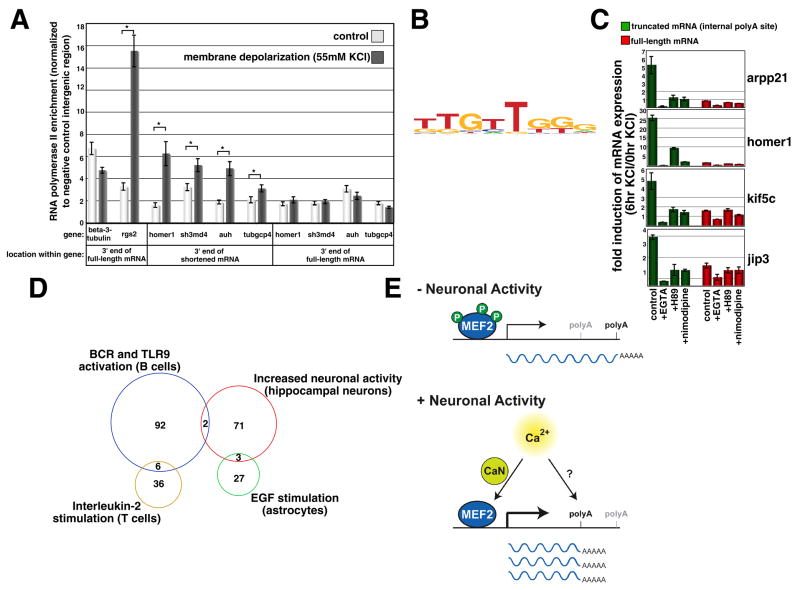
Several additional lines of evidence support this conclusion. First, using an RT-PCR-based approach, we verified that the distinct mRNA isoforms produced by these genes while they differ in their 3′ ends share the same 5′ exons (Fig. 4F). This demonstrates that the shortened mRNAs that are present after stimulation belong to the same transcriptional units as the full-length mRNAs that are present prior to membrane depolarization. We also investigated RNA polymerase II pausing at the activity-regulated genes that display activity-dependent polyA site switching. Recent studies have revealed that the RNA polymerase II complex pauses after polyA hexamers at the 3′ ends of genes, leading to transcription termination, mRNA cleavage and polyA tail addition (Gromak et al., 2006). Consistent with these results, we detect an enrichment of RNA polymerase II at the 3′ end of neuron-specific gene beta-3-tubulin and an activity-dependent increase in RNA polymerase II occupancy at the 3′ end of the activity-regulated gene rgs2 (Fig. 5A). However, at homer1 and other activity-regulated genes that our analyses indicate display polyA site switching, activity-dependent RNA polymerase II pausing was detected at the 3′ end of the short mRNA rather than the 3′ end of the full-length mRNA (Fig. 5A). This indicates that increased neuronal activity leads to RNA polymerase II pausing in genomic regions surrounding the sites of activity-dependent polyA tail addition, which likely leads to the cleavage of pre-mRNAs in this region, followed by the addition of a polyA tail. Taken together with the analyses described above, these data provide strong evidence that neuronal activity leads to polyA site switching at a number of activity-regulated genes, including many MEF2 target genes.
Figure 5. Features of polyA regions that display activity-dependent use.
(A) Data from two independent ChIP experiments conducted with an antibody against the elongating form of RNA polymerase II are shown as levels of RNA polymerase II enrichment at the indicated locations in the mouse genome, normalized to a negative control intergenic region that does not display RNA polymerase II occupancy. Note that analyses of microarray data (Fig. 1) indicate activity-dependent polyA site switching at homer1, while polyA site switching at sh3 multiple domains 4 (sh3md4), AU RNA binding protein/enoyl-Coenzyme A Hydratase (auh) and tubulin, gamma complex associated protein 4 (tubgcp4) is indicated by additional expression analyses (data not shown). Data are shown as means ± SEM; asterisk indicates statistically significant increase in RNA polymerase II occupancy for a given genomic region (p<0.01, Two-Factor ANOVA). (B) Logo of a novel motif that was identified as highly enriched in the regions surrounding the truncated polyA regions. (C) RT-qPCR analysis of mRNA expression of the indicated truncated and full-length mRNAs. The fold induction of mRNA expression was analyzed in response to six hours of membrane depolarization in the absence or presence of the indicated inhibitors. Data from one representative experiment are shown as means ± SEM. (D) Sets of genes identified through microarray analysis as displaying polyA site switching in response to different extracellular stimuli. Note that there is almost no overlap between these different sets of genes. (E) Model for activity-regulated polyadenylation site switching. At several MEF2 target genes, and other genes that display activity-dependent transcription, neuronal activity promotes both increased levels of transcription and increased usage of internal polyadenylation sites, which leads to a robust increase in the abundance of short mRNAs that differ from the full-length mRNAs produced in the absence of neuronal activity. The calcium-dependent activation of MEF2 is mediated in large part through the calcineurin (CaN)-dependent dephosphorylation of MEF2 proteins, while calcium-dependent polyadenylation site switching is mediated through signal transduction pathways that remain unknown. Note that although these processes are coupled at individual genes (like the gene depicted in the model), the signaling pathways that lead to activity-dependent transcription and polyA site switching might be completely independent.
Molecular mechanisms underlying activity-dependent polyA site switching
We took several approaches to begin to identify the molecular mechanisms by which neuronal activity triggers polyA site switching. Although our bioinformatic analyses indicated that the genomic regions surrounding the sites of activity-regulated polyA tail addition have features of typical sites of polyA tail addition (including the polyA signal hexamer, AATAAA and closely related sequences; Fig. 4D–E), we performed an unbiased search for additional nucleotide sequence motifs that were specifically enriched within the genomic regions where activity-dependent mRNA cleavage and polyA tail addition occur. This analysis resulted in the identification of a novel motif that was present in many of these genomic regions (42%; TTGTTGGG; Fig. 5B), but was only rarely found in the genomic regions that surround non-activity-regulated sites of polyA tail addition (8%). While the function of this motif is not yet clear, our bioinformatic analyses suggest that this sequence motif may play an important role in directing activity-dependent polyadenylation to specific sites, perhaps by promoting activity-dependent pausing of RNA polymerase II in these specific genomic regions, leading to mRNA cleavage and polyA tail addition.
To determine how neuronal activity might trigger RNA polymerase II pausing and polyA tail addition at these genes, we monitored the expression of both the short and full-length mRNAs produced by several of the genes that display activity-dependent polyA site switching (including several MEF2 target genes) and performed a pharmacological screen to identify agents that altered the activity-dependent expression of these mRNAs. None of the pharmacological agents applied (15 total) successfully dissociated the process of activity/MEF2-dependent transcription from the process of activity/MEF2-dependent polyA tail addition at any of the genes examined (Fig. 5C and data not shown). This result suggests that these two processes might be coupled. However, we note that MEF2 is not the only transcription factor that can couple activity-dependent transcription to activity-dependent polyA site switching, as 41 activity-regulated genes that are not regulated by MEF2 also display a switch in polyA site usage in response to membrane depolarization. Although we were unable to dissociate the processes of activity-dependent transcriptional intiation and polyA site switching, we were able to identify several agents (the calcium chelator EGTA, the L-type voltage-gated calcium channel (L-VGCC) antagonist nimodipine, and the cAMP-regulated protein kinase (PKA) inhibitor H89) that prevented the activity-dependent transcription/polyadenylation of each of the short mRNAs that were examined. These data indicate that the activity-dependent induction of truncated mRNAs is due to the coupling of activity-dependent transcription and activity-dependent polyA site switching at select genes, and requires calcium influx through L-VGCCs, as well as the activation of PKA (Fig. 5C).
PolyA site switching in other transcriptional programs
In considering the potential functions of activity-dependent polyA site switching, we reasoned that this mechanism might exist to promote the expression of short mRNA transcripts that are important for activity- and MEF2-dependent synapse development. Consistent with this idea, when we monitored the expression of several of these short mRNAs, we found that application of other extracellular factors that do not promote activity- and MEF2-dependent synapse development in hippocampal neurons (such as BDNF, NT-4, EGF and forskolin) does not lead to polyA site switching at the same set of genes that display this switch in response to increased neuronal activity (data not shown).
Nevertheless, we reasoned that extracellular stimulus-dependent polyA site switching might be a general mechanism used by cells in a variety of different contexts to alter gene function in response to changes in their environment. To determine whether application of other extracellular factors to cells leads to polyA site switching at genes that do not display this switch in response to neuronal activity, we analyzed a number of publicly available microarray data sets. We were able to identify genes that displayed polyA site switching in response to: (1) EGF application to astrocytes, (2) interleukin-2 application to T cells and (3) anti-IgM and CpG co-application to B cells. Furthermore, while this paper was under review, a separate study showed that stimulation of several types of immune cells also causes polyA site switching, which leads to the production of shortened mRNAs (Sandberg et al., 2008). The sets of genes that display polyA site switching in these different cellular contexts are almost completely non-overlapping (Fig. 5D). These data indicate that a wide variety of extracellular factors can trigger polyA site switching, but the specific genes that display this switch vary depending on the cell type and the extracellular stimulus.
Functional consequences of activity-dependent polyA site switching
Activity-dependent polyA site switching is likely to play a widespread role in the regulation of neuronal gene expression (see Table 1 for full list of genes that display this switch). This gene regulatory mechanism significantly alters the functions of ~10% of the mRNAs that are transcribed from MEF2 target genes in response to increased neuronal activity. The shortened, activity-regulated mRNAs are often predicted to encode proteins that are functionally distinct from the proteins encoded by the full-length mRNAs produced by the same gene. For instance, these mRNAs are predicted to encode scaffolding proteins that lack one or more of their C-terminal protein-protein interaction domains (homer1, jip3), signaling enzymes that lack C-terminal regulatory domains (casein kinase I epsilon, arhgef9) and transport proteins that lack their ability to transport dendritic mRNAs (kif5c; (Kanai et al., 2004)). In the case of homer1, the shortened mRNA encodes a dominant-negative form of homer1 that disrupts full-length homer1 function and restricts excitatory synapse number (Sala et al., 2003).
Table 1. Genes that display a neuronal activity-dependent switch in polyadenylation site usage.
These genes are divided into two groups. The 18 MEF2- and activity-regulated genes listed were included among the 182 MEF2 target genes that we identified, while the 41 activity-regulated genes (not MEF2-regulated) that are listed were included among the 643 activity-regulated genes identified in our study, but did not display evidence of MEF2-dependent regulation.
| MEF2- and activity-regulated genes | |
|---|---|
| Common Name | Description |
| JSAP1 | JNK/SAPK-associated protein 1 |
| Odz2 | odd Oz/ten-m homolog 2 (Drosophila) |
| Arhgef9 | Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| Csnk1e | Casein kinase 1, epsilon |
| Vesl-1; HOMER1F | homer homolog 1 (Drosophila) |
| Pcdh9_predicted | Protocadherin 9 (predicted) |
| Omp25 | synaptojanin 2 binding protein |
| Pkib | protein kinase (cAMP dependent, catalytic) inhibitor beta |
| LOC299907 | similar to Ext1 |
| LOC498145 | similar to RIKEN cDNA 2810453I06 |
| Septin 11 | Septin 6 (predicted) |
| Rybp_predicted | RING1 and YY1 binding protein (predicted) |
| Rinzf | zinc finger and BTB domain containing 10 |
| RGD1566117_predicted | similar to hypothetical protein FLJ23033 |
| RGD1311381_predicted | similar to hypothetical protein FLJ20037 (predicted) |
| Klf6 | Core promoter element binding protein |
| RGD1308432_predicted | similar to cDNA sequence BC020002 (predicted) |
| Tcf4 | Transcription factor 4 |
| Activity-regulated genes (not MEF2-regulated) | |
| Common Name | Description |
| Usp9x_predicted | similar to ubiquitin specific protease 9, X-linked (fat facets-like, Drosophila) |
| Syncrip | synaptotagmin binding, cytoplasmic RNA interacting protein (predicted) |
| Dab1 | disabled homolog 1 (Drosophila) |
| Arih1 | ariadne ubiquitin-conjugating enzyme E2 binding protein homolog 1 (Drosophila) (predicted) |
| HSPG | syndecan 2 |
| Camk2d | Calcium/calmodulin-dependent protein kinase II, delta |
| Rnf153 | ring finger protein 153 (predicted) |
| Rab21 | RAB21, member RAS oncogene family |
| Impad1 | inositol monophosphatase domain containing 1 |
| Kif5c_predicted | Kinesin family member 5C (predicted) |
| Mbd1 | Methyl-CpG binding domain protein 1 (predicted) |
| LOC363153 | Similar to protein phosphatase 1, regulatory(inhibitory) subunit 1C; thymocyte ARPP; |
| RGD1562438_predicted | similar to amyloid beta (A4) precursor protein- binding, family B, member 2 |
| Gpr177 | CG6210-like |
| Ppp1r2 | protein phosphatase 1, regulatory (inhibitor) subunit 2 |
| Zfp68_predicted | zinc finger protein 68 (predicted) |
| Idb4 | inhibitor of DNA binding 4 |
| Pabpn1 | poly(A) binding protein, nuclear 1 |
| Usp19 | ubiquitin specific protease 19 |
| Ssr1 | signal sequence receptor, alpha (predicted) |
| Txnl; MGC114276 | thioredoxin-like (32kD) |
| Pofut2_predicted | protein O-fucosyltransferase 2 (predicted) |
| Spred1 | sprouty protein with EVH-1 domain 1, related sequence (predicted) |
| Csrp2bp_predicted | cysteine and glycine-rich protein 2 binding protein (predicted) |
| Vcip135 | valosin-containing protein (p97)/p47 complex- interacting protein 135 |
| Cachd1_predicted | cache domain containing 1 (predicted) |
| Josd3 | Josephin domain containing 3 |
| Pafah1b1 | platelet-activating factor acetylhydrolase, isoform Ib, alpha subunit 45kDa |
| Ahctf1_predicted | AT hook containing transcription factor 1 (predicted) |
| Degs | degenerative spermatocyte homolog (Drosophila) |
| Cggbp1_predicted | CGG triplet repeat binding protein 1 (predicted) |
| AIP1; Alix | programmed cell death 6 interacting protein |
| Rptpk | Protein tyrosine phosphatase, receptor type, K, extracellular region |
| Ppp2r2a | Protein phosphatase 2 (formerly 2A), regulatory subunit B (PR 52), alpha isoform |
| Rnpc2 | RNA-binding region (RNP1, RRM) containing 2(predicted) |
| similar to DIP13 alpha (predicted) | |
| RGD1307879_predicted | similar to hypothetical protein D10Ertd438e (predicted) |
| RGD1309054_predicted | similar to FKSG26 protein (predicted) |
| RGD1559923_predicted | similar to chromosome 14 open reading frame 35 |
| proteasome (prosome, macropain) activator subunit 4 | |
| RGD1310651_predicted | Similar to hypothetical protein MGC20460(predicted) |
For other genes, the only difference between the shortened, activity-regulated mRNAs and the full-length mRNAs is that the shortened mRNAs lack a portion of their 3′UTRs. Interestingly, a recent study showed that the truncation of 3′ UTRs as a result of polyA site switching removes microRNA binding sites that repress mRNA translation (Sandberg et al., 2008). Thus, while these mRNAs are predicted to encode the same proteins as the full-length mRNAs, they likely lack elements within their 3′ UTRs that might specify where and when the mRNAs will be translated.
Based on the homer1 example, as well as our analyses of the sequences of the other newly identified truncated mRNAs, we hypothesize that stimulus-dependent polyA site switching might be a general mechanism used to alter gene function in response to changes in a cell’s extracellular environment. It is possible that stimulus-regulated transcriptional programs such as the activity-dependent MEF2 genetic program commonly use polyA site switch mechanisms to interfere with or alter the function of pre-existing proteins. Because new gene transcription does not necessarily lead to the disappearance of stably expressed proteins, the production of shortened, dominantly interfering or activating forms of these proteins may provide an efficient way to acutely disrupt or alter the function of existing proteins. Given that the remodeling of synapses likely requires the destabilization of existing structures (through activity-regulated genes like homer1a), the polyA site switch mechanism may have evolved to generate truncated proteins and/or mRNAs that can acutely reverse cellular processes such as synapse development.
MEF2 target genes that function at synapses to control synaptic remodeling
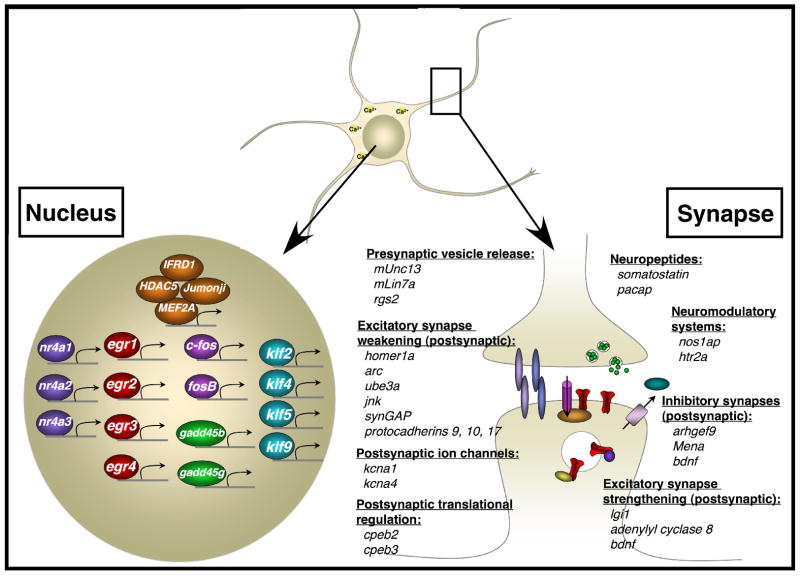
A striking feature of the group of MEF2 target genes that we have identified is that this set of genes displays enriched expression specifically within the CNS (based on comparisons to GNF Atlas 2 expression data; (Su et al., 2004); Table S3) and a large number of these genes encode proteins that function at neuronal synapses (p=2.14 ×10−7, enrichment for Synaptic Transmission GO category, Ingenuity Pathway Analysis; Fig. 6). In support of the recently described function of MEF2 in restricting the number of excitatory synapses that form on neurons, many of the MEF2 targets we have identified are known to contribute to the weakening/loss of excitatory synapses by regulating different aspects of signal transduction on the postsynaptic side of the synapse: Homer1a is involved in the disassembly of postsynaptic scaffolding complexes (Sala et al., 2003), while Activity-regulated cytoskeletal protein (Arc) and c-jun N-terminal kinase (Jnk) promote AMPAR removal from the postsynaptic membrane (Chowdhury et al., 2006; Zhu et al., 2005). The MEF2 target genes also include three members of the delta subfamily of protocadherins (protocadherins 9, 10, and 17), which have recently been shown to restrict the number of synapses formed onto neurons by promoting the internalization of N-cadherin, a synaptic adhesion molecule that stabilizes excitatory synapses (Yasuda et al., 2007). Finally, increased expression of the postsynaptically localized potassium channels encoded by the MEF2 target genes kcna1 and kcna4 would also be predicted to decrease excitatory postsynaptic strength, as dendritic areas of increased potassium channel conductance display more limited excitatory postsynaptic potential (EPSP) summation (Lujan et al., 2003; Raab-Graham et al., 2006). The finding that each of these genes is a MEF2 target indicates that MEF2 restricts synapse number through multiple mechanisms that together contribute to synapse weakening and synapse elimination.
Figure 6. MEF2 target genes that function at the synapse and in the nucleus.
The list of newly identified MEF2 target genes was enriched for genes that encode transcriptional regulators and for genes that encode proteins that function at the synapse. Several examples of these types of genes are shown. See main text for additional details.
The application of genomic strategies to identify MEF2 targets at the time that synapses are forming has also revealed new and unexpected roles for MEF2 in synapse biology. Unexpectedly, several of the MEF2 target genes encode proteins known to localize to the postsynaptic side of excitatory synapses and promote synaptic strengthening: Leucine-rich glioma-inactivated 1 (Lgi1) promotes AMPAR insertion into the postsynaptic membrane by binding to its transmembrane receptor (Fukata et al., 2006); adenylyl cyclase 8 mediates calcium-dependent cAMP production necessary for calcium-dependent synaptic strengthening (Wang et al., 2003); and Bdnf is known to enhance excitatory synaptic strength though a variety of mechanisms, for instance by acting as a retrograde signal that enhances presynaptic release (Zhang and Poo, 2002). One possible explanation for the coordinated expression of MEF2 target genes with antagonistic functions (synaptic weakening vs. strengthening) is that the activation of MEF2 might lead to a decrease in the total number of synapses formed onto cells through one group of target genes (e.g. homer1a, arc, kcna1, etc.) and at the same time lead to the strengthening of a separate subset of synapses through a separate group of target genes (e.g. adenylyl cyclase 8, bdnf, etc.). In fact, it is known that synapse elimination and synaptic potentiation (of other synapses) occur concurrently within individual cells during synapse development (Chen and Regehr, 2000). Further experiments will be necessary to determine the specific synapses at which each of these different MEF2 target genes functions.
Several of the genes identified in our screen for MEF2 target genes encode proteins that function postsynaptically at inhibitory synapses, providing the first evidence that MEF2 regulates inhibitory synaptic function. The Cdc42 guanine nucleotide exchange factor (GEF) Arhgef9 and the actin organizing molecule Mena both regulate the actin cytoskeleton in order to promote the clustering of gephyrin, a scaffolding protein that is specifically found at inhibitory synapses (Giesemann et al., 2003; Papadopoulos et al., 2007), while Bdnf is critical for activity-dependent inhibitory synapse formation (Rutherford et al., 1997). The finding that these genes are also included among the MEF2 target genes indicates that MEF2 may play a positive role in promoting inhibitory synapse development in addition to its effects on excitatory synapse development. Consistent with this idea, recent studies from our laboratory have identified an activity-regulated transcription factor, NPAS4 (not present on the RAE230.2 microarrays used in this study), that plays a key role in the formation of inhibitory synapses onto excitatory neurons and appears to be a direct target of MEF2 ((Lin et al., 2008); and data not shown).
The activity-dependent activation of MEF2 within a neuron also triggers the induction of target genes that regulate the presynaptic outputs of the neuron. For example, mUNC13, mLin7A, and rgs2 each encode proteins that enhance synaptic vesicle release. Moreover, the neuropeptides Somatostatin and adenylate cyclase activating polypeptide 1 (ADCYAP1, or PACAP), both encoded by activity-regulated MEF2 target genes, are known to be secreted from presynaptic sites and to modulate excitatory synaptic transmission (Boehm and Betz, 1997; Roberto et al., 2001).
These data demonstrate that in response to neuronal activity MEF2 activates target genes that regulate multiple aspects of neural circuit development and function. In addition to activating genes that weaken excitatory synaptic inputs formed onto the MEF2-expressing neuron, MEF2 also activates additional groups of target genes that strengthen select excitatory synaptic inputs, remodel inhibitory synaptic inputs and regulate the presynaptic outputs of the MEF2-expressing neuron. Further experiments will be necessary to determine if these MEF2 targets are all expressed together in the same neurons or, alternatively, if distinct subsets of these target genes are expressed in different subsets of neurons (e.g. CA3 versus CA1 pyramidal neurons). Nevertheless, these findings indicate that in response to an increase in neural circuit activity, the activation of MEF2 within individual neurons in the circuit leads to the induction of an extensive genetic program that likely plays a central role in the process of experience-dependent synaptic development, refinement and plasticity.
MEF2 target genes that function within the nucleus
Another group of genes that is statistically over-represented among the MEF2 target genes encode proteins that function within the nucleus to regulate transcription (p=1.75 × 10−31, enrichment for Transcription GO category, Ingenuity; Fig. 6). Among these transcriptional regulators are several molecules (e.g. c-fos, fosB, early growth response (egr1), egr3, nur77, histone deacetylase 5 (hdac5) and mef2a itself) that have been documented to play a critical role in synapse development and plasticity, although for the most part the target genes of these transcription factors remain to be identified (Flavell et al., 2006; Fleischmann et al., 2003; Hiroi et al., 1997; Jones et al., 2001; Li et al., 2007; Renthal et al., 2007; Shalizi et al., 2006). The identification of these genes as MEF2 targets suggests that MEF2 induces the expression of a set of regulatory factors that work together with MEF2 to control synaptic function. Consistent with this idea, a number of the MEF2 targets are known to enhance MEF2 activity (IFRD1 and Mef2a; (Micheli et al., 2005)) whereas other targets interact with MEF2 and inhibit its transcriptional activity (the putative histone demethylase Jumonji and the histone deacetylase HDAC5; (Kim et al., 2005; McKinsey et al., 2000)).
MEF2 target genes in neurological disease
The importance of the MEF2 transcriptional program for neural circuit development and function in humans is highlighted by the fact that mutations in several of the MEF2 targets in humans give rise to neurological disorders. Mutations in the human orthologs of the MEF2 target genes lgi1, kcna1 and arhgef9 result in an imbalance between synaptic excitation and inhibition that leads to epilepsy (Table 2 and references therein). In addition, mutations in the human orthologs of the MEF2 target genes ube3a and solute carrier 9, member 6 (slc9a6) are known to cause Angelman syndrome, a disorder characterized by mental retardation and increased susceptibility to seizures. Finally, two of five mutations that were recently shown to give rise to autism and epilepsy in families in which parents share ancestors occur within the putative regulatory regions of protocadherin 10 and c3orf58 (Morrow et al., 2008). Protocadherin 19, which is closely related to protocadherin 10, has also recently been linked to epilepsy and mental retardation in several families (Dibbens et al., 2008).
Table 2.
MEF2 target genes related to epilepsy and autism-spectrum disorders in humans.
| Disease | Gene Name | Gene Symbol | Reference (Pubmed ID) |
|---|---|---|---|
| Epilepsy | leucine-rich, glioma inactivated 1 | lgi1 | 11810107 |
| Epilepsy | potassium voltage-gated channel, shaker-related subfamily, member 1 | kcna1 | 10414318 |
| Epilepsy | Cdc42 guanine nucleotide exchange factor (GEF) 9 | arhgef9 | 15215304 |
| Angelman syndrome | ubiquitin protein ligase E3A | ube3a | 8988171 |
| Angelman syndrome | solute carrier family 9 (sodium/hydrogen exchanger), member 6 | slc9a6 | 18342287 |
| Autism/Epilepsy | protocadherin 10 | Pcdh10 | 18621663 |
| Autism/Epilepsy | chromosome 3, open reading frame 58 | c3orf58 | 18621663 |
The symptoms of epilepsy, Angelman syndrome and autism are typically manifested in the first years of life during which sensory experience regulates synapse development and function. Thus, our finding that MEF2, a transcription factor activated by sensory experience, regulates activity-dependent synaptic development and induces the expression of a set of genes, several of which are mutated in disorders of human cognition, suggests that a common mechanism of these disorders may be the disruption of the activity-dependent MEF2-regulated gene expression program. This also raises the possibility that human genetic studies combined with the characterization of cell type-specific transcriptional programs that regulate synapse development and maturation (like the activity-regulated MEF2 transcription program) may provide an enhanced understanding of the molecular basis of epilepsy and autism.
CONCLUDING REMARKS
In the CNS, sensory stimulation leads to the activation of MEF2 and several other activity-regulated transcription factors that in turn activate programs of gene expression necessary for synapse development and remodeling (Flavell and Greenberg, 2008). By performing a series of genome-wide analyses in hippocampal neurons at the time that synapses are developing, we were able to effectively identify over 150 activity-regulated MEF2 target genes that are primarily expressed in the CNS. While previous studies have utilized loss- and gain-of-function experiments to suggest that MEF2 regulates synapse development, it has been difficult to distinguish between the direct functions of MEF2 (mediated by its target genes) and the homeostatic changes that occur in cells where the expression of MEF2 has been altered for several days. The set of MEF2 targets identified in our study is enriched for genes whose protein products are localized to synapses and these genes control multiple aspects of synaptic function. Thus, the identification of this set of MEF2 targets provides evidence that MEF2 plays a key role in mediating many aspects of synapse and neural circuit development.
One issue left unresolved by this study is which of the newly identified MEF2 target genes that have not yet been shown to play a critical role in synapse development are actually involved in this process. To address this issue, each of the MEF2-regulated genes will need to be individually tested for their role in synapse development. The MEF2 target genes that are known to harbor mutations and polymorphisms in the human population that lead to an increased susceptibility to neurological disorders such as epilepsy and autism-spectrum disorders (lgi1, arhgef9, kcna1, ube3a, slc9a6, pcdh10, c3orf58) are especially important to test for their potential roles in synapse development. The observation that several activity-dependent MEF2 target genes are mutated in cases of epilepsy and autism suggests that these disorders may be caused at least in part by the disruption of important components of the activity-regulated MEF2 genetic program that controls neural circuit development.
Our approach to MEF2 target gene identification also revealed a novel feature of the activity-dependent genetic program. Neuronal activity promotes the use of alternative sites of polyA addition at many of the MEF2 target genes, which results in the activity-dependent production of truncated mRNAs and in some cases truncated proteins (Fig. 5E). In cases where activity-dependent polyA site switching leads to truncated proteins, these proteins are frequently predicted to act dominantly to alter the function of pre-existing full-length proteins. In other cases where activity-dependent polyA site switching leads to shortened mRNAs that contain full-length open reading frames but shortened 3′ UTRs, this switch will not alter protein function but instead will likely alter when and where the mRNAs are translated. We hypothesize that stimulus-dependent changes in polyadenylation site usage may thus provide a mechanism to acutely alter gene function in response to changes in a cell’s extracellular environment.
It is likely that the genomic approaches that we have used to study MEF2 function in neurons can be generally applied to elucidate the gene expression programs that control different aspects of neural development. In addition, the further characterization of stimulus-regulated transcriptional programs in neurons and other cell types should provide new insights into the importance of polyA site switching in stimulus-dependent transcriptional control and should more generally reveal the molecular mechanisms by which extracellular factors induce complex cellular responses that allow multi-cellular organisms to adapt to changes in their environment.
EXPERIMENTAL PROCEDURES
Cell Culture
Hippocampal neurons from E18 Long-Evans rats were isolated as previously described (Xia et al., 1996). Detailed information about cell culture conditions, lentiviral vectors and pharmacological agents is available in Supplemental Data.
Plasmids
MEF2A and MEF2D shRNA constructs were previously described (Flavell et al., 2006). Descriptions of the plasmids encoding MEF2-VP16-ER and the bdnf luciferase reporter plasmids are available in the Supplemental Data.
Exposure of rats to novel environment
Novel environment experiments were conducted using standard procedures. Details are available in Supplemental Data.
Mouse visual cortex stimulation
Detailed protocols for mouse visual stimulation and visual cortex isolation are available in Supplemental Data.
mRNA profiling microarray analyses
For expression profiling of cultured neurons, three biological replicates of each experiment were performed. For the novel environment experiment, RNA was collected from six animals for each experimental condition. Arrays from all experimental conditions (and all replicates) of a given experiment were normalized to one another and all subsequent analyses were conducted using GeneSpring GX software (Agilent, Santa Clara, CA). Additional details about RNA isolation, sample preparation and array analysis methods are available in Supplemental Data.
Identification of multiple probe sets mapping to individual genes
To identify cases where multiple probe sets mapping to the same gene showed different expression profiles, we used standard procedures that are described in detail in Supplemental Data.
Statistical Analyses and Comparisons
Statistical methods used to investigate MEF2 target gene expression in different tissues (through GNF Atlas 2 data), as well as details about Ingenuity Pathway analyses, are available in Supplemental Data.
Reverse Transcription-quantitative PCR (RT-qPCR)
All RT-qPCR experiments were performed essentially as previously described (Flavell et al., 2006). Details are available in Supplemental Data.
Luciferase Assays
All luciferase assays were performed essentially as previously described (Flavell et al., 2006). Details are available in Supplemental Data.
Antibodies
The antibodies used in all experiments are described in Supplemental Data.
Chromatin-immunoprecipitation (ChIP)
Typically 20–40 million hippocampal neurons were used for a single ChIP experiment. A detailed description of the ChIP procedure is available in Supplemental Data.
Quantitative-PCR (qPCR) using ChIP samples
ChIP analysis of all individual genomic regions was conducted using qPCR and is described in Supplemental Data.
ChIP-chip
The arrays used for ChIP-chip were manufactured by Nimblegen Systems, Inc (Madison, WI). We performed two biological replicates of each ChIP-chip experiment, which included three different stimulation conditions (unstimulated, 20 and 140 min after KCl depolarization, which was conducted as described above). Furthermore, each ChIP experiment was paired with a negative control ChIP reaction that was performed with a negative control antibody. For MEF2D ChIP, the negative control antibody was a MEF2D antibody that was pre-blocked with antigen peptide. For RNA Polymerase II ChIP, mouse IgG mix (Santa Cruz Biotechnology, Inc) was used as a negative control. Details about microarrays and ChIP-chip library preparation are available in Supplemental Data.
ChIP-chip data analysis
Signal intensity data was extracted from the scanned images of each array using NimbleScan, NimbleGen’s data extraction software. Subsequently, scaled log2-ratio values from each probe were obtained (log2[ChIP signal intensity/input signal intensity]). The log2-ratio was computed and scaled to center these values around zero. Scaling was performed by subtracting the bi-weight mean for the log2-ratio values for all features on the array from each log2-ratio value. We then identified regions of MEF2D and RNA polymerase II occupancy using a custom-written ChIP-chip data analysis program that was implemented using the R language and is composed of three parts that are described in Supplemental Data.
Bioinformatic analyses
Methods used to investigate evolutionary conservation and sequence motifs within the rodent genome are described in Supplemental Data.
Supplementary Material
01
Acknowledgments
We thank members of the Greenberg lab for their helpful suggestions and discussions and for critical reading of the manuscript. In particular, we thank J. Zieg for assistance with figures and S. Vasquez for preparing primary neuronal cell cultures. We thank the Molecular Genetics Core Facility at Children’s Hospital Boston for assistance with microarrays. We also thank Soren Impey and Richard H. Goodman for their early guidance concerning genome-wide applications of ChIP, and Gabriel Kreiman for bioinformatics help. M.E.G. acknowledges the generous support of the Nancy Lurie Marks Family Foundation and the generous support of the F.M. Kirby Foundation to the F.M. Kirby Neurobiology Center of Children’s Hospital Boston. This work was supported by a Jane Coffin Childs Memorial Fund postdoctoral fellowship (T.K.K), a Lefler postdoctoral fellowship (T.K.K.), a Helen Hay Whitney postdoctoral fellowship (J.M.G.), a National Library of Medicine training fellowship in Biomedical Informatics (D.A.H.), Developmental Disability Research Center grant HD18655 and National Institutes of Health grant NS028829 (M.E.G.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aizawa H, Hu SC, Bobb K, Balakrishnan K, Ince G, Gurevich I, Cowan M, Ghosh A. Dendrite development regulated by CREST, a calcium-regulated transcriptional activator. Science. 2004;303:197–202. doi: 10.1126/science.1089845. [DOI] [PubMed] [Google Scholar]
- Barbosa AC, Kim MS, Ertunc M, Adachi M, Nelson ED, McAnally J, Richardson JA, Kavalali ET, Monteggia LM, Bassel-Duby R, Olson EN. MEF2C, a transcription factor that facilitates learning and memory by negative regulation of synapse numbers and function. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0802679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barco A, Alarcon JM, Kandel ER. Expression of constitutively active CREB protein facilitates the late phase of long-term potentiation by enhancing synaptic capture. Cell. 2002;108:689–703. doi: 10.1016/s0092-8674(02)00657-8. [DOI] [PubMed] [Google Scholar]
- Boehm S, Betz H. Somatostatin inhibits excitatory transmission at rat hippocampal synapses via presynaptic receptors. J Neurosci. 1997;17:4066–4075. doi: 10.1523/JNEUROSCI.17-11-04066.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Regehr WG. Developmental remodeling of the retinogeniculate synapse. Neuron. 2000;28:955–966. doi: 10.1016/s0896-6273(00)00166-5. [DOI] [PubMed] [Google Scholar]
- Chowdhury S, Shepherd JD, Okuno H, Lyford G, Petralia RS, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron. 2006;52:445–459. doi: 10.1016/j.neuron.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibbens LM, Tarpey PS, Hynes K, Bayly MA, Scheffer IE, Smith R, Bomar J, Sutton E, Vandeleur L, Shoubridge C, et al. X-linked protocadherin 19 mutations cause female-limited epilepsy and cognitive impairment. Nat Genet. 2008;40:776–781. doi: 10.1038/ng.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etkin A, Alarcon JM, Weisberg SP, Touzani K, Huang YY, Nordheim A, Kandel ER. A role in learning for SRF: deletion in the adult forebrain disrupts LTD and the formation of an immediate memory of a novel context. Neuron. 2006;50:127–143. doi: 10.1016/j.neuron.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, Griffith EC, Hu LS, Chen C, Greenberg ME. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science. 2006;311:1008–1012. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Greenberg ME. Signaling Mechanisms Linking Neuronal Activity to Gene Expression and Plasticity of the Nervous System. Annu Rev Neurosci. 2008;31:563–590. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann A, Hvalby O, Jensen V, Strekalova T, Zacher C, Layer LE, Kvello A, Reschke M, Spanagel R, Sprengel R, et al. Impaired long-term memory and NR2A-type NMDA receptor-dependent synaptic plasticity in mice lacking c-Fos in the CNS. J Neurosci. 2003;23:9116–9122. doi: 10.1523/JNEUROSCI.23-27-09116.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata Y, Adesnik H, Iwanaga T, Bredt DS, Nicoll RA, Fukata M. Epilepsy-related ligand/receptor complex LGI1 and ADAM22 regulate synaptic transmission. Science. 2006;313:1792–1795. doi: 10.1126/science.1129947. [DOI] [PubMed] [Google Scholar]
- Gaudilliere B, Konishi Y, de la Iglesia N, Yao G, Bonni A. A CaMKII-NeuroD signaling pathway specifies dendritic morphogenesis. Neuron. 2004;41:229–241. doi: 10.1016/s0896-6273(03)00841-9. [DOI] [PubMed] [Google Scholar]
- Giesemann T, Schwarz G, Nawrotzki R, Berhorster K, Rothkegel M, Schluter K, Schrader N, Schindelin H, Mendel RR, Kirsch J, Jockusch BM. Complex formation between the postsynaptic scaffolding protein gephyrin, profilin, and Mena: a possible link to the microfilament system. J Neurosci. 2003;23:8330–8339. doi: 10.1523/JNEUROSCI.23-23-08330.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromak N, West S, Proudfoot NJ. Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol Cell Biol. 2006;26:3986–3996. doi: 10.1128/MCB.26.10.3986-3996.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroi N, Brown JR, Haile CN, Ye H, Greenberg ME, Nestler EJ. FosB mutant mice: loss of chronic cocaine induction of Fos-related proteins and heightened sensitivity to cocaine’s psychomotor and rewarding effects. Proc Natl Acad Sci U S A. 1997;94:10397–10402. doi: 10.1073/pnas.94.19.10397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua JY, Smith SJ. Neural activity and the dynamics of central nervous system development. Nat Neurosci. 2004;7:327–332. doi: 10.1038/nn1218. [DOI] [PubMed] [Google Scholar]
- Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeney S, Dunn JJ, Mandel G, Goodman RH. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- Ince-Dunn G, Hall BJ, Hu SC, Ripley B, Huganir RL, Olson JM, Tapscott SJ, Ghosh A. Regulation of thalamocortical patterning and synaptic maturation by NeuroD2. Neuron. 2006;49:683–695. doi: 10.1016/j.neuron.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Jones MW, Errington ML, French PJ, Fine A, Bliss TV, Garel S, Charnay P, Bozon B, Laroche S, Davis S. A requirement for the immediate early gene Zif268 in the expression of late LTP and long-term memories. Nat Neurosci. 2001;4:289–296. doi: 10.1038/85138. [DOI] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Kim TG, Jung J, Mysliwiec MR, Kang S, Lee Y. Jumonji represses alpha-cardiac myosin heavy chain expression via inhibiting MEF2 activity. Biochem Biophys Res Commun. 2005;329:544–553. doi: 10.1016/j.bbrc.2005.01.154. [DOI] [PubMed] [Google Scholar]
- Li L, Yun SH, Keblesh J, Trommer BL, Xiong H, Radulovic J, Tourtellotte WG. Egr3, a synaptic activity regulated transcription factor that is essential for learning and memory. Mol Cell Neurosci. 2007;35:76–88. doi: 10.1016/j.mcn.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Bloodgood BL, Hauser JL, Lapan AD, Koon AC, Kim TK, Hu LS, Malik AN, Greenberg ME. Activity-dependent regulation of inhibitory synapse development by Npas4. Nature. 2008 doi: 10.1038/nature07319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan R, de Cabo de la Vega C, Dominguez del Toro E, Ballesta JJ, Criado M, Juiz JM. Immunohistochemical localization of the voltage-gated potassium channel subunit Kv1.4 in the central nervous system of the adult rat. J Chem Neuroanat. 2003;26:209–224. doi: 10.1016/j.jchemneu.2003.07.006. [DOI] [PubMed] [Google Scholar]
- Majdan M, Shatz CJ. Effects of visual experience on activity-dependent gene regulation in cortex. Nat Neurosci. 2006;9:650–659. doi: 10.1038/nn1674. [DOI] [PubMed] [Google Scholar]
- Malinow R, Malenka RC. AMPA receptor trafficking and synaptic plasticity. Annu Rev Neurosci. 2002;25:103–126. doi: 10.1146/annurev.neuro.25.112701.142758. [DOI] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Lu J, Olson EN. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408:106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheli L, Leonardi L, Conti F, Buanne P, Canu N, Caruso M, Tirone F. PC4 coactivates MyoD by relieving the histone deacetylase 4-mediated inhibition of myocyte enhancer factor 2C. Mol Cell Biol. 2005;25:2242–2259. doi: 10.1128/MCB.25.6.2242-2259.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow EM, Yoo SY, Flavell SW, Kim TK, Lin Y, Hill RS, Mukaddes NM, Balkhy S, Gascon G, Hashmi A, et al. Identifying autism Loci and genes by tracing recent shared ancestry. Science. 2008;321:218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nithianantharajah J, Hannan AJ. Enriched environments, experience-dependent plasticity and disorders of the nervous system. Nat Rev Neurosci. 2006;7:697–709. doi: 10.1038/nrn1970. [DOI] [PubMed] [Google Scholar]
- Papadopoulos T, Korte M, Eulenburg V, Kubota H, Retiounskaia M, Harvey RJ, Harvey K, O’Sullivan GA, Laube B, Hulsmann S, et al. Impaired GABAergic transmission and altered hippocampal synaptic plasticity in collybistin-deficient mice. Embo J. 2007;26:3888–3899. doi: 10.1038/sj.emboj.7601819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development. 2007;134:4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- Pulipparacharuvil S, Renthal W, Hale CF, Taniguchi M, Xiao G, Kumar A, Russo SJ, Sikder D, Dewey CM, Davis MM, et al. Cocaine regulates MEF2 to control synaptic and behavioral plasticity. Neuron. 2008;59:621–633. doi: 10.1016/j.neuron.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab-Graham KF, Haddick PC, Jan YN, Jan LY. Activity- and mTOR-dependent suppression of Kv1.1 channel mRNA translation in dendrites. Science. 2006;314:144–148. doi: 10.1126/science.1131693. [DOI] [PubMed] [Google Scholar]
- Ramanan N, Shen Y, Sarsfield S, Lemberger T, Schutz G, Linden DJ, Ginty DD. SRF mediates activity-induced gene expression and synaptic plasticity but not neuronal viability. Nat Neurosci. 2005;8:759–767. doi: 10.1038/nn1462. [DOI] [PubMed] [Google Scholar]
- Renthal W, Maze I, Krishnan V, Covington HE, 3rd, Xiao G, Kumar A, Russo SJ, Graham A, Tsankova N, Kippin TE, et al. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron. 2007;56:517–529. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- Roberto M, Scuri R, Brunelli M. Differential effects of PACAP-38 on synaptic responses in rat hippocampal CA1 region. Learn Mem. 2001;8:265–271. doi: 10.1101/lm.40501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford LC, DeWan A, Lauer HM, Turrigiano GG. Brain-derived neurotrophic factor mediates the activity-dependent regulation of inhibition in neocortical cultures. J Neurosci. 1997;17:4527–4535. doi: 10.1523/JNEUROSCI.17-12-04527.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala C, Futai K, Yamamoto K, Worley PF, Hayashi Y, Sheng M. Inhibition of dendritic spine morphogenesis and synaptic transmission by activity-inducible protein Homer1a. J Neurosci. 2003;23:6327–6337. doi: 10.1523/JNEUROSCI.23-15-06327.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalizi A, Gaudilliere B, Yuan Z, Stegmuller J, Shirogane T, Ge Q, Tan Y, Schulman B, Harper JW, Bonni A. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 2006;311:1012–1017. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- Simon DJ, Madison JM, Conery AL, Thompson-Peer KL, Soskis M, Ruvkun GB, Kaplan JM, Kim JK. The microRNA miR-1 regulates a MEF-2-dependent retrograde signal at neuromuscular junctions. Cell. 2008;133:903–915. doi: 10.1016/j.cell.2008.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Pineda VV, Chan GC, Wong ST, Muglia LJ, Storm DR. Type 8 adenylyl cyclase is targeted to excitatory synapses and required for mossy fiber long-term potentiation. J Neurosci. 2003;23:9710–9718. doi: 10.1523/JNEUROSCI.23-30-09710.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z, Dudek H, Miranti CK, Greenberg ME. Calcium influx via the NMDA receptor induces immediate early gene transcription by a MAP kinase/ERK-dependent mechanism. J Neurosci. 1996;16:5425–5436. doi: 10.1523/JNEUROSCI.16-17-05425.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda S, Tanaka H, Sugiura H, Okamura K, Sakaguchi T, Tran U, Takemiya T, Mizoguchi A, Yagita Y, Sakurai T, et al. Activity-induced protocadherin arcadlin regulates dendritic spine number by triggering N-cadherin endocytosis via TAO2beta and p38 MAP kinases. Neuron. 2007;56:456–471. doi: 10.1016/j.neuron.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn HD, Chatila TA, Liu JO. Integration of calcineurin and MEF2 signals by the coactivator p300 during T-cell apoptosis. Embo J. 2000;19:4323–4331. doi: 10.1093/emboj/19.16.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Poo MM. Localized synaptic potentiation by BDNF requires local protein synthesis in the developing axon. Neuron. 2002;36:675–688. doi: 10.1016/s0896-6273(02)01023-1. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Pak D, Qin Y, McCormack SG, Kim MJ, Baumgart JP, Velamoor V, Auberson YP, Osten P, van Aelst L, et al. Rap2-JNK removes synaptic AMPA receptors during depotentiation. Neuron. 2005;46:905–916. doi: 10.1016/j.neuron.2005.04.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01